Abstract
Methylation and acetylation of DNA and histone proteins are the chemical basis for epigenetics. From bacteria to humans, methylation and acetylation are sensitive to cellular metabolic status. Modification rates depend on the availability of one-carbon and two-carbon substrates (S-adenosylmethionine, acetyl-CoA, and in bacteria also acetyl-phosphate). In addition, they are sensitive to demodification enzyme cofactors (α-ketoglutarate, NAD+) and structural analog metabolites that function as epigenetic enzyme inhibitors (e.g., S-adenosylhomocysteine, 2-hydroxyglutarate). Methylation and acetylation likely initially evolved to tailor protein activities in microbes to their metabolic milieu. While the extracellular environment of mammals is more tightly controlled, the combined impact of nutrient abundance and metabolic enzyme expression impacts epigenetics in mammals sufficiently to drive important biological outcomes such as stem cell fate and cancer.
In classical kinase signal transduction cascades, a chemical trigger (such as hormone-receptor binding) leads to a rapid (~ 1 min) series of phosphorylation events that augment the signal and drive downstream effector functions such as gene transcription. While requiring ATP, such cascades are generally independent of metabolic status, as the cellular concentration of ATP (~ 10 mM) dwarfs the amount needed to saturate kinase active sites (~ 0.002-0.1 mM) and is also sufficient to outcompete related metabolites like ADP [1]. Thus, with the exception of kinases like AMPK and TOR that are specifically designed to sense metabolites, metabolism and kinase signaling can be reasonably viewed as distinct biochemical networks.
In contrast, other important protein covalent modifications occur on slower timescales and are tightly linked to cellular metabolite abundances. Foremost among these are methylation and acetylation. For these reactions, the physiological substrate concentrations are lower than ATP. Moreover, the reaction products, or other related endogenous metabolites, are often competitive inhibitors of substrate binding [2]. The tight binding (low Ki) of these inhibitors renders reaction rates sensitive to substrate concentration, even when substrate is nominally sufficient to saturate the enzyme ([substrate] > Km):
Although more work on the underlying enzymology is needed, much of metabolic control of methylation and acetylation seems to rely on such active site competition.
Acetylation and deacetylation
In bacteria, acetylation can be driven by acetyl-CoA or acetyl-phosphate. Based on recent experiments in E. coli, acetyl-phosphate, which reacts spontaneously with protein lysines, is thought to predominate. Manipulations that increase acetyl-phosphate, such as deletion of acetate kinase or nitrogen limitation, increase protein lysine acetylation. In contrast, knockout of phosphotransacetylase, which converts acetyl-CoA into acetyl-phosphate, decreases protein acetylation. While E. coli encodes a homolog of the classical eukaryotic histone acetylation enzyme Gcn5 (YfiQ), its knockout does not broadly alter protein acetylation. Thus, in bacteria, acetyl-phosphate levels are likely the primary determinant of protein acetylation rates [3].
Most eukaryotes are not known to make acetyl-phosphate and the only known substrate for acetylation is acetyl-CoA. Based on analysis of isolated mitochondria, their acetyl-CoA concentration is estimated to be 0.1-1.5 mM, [4]. The combination of abundant acetyl-CoA and high pH (which enhances the fraction of lysine residues in their neutral and thus nucleophilic form), results in substantial spontaneous mitochondrial protein lysine acetylation [5]. Such nonenzymatic protein acetylation may also happen outside mitochondria, facilitated by basic amino acid residues just upstream of the critical lysine in the protein sequence [6]. Nevertheless, due to lower acetyl-CoA levels (0.002-0.013 mM) [7] and pH, most acetylation outside mitochondria, including in the nucleus where histones reside, depends on specific modification enzymes such as Gcn5, MYST, and p300/CBP. Histone acetylation generally promotes associated gene transcription.
Acetyl-CoA can be made in mitochondria via catabolism of pyruvate, amino acids, or fatty acids (Figure 1). Transport of acetyl-CoA into the cytosol involves an ATP-driven metabolic cycle, where mitochondrial acetyl-CoA condenses with oxaloacetate to form citrate, which is transported into the cytosol and cleaved by ATP citrate lyase [8]. Activity of this cycle, which is induced by signals including insulin and Akt [7,9], impacts cytosolic acetyl-CoA levels. In hypoxia, pyruvate dehydrogenase is inhibited and acetate becomes a major source of cytosolic acetyl-CoA. The ligation of acetate and CoA, at the expense of ATP, is catalyzed by the enzyme acetyl-CoA synthetase 2 (ACSS2) in mammals. Hypoxic cancer cells in culture derive nearly half of cytosolic acetyl-CoA from acetate [10], and significant expression of ACSS2 has been found in certain breast, ovarian, and lung tumors [11]. Recently it has been reported that pyruvate dehydrogenase complex can be translocated from the mitochondria to the nucleus [12]. This putatively enables direct conversion of nuclear pyruvate into acetyl-CoA for histone acetylation.
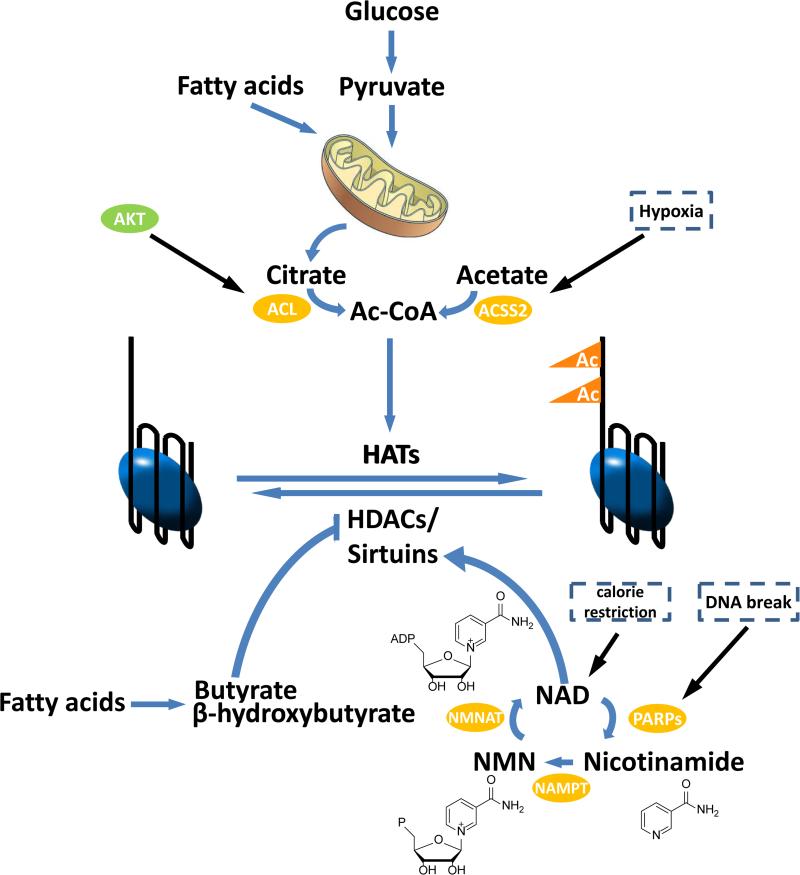
Figure 1.
Metabolic pathways contributing to histone acetylation and deacetylation. Acetyl-CoA is the substrate of histone acetyltransferase (HATs). Glucose derived pyruvate and fatty acids feed into mitochondria to produce acetyl-CoA and subsequently citrate. Mitochondrial citrate can be exported and converted to cytosolic acetyl-CoA by citrate-ATP lyase (ACL). AKT activates ACL by phosphorylation. Alternatively, cytosolic acetyl-CoA can be generated from acetate, which is the primary production route under hypoxia. Two classes of enzymes remove the histone acetylation marks, HDACs and sirtuins. Sirtuins use NAD+ as the substrate for deacetylation, generating nicotinamide and O-acetyl-ADP-ribose as the products. Nicotinamide is a sirtuin inhibitor. Calorie restriction or supplementation of NAD biosynthetic precursors enhance NAD+ levels and thus sirtuin activity. Poly(ADP-ribose) polymerases (PARPs) use NAD+ as substrate and deplete NAD under DNA damage conditions. Other HDACs have no co-substrate requirement, but can be inhibited by β-hydroxybutyrate.
Although lysine acetylation is chemically stable, the removal of lysine acetylation can be achieved by straightforward amide hydrolysis by water. Three different phylogenetic classes of histone deacetylases (HDACs) carry out this reaction in a Zn2+-dependent manner: Class I (human HDAC1–3 and HDAC8), II (human HDAC4–7 and HDAC9–10), and IV (human HDAC11). In contrast, Class III HDACs, also known as sirtuins (human SirT1-7), carry out an alternative reaction with NAD+ as a co-substrate. Thus, NAD+ levels, relative to NADH and nicotinamide, are important regulators of deacetylation. Although straightforward deacetylases are found in some eubacteria, the ties between NAD+ and protein deacetylation are evolutionarily conserved, with deacetylation in E. coli carried out by the NAD+-dependent deacetylase CobB [13]. Thus, protein acetylation and deacetylation depend on the concentrations of two of the most important central metabolic cofactors: acetyl-CoA and NAD+.
Methylation and demethylation
The substrate for methylation is the activated one-carbon donor S-adenosylmethionine (SAM), which is transferred to DNA and protein targets via specific modification enzymes in both prokaryotes and eukaryotes (Figure 2). DNA methylation in prokaryotes occurs at the 6-position of adenosine residues (6mA), whereas in eukaryotes it typically occurs at the 5-position of cytosine residues (5mC). In both cases, DNA methylation is generally transcription inhibitory [14,15].
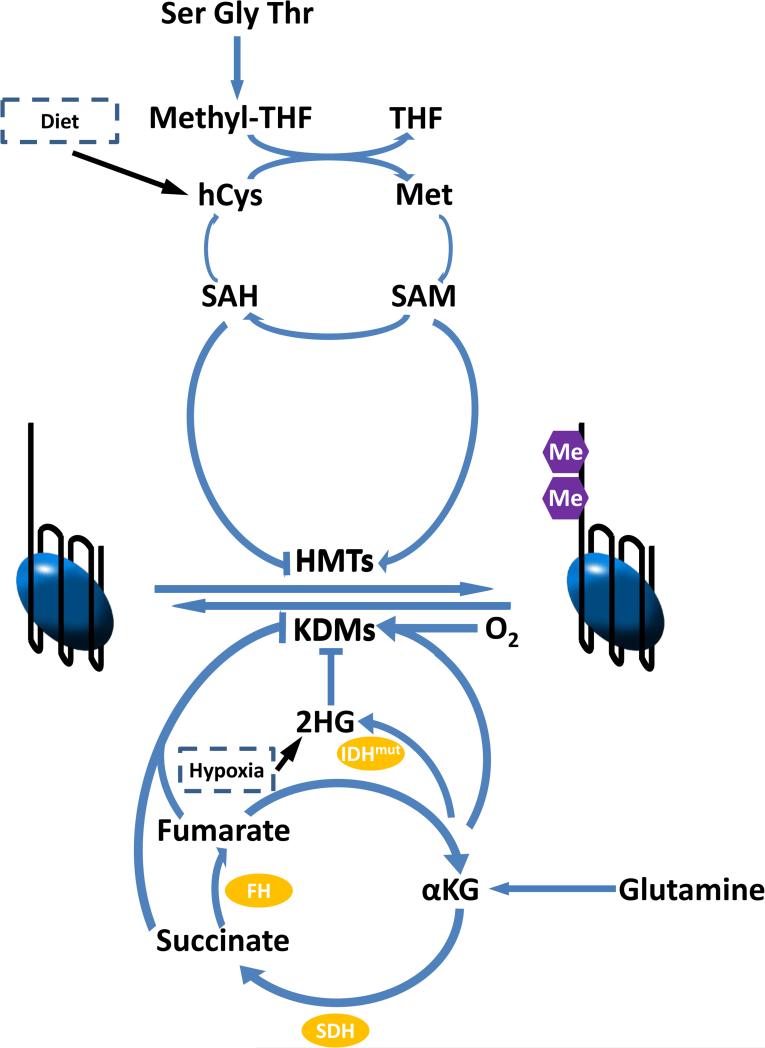
Figure 2.
Metabolic pathways contributing to histone and DNA methylation and demethylation. Histone and DNA methyltransferases use SAM as substrate and produce SAH as product. SAH is a competitive inhibitor of the methyltransferases, therefore the SAM:SAH ratio dictates the activity of the transferases. SAH can be removed by S-adenosylhomocysteine hydrolase, producing homocysteine, which can be remethylated to form methionine. This set of reactions is called the methionine cycle. The remethylation uses methyl-THF and vitamin B12. Serine, glycine and (in mouse but not human) threonine can all contribute to the methyl-THF pool. S-adenosylmethionine synthetase converts methionine and ATP into SAM. DNA demethylation and most histone demethylation depend on both O2 and α-ketoglutarate. Other dicarboxylic acids, such as succinate, fumarate and 2-hydroxyglutarate (2HG), are inhibitors of the α-ketoglutarate-dependent demethylases. Oncogenic mutations in isocitrate dehydrogenase (IDH1 R132H and IDH2 R172K) result in D-2HG production. Loss of succinate dehydrogenase or fumarase can similarly cause cancer due to accumulation of succinate and fumarate. All these mutations are oncogenic due to the inhibition of DNA and histone demethylation.
On histones, methylation can occur on arginines or lysines. Each individual lysine residue can be mono-, di-, or tri-methylated. While the residue is cationic in any of these forms, methylation enhances hydrophobicity and thereby alters binding of important gene-expression regulating proteins. Methyl-binding domains frequently recognize the modifications via aromatic cages. One class of histone methyltransferase, DOT1, targets the core histone body to regulate transcriptional activation and elongation. Chromosomal rearrangements that activate DOT1 are the leading causes of infant acute leukemias [16]. Another class, involving a highly conserved catalytic protein domain called SET, targets the histone tail where many regulatory proteins bind. While SET domain expressing proteins have the conserved chemical function of histone methylation, the gene expression outcome varies depending on the specific tail site. Typically, trimethylation of lysine 27 on histone H3 (H3K27me2/3) that is mediated by polycomb-group complex 2 (PRC2) represses gene expression, whereas trimethylation of lysine 4 of histone H3 (H3K4me3) that is mediated by Trithorax complex activates transcription.
While all methylation enzymes share the common substrate of SAM, the impact of SAM concentrations can be histone site specific. This can be achieved by differences in histone methyltransferase's SAM affinity. For example, in yeast, core histone methylation catalyzed by Dot1 is less sensitive to disrupted methionine and SAM biosynthesis than histone tail methylation catalyzed by SET enzymes. This reflects the lower Km of Dot1 for SAM, and can be reversed by mutants that increase this Km [17]. In general, H3K4me2/3, which is methylated by the yeast enzyme Set1, is the most sensitive site to SAM [18-20]. Set1 was recently shown to interact with a metabolic enzyme complex comprising serine biosynthetic enzymes, serine hydroxymethyltransferase and methionine adenosyltransferases [21]. The higher sensitivity of H3K4 methylation to one-carbon status may arise due to this complex, the Km of Set1 for SAM, and/or H3K4 being more exposed than other histone sites to demethylation. Consistent with complexes between metabolic enzymes and histone methyl transferases being functionally important, in Caenorhabditis elegans, different methionine adenosyltransferases appear to impact differentially histone methylation. Worms lacking methionine adenosyltransferase 1 (sams-1) are more sensitive to Pseudomonas, due to a reduction in H3K4me3, which is required to induce infection response gene transcription [22]. Knockdown of methionine adenosyltransferase 3 (sams-3) has no effect on H3K4me3, but decreases methylation of other sites (H3K9, H3K27, H3K36) [23]. Thus, histone methyltransferases may depend on local SAM production by specific associated methionine adenosyltransferase enzymes.
Removal of methylation is chemically challenging and involves diverse metabolic inputs. In bacteria, the removal of DNA methylation marks is usually achieved by two rounds of DNA replication. On the other hand, DNA alkylation damage including 1mA and 3mC can be removed by the enzyme AlkB. AlkB is a member of the chemically versatile class of enzymes known as Fe2+/α-ketoglutarate-dependent oxygenases [24]. These enzymes couple iron-catalyzed oxidative decomposition of the co-substrate α-ketoglutarate (forming CO2 and succinate) to the hydroxylation of the primary substrate. Their function in both DNA and protein demethylation is conserved from bacteria to humans. Although the closest eukaryotic homologs of AlkB function in DNA and RNA alkylation repair, the same catalytic strategy is used by the main family of eukaryotic DNA demethylases (TET) and histone demethylases (JMJC). Another family of histone demethylases, LSD1, while still consuming oxygen, uses FAD as a cofactor and does not require any co-substrate. Importantly, only the α-ketoglutarate-dependent JMJC enzymes can remove lysine tri-methylation.
The dependence of TET and JMJC enzymes on oxygen and α-ketoglutarate as co-substrates renders demethylation rates sensitive to both oxygenation and TCA cycle metabolism. Among TCA cycle metabolites, α-ketoglutarate promotes demethylation, whereas succinate and fumarate act as competitive inhibitors. In addition, α-ketoglutarate can undergo two-electron reduction to produce 2-hydroxyglutarate (2HG), which also competitively inhibits demethylation. Thus, methylation status is broadly sensitive to oxygen, one-carbon, and TCA-related metabolism.
Physiological function of acetylation and methylation in microorganisms
In E. coli, when carbon availability exceeds nitrogen availability, acetate and acetyl-phosphate accumulate, increasing protein acetylation. In complete media, deletion of the main enzymes making and consuming acetyl-phosphate (ackAΔ and ptaΔ) result in 10-fold changes in overall protein acetylation, without markedly impacting growth rate. This suggests that any functional role of protein acetylation is subtle and/or context dependent [3]. In eukaryotes, metabolism is rewired to eliminate acetyl-phosphate, at the expense of reducing the ATP yield of glycolysis from 4 ATP to 2 ATP per glucose. It is possible that such rewiring evolved in part to minimize spontaneous protein acetylation, which in turn allowed broader use of enzymatic acetylation for regulatory purposes.
Consistent with this, acetyl-CoA plays an important role in coordinating metabolism and transcription in yeast. When yeast are limited for glucose in continuous culture, they undergo rounds of synchronized metabolic cycling, which temporally separate expression of growth genes (such as ribosomes) from protective genes (such as antioxidant enzymes). A critical determinant of this metabolic cycling is intracellular acetyl-CoA concentration [25]. Acetyl-CoA levels are high coincident with the expression of growth genes, both in metabolically cycling yeast and in standard batch cultures. In glucose-limited metabolically cycling yeast, addition of acetate will induce growth gene expression. These effects are mediated through the histone acetyltransferase Gcn5, part of the SAGA complex. Gcn5 preferentially acetylates histone lysine residue H3K9 at growth genes [26]. Remarkably, roughly two-fold changes in acetyl-CoA are sufficient to alter histone acetylation and gene expression several-fold. This may reflect acetyl-CoA cooperatively regulating Gcn5 activity, both through acetylation of other SAGA subunits and through its direct role as the histone acetylation substrate. The net effect is that acetyl-CoA ties carbon status to growth gene transcription [27].
While histone acetylation is important for cell growth, deacetylation is important for transcriptional repression at certain loci. Yeast cells that fail to repress certain genes, including ribosomal RNA, have short replicative life span [28]. Overexpression of the sirtuin family histone deacetylate Sir2p extends the life span of yeast cells. Alternatively, the life span of yeast can be extended by mild carbon limitation, which activates Sir2p by decreasing NADH and increasing the NAD+/NADH ratio [29]. Hypoxia is a powerful metabolic manipulation that markedly decreases the NAD+/NADH ratio, and may accordingly be expected to inhibit sirtuin activity. Further investigation of the impact of hypoxia on microbial acetylation status is warranted.
Similarly, environmental factors can impact concentrations of metabolites involved in methylation and demethylation. While existing literature in microbes is limited, one can envision the environmental concentrations of folate, formate, serine, or methionine feeding into SAM levels and thus methylation rates. In terms of demethylation, in both E. coli and yeast, α-ketoglutarate responds the most strongly of any metabolite to nitrogen limitation. In E. coli, it coordinates carbon and nitrogen metabolism, suppressing glucose uptake and inhibiting transcription of alternative carbon source genes [30,31]. In yeast, it regulates the expression of nitrogen assimilation genes through the PII signal transduction proteins. It seems likely that the dependence of demethylases on α-ketoglutarate as a cofactor evolved in part to couple demethylation to nitrogen status. Thus, in addition to the well-established connection between acetyl-CoA, NAD+, and growth gene transcription in yeast, there are likely other functionally important connections between metabolism, acetylation, and methylation in microbes.
Impact of metabolic milieu on mammalian epigenetics
In mammals, cells are fed through the circulation, with extracellular metabolite concentrations maintained in a relatively narrow range through systemic homeostatic mechanisms. Nevertheless, the metabolic environment can impact acetylation and methylation. Acetyl-CoA can be made from each major category of mammalian nutrients (carbohydrates, fats, protein). In mouse liver, free CoA is higher under fasting conditions, while acetyl-CoA is relatively stable [32]. To date, we are unaware of studies adequately investigating the link between dietary nutrient consumption, acetyl-CoA levels, and in vivo acetylation rates. Deacetylation by sirtuins, however, is known to be influenced by levels of their co-substrate NAD+ (relative to competitive inhibitors). Moreover, even the deacetylases that use water as the sole substrate are subject to allosteric regulation by circulating β-hydroxybutyrate.
Both calorie restriction and high-fat diet may impact histone acetylation via NAD+ levels, with calorie restriction potentially promoting high NAD+/NADH and thus deacetylation [33]. High-fat diet has the converse effect, decreasing NAD+ and thus sirtuin activity [34,35]. In high-fat diet-induced diabetic mice, the activity of nicotinamide phosphoribosyltransferase (NAMPT), a key enzyme in mammalian NAD+ synthesis, is severely reduced. Administration of nicotinamide mononucleotide or riboside restores NAD+, activates SirT1 and SirT3, and enhances insulin sensitivity [36,37]. The NAD+-consuming enzyme nicotinamide N-methyltransferase (NNMT) is up-regulated in mouse models of obesity and insulin resistance [38] and is overexpressed in many tumors [39]. This enzyme has dual epigenetic effects, simultaneously depleting SAM and NAD+, and thereby impairing methylation and deacetylation [38].
Although other HDACs have no co-factor requirement, their activity is nevertheless coupled to metabolism via the “ketone body” β-hydroxybutyrate, which inhibits class I HDACs, leading to increased H3K9 and H3K14 acetylation [40]. During fasting, the liver switches to fatty acid oxidation and circulating D-β-hydroxybutyrate levels can rise to above 1 mM. β-hydroxybutyrate increases H3K9 acetylation at the promoter of FOXO3A, a transcription factor for oxidative stress resistance genes [40], and diets that are low in carbohydrate promote ketogenesis and protect neurons from oxidative damage [41]. Altered acetylation does not, however, correlate with seizure control by ketogenic diet [42]. Thus, high calorie diets that include carbohydrates tend to inhibit deacetylation by sirtuins, whereas ketogenic conditions inhibit deacetylation by other HDACs.
The impact of diet on methylation rate is increasingly well established. Circulating homocysteine levels correlate inversely with cellular SAM:S-adenosylhomocysteine (SAH) ratio. Homocysteine levels tend to increase with age and disease, including atherosclerosis and thrombosis, although a causative role for homocysteine in these conditions has not been established [43]. Diet that is high in methionine and low in folate and cobalamin causes high circulating homocysteine [44]. In rodent studies, either methionine restriction (0.12% methionine diet in mice) or excess (regular diet plus 1% methionine in rats) has been reported to lower the SAM:SAH ratio and H3K4me3 [18,45]. In humans, vegetarians, who have a low intake of cobalamin (the cofactor for converting homocysteine into methionine), tend to have high SAH correlated with low whole-genome methylation [46]. Overall, dietary factors have been estimated to account for 30% of serum methionine variation [18]. Thus, both amino acid and vitamin intake can propagate through metabolism to impact methylation rates.
A primary environmental factor impacting demethylation is hypoxia, which occurs in a variety of physiological and pathological settings, including intense exercise, high altitude, cancer, and atherosclerosis. Elevated NADH due to hypoxia may potentially inhibit sirtuin activity. Ties between low oxygen and demethylation are better established, with low oxygen inhibiting demethylation through two mechanisms: lack of oxygen as a substrate for demethylase enzymes and increased (likely via elevated NADH) synthesis of L-2HG, which competitively inhibits α-ketoglutarate-dependent demethylases [47]. The catalytic mechanism of α-ketoglutarate-dependent demethylases is similar to that of the classical oxygen sensors HIF prolyl hydroxylases, with human JMJC demethylase KDM4E having a saturating concentration of oxygen above the atmospheric concentration. Nevertheless, H3K9me3 enhancement in hypoxia is dependent on elevated L-2HG [48,49].
Collectively, these examples highlight the potential for physiological conditions to impact epigenetics via metabolism. Further work is needed, however, to establish the extent to which transcriptional adaptation to nutrient and oxygen availability occurs via acetylation and methylation versus other mechanisms. One appealing possibility is that metabolism-specific signaling machinery like insulin and HIF play a predominant role in short-term adaptation, whereas acetylation and methylation contribute substantially to long-term adaptation.
Control of stem cell fate by epigenetic regulatory metabolites
The maintaining of pluripotency requires an open and accessible chromatin structure. Histone acetylation contributes to the openness of chromatin and stem cell pluripotency. Human embryonic stem cells differentiate upon withdraw of basic fibroblast growth factor (bFGF), with concomitant decreases in the expression of pyruvate dehydrogenase, ATP citrate lyase, and ACCS2, and depletion of acetyl-CoA. Addition of acetate promoted H3K9/K27 acetylation, complementing bFGF in maintaining pluripotency [50]. While ties between deacetylation and stem cell maintenance are less well established, nicotinamide can facilitate human stem cell maintenance, possibly via sirtuins [51].
Open chromatin structure in stem cells is also dependent on both methylation and demethylation. H3K4me3 is transcriptionally activating and a crucial ESC self-renewal signal [52], and SAM production is important in maintaining the pluripotency of mouse embryonic stem cells (mESC). mESC specifically express threonine dehydrogenase [53], which oxidizes threonine into acetyl-CoA and glycine, a one-carbon donor. When the mouse embryonic fibroblasts are reprogrammed into induced pluripotent stem cells, SAM levels increase, along with H3K4 methylation, driven by threonine-derived methyl groups [19]. In humans, however, threonine dehydrogenase is a nonfunctional pseudogene, and SAM production relies on methionine. Methionine withdrawal or inhibition of methionine adenosyltransferase inhibits human stem cell growth [20]. On the demethylation side, mESC differ from most cultured cells in having high flux from glucose to α-ketoglutarate. This both enables growth in the absence of glutamine, and maintains a high intracellular ratio of α-ketoglutarate to succinate, which facilitates demethylation. mESC cells have low levels of H3K27me3 and low DNA methylation. Supplementation with α-ketoglutarate favors mESC self-renewal, whereas succinate promotes differentiation [54]. Thus, in addition to responding to their nutrient environment, cells can modulate their metabolic network through changing enzyme levels to maintain particular epigenetic states.
Metabolism and epigenetics in cancer
Cancer cells resemble stem cells in many ways. Abnormal metabolism not only fuels cancer cell growth, but also shapes their epigenetics to favor proliferation. Oncogenic AKT and Myc promote ATP-citrate lyase-dependent histone acetylation [7,55], and pAKT(S473) correlates with histone acetylation in human gliomas and prostate tumors [7]. Butyrate is one of the bacterial fermentation products of dietary fiber in colon, and has been shown to reduce the risk of colorectal cancer [56]. In normal cells, butyrate goes through β-oxidation to generate acetyl-CoA. In cancerous colonocytes, butyrate functions as a HDAC inhibitor and is growth inhibitory. In both cases butyrate favors histone acetylation and activates transcription. For example, the upregulation of the tumor suppressor TES relies on butyrate-derived acetyl-CoA, whereas the upregulation of apoptosis related genes depends on the HDAC inhibition by butyrate [57].
Changes in methylation are prevalent in cancer genomes, with 2000-3000 promoters typically are aberrantly methylated per cancer cell and 70% of all the histone and DNA modification genes known to be mutated in human cancer [58]. In contrast, only a few of the ~ 2000 metabolic enzymes in the human genome are systematically mutated in cancer. Remarkably, each known oncogenic mutation in metabolic enzymes affects methylation. The most common and important of these are active site mutations in isocitrate dehydrogenase (IDH) 1 and 2, which lead to cancer through production of D-2HG [59-61]. Almost all cases of AML involve in activation of the DNA methylase TET2, which can occur through homozygous TET2 deletion or single-copy point mutation of IDH [62]. Loss-of-function mutations in fumarate hydratase and succinate dehydrogenase subunits cause the accumulation of fumarate or succinate and thereby also result in cancer through demethylase inhibition [63,64]. The discovery of these oncometabolites offers new therapeutic opportunities. A small molecule inhibitor of mutant IDH1 inhibitor reduces histone H3K9me3 and promotes glioma cells differentiation [65].
In addition to genetic lesions to metabolic enzymes, cancer epigenetics can be influenced by the tumor metabolic status. While the tumor environment is typically nutrient poor and low in serine, overall cancer genomes are hypermethylated. One contributor may be phosphoglycerate dehydrogenase, which diverts glycolytic intermediate 3-phosphglycerate into serine de novo synthesis and is genomically amplified in breast cancer and melanoma [66]. In addition, genes to convert serine into one-carbon units are strongly overexpressed in a broad spectrum of tumors and show correlation between expression and cancer progression [67]. On the demethylation side, one factor that could contribute to cancer genome hypermethylation is hypoxia.
Beyond the inhibition of specific metabolic oncogenes, it is intriguing to speculate that metabolic manipulations could, through epigenetic mechanisms, help to treat a larger subset of cancers. Methotrexate, an antifolate, can increases Fas death receptor expression by reducing promoter methylation [68]. The NAMPT inhibitor FK866 lowers cellular NAD+ levels and increases p53 acetylation [69]. Further investigation is merited to identify the optimal combination of diet, metabolic inhibition, and epigenetic therapy to stop cancer.
Highlights.
■ Epigenetic methylation and acetylation are sensitive to cellular metabolic status
■ Diet and environment conditions shape epigenetics through metabolism
■ Stem cells rewire metabolism to maintain open chromatin structure
■ Oncometabolites cause cancer by interfering with DNA and histone demethylation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Knight ZA, Shokat KM. Features of selective kinase inhibitors. Chem Biol. 2005;12:621–637. doi: 10.1016/j.chembiol.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 2.Fan J, Krautkramer KA, Feldman JL, Denu JM. Metabolic regulation of histone post-translational modifications. ACS Chem Biol. 2015;10:95–108. doi: 10.1021/cb500846u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3•.Weinert BT, Iesmantavicius V, Wagner SA, Scholz C, Gummesson B, Beli P, Nystrom T, Choudhary C. Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol Cell. 2013;51:265–272. doi: 10.1016/j.molcel.2013.06.003. [This paper shows that the main substrate driving lysine acetylation in E. coli is acetyl-phosphate, whose concentration impacts the overall extent of lysine acetylation. The NAD+-dependent deacetylase cobB removes the lysine acetylation.] [DOI] [PubMed] [Google Scholar]
- 4.Hansford RG. The control of tricarboxylate-cycle of oxidations in blowfly flight muscle. The steady-state concentrations of coenzyme A, acetyl-coenzyme A and succinyl-coenzyme A in flight muscle and isolated mitochondria. Biochem J. 1974;142:509–519. doi: 10.1042/bj1420509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wagner GR, Payne RM. Widespread and enzyme-independent Nepsilon-acetylation and Nepsilon-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J Biol Chem. 2013;288:29036–29045. doi: 10.1074/jbc.M113.486753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olia AS, Barker K, McCullough CE, Tang HY, Speicher DW, Qiu J, LaBaer J, Marmorstein R. Nonenzymatic Protein Acetylation Detected by NAPPA Protein Arrays. ACS Chem Biol. 2015;10:2034–2047. doi: 10.1021/acschembio.5b00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7••.Lee JV, Carrer A, Shah S, Snyder NW, Wei S, Venneti S, Worth AJ, Yuan ZF, Lim HW, Liu S, et al. Akt-dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell Metab. 2014;20:306–319. doi: 10.1016/j.cmet.2014.06.004. [This study shows that Akt activates ATP-citrate lyase and thereby promotes histone acetylation. pAKT(Ser473) significantly correlates with histone acetylation in tumor samples.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swergold GD, Rosen OM, Rubin CS. Hormonal regulation of the phosphorylation of ATP citrate lyase in 3T3-L1 adipocytes. Effects of insulin and isoproterenol. J Biol Chem. 1982;257:4207–4215. [PubMed] [Google Scholar]
- 10.Kamphorst JJ, Chung MK, Fan J, Rabinowitz JD. Quantitative analysis of acetyl-CoA production in hypoxic cancer cells reveals substantial contribution from acetate. Cancer Metab. 2014;2:23. doi: 10.1186/2049-3002-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11•.Comerford SA, Huang Z, Du X, Wang Y, Cai L, Witkiewicz AK, Walters H, Tantawy MN, Fu A, Manning HC, et al. Acetate dependence of tumors. Cell. 2014;159:1591–1602. doi: 10.1016/j.cell.2014.11.020. [This study shows that the cytosolic acetyl-CoA synthetase ACSS2, which is expressed at minimal levels in normal tissues, is more highly expressed across a broad spectrum of tumor samples. A quinoxaline ACSS2 inhibitor has been shown to decrease labeling of histones from labeled acetate.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12••.Sutendra G, Kinnaird A, Dromparis P, Paulin R, Stenson TH, Haromy A, Hashimoto K, Zhang N, Flaim E, Michelakis ED. A nuclear pyruvate dehydrogenase complex is important for the generation of acetyl-CoA and histone acetylation. Cell. 2014;158:84–97. doi: 10.1016/j.cell.2014.04.046. [This paper shows that pyruvate dehydrogenase complex, which is his classically only found in mitochondria, can translocate to the nucleus and make acetyl-CoA, thereby contributing to histone acetylation. The extent of translocation is responsive to growth factors.] [DOI] [PubMed] [Google Scholar]
- 13.Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 14.Casadesus J, Low D. Epigenetic gene regulation in the bacterial world. Microbiol Mol Biol Rev. 2006;70:830–856. doi: 10.1128/MMBR.00016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhutani N, Burns DM, Blau HM. DNA demethylation dynamics. Cell. 2011;146:866–872. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen AT, Zhang Y. The diverse functions of Dot1 and H3K79 methylation. Genes Dev. 2011;25:1345–1358. doi: 10.1101/gad.2057811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sadhu MJ, Guan Q, Li F, Sales-Lee J, Iavarone AT, Hammond MC, Cande WZ, Rine J. Nutritional control of epigenetic processes in yeast and human cells. Genetics. 2013;195:831–844. doi: 10.1534/genetics.113.153981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18•.Mentch SJ, Mehrmohamadi M, Huang L, Liu X, Gupta D, Mattocks D, Gomez Padilla P, Ables G, Bamman MM, Thalacker-Mercer AE, et al. Histone Methylation Dynamics and Gene Regulation Occur through the Sensing of One-Carbon Metabolism. Cell Metab. 2015;22:861–873. doi: 10.1016/j.cmet.2015.08.024. [This study shows that methionine restriction is sufficient to change histone methylation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19••.Shyh-Chang N, Locasale JW, Lyssiotis CA, Zheng Y, Teo RY, Ratanasirintrawoot S, Zhang J, Onder T, Unternaehrer JJ, Zhu H, et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013;339:222–226. doi: 10.1126/science.1226603. [This paper shows that threonine is metabolized into acetyl-CoA and glycine in mouse stem cells, where glycine serves as an important source of one-carbon units, increasing SAM levels and H3K4me3.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20•.Shiraki N, Shiraki Y, Tsuyama T, Obata F, Miura M, Nagae G, Aburatani H, Kume K, Endo F, Kume S. Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab. 2014;19:780–794. doi: 10.1016/j.cmet.2014.03.017. [This paper shows that human stem cells require high amounts of methionine. Methionine deprivation lowers SAM level and causes rapid DNA and histone demethylation.] [DOI] [PubMed] [Google Scholar]
- 21•.Li S, Swanson SK, Gogol M, Florens L, Washburn MP, Workman JL, Suganuma T. Serine and SAM Responsive Complex SESAME Regulates Histone Modification Crosstalk by Sensing Cellular Metabolism. Mol Cell. 2015;60:408–421. doi: 10.1016/j.molcel.2015.09.024. [This paper provides evidence for a multi-protein complex in yeast that comprises pyruvate kinase, serine metabolic enzymes, and SAM synthetases, binds the histone methyl transferase Set1, and contributes to H3T11 phosphorylation and H3K4 methylation.] [DOI] [PubMed] [Google Scholar]
- 22•.Ding W, Smulan LJ, Hou NS, Taubert S, Watts JL, Walker AK. s-Adenosylmethionine Levels Govern Innate Immunity through Distinct Methylation-Dependent Pathways. Cell Metab. 2015 doi: 10.1016/j.cmet.2015.07.013. [This paper shows that the induction of Pseudomonas infection response genes in C. elegans depends on SAM production by SAM synthetase-1 and H3K4 trimethylation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Towbin BD, Gonzalez-Aguilera C, Sack R, Gaidatzis D, Kalck V, Meister P, Askjaer P, Gasser SM. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell. 2012;150:934–947. doi: 10.1016/j.cell.2012.06.051. [DOI] [PubMed] [Google Scholar]
- 24.Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature. 2002;419:174–178. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- 25.Tu BP, Mohler RE, Liu JC, Dombek KM, Young ET, Synovec RE, McKnight SL. Cyclic changes in metabolic state during the life of a yeast cell. Proc Natl Acad Sci U S A. 2007;104:16886–16891. doi: 10.1073/pnas.0708365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26••.Cai L, Sutter BM, Li B, Tu BP. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol Cell. 2011;42:426–437. doi: 10.1016/j.molcel.2011.05.004. [This paper shows that addition of acetate to slow-growing yeast triggers expression of growth genes. The mechanism of gene regulation is activation Gcn5-mediated histone acetylation by acetyl-CoA.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi L, Tu BP. Acetyl-CoA and the regulation of metabolism: mechanisms and consequences. Curr Opin Cell Biol. 2015;33:125–131. doi: 10.1016/j.ceb.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin SJ, Ford E, Haigis M, Liszt G, Guarente L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 2004;18:12–16. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doucette CD, Schwab DJ, Wingreen NS, Rabinowitz JD. alpha-Ketoglutarate coordinates carbon and nitrogen utilization via enzyme I inhibition. Nat Chem Biol. 2011;7:894–901. doi: 10.1038/nchembio.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.You C, Okano H, Hui S, Zhang Z, Kim M, Gunderson CW, Wang YP, Lenz P, Yan D, Hwa T. Coordination of bacterial proteome with metabolism by cyclic AMP signalling. Nature. 2013;500:301–306. doi: 10.1038/nature12446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao L, Chiou W, Tang H, Cheng X, Camp HS, Burns DJ. Simultaneous quantification of malonyl-CoA and several other short-chain acyl-CoAs in animal tissues by ion-pairing reversed-phase HPLC/MS. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;853:303–313. doi: 10.1016/j.jchromb.2007.03.029. [DOI] [PubMed] [Google Scholar]
- 33.Michan S. Calorie restriction and NAD(+)/sirtuin counteract the hallmarks of aging. Front Biosci (Landmark Ed) 2014;19:1300–1319. doi: 10.2741/4283. [DOI] [PubMed] [Google Scholar]
- 34.Cheng Z, Guo S, Copps K, Dong X, Kollipara R, Rodgers JT, Depinho RA, Puigserver P, White MF. Foxo1 integrates insulin signaling with mitochondrial function in the liver. Nat Med. 2009;15:1307–1311. doi: 10.1038/nm.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Braidy N, Guillemin GJ, Mansour H, Chan-Ling T, Poljak A, Grant R. Age related changes in NAD+ metabolism oxidative stress and Sirt1 activity in wistar rats. PLoS One. 2011;6:e19194. doi: 10.1371/journal.pone.0019194. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Yoshino J, Mills KF, Yoon MJ, Imai S. Nicotinamide mononucleotide, a key NAD(+) intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab. 2011;14:528–536. doi: 10.1016/j.cmet.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Canto C, Houtkooper RH, Pirinen E, Youn DY, Oosterveer MH, Cen Y, Fernandez-Marcos PJ, Yamamoto H, Andreux PA, Cettour-Rose P, et al. The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab. 2012;15:838–847. doi: 10.1016/j.cmet.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38•.Kraus D, Yang Q, Kong D, Banks AS, Zhang L, Rodgers JT, Pirinen E, Pulinilkunnil TC, Gong F, Wang YC, et al. Nicotinamide N-methyltransferase knockdown protects against diet-induced obesity. Nature. 2014;508:258–262. doi: 10.1038/nature13198. [The study shows NNMT regulates the availability of both SAM and NAD+ in adipocytes. NNMT knockdown activates polyamine biosynthesis gene expression by increasing H3K4 methylation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ulanovskaya OA, Zuhl AM, Cravatt BF. NNMT promotes epigenetic remodeling in cancer by creating a metabolic methylation sink. Nat Chem Biol. 2013;9:300–306. doi: 10.1038/nchembio.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40•.Shimazu T, Hirschey MD, Newman J, He W, Shirakawa K, Le Moan N, Grueter CA, Lim H, Saunders LR, Stevens RD, et al. Suppression of oxidative stress by beta-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science. 2013;339:211–214. doi: 10.1126/science.1227166. [This study shows that β-hydroxybutyrate is a specific inhibitor of Class I HDACs, and thereby proposes a mechanism by which ketosis can impact epigenetics.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim do Y, Davis LM, Sullivan PG, Maalouf M, Simeone TA, van Brederode J, Rho JM. Ketone bodies are protective against oxidative stress in neocortical neurons. J Neurochem. 2007;101:1316–1326. doi: 10.1111/j.1471-4159.2007.04483.x. [DOI] [PubMed] [Google Scholar]
- 42.Chang P, Zuckermann AM, Williams S, Close AJ, Cano-Jaimez M, McEvoy JP, Spencer J, Walker MC, Williams RS. Seizure control by derivatives of medium chain fatty acids associated with the ketogenic diet show novel branching-point structure for enhanced potency. J Pharmacol Exp Ther. 2014;352:43–52. doi: 10.1124/jpet.114.218768. [DOI] [PubMed] [Google Scholar]
- 43.Selhub J. The many facets of hyperhomocysteinemia: studies from the Framingham cohorts. J Nutr. 2006;136:1726S–1730S. doi: 10.1093/jn/136.6.1726S. [DOI] [PubMed] [Google Scholar]
- 44.Mandaviya PR, Stolk L, Heil SG. Homocysteine and DNA methylation: a review of animal and human literature. Mol Genet Metab. 2014;113:243–252. doi: 10.1016/j.ymgme.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 45.Jiang Y, Sun T, Xiong J, Cao J, Li G, Wang S. Hyperhomocysteinemia-mediated DNA hypomethylation and its potential epigenetic role in rats. Acta Biochim Biophys Sin (Shanghai) 2007;39:657–667. doi: 10.1111/j.1745-7270.2007.00327.x. [DOI] [PubMed] [Google Scholar]
- 46.Geisel J, Schorr H, Bodis M, Isber S, Hubner U, Knapp JP, Obeid R, Herrmann W. The vegetarian lifestyle and DNA methylation. Clin Chem Lab Med. 2005;43:1164–1169. doi: 10.1515/CCLM.2005.202. [DOI] [PubMed] [Google Scholar]
- 47•.Intlekofer AM, Dematteo RG, Venneti S, Finley LW, Lu C, Judkins AR, Rustenburg AS, Grinaway PB, Chodera JD, Cross JR, et al. Hypoxia Induces Production of L-2-Hydroxyglutarate. Cell Metab. 2015;22:304–311. doi: 10.1016/j.cmet.2015.06.023. [The study shows that L-2HG is produced from glutamine-derived α-ketoglutarate by LDHA, and that such production is enhanced in hypoxia. Malate dehydrogenase is a minor producer of L-2HG.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, Leung IK, Li XS, Woon EC, Yang M, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12:463–469. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Xiao MT, Liu LX, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50•.Moussaieff A, Rouleau M, Kitsberg D, Cohen M, Levy G, Barasch D, Nemirovski A, Shen-Orr S, Laevsky I, Amit M, et al. Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab. 2015;21:392–402. doi: 10.1016/j.cmet.2015.02.002. [This paper shows that glucose-derived acetyl-CoA is crucial for histone acetylation and maintaining pluripotency. Acetyl-CoA levels quickly decrease during early differentiation. The primary acetyl-CoA production route is through ATP-citrate lyase. Alternatively, acetate has the ability to delay differentiation by increasing acetyl-CoA availability.] [DOI] [PubMed] [Google Scholar]
- 51.Son MJ, Son MY, Seol B, Kim MJ, Yoo CH, Han MK, Cho YS. Nicotinamide overcomes pluripotency deficits and reprogramming barriers. Stem Cells. 2013;31:1121–1135. doi: 10.1002/stem.1368. [DOI] [PubMed] [Google Scholar]
- 52.Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K, Ding J, Ge Y, Darr H, Chang B, et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145:183–197. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang J, Alexander P, Wu L, Hammer R, Cleaver O, McKnight SL. Dependence of mouse embryonic stem cells on threonine catabolism. Science. 2009;325:435–439. doi: 10.1126/science.1173288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54••.Carey BW, Finley LW, Cross JR, Allis CD, Thompson CB. Intracellular alpha-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature. 2015;518:413–416. doi: 10.1038/nature13981. [This study shows reprogramming of glucose and glutamine metabolism helps to maintain pluripotency through increased α-ketoglutarate levels. Exogenous α-ketoglutarate promotes, whereas succinate inhibits, murine stem cell pluripotency.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morrish F, Noonan J, Perez-Olsen C, Gafken PR, Fitzgibbon M, Kelleher J, VanGilst M, Hockenbery D. Myc-dependent mitochondrial generation of acetyl-CoA contributes to fatty acid biosynthesis and histone acetylation during cell cycle entry. J Biol Chem. 2010;285:36267–36274. doi: 10.1074/jbc.M110.141606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goncalves P, Martel F. Butyrate and colorectal cancer: the role of butyrate transport. Curr Drug Metab. 2013;14:994–1008. doi: 10.2174/1389200211314090006. [DOI] [PubMed] [Google Scholar]
- 57•.Donohoe DR, Collins LB, Wali A, Bigler R, Sun W, Bultman SJ. The Warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Mol Cell. 2012;48:612–626. doi: 10.1016/j.molcel.2012.08.033. [This paper shows that butyrate can turn on histone acetylation and gene expression by producing acetyl-CoA. In cancerous cells, however, Warburg effect causes more glucose usage and less butyrate consumption, increasing histone acetylation by inhibiting HDACs. Two conditions activate different subset of genes, reflecting the different targets of HATs and HDACs.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58•.Plass C, Pfister SM, Lindroth AM, Bogatyrova O, Claus R, Lichter P. Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nat Rev Genet. 2013;14:765–780. doi: 10.1038/nrg3554. [This is an excellent review of epigenetic regulators in cancer.] [DOI] [PubMed] [Google Scholar]
- 59.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Letouze E, Martinelli C, Loriot C, Burnichon N, Abermil N, Ottolenghi C, Janin M, Menara M, Nguyen AT, Benit P, et al. SDH mutations establish a hypermethylator phenotype in paraganglioma. Cancer Cell. 2013;23:739–752. doi: 10.1016/j.ccr.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 64.Xiao M, Yang H, Xu W, Ma S, Lin H, Zhu H, Liu L, Liu Y, Yang C, Xu Y, et al. Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 2012;26:1326–1338. doi: 10.1101/gad.191056.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C, Tsoi J, Clark O, Oldrini B, Komisopoulou E, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340:626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43:869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer. 2013;13:572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nihal M, Wu J, Wood GS. Methotrexate inhibits the viability of human melanoma cell lines and enhances Fas/Fas-ligand expression, apoptosis and response to interferon-alpha: rationale for its use in combination therapy. Arch Biochem Biophys. 2014;563:101–107. doi: 10.1016/j.abb.2014.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thakur BK, Dittrich T, Chandra P, Becker A, Lippka Y, Selvakumar D, Klusmann JH, Reinhardt D, Welte K. Inhibition of NAMPT pathway by FK866 activates the function of p53 in HEK293T cells. Biochem Biophys Res Commun. 2012;424:371–377. doi: 10.1016/j.bbrc.2012.06.075. [DOI] [PubMed] [Google Scholar]