Abstract
High throughput screening has historically been used for drug discovery almost exclusively by the pharmaceutical industry. Due to a significant decrease in costs associated with establishing a high throughput facility and an exponential interest in discovering probes of development and disease associated biomolecules, HTS core facilities have become an integral part of most academic and non-profit research institutions over the past decade. This major shift has led to the development of new HTS methodologies extending beyond the capabilities and target classes used in classical drug discovery approaches such as traditional enzymatic activity-based screens. In this brief review we describe some of the most interesting developments in HTS technologies and methods for chemical probe discovery.
Building a Chemical Toolkit for Probing Biological Networks
Quantifying biomolecules in their natural setting can give a portrait of a biological system; but to elucidate the connectivity in these systems—the intricate relationships between nodes in a network—and to understand the spatial and temporal dynamics of these relationships, we need a means to precisely disrupt the system. Chemical probes, including small molecule modulators of biological function, allow us to selectively perturb individual members of a biological system in a virtually natural state of the system. Using these tools, we can further our understanding of not only the function(s) of our target biomolecule, but also the connectivity of the system as a whole.[1–3]
Depending on the biological question at hand, the search for a small molecule probe generally follows one of two general approaches: reverse chemical genetics or forward chemical genetics.[4] Reverse chemical genetics places emphasis on target engagement and typically involves biochemical or biophysical methods. These types of binding assays assume that a considerable number of compounds that directly bind the target will have some functional effects, which will be confirmed and characterized downstream. Forward chemical genetics places an emphasis on a functional effect or phenotypic consequence in a cellular setting, with the knowledge that the target(s) and mechanism(s) of action of the small molecule will be determined in follow-up studies.
While HTS methods for both target-based and phenotype-based approaches have been around for many years, in the past decade, we have seen the fruits of those technological developments, as well as their application to new target classes. Recent efforts in HTS assay development for chemical probe discovery show a significant effort towards designing more robust assays as well as combinations of well-established HTS principles.[5] Recent efforts in HTS development have not been focused merely on pushing the limits of screening throughput or format miniaturization, but increasing the quality, quantity, and scope of measurable content. Here we discuss some of the most recent probe discovery efforts using novel biochemical and cell-based assay approaches.
Biochemical and Biophysical Assays
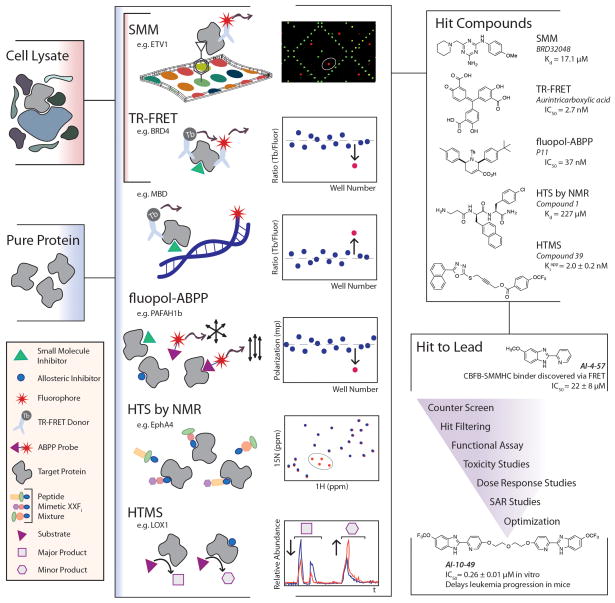
Target-focused screening methods are ideal for discovering small molecule inhibitors of a defined enzymatic activity or a specific bimolecular interaction. These assays typically involve highly purified and structurally stable proteins, but recent efforts show adaptations of these assay formats to accommodate screening target molecules in complex settings, including cell lysate (Figure 1).
Figure 1. Chemical Probe Discovery via High-Throughput Biochemical and Biophysical Assays.
The primary assays for this approach typically require pure protein, but some have been recently modified to accommodate cell lysates to detect small molecules that directly bind a target protein (SMM, TR-FRET Stabilization Assay, HTS by NMR, HTMS) or disrupt bimolecular interactions (TR-FRET, fluopol-ABPP). A schematic representing a molecular view of an active compound (“hit”) in the types of assays outlined in this review and how this event would be distinguished from other events in the data is shown in the center panel. Once hits are identified, the reduction of a large list of primary assay hits through the development of one or more lead probe compounds generally follows the workflow outlined in the bottom-right panel, where the story of AI-10–49 from Illendula et al. serves as a recent and relevant example.
Fluorescence polarization (FP) assays have been around for decades and are highly amenable to high throughput screening for a variety of target classes. Several years ago, this venerated principle for high-throughput binding detection was leveraged to detect functional outcomes, namely inhibition of enzymatic activity through fluorescence polarization-activity-based protein profiling (fluopol-ABPP). [6,7] In what is essentially a high-throughput competition assay, purified, active enzymes are screened against potential inhibitors in the presence of a fluorescence-conjugated natural ligand mimetic, which acts as a competitor molecule and provides an increased FP signal when enzyme activity is uninhibited. Recently, Chang et al., applied this approach in the development of P11 as an inhibitor of two different yet structurally similar platelet-activating factor acetylhydrolases (PAFAH1b2 and PAFAH1b3), which are implicated but relatively uncharacterized in a variety of disease states.[8] Using recombinant mouse PAFAH1b2 and a serine hydrolase specific ABPP probe, a rhodamine-conjugated fluorophosphonate that covalently binds to serine residues in the active site of the enzyme, the authors screened over 300,000 compounds from an NIH curated library with a 0.37% hit rate. After filtering out hits with reported activity in literature and removing false positives identified by using the same ABPP probe in a gel based competition assay, a group of tetrahydropyridines emerged, with one compound, P11, exhibiting the highest potency with an IC50 of 0.8 μM. Follow on studies demonstrate the molecule as an inhibitor of PAFAH1b2 and PAFAH1b3 exclusively and that P11 inhibition of PAFAH1 activity in a cancer cell model results in significantly decreased cell survival.[8] While the exploration of PAFAH1 inhibition as a therapeutic strategy is ongoing, this work demonstrates the incredible scale and utility of the fluopol-ABPP for novel enzymatic probe discovery.
Förster resonance energy transfer (FRET) assays are powerful high throughput techniques used to discover inhibitors or stabilizers of biomolecular interactions.[9] In a recent example, a FRET assay involving purified, full length CBFβ-SMMHC and the purified Runt domain of RUNX1, Illendula et. al. identified the compound AI-4–57, which disrupts the protein-protein interaction (PPI) of these molecules with an IC50 of 22 μM and prevents the acute myeloid leukemia-associated fusion protein CBFβ-SMMHC from modulating the activity of the RUNX1 transcription factor.[10] Further study of this molecule lead to the development of AI-10–49, a multivalent analog that restores normal RUNX1 transcriptional activity and delays leukemia in mice, demonstrating both the power of a chemical probe in testing therapeutic strategies and the utility of a high throughput FRET assay to discover modulators of PPIs with relevant in vivo activity. More recently, FRET methodology has been coupled with time resolved fluorometry in a technique called time-resolved FRET (TR-FRET), which uses the long-lived fluorescence of lanthanide metals as FRET donors to increase signal-to-noise ratio in a FRET assay. Whys et al. use the TR-FRET approach for screening methyl binding domains in order to identify molecules that could interfere with the DNA binding function.[11] Using a purified, His-tagged methyl binding domain of MBD2 (MBD2-MBD) and a terbium-labeled anti-His-tag antibody as the FRET donor and a FAM labeled hairpin-forming C-methylated oligonucleotide as the FRET acceptor, the authors returned 4 hits with IC50 values in the 10–100 nM range from a screen of 1280 commercially available compounds. The authors adapted this screen to serve as a counter screen for specificity by using purified, His-tagged SP1 (a transcription factor) and unmethylated oligos, which showed that the four initial hits, two DNA intercalators and two other bioactives, nonspecifically block protein-oligo interactions. Though no true probes of MBD2-MBD were found in this pilot screen, this proof of concept work demonstrates high-throughput approaches for discovery of probes against a new class of epigenetic regulators through TR-FRET. Recently Schulze et al. have devised a novel use of TR-FRET reagents for a high-throughput protein stabilization assay in cell lysate with another epigenetic systems protein, BRD4, demonstrating proof of concept with a well characterized probe of the protein, JQ1.[12,13] The assay uses the principle that small-molecule bound protein will have increased stability to indirectly detect binding events in live cells through the identification of increased intracellular protein concentration levels. Compound-treated cell lines overexpressing N- and C-terminal epitope tagged BRD4 are lysed with buffer containing TR-FRET antibodies against the N- and C-terminal epitope tags of the protein to enable protein quantification without protein purification, an added advantage for unbiased probe discovery against targets which are difficult to purify.[12]
Small molecule microarrays (SMMs) are another HTS assay equally suited for purified targets and targets in cell lysate, which allows for the screening of a range of targets from the traditional to the challenging. We point the reader to a review of recent successful applications of this technology extensively used in our laboratory for concrete examples, but, will highlight that this approach is especially attractive for screening compounds against difficult targets, such as proteins residing in complexes and intrinsically disordered proteins.[14] For example, transcription factors, many of which are intrinsically disordered when purified away from interacting partners, are attractive as potential therapeutic targets and have been considered ‘undruggable.’ Recently, BRD32048, an inhibitor of a prostate cancer-associated transcription factor, ETV1, was discovered using SMMs screened against cell lysates containing epitope-tagged ETV1.[15,16] Mechanistic studies involving BRD32048 suggested a new avenue for modulating the function of recalcitrant transcriptional targets through inhibition of vital posttranslational events such as acetylation. This story demonstrates that unbiased binding screens involving proteins in cell lysate can give rise to compounds with novel or unexpected mechanisms of modulation from a single screen.
To this point, we have focused on chemical screening approaches that rely on some aspect of fluorescence for detection of binding events. While these approaches are easily scalable to test compound libraries over 106 in size, follow-up assays often reveal significant false positive rates.[17] In reaction to this reality, some have turned to fragment-based screening methods, where hundreds of chemical fragments are screened in a high fidelity biophysical assay, such as nuclear magnetic resonance spectroscopy (NMR). Though fragment-based NMR screening is much lower in throughput and requires considerable downstream chemistry, this technique gives true positives for target binding even for weak binders and, most importantly, continues to see great success in both drug and probe discovery against many diverse targets.[18–20] However, only a handful of years ago, the limits of throughput in a fragment-based screening workflow were challenged in what Wu et. al. term HTS by NMR.[21] In this workflow for discovering antagonists of protein-protein interactions, a peptide mimetic library for screening is assembled based on positional scanning to greatly reduce the amount of NMR runs necessary to test a large number of fragment combinations. The details on the theory behind this approach can be found in the 2013 paper, but, briefly, mixtures of diverse compounds of equal fragment length which share one common fragment in a given position are assembled and tested together in a single reaction mixture, under the assumption that the fixed position fragment is responsible for the majority of the chemical shift detected through NMR. In their proof of concept, this allows a library of 100 fragments representing 106 potential structures to be tested robustly by NMR using just 300 compounds. This technique has the added advantages of reducing the burden of downstream combinatorial chemistry found in typical small molecule fragment library screening campaigns due to the common backbone of fragments in a peptide-mimetic library. Most importantly, the authors demonstrate the utility of this workflow for de novo discovery of three novel, selective, and cellularly active EphA4 receptor inhibitors with IC50 values in the single to double-digit micromolar range using purified EphA4 LBD in an HTS by NMR workflow.
Mass spectrometry (MS) is another label-free technique that has the added advantages of quantitation and identification of target peptides, chemical species, and the substrates and products of peptides under scrutiny. Historically, MS has been reserved for later stages of probe development pipelines, as sample runs typically require liquid chromatography (LC) separation of reaction mixtures and are time intensive processes. Recently, though, MS has been pushed to the front of the discovery pipeline through the availability of new sample processing technology platforms, like RapidFire Solid Phase Extraction (SPE) from Agilent, which eliminates the need for liquid chromatography and is compatible with plate-based formats with rapid sample processing time.[22,23] These high throughput mass spectrometry (HTMS) approaches are gaining more traction in the field where established primary assays are resource intensive or detection limited. For example, while chemical screening of protein methyltransferases (PMTs), an attractive therapeutic target class, through radio filter-binding assays requires hazardous radioactive materials and is unable to provide information related to methylated products (i.e. monomethylated or dimethylated peptides), a recently developed HTMS workflow using RapidFire SPE-MS to analyze reaction mixtures of full length, purified PRMT5 (a PMT), MEP50 (a binding complex partner), and H4 (histone substrate) performs similarly to the RFA gold standard with the added advantage of identifying methylated product formation and quantifying levels of methyl donor states.[24] Obviously, the use of an HTMS platform of this type is not accessible for all groups due to substantial equipment and training investments, but many in the field have devised other ways for overcoming the difficulties of LC in an HTMS workflow. Recently, one group has demonstrated the use of an acid cleavable detergent (sodium 3-(4-(1,1-bis(hexyloxy)ethyl)pyridinium-1-yl)propane-1-sulfonate, or PPS) to enable an HPLC-MS approach for finding lipoxygenase (LOX) inhibitors in a 96-well format. This workflow not only provides greater sensitivity for potent inhibitors below the limit of detection in standard UV assays, but allows monitoring of product formations to enable a directed search for allosteric inhibitors, defined as compounds which force a dose-dependent shift in product:substrate or product:alternative product ratios, which was validated using known allosteric inhibitors of 15-LOX-1 and 15-LOX-2.[25]
Cell-Based Phenotypic Assays
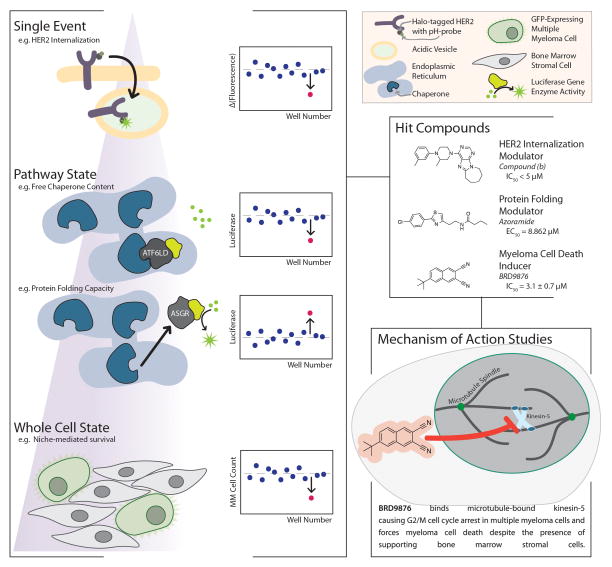
Cell based high-throughput assays evaluate compounds based on their modulation of a precisely defined phenotype often using a luminescence or fluorescence readout (Figure 2). Examples of commonly assayed phenotypes include gene expression (e.g. luciferase reporter assays, Luminex L1000, or fluorescent protein-gene product fusions),[26–28] protein localization (e.g. intracellular vs. extracellular or cytoplasm vs. nucleus),[29–31] or cell morphology (e.g. changes in organelles or the overall cell).[32–35] All of these approaches have enabled identification of small molecules that modulate a given phenotype, but do not directly provide insight into the mechanism of action for a molecule or even the precise target(s) the molecule acts upon. As these approaches are typically target-agnostic, complex studies to identify the direct target(s) and mechanism(s) of action are often required for cell-based HTS assay hits.[36,37] Despite this drawback, these assays provide a powerful means for discovering probe molecules of disease phenotypes and physiological pathways with cellular activity, even without a hypothesis of which nodes in a network should be targeted to induce the phenotype of interest.
Figure 2. Chemical Probe Discovery via High-Throughput Cell-based and Phenotypic Assays.
The primary assays in this approach require a clear definition of phenotype for observation. These phenotypes can range in complexity and are often monitored via fluorescence, as shown in the left panel where a schematic of the phenotype for the assay is shown alongside a depiction of the way in which an active compound in the assay could be distinguished from other events in the data. Once hits are identified, their mechanism of action (the cellular targets they interact with and the consequences of those interactions) must be uncovered and proven in what is typically a lengthy experimental campaign. In the bottom-right panel, we show one example of a proven mechanism of action from a phenotypic screen demonstrated by Chattopadhyay et. al. from their work highlighted in the main text of this review.
Isa et al. recently described a novel HTS assay that exploits a well established biological mechanism of HER2 regulation, the internalization of HER2 for sorting to acidic endosomes and lysosomes for its degradation.[29] The authors used the emerging HaloTag labeling technology in combination with an acidic pH-activatable probe to search from compounds that induce internalization of the receptor. Using 17-AAG, a HSP-90 inhibitor known to induce HER2 internalization, and NH4Cl, which reverses signal of the probe through intravesicular alkalization, the authors validated the assay format and demonstrated minimal nonspecific signal. The assay monitored the change in fluorescence signal between internalized HER2 signal and NH4Cl treatment (ΔF = Fbefore − Fafter) providing an excellent Z factor (>0.5) and high signal-to-noise ratio (44.6). Using a 384-well format the authors screened a library of ~155,000 small molecules to identify small molecules that induced internalization and degradation. They chose to retest the top ~0.1% compounds in quadruplicate, resulting in 16 hits with >15% activity. One compound led to a 50% decrease of HER2 in 1h at 5 μM. This robust assay format can potentially be adapted to other receptors with similar internalization mechanisms and could be considered as a complement to the current antibody-based campaigns seen in drug discovery campaigns.
In another recent HTS report, Tang et al.[38] developed a high-content cell based assay to identify small molecules antagonists of AXL. This assay relies on antibody labeling to quantify pAKT in the presence of compound after stimulation with either GAS6 ligand or MAB154 agonist, screening this receptor tyrosine kinase activity in a cell-based format may present advantages over a biophysical format using purified components, as AXL has unique activation behaviors dependent on spatial presentation of GAS6.[39] The sensitivity of the assay is relatively strong (with a 4-fold pAKT induction in H1299 cell line) and the parallel mechanisms of receptor activation can aid in identifying the hits that inhibit AXL kinase activity rather than GAS6 binding. This screening approach may have the potential to identify novel and specific AXL pathway inhibitors by preventing AXL activation.
Fu et al. demonstrate the value of thoroughly understanding the biological mechanisms governing a phenotypic state when designing a cell-based assay with their recent work related to endoplasmic reticulum (ER) stress.[30] The authors sought to discover small molecules that are able to correct ER stress, a phenomena believed to be involved in a number of human pathologies ranging from diabetes and obesity to cancer and inflammation, by exploiting the trafficking of two ER-associated proteins. In a normal, low stress state ATF6, a transcription factor that controls chaperone gene expression, is repressed by abundant chaperone binding, whereas ASGR1, a membrane-associated protein, is highly produced and secreted. Inversely, a high stress state (reduced folding capacity in the ER) traffics ATF6 out of ER through the Golgi and retains ASGR1 in the ER until chaperone levels are restored. The authors designed two complementary assays where they fused Luciferase to the luminal domain of AFL6 or to the truncated ASRG1 lacking the membrane-anchoring domain, using a second luciferase in both assays as an internal control. Using these two systems to monitor ER stress through luciferase activity, they have identified azoramide, which protects cells from induced ER stress. The authors determined the IC50 for the AFL6 system to be ~1.3 μM and the EC50 for the secreted ASRG1 to be 8.8 μM. This screening campaign was performed in 96-well format, but it should be easily scalable to 384-well format since the amount of media needed for luciferase measurements is relatively small (10 μL).
Finally a recent HTS report uses a niche-based, two-cell culture system to screen compounds against multiple myeloma (MM) cells to identify compounds that overcome stromal resistance, a term used to describe the survival benefit conferred to multiple myeloma cells by neighboring bone marrow stromal cells (BMSC) resulting in resistance to standard chemotherapy.[40] The authors designed a high-throughput assay using GFP-expressing MOLP5 cells co-cultured with primary BMSC. The readout involved monitoring cell viability by measuring changes in fluorescence intensity. Screening a library of ~25,000 compounds they identified BRD9876 (IC50 ~2.2 μM in MM1S cells) that inhibits various MM cell lines, overcomes BMSC-conferred resistance and displays selectivity over hematopoietic progenitors. The authors then employed an extraordinary battery of functional genomic and cellular assay that identified Kinesin-5 (Eg5) as the primary target of BRD9876, which interestingly is inhibitory only on microtubule-bound Eg5. This article is a great example of how cell-based HTS assays need to (i) exploit a defined physiologic phenotype, (ii) have a robust readout parameter, (iii) be complemented by counter screen assays to judge selectivity, and (iv) be followed by target identification studies and mechanism of action studies.
New Frontiers for HTS in Probe Discovery
A successful high-throughput screening campaign can yield multiple high quality hits, but this outcome is largely dependent on the content of the chemical library being screened, therefore placing a high level of importance on library design and selection.[41–43] While there have been many advances in high-throughput screening methods, fewer advances have been made to increase scale and economy for diverse and novel synthetic chemistry for library generation. A noteworthy report published by Santanilla et al. demonstrates the automated use of Rapid MISER (multiple injections in a single run) HPLC-MS to identify optimal reaction conditions for metal-catalyzed coupling of complex electrophiles to highly polar nucleophilic building blocks. The technology enables 1500 nanomole-scale reactions per day.[44]
Selected high-value assay positives identified using HTS can be promoted for development into a high quality probe with sufficient potency and selectivity.[1] This development stage can be cumbersome with the difficulty closely related to target biology and the chemical features of the small molecule. Moreover understanding the mechanism of action is a vital component in understanding the biological outcome. Therefore probe development campaigns are often lengthy and resource intensive. To overcome some of these obstacles, the HTS field must continue the development of HTS assays with higher information content, including those that may combine defined binding events with robust measurable functional activity. Development of physiologically relevant cell-based or biochemical assays will benefit tremendously from the deep understanding of biological activity of the target or pathway in question as well as the fine characterization of a unique disease-specific phenotype. Finally, technologies to assist demonstration of target engagement in cells, evaluating cellular specificity profiles, and uncovering precise mechanism of action are needed to define probe quality and to leverage the probe for systems-level studies.
As new genetic tools continue to develop and large gene expression data sets are being constructed, we witness an exponential increase in using computational means to link a cellular response of a novel molecule to the cellular profiles of known molecules. Using such powerful comparisons will certainly reduce the number of potential targets, prioritize hypothesis testing of mechanism of action and add valuable information related to specificity and potential off-target toxicity.
To further improve the power of primary HTS assays using pure proteins or cell lysates, it would behoove us to explore how we might leverage innovative methods for labeling proteins and other biomolecules using nucleic acid sequences and/or peptide sequences [45–48] possibly allowing a multiplexed screening campaign to detect unique binding events through high fidelity target identification when screening compound collections in the presence of multiple relevant targets.
For phenotypic assays, thinking of integrating target ID into a primary screen might involve bridging compound treatments directly to mass spectrometry (MS) to identify targets engaged by phenotypically active molecules. Chemical moieties for covalent tethering of a molecule to its target or for target pull down upstream of MS analysis are commonly coupled to HTS hit compounds in follow-up studies, but devising clever ways to functionalize a relatively large library with these moieties in a manner that does not bias hits towards more reactive species or prevent target engagement through steric hindrance are worth exploring in high-throughput assay development. [49,50]
In closing, the chemical biology community has made tremendous advances in the area of high throughput screening for chemical probe discovery. We are encouraged by the combinations of established principles to provide higher-content information in primary assays and the adaptations of venerated platforms to accommodate diverse target classes. We expect to see a further increase in the development of higher-content HTS assays. This will significantly influence our campaigns for probe discovery and will further our understanding of unexamined biology for therapeutic development.
HIGHLIGHTS.
We review selected recent developments in chemical high-throughput screening (HTS)
Target-based HTS assays have recently been adapted for new target classes
Cell-based HTS assays rely on the ability to define and quantify complex phenotypes
Both strategies have yielded effective probes for studying uncharacterized biology
Acknowledgments
We thank members of the Koehler lab for useful comments and discussions. This material is based upon work supported by the National Science Foundation Graduate Research Fellowship under Grant No. 1122374. Any opinion, findings, and conclusions or recommendations expressed in this material are those of the authors(s) and do not necessarily reflect the views of the National Science Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schreiber SL, Kotz JD, Li M, Aubé J, Austin CP, Reed JC, Rosen H, White EL, Sklar LA, Lindsley CW, et al. Advancing Biological Understanding and Therapeutics Discovery with Small-Molecule Probes. Cell. 2015;161:1252–1265. doi: 10.1016/j.cell.2015.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huston A, Arrowsmith CH, Knapp S, Schapira M. Probing the epigenome. Nat Chem Biol. 2015;11:542–545. doi: 10.1038/nchembio.1871. [DOI] [PubMed] [Google Scholar]
- 3.Macarron R, Banks MN, Bojanic D, Burns DJ, Cirovic DA, Garyantes T, Green DVS, Hertzberg RP, Janzen WP, Paslay JW, et al. Impact of high-throughput screening in biomedical research. 2011 doi: 10.1038/nrd3368. [DOI] [PubMed] [Google Scholar]
- 4.Stockwell BR. Chemical genetics: ligand-based discovery of gene function. Nat Rev Genet. 2000;1:116–125. doi: 10.1038/35038557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mayr LM, Bojanic D. Novel trends in high-throughput screening. Current Opinion in Pharmacology. 2009;9:580–588. doi: 10.1016/j.coph.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 6.Simon GM, Cravatt BF. Activity-based proteomics of enzyme superfamilies: serine hydrolases as a case study. J Biol Chem. 2010;285:11051–11055. doi: 10.1074/jbc.R109.097600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bachovchin DA, Brown SJ, Rosen H, Cravatt BF. Identification of selective inhibitors of uncharacterized enzymes by high-throughput screening with fluorescent activity-based probes. Nat Biotechnol. 2009;27:387–394. doi: 10.1038/nbt.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8**.Chang JW, Zuhl AM, Speers AE, Niessen S, Brown SJ, Mulvihill MM, Fan YC, Spicer TP, Southern M, Scampavia L, et al. Selective inhibitor of platelet-activating factor acetylhydrolases 1b2 and 1b3 that impairs cancer cell survival. ACS Chem Biol. 2015;10:925–932. doi: 10.1021/cb500893q. The authors demonstrate a fluorescence polarization assay that uses an activity-based protein profiling (ABPP) probe for serine hydrolases as a competitor molecule to discover small molecules that can inhibit ABPP probe binding at the enzyme active site of purified PAFAH1b2 (a platelet-activating factor acetylhydrolase). The most potent hit from the screen proved selective for PAFAH1b2 and PAFAH1b3 and allowed the authors to suggest a functional significance of these cancer-associated genes by showing decreased tumor survival following compound treatment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song Y, Madahar V, Liao J. Development of FRET Assay into Quantitative and High-throughput Screening Technology Platforms for Protein–Protein Interactions. Ann Biomed Eng. 2010;39:1224–1234. doi: 10.1007/s10439-010-0225-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Illendula A, Pulikkan JA, Zong H, Grembecka J, Xue L, Sen S, Zhou Y, Boulton A, Kuntimaddi A, Gao Y, et al. Chemical biology. A small-molecule inhibitor of the aberrant transcription factor CBFβ-SMMHC delays leukemia in mice. Science. 2015;347:779–784. doi: 10.1126/science.aaa0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11*.Wyhs N, Walker D, Giovinazzo H, Yegnasubramanian S, Nelson WG. Time-Resolved Fluorescence Resonance Energy Transfer Assay for Discovery of Small-Molecule Inhibitors of Methyl-CpG Binding Domain Protein 2. J Biomol Screen. 2014;19:1060–1069. doi: 10.1177/1087057114526433. The authors adapt a bimolecular TR-FRET assay to study an epigenetic protein target, methyl-CpG binding domain protein 2 (MBD2), using purified MBD2-MBD and fluorescently labeled methylated oligonucleotide. This proof of concept paper demonstrates outstanding practices in assay development and the utility of an appropriately designed counter screen. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12*.Schulze J, Moosmayer D, Weiske J, Fernandez-Montalvan A, Herbst C, Jung M, Haendler B, Bader B. Cell-Based Protein Stabilization Assays for the Detection of Interactions between Small-Molecule Inhibitors and BRD4. J Biomol Screen. 2015;20:180–189. doi: 10.1177/1087057114552398. The authors demonstrate the use of TR-FRET reagents to quantify increases in cellular protein concentration caused by increased stability through the binding of small-molecules. This assay provides a high-throughput mechanism for finding small molecule binders of proteins that engage their target in live cells. [DOI] [PubMed] [Google Scholar]
- 13.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong JA, Neel DV, Wassaf D, Caballero F, Koehler AN. Recent discoveries and applications involving small-molecule microarrays. Curr Opin Chem Biol. 2014;18:21–28. doi: 10.1016/j.cbpa.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pop MS, Stransky N, Garvie CW, Theurillat J-P, Hartman EC, Lewis TA, Zhong C, Culyba EK, Lin F, Daniels DS, et al. A small molecule that binds and inhibits the ETV1 transcription factor oncoprotein. Mol Cancer Ther. 2014;13:1492–1502. doi: 10.1158/1535-7163.MCT-13-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pop MS, Wassaf D, Koehler AN. Probing small-molecule microarrays with tagged proteins in cell lysates. Curr Protoc Chem Biol. 2014;6:209–220. doi: 10.1002/9780470559277.ch140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inglese J, Johnson RL, Simeonov A, Xia M, Zheng W, Austin CP, Auld DS. High-throughput screening assays for the identification of chemical probes. Nat Chem Biol. 2007;3:466–479. doi: 10.1038/nchembio.2007.17. [DOI] [PubMed] [Google Scholar]
- 18.Barile E, Pellecchia M. NMR-Based Approaches for the Identification and Optimization of Inhibitors of Protein–Protein Interactions. Chem Rev. 2014;114:4749–4763. doi: 10.1021/cr500043b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garavís M, López-Méndez B, Somoza A, Oyarzabal J, Dalvit C, Villasante A, Campos-Olivas R, González C. Discovery of Selective Ligands for Telomeric RNA G-quadruplexes (TERRA) through 19F-NMR Based Fragment Screening. ACS Chem Biol. 2014;9:1559–1566. doi: 10.1021/cb500100z. [DOI] [PubMed] [Google Scholar]
- 20.Lee M-K, Bottini A, Kim M, Bardaro MF, Zhang Z, Pellecchia M, Choi B-S, Varani G. A novel small-molecule binds to the influenza A virus RNA promoter and inhibits viral replication. Chem Commun (Camb) 2014;50:368–370. doi: 10.1039/c3cc46973e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu B, Zhang Z, Noberini R, Barile E, Giulianotti M, Pinilla C, Houghten RA, Pasquale EB, Pellecchia M. HTS by NMR of Combinatorial Libraries: A Fragment-Based Approach to Ligand Discovery. Chem Biol. 2013;20:19–33. doi: 10.1016/j.chembiol.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adam GC, Meng J, Rizzo JM, Amoss A, Lusen JW, Patel A, Riley D, Hunt R, Zuck P, Johnson EN, et al. Use of High-Throughput Mass Spectrometry to Reduce False Positives in Protease uHTS Screens. J Biomol Screen. 2015;20:212–222. doi: 10.1177/1087057114555832. [DOI] [PubMed] [Google Scholar]
- 23.Leveridge M, Buxton R, Argyrou A, Francis P, Leavens B, West A, Rees M, Hardwicke P, Bridges A, Ratcliffe S, et al. Demonstrating Enhanced Throughput of RapidFire Mass Spectrometry through Multiplexing Using the JmjD2d Demethylase as a Model System. J Biomol Screen. 2014;19:278–286. doi: 10.1177/1087057113496276. [DOI] [PubMed] [Google Scholar]
- 24.Maegley KA, Krivacic C, Bingham P, Liu W, Brooun A. Comparison of a High-Throughput Mass Spectrometry Method and Radioactive Filter Binding to Assay the Protein Methyltransferase PRMT5. Assay Drug Dev Technol. 2015;13:235–240. doi: 10.1089/adt.2015.640. [DOI] [PubMed] [Google Scholar]
- 25.Jameson JB, II, Kenyon V, Holman TR. A high-throughput mass spectrometric assay for discovery of human lipoxygenase inhibitors and allosteric effectors. Analytical Biochemistry. 2015;476:45–50. doi: 10.1016/j.ab.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagiec MM, Skepner AP, Negri J, Eichhorn M, Kuperwasser N, Comer E, Muncipinto G, Subramanian A, Clish C, Musunuru K, et al. Modulators of Hepatic Lipoprotein Metabolism Identified in a Search for Small-Molecule Inducers of Tribbles Pseudokinase 1 Expression. PLoS ONE. 2015;10:e0120295–26. doi: 10.1371/journal.pone.0120295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Radomska HS, Jernigan F, Nakayama S, Jorge SE, Sun L, Tenen DG, Kobayashi SS. A Cell-Based High-Throughput Screening for Inducers of Myeloid Differentiation. J Biomol Screen. 2015 doi: 10.1177/1087057115592220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bernardo SM, Allen CP, Waller A, Young SM, Oprea T, Sklar LA, Lee SA. An automated high-throughput cell-based multiplexed flow cytometry assay to identify novel compounds to target Candida albicans virulence-related proteins. PLoS ONE. 2014;9:e110354. doi: 10.1371/journal.pone.0110354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29*.Isa M, Asanuma D, Namiki S, Kumagai K, Kojima H, Okabe T, Nagano T, Hirose K. High-throughput screening system to identify small molecules that induce internalization and degradation of HER2. ACS Chem Biol. 2014;9:2237–2241. doi: 10.1021/cb500654q. The authors demonstrate the use of HaloTag technology to link a pH-sensitive fluorophore to cell surface receptor HER2 to monitor the receptor’s internalization to acidic vesicles in live cells. The assay provides an excellent window for detection demonstrated with a known HER2 internalization agonist and an alkalizing agent to influence the level of fluorescence from the acid-sensitive pH-probe. [DOI] [PubMed] [Google Scholar]
- 30**.Fu S, Yalcin A, Lee GY, Li P, Fan J, Arruda AP, Pers BM, Yilmaz M, Eguchi K, Hotamisligil GS. Phenotypic assays identify azoramide as a small-molecule modulator of the unfolded protein response with antidiabetic activity. Sci Transl Med. 2015;7:292ra98–292ra98. doi: 10.1126/scitranslmed.aaa9134. The authors harness detailed knowledge of endoplasmic reticulum (ER) biology to design luciferase fusion constructs, which allow for the monitoring of ER stress states in live cells through chemiluminescence. This assay provides a high-throughput mechanism for discovering modulators of ER function in live cells through two distinct, but complementary assays, which are detailed and robustly tested in this proof of concept paper. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burns SM, Vetere A, Walpita D, Dančík V, Khodier C, Perez J, Clemons PA, Wagner BK, Altshuler D. High-Throughput Luminescent Reporter of Insulin Secretion for Discovering Regulators of Pancreatic Beta-Cell Function. Cell Metabolism. 2015;21:126–137. doi: 10.1016/j.cmet.2014.12.010. [DOI] [PubMed] [Google Scholar]
- 32.Hartwell KA, Miller PG, Mukherjee S, Kahn AR, Stewart AL, Logan DJ, Negri JM, Duvet M, sMJARA, Puram R, et al. Niche-based screening identifies small-molecule inhibitors of leukemia stem cells. Nat Chem Biol. 2013;9:834–839. doi: 10.1038/nchembio.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chattopadhyay S, Stewart AL, Mukherjee S, Huang C, Hartwell KA, Miller PG, Subramanian R, Carmody LC, Yusuf RZ, Sykes DB, et al. Niche-Based Screening in Multiple Myeloma Identifies a Kinesin-5 Inhibitor with Improved Selectivity over Hematopoietic Progenitors. Cell Reports. 2015;10:755–770. doi: 10.1016/j.celrep.2015.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stanley SA, Barczak AK, Silvis MR, Luo SS, Sogi K, Vokes M, Bray M-A, Carpenter AE, Moore CB, Siddiqi N, et al. Identification of host-targeted small molecules that restrict intracellular Mycobacterium tuberculosis growth. PLoS Pathog. 2014;10:e1003946. doi: 10.1371/journal.ppat.1003946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang YM, Gupta SK, Kim KJ, Powers BE, Cerqueira A, Wainger BJ, Ngo HD, Rosowski KA, Schein PA, Ackeifi CA, et al. A Small Molecule Screen in Stem-Cell-Derived Motor Neurons Identifies a Kinase Inhibitor as a Candidate Therapeutic for ALS. Stem Cell. 2013;12:713–726. doi: 10.1016/j.stem.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schenone M, Dančík V, Wagner BK, Clemons PA. Target identification and mechanism of action in chemical biology and drug discovery. Nat Chem Biol. 2013;9:232–240. doi: 10.1038/nchembio.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bunnage ME, Chekler ELP, Jones LH. Target validation using chemical probes. Nature Publishing Group. 2013;9:195–199. doi: 10.1038/nchembio.1197. [DOI] [PubMed] [Google Scholar]
- 38.Tang H, Yang J, Shen DR, Calambur D, Witmer M, Wu S, Carpenter B, Zhang Y, Gao M, Constantine K, et al. High-throughput high-content imaging assays for identification and characterization of selective AXL pathway inhibitors. Assay Drug Dev Technol. 2014;12:80–86. doi: 10.1089/adt.2013.540. [DOI] [PubMed] [Google Scholar]
- 39.Meyer AS, Zweemer AJM, Lauffenburger DA. The AXL Receptor Is a Sensor of Ligand Spatial Heterogeneity. Cell Systems. 2015;1:25–36. doi: 10.1016/j.cels.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson KC. Targeted therapy of multiple myeloma based upon tumor-microenvironmental interactions. Exp Hematol. 2007;35:155–162. doi: 10.1016/j.exphem.2007.01.024. [DOI] [PubMed] [Google Scholar]
- 41.Nielsen TE, Schreiber SL. Towards the Optimal Screening Collection: A Synthesis Strategy. Angew Chem Int Ed. 2008;47:48–56. doi: 10.1002/anie.200703073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dandapani S, Marcaurelle LA. Accessing new chemical space for “undruggable” targets. Nat Chem Biol. 2010;6:861–863. doi: 10.1038/nchembio.479. [DOI] [PubMed] [Google Scholar]
- 43.Wawer MJ, Li K, Gustafsdottir SM, Ljosa V, Bodycombe NE, Marton MA, Sokolnicki KL, Bray M-A, Kemp MM, Winchester E, et al. Toward performance-diverse small-molecule libraries for cell-based phenotypic screening using multiplexed high-dimensional profiling. Proc Natl Acad Sci US A. 2014;111:10911–10916. doi: 10.1073/pnas.1410933111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buitrago Santanilla A, Regalado EL, Pereira T, Shevlin M, Bateman K, Campeau L-C, Schneeweis J, Berritt S, Shi Z-C, Nantermet P, et al. Organic chemistry. Nanomole-scale high-throughput chemistry for the synthesis of complex molecules. Science. 2015;347:49–53. doi: 10.1126/science.1259203. [DOI] [PubMed] [Google Scholar]
- 45.Gu L, Li C, Aach J, Hill DE, Vidal M, Church GM. Multiplex single-molecule interaction profiling of DNA-barcoded proteins. Nature. 2014;515:554–557. doi: 10.1038/nature13761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scheuermann J, Neri D. Dual-pharmacophore DNA-encoded chemical libraries. Curr Opin Chem Biol. 2015;26:99–103. doi: 10.1016/j.cbpa.2015.02.021. [DOI] [PubMed] [Google Scholar]
- 47.Blakskjaer P, Heitner T, Hansen NJV. Fidelity by design: Yoctoreactor and binder trap enrichment for small-molecule DNA-encoded libraries and drug discovery. Curr Opin Chem Biol. 2015;26:62–71. doi: 10.1016/j.cbpa.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 48.Zambaldo C, Barluenga S, Winssinger N. PNA-encoded chemical libraries. Curr Opin Chem Biol. 2015;26:8–15. doi: 10.1016/j.cbpa.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 49.Kathman SG, Xu Z, Statsyuk AV. A Fragment-Based Method to Discover Irreversible Covalent Inhibitors of Cysteine Proteases. J Med Chem. 2014;57:4969–4974. doi: 10.1021/jm500345q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Friedman Ohana R, Kirkland TA, Woodroofe CC, Levin S, Uyeda HT, Otto P, Hurst R, Robers MB, Zimmerman K, Encell LP, et al. Deciphering the Cellular Targets of Bioactive Compounds Using a Chloroalkane Capture Tag. ACS Chem Biol. 2015 doi: 10.1021/acschembio.5b00351. [DOI] [PubMed] [Google Scholar]