Abstract
A large number of pharmaceuticals, endogenous metabolites, and environmental chemicals act through covalent mechanisms with protein targets. Yet, their specific interactions with the proteome still remain poorly defined for most of these reactive chemicals. Deciphering direct protein targets of reactive small-molecules is critical in understanding their biological action, off-target effects, potential toxicological liabilities, and development of safer and more selective agents. Chemoproteomic technologies have arisen as a powerful strategy that enable the assessment of proteome-wide interactions of these irreversible agents directly in complex biological systems. We review here several chemoproteomic strategies that have facilitated our understanding of specific protein interactions of irreversibly-acting pharmaceuticals, endogenous metabolites, and environmental electrophiles to reveal novel pharmacological, biological, and toxicological mechanisms.
Introduction
We are exposed to a large number of chemicals that act through covalent mechanisms. These chemicals include pharmaceutical agents that irreversibly inhibit their respective protein targets to treat human diseases, such as Alzheimer’s disease, obesity, pain, and cancer [1–5]. Also included are reactive endogenous metabolites that are formed through metabolism, such as lipid aldehydes and various forms of reactive oxygen species or nitrogen stress. Many pesticides, environmental contaminants, and household chemicals also act through covalent mechanisms [6–10]. While most of these chemicals have undergone standard toxicological testing, the reactivity of these chemicals across the proteome still remains poorly defined. Understanding the selectivity of these reactive agents is of paramount importance in comprehending the mechanisms underlying their biological or therapeutic action, identifying off-target effects that may lead to “idiosyncratic” toxicities, and informing the development of safer and more selective agents.
Over the past several years, there have been major advancements in the development and use of chemoproteomic platforms to determine the proteome-wide interactions of irreversible small-molecule tool compounds, therapeutics, endogenous electrophiles, and environmental chemicals. In this review, we will describe how chemoproteomic technologies have been used to assess both the selectivity of therapeutic agents and the toxicological mechanisms of environmental chemicals.
Chemoproteomic Profiling to Assess Selectivity of Therapeutic Irreversible Small-Molecule Inhibitors
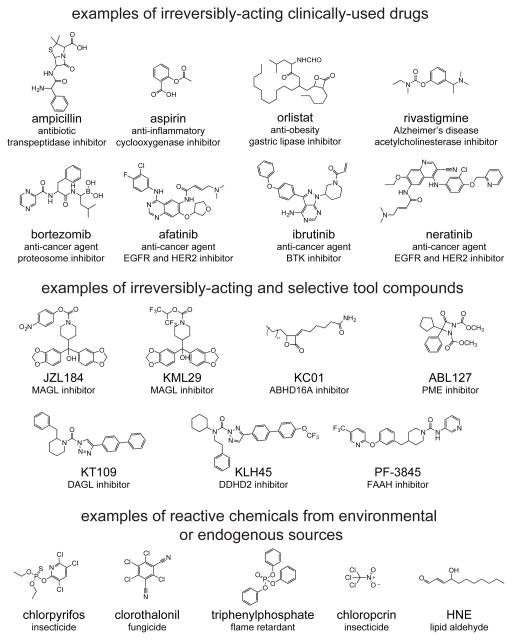
Pharmaceutical companies have historically shied away from pursuing covalent inhibitors due to risks of haptenization and immunologic reactions that may occur through non-specific covalent modification of small-molecules with protein targets [11]. Nonetheless, many irreversible or pseudo-irreversible inhibitors have been successfully developed as well-tolerated drugs in the clinic. Examples include the anti-inflammatory drug aspirin, the broad class of antibacterial beta-lactam antibiotics such as penicillin, drugs that require metabolic bioactivation including the proton pump inhibitor omeprazole, the Alzheimer’s drug rivastigmine that inhibits acetylcholinesterase, the cancer therapy bortezomib (Velcade) that targets the proteosome, and the anti-obesity drug tetrahydrolipstatin (Orlistat) that inhibits gastric lipase [1–3,5]. In recent years, there has been resurgence in developing covalent and irreversible inhibitors, including several acrylamide-based inhibitors that act through Michael addition with a cysteine in the ATP binding pocket of oncogenic kinases for cancer therapy. Some examples include PCI-32765 (ibrutinib), a Bruton’s tyrosine kinase (BTK) inhibitor now FDA approved for mantle cell lymphoma and chronic lymophoblastic leukemia; BIBW-2992 (afatinib) and HKI-272 (neratinib) that dually inhibit human epidermal growth factor receptor 2 (HER2) and epidermal growth factor receptor (EGFR), both of which are approved or in development for non-small cell lung cancer (NSCLC) and breast cancer, respectively; and CO1686 (Rociletinib) that specifically inhibits the mutant T790M form of EGFR and is also currently in development for NSCLC [4].
Although it may be counterintuitive to develop selective inhibitors through reactive and covalent mechanisms, irreversible inhibitors as therapeutics in the modern era of drug discovery and chemical biology afford many benefits. First, covalent inhibitors can provide extended target engagement without the need to maintain high levels of drug. Second, the electrophilicity of the inhibitor can be fine-tuned with the affinity of the small-molecule for a particular binding pocket of a specific protein target, such that the reaction occurs selectively with minimal off-target liabilities. Third, various modern chemoproteomic approaches can be utilized to confirm target engagement and proteome-wide selectivity of covalent inhibitors in situ and in vivo, which can, in-turn, inform further medicinal chemistry efforts to optimize inhibitor properties or to confirm the safety and specificity of lead molecules. We will discuss several examples showcasing the utility of chemoproteomic platforms to define the selectivity of irreversible small-molecule inhibitors and drugs.
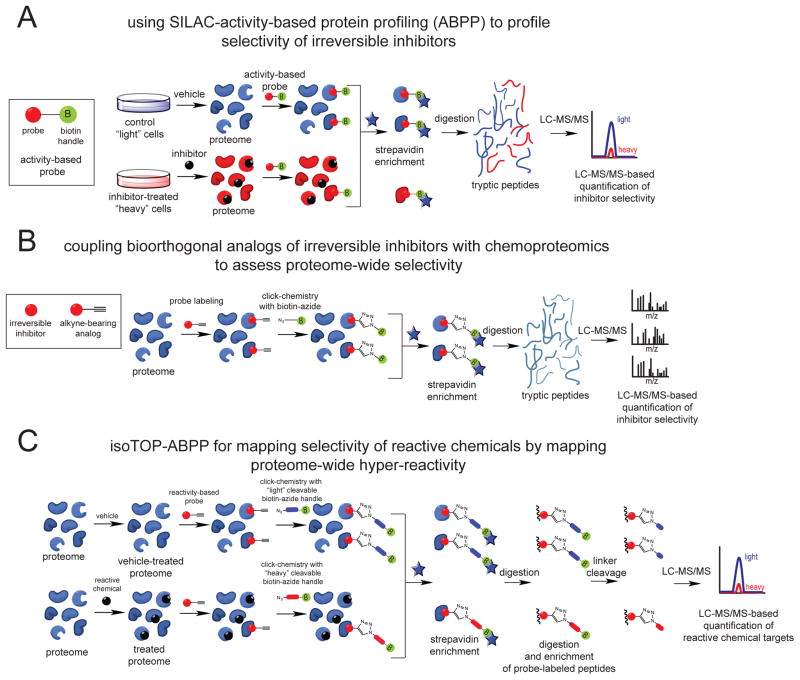
One chemoproteomic platform that has been successfully used to develop selective inhibitors against many protein targets is activity-based protein profiling (ABPP). ABPP uses activity-based chemical probes that directly bind to the active sites of large numbers of enzymes, thus providing a functional readout of enzyme activities en masse directly in complex proteomes [12,13]. Because these activity-based probes bind to the active-sites of enzymes, small-molecule inhibitors can be competed against probe-binding, therefore enabling the development of small-molecules for both characterized and uncharacterized enzymes. Since the activity-based probes evaluate enzyme activities across an entire enzyme class, the proteome-wide selectivity of the small-molecule inhibitor can be assessed within that particular enzyme class. While the ABPP platform has been used to develop selective reversible and irreversible inhibitors of enzymes [14–21], this approach has been particularly useful for testing the efficacy and selectivity of irreversible inhibitors. Target engagement and proteome-wide selectivity can be confirmed for irreversible inhibitors by comparing ex vivo labeling of vehicle and inhibitor-treated proteomes [14,17,18]. The ABPP platform has also been adapted to be compatible with modern quantitative proteomic approaches through stable isotope labeling of cells (SILAC), in which vehicle-treated “light” cells and inhibitor-treated “heavy” cells are combined after labeling with activity-based probes and, subsequently, analyzed for their selectivity by SILAC ratios [20].
Several highly potent, selective, and in vivo active irreversible small-molecule inhibitors that show potential therapeutic benefit have been developed using the ABPP platform. These include monoacylglycerol lipase (MAGL) and diacylglycerol lipase (DAGL) inhibitors that hydrolyze or generate, respectively, the endocannabinoid signaling lipid 2-arachidonoylglycerol (2-AG), and also control arachidonic acid release for pro-inflammatory prostaglandin synthesis [22,23]. The development of selective and in vivo efficacious irreversible MAGL inhibitors, such as JZL184, KML29, and MJN110, have led to the discovery that MAGL blockade leads to heightened 2-AG levels, cannabinoid receptor stimulation, and lower arachidonic acid and pro-inflammatory prostaglandin levels in the brain, thus providing antinociceptive, anti-inflammatory, anxiolytic, and neuroprotective effects [17,24,25]. Hsu et al. developed DAGL inhibitors, such as KT172 and KT109, which have been used to show that DAGL blockade leads to depletion in 2-AG, arachidonic acid, and pro-inflammatory prostaglandin levels to suppress inflammatory cytokine release from macrophages [22].
Hoover et al. used ABPP platforms to show that the obesity drug tetrahydrolipstatin inhibits multiple metabolic enzyme targets in brain, including ABHD12, TPP2, BAT5, and PLA2G7 [3]. Inloes et al. recently discovered that the previously uncharacterized enzyme DDHD2, which is linked to the genetic disorder hereditary spastic paraplegia, was the primary triacylglycerol hydrolase in brain. Using ABPP platforms, the authors developed a selective, in vivo efficacious and irreversible DDHD2 inhibitor, KLH45 [26]. Using this inhibitor alongside genetic DDHD2 knockout mouse models, the authors showed that DDHD2 blockade led to striking accumulations in triacylglycerol levels in the brains of these mice, potentially explaining the metabolic mechanisms underlying the associated neurological disorder. Using ABPP platforms, Kamat et al. developed a highly selective inhibitor, KC01, for a previously uncharacterized enzyme, ABHD16A. They then used this inhibitor to characterize ABHD16A as a phosphatidylserine hydrolase that generates lysophosphatidylserine (LPS) that, in-turn, fuels a neuroinflammatory response [27]. Previous studies showed that another formerly uncharacterized hydrolase ABHD12 is mutationally inactivated in a neurodegenerative disease known as Polyneuropathy, Hearing loss, Ataxia, Retinitis Pigmentosa, and Cataracts (PHARC), leading to accumulation of brain LPS and neuroinflammation [28]. Kamat et al. showed that KC01 lowers the high levels of LPS found in ABHD12-deficient macrophages, leading to suppression of inflammatory cytokine release, indicating that ABHD16A inhibitors may act as anti-inflammatory agents through modulating LPS signaling [27]. Thus, ABPP has been used successfully to develop irreversible small-molecule inhibitors against both characterized and uncharacterized enzymes to further our understanding of the biological and potential therapeutic functions of these enzymes.
Many studies have also developed “clickable” analogs of lead small-molecule therapies or inhibitors of therapeutic targets bearing either alkyne or azide handles for chemoproteomic profiling to confirm the small-molecule’s selectivity or identify any off-targets. Lanning et al. used alkyne-bearing analogs of cysteine-reactive irreversible kinase inhibitors, ibrutinib and PF-6274484, that target BTK and EGFR, respectively, to assess their selectivity in cancer cells using click-chemistry-based chemoproteomic approaches [29]. Cheng et al. developed an alkyne-bearing analog of the widely used inhibitor, C75, of the cancer therapy target fatty acid synthase (FASN), and showed that it possessed many off-targets including CPT1A, GAPDH, and 13 other enzymes beyond FASN, which may explain the high level of toxicity associated with C75 [30]. Bateman et al. developed an aspirin-alkyne probe and coupled the labeling of this probe with chemoproteomic profiling to identify 120 protein targets of aspirin, 112 of which had not been previously reported to be acetylated by aspirin in cellular or in vivo contexts [31]. The authors showed that aspirin-alkyne modified core histone proteins, thus implicating aspirin as a potential chemical-regulator of transcription [31]. The proteome-wide selectivity of many of the serine hydrolase inhibitors that have been developed using ABPP platforms have also been confirmed for their selectivity outside the serine hydrolase family through the development of alkyne-bearing analogs. Examples include the FAAH inhibitor PF-3845 and PF-04457845, the PME inhibitor ABL127, and the MAGL inhibitor MJN110 [24,32–34].
Chemoproteomics has also been used to identify the targets of various natural products through the synthesis of reporter-bearing analogs. Using a Wortmannin analog bearing a tetramethylrhodamine, the conjugate AX7503 was shown to not only bind phosphoinositide 3-kinase (PI3K) and PI3K-related kinases, but also Polo-like kinase 1 (PLK1) [35]. Stephan Sieber’s group synthesized a series of alkyne-bearing β-lactam antibiotic analogs of penicillin, aztreonam, and cephalosporin to label diverse penicillin binding proteins [36]. The authors also synthesized a series of additional β-lactam probes, which labeled and inhibited a selection of penicillin-binding proteins as well as unrelated bacterial targets, including the virulence-associated enzyme ClpP and resistance-associated β-lactamase [36]. Yang et al. synthesized an alkyne-bearing analog of tetrahydrolipstatin (Orlistat), an FDA-approved anti-obesity drug with potential antitumor activities, and identified 8 novel targets of orlistat beyond FASN, including Hsp90AB1, GAPDH, Annexin A2, RPL7a, and RPS9 [37]. In another example, Abegg et al. used an ethynyl benziodoxolones cysteine-reactive probe, JW-RF-010, to identify biological targets of the potential anti-cancer therapy curcumin. The authors identified 42 additional targets of curcumin, only one of which was previously known [38].
Chemoproteomic Profiling of Reactive Environmental Chemicals and Endogenous Reactive Metabolites to Understand Toxicological Mechanisms
We are exposed to countless chemicals, many of which have been linked to adverse health effects, and most of which have not been characterized in terms of their toxicological potential or mechanisms. Of particular concern among chemicals in our environment are reactive electrophiles that we are directly exposed to or those that form through bioactivation, which have the potential to covalently and cumulatively react with nucleophilic amino acid hotspots within the proteome, leading to potential protein dysfunction and pathophysiological effects. Understanding the direct chemical-protein interactions of these reactive agents informs our understanding of downstream molecular, metabolic, and pathophysiological effects that may arise from chemical exposure, and provides a more direct approach towards identifying toxicological drivers of human disease. We will discuss several chemoproteomic approaches that have been successfully applied to understand unique and novel toxicological mechanisms for both environmental chemicals and endogenous reactive metabolites.
ABPP platforms have been used to identify off-targets of widely used organophosphorus (OP) and carbamate pesticides in vivo. These pesticides act as insecticides through inhibiting acetylcholinesterase, but there have been toxicological effects associated with exposure to these agents that cannot be explained by acetylcholinesterase inhibition alone, indicating possible off-targets. Using the serine hydrolase-directed activity-based probe, Nomura et al. and Medina-Cleghorn et al. identified many in vivo off-targets of these pesticides that are inhibited in mice, leading to downstream biochemical effects. For example, several studies have shown that OP pesticides inhibit MAGL and fatty acid amide hydrolase (FAAH) in mouse brain causing elevations in endocannabinoid signaling lipids, 2-AG, and anandamide, all of which lead to downstream cannabinoid-like behavioral effects [6,8,9].
Wang et al. used an elegant quantitative chemoproteomic strategy, termed isotopic tandem orthogonal proteolysis activity-based protein profiling (isoTOP-ABPP), for mapping cysteine reactivity to investigate direct targets and site-of-modifications of lipid aldehydes generated during lipid peroxidation through competition of lipid aldehydes against the cysteine-reactive iodoacetamide-alkyne (IAyne) reactivity-based probe [39,40]. Probe-labeled control and treated proteomes were appended to a biotin-azide analytical handle bearing a light or heavy valine and TEV protease cleavage site using click-chemistry, control and probe labeled proteomes were combined, and probe-labeled tryptic peptides were subsequently enriched and analyzed by quantitative proteomic platforms. Surprisingly, the authors showed that 4-hydroxy-2-nonenal (HNE) interacts with a select set of proteins that constitute hotspots for modifications by various lipid-derived electrophiles, rather than non-specifically reacting with cysteines. For example, they showed that HNE specifically reacts with an active-site proximal cysteine on sterile alpha motif and leucine zipper containing kinase (ZAK), leading to enzyme inhibition creating a negative feedback mechanism that can suppress the activation of c-Jun N-terminal kinase (JNK) pathways induced by oxidative stress [39].
The toxic mechanisms of alkylation by lipid aldehydes were also explored with alkyne-bearing analogs of HNE and 4-oxo-2-nonenal (ONE) coupled with chemoproteomic approaches. The authors showed that HNE and ONE show particular susceptibility towards alkylating protein targets mapping to networks involved in cytoskeletal regulation with low susceptibility towards proteins involved in protein synthesis and turnover. The authors then postulated that the differential sensitivity of protein targets to lipid aldehyde alkylation may protect cells from cytotoxicity as a result of moderate levels of lipid aldehydes [41].
Medina-Cleghorn et al. recently used ABPP approaches to map direct biological targets of several reactive environmental chemicals, including the fungicide chlorothalonil (CTN), the environmental contaminant monomethylarsenous acid (MMA), and a broad-spectrum insecticide chloropicrin [10]. The authors performed in vitro competition of these agents against the cysteine-reactive IAyne reactivity-based probe directly in mouse liver proteomes and found that CTN, MMA, and chloropicrin commonly inhibit several metabolic enzymes involved in fatty acid metabolism and energetic enzymes. The authors further delved into the mechanisms underlying previously reported kidney-specific toxicity associated with CTN through in vivo profiling of CTN targets, and subsequent ex vivo labeling with an alkyne-bearing CTN analog for chemoproteomic discovery of in vivo CTN targets in kidney. The authors showed that CTN inhibits fatty acid transport proteins, fatty acid oxidation enzymes, and glycolytic enzymes in vivo, leading to alterations in kidney lipid metabolism, thus revealing a novel mechanism of toxicity underlying this major fungicide [10].
In another study, Morris et al. used chemoproteomic profiling approaches to comprehensively identify the biological targets of the widely-used flame retardant chemical triphenylphosphate (TPP) by using an alkyne-bearing TPP analog. The authors showed that specific liver carboxylesterases (CES), in particular CES1G, were inhibited by TPP leading to heightened DAG levels and protein kinase C stimulation in liver and serum hypertriglyceridemia [7].
Conclusion
We provide here several examples of chemoproteomic platforms and their applications to assess the selectivity or off-target profiles of tool compounds, therapeutics, and environmental chemicals that act through irreversible mechanisms. Historically, small-molecule agents that act through covalent mechanisms have been feared to cause non-specific adducts on proteins, which, in-turn, may lead to non-specific toxicities and potential haptenization or other types of idiosyncratic toxicities. Certainly, there have been historical examples of highly reactive agents or reactive metabolites that have caused these types of toxicities [11]. However, modern chemoproteomic technologies have provided a more precise and deeper understanding of how reactive chemicals interact with the proteome.
There are indeed reactive chemicals that show large numbers of off-targets. However, chemoproteomic studies have shown that even highly reactive chemicals, such as lipid aldehydes, show relatively selective interactions with specific subsets of targets over others and that these interactions occur oftentimes at hyper-reactive and functional sites on protein targets, rather than non-specific alkylation events. Chemoproteomic profiling of covalently-acting and clinically approved drugs, such as ibrutinib, have revealed potentially large numbers of off-targets at high concentrations. However, studies have shown that these off-target liabilities can be greatly minimized upon even slight chemical modifications of a drug. There have also been a substantial number of highly selective irreversibly-acting small-molecule inhibitors that have been developed in conjunction with technologies such as activity-based proteomics or click-chemistry based chemoproteomic approaches.
Thus, while many pharmaceutical and environmental electrophilic chemicals show potential off-target liabilities, a high degree of selectivity and specificity can still be achieved with irreversible compounds through medicinal chemistry efforts, especially when optimization efforts are coupled with chemoproteomic profiling. Irreversible inhibitors coupled with chemoproteomic platforms also affords substantial advantages for confirming target engagement and in vivo selectivity profiling, which is oftentimes difficult with reversible inhibitors. Thus, the aim of this review is not to disparage the development of irreversible inhibitors, but instead to promote the development of irreversible inhibitors coupled with the application of chemoproteomic platforms to facilitate the development of highly selective and covalent therapeutics or even agrochemicals.
Figure 1.
Examples of irreversibly-acting drugs, tool compounds, environmental chemicals, and endogenous electrophiles.
Figure 2. Chemoproteomic platforms for assessing proteome-wide targets of irreversibly-acting chemicals.
(A) SILAC-ABPP uses active-site directed chemical probes to assess the functional state of large numbers of enzymes directly in complex proteomes. Small-molecule inhibitors can be competed against the binding of activity-based probes to enzymes to assess enzyme class-wide selectivity. Cells can be labeled with light or heavy isotopic amino acids for quantitative proteomic analysis. (B) Analogs of these inhibitors bearing a bioorthogonal handle (e.g. alkyne) can be used to assess proteome-wide selectivity of small-molecule inhibitors using chemoproteomic approaches. (C) Isotopic Tandem Orthogonal Proteolysis-ABPP (isoTOP-ABPP) can be used to map hyper-reactive and functional sites across the proteome using reactivity-based chemical probes bearing bioorthogonal handles (e.g. alkyne). Reactive electrophiles can be competed against probe binding to hyper-reactive sites to map protein targets of these reactive agents. Probe-labeled peptides can be identified through subsequent appending of a biotin-azide analytical handle bearing a TEV protease recognition sequence and heavy or light isotopic valine tag using copper-catalyzed click chemistry. Upon mixing control and treated proteomes, probe-labeled proteins can be avidin-enriched, tryptically digested, and probe-labeled peptides can be subsequently enriched and released by TEV protease for subsequent quantitative proteomic analysis.
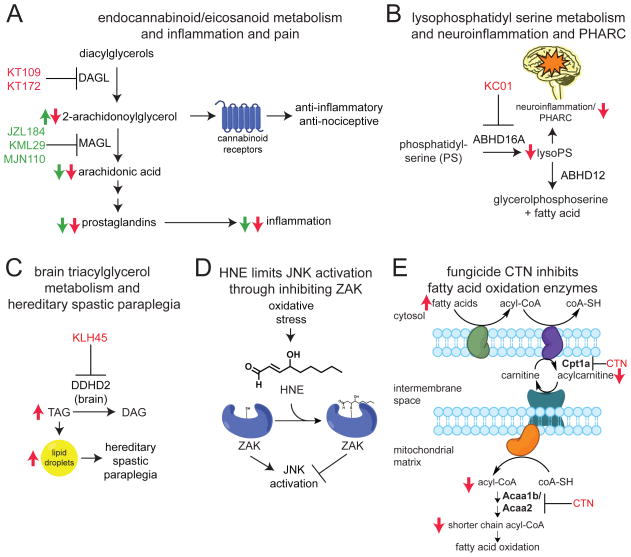
Figure 3. Biological insights gained from using chemoproteomic platforms.
(A) ABPP has been successfully used to develop selective small-molecule inhibitors against enzymes involved the synthesis and degradation of the endocannabinoid 2-arachidonoylglycerol (2-AG). Selective DAGL inhibitors KT109 and KT172 and selective MAGL inhibitors JZL184, KML29, and MJN110 have been used to not only identify that DAGL and MAGL regulate endocannabioid metabolism and signaling, but also to show that this pathway controls arachidonic metabolism that fuels pro-inflammatory prostaglandin synthesis. (B) ABPP was used to develop the selective DDHD2 inhibitor KLH45, which was used to show that the previously uncharacterized enzyme DDHD2 was the primary triacylglycerol (TAG) hydrolase in the brain, and that inhibiting this enzyme led to accumulation in brain triacylglycerol levels and accumulation of lipid droplets. (C) ABPP was used to develop the selective ABHD16A inhibitor KC01 to determine that ABHD16A was the primary phosphatidylserine (PS) hydrolase that generates the pro-inflammatory signaling lipid lyso-PS, which is in-turn hydrolyzed by ABHD12. Previous studies had shown that ABHD12 inactivation caused a neurodegenerative disease known as PHARC. ABHD16A inhibition protected mice from the neurotoxicological markers associated with PHARC. (D) Lipid aldehydes such as HNE were competed against the cysteine-reactive iodoacetamide-alkyne (IAyne) probe and coupled to the isoTOP-ABPP platform to map the direct protein targets of HNE. HNE showed selective interactions with certain sites such as the active-site proximal cysteine of ZAK, leading to ZAK inhibition and JAK inactivation. (E) Reactive environmental chemicals, such as the fungicide chlorothalonil (CTN) were competed against IAyne to map direct protein targets of these chemicals, leading to the discovery that CTN binds to and inhibits multiple enzymes involved in fatty acid oxidation.
Highlights.
Many pharmaceuticals and environmental chemicals act through covalent mechanisms.
Deciphering direct targets of reactive chemicals is important in mitigating toxicity
Chemoproteomics enables selectivity assessment of irreversibly-acting chemicals
Acknowledgments
This work was supported by the Searle Scholar Award, the Center for Environmental Research on Toxics, the National Institutes of Health (P42ES004705; R01CA172667), the American Cancer Society Research Scholar Award (RSG-14-242-01-TBE), the DOD Breakthroughs Award (CDMRP W81XWH-15-1-0050), and the NSF Graduate Fellowship Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Beck P, Dubiella C, Groll M. Covalent and non-covalent reversible proteasome inhibition. Biological chemistry. 2012;393(10):1101–1120. doi: 10.1515/hsz-2012-0212. [DOI] [PubMed] [Google Scholar]
- 2.Colovic MB, Krstic DZ, Lazarevic-Pasti TD, Bondzic AM, Vasic VM. Acetylcholinesterase inhibitors: Pharmacology and toxicology. Current neuropharmacology. 2013;11(3):315–335. doi: 10.2174/1570159X11311030006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoover HS, Blankman JL, Niessen S, Cravatt BF. Selectivity of inhibitors of endocannabinoid biosynthesis evaluated by activity-based protein profiling. Bioorganic & medicinal chemistry letters. 2008;18(22):5838–5841. doi: 10.1016/j.bmcl.2008.06.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Q, Sabnis Y, Zhao Z, Zhang T, Buhrlage SJ, Jones LH, Gray NS. Developing irreversible inhibitors of the protein kinase cysteinome. Chemistry & biology. 2013;20(2):146–159. doi: 10.1016/j.chembiol.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Potashman MH, Duggan ME. Covalent modifiers: An orthogonal approach to drug design. Journal of medicinal chemistry. 2009;52(5):1231–1246. doi: 10.1021/jm8008597. [DOI] [PubMed] [Google Scholar]
- 6.Medina-Cleghorn D, Heslin A, Morris PJ, Mulvihill MM, Nomura DK. Multidimensional profiling platforms reveal metabolic dysregulation caused by organophosphorus pesticides. ACS chemical biology. 2014;9(2):423–432. doi: 10.1021/cb400796c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morris PJ, Medina-Cleghorn D, Heslin A, King SM, Orr J, Mulvihill MM, Krauss RM, Nomura DK. Organophosphorus flame retardants inhibit specific liver carboxylesterases and cause serum hypertriglyceridemia. ACS chemical biology. 2014;9(5):1097–1103. doi: 10.1021/cb500014r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nomura DK, Blankman JL, Simon GM, Fujioka K, Issa RS, Ward AM, Cravatt BF, Casida JE. Activation of the endocannabinoid system by organophosphorus nerve agents. Nature chemical biology. 2008;4(6):373–378. doi: 10.1038/nchembio.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nomura DK, Casida JE. Activity-based protein profiling of organophosphorus and thiocarbamate pesticides reveals multiple serine hydrolase targets in mouse brain. Journal of agricultural and food chemistry. 2011;59(7):2808–2815. doi: 10.1021/jf101747r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **10.Medina-Cleghorn D, Bateman LA, Ford B, Heslin A, Fisher KJ, Dalvie ED, Nomura DK. Mapping proteome-wide targets of environmental chemicals using reactivity-based chemoproteomic platforms. Chemistry & biology. 2015;22(10):1394–1405. doi: 10.1016/j.chembiol.2015.09.008. This paper uses reactivity-based chemoproteomic strategies to map proteome-wide targets of highly used reactive environmental chemicals and contaminants and shows that metabolic enzymes involved in fatty acid metabolism are directly adducted on hyper-reactive cysteines, inhibit their activity, and cause abberant lipid metabolism in vivo. This study shows the utility of using chemoproteomic platforms to identify novel toxicological mechanisms of environmental chemicals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uetrecht J. Idiosyncratic drug reactions: Past, present, and future. Chemical research in toxicology. 2008;21(1):84–92. doi: 10.1021/tx700186p. [DOI] [PubMed] [Google Scholar]
- 12.Medina-Cleghorn D, Nomura DK. Exploring metabolic pathways and regulation through functional chemoproteomic and metabolomic platforms. Chemistry & biology. 2014;21(9):1171–1184. doi: 10.1016/j.chembiol.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nomura DK, Dix MM, Cravatt BF. Activity-based protein profiling for biochemical pathway discovery in cancer. Nature reviews Cancer. 2010;10(9):630–638. doi: 10.1038/nrc2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang JW, Nomura DK, Cravatt BF. A potent and selective inhibitor of kiaa1363/aadacl1 that impairs prostate cancer pathogenesis. Chemistry & biology. 2011;18(4):476–484. doi: 10.1016/j.chembiol.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang JW, Zuhl AM, Speers AE, Niessen S, Brown SJ, Mulvihill MM, Fan YC, Spicer TP, Southern M, Scampavia L, Fernandez-Vega V, et al. Selective inhibitor of platelet-activating factor acetylhydrolases 1b2 and 1b3 that impairs cancer cell survival. ACS chemical biology. 2015;10(4):925–932. doi: 10.1021/cb500893q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leung D, Hardouin C, Boger DL, Cravatt BF. Discovering potent and selective reversible inhibitors of enzymes in complex proteomes. Nature biotechnology. 2003;21(6):687–691. doi: 10.1038/nbt826. [DOI] [PubMed] [Google Scholar]
- 17.Long JZ, Li W, Booker L, Burston JJ, Kinsey SG, Schlosburg JE, Pavon FJ, Serrano AM, Selley DE, Parsons LH, Lichtman AH, et al. Selective blockade of 2-arachidonoylglycerol hydrolysis produces cannabinoid behavioral effects. Nature chemical biology. 2009;5(1):37–44. doi: 10.1038/nchembio.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long JZ, Nomura DK, Vann RE, Walentiny DM, Booker L, Jin X, Burston JJ, Sim-Selley LJ, Lichtman AH, Wiley JL, Cravatt BF. Dual blockade of faah and magl identifies behavioral processes regulated by endocannabinoid crosstalk in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(48):20270–20275. doi: 10.1073/pnas.0909411106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adibekian A, Martin BR, Chang JW, Hsu KL, Tsuboi K, Bachovchin DA, Speers AE, Brown SJ, Spicer T, Fernandez-Vega V, Ferguson J, et al. Probe reports from the nih molecular libraries program. Bethesda (MD): 2010. Characterization of a selective, reversible inhibitor of lysophospholipase 2 (lypla2) [PubMed] [Google Scholar]
- 20.Adibekian A, Martin BR, Wang C, Hsu KL, Bachovchin DA, Niessen S, Hoover H, Cravatt BF. Click-generated triazole ureas as ultrapotent in vivo-active serine hydrolase inhibitors. Nature chemical biology. 2011;7(7):469–478. doi: 10.1038/nchembio.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bachovchin DA, Ji T, Li W, Simon GM, Blankman JL, Adibekian A, Hoover H, Niessen S, Cravatt BF. Superfamily-wide portrait of serine hydrolase inhibition achieved by library-versus-library screening. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(49):20941–20946. doi: 10.1073/pnas.1011663107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu KL, Tsuboi K, Adibekian A, Pugh H, Masuda K, Cravatt BF. Daglbeta inhibition perturbs a lipid network involved in macrophage inflammatory responses. Nature chemical biology. 2012;8(12):999–1007. doi: 10.1038/nchembio.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nomura DK, Morrison BE, Blankman JL, Long JZ, Kinsey SG, Marcondes MC, Ward AM, Hahn YK, Lichtman AH, Conti B, Cravatt BF. Endocannabinoid hydrolysis generates brain prostaglandins that promote neuroinflammation. Science. 2011;334(6057):809–813. doi: 10.1126/science.1209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niphakis MJ, Johnson DS, Ballard TE, Stiff C, Cravatt BF. O-hydroxyacetamide carbamates as a highly potent and selective class of endocannabinoid hydrolase inhibitors. ACS chemical neuroscience. 2012;3(5):418–426. doi: 10.1021/cn200089j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang JW, Niphakis MJ, Lum KM, Cognetta AB, 3rd, Wang C, Matthews ML, Niessen S, Buczynski MW, Parsons LH, Cravatt BF. Highly selective inhibitors of monoacylglycerol lipase bearing a reactive group that is bioisosteric with endocannabinoid substrates. Chemistry & biology. 2012;19(5):579–588. doi: 10.1016/j.chembiol.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inloes JM, Hsu KL, Dix MM, Viader A, Masuda K, Takei T, Wood MR, Cravatt BF. The hereditary spastic paraplegia-related enzyme ddhd2 is a principal brain triglyceride lipase. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(41):14924–14929. doi: 10.1073/pnas.1413706111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *27.Kamat SS, Camara K, Parsons WH, Chen DH, Dix MM, Bird TD, Howell AR, Cravatt BF. Immunomodulatory lysophosphatidylserines are regulated by abhd16a and abhd12 interplay. Nature chemical biology. 2015;11(2):164–171. doi: 10.1038/nchembio.1721. This paper uses ABPP platforms to develop selective inhibitors against an uncharacterized enzyme ABHD16A. Using this selective inhibitor, the authors discover that ABHD16A is a phosphatidyl serine hydrolase and that inhibiting ABHD16A protects against the neuroinflammatory insults conferred by inactivation of ABHD12, a lysophsohatidyl serine hydrolase. This paper is a recent example of how ABPP can be used to develop selective inhibitors of enzyme targets. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blankman JL, Long JZ, Trauger SA, Siuzdak G, Cravatt BF. Abhd12 controls brain lysophosphatidylserine pathways that are deregulated in a murine model of the neurodegenerative disease pharc. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(4):1500–1505. doi: 10.1073/pnas.1217121110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **29.Lanning BR, Whitby LR, Dix MM, Douhan J, Gilbert AM, Hett EC, Johnson TO, Joslyn C, Kath JC, Niessen S, Roberts LR, et al. A road map to evaluate the proteome-wide selectivity of covalent kinase inhibitors. Nature chemical biology. 2014;10(9):760–767. doi: 10.1038/nchembio.1582. This paper uses a bioorthogonal chemoproteomic strategy to show that clinically used covalent kinase inhibitors such as ibrutinib show substantial off-target reactivity at higher concentrations. This study also shows that minor structural modifications confer substantially improved selectivity, indicating that selectivity can be achieved with covalent inhibitors when medicinal chemistry efforts are combined with chemoproteomic profiling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng X, Li L, Uttamchandani M, Yao SQ. In situ proteome profiling of c75, a covalent bioactive compound with potential anticancer activities. Organic letters. 2014;16(5):1414–1417. doi: 10.1021/ol500206w. [DOI] [PubMed] [Google Scholar]
- 31.Bateman LA, Zaro BW, Miller SM, Pratt MR. An alkyne-aspirin chemical reporter for the detection of aspirin-dependent protein modification in living cells. Journal of the American Chemical Society. 2013;135(39):14568–14573. doi: 10.1021/ja408322b. [DOI] [PubMed] [Google Scholar]
- 32.Ahn K, Johnson DS, Mileni M, Beidler D, Long JZ, McKinney MK, Weerapana E, Sadagopan N, Liimatta M, Smith SE, Lazerwith S, et al. Discovery and characterization of a highly selective faah inhibitor that reduces inflammatory pain. Chemistry & biology. 2009;16(4):411–420. doi: 10.1016/j.chembiol.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahn K, Smith SE, Liimatta MB, Beidler D, Sadagopan N, Dudley DT, Young T, Wren P, Zhang Y, Swaney S, Van Becelaere K, et al. Mechanistic and pharmacological characterization of pf-04457845: A highly potent and selective fatty acid amide hydrolase inhibitor that reduces inflammatory and noninflammatory pain. The Journal of pharmacology and experimental therapeutics. 2011;338(1):114–124. doi: 10.1124/jpet.111.180257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bachovchin DA, Mohr JT, Speers AE, Wang C, Berlin JM, Spicer TP, Fernandez-Vega V, Chase P, Hodder PS, Schurer SC, Nomura DK, et al. Academic cross-fertilization by public screening yields a remarkable class of protein phosphatase methylesterase-1 inhibitors. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(17):6811–6816. doi: 10.1073/pnas.1015248108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, Shreder KR, Gai W, Corral S, Ferris DK, Rosenblum JS. Wortmannin, a widely used phosphoinositide 3-kinase inhibitor, also potently inhibits mammalian polo-like kinase. Chemistry & biology. 2005;12(1):99–107. doi: 10.1016/j.chembiol.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 36.Staub I, Sieber SA. Beta-lactams as selective chemical probes for the in vivo labeling of bacterial enzymes involved in cell wall biosynthesis, antibiotic resistance, and virulence. Journal of the American Chemical Society. 2008;130(40):13400–13409. doi: 10.1021/ja803349j. [DOI] [PubMed] [Google Scholar]
- 37.Yang PY, Liu K, Ngai MH, Lear MJ, Wenk MR, Yao SQ. Activity-based proteome profiling of potential cellular targets of orlistat--an fda-approved drug with anti-tumor activities. Journal of the American Chemical Society. 2010;132(2):656–666. doi: 10.1021/ja907716f. [DOI] [PubMed] [Google Scholar]
- 38.Abegg D, Frei R, Cerato L, Prasad Hari D, Wang C, Waser J, Adibekian A. Proteome-wide profiling of targets of cysteine reactive small molecules by using ethynyl benziodoxolone reagents. Angewandte Chemie. 2015;54(37):10852–10857. doi: 10.1002/anie.201505641. [DOI] [PubMed] [Google Scholar]
- **39.Wang C, Weerapana E, Blewett MM, Cravatt BF. A chemoproteomic platform to quantitatively map targets of lipid-derived electrophiles. Nature methods. 2014;11(1):79–85. doi: 10.1038/nmeth.2759. This manuscript uses isoTOP-ABPP platforms to map proteome-wide interactions of reactive lipid aldehydes. The study shows that lipid aldehydes such as HNE specifically modify hyper-reactive cysteines on specific protein targets such as ZAK to modulate its activity and downstream JNK signaling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MB, Bachovchin DA, Mowen K, Baker D, Cravatt BF. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468(7325):790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Codreanu SG, Ullery JC, Zhu J, Tallman KA, Beavers WN, Porter NA, Marnett LJ, Zhang B, Liebler DC. Alkylation damage by lipid electrophiles targets functional protein systems. Molecular & cellular proteomics : MCP. 2014;13(3):849–859. doi: 10.1074/mcp.M113.032953. [DOI] [PMC free article] [PubMed] [Google Scholar]