Abstract
Dun is a wild-type coat color in horses characterized by pigment dilution with a striking pattern of dark areas termed primitive markings. Here we show that pigment dilution in Dun horses is due to radially asymmetric deposition of pigment in the growing hair caused by localized expression of the T-box 3 (TBX3) transcription factor in hair follicles, which in turn determines the distribution of hair follicle melanocytes. Most domestic horses are non-dun, a more intensely pigmented phenotype caused by regulatory mutations impairing TBX3 expression in the hair follicle, resulting in a more circumferential distribution of melanocytes and pigment granules in individual hairs. We identified two different alleles (non-dun1 and non-dun2) causing non-dun color. non-dun2 is a recently derived allele, whereas the Dun and non-dun1 alleles are found in ancient horse DNA, demonstrating that this polymorphism predates horse domestication. These findings uncover a new developmental role for T-box genes and new aspects of hair follicle biology and pigmentation.
The Dun coat color phenotype in horses is characterized by pigmentary dilution affecting most of the body hair, leaving areas with undiluted pigment in a variable pattern, with the most common feature being a dark dorsal stripe. This stripe and other Dun pattern elements are termed primitive markings (Fig. 1a, Online Methods and Supplementary Fig. 1). Most domestic horses, including the individual used for the genome assembly1, are non-dun, with little or no pigment dilution and a faint or absent dorsal stripe. The Dun coat color is presumed to be wild type, as the Przewalski’s horse, a close relative of the ancestor of domestic horses2,3, exhibits Dun color, as do other wild equids—the kiang, onager and African wild ass, as well as the quagga, a now extinct subspecies of plains zebra. The phylogenetic distribution of the Dun phenotype and the reduced pigment intensity of Dun horses (Supplementary Fig. 1) suggest that Dun coloring serves an important camouflage role in equids.
Figure 1.
Phenotypic characterization. (a) Three horses with different genotypes at the Dun g locus on a similar pigmentary (E/−; a/a) background, including a blue Dun (D/−) horse, a black horse with primitive markings (d1/d1) and a black horse without primitive markings (d2/d2). Photographs by Freyja Imsland and Páll Imsland. (b) Cross-sections of hairs from the croup of the three horses in a. (c) Skin and hair section stained with hematoxylin and eosin from a Dun horse. (d) Hair sections stained with hematoxylin and eosin from Dun and non-dun horses. (e) Color intensity differences across the diameter of the hair cortex (means ± s.e.m.) as shown in the inset for the croup and dorsal midline in each phenotype (n = 6 Dun and n = 6 non-dun). **P < 0.01 for Dun versus non-dun croup, two-tailed t test. (f) Cladogram of the Equidae family (based on ref. 10); species with hair histology in g are shown in bold. (g) Photographs of Przewalski’s horses (Equus ferus przewalskii) and Somali wild ass (Equus africanus somaliensis) (left) and photomicrographs of transverse sections through dilute-colored flank hairs (right). Photographs by Waltraut Zimmerman and the St. Louis Zoo. Scale bars, 35 μm (b), 100 μm (c), 35 μm (d), 10 μm (e) and 50 μm (g).
Dun (D) is fully dominant over non-dun (d) (ref. 4). However, the corresponding phenotypes are sometimes misclassified because some non-dun horses exhibit faint primitive markings and may appear superficially similar to Dun horses, especially if mutations in other pigment dilution genes are present4 (Fig. 1a and Supplementary Fig. 2).
Here we show that in horses the TBX3 gene (encoding the T-box 3 transcription factor) is normally expressed in a pattern resulting in the Dun phenotype and that regulatory mutations specifically impairing TBX3 expression in the hair follicle cause non-dun coat color. In humans, heterozygosity for loss-of-function mutations in TBX3 causes a well-recognized pattern of developmental defects, ulnar-mammary syndrome, with abnormalities in limb, apocrine gland, tooth and genital development5. Experimental studies of Tbx3 in mice have provided insight into the mechanism of these abnormalities6,7, but TBX3 has not previously been implicated in pigmentation.
RESULTS
Dun color is caused by asymmetric deposition of hair pigment
Microscopic examination of dilute-colored hairs from the dorsal hindquarters (croup; Supplementary Fig. 2a) of Dun horses showed a striking reduction in pigment in a stereotyped, radially asymmetric pattern (Fig. 1b–e). In sections perpendicular to the hair shaft, pigment granules in dilute hairs from the croup were limited to approximately 25–50% of the cortex (Fig. 1b, left). By contrast, pigment granules in dorsal stripe hairs from Dun individuals (Supplementary Fig. 2a) and in both croup and dorsal midline hairs from non-dun individuals (Fig. 1b and Supplementary Fig. 2a) are more evenly dispersed throughout the hair cortex. A similar observation was described by Gremmel8 more than 75 years ago as pigment granule crowding or clumping but has not been otherwise investigated with regard to the underlying mechanisms.
Asymmetric pigment distribution in dilute hairs was also apparent in histological sections of skin, with the most intensely pigmented area lying on the outward-facing side of the hair (Fig. 1c). Furthermore, examination of longitudinal sections of anagen hair follicles showed that the asymmetry in pigmentation begins in the hair bulb (Fig. 1d) and therefore arises during or before melanin synthesis rather than after pigment deposition.
We also examined pigment distribution in hairs from other equids (Fig. 1f,g and Supplementary Table 1). Przewalski’s horse exhibits a Dun phenotype with a dilute coat color and primitive markings, including a dark dorsal stripe. As in Dun domestic horses, dilute hairs from Przewalski’s horses exhibit asymmetric pigmentation, whereas dorsal stripe hairs are uniformly pigmented. The African wild ass, which diverged from the domestic horse more than 4 million years ago2, also has a Dun phenotype with especially prominent primitive markings on the legs and asymmetric hair pigmentation (Fig. 1a,g).
non-dun is caused by noncoding TBX3 mutations
We first mapped the Dun locus to a region on horse chromosome 8 (chr. 8: 18,061,745–18,482,196) using microsatellite markers and then fine-mapped the locus with a 27-SNP panel to a 200-kb region containing only one gene, TBX3 (Fig. 2a). In a recent study of the Gait-keeper mutation in horses9, we used a non-dun and a heterozygous Dun horse for whole-genome resequencing, with each horse sequenced to about 30× coverage with paired-end reads. A careful examination of the associated region identified partially aligned (soft-clipped) and unpaired reads in the Dun individual in an area where the reads from the non-dun individual aligned well (Fig. 2b), indicating the presence of a structural variant about 5 kb downstream of TBX3. PCR amplification and Sanger sequencing showed that the Dun haplotype has 1,617 bp that is missing from the EquCab2 assembly, which is based on a non-dun horse. The missing sequence consists of nearly contiguous 1,609-bp and 8-bp segments (Fig. 2c). The Dun-specific sequence shows 86% identity to the corresponding human sequence. Thus, the Dun allele is representative of the ancestral state, and the deletion present on the sequenced non-dun chromosomes, as well as in the reference assembly, is derived.
Figure 2.
Genetic analysis. (a) Association analysis (χ2) of the Dun phenotype with SNP genotypes at chr. 8: 18,061,745–18,482,196. (b) Read alignments from whole-genome sequencing of a Dun heterozygote and a non-dun homozygote. Red borders denote a read pair where one of the reads is unmapped; blue segments represent soft-clipped parts of reads where part of the read cannot be aligned. The position of the deletion downstream of TBX3 at chr. 8: 18,227,267–18,227,279 is indicated, and the extent of sequence conservation is illustrated using human genome annotation. (c) Alignment of the Dun, non-dun1 and non-dun2 alleles at the deletion breakpoints, showing how the deletion breakpoint closer to TBX3 is flanked by an additional deletion of 8 bp in non-dun2; there is a 1-bp indel polymorphism between Dun and non-dun1. (d) SNP association analysis (−log10 (P value), two-tailed Fisher’s exact test) between Dun and non-dun1 haplotypes, including both domestic and Przewalski’s horses. SNP1 and SNP2 are marked with a black arrow. Red dots mark candidate SNPs selected on the basis of allele frequencies of more than 50% in Dun individuals (dominant trait) and less than 5% in non-dun individuals (erroneous calls) (supplementary table 3). The location of the deletion is marked with a red arrow in d and f. (e) Alignment of six sequence polymorphisms associated with Dun and non-dun1 haplotypes from domestic horses. SNP1 and SNP2 are shown in bold. (f) Nucleotide diversity at polymorphic sites estimated in sliding windows of 100 SNPs for Dun (D), non-dun1 (d1) and non-dun2 (d2) chromosomes among domestic horses and Przewalski’s horses.
We genotyped a large number of Dun and non-dun horses from various breeds for the deletion and concluded that it was not present in the homozygous state in any of the 529 Dun horses, as expected if it is a causal mutation for the recessive non-dun phenotype (Table 1). However, only two-thirds of the non-dun horses were homozygous for the deletion. Careful inspection of a subset of non-dun horses demonstrated that horses homozygous for the deletion did not show primitive markings, whereas many horses homozygous or heterozygous for the non-deleted allele did (P = 4.3 × 10−17, Fisher’s exact test; Fig. 1a, Table 1 and Supplementary Fig. 2). Thus, the non-dun phenotype is caused by either of two alleles: non-dun1 (d1; associated with markings) or non-dun2 (d2; not associated with markings, containing the derived 1.6-kb deletion). As described further below (Online Methods and Supplementary Fig. 2), the non-dun1 allele has a weaker effect than the non-dun2 allele with regard to both gross and microscopic pigmentary phenotypes.
Table 1.
Association between TBX3 sequence variation and Dun phenotypes
| Genotype frequency for the 1.6-kb deletion
| ||||
|---|---|---|---|---|
| Phenotypea | Deletion
|
|||
| WT/WT | WT/del | del/del | Total | |
| Dun | 222 | 307 | 0 | 529 |
| non-dun, dorsal stripe status unknown | 108 | 280 | 803 | 1,191 |
| non-dun, with dorsal stripe | 28 | 44 | 0 | 72 |
| non-dun, without dorsal stripe | 0 | 3 | 19 | 22 |
| Total | 358 | 634 | 822 | 1,814 |
| Genotype frequency for causal TBX3 variants
| ||||||
|---|---|---|---|---|---|---|
|
TBX3 genotypes
|
Dunb | Phenotype
|
||||
| SNP2 | SNP1 | Deletion | Dun | non-dun | Total | |
| G/G | G/G | WT/WT | D/D | 152 | 0 | 152 |
| A/A | G/G | WT/WT | D/D | 2 | 0 | 2 |
| A/G | G/T | WT/WT | D/d1 | 66 | 0 | 66 |
| A/A | G/T | WT/WT | D/d1 | 2 | 0 | 2 |
| A/G | G/− | WT/del | D/d2 | 303 | 0 | 303 |
| A/A | G/− | WT/del | D/d2 | 4 | 0 | 4 |
| A/A | T/T | WT/WT | d1/d1 | 0 | 136 | 136 |
| A/A | T/− | WT/del | d1/d2 | 0 | 327 | 327 |
| A/A | −/− | del/del | d2/d2 | 0 | 822 | 822 |
| 529 | 1,285 | 1,814 | ||||
Animals representing more than 45 breeds were used (supplementary table 4). Del, deletion; WT, wild type.
Of the 1,285 non-dun horses tested for the deletion, 94 were carefully examined for the presence of a dorsal stripe.
Genotype inferred from TBX3 genotypes and Dun versus non-dun phenotype.
To further explore genetic relationships between the Dun, non-dun1 and non-dun2 alleles, we identified SNPs from the TBX3 region using resequencing data from 21 modern domestic horses, a modern Przewalski’s horse and two ancient horses, one ~4,400 years old from Yakutia and one ~42,700 years old from the Taymyr peninsula, both of which are in Russia10 (Supplementary Table 2). We designed a 384-SNP panel covering ~375 kb with a SNP density in the core 200-kb region of approximately 1 SNP/550 bp, including three SNPs within the region deleted in non-dun2. Genotyping data were combined with resequencing data (in total, for 71 Dun domestic horses, 152 non-dun domestic horses, 15 Przewalski’s horses and one domestic horse–Przewalski’s horse hybrid; Supplementary Table 2) to analyze genotype-phenotype relationships. Only two SNPs showed complete concordance with the Dun versus non-dun phenotype when Przewalski’s horses were included in the genetic analysis (Fig. 2d and Supplementary Table 3). One of these, SNP1 (G in Dun, T in non-dun1), is located within the region deleted in non-dun2, 1,067 bp downstream of the deletion breakpoint at chr. 8: 18,227,267. The second, SNP2 (G in Dun, A in non-dun1 and non-dun2), is located 362 bp upstream of the deleted region in non-dun2 at chr. 8: 18,226,905. We analyzed resequencing data from modern horses, Przewalski’s horses, the two ancient horses and 13 other equids, for a total of 87 equids (Supplementary Table 2), to comprehensively investigate potential causal SNPs for non-dun1. This analysis identified three additional SNPs and a 1-bp indel, all within the deleted region in non-dun2, showing nearly complete linkage disequilibrium with the Dun and non-dun1 alleles in domestic horses (Fig. 2e). However, these four variants could be excluded as causal for the non-dun phenotype because they were variable in Przewalski’s horses, which are always Dun.
A total of 1,814 horses of known phenotype, representing more than 45 breeds (including Przewalski’s horses), were genotyped for SNP1 and SNP2 and for the presence of the 1.6-kb deletion. We observed three common haplotypes: GG (associated with Dun), AT (associated with non-dun1) and A-del (associated with non-dun2), where genotypes are given for SNP2 and SNP1 or for SNP2 and the deletion. These haplotypes showed complete concordance with phenotype across this diverse set of horses (Table 1 and Supplementary Table 4). In addition, a rare AG haplotype (associated with Dun) was found in Estonian native horses, suggesting that SNP2 is not required for the Dun phenotype. Taken together with the resequencing data (Fig. 2e), these results indicate that SNP1 is sufficient to cause a non-dun1 phenotype and that the 1.6-kb deletion is the causal mutation for the non-dun2 phenotype.
Bioinformatic analysis11 predicted binding sites in the region deleted in non-dun2 for ALX4 and MSX2, two transcription factors with an established role in hair follicle development12–15. Furthermore, SNP1 is predicted to affect binding of the CCAT box–binding transcription factors, NF-Y and NF-I.
We used the pattern of sequence variation in the TBX3 region in equids to explore the evolutionary history of the Dun and non-dun haplotypes (Fig. 2e and Supplementary Tables 3 and 5). There was almost no diversity among non-dun2 chromosomes, as expected for a recently derived allele (Fig. 2f). In contrast, non-dun1 chromosomes showed extensive diversity across the entire region (Fig. 2f and Supplementary Table 5), demonstrating that non-dun1 cannot be a recently derived allele. Surprisingly, Dun chromosomes in domestic horses were associated with as little nucleotide diversity as the non-dun2 allele. This relative lack of diversity likely represents either a founder effect of horse domestication or a subsequent bottleneck, as Dun chromosomes from Przewalski’s horses showed extensive nucleotide diversity (Fig. 2f).
Interestingly, the 4,400-year-old ancient horse could be deduced to be homozygous non-dun1, whereas the 42,700-year-old horse was heterozygous Dun/non-dun1. Thus, the divergence of Dun and non-dun1 haplotypes predates domestication. All other equids, including three species of asses and three species of zebras, could be deduced to be homozygous Dun on the basis of SNP1 and SNP2 genotypes (Fig. 2e). Thus, non-dun1 is a derived allele that arose over 40,000 years ago.
Differential gene expression in skin
To gain insight into the mechanism of pigmentary dilution, we generated and analyzed transcriptome data from the croup skin of seven Dun and 11 non-dun horses. We identified 57 differentially expressed genes (Fig. 3a and Supplementary Table 6), including TBX3, which was downregulated by 1.6-fold in non-dun skin. Quantitative RT-PCR (RT-qPCR) confirmed downregulation of TBX3 in both non-dun1 (3.5-fold; n = 3) and non-dun2 (2.5-fold; n = 6) homozygotes; notably, the neighboring gene, TBX5, did not show differential expression (Supplementary Fig. 3a). The RNA sequencing (RNA-seq) results also showed upregulation of eight genes with well-established roles in melanocyte-specific pigment production (TYR, DCT, MC1R, TRPM1, SLC24A5, MLANA, KIT and OCA2) in non-dun croup skin, consistent with increased pigmentation intensity in non-dun hair (Fig. 1b,d). ASIP, EDN3 and KITLG are three genes with known regulatory roles in animal color patterns; of these, only KITLG exhibited differential expression. We also examined transcriptome data from the dorsal stripe skin of Dun horses and skin from the corresponding location in non-dun horses and did not observe differential expression of melanogenic or melanocyte regulatory genes (Supplementary Table 6).
Figure 3.
Differential gene and protein expression in the croup skin of Dun and non-dun horses. (a) Transcript levels (normalized gene counts) plotted as a function of differential expression (log2-transformed fold change) in Dun (D/−, n = 7) versus non-dun (d1/d1, n = 3; d2/d2, n = 6; d1/d2, n = 2) samples. The 57 genes demonstrating significant (false discovery rate (FDR) < 0.1) differential expression are shown in dark yellow (higher expression in Dun) or red (lower expression in Dun; Online Methods and Supplementary Table 6); seven additional pigmentation-related genes that are not differentially expressed are shown in blue. (b–e) Immunofluorescence for TBX3 (green) (b), MITF (green) (c), KIT (green) (d) and KITLG (green) (e) in sections of anagen hair follicles from the croup of Dun/−, non-dun1/non-dun1 and non-dun2/non-dun2 horses. Pigment is pseudocolored in b (red). Corresponding bright-field images are on the right in c–e. DAPI staining is in blue in c–e, and white lines indicate the basement membrane. Each photomicrograph is representative of at least two individuals of each genotype. Scale bars, 100 μm (b–e).
TBX3 is asymmetrically expressed in Dun hair follicles
Immunohistochemistry of sections from Dun croup skin displayed an unusual pattern of TBX3 expression in hair follicles mirroring the pattern of pigment deposition (Fig. 3b and Supplementary Fig. 4a). In sequential sections of anagen hair follicles, TBX3 immunostaining was localized to a cluster of keratinocytes in the developing hair cortex (Supplementary Fig. 4a). TBX3 was not expressed in hair cortex keratinocytes from non-dun croup skin (Fig. 3b) nor from the dark dorsal midline of Dun and non-dun horses (Supplementary Fig. 4b) but was expressed in the outer cuticular layer of hair follicles in all samples (Fig. 3b and Supplementary Fig. 4). These observations suggest that TBX3 operates in a specific subset of hair cortex keratinocytes to inhibit pigment synthesis in the dilute hair of Dun horses.
We used immunohistochemistry for MITF and KIT, markers of mature pigment cells16, to investigate the distribution of melanocytes in croup skin biopsies from Dun and non-dun horses. MITF was detected in the pigmented epidermis of Dun and non-dun horses and showed symmetrical immunostaining in hair follicles of non-dun horses (Fig. 3c and Supplementary Fig. 5a). In contrast, asymmetric immunostaining of MITF and KIT was observed in hair follicles from the croup skin of Dun horses, corresponding to the pattern of pigment deposition (Fig. 3c,d and Supplementary Fig. 5a). Thus, both the histological and transcriptomic differences between Dun and non-dun horses arise from differences in melanocyte distribution within the hair follicle.
In the RNA-seq data, we observed no difference between non-dun and Dun skin in the expression of MITF, likely owing to the presence of cells in the dermis whose expression of MITF was not affected by Dun genotype. By contrast, the RNA-seq data showed a significant increase in the expression of KIT and, notably, KITLG (encoding KIT ligand) in non-dun skin in comparison to Dun skin (Fig. 3a), a finding confirmed by RT-qPCR experiments (Supplementary Fig. 3b). Previous studies have identified KITLG as a keratinocyte-derived molecule necessary for melanocyte migration and survival in the skin and hair follicle17,18. Immunohistochemistry showed that KITLG-positive cells were uniformly distributed in the hair bulbs of non-dun animals and in the pigmented epidermis of all animals examined (Fig. 3e and Supplementary Fig. 5b). However, KITLG showed asymmetric expression in hair bulb keratinocytes from Dun croup skin, consistent with the asymmetry of melanocyte distribution and pigment deposition. Apparent suppression of KITLG expression occurred in a similar but not identical location to where asymmetric expression of TBX3 occurred in Dun croup skin (Fig. 3e and Supplementary Fig. 5b).
TBX3 and hair follicle symmetry and differentiation
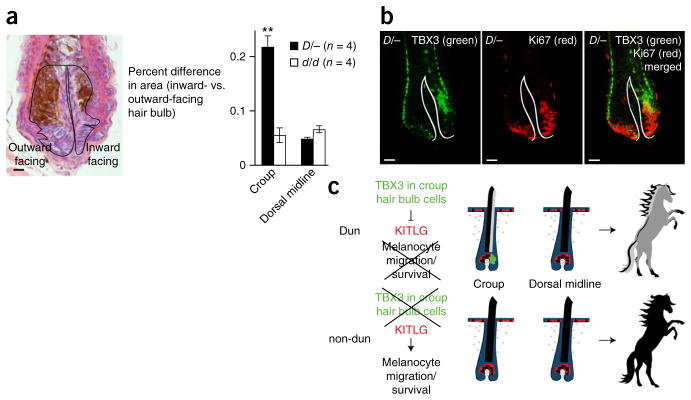
To further investigate the mechanism by which TBX3 affects KITLG expression and melanocyte localization, we examined the morphology and differentiation of croup hair follicles in Dun horses in comparison to those in non-dun horses. In addition to asymmetry in pigment deposition, we observed that croup hair follicles from Dun horses exhibited an apparent asymmetry in hair bulb volume. To assess this difference quantitatively, we measured hair bulb volume in serial hair follicle sections, using the center of the dermal papilla as a central boundary, and expressed the extent of asymmetry as the percent difference in volume between the outward-facing and inward-facing halves of each hair bulb (Fig. 4a). Croup hair follicles from Dun horses (n = 4) exhibited significantly greater asymmetry in bulb volume than those from non-dun horses (n = 4). In contrast, no difference between Dun and non-dun horses was observed for dorsal midline follicles (Fig. 4a).
Figure 4.
Relationship between TBX3 expression and hair follicle symmetry and differentiation. (a) Left, hair follicle section stained with hematoxylin and eosin from Dun croup skin. Black lines outline the outward-facing and inward-facing halves of the hair bulb. Right, the percent difference in area (mean ± s.e.m.) between the outward- and inward-facing halves of the hair bulb was calculated for the croup and dorsal midline from Dun (D/−; n = 4) and non-dun (d/d; n = 4) horses. Bulb area was measured from at least two serial sections (range of 2–12 sections/follicle) from at least two follicles (range of 2–7 follicles) per anatomical location. **P = 0.004 for Dun versus non-dun croup, two-tailed t test. (b) Immunofluorescence for TBX3 (green; left) and Ki67 (red; center) in a croup hair follicle from a Dun horse. The merged image is on the right; white lines delineate the basement membrane. Scale bars, 25 μm (a) and 50 μm (b). (c) Asymmetric expression of TBX3 (green) in hair bulb keratinocytes of Dun croup hairs impairs the expression of KITLG (red) and melanocyte survival in one-half of the follicle. Dun croup hairs are asymmetrically pigmented and appear lighter colored than hairs from the dorsal stripe or from non-dun horses.
To better refine the location of hair bulb TBX3 expression, we examined its distribution relative to established hair follicle differentiation markers, including AE13 (cortex, hair cuticle), Ki67 (proliferative cells), AE15 (medulla, inner root sheath) and keratin 6 (KRT6; companion cell layer). The patterns of AE15 and KRT6 immunostaining were very different from that of TBX3 (Supplementary Fig. 6a). TBX3-expressing cells lay in close proximity to the developing hair cortex (Fig. 3b and Supplementary Fig. 4a). We observed no colocalization of TBX3 and AE13 (Supplementary Fig. 6b), but double immunostaining demonstrated that a subset of Ki67-positive cells also expressed TBX3 (Fig. 4b). Thus, TBX3 expression occurs during early differentiation of croup hair follicle keratinocytes but does not correspond to a known lineage or compartment. Instead, TBX3 appears to shift the balance between dividing and differentiating cells, leading to reduced hair bulb volume. Taken together, our results demonstrate that asymmetric TBX3 expression in hair bulb keratinocytes from Dun horses leads to asymmetric pigmentation via altered KITLG expression and that the mutations in non-dun horses disrupt this mechanism (Fig. 4c).
DISCUSSION
The genetics of animal coat color is a longstanding model system for studying fundamental aspects of gene action and interaction. As shown here, equine Dun coloration exemplifies a new aspect of pigmentary variation with implications for epithelial biology, mammalian evolution and the history of animal domestication.
In contrast to the numerous loss-of-function mutations causing dilute pigmentation in mice and other vertebrates as a result of defects in the pigmentary machinery, dilute pigmentation in equids represents the wild-type state and maintains intact melanogenesis. The mutations in non-dun horses disrupt this process and, in doing so, uncover an unexpected mechanism whereby TBX3 regulates pigment deposition along the radial axis of hair follicles (Fig. 4c). Regulation of pigment deposition along the longitudinal axis of hair follicles, controlled by the ASIP gene, underlies color differences between body regions in many species of mammals. Whereas ASIP variation affects coat color in horses (Supplementary Fig. 1a), the longitudinal hair banding mediated by ASIP is largely absent from equids. Instead, differential expression of TBX3 and its localized effect on pigment deposition provide an alternative and unexpected mechanism modulating the appearance of individual hairs in different regions of the body. From this perspective, TBX3 expression seems to mark a previously unappreciated hair follicle compartment that is sensitive to both the microscopic position of cells within individual follicles (radial asymmetry) and the macroscopic region of the body (for example, croup versus dorsal stripe).
Hair follicle cells that express TBX3 include keratinocytes of the cuticle and the hair bulb cortex, but only the latter group is sensitive to position and controls Dun coloration. These TBX3-expressing cells in the hair bulb cortex do not correspond to a known cell lineage or differentiation compartment. Instead, they are likely to respond to positional signals correlated with the direction of hair follicle downgrowth and bending19, as TBX3 is expressed in the inward-facing half of the hair follicle where KITLG expression is extinguished and melanocytes are excluded. However, several observations suggest that the connection between expression of TBX3 and KITLG in the hair bulb is likely to be indirect. First, the location of TBX3 immunostaining within the hair bulb only partially overlap with the location in which KITLG immunostaining is absent. Second, our histological and morphometric observations point to altered distributions of hair bulb cell composition and differentiation rather than just alterations in gene expression. Finally, existing chromatin immunoprecipitation and sequencing (ChIP-seq) data for TBX3 do not include potential regulatory regions for KITLG20. Nonetheless, our data suggest that the most striking effect of TBX3 expression in hair bulb keratinocytes concerns hair pigmentation, and, thus, most of the differentially expressed genes we identified represent differences in melanocyte abundance.
TBX3 is expressed in many cell types and has a critical role during development5. Our data indicate that the non-dun2 deletion as well as the non-dun1 allele disrupt the function of a transcriptional enhancer regulating TBX3 expression in a specific subset of hair bulb keratinocytes during hair growth. This conclusion is based on the clear difference in TBX3 expression between Dun and non-dun horses, the high sequence conservation across mammals in the region and the prediction of transcription factor binding sites affected by the non-dun mutations. Furthermore, the horse TBX3 mutations appear to have tissue-specific effects, as neither Dun nor non-dun horses show any recognized differences in limb development or other pleiotropic effects outside the hair follicle.
The ability of TBX3 to influence pigmentation by altering hair bulb keratinocyte differentiation and KITLG expression could be limited to equids with a last common ancestor present 4.0–4.5 million years ago. Alternatively, a role for TBX3 in hair color may have a more ancient mammalian origin. Various pigment markings and patterned coat color dilutions are widely distributed among Ruminantia (with an origin ~55 million years ago). Animals such as the wildebeest, eland, okapi, fallow deer and wild goat display some aspects of coat patterning reminiscent of Dun coloring, and it will be interesting to investigate the developmental and genetic basis of these markings. A closely related question within equids has to do with the potential role of TBX3 in zebra stripes, which could be viewed as a modification of the Dun phenotype where primitive markings extend over the entire body and the dilution has become so pronounced that the hairs are unpigmented. From this perspective, understanding the spatial control of TBX3 expression in horse skin, that is, dorsal stripe versus croup, may provide insight into the etiology of zebra stripes.
The non-dun2 deletion allele has a stronger phenotypic effect than the non-dun1 allele, exhibits very low nucleotide diversity in flanking DNA sequences and was not observed outside of domestic horses, suggesting a relatively recent origin, probably during the last several thousand years. In contrast, the non-dun1 allele exhibits far greater nucleotide diversity and is much older. Haplotype comparisons and breed distribution suggest that the non-dun2 deletion arose on a non-dun1 chromosome and then increased in frequency, a history consistent with persistent selection during and after horse domestication for the non-dun phenotype; in fact, selection against camouflage color is likely an important reason for changes in coat color during animal domestication21.
Additional studies of ancient horse DNA should help to more precisely date the origin of the non-dun1 allele, but its presence in a ~43,000-year-old sample is consistent with other work inferring that multiple color morphs existed in predomestic horses22, pointing toward a hybrid origin of the domestic horse. The major contribution to horse domestication may have occurred from a population carrying the non-dun1 allele at high frequency, potentially the same population as the now extinct tarpan (Equus ferus ferus). Indeed, the ~43,000-year-old horse from the Taymyr peninsula carrying non-dun1 belonged to a population that contributed substantially to the domestic horse lineage10. Conversely, a predominantly Dun population, the ancestor of Przewalski’s horse or a close relative, could have made a minor contribution to domestic horses, as suggested by the low diversity observed among domestic horse Dun chromosomes. This idea can be further explored with additional ancient horse DNA samples spanning a large temporal and geographic range.
URLs
Picard, http://broadinstitute.github.io/picard/; Genome Analysis Toolkit (GATK) wiki, http://www.broadinstitute.org/gsa/wiki/index.php/The_Genome_Analysis_Toolkit; ImageJ, http://imagej.nih.gov/ij/.
ONLINE METHODS
Animals and phenotyping
We obtained extracted DNA, hair or blood samples collected by authors and collaborators, provided by horse owners, with permission from the St. Louis Zoo, or archived at the Veterinary Genetics Laboratory (University of California, Davis), the Animal Genetics Laboratory (Swedish University of Agricultural Sciences, Sweden) or the Smithsonian National Museum of Natural History. DNA was extracted from samples following standard methods. DNA from the CITES protected Przewalski’s horse was transferred from the Zoological Society of San Diego to the Centre for GeoGenetics to perform high-throughput sequencing analyses under biomaterial request BR2013045.
Skin plug biopsies were collected from horses in Iceland by practicing veterinarians. Local anesthetic was given to the horses before biopsy extraction. Punch biopsies were taken from the dorsal midline on the hindquarters, sampling the dorsal stripe if present, and from the croup a few centimeters lateral to the sampling on the dorsal midline, beyond the area in which a dorsal stripe is present in Dun horses. The samples from beyond the dorsal midline are referred to as croup samples. Anatomical locations of sampling are indicated in Supplementary Figure 2a.
Dun phenotypes were assigned by visual assessment of live individuals or photographs; in some instances, Dun phenotypes were assigned on the basis of owner assessment and/or breed criteria. The Dun coat color is characterized by specific patterns of intensely pigmented markings on a diluted background. These specific patterns can be broken down into elements, which can occur in different combinations. Some elements are rare in general but common in certain breeds. Other elements, such as the dorsal stripe, are nearly universal. The dorsal stripe is an intensely pigmented stripe extending from the forehead, along the dorsal midline all the way down to the tip of the horse’s tail. A non-exhaustive list of other Dun pattern elements includes ear markings, dark legs, leg barring, shoulder crosses, neck or shoulder shadows, dark facial masks, eye shadows, darkly pigmented areas around vibrissae, dorsal barbs, cobwebbing on the forehead, and netting on the neck, shoulder and flank (Supplementary Fig. 1b).
non-dun horses carrying the non-dun1 allele often show primitive markings in the absence of marked color dilution. As the body hair color of non-dun horses is much darker than that of Dun horses, these markings can be faint. Furthermore, the base color of the individual horse affects the visibility of primitive markings. A horse with primarily red body color, such as a chestnut or a red bay, will show such markings more readily than a horse with primarily black body pigmentation, such as a black bay or black horse, as the visual intensity of black pigment is greater than that of red.
Dun is dominant to non-dun1 and to non-dun2, that is, Dun/Dun, Dun/non-dun1 and Dun/non-dun2 horses are phenotypically identical. However, the effects of the non-dun1 allele are not as strong as those of the non-dun2 allele. Cross-sections of croup hairs from non-dun1/non-dun1 or non-dun1/non-dun2 horses exhibit some asymmetry in pigment deposition, although not nearly as great as in Dun/− horses (Fig. 1a,b and Supplementary Fig. 2a). Likewise, primitive markings in non-dun1/non-dun1 horses are usually more pronounced than in non-dun1/non-dun2 horses (Supplementary Fig. 2b,c).
Initial SNP association analysis
Twenty-seven SNPs were genotyped across the region ECA8: 18,061,745–18,482,196 in 366 individuals from 19 breeds, including Przewalski’s horses. Ninety-six individuals were homozygous Dun, 111 were heterozygous Dun/non-dun and 159 were homozygous non-dun. Genotyping was performed on the Sequenom platform. Primers and probes were designed in a multiplex format using SpectroDESIGNER software (Sequenom). Assay amplification was performed as previously described23. Statistical evaluation of data was carried out using Haploview24.
Targeted resequencing
Two Dun individuals, an Icelandic horse and a Quarter Horse, were chosen for massively parallel resequencing of the Dun region. Both were known to be homozygous for Dun on the basis of offspring data. Five non-dun horses were also included: an Appaloosa, an Arabian, a Knabstrupper, a Lipizzaner and a Noriker. NimbleGen Sequence Capture Arrays (Roche NimbleGen) were designed to target the 200-kb region on horse chromosome 8 harboring the Dun locus. Targeted resequencing was performed as described25. Subsequent to the discovery of the non-dun2 deletion, sequence for the deletion was obtained via Sanger sequencing for these seven individuals. Primer sequences are given in Supplementary Table 7.
Genome resequencing
DNA samples from two Icelandic horses, one Dun heterozygote and one non-dun homozygote, were prepared for whole-genome sequencing as part of our previous study on the Gait-keeper mutation9. Initially, these two samples were aligned to the horse reference assembly (EquCab2.0). Following the detection of the deletion in non-dun2, we added Sanger-derived sequence from a Dun chromosome (GenBank, KT896508–KT896515) to the EquCab2 chromosome 8 sequence for bases chr. 8: 18,227,267–18,227,279, thereby creating an alternative assembly in silico (EquCab2.0-Dun), which was used for all subsequent alignment steps.
Whole-genome sequencing was carried out using 2 × 100-bp paired-end libraries on an Illumina HiSeq instrument at the SciLifeLab Illumina platform, with insert sizes ranging between approximately 200 and 600 bases, to generate approximately 6–10× coverage. The reads from both targeted resequencing and whole-genome sequencing (BioSample accession numbers SAMN03396927–SAMN03396975; Supplementary Table 2) were mapped to EquCab2.0-Dun using Burrows-Wheeler Aligner (BWA)26. PCR duplicates were removed using Picard. SNPs and small insertion-deletions were called from mapping data using the Genome Analysis Toolkit (GATK)27 UnifiedGenotyper command after having performed realignment around indels. Sequence variants were collected for EquCab2.0-Dun chromosome 8: 18,120,000–18,322,000. The variant calls were subjected to recommended filters for SNPs listed on the GATK wiki page.
Sequences with BioSample accession numbers SAMN04002334– SAMN04002382 (Supplementary Table 2) had their reads mapped to a segment of chromosome 8 of the EquCab2.0-Dun hybrid assembly, using the PALEOMIX pipeline28, omitting mapping quality checks and PCR duplicate filtering. Sequencing reads were first trimmed using AdapterRemoval v.1 (ref. 29). All mapped reads were then collected and converted from BAM records to FASTQ reads. Next, the reads were remapped against the full EquCab2.0-Dun genome using the PALEOMIX pipeline28, filtering for mapping quality (MAPQ ≥25), removing PCR duplicates and realigning around indels. Paired mates were kept in pairs if both mates were found among the alignments and orphaned mates were treated as single-end reads during remapping. Finally, all hits mapping to the region EquCab2.0-Dun chr. 8: 18,120,000–18,322,000 were collected. This procedure, which first identifies candidate hits against the target region and then checks whether better alignment exists somewhere else in the genome, was devised to avoid remapping the whole set of reads (comprising several billion reads per individual). The software ANNOVAR30 was used to annotate SNPs in relation to Ensembl genes.
SNP genotyping using BeadXpress
A VeraCode GoldenGate assay (Illumina) was designed targeting 384 SNPs that were called in the resequencing samples. A total of 200 domestic horses, one Przewalski’s horse and five other equids were genotyped using the standard protocol. The GoldenGate assays were read using a BeadXpress Reader (Illumina), and data were analyzed on GenomeStudio v2011.1 software (Illumina). SNPs located outside the non-dun2 deletion with a call rate of less than 80% (17 SNPs) or invariable in typed individuals (one SNP) were excluded from the analysis. The three SNPs within the deleted region had a call rate of 67–68%, consistent with one-third of the samples being homozygous for the non-dun2 deletion. All samples used had call rates of no less than 97% (domestic horses) or 90% (other equids).
In-depth SNP association analysis
Phenotypic association tests assuming dominant inheritance of the Dun phenotype were carried out with PLINK31 using Fisher’s exact test. SNP genotypes from the GoldenGate assay were combined with genotypes for those particular SNPs from the resequencing data. Candidate SNPs were selected on the basis of allele frequencies of more than 50% in Dun individuals (dominant trait) and less than 5% in non-dun individuals (erroneous calls) (Supplementary Table 3).
A similar analysis was performed on SNP calls from the resequencing experiments. Over 5,000 SNPs from the interval chr. 8: 18,119,043–18,321,208 passed quality filtering criteria of quality over 100, genotype quality higher than 9 and proportion of missing genotypes across samples less than 0.2. SNPs at positions 18,276,126, 18,276,338 and 18,277,478 on chromosome 8 showed strong association with the Dun phenotype in the resequencing data. The SNPs at 18,276,338 and 18,277,478 on chromosome 8 were included in the GoldenGate assay (Supplementary Table 3). All three SNPs were genotyped via Sanger sequencing for a family of three recombinant Dun Icelandic horses, which excluded them as causal. Primer sequences are given in Supplementary Table 7.
Haplotype diversity analysis
To calculate diversity, all screened SNPs from the GoldenGate assay were phased with SHAPEIT2 (ref. 32; using 100 burn-in iterations, 100 pruning iterations, 100 main iterations and 1,000 conditioning states per SNP, effective population size of 100 and recombination parameter rho of 0.001) and had haplotypes inferred according to genotype at SNP1, SNP2 and the deletion. Average nucleotide diversity per polymorphic site was calculated for each of the Dun, non-dun1 and non-dun2 haplotypes as 2 × f1 × f2 × n/(n − 1), where f denotes allele frequency and n the total number of alleles.
Genotyping of Dun, non-dun1 and non-dun2 alleles
DNA samples from horses of various breeds were genotyped for the Dun, non-dun1 and non-dun2 alleles. The non-dun2 deletion was typed by standard agarose or capillary (ABI 3730 DNA Analyzer, Applied Biosystems) electrophoresis for detection of the amplification length polymorphism. Betaine or DMSO additives were required for successful PCR amplification. SNP2 was genotyped by pyrosequencing on PSQ 96MA 2.1 and PyroMark Q96 MD instruments (Qiagen) according to the manufacturer’s recommendations. SNP1 was genotyped with pyrosequencing or with a TaqMan allelic discrimination assay on a 7900HT Fast Real-Time PCR system (Applied Biosystems) according to the manufacturer’s instructions. Primer sequences are given in Supplementary Table 7.
Expression analysis
Punch biopsies of skin from the croup and dorsal midline of 18 horses (seven Dun and 11 non-dun) were collected. The samples were preserved in RNAlater and stored at −80 °C before processing. Total RNA was isolated from skin biopsies using the RNeasy Fibrous Tissue Mini kit (Qiagen). RNA quality was assessed with an Agilent Bioanalyzer instrument. All samples had an RNA integrity number (RIN) greater than 7.0. cDNA libraries were further prepared for each sample using Illumina’s TruSeq RNA Sample Preparation Kit v2. Individual libraries were multiplexed, six per lane, and sequenced as single-end 50-bp reads on an Illumina HiSeq 2000 instrument at the Genome Sequencing Laboratory of the HudsonAlpha Institute.
RNA-seq reads obtained for each sample were aligned against the horse reference genome with TopHat2 software (v. 2.0.12), using genomic sequence and transcript annotations obtained from Ensembl (release 75). Gene counts were computed from read alignments with GenomicRanges and GenomicFeatures packages from Bioconductor (release 2.14) and then used to test for differential gene expression between Dun and non-dun skin samples with the DESeq2 package from Bioconductor. DESeq2 relies on negative binomial generalized linear models to determine whether the number of counts for a transcript or gene is significantly different across a range of experimental conditions. To account for expression changes related to differences in the base coat color (black, bay or chestnut; Supplementary Fig. 1a) of different samples rather than the presence of the Dun allele, we included base coat color as a covariate in the generalized linear model.
RT-qPCR for TBX3, TBX5 and KITLG
Total RNA for RT-qPCR was isolated from skin biopsies using TRIzol (Invitrogen Life Technologies), purified using the PureLink RNA Mini kit (Ambion) and treated with DNase I (Invitrogen Life Technologies) before reverse transcription with Superscript III (Invitrogen Life Technologies). cDNA was amplified using the LightCycler FastStart DNA Master Plus SYBR Green I System (Roche Diagnostics). β2-microglobulin (B2M) was used to compare relative levels of mRNA. Primer sequences are given in Supplementary Table 7.
Histology and immunofluorescence
Skin samples were fixed in 4% para-formaldehyde, washed in PBS, dehydrated and embedded in paraffin. Five-micrometer sections of hair, croup skin and dorsal midline skin were stained with hematoxylin and eosin. ImageJ was used to measure hair bulb area and hair color intensity. Color intensity was measured at six points across the diameter of the hair cortex and averaged for each hair. Hair color intensity measurements were taken from at least five hairs from each genotype. Immunofluorescence was carried out with goat antibody to TBX3 (Santa Cruz Biotechnology, sc-17871), mouse antibody to MITF (Abcam, ab12039), rabbit antibody to SCF (KITLG; Abcam, ab64677), mouse antibody to AE13 (Abcam, ab16113), rabbit antibody to Ki67 (Abcam, ab15580), mouse antibody to AE15 (Abcam, ab58755), rabbit antibody to KRT6 (Abcam, ab24646) and rabbit antibody to KIT (Dako, A4502) after antigen retrieval using Tris-EDTA, pH 9 (TBX3) or 0.01 M citrate buffer, pH 6 (MITF, KITLG, AE13, Ki67, AE15, KRT6 and KIT) in a pressure cooker. All photomicrographs are representative of at least two animals of each genotype and anatomical location.
Supplementary Material
Acknowledgments
We thank the numerous horse owners who provided samples, T. Raudsepp and I. Randlaht for samples from Estonian native horses, C. Asa and M. Fischer (St. Louis Zoo) and F. Marshall for providing hair samples from the Somali wild ass, W. Zimmerman for photographs of Przewalski’s horse, S. Fard, H. Ring and F. Hallböök for advice on histological characterization, O. Ryder, L. Chemnick and C. Steiner for delivering DNA extracts from Przewalski’s horses, C. Der Sarkissian and L. Ermini for assistance in whole-genome resequencing at the Centre for GeoGenetics, Denmark, and the HudsonAlpha Genomic Services Laboratory for RNA-seq. This work was supported by grants from the Knut and Alice Wallenberg foundation (to L.A.) and the US National Institutes of Health (to G.S.B.), as well as by an Erasmus Mundus fellowship within the framework of the European Graduate School of Animal Breeding and Genetics (to D.S.). Sequencing was performed by the SNP&SEQ Technology Platform, supported by Uppsala University and Hospital, the Science for Life Laboratory and the Swedish Research Council (80576801 and 70374401).
Footnotes
Accession codes. Data from sequence capture and whole-genome resequencing have been deposited in GenBank under BioProject PRJNA277815. Sanger sequences of the non-dun2 deletion have been submitted to GenBank with accessions KT896508–KT896515.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
L.A. led the genetic characterization and G.S.B. led the RNA-seq and immunohistochemistry studies. F.I., P.I., K.M. and M.C.T.P. did the sampling. F.I. was responsible for phenotyping and carried out genotyping together with U.G., L.M. and M.C.T.P. K.M. performed immunohistochemistry analysis. C.H. performed RNA-seq analysis. C.-J.R., F.I., J.B., M.S. and L.O. were responsible for genome sequence analysis. E.S. analyzed transcription factor binding sites. D.S. contributed to TBX3 expression analysis. F.I., L.A., M.C.T.P., K.L.-T., G.L., S.M. and C.W. took part in the initial mapping of Dun. L.A., F.I., K.M., C.H. and G.S.B. wrote the manuscript with input from other authors. All authors approved the manuscript before submission.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Wade CM, et al. Genome sequence, comparative analysis, and population genetics of the domestic horse. Science. 2009;326:865–867. doi: 10.1126/science.1178158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orlando L, et al. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature. 2013;499:74–78. doi: 10.1038/nature12323. [DOI] [PubMed] [Google Scholar]
- 3.Bowling AT, Ryder OA. Genetic studies of blood markers in Przewalski’s horses. J Hered. 1987;78:75–80. doi: 10.1093/oxfordjournals.jhered.a110340. [DOI] [PubMed] [Google Scholar]
- 4.Adalsteinsson S. Inheritance of yellow dun and blue dun in the Icelandic toelter horse. J Hered. 1978;69:146–148. doi: 10.1093/oxfordjournals.jhered.a108913. [DOI] [PubMed] [Google Scholar]
- 5.Bamshad M, et al. Mutations in human TBX3 alter limb, apocrine and genital development in ulnar-mammary syndrome. Nat Genet. 1997;16:311–315. doi: 10.1038/ng0797-311. [DOI] [PubMed] [Google Scholar]
- 6.Frank DU, Emechebe U, Thomas KR, Moon AM. Mouse TBX3 mutants suggest novel molecular mechanisms for Ulnar-mammary syndrome. PLoS One. 2013;8:e67841. doi: 10.1371/journal.pone.0067841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar PP, et al. TBX3 regulates splicing in vivo: a novel molecular mechanism for Ulnar-mammary syndrome. PLoS Genet. 2014;10:e1004247. doi: 10.1371/journal.pgen.1004247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gremmel F. Coat colors in horses. J Hered. 1939;30:437–445. [Google Scholar]
- 9.Andersson LS, et al. Mutations in DMRT3 affect locomotion in horses and spinal circuit function in mice. Nature. 2012;488:642–646. doi: 10.1038/nature11399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schubert M, et al. Prehistoric genomes reveal the genetic foundation and cost of horse domestication. Proc Natl Acad Sci USA. 2014;111:E5661–E5669. doi: 10.1073/pnas.1416991111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas-Chollier M, et al. Transcription factor binding predictions using TRAP for the analysis of ChIP-seq data and regulatory SNPs. Nat Protoc. 2011;6:1860–1869. doi: 10.1038/nprot.2011.409. [DOI] [PubMed] [Google Scholar]
- 12.Ma L, et al. ‘Cyclic alopecia’ in Msx2 mutants: defects in hair cycling and hair shaft differentiation. Development. 2003;130:379–389. doi: 10.1242/dev.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kayserili H, et al. ALX4 dysfunction disrupts craniofacial and epidermal development. Hum Mol Genet. 2009;18:4357–4366. doi: 10.1093/hmg/ddp391. [DOI] [PubMed] [Google Scholar]
- 14.Reginelli AD, Wang YQ, Sassoon D, Muneoka K. Digit tip regeneration correlates with regions of Msx1 (Hox 7) expression in fetal and newborn mice. Development. 1995;121:1065–1076. doi: 10.1242/dev.121.4.1065. [DOI] [PubMed] [Google Scholar]
- 15.Satokata I, et al. Msx2 deficiency in mice causes pleiotropic defects in bone growth and ectodermal organ formation. Nat Genet. 2000;24:391–395. doi: 10.1038/74231. [DOI] [PubMed] [Google Scholar]
- 16.Steingrímsson E, Copeland NG, Jenkins NA. Melanocytes and the microphthalmia transcription factor network. Annu Rev Genet. 2004;38:365–411. doi: 10.1146/annurev.genet.38.072902.092717. [DOI] [PubMed] [Google Scholar]
- 17.Kunisada T, et al. Transgene expression of steel factor in the basal layer of epidermis promotes survival, proliferation, differentiation and migration of melanocyte precursors. Development. 1998;125:2915–2923. doi: 10.1242/dev.125.15.2915. [DOI] [PubMed] [Google Scholar]
- 18.Yoshida H, et al. Review: melanocyte migration and survival controlled by SCF/c-kit expression. J Investig Dermatol Symp Proc. 2001;6:1–5. doi: 10.1046/j.0022-202x.2001.00006.x. [DOI] [PubMed] [Google Scholar]
- 19.St-Jacques B, et al. Sonic hedgehog signaling is essential for hair development. Curr Biol. 1998;8:1058–1068. doi: 10.1016/s0960-9822(98)70443-9. [DOI] [PubMed] [Google Scholar]
- 20.van den Boogaard M, et al. Genetic variation in T-box binding element functionally affects SCN5A/SCN10A enhancer. J Clin Invest. 2012;122:2519–2530. doi: 10.1172/JCI62613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang M, Larson G, Ribeiro HS, Li N, Andersson L. Contrasting mode of evolution at a coat color locus in wild and domestic pigs. PLoS Genet. 2009;5:e1000341. doi: 10.1371/journal.pgen.1000341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pruvost M, et al. Genotypes of predomestic horses match phenotypes painted in Paleolithic works of cave art. Proc Natl Acad Sci USA. 2011;108:18626–18630. doi: 10.1073/pnas.1108982108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gabriel SB, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 24.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 25.Sundström E, et al. Copy number expansion of the STX17 duplication in melanoma tissue from Grey horses. BMC Genomics. 2012;13:365. doi: 10.1186/1471-2164-13-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schubert M, et al. Characterization of ancient and modern genomes by SNP detection and phylogenomic and metagenomic analysis using PALEOMIX. Nat Protoc. 2014;9:1056–1082. doi: 10.1038/nprot.2014.063. [DOI] [PubMed] [Google Scholar]
- 29.Lindgreen S. AdapterRemoval: easy cleaning of next-generation sequencing reads. BMC Res Notes. 2012;5:337. doi: 10.1186/1756-0500-5-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2012;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.