Abstract
Background
In the burn-injured patient, older age, larger percent total body surface area (%TBSA) burned, and inhalation injury are established risk factors for death, which typically results from multisystem organ failure and sepsis, implicating burn-induced immune dysregulation as a contributory mechanism. We sought to identify early transcriptomic changes in circulating leukocytes underlying increased mortality associated these three risk factors.
Methods
We performed a retrospective analysis of the Glue Grant database. From 2003–2010, 324 adults with ≥20% TBSA burned were prospectively enrolled at five US burn centers, and 112 provided blood samples within one week post-burn. RNA was extracted from pooled leukocytes for hybridization onto Affymetrix HU133 Plus 2.0 GeneChips. A multivariate regression model was constructed to determine risk factors for mortality. Testing for differential gene association associated with age, burn size, and inhalation injury was based on linear models using a fold-change threshold of 1.5 and false-discovery rate of 0.05.
Results
After adjusting for potential confounders, age >60 (RR 4.53; 95% CI: 2.93–6.99), burn size >40% TBSA (RR 4.24; 95% CI: 2.61–6.91), and inhalation injury (RR 2.08; 95% CI: 1.35–3.21) were independently associated with mortality. No genes were differentially expressed in association with age >60 or inhalation injury. Fifty-one probe sets representing thirty-nine unique genes were differentially expressed in leukocytes from patients with burn size >40% TBSA; these genes were associated with platelet activation and degranulation/exocytosis, and gene-set enrichment analysis suggested increased cellular proliferation and down-regulation of pro-inflammatory cytokines.
Conclusions
Among adults with large burns, older age, increasing burn size, and inhalation injury have a modest effect on the leukocyte transcriptome in the context of the “genomic storm” induced by a ≥20% TBSA burn. The 39-gene signature we identified may provide novel targets for the development of therapies to reduce morbidity and mortality associated with burns >40% TBSA.
Level of Evidence
Epidemiologic study, level III.
Keywords: Burn mortality, inflammatory response to injury, genomic storm, transcriptome profiling
INTRODUCTION
Breakthroughs in modern burn care including the advent of topical antimicrobials, formalized fluid-resuscitation, and early burn-wound excision and grafting have dramatically improved burn survival during the past century (1). However, there are still over 300,000 deaths from burn-related injuries each year worldwide (2). Beginning in 1949 with Bull and Squire (3), dozens of investigators have developed prognostic scores and statistical models to help predict mortality after burn injury (4). Over time, three factors have remained consistently associated with increased risk of mortality after burns: increased age, burn size (% total body surface area [TBSA] burned), and presence of inhalation injury (5). Understanding how these factors contribute to increased mortality risk will be critical in continuing to improve outcomes for patients with severe burn injuries.
Whereas deaths within the first few days post-burn are generally due to burn shock, multisystem organ failure and sepsis account for the majority of later deaths (6–9). Each of these major causes of death implicates immune dysregulation as a contributing factor: a massive inflammatory response to burn injury is thought to contribute to burn shock and hypermetabolism leading to multisystem organ failure, and impaired immunity against opportunistic organisms likely predisposes to sepsis (10, 11). Burn injury is known to induce a “genomic storm” of early, pervasive changes in the leukocyte transcriptome (14) and all three major risk factors for mortality have been associated with altered levels of circulating cytokines in studies examining small sets of candidate molecules (10, 12, 13). Thus, we hypothesized that early transcriptomic changes in circulating leukocytes would underlie increased mortality associated with age, %TBSA burned, and inhalation injury.
MATERIALS AND METHODS
Study design, population, and setting
We performed a retrospective analysis of data from the Inflammation and Host Response to Injury “Glue Grant”, a large-scale multicenter prospective cohort study of genomic responses to injury. Patients were enrolled from May 2003 to February 2010 at five burn centers following approval by their respective institutional review boards: Loyola University, Chicago, IL; Massachusetts General Hospital, Boston, MA; University of Texas Medical Branch, Galveston, TX; University of Texas Southwestern, Dallas, TX; and University of Washington, Seattle, WA. Enrolled subjects met the following inclusion criteria: age <90 years, admission to a participating center within 96 hours of injury, burn size ≥20% total body surface area (%TBSA), and at least one surgical procedure (15). Exclusion criteria included chemical or electrical burns, multiple injuries with injury severity score (ISS) ≥25, significant premorbid conditions, or decision not to treat (15). In this study, we limited our analysis to adults (age ≥18 years) because both mortality risk associated with burn size and inhalation injury (5) and burn-induced plasma cytokine profiles (10) are known to differ considerably between adults and children. Patients were treated according to standard operating procedures defined by the Inflammation and the Host Response to Injury Burn Core (16), and data on patient- and injury characteristics and clinical course were recorded prospectively for all subjects. A subset of subjects provided blood and/or tissue samples for genomic profiling (15).
Exposures and outcomes
Our exposures of interest were age >60 years, burn size >40% TBSA, and inhalation injury, as these three factors were previously shown to be highly predictive of mortality after burn injury (17). The diagnosis of inhalation injury was made either bronchoscopically or by clinical judgment, as detailed in the standard operating procedures (16). We examined two outcomes: death within one year of burn injury and differential leukocyte gene-expression.
Gene-expression data
RNA was isolated from whole-blood leukocytes and processed as previously described (18, 19). Purified RNA samples were hybridized to Human Genome U133 Plus 2.0 GeneChip microarrays (Affymetrix, Santa Clara, CA), and CEL files containing raw probe intensities were uploaded to the Trauma Research Database. We downloaded CEL files from RNA samples with quality scores ≥2 isolated from burned adults within 7 days of injury. For patients with multiple buffy-coat samples within this time frame, we used data from the earliest available sample. In preparation for differential-expression analysis, we calculated robust multichip average (RMA) expression measures from probe-level data after RMA background-correction and quantile-normalization (20) and corrected for batch effects using an empirical Bayes framework (21).
Statistical analysis
Descriptive data are summarized as number (percent) for categorical variables and median (interquartile range [IQR]) for continuous variables. To estimate the relative risk of mortality associated with our exposures of interest, we analyzed all burned adults with complete clinical data using Poisson regression with robust variance estimates. We favored Poisson over logistic regression because mortality was common (>10%) in the analyzed cohort, in which case estimated odds-ratios would fail to approximate relative risks. In univariate analysis, one model was fit for each of our three exposures of interest (dummy-variable coded as follows: age >60 or ≤60, burn size >40% TBSA or ≤40%, and inhalation injury present or absent). In multivariate analysis, all three exposures of interest were included in a single model along with the following potential confounders: sex, body mass index, burn mechanism, and self-identified race and ethnicity. In Poisson regression analyses, statistical significance was defined as Wald test P-value <0.05.
We modeled gene expression as a linear combination of our three exposures of interest and based statistical inference on their estimated regression coefficients by calculating moderated t-statistics through an empirical Bayes method (22). This approach allowed us to examine the individual effect of each exposure on gene expression while adjusting for the other two exposures. We defined differential expression as |fold change| >1.5 and false discovery rate (FDR)-adjusted P-value <0.05 (23). To visualize patterns of gene expression, we plotted differentially expressed probe sets as a heat map of log-transformed expression values.
We entered differentially expressed genes into the GeneMANIA Cytoscape plugin (24) in order identify related genes, construct a network based on predicted functional relationships, and identify Gene Ontology (GO) terms (25) significantly enriched (FDR-adjusted P <0.05) among the genes in the network. This method of pathway analysis follows the over-representation analysis (ORA) approach, which is based on large changes in a small number of differentially expressed genes (26). Since smaller, coordinated changes in larger sets of genes can also significantly impact cellular function, we used LRpath (27) to conduct a complementary pathway-analysis through a functional class scoring (FCS) approach. We uploaded both P-values and log fold change values for all genes in order to use LRpath to perform directional testing of GO Biological Process gene concepts. As before, we defined statistical significance as FDR-adjusted P <0.05.
Software
Except where noted, all analyses were performed in the open-source R software environment (28) version 3.1.2 with the following additional packages, all of which are freely available through Bioconductor (29): affy (for probe summarization), sva (for batch-effect correction), limma (for differential-expression analysis), and pheatmap (for heat map generation).
RESULTS
Risk factors for mortality after burns
Over 8 years, 324 adult burn patients were enrolled. These patients were predominantly middle-aged White males with large flame burns, and the majority died from infectious causes or multiple organ failure (Table 1), consistent with previous reports (6–8). In univariate analysis, age >60 years, burn size >40% TBSA, and inhalation injury were each associated with increased risk of mortality, and in a multivariate model including several potential confounders, the associations with each of these three factors remained highly significant (Table 2). These results agreed with a recently published analysis of the same dataset by Jeschke et al. demonstrating that burn size >40% TBSA was significantly associated increased risk of mortality as well as infectious complications and multiple organ failure (30). In addition, our results confirm those of Ryan et al., who retrospectively identified and prospectively validated these three factors as being highly predictive of mortality (17).
TABLE 1.
Patient demographics, injury characteristics, and causes of death in the overall cohorta (N = 324).
| Age (years)b | 41 (28–52) |
| Male sex | 242 (75%) |
| Body mass index (kg/m2)c | 27 (23–31) |
| Hispanic ethnicityd | 48 (15%) |
| Racee | |
| White | 230 (73%) |
| Black/African American | 48 (15%) |
| Otherf | 39 (12%) |
| Burn Mechanism | |
| Flame | 270 (83%) |
| Flash | 22 (7%) |
| Scald | 17 (5%) |
| Other | 15 (5%) |
| Burn size (%TBSA)b | 36 (27–51) |
| Inhalation injury | 127 (39%) |
| Deathg | 64 (20%) |
| Sepsis/infection | 18 (28%) |
| Multisystem organ failure | 15 (23%) |
| Withdrawal of therapy | 8 (13%) |
| Cardiac dysfunction/arrest | 8 (13%) |
| Brain death | 5 (8%) |
| Respiratory failure/arrest | 5 (8%) |
| Other/unknownh | 5 (8%) |
Data presented as number (%), except where indicated.
Reported as median (interquartile range).
Missing data for 5 patients.
Missing data for 1 patient.
Missing data for 2 patients.
Includes 10 self-identifying as Asian, 5 as Native American, 1 as Pacific Islander, and 23 as Other.
Percentages for individual causes of death do not sum to 100% due to rounding error.
Includes 1 who died of renal failure, 1 who died of hypovolemic shock, and 3 for whom the specific cause of death was unknown.
TBSA, total body surface area burned.
TABLE 2.
Estimated relative risk of mortality associated with age, burn size, and inhalation injury (N = 317)a.
| Risk Factor | Crude
|
Adjusted
|
||||
|---|---|---|---|---|---|---|
| RR | 95% CI | P | RRadjb | 95% CI | P | |
| Age >60 years | 3.53 | (2.33–5.36) | <0.001 | 4.53 | (2.93–6.99) | <0.001 |
| Burn size >40% TBSA | 4.21 | (2.49–7.11) | <0.001 | 4.24 | (2.61–6.91) | <0.001 |
| Inhalation injury | 2.61 | (1.63–4.16) | <0.001 | 2.08 | (1.35–3.21) | 0.001 |
Seven subjects were excluded due to missing data for one or more adjustment covariates.
The estimate for each of the three risk factors is adjusted for the other factors as well as sex, body mass index, race, ethnicity, and burn mechanism.
RR, relative risk; RRadj, adjusted relative risk; CI, confidence interval; TBSA, total body surface area burned.
Early gene-expression associated with risk factors for mortality
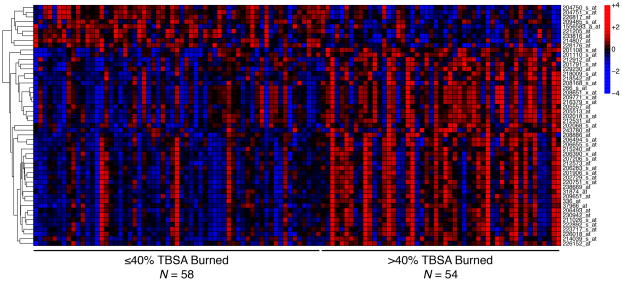
A subset of 112 patients had pooled-leukocyte samples from within 7 days post-burn available for differential-expression analysis (Supplemental Digital Content 1); their characteristics and outcomes were highly consistent with those of the overall cohort (Table 1). The median time from burn injury to blood sampling for RNA isolation was 26 hours (IQR: 15–55 hours). We did not identify any differentially expressed probe sets associated with age >60 years or inhalation injury, but burn size >40% TBSA was independently associated with differential expression of 51 probe sets, of which 42 were up-regulated (Figure 1 and Supplemental Digital Content 2). These 51 differentially expressed probe sets represented 39 unique genes (Table 3). Functional prediction illustrated a high degree of known association among this gene set, based predominantly on co-expression but also on physical interactions and co-localization identified in previous studies (Figure 2).
FIGURE 1.
Heat map of 51 probe sets differentially expressed in whole-blood leukocytes isolated from patients with burn size >40% total body surface area (TBSA). Columns represent individual patient samples, ordered from left to right by increasing %TBSA burned. Each row represents one probe set that was differentially expressed in association with >40% TBSA burn after adjusting for age and inhalation injury; rows were ordered by hierarchical clustering. Expression values are scaled in the row direction, with red indicating relatively high expression and blue relatively low expression.
TABLE 3.
Genes differentially expresseda in whole-blood leukocytes isolated from patients with >40% TBSA burn.
| Symbol | Name | Fold Change | Padj |
|---|---|---|---|
| LCN2 | lipocalin 2 | 2.63 | 0.043 |
| LTF | lactotransferrin | 2.39 | 0.029 |
| THBS1b | thrombospondin 1 | 2.21 | 0.028 |
| ITGA2Bb | integrin, alpha 2b | 2.05 | 0.032 |
| CD24b | cluster of differentiation 24 molecule | 2.01 | 0.041 |
| LAPTM4B | lysosomal protein transmembrane 4 beta | 1.93 | 0.023 |
| TCN1 | transcobalamin I | 1.93 | 0.029 |
| BPI | bactericidal/permeability-increasing protein | 1.92 | 0.029 |
| SLC51A | solute carrier family 51, alpha subunit | 1.81 | 0.029 |
| PF4 | platelet factor 4 | 1.76 | 0.045 |
| TGFB1I1 | TGF beta 1 induced transcript 1 | 1.74 | 0.023 |
| MTURN | maturin, neural progenitor differentiation regulator homolog | 1.72 | 0.014 |
| ALOX12 | arachidonate 12-lipoxygenase | 1.71 | 0.027 |
| ACRBP | acrosin binding protein | 1.68 | 0.028 |
| TAL1 | T-cell acute lymphocytic leukemia 1 | 1.68 | 0.031 |
| LTBP1 | latent TGF beta binding protein 1 | 1.62 | 0.016 |
| CTDSPL | CTD small phosphatase-like | 1.61 | 0.023 |
| PTGS1 | prostaglandin-endoperoxide synthase 1 | 1.61 | 0.023 |
| CHIT1 | chitinase 1 | 1.61 | 0.026 |
| ENDOD1 | endonuclease domain containing 1 | 1.58 | 0.025 |
| MGLL | monoglyceride lipase | 1.58 | 0.042 |
| GAS2L1 | growth arrest-specific 2 like 1 | 1.57 | 0.031 |
| LDLR | low density lipoprotein receptor | 1.57 | 0.026 |
| CEP55 | centrosomal protein 55kDa | 1.55 | 0.027 |
| TMEM40 | transmembrane protein 40 | 1.54 | 0.029 |
| TTC7B | tetratricopeptide repeat domain 7B | 1.54 | 0.023 |
| CMTM5 | CKLF-like MARVEL transmembrane domain containing 5 | 1.53 | 0.040 |
| PRC1 | protein regulator of cytokinesis 1 | 1.53 | 0.027 |
| H1F0 | H1 histone family, member 0 | 1.53 | 0.016 |
| DHCR7 | 7-dehydrocholesterol reductase | 1.52 | 0.012 |
| RPS6KA2 | ribosomal protein S6 kinase, 90kDa, polypeptide 2 | 1.52 | 0.033 |
| ITGB3 | integrin, beta 3 | 1.52 | 0.031 |
| TBXA2R | thromboxane A2 receptor | 1.51 | 0.016 |
| PARVB | parvin, beta | 1.51 | 0.027 |
| FAXDC2 | fatty acid hydroxylase domain containing 2 | 1.50 | 0.027 |
| SLC8A1 | solute carrier family 8, member 1 | −1.51 | 0.049 |
| S1PR3 | sphingosin×10 -phosphate receptor 3 | −1.52 | 0.026 |
| OSBPL1A | oxysterol binding protein-like 1A | −1.66 | 0.023 |
| DSC2b | desmocollin 2 | −1.83 | 0.016 |
Defined as |fold change| >1.5 and Padj <0.05.
For genes with multiple significant probe sets, data are for the probe set with the greatest |fold change|. CKLF, chemokine-like factor; CTD, carboxy-terminal domain; kDa, kilodalton; Padj, false-discovery-rate-adjusted P-value; TBSA, total body surface area; TGF, transforming growth factor.
FIGURE 2.
Gene network based on the 39 unique genes differentially regulated in whole-blood leukocytes isolated from patients with burn size >40% total body surface area (TBSA). Nodes represent gene products, with black circles indicating genes identified in our differential-expression analysis and gray circles indicating related genes. For related genes (gray circles), node size is proportional to the gene score assigned by GeneMANIA. Line thickness is proportional to the weight of the link, and line color indicates the type of link: purple, co-expression; pink, physical interaction; blue, co-localization; and cyan, participation in a common reaction within a pathway.
Biological pathways associated with burn size >40% TBSA
In order to identify cellular processes differentially regulated in association with burn size >40% TBSA, we performed pathway analysis using two approaches. Testing for GO terms significantly enriched among the gene network constructed from the 39 differentially expressed genes (Figure 2) yielded 36 terms with FDR-adjusted P <0.05 (Table 4). Significantly associated GO terms pertained to platelet activation, cellular exocytosis and degranulation, tumor necrosis factor (TNF) production, and cellular adhesion and migration, consistent with expected cellular responses to an inflammatory stimulus.
TABLE 4.
GO terms significantlya enriched among 39 genes differentially expressed in whole-blood leukocytes isolated from patients with >40% TBSA burn.
| ID | Name | Padj | # Genesb |
|---|---|---|---|
| GO:0030168 | Platelet activation | 2.33×10−8 | 12 (211) |
| GO:0002576 | Platelet degranulation | 2.47×10−8 | 9 (82) |
| GO:0031091 | Platelet alpha granule | 6.69×10−8 | 8 (61) |
| GO:0030141 | Secretory granule | 1.23×10−7 | 10 (152) |
| GO:0060205 | Cytoplasmic membrane-bounded vesicle lumen | 9.06×10−6 | 7 (76) |
| GO:0031983 | Vesicle lumen | 9.06×10−6 | 7 (76) |
| GO:0006887 | Exocytosis | 1.45×10−5 | 9 (191) |
| GO:0031093 | Platelet alpha granule lumen | 1.51×10−5 | 6 (48) |
| GO:0034774 | Secretory granule lumen | 6.50×10−5 | 6 (62) |
| GO:0030198 | Extracellular matrix organization | 3.45×10−4 | 9 (290) |
| GO:0043062 | Extracellular structure organization | 3.45×10−4 | 9 (291) |
| GO:0030099 | Myeloid cell differentiation | 1.40×10−2 | 6 (164) |
| GO:0034329 | Cell junction assembly | 1.40×10−2 | 6 (164) |
| GO:0045637 | Regulation of myeloid cell differentiation | 1.40×10−2 | 5 (96) |
| GO:0050431 | Transforming growth factor beta binding | 1.47×10−2 | 3 (15) |
| GO:0006875 | Cellular metal ion homeostasis | 1.62×10−2 | 7 (264) |
| GO:0007160 | Cell-matrix adhesion | 1.62×10−2 | 5 (103) |
| GO:0034330 | Cell junction organization | 1.80×10−2 | 6 (181) |
| GO:0032640 | Tumor necrosis factor production | 2.00×10−2 | 4 (54) |
| GO:0032680 | Regulation of tumor necrosis factor production | 2.00×10−2 | 4 (54) |
| GO:0030003 | Cellular cation homeostasis | 2.01×10−2 | 7 (284) |
| GO:0055065 | Metal ion homeostasis | 2.01×10−2 | 7 (288) |
| GO:0019955 | Cytokine binding | 2.01×10−2 | 4 (56) |
| GO:0006873 | Cellular ion homeostasis | 2.10×10−2 | 7 (292) |
| GO:0071706 | TNF superfamily cytokine production | 2.27×10−2 | 4 (59) |
| GO:0032369 | Negative regulation of lipid transport | 2.44×10−2 | 3 (21) |
| GO:0008201 | Heparin binding | 2.54×10−2 | 4 (63) |
| GO:0070161 | Anchoring junction | 2.54×10−2 | 5 (125) |
| GO:0010634 | Positive regulation of epithelial cell migration | 2.54×10−2 | 4 (63) |
| GO:0010631 | Epithelial cell migration | 3.39×10−2 | 5 (135) |
| GO:0050900 | Leukocyte migration | 4.01×10−2 | 6 (230) |
| GO:0042383 | Sarcolemma | 4.31×10−2 | 3 (27) |
| GO:0010743 | Regulation of foam cell differentiation | 4.40×10−2 | 3 (28) |
| GO:0001525 | Angiogenesis | 4.40×10−2 | 6 (237) |
| GO:0006869 | Lipid transport | 4.40×10−2 | 5 (147) |
| GO:0031589 | Cell-substrate adhesion | 4.96×10−2 | 5 (152) |
Defined as Padj <0.05.
Number of identified genes (vs. total number of known genes) associated with GO term.
GO, Gene Ontology; Padj, false-discovery-rate-adjusted P-value; TBSA, total body surface area.
Taking a complementary approach, we tested 5,389 GO Biological Process concepts (sets of genes associated with specific GO Biological Process terms) for coordinated changes in expression of multiple genes. We identified 113 GO concepts that were significantly enriched with up-regulated genes (Supplemental Digital Content 3) and 51 significantly enriched with down-regulated genes (Supplemental Digital Content 4) in association with burn size >40% TBSA. Nearly all of the GO concepts most significantly enriched with up-regulated genes related to cellular division (Supplemental Digital Content 3), suggesting increased leukocyte proliferation in response to inflammatory cytokines released by burned tissue; in addition, a few related to activation of the clotting cascade, consistent with results of our pathway analysis based on significantly differentially expressed genes (Table 4). Some of the GO concepts most significantly enriched with down-regulated genes pertained to regulation and production of cytokines, specifically IL-1b and IL-6 (Supplemental Digital Content 4). Thus, we examined expression of IL1B and IL6 and confirmed that both were modestly down-regulated in association with >40% TBSA burn after adjusting for age and inhalation injury: fold change = −1.44, unadjusted P = 0.019 for IL1B; and fold change = −1.06, unadjusted P = 0.046 for IL6.
DISCUSSION
In the burn-injured patient, older age, larger burn, and inhalation injury are well established risk factors for death, which has historically resulted from burn shock, multisystem organ failure, and sepsis, implicating burn-induced immune dysregulation as a contributory mechanism. Analyzing data from a recent multicenter prospective cohort study, we have confirmed that in the modern era of burn care, these three factors remain highly associated with risk of mortality and that multisystem organ failure and infectious complications persist as the most common causes of death among severely burned adults. These observations underscore the importance of understanding the effect of age, burn size, and inhalation injury on the host inflammatory response to burn injury. We report the first published analysis of transcriptomic changes associated with the three major risk factors for burn mortality.
There were relatively few significant changes in expression of individual genes associated with the three risk factors of interest. Since only 13 (12%) patients included in our genomic analysis were older than 60 years, it is possible that our analysis was underpowered to detect differential expression associated with advanced age. Although nearly half of the included patients had an inhalation injury, some patients were diagnosed based on clinical suspicion alone (16), predisposing to potential differential misclassification that would bias estimates of association toward the null. However, when analyzing gene expression associated with burn size, which benefitted from both more direct and objective exposure-measurement and a near-equal distribution of exposed vs. unexposed patients (Figure 1), we identified only 39 differentially expressed genes. In contrast, a previous analysis comparing patients with >20% TBSA burned to uninjured controls and applying even more stringent significance criteria than used in our present analysis identified 3,250 differentially expressed genes (31). As all of the patients in our study had burns >20% TBSA, we hypothesize that this “genomic storm” induced by a >20% TBSA burn may saturate the leukocyte transcriptomic response, making it more difficult to detect more subtle perturbations in leukocyte gene-expression induced by other factors. Studying a patient population with smaller burns (to increase the signal-to-noise ratio) or a much larger population of patients with large burns (to increase power to detect more subtle gene-expression changes) would likely facilitate future studies of the transcriptomic changes associated with older age and inhalation injury in burn patients.
The genes and biological processes we identified in association with burn size >40% TBSA may help elucidate the increased risk of morbidity and mortality in this group (30). GO-term enrichment analysis based on the set of 39 differentially expressed genes highlighted platelet activation and leukocyte degranulation/exocytosis as being over-represented (Table 5), suggesting endothelial damage and thrombosis as contributory pathophysiologic mechanisms. Consistent with this notion are clinical observations that greater burn size is associated with higher incidence of thromboembolic complications (32, 33). In addition, burn size >40% TBSA has been associated with increased hypercoagulability and disseminated intravascular coagulation (DIC) contributing to multisystem organ failure (34, 35), the most common cause of death among patients included in our genomic analysis (Table 3). Since anti-thrombin administration has been shown to decrease multisystem organ failure and mortality in a small randomized trial of severely burned patients (36), pro-thrombotic genes identified in this study (Table 4), such as platelet factor 4 (PF4) and prostaglandin-endoperoxide synthase 1 (PTGS1, also known as cyclooxygenase 1), may represent novel therapeutic targets for preventing multisystem organ failure and death after burn injury.
Sepsis was a common cause of mortality (Table 1) in our study, and burn size >40% TBSA was associated with increased incidence of sepsis as well as pneumonia and burn wound infections in a recent analysis of the same cohort (30). IL-1β is a pro-inflammatory cytokine secreted by macrophages that has been demonstrated to be elevated in plasma following burns (11, 37) and in association with sepsis (38). Interestingly, the top GO concepts significantly enriched with down-regulated genes in association with burn size >40% TBSA were largely related to immune-cell activation and production of pro-inflammatory cytokines, particularly IL-1β (Supplemental Digital Content 4), and we also found that IL1B transcription was down-regulated. Since transcriptional changes do not necessarily correlate with protein levels, the significance of these findings is unclear. Down-regulation of IL1B at the mRNA level may correspond to decreased IL-1β protein levels, or it may represent a homeostatic negative-feedback response to increased levels of IL-1β protein. Hence, our results call for future studies to investigate the potential role of leukocyte IL-1β production in sepsis risk associated with larger burns.
In addition to increased risk of morbidity and mortality in the immediate post-burn period, larger burns are also associated with increased severity of hypertrophic scarring (39), a common long-term sequela of burn injury that results in stiff, raised, contracted scars associated with disfigurement, functional limitation, and decreased quality of life (40). The mechanism connecting burn size to increased scar severity is currently unknown, and a wide range of circulating immune cells and factors have been implicated in HTS formation (41). Thus, the differential gene-expression profile we have identified in association with burn size >40% TBSA may help to elucidate HTS pathobiology. For instance, thrombospondin 1 was one of the top up-regulated genes in association with burn size >40% TBSA (Table 4), and it is known to activate transforming growth factor β1, a key mediator of fibrogenesis and hypertrophic scarring (42). Although thrombospondin 1 has long been implicated in wound healing and scarring, this is to our knowledge the first report of increased thrombospondin 1 expression by circulating leukocytes in patients with large burns. Hence, further studies examining leukocyte expression and plasma levels of thrombospondin 1 (Table 4) in relation to scar outcomes may identify novel therapeutic targets.
Since we examined gene expression of whole-blood leukocytes, it is not clear which cell type(s) may be driving the observed changes nor whether observed gene-expression changes actually reflect differences in relative concentrations of different leukocyte populations. Further studies using leukocyte separations will be required to clarify this issue. In addition, to maximize the number of samples available for analysis, we included patients with samples available at any time within 7 days post-burn. In doing so, we could have failed to detect biologically relevant differences in gene expression occurring only at specific time points within the first week. Finally, changes in gene expression may not reflect changes in expression at the protein level, and thus measuring protein expression will be a necessary first step in further studies of gene targets we have identified.
In summary, age >60 years, burn size >40% TBSA, and inhalation injury continue to be strongly associated with risk of death in burned adults. Although our transcriptomic analyses suggest that changes in leukocyte gene expression related to these factors are modest in comparison to the “genomic storm” induced by >20% TBSA burn, we were able to identify differentially regulated genes and pathways associated with burn size >40% TBSA. Understanding these associations may help elucidate the biological mechanisms linking burn size to increased morbidity and mortality.
Supplementary Material
Patient demographics, injury characteristics, and causes of death among patients with pooled-leukocyte microarray data obtained within 7 days of burn injury (N = 112).
Probe sets differentially expressed by whole-blood leukocytes isolated from patients with >40% TBSA burn.
Gene Ontology (GO) Biological Process Concepts signficantly enriched with up-regulated genes in whole-blood leukocytes isolated from patients with >40% TBSA burn.
Gene Ontology (GO) Biological Process Concepts signficantlya enriched with down-regulated genes in whole-blood leukocytes isolated from patients with >40% TBSA burn.
Acknowledgments
National Institutes of Health funding support: U54 GM6211904 and T32 GM007037.
We would like to thank our Glue Grant collaborators for collecting data, processing samples, and maintaining the Trauma Research Database.
Footnotes
Disclosure:
The authors declare no conflicts of interest.
Presented at the American College of Surgeons Clinical Congress Surgical Forum on October 29, 2014 in San Francisco, California.
AUTHOR CONTRIBUTIONS
R.F.S. conceptualized and designed the study, performed the statistical analysis, interpreted the data, drafted the article, and approved the final version of the article as submitted. N.S.G. conceptualized and designed the study, interpreted the data, drafted the article, and approved the final version of the article as submitted. B.D.A. reviewed the analysis, revised the article, and approved the final version of the article as submitted. R.L.G. reviewed the analysis, revised the article, and approved the final version of the article as submitted. D.N.H. reviewed the analysis, revised the article, and approved the final version of the article as submitted. R.G.T reviewed the analysis, revised the article, and approved the final version of the article as submitted.
References
- 1.Branski LK, Herndon DN, Barrow RE. A brief history of acute burn care management. In: Herndon DN, editor. Total Burn Care. 4. Philadelphia, PA: Saunders; 2012. pp. 1–9. [Google Scholar]
- 2.Peck MD. Epidemiology of burns throughout the world. Part I: Distribution and risk factors. Burns. 2011;37(7):1087–100. doi: 10.1016/j.burns.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 3.Bull JP, Squire JR. A Study of Mortality in a Burns Unit: Standards for the Evaluation of Alternative Methods of Treatment. Ann Surg. 1949;130(2):160–73. doi: 10.1097/00000658-194908000-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hussain A, Choukairi F, Dunn K. Predicting survival in thermal injury: a systematic review of methodology of composite prediction models. Burns. 2013;39(5):835–50. doi: 10.1016/j.burns.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 5.Taylor SL, Lawless M, Curri T, Sen S, Greenhalgh DG, Palmieri TL. Predicting mortality from burns: the need for age-group specific models. Burns. 2014;40(6):1106–15. doi: 10.1016/j.burns.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Latenser BA, Miller SF, Bessey PQ, Browning SM, Caruso DM, Gomez M, Jeng JC, Krichbaum JA, Lentz CW, Saffle JR, Schurr MJ, Greenhalgh DG, Kagan RJ. National Burn Repository 2006: a ten-year review. J Burn Care Res. 2007;28(5):635–58. doi: 10.1097/BCR.0B013E31814B25B1. [DOI] [PubMed] [Google Scholar]
- 7.Bloemsma GC, Dokter J, Boxma H, Oen IM. Mortality and causes of death in a burn centre. Burns. 2008;34(8):1103–7. doi: 10.1016/j.burns.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 8.Brusselaers N, Monstrey S, Vogelaers D, Hoste E, Blot S. Severe burn injury in Europe: a systematic review of the incidence, etiology, morbidity, and mortality. Crit Care. 2010;14(5):R188. doi: 10.1186/cc9300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swanson JW, Otto AM, Gibran NS, Klein MB, Kramer CB, Heimbach DM, Pham TN. Trajectories to death in patients with burn injury. J Trauma Acute Care Surg. 2013;74(1):282–8. doi: 10.1097/TA.0b013e3182788a1c. [DOI] [PubMed] [Google Scholar]
- 10.Finnerty CC, Jeschke MG, Herndon DN, Gamelli R, Gibran N, Klein M, Silver G, Arnoldo B, Remick D, Tompkins RG Investigators of the I, the Host Response Glue G. Temporal cytokine profiles in severely burned patients: a comparison of adults and children. Mol Med. 2008;14(9–10):553–60. doi: 10.2119/2007-00132.Finnerty. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeschke MG, Chinkes DL, Finnerty CC, Kulp G, Suman OE, Norbury WB, Branski LK, Gauglitz GG, Mlcak RP, Herndon DN. Pathophysiologic response to severe burn injury. Ann Surg. 2008;248(3):387–401. doi: 10.1097/SLA.0b013e3181856241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeschke MG, Mlcak RP, Finnerty CC, Norbury WB, Gauglitz GG, Kulp GA, Herndon DN. Burn size determines the inflammatory and hypermetabolic response. Crit Care. 2007;11(4):R90. doi: 10.1186/cc6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis CS, Janus SE, Mosier MJ, Carter SR, Gibbs JT, Ramirez L, Gamelli RL, Kovacs EJ. Inhalation injury severity and systemic immune perturbations in burned adults. Ann Surg. 2013;257(6):1137–46. doi: 10.1097/SLA.0b013e318275f424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao W, Mindrinos MN, Seok J, Cuschieri J, Cuenca AG, Gao H, Hayden DL, Hennessy L, Moore EE, Minei JP, Bankey PE, Johnson JL, Sperry J, Nathens AB, Billiar TR, West MA, Brownstein BH, Mason PH, Baker HV, Finnerty CC, Jeschke MG, Lopez MC, Klein MB, Gamelli RL, Gibran NS, Arnoldo B, Xu W, Zhang Y, Calvano SE, McDonald-Smith GP, Schoenfeld DA, Storey JD, Cobb JP, Warren HS, Moldawer LL, Herndon DN, Lowry SF, Maier RV, Davis RW, Tompkins RG Inflammation, Host Response to Injury Large-Scale Collaborative Research P. A genomic storm in critically injured humans. J Exp Med. 2011;208(13):2581–90. doi: 10.1084/jem.20111354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klein MB, Silver G, Gamelli RL, Gibran NS, Herndon DN, Hunt JL, Tompkins RG Inflammation, the Host Response to Injury I. Inflammation and the host response to injury: an overview of the multicenter study of the genomic and proteomic response to burn injury. J Burn Care Res. 2006;27(4):448–51. doi: 10.1097/01.BCR.0000227477.33877.E6. [DOI] [PubMed] [Google Scholar]
- 16.Silver GM, Klein MB, Herndon DN, Gamelli RL, Gibran NS, Altstein L, McDonald-Smith GP, Tompkins RG, Hunt JL Inflammation, the Host Response to Trauma CRP. Standard operating procedures for the clinical management of patients enrolled in a prospective study of Inflammation and the Host Response to Thermal Injury. J Burn Care Res. 2007;28(2):222–30. doi: 10.1097/BCR.0B013E318031AA44. [DOI] [PubMed] [Google Scholar]
- 17.Ryan CM, Schoenfeld DA, Thorpe WP, Sheridan RL, Cassem EH, Tompkins RG. Objective estimates of the probability of death from burn injuries. N Engl J Med. 1998;338(6):362–6. doi: 10.1056/NEJM199802053380604. [DOI] [PubMed] [Google Scholar]
- 18.Feezor RJ, Baker HV, Mindrinos M, Hayden D, Tannahill CL, Brownstein BH, Fay A, MacMillan S, Laramie J, Xiao W, Moldawer LL, Cobb JP, Laudanski K, Miller-Graziano CL, Maier RV, Schoenfeld D, Davis RW, Tompkins RG Inflammation Host Response to Injury L-SCRP. Whole blood and leukocyte RNA isolation for gene expression analyses. Physiol Genomics. 2004;19(3):247–54. doi: 10.1152/physiolgenomics.00020.2004. [DOI] [PubMed] [Google Scholar]
- 19.Cobb JP, Mindrinos MN, Miller-Graziano C, Calvano SE, Baker HV, Xiao W, Laudanski K, Brownstein BH, Elson CM, Hayden DL, Herndon DN, Lowry SF, Maier RV, Schoenfeld DA, Moldawer LL, Davis RW, Tompkins RG, Baker HV, Bankey P, Billiar T, Brownstein BH, Calvano SE, Camp D, Chaudry I, Cobb JP, Davis RW, Elson CM, Freeman B, Gamelli R, Gibran N, Harbrecht B, Hayden DL, Heagy W, Heimbach D, Herndon DN, Horton J, Hunt J, Laudanski K, Lederer J, Lowry SF, Maier RV, Mannick J, McKinley B, Miller-Graziano C, Mindrinos MN, Minei J, Moldawer LL, Moore E, Moore F, Munford R, Nathens A, O’Keefe G, Purdue G, Rahme L, Remick D, Sailors M, Schoenfeld DA, Shapiro M, Silver G, Smith R, Stephanopoulos G, Stormo G, Tompkins RG, Toner M, Warren S, West M, Wolfe S, Xiao W, Young V Inflammation, Host Response to Injury Large-Scale Collaborative Research P. Application of genome-wide expression analysis to human health and disease. Proc Natl Acad Sci U S A. 2005;102(13):4801–6. doi: 10.1073/pnas.0409768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 21.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–27. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 22.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 23.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995:289–300. [Google Scholar]
- 24.Montojo J, Zuberi K, Rodriguez H, Kazi F, Wright G, Donaldson SL, Morris Q, Bader GD. GeneMANIA Cytoscape plugin: fast gene function predictions on the desktop. Bioinformatics. 2010;26(22):2927–8. doi: 10.1093/bioinformatics/btq562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khatri P, Sirota M, Butte AJ. Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput Biol. 2012;8(2):e1002375. doi: 10.1371/journal.pcbi.1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim JH, Karnovsky A, Mahavisno V, Weymouth T, Pande M, Dolinoy DC, Rozek LS, Sartor MA. LRpath analysis reveals common pathways dysregulated via DNA methylation across cancer types. BMC Genomics. 2012;13:526. doi: 10.1186/1471-2164-13-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Team RC. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2014. [Google Scholar]
- 29.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeschke MG, Pinto R, Kraft R, Nathens AB, Finnerty CC, Gamelli RL, Gibran NS, Klein MB, Arnoldo BD, Tompkins RG, Herndon DN Inflammation, the Host Response to Injury Collaborative Research P. Morbidity and survival probability in burn patients in modern burn care. Crit Care Med. 2015;43(4):808–15. doi: 10.1097/CCM.0000000000000790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, Richards DR, McDonald-Smith GP, Gao H, Hennessy L, Finnerty CC, Lopez CM, Honari S, Moore EE, Minei JP, Cuschieri J, Bankey PE, Johnson JL, Sperry J, Nathens AB, Billiar TR, West MA, Jeschke MG, Klein MB, Gamelli RL, Gibran NS, Brownstein BH, Miller-Graziano C, Calvano SE, Mason PH, Cobb JP, Rahme LG, Lowry SF, Maier RV, Moldawer LL, Herndon DN, Davis RW, Xiao W, Tompkins RG Inflammation Host Response to Injury LSCRP. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2013;110(9):3507–12. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mullins F, Mian MA, Jenkins D, Brandigi C, Shaver JR, Friedman B, Alam B, Schwartz M, Hassan Z. Thromboembolic complications in burn patients and associated risk factors. J Burn Care Res. 2013;34(3):355–60. doi: 10.1097/BCR.0b013e31827819a1. [DOI] [PubMed] [Google Scholar]
- 33.Harrington DT, Mozingo DW, Cancio L, Bird P, Jordan B, Goodwin CW. Thermally injured patients are at significant risk for thromboembolic complications. J Trauma. 2001;50(3):495–9. doi: 10.1097/00005373-200103000-00014. [DOI] [PubMed] [Google Scholar]
- 34.Garcia-Avello A, Lorente JA, Cesar-Perez J, Garcia-Frade LJ, Alvarado R, Arevalo JM, Navarro JL, Esteban A. Degree of hypercoagulability and hyperfibrinolysis is related to organ failure and prognosis after burn trauma. Thromb Res. 1998;89(2):59–64. doi: 10.1016/s0049-3848(97)00291-0. [DOI] [PubMed] [Google Scholar]
- 35.Lippi G, Ippolito L, Cervellin G. Disseminated intravascular coagulation in burn injury. Semin Thromb Hemost. 2010;36(4):429–36. doi: 10.1055/s-0030-1254051. [DOI] [PubMed] [Google Scholar]
- 36.Lavrentieva A, Kontakiotis T, Bitzani M, Parlapani A, Thomareis O, Scourtis H, Tsotsolis N, Lazaridis L, Giala MA. The efficacy of antithrombin administration in the acute phase of burn injury. Thromb Haemost. 2008;100(2):286–90. [PubMed] [Google Scholar]
- 37.Jeschke MG, Gauglitz GG, Kulp GA, Finnerty CC, Williams FN, Kraft R, Suman OE, Mlcak RP, Herndon DN. Long-term persistance of the pathophysiologic response to severe burn injury. PLoS One. 2011;6(7):e21245. doi: 10.1371/journal.pone.0021245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pierrakos C, Vincent JL. Sepsis biomarkers: a review. Crit Care. 2010;14(1):R15. doi: 10.1186/cc8872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sood RF, Hocking AM, Muffley LA, Ga M, Honari S, Reiner AP, Rowhani-Rahbar A, Gibran NS. Race and Melanocortin 1 Receptor Polymorphism R163Q are Associated with Post-Burn Hypertrophic Scarring: A Prospective Cohort Study. J Invest Dermatol. 2015 doi: 10.1038/jid.2015.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lawrence JW, Mason ST, Schomer K, Klein MB. Epidemiology and impact of scarring after burn injury: a systematic review of the literature. J Burn Care Res. 2012;33(1):136–46. doi: 10.1097/BCR.0b013e3182374452. [DOI] [PubMed] [Google Scholar]
- 41.Bloemen MC, van der Veer WM, Ulrich MM, van Zuijlen PP, Niessen FB, Middelkoop E. Prevention and curative management of hypertrophic scar formation. Burns. 2009;35(4):463–75. doi: 10.1016/j.burns.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 42.Leask A, Abraham DJ. TGF-beta signaling and the fibrotic response. FASEB J. 2004;18(7):816–27. doi: 10.1096/fj.03-1273rev. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Patient demographics, injury characteristics, and causes of death among patients with pooled-leukocyte microarray data obtained within 7 days of burn injury (N = 112).
Probe sets differentially expressed by whole-blood leukocytes isolated from patients with >40% TBSA burn.
Gene Ontology (GO) Biological Process Concepts signficantly enriched with up-regulated genes in whole-blood leukocytes isolated from patients with >40% TBSA burn.
Gene Ontology (GO) Biological Process Concepts signficantlya enriched with down-regulated genes in whole-blood leukocytes isolated from patients with >40% TBSA burn.