Abstract
We report that the mammalian 5-methylcytosine (5mC) oxidase Tet3 exists as three major isoforms and characterized the full-length isoform containing an N-terminal CXXC domain (Tet3FL). This CXXC domain binds to unmethylated CpGs but unexpectedly its highest affinity is towards 5-carboxylcytosine (5caC). We determined the crystal structure of the CXXC domain - 5caC-DNA complex revealing the structural basis of the binding specificity of this domain as a reader of CcaCG sequences. Mapping of Tet3FL in neuronal cells shows that Tet3FL is localized precisely at the transcription start sites (TSS) of genes involved in lysosome function, mRNA processing and key genes of the base excision repair pathway. Thus, Tet3FL may function as a regulator of 5caC removal by base excision repair. Active removal of accumulating 5mC from the TSS of genes coding for lysosomal proteins by Tet3FL in postmitotic neurons of the brain may be important for preventing neurodegenerative diseases.
INTRODUCTION
5-methylcytosine (5mC) is a modified cytosine base implicated in gene control and has long been thought to be the only modified base naturally present in mammalian DNA (Klose and Bird, 2006). Only recently, 5-hydroxymethylcytosine (5hmC) has also been identified (Kriaucionis and Heintz, 2009; Tahiliani et al., 2009). 5hmC is formed enzymatically by the Tet family of 5mC oxidases (Tahiliani et al., 2009; Ito et al., 2010) and is now thought to be a stable component of the epigenetic code (Koh and Rao, 2013; Pfeifer et al., 2013; Wu and Zhang, 2014). Alternatively, 5hmC has been viewed as an intermediate base in developmentally controlled DNA demethylation reactions. The two proposed functions of 5hmC are not necessarily mutually exclusive (Hahn et al., 2014).
Levels of 5hmC are particularly high in neuronal cells where they reach up to ~1% of all cytosines, for example in the human brain (Münzel et al., 2010). The function of this base in neurons is still unclear. Mapping studies have shown that 5hmC is prominently localized within transcribed sequences of a number of neuronal function-related genes (Szulwach et al., 2011; Hahn et al., 2013). In the embryonic mouse brain, 5hmC formation along important neuron-specific genes parallels neuronal differentiation, and depletion of Tet2 and Tet3 leads to a block of neuron migration suggesting that 5hmC is important for brain development (Hahn et al., 2013). Substantial levels of 5hmC also occur at regions near promoters and enhancers, for example in embryonic stem (ES) cells (Kriaucionis and Heintz, 2009; Ficz et al., 2011; Williams et al., 2011). 5mC oxidation appears to be important in keeping such sequences in an unmethylated state (Williams et al., 2011; Hahn et al., 2014). During other developmental phases, when DNA methylation is erased globally, 5hmC can be viewed as a transiently existing base that promotes DNA demethylation, e.g. in zygotes and in primordial germ cells (Gu et al., 2011; Iqbal et al., 2011; Wossidlo et al., 2011; Hackett et al., 2013). Recent studies have dissected the contribution of Tet3 to DNA demethylation in zygotes and concluded that there are Tet3-dependent and Tet3-independent but replication-associated DNA demethylation events in the paternal pronucleus of the zygote (Guo et al., 2014; Peat et al., 2014; Shen et al., 2014).
The three related mammalian proteins Tet1, Tet2, and Tet3 all possess 5mC oxidase activity but they differ in terms of domain architecture and tissue specificity of their expression levels (Tahiliani et al., 2009; Ito et al., 2010). For example, while Tet1 and Tet2 mRNA levels are abundant in embryonic stem (ES) cells and in primordial germ cells (Ito et al., 2010; Ficz et al., 2011; Gu et al., 2011; Hackett et al., 2013; Yamaguchi et al., 2013; Huang et al., 2014), Tet3 is the only Tet gene expressed at substantial levels in oocytes and zygotes (Gu et al., 2011; Iqbal et al., 2011; Wossidlo et al., 2011). Presumably, the Tet proteins also show functional differences but their specific properties are currently not understood. The Tet1 and Tet3 5mC oxidases are characterized by two conserved domains, an N-terminal CXXC domain, which binds to CpG dinucleotides, and a C-terminal Fe(II) and 2-ketoglutarate-dependent catalytic domain which progressively converts 5mC to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and terminally to 5-carboxylcytosine (5caC) resulting in active or passive DNA demethylation (Tahiliani et al., 2009; Ito et al., 2010; He et al., 2011; Ito et al., 2011; Shen et al., 2013; Hashimoto et al., 2014; Hu et al., 2014). Passive DNA demethylation is achieved by the inability of maintenance DNA methyltransferase Dnmt1 to copy the CpG methylation pattern at sequences that contain 5hmC (Valinluck and Sowers, 2007; Hashimoto et al., 2012). Active demethylation can be accomplished by removal of 5fC or 5caC through thymine DNA glycosylase (TDG) initiated base excision repair (BER) (He et al., 2011), or perhaps alternatively through a yet unidentified 5caC decarboxylase activity.
Here, we have focused on mouse Tet3 and characterized Tet3 function with special emphasis on its long isoform that contains an N-terminal CXXC domain. We found that the CXXC domain of Tet3 has the capacity to bind to unmethylated and carboxylated cytosines at CpG sequences and that full-length Tet3 has a very restricted genomic localization pattern with a preference for transcription start sites of a specific set of important genes in neuronal cell populations.
RESULTS
Three different Tet3 transcript isoforms
Tet1 has a single transcript containing all conserved domains. Tet2, however, has lost its N-terminal CXXC domain, which is now encoded by a separate, neighboring gene named Idax/Cxxc4 (Ko et al., 2013). Idax/Cxxc4 forms a complex with Tet2 negatively regulating its activity. Tet3 possesses two known isoforms, one containing the CXXC domain, which we refer to here as Tet3-full-length (Tet3FL), and one lacking a CXXC domain, Tet3-short (Tet3s) (Fig. 1A). A recent study indicated that the CXXC domain of Xenopus tropicalis Tet3 (xtTet3) functions via binding to CpG dinucleotides in either unmodified or methylated states (Xu et al., 2012). Interestingly, the level of Xenopus Tet3 mRNA is very low in oocytes, and this may explain the lack of genome-wide DNA demethylation in fertilized Xenopus eggs, which in mammals involves Tet3-dependent oxidation of 5-methylcytosines in the paternal genome after fertilization (Gu et al., 2011; Iqbal et al., 2011; Wossidlo et al., 2011). In contrast, upregulation of mouse Tet3 and the accompanying increase of 5hmC levels during neuronal differentiation affects specific gene targets rather than the genome globally suggesting that Tet3 activity can be modulated based on cellular and genomic context (Hahn et al., 2013).
Figure 1. Functional characterization of different Tet3 isoforms.
(A) Schematic representation of the three Tet3 isoforms.
(B) Anti-V5 Western blot of transfected Tet3 protein isoforms.
(C) Immunofluorescence detection of 5hmC in cells transfected with different Tet3 isoforms. Cells were co-immunostained with anti-V5 tag antibody (green) and with anti-5hmC antibody (red). Scale bar, 20 μm.
(D) 5hmC production by Tet3 isoforms as analyzed by anti-5hmC dot blot. Tet3FL-CXXC-DM, Tet3FL with CXXC domain double mutation (C60A and C72A) deficient in DNA binding.
(E) Reactivation of a methylation-silenced luciferase construct by Tet3 isoforms and by Tet3FL containing the CXXC domain DNA binding mutations (Tet3FLCXXC-DM) in absence or presence of sodium ascorbate.
(F) An unmethylated luciferase vector was used as a control for panel E.
In order to characterize mammalian Tet3, we examined the transcripts of Tet3FL and Tet3s using RT-qPCR with primer pairs spanning the isoform-specific exons at the 5′ end and measured the total levels of Tet3 transcripts with primers spanning the conserved 3′ exons (Fig. S1; Table S1). Total levels of Tet3 were highly abundant in oocytes (Fig. S1D) and were upregulated during retinoic acid-induced differentiation of mouse ES cells towards the neuronal lineage (Fig. S1E,F), consistent with previous results (Ko et al., 2010; Iqbal et al., 2011). Analysis of neuronal markers confirmed the progression of differentiation (Fig. S1G). Using 5′-RACE with oocyte mRNA as well as in silico searches of mouse oocyte EST databases, we identified an undocumented variant of Tet3 in mouse oocytes. This transcript is created by alternative promoter usage and contains an additional N-terminal exon coding for 11 amino acids (MFLPETPQQYA) (Fig. S1A). The promoter is located approximately 5 kb upstream of the start codon of the mouse Tet3FL isoform but lacks the CXXC domain via alternative splicing (Fig. 1A and Fig. S1A). This Tet3 isoform was named Tet3o. Using RT-PCR, we confirmed that there are no additional major spliced Tet3 isoforms other than the three N-terminal variants, Tet3s, Tet3FL, and Tet3o in oocytes or neuronal cells (Fig. S1B,C). A Tet3FL isoform lacking both exons 1 and 2, as described by Liu et al in a neural stem cell line (Liu et al., 2013a), was present at less than 5% of total Tet3FL transcripts in E15.5 mouse embryo brain. Tet3s and Tet3FL were upregulated during ES cell differentiation towards the neuronal lineage (Fig. S1F). Tet3o was not expressed in ES cells or in any other cell type or adult mouse tissue tested (Fig. S1F and data not shown). Together these data show that Tet3o is an oocyte-specific isoform of Tet3 that is predicted to be involved in preferential global oxidation of the paternal genome in mouse zygotes (Gu et al., 2011; Iqbal et al., 2011; Wossidlo et al., 2011) suggesting different functions of Tet3 during early embryonic development and in the neuronal lineage.
Characterization of Tet3 isoforms
We next examined the subcellular localization and activity of each Tet3 isoform in HEK293 cells using transient transfection and immunofluorescence staining with anti-tag (V5) and anti-5hmC antibodies (Fig. 1B,C). The full-length V5-epitope tagged variants of Tet3 were localized with increased 5hmC levels in the nucleus, indicating that they all function as nuclear 5mC oxidases (Fig. 1C). Tet3 isoform-induced genome-wide 5mC oxidation activity was confirmed by measurement of 5hmC levels in genomic DNA using immuno-dot blots (Fig. 1D). In these assays, Tet3FL had the lowest activity. We then used a reporter assay with a 5mC-silenced luciferase vector where repression can be released due to oxidation of 5mC via overexpressed Tet proteins in a vitamin C dose-dependent manner (Fig. 1E,F) (Blaschke et al., 2013; Minor et al., 2013; Yin et al., 2013). This in vivo assay showed that all variants are functionally active, and the oxidase activity of Tet3o and Tet3s can significantly relieve 5mC-induced repression of the luciferase reporter. However, Tet3FL was substantially less effective than Tet3o and Tet3s (Fig. 1E). The different assays suggest that the CXXC domain of Tet3 restricts its genome-wide 5mC oxidation capacity. Our data are in apparent contrast to a previous report (Liu et al., 2013a). Liu et al used an N-terminal GFP-tagged version of Tet3FL. Our construct had a C-terminal V5 tag. In their study, Liu et al showed that direct N-terminal tagging of the Tet1 CXXC domain interferes with DNA binding. Since the CXXC domain of Tet3 is directly at the N-terminus, their tagging approach presents a possible explanation for the differences observed.
Our findings prompted us to investigate the biological properties of Tet3’s CXXC domain. Two previous reports regarding the role of other CXXC domains have indicated that (i) the CXXC domain of DNMT1 can inhibit DNMT1 activity by direct interaction with the catalytic domain of DNMT1 (Song et al., 2011) and (ii) overexpression of Idax/Cxxc4 is capable of inducing caspase-dependent degradation of its binding partner Tet2 (Ko et al., 2013). To test if the first of these CXXC domain-dependent mechanisms could play a role in modulation of Tet3 activity, we incubated the recombinant GST-tagged CXXC domain of Tet3 with a HEK293 cell lysate expressing the catalytic domain of Tet3, but no direct interaction between the CXXC domain and the catalytic domain was observed (data not shown) although we cannot exclude the possibility that such interactions may occur in vivo. To test the second mechanism, caspase inhibitors were added alongside overexpressed Tet3 variants, but this had no specific effect on protein quantity of the Tet3FL isoform (not shown). We therefore hypothesized that the CXXC DNA binding domain restricts the ability of the catalytic domain of Tet3FL to oxidize 5mC globally by limiting its mobility in the nucleus. To abolish the DNA binding, we generated CXXC-domain mutations (C60A and C72A) (Fig. 2A) based on Tet1-CXXC mutations shown to cause a loss of DNA binding (Xu et al., 2011). The assays revealed that loss of DNA binding of the CXXC domain increases the catalytic activity of Tet3FL (Fig. 1D,E). Together, these data suggest that the CXXC domain of mouse Tet3 restricts the in vivo activity of Tet3FL by DNA binding and genomic targeting.
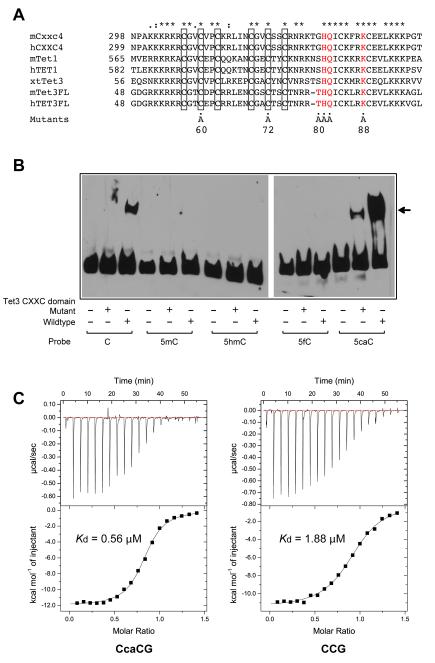
Figure 2. The CXXC domain of Tet3 is a specific reader of 5-carboxylcytosine at 5′CCG sequences.
(A) Sequence of the CXXC domains of Tet-associated CXXC domains from mouse (m), human (h) and Xenopus tropicalis (xt). Cxxc4 is a separate protein associated with Tet2. Amino acid positions and mutants are indicated. Asterisks and dots indicate identical and similar residues, respectively.
(B) Gel mobility shift assay of the mouse Tet3 CXXC domain and its mutant (C60A and C72A) with oligonucleotides containing different cytosine modifications.
(C) Isothermal titration calorimetry (ITC) of the Tet3 CXXC domain and oligonucleotides containing 5caC or C at a CCG sequence.
See also Fig. S2.
DNA binding properties of Tet3’s CXXC domain
To characterize the DNA binding activity of the mouse Tet3 CXXC domain, we prepared wild type and mutant (C60A and C72A) 182 amino acid (aa) recombinant GST-CXXC proteins and performed gel mobility shift assays using oligonucleotides containing all known cytosine C5 modifications (C, 5mC, 5hmC, 5fC, and 5caC). Unexpectedly, we observed that the Tet3 CXXC domain binds more strongly to the oligonucleotide containing 5caC than to normal cytosine (Fig. 2B). Binding in mobility shift assays was not detectable with 64-mer sequences containing 5mC, 5hmC, or 5fC. Next, to quantify the binding affinities of Tet3’s CXXC domain with both the unmodified DNA and the 5caC-modified DNA, we used isothermal titration calorimetry (ITC) to measure binding of the mouse Tet3 CXXC domain (mTet3-CXXC, residues 51-96) to a 12-mer palindromic DNA duplex, centered around an unmodified CpG dinucleotide (CCG DNA: GAATCCGGATTC) or a 5caCpG dinucleotide (CcaCG DNA: GAATCcaCGGATTC, caC = 5-carboxyldeoxycytidine) on both strands. The ITC analysis indicated that the mTet3-CXXC domain binds to the unmodified CCG DNA with a Kd of 1.88 μM but binds to the CcaCG DNA with much higher affinity (Kd of 0.56 μM) (Fig. 2C). This 3.4-fold binding preference for 5caC over unmethylated DNA is greater than, for example, the two-fold binding preference of the UHRF1 SRA domain for hemi- over unmethylated DNA (Rottach et al., 2010). Then, we measured binding of the mTet3-CXXC domain to a 12-mer CmCG, CfCG or ChmCG DNA duplex that shows identical sequence to the aforementioned CcaCG DNA except for the replacement of 5caC with 5mC, 5fC or 5hmC. Our results showed that the mTet3-CXXC domain binds to CmCG and CfCG with a Kd of 5.59 μM and 6.13 μM, respectively, and even more weakly to ChmCG (Fig. S2A,B). Notably, when 5caC on one of the DNA strands was replaced with unmodified C, resulting in a hemi-carboxylated DNA (hemi-CcaCG), we observed slightly reduced binding affinity for mTet3-CXXC, with a Kd of 1.57 μM (Fig. S2A,B). Replacement of the unmodified C preceding 5caC with A, T or G also led to a reduced binding affinity (Fig. S2C). Together, these observations establish that the Tet3 CXXC domain is a specific reader for 5caC DNA within the CcaCG sequence. We examined the presence of 5caC at 5′CCG sequences in vivo using published data sets in which 5caC was mapped at single base resolution (Lu et al., 2015). In this study, 48.5% of the 5caC bases were at 5′ACG, 10.6% were at 5′TCG, 12.5% were at 5′GCG and 28.3% were at 5′CCG. Therefore, CCG accounts for a substantial fraction of 5caC. This particular sequence context is overrepresented at CpG islands.
We also performed in vitro binding experiments with Tet3FL or Tet3CD proteins purified from HEK293T cell. We observed that purified Tet3FL and Tet3CD strongly bind to DNA irrespectively of modifications (data not shown), most likely due to the inherent DNA binding properties of the catalytic domain as shown previously for Tet2 (Hu et al., 2013). The DNA binding property of Tet3’s CXXC domain to 5caC prompted us to test a possible decarboxylase activity of this domain that could directly remove carboxyl groups from 5caC. However, we did not observe any ability of Tet3-CXXC to function as an “eraser” of 5-carboxyl groups on 5caC DNA using in vivo and in vitro studies (Fig. S2D,E). In addition, presence of the CXXC domain of Tet3 did not affect the cleavage activity of the DNA glycosylase TDG towards 5caC (Fig. S2F), a preferred substrate of this DNA glycosylase (He et al., 2011; Maiti and Drohat, 2011; Zhang et al., 2012), nor did we find any direct interaction between the Tet3 CXXC domain or Tet3FL and TDG (data not shown).
Structure of the Tet3 CXXC domain in complex with 5caC
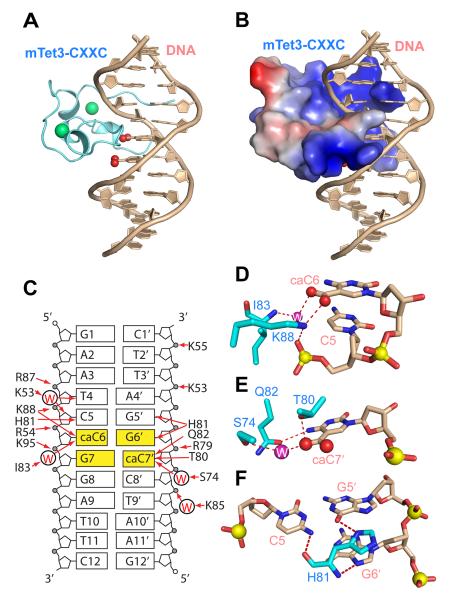
We then determined the crystal structure of mTet3-CXXC bound to the 12-mer CcaCG DNA, at 1.3 Å resolution (Fig. 3; Table S2). The structure reveals that the mTet3 CXXC domain, with a crescent-like fold and two Cys4-type zinc clusters, interacts with the CcaCG DNA from both major and minor grooves (Fig. 3A,B). Association of mTet3-CXXC with CcaCG DNA involves extensive protein-DNA contacts (Fig. 3C). Of particular note, we observed a number of interactions targeting the carboxylate group of the 5caC nucleotides from both DNA strands: The carboxylate of caC6 forms direct and water-mediated hydrogen bonds with the side chain amino group of mTet3 Lys88 and the backbone amide of mTet3 Ile83, respectively (Fig. 3D), while the carboxylate of caC7′ forms direct and water-mediated hydrogen bonds with the side chain hydroxyl groups of mTet3 Thr80 and Ser74, respectively (Fig. 3E). Other CcaCG base-specific interactions include the hydrogen bond between the side chain carbonyl oxygen of mTet3 Gln82 and the N4 atom of caC7′ (Fig. 3E), and the hydrogen bonds between the backbone carbonyl oxygen, amide nitrogen and side chain Nδ1 atom of mTet3 His81 and the N4 atom of C5, N7 atom of G5′, and O6 atom of G6′, respectively (Fig. 3F). Note that residues His81, Gln82 and Lys88 of mTet3 are highly conserved throughout evolution (Fig. 2A, red color), suggesting that the specific 5caC binding is a common feature of the Tet-associated subfamily of CXXC domains. The binding of mTet3-CXXC to CcaCG DNA is further supported by a number of intermolecular interactions between the CXXC domain and the phosphate backbone and sugar moieties of DNA (Fig. S3A,B).
Figure 3. Complex of the Tet3 CXXC domain with DNA containing 5caC.
X-ray crystallography was used for structure determination.
(A) Ribbon representation of the mTet3 CXXC domain (light blue) bound to CcaCG DNA (wheat). The zinc ions in the CXXC domain and the carboxylates in DNA are shown as green and red spheres, respectively.
(B) Electrostatic surface representation of the mTet3 CXXC domain bound to CcaCG DNA, with positively charged, neutral and negatively charged areas painted in blue, grey and red, respectively.
(C) Schematic view of intermolecular interactions between the Tet3 CXXC domain and CcaCG DNA, with intermolecular hydrogen bonds shown by red arrows.
(D-F) Specific hydrogen bonding interactions between side chains of the Tet3 CXXC domain and the CcaC step in the DNA. The nitrogen, oxygen and phosphorous atoms are shown in dark blue, red and yellow, respectively. Water molecules are shown in light purple.
The CXXC domain of Xenopus tropicalis Tet3 (XtTet3) has recently been shown to bind to unmodified and methylated CpG sites in a different manner. The mTet3 His81 equivalent residue in XtTet3 interacts with the central CpG dinucleotide of the unmodified DNA (ACG DNA), but shifts its binding to the preceding C•G pair when the cytosine in the CpG dinucleotide becomes methylated (CmCG DNA) (Xu et al., 2012). Consistent with that, our ITC analysis indicates that mTet3-CXXC binds to the CmCG DNA sequence with a lower affinity than that for CCG and CcaCG DNA sequences (Fig S2A,B). To understand the binding specificity of Tet3-CXXC towards 5caC DNA, we compared the structure of mTet3-CXXC - CcaCG DNA with those of XtTet3-CXXC - ACG DNA and XtTet3-CXXC - CmCG DNA (Fig. S3C,D). The analysis indicates that the mTet3 His81 equivalent residues from mTet3-CXXC – CcaCG and XtTet3-CXXC – CmCG complexes both interact with the C•G pair preceding the central (ca/m)CpG dinucleotides, thereby positioning the DNA molecules in a similar fashion. However, it is apparent that the central (ca/m)CpG dinucleotides are differently recognized in the two complexes: In the mTet3-CXXC - CcaCG complex, the central caCpG dinucleotides are recognized by mTet3 Thr80, Gln82 and Lys88 through hydrogen bond interactions; by contrast, there is no appreciable base-specific interaction involving the central mCpG dinucleotides in the XtTet3-CXXC – CmCG complex, and the corresponding protein residues are largely disordered (Xu et al., 2012). Through superimposition of the structures of mTet3-CXXC – CcaCG and XtTet3-CXXC – CmCG complexes, we generated a model of an XtTet3-CXXC – CcaCG complex (Fig. S3D). The strictly conserved residues Q91 and K97, equivalent to mTet3 Q82 and K88, respectively, are both engaged in the interactions with the caC nucleotides, while the mTet3 His81 equivalent residue recognizes the preceding C•G pair (Fig. S3D). Consistent with the modeling analysis, our ITC assays indicate that the XtTet3-CXXC domain also binds to CcaCG strongly, with a Kd of 1.30 μM (Fig. S4). By contrast, it binds to CCG and CmCG with a Kd of 2.58 μM and 9.00 μM, respectively, which is in good agreement with what was reported in a previous study (Xu et al., 2012). In addition, we show that the XtTet3-CXXC domain binds to CfCG and ChmC with a Kd of 10.4 μM and 18.6 μM, respectively. (Fig. S4). Therefore, we conclude that the Tet3 CXXC domains across species strongly bind to CcaCG DNA, with a binding mode distinct from their binding to either unmodified or other modified forms of DNA. In fact, it is conceivable that 5caC recognition is a conserved feature of the Tet-associated CXXC domains given the fact that the 5caC-interacting residues of mTet3, including Q82 and K88, are highly conserved within this family of CXXC domains (Fig. 2A). It is worth noting that 5caC recognition by the Tet3 CXXC domain is also distinct from what was observed for the TDG-5caC interaction (Zhang et al., 2012; Hashimoto et al., 2013), in which the 5caC base is flipped out of the DNA helix and inserted into a pocket formed by TDG residues.
Mutations in the CXXC domain
We then selected a number of CcaCG-interacting residues for mutagenesis and monitored their impact on the binding affinities of mTet3-CXXC towards both CCG and CcaCG DNA by ITC (Fig. 4). First, we observed that mutation of His81 to alanine abolished the binding of the domain to both CCG and CcaCG DNA, in agreement with our structural observation as well as the pervious report that this residue is essential for DNA binding (Xu et al., 2012). Second, the K88A mutation led to >27-fold weaker binding toward CcaCG DNA (Kd > 15 μM), but only a 3-fold weaker binding toward CCG DNA (Kd = 5.32 μM) (Fig. 4A-C). These observations reinforce the notion that residue Lys88, through direct interaction with 5caC (Fig. 3C), plays a key role in specific recognition between mTet3-CXXC and CcaCG DNA. Third, the mTet3-CXXC T80A and Q82A mutations both led to a modest decrease in binding affinity toward CcaCG DNA (Kd = 0.91 μM and 0.97 μM, respectively), but an even more negligible change in binding affinity toward CCG DNA (Kd = 1.79 μM and 2.38 μM, respectively), consistent with the structural observations that both residues contribute to binding to CcaCG DNA. We observed that Tet3 activity to reactivate a methylated luciferase reporter gene was only modestly (~20%) increased when a Tet3 construct with K88A mutation was used (data not shown). However, one needs to take into account that only very low levels of 5caC are formed under these conditions. Together, our mutational studies and ITC assays lend strong support to our structural observations for the mTet3-CXXC – CcaCG complex.
Fig. 4. Isothermal titration calorimetry analysis of different CXXC domain mutants.
(A) Interaction of mutants with DNA containing CCG.
(B) Interaction of mutants with DNA containing CcaCG.
(C) Summary of ITC data. N/A, Not Available.
See also Fig. S4.
In vivo mapping of Tet3FL
To elucidate the role of Tet3FL in vivo, we developed a specific antibody against its CXXC domain (Fig. S5). This antibody does not recognize the shorter isoforms of Tet3 (Fig. S5A,B) and detects a single band in neuronal cells and other mouse tissues. Comparison with an antibody directed against the C-terminus of Tet3 (Active Motif) that recognizes all Tet3 isoforms shows that Tet3FL, owing to its slightly lower mobility is likely the predominant Tet3 isoform in neuronal cells and in other mouse tissues analyzed (Fig. S5B,C). Since Tet3FL is relatively abundant in neuronal cells derived from differentiating mouse ES cells, we first mapped the distribution of Tet3FL in these cells by ChIP-seq using our in house antibody (Fig. 5). The C-terminal antibody did not work in ChIP-seq. One third of the peaks identified in neuronal progenitor cells (NPC) mapped very close to transcription start sites (TSS; −0.5 to +0.5 kb) (Fig. 5A; Table S3). These TSS peaks were strong, whereas peaks in intra- or inter-genic regions were much weaker and did not occur at CpG islands. To obtain additional support for Tet3FL localization in an in vivo setting, we mapped Tet3FL in E15.5 embryonic mouse brain (containing cortex, ganglionic eminence and hippocampus). This tissue contains mostly neurons and few glial cells. Similar TSS peaks were observed in NPC and in embryonic brain (Fig. 6; Fig. 6S) but their distribution was even more biased towards the TSS in brain-derived neurons (Fig. 5A; Table S3). About 42% of the TSS peaks found in brain were also scores in NPC. Almost half of the genes having Tet3FL peaks near the TSS in brain tissue are preferentially expressed in brain. The Tet3FL peaks at the TSS strongly overlapped with H3K4me3 peaks in embryonic mouse brain (Fig. 5B). Histograms and composite profile analysis show that the Tet3FL peaks are very precisely targeted to the TSS (Fig. 5C,D). Tet3FL peaks overlap with the distribution of H3K4me3 and appear to center within a nucleosome-free region as suggested by the dip in the H3K4me3 profile at the TSS (Fig. 5D). Genes with Tet3FL peak at the TSS have a higher expression level than genes without Tet3FL peak (Fig. 5G) although there are also many highly expressed genes with no Tet3FL peak.
Figure 5. Mapping of Tet3FL in mouse neural progenitor cells and embryonic brain-derived neurons.
(A) Distribution of peaks in the genome. TSS peaks were defined as peaks between −0.5 and +0.5 kb relative to the TSS (green). Extended promoter peaks are located outside of the TSS and are between 0.5 kb and 2.5 kb up- or down-stream of the TSS (purple).
(B) Overlap of Tet3FL peaks with H3K4me3 and H3K27me3.
(C) Histogram of E15.5 brain Tet3FL distribution near the TSS. The histogram shows the TSS +/− 1 kb on each side with 100 bp in each bin. The TSS is at the center.
(D) Composite profile of Tet3FL (red), H3K4me3 (blue) and H3K27me3 (green) for mouse embryo brain. The profile shows the gene body +/− 5 kb. The gene body was separated into 250 bins, while the upstream and downstream 5 kb was separated into 125 bins each.
(E) Gene ontology analysis of genes with Tet3FL peaks near the TSS in E15.5 brain. Shown are MSigDB analysis results using GREAT.
(F) Enrichment of TFEB sequence motifs at Tet3FL peaks.
(G) Expression levels of peaks with and without Tet3FL peaks at the TSS in E15.5 embryonic brain. Genes with Tet3FL peaks are expressed at higher levels (P<2.2×10−e16).
Fig. 6. Transcription start site-centered Tet3FL peaks.
ChIP-sequencing data show Tet3FL peaks at the transcription start sites (TSS) in NPCs (light blue) and mouse brain (dark blue). The 5mC patterns (shown as vertical bars representing 0-100% methylation at individual CpG sites) were obtained from whole genome bisulfite sequencing data of mouse ESC (red) and NPC (blue) (Stadler et al., 2011).
(A) Genes related to the base excision repair pathway.
(B) Genes important for lysosome structure and function.
Gene ontology analysis using GREAT (McLean et al., 2010) revealed that genes having Tet3FL peaks near the TSS in brain are significantly enriched in lysosome function, mRNA processing, splicing, and base excision repair (Fig. 5E; Tables S3 and S4). Figure 6B and Figure S6A provide genome browser views of several of the Tet3FL-targeted genes from the most prominently enriched lysosome functional category. Strikingly, detailed analysis in Table S3 shows that 68 of 220 genes (31%) with Tet3FL peaks at the TSS in brain-derived neurons are genes implicated in lysosome function, protein degradation and autophagy.
Whereas the exact roles of Tet3FL in regulating mRNA processing and in the function of lysosomes are currently unknown, the selective targeting of base excision repair genes by Tet3FL provides another important clue suggesting that active DNA demethylation is mediated by the Tet3FL/BER pathway. Promoters with strong Tet3FL peaks included several genes of the core base excision repair pathway such as Apex1 (AP endonuclease 1), Parp1 (Poly-ADP-ribose polymerase 1), and Tdg (Fig. 6A; Table S4). We also found peaks at the TSS of Sae1, an E1-activating enzyme as well as at Pias4, which is a SUMO ligase potentially involved in SUMO-dependent activation of Tdg (Fig. 6A, Table S3) (Hardeland et al., 2002; Coey et al., 2014) as well as Rad9, another binding partner and activator of Tdg (Guan et al., 2007). In addition, we found a strong Tet3FL peak at the TSS of Stub1/ChIP, an E3 ubiquitin ligase that regulates the function of the base excision repair proteins XRCC1 and DNA polymerase beta (Parsons et al., 2008; Sobol, 2008). The Tdg-initiated base excision repair pathway is known to be critical for removal of 5caC from DNA in the 5mC oxidation-demethylation cycle (He et al., 2011). These Tet3FL-marked genes were upregulated upon differentiation of mESC towards the neuronal lineage (Fig. S6B). During that transition, the total levels of 5caC are close to undetectable (data not shown) making it unlikely that the Tet3FL peaks we observed were at 5caC sites. Also, using anti-5caC IP followed by PCR, we did not detect any enrichment of 5caC at sequences with Tet3FL peaks near the TSS of these genes (not shown) suggesting that – if 5caC is formed there – it is too transient to be detectable. In mouse brain, steady-state levels of 5caC are close to undetectable as well (Liu et al., 2013b). Whole genome bisulfite sequencing data of differentiating mESC (Stadler et al., 2011) showed that genomic sequences near Tet3FL peaks are largely unmethylated in ES cells and NPC (Fig. 6). Interestingly, however, we observed an encroachment of 5mC at sequences immediately surrounding the TSS and Tet3FL peaks in NPC (Fig. 6).
We used a motif finding algorithm to identify sequence motifs within Tet3FL peaks near the TSS. This motif search identified a strong enrichment for a sequence recognized by the transcription factor EB (TFEB), 5′TCACGTGA (Fig. 5F) (P<6.5e-06). TFEB motifs were found within 55% of all Tet3FL TSS peaks and were centered at those peaks. This finding is consistent with the identification of the lysosome pathway as most significantly enriched for Tet3FL binding (Fig. 5E) because TFEB is viewed as the master regulator of autophagy and lysosomal genes (Palmieri et al., 2011; Settembre et al., 2011).
DISCUSSION
In this study, we identified and characterized three different isoforms of the mammalian 5mC oxidase Tet3. Transcription of the oocyte-specific isoform is initiated at a unique promoter and CpG island. This CpG island contains several binding sites for factors involved in oocyte-specific transcription including NBE and E-box (Tsunemoto et al., 2008). Tet3o lacks an N-terminal CXXC domain and therefore is expected to be less targeted to specific genomic sequences, e.g. to CpG islands. Rather, this isoform may participate in preferential genome-scale oxidation of paternal pronuclear DNA in zygotes (Gu et al., 2011; Iqbal et al., 2011; Wossidlo et al., 2011). Specific knockout of this oocyte-specific isoform in the female germ line will be required to prove its key function in zygotic epigenome remodeling.
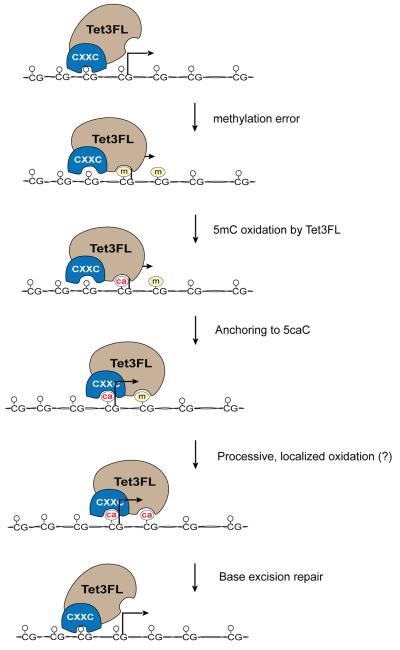
Tet proteins effectively produce 5hmC. There are many unresolved issues, for example how and under what conditions do Tet proteins proceed beyond 5hmC formation to produce 5caC resulting in DNA demethylation? Importantly, this study has identified the CXXC domain of Tet3 as a specific reader for 5caC. Our luciferase assays further suggest that binding of the CXXC domain of Tet3FL to 5caC or C restricts its genome-wide 5mC oxidation capacity. Based on the data presented, we propose a model for Tet3FL function (Fig. 7). Constitutive binding of Tet3FL near the TSS of unmethylated CpG islands suggests a housekeeping function to prevent methylation of critical CpG sites near the transcription start site (Fig. 6). Upon inadvertent introduction of 5mC by a DNA methyltransferase, Tet3FL oxidizes 5mC to 5caC. The high affinity of the CXXC domain of Tet3FL to 5caC causes Tet3FL to become even more tightly bound to genomic regions in which 5mC has been oxidized to 5caC. Through this anchoring mechanism, the catalytic domain of Tet3 is then able to rapidly oxidize additional 5mC bases in the immediate vicinity of the first oxidized base resulting in a highly effective localized 5mC oxidation reaction to preserve the integrity of critical unmethylated CpG islands at TSS regions (Fig. 7). Our model describes an anchoring mechanism where two different domains of the same protein can be engaged (the catalytic domain and the CXXC domain, which binds to the reaction product of Tet3FL activity). This theme is becoming quite common in chromatin regulators, which often contain writer and reader domains either within the same polypeptide or within a complex (Hassan et al., 2002; Lan et al., 2007; Hansen et al., 2008; Zhang et al., 2008; Margueron et al., 2009; Klein et al., 2014; Dumesic et al., 2015; Liu et al., 2015; Torres et al., 2015). The purpose of these dual domains generally is viewed as anchoring the modifier to its preferred sites of activity.
Fig. 7. Model for the biological role of Tet3FL in maintaining the unmethylated state of CpG islands through anchoring to 5caC.
Tet3FL binds to unmethylated CpG sites at the TSS of CpG islands of specific target genes. Tethering of Tet3FL to transcription start sites maintains these critical sequences in a methylation-free state. Binding of Tet3FL to its reaction product 5-carboxylcytosine (ca) allows for highly localized 5mC (m) oxidation in which Tet3 stays bound to 5caC through the CXXC domain promoting rapid oxidation of adjacent erroneously introduced methyl groups.
Gene ontology analysis suggests that Tet3FL preferentially targets genes functioning in lysosome pathways, mRNA processing and base excision repair (Fig. 5E). The limited number and selective nature of the Tet3FL peaks we observed suggests that Tet3FL is targeted to these loci specifically either based on DNA sequence preference, chromatin environment or interaction with transcription factors. A fraction of Tet3 in neurons may not contain the CXXC domain. A recent study described an interaction of the short form of Tet3 (Tet3s) with certain transcription factors including REST (Perera et al., 2015), although the genomic location of Tet3s was not mapped in that study. We show here that Tet3FL is the predominant Tet3 protein isoform in neurons (Fig. S5B). The exact targeting mechanisms of Tet3FL remain to be determined. For the lysosomal and autophagy genes, our motif analysis of Tet3FL peaks uncovered TFEB as a likely factor involved in Tet3FL recruitment (Fig. 5F).
Of particular significance, binding of Tet3FL was observed preferentially at transcription start sites of genes functioning in base excision repair. This fact suggests a regulatory function of Tet3FL in promoting the expression of factors that process the final DNA oxidation product of Tet activity, 5caC, including Tdg and AP endonuclease, which implies the existence of a Tet3FL-dependent regulatory feed-forward pathway for active, base excision repair-associated DNA demethylation.
Interestingly, mapping of Myc-tagged Idax/Cxxc4, which presumably is in a complex with Tet2, revealed partially similar GO categories with mRNA splicing as the most significantly enriched one (Ko et al., 2013). The exact role of Tet proteins and/or their associated 5mC oxidation products in RNA splicing is currently not understood although it has been found in some studies that 5hmC is enriched at exon-intron boundaries suggesting a mechanistic link between 5hmC and splicing (Khare et al., 2012; Wen et al., 2014).
The association of Tet3FL with genes regulating lysosome function is particularly intriguing and is of potential significance for neurodegenerative diseases. A large fraction (~31%) of all Tet3FL peaks in brain-derived neurons was localized at the TSS of genes functioning in lysosomal and autophagy pathways (Table S3). The genes identified in the category “lysosome” included Ctsd (cathepsin D), Gba (glucocerebrosidase), Hexa (hexosaminidase A), Lamp1 (lysosomal-associated membrane protein 1), Sumf1 (sulfatase modifying factor 1), Tpp1 (tripeptidyl peptidase I), Cln3 (ceroid-lipofuscinosis, neuronal 3), Atg3 (autophagy related 3) and a number of others (Fig. 6; Fig. S6; Table S3, S4). Several of these genes are commonly mutated in neuronal ceroid lipofuscinoses (a subgroup of lysosome storage diseases), a set of brain disorders caused by defects in either soluble enzymes or membrane-associated structural proteins that result in dysfunction of lysosomes (Shacka, 2012). Mutations in these genes are often associated with neurodegeneration due to failure to degrade sphingolipids and specific neuropeptides. Lysosomal activities are rate-limiting for degradation of aged or damaged proteins and diminished lysosome function has been linked to other neurodegenerative diseases including Parkinson’s disease (Schneider and Zhang, 2010; Gan-Or et al., 2015). Heterozygous mutations in beta-glucocerebrosidase (GBA) are the strongest genetic risk factor for Parkinson’s disease known to date (Sidransky et al., 2009; Schondorf et al., 2014). Aging brains exhibit decreased lysosomal function (Cuervo and Dice, 2000). Further studies are required to understand why Tet3FL is particularly prevalent at the TSS of this unique group of genes, whether Tet3FL maintains long-term expression of these genes by repairing methylation errors and how this control might be disabled during organismal aging potentially contributing to neurodegenerative diseases. The existence of a Tet3FL-mediated active 5mC removal process may be of special significance for post-mitotic neurons, in which replication-dependent dilution of erroneously deposited 5mC is not an option.
EPERIMENTAL PROCEDURES
The Tet3 isoforms were identified through RT-qPCR, and their activities were examined using transfection assays, luciferase reporter assays and dot blots. The interaction of Tet3-CXXC with DNA was determined using EMSA and ITC assays. The structure of mTet3-CXXC in complex with 5caC DNA was determined using X-ray crystallography. Mapping of Tet3FL in mouse neural progenitor cells and embryonic brain-derived neurons was performed using the Chip-sequencing method.
Detailed Experimental Procedures are available as Supplemental Material.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH grants CA160965 to G.P.P, GM064378 to P.E.S., NS075393 to Q.L., and MH094599 to Q.L. and G.P.P., Kimmel Scholar Award from Sidney Kimmel Foundation for Cancer Research, University of California Cancer Research Coordination Committee Award (CRC-15-380558), UCR Regent Fellowship and March of Dimes Foundation (1-FY15-345) to J.S., and by the City of Hope – UC Riverside Biomedical Research Initiative to G.P.P and J.S. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
ChIP-seq data were deposited into the GEO database (accession number GSE56821). Structural data were deposited in the Protein Data Bank under accession number 5EXH.
Supplemental Information includes six Figures, four Tables and Supplemental Experimental Procedures and can be found with this article online.
AUTHOR CONTRIBUTION
S-G.J. characterized Tet3 isoforms, developed the anti-Tet3FL antibody and performed ChIP-sequencing, T.L.D. prepared recombinant proteins and performed gel mobility shift assays, J.S. and Z-M.Z. performed crystallographic experiments, J.S., Z-M.Z. and M.R.H. conducted ITC and EMSA assays, J.J. conducted Western blot and Tet3 binding experiments, X.W. analyzed ChIP-sequencing data, J.L., Q.L. and P.E.S. isolated mouse brain cells and oocytes, Z.L. and G.-L.X. purified Tet3 and conducted EMSAs, J.S. and G.P.P. designed and organized the study, S-G.J., G.P.P. and J.S. prepared the manuscript.
REFERENCES
- Blaschke K, Ebata KT, Karimi MM, Zepeda-Martinez JA, Goyal P, Mahapatra S, Tam A, Laird DJ, Hirst M, Rao A, et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 2013;500:222–226. doi: 10.1038/nature12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coey CT, Fitzgerald ME, Maiti A, Reiter KH, Guzzo CM, Matunis MJ, Drohat AC. E2-mediated Small Ubiquitin-like Modifier (SUMO) Modification of Thymine DNA Glycosylase Is Efficient but Not Selective for the Enzyme-Product Complex. J. Biol. Chem. 2014;289:15810–15819. doi: 10.1074/jbc.M114.572081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM, Dice JF. When lysosomes get old. Exp. Gerontol. 2000;35:119–131. doi: 10.1016/s0531-5565(00)00075-9. [DOI] [PubMed] [Google Scholar]
- Dumesic PA, Homer CM, Moresco JJ, Pack LR, Shanle EK, Coyle SM, Strahl BD, Fujimori DG, Yates JR, 3rd, Madhani HD. Product binding enforces the genomic specificity of a yeast polycomb repressive complex. Cell. 2015;160:204–218. doi: 10.1016/j.cell.2014.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- Gan-Or Z, Dion PA, Rouleau GA. Genetic perspective on the role of the Autophagy-Lysosome Pathway in Parkinson disease. Autophagy. 2015;11:1443–1457. doi: 10.1080/15548627.2015.1067364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- Guan X, Madabushi A, Chang DY, Fitzgerald ME, Shi G, Drohat AC, Lu AL. The human checkpoint sensor Rad9-Rad1-Hus1 interacts with and stimulates DNA repair enzyme TDG glycosylase. Nucleic Acids Res. 2007;35:6207–6218. doi: 10.1093/nar/gkm678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Li X, Liang D, Li T, Zhu P, Guo H, Wu X, Wen L, Gu TP, Hu B, et al. Active and passive demethylation of male and female pronuclear DNA in the Mammalian zygote. Cell stem cell. 2014;15:447–458. doi: 10.1016/j.stem.2014.08.003. [DOI] [PubMed] [Google Scholar]
- Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, Surani MA. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science. 2013;339:448–452. doi: 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MA, Qiu R, Wu X, Li AX, Zhang H, Wang J, Jui J, Jin SG, Jiang Y, Pfeifer GP, et al. Dynamics of 5-hydroxymethylcytosine and chromatin marks in Mammalian neurogenesis. Cell Rep. 2013;3:291–300. doi: 10.1016/j.celrep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MA, Szabo PE, Pfeifer GP. 5-Hydroxymethylcytosine: A stable or transient DNA modification? Genomics. 2014;104:314–323. doi: 10.1016/j.ygeno.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K. A model for transmission of the H3K27me3 epigenetic mark. Nat. Cell Biol. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- Hardeland U, Steinacher R, Jiricny J, Schar P. Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J. 2002;21:1456–1464. doi: 10.1093/emboj/21.6.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, Zhang X, Cheng X. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res. 2012;40:4841–4849. doi: 10.1093/nar/gks155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Pais JE, Zhang X, Saleh L, Fu ZQ, Dai N, Correa IR, Jr., Zheng Y, Cheng X. Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA. Nature. 2014;506:391–395. doi: 10.1038/nature12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Zhang X, Cheng X. Activity and crystal structure of human thymine DNA glycosylase mutant N140A with 5-carboxylcytosine DNA at low pH. DNA repair. 2013;12:535–540. doi: 10.1016/j.dnarep.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002;111:369–379. doi: 10.1016/s0092-8674(02)01005-x. [DOI] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Li Z, Cheng J, Rao Q, Gong W, Liu M, Shi YG, Zhu J, Wang P, Xu Y. Crystal structure of TET2-DNA complex: insight into TET-mediated 5mC oxidation. Cell. 2013;155:1545–1555. doi: 10.1016/j.cell.2013.11.020. [DOI] [PubMed] [Google Scholar]
- Hu X, Zhang L, Mao SQ, Li Z, Chen J, Zhang RR, Wu HP, Gao J, Guo F, Liu W, et al. Tet and TDG Mediate DNA Demethylation Essential for Mesenchymal-to-Epithelial Transition in Somatic Cell Reprogramming. Cell Stem Cell. 2014;14:512–522. doi: 10.1016/j.stem.2014.01.001. [DOI] [PubMed] [Google Scholar]
- Huang Y, Chavez L, Chang X, Wang X, Pastor WA, Kang J, Zepeda-Martinez JA, Pape UJ, Jacobsen SE, Peters B, et al. Distinct roles of the methylcytosine oxidases Tet1 and Tet2 in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2014;111:1361–1366. doi: 10.1073/pnas.1322921111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal K, Jin SG, Pfeifer GP, Szabo PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc. Natl. Acad. Sci. USA. 2011;108:3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare T, Pai S, Koncevicius K, Pal M, Kriukiene E, Liutkeviciute Z, Irimia M, Jia P, Ptak C, Xia M, et al. 5-hmC in the brain is abundant in synaptic genes and shows differences at the exon-intron boundary. Nat. Struct. Mol. Biol. 2012;19:1037–1043. doi: 10.1038/nsmb.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein BJ, Piao L, Xi Y, Rincon-Arano H, Rothbart SB, Peng D, Wen H, Larson C, Zhang X, Zheng X, et al. The histone-H3K4-specific demethylase KDM5B binds to its substrate and product through distinct PHD fingers. Cell Rep. 2014;6:325–335. doi: 10.1016/j.celrep.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Ko M, An J, Bandukwala HS, Chavez L, Aijo T, Pastor WA, Segal MF, Li H, Koh KP, Lahdesmaki H, et al. Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature. 2013;497:122–126. doi: 10.1038/nature12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh KP, Rao A. DNA methylation and methylcytosine oxidation in cell fate decisions. Curr. Opin. Cell Biol. 2013;25:152–161. doi: 10.1016/j.ceb.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F, Collins RE, De Cegli R, Alpatov R, Horton JR, Shi X, Gozani O, Cheng X, Shi Y. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007;448:718–722. doi: 10.1038/nature06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Wang M, Deng W, Schmidt CS, Qin W, Leonhardt H, Spada F. Intrinsic and extrinsic connections of Tet3 dioxygenase with CXXC zinc finger modules. PloS One. 2013a;8:e62755. doi: 10.1371/journal.pone.0062755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Zhang Z, Wu H, Jiang Y, Meng L, Xiong J, Zhao Z, Zhou X, Li J, Li H, et al. Recognition of H3K9 methylation by GLP is required for efficient establishment of H3K9 methylation, rapid target gene repression, and mouse viability. Genes Dev. 2015;29:379–393. doi: 10.1101/gad.254425.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Wang J, Su Y, Guerrero C, Zeng Y, Mitra D, Brooks PJ, Fisher DE, Song H, Wang Y. Quantitative assessment of Tet-induced oxidation products of 5-methylcytosine in cellular and tissue DNA. Nucl. Acids Res. 2013b;41:6421–6429. doi: 10.1093/nar/gkt360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Han D, Zhao BS, Song CX, Zhang LS, Dore LC, He C. Base-resolution maps of 5-formylcytosine and 5-carboxylcytosine reveal genome-wide DNA demethylation dynamics. Cell Res. 2015;25:386–389. doi: 10.1038/cr.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J. Biol. Chem. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor EA, Court BL, Young JI, Wang G. Ascorbate induces ten-eleven translocation (Tet) methylcytosine dioxygenase-mediated generation of 5-hydroxymethylcytosine. J. Biol. Chem. 2013;288:13669–13674. doi: 10.1074/jbc.C113.464800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münzel M, Globisch D, Bruckl T, Wagner M, Welzmiller V, Michalakis S, Muller M, Biel M, Carell T. Quantification of the sixth DNA base hydroxymethylcytosine in the brain. Angew. Chem. Int. Ed. Engl. 2010;49:5375–5377. doi: 10.1002/anie.201002033. [DOI] [PubMed] [Google Scholar]
- Palmieri M, Impey S, Kang H, di Ronza A, Pelz C, Sardiello M, Ballabio A. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum. Mol. Genet. 2011;20:3852–3866. doi: 10.1093/hmg/ddr306. [DOI] [PubMed] [Google Scholar]
- Parsons JL, Tait PS, Finch D, Dianova II, Allinson SL, Dianov GL. CHIP-mediated degradation and DNA damage-dependent stabilization regulate base excision repair proteins. Mol. Cell. 2008;29:477–487. doi: 10.1016/j.molcel.2007.12.027. [DOI] [PubMed] [Google Scholar]
- Peat JR, Dean W, Clark SJ, Krueger F, Smallwood SA, Ficz G, Kim JK, Marioni JC, Hore TA, Reik W. Genome-wide bisulfite sequencing in zygotes identifies demethylation targets and maps the contribution of TET3 oxidation. Cell Rep. 2014;9:1990–2000. doi: 10.1016/j.celrep.2014.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera A, Eisen D, Wagner M, Laube SK, Kunzel AF, Koch S, Steinbacher J, Schulze E, Splith V, Mittermeier N, et al. TET3 Is Recruited by REST for Context-Specific Hydroxymethylation and Induction of Gene Expression. Cell Rep. 2015;11:283–294. doi: 10.1016/j.celrep.2015.03.020. [DOI] [PubMed] [Google Scholar]
- Pfeifer GP, Kadam S, Jin SG. 5-hydroxymethylcytosine and its potential roles in development and cancer. Epigenetics Chromatin. 2013;6:10. doi: 10.1186/1756-8935-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottach A, Frauer C, Pichler G, Bonapace IM, Spada F, Leonhardt H. The multi-domain protein Np95 connects DNA methylation and histone modification. Nucleic Acids Res. 2010;38:1796–1804. doi: 10.1093/nar/gkp1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider L, Zhang J. Lysosomal function in macromolecular homeostasis and bioenergetics in Parkinson's disease. Mol. Neurodegener. 2010;5:14. doi: 10.1186/1750-1326-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schondorf DC, Aureli M, McAllister FE, Hindley CJ, Mayer F, Schmid B, Sardi SP, Valsecchi M, Hoffmann S, Schwarz LK, et al. iPSC-derived neurons from GBA1-associated Parkinson's disease patients show autophagic defects and impaired calcium homeostasis. Nat. Commun. 2014;5:4028. doi: 10.1038/ncomms5028. [DOI] [PubMed] [Google Scholar]
- Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shacka JJ. Mouse models of neuronal ceroid lipofuscinoses: useful pre-clinical tools to delineate disease pathophysiology and validate therapeutics. Brain. Res. Bull. 2012;88:43–57. doi: 10.1016/j.brainresbull.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Shen L, Inoue A, He J, Liu Y, Lu F, Zhang Y. Tet3 and DNA replication mediate demethylation of both the maternal and paternal genomes in mouse zygotes. Cell Stem Cell. 2014;15:459–470. doi: 10.1016/j.stem.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Wu H, Diep D, Yamaguchi S, D'Alessio AC, Fung HL, Zhang K, Zhang Y. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell. 2013;153:692–706. doi: 10.1016/j.cell.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidransky E, Nalls MA, Aasly JO, Aharon-Peretz J, Annesi G, Barbosa ER, Bar-Shira A, Berg D, Bras J, Brice A, et al. Multicenter analysis of glucocerebrosidase mutations in Parkinson's disease. N. Engl. J. Med. 2009;361:1651–1661. doi: 10.1056/NEJMoa0901281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobol RW. CHIPping away at base excision repair. Molecular cell. 2008;29:413–415. doi: 10.1016/j.molcel.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Song J, Rechkoblit O, Bestor TH, Patel DJ. Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation. Science. 2011;331:1036–1040. doi: 10.1126/science.1195380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, Irier H, Upadhyay AK, Gearing M, Levey AI, et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nature Neuroscience. 2011;14:1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres IO, Kuchenbecker KM, Nnadi CI, Fletterick RJ, Kelly MJ, Fujimori DG. Histone demethylase KDM5A is regulated by its reader domain through a positive-feedback mechanism. Nat. Commun. 2015;6:6204. doi: 10.1038/ncomms7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunemoto K, Anzai M, Matsuoka T, Tokoro M, Shin SW, Amano T, Mitani T, Kato H, Hosoi Y, Saeki K, et al. Cis-acting elements (E-box and NBE) in the promoter region of three maternal genes (Histone H1oo, Nucleoplasmin 2, and Zygote Arrest 1) are required for oocyte-specific gene expression in the mouse. Mol. Reprod. Dev. 2008;75:1104–1108. doi: 10.1002/mrd.20863. [DOI] [PubMed] [Google Scholar]
- Valinluck V, Sowers LC. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res. 2007;67:946–950. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
- Wen L, Li X, Yan L, Tan Y, Li R, Zhao Y, Wang Y, Xie J, Zhang Y, Song C, et al. Whole-genome analysis of 5-hydroxymethylcytosine and 5-methylcytosine at base resolution in the human brain. Genome Biol. 2014;15:R49. doi: 10.1186/gb-2014-15-3-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, Arand J, Nakano T, Reik W, Walter J. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat. Commun. 2011;2:241. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, Barbera AJ, Zheng L, Zhang H, Huang S, et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol. Cell. 2011;42:451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Xu C, Kato A, Tempel W, Abreu JG, Bian C, Hu Y, Hu D, Zhao B, Cerovina T, et al. Tet3 CXXC domain and dioxygenase activity cooperatively regulate key genes for Xenopus eye and neural development. Cell. 2012;151:1200–1213. doi: 10.1016/j.cell.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Shen L, Liu Y, Sendler D, Zhang Y. Role of Tet1 in erasure of genomic imprinting. Nature. 2013;504:460–464. doi: 10.1038/nature12805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin R, Mao SQ, Zhao B, Chong Z, Yang Y, Zhao C, Zhang D, Huang H, Gao J, Li Z, et al. Ascorbic acid enhances Tet-mediated 5-methylcytosine oxidation and promotes DNA demethylation in mammals. J. Am. Chem. Soc. 2013;135:10396–10403. doi: 10.1021/ja4028346. [DOI] [PubMed] [Google Scholar]
- Zhang K, Mosch K, Fischle W, Grewal SI. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat. Struct. Mol. Biol. 2008;15:381–388. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
- Zhang L, Lu X, Lu J, Liang H, Dai Q, Xu GL, Luo C, Jiang H, He C. Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nat. Chem. Biol. 2012;8:328–330. doi: 10.1038/nchembio.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.