Abstract
Protein fatty-acylation in eukaryotes has been associated with many fundamental biological processes. However, the diversity, abundance and regulatory mechanisms of protein fatty-acylation in vivo remain to be explored. Herein, we review the proteomic analysis of fatty-acylated proteins, with a focus on N-myristoylation and S-palmitoylation. We then highlight major challenges and emerging methods for direct site identification, quantitation, and lipid structure characterization to understand the functions and regulatory mechanisms of fatty-acylated proteins in physiology and disease.
Graphic abstract
Introduction
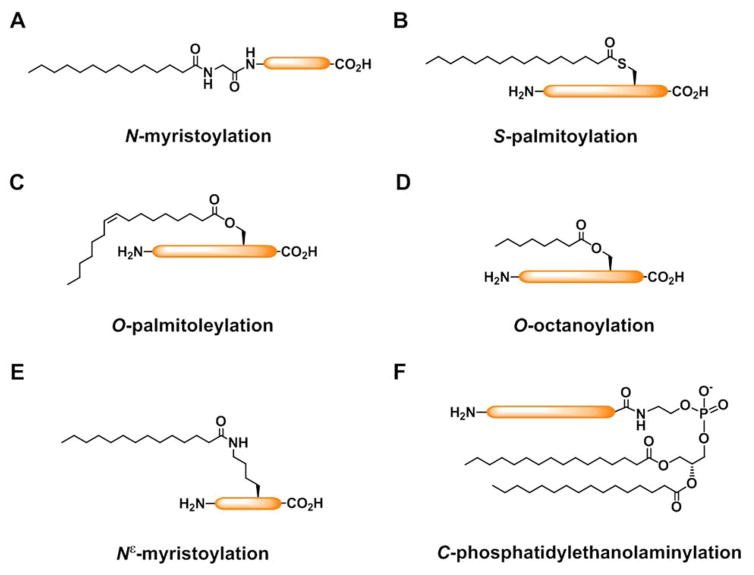
Protein fatty-acylation describes the covalent attachment of diverse fatty acids onto a variety of amino acid residues on proteins, including N-myristoylation, S-palmitoylation (or called S-fatty-acylation), Nε-fatty-acylation, O-fatty-acylation (e.g., O-palmitoleylation and O-octanoylation), glycosylphosphatidylinositol (GPI)-anchor modification, and C-phosphatidylethanolaminylation (Figure 1) [1]. N-myristoylation and S-palmitoylation are the two most prominent forms of protein fatty-acylation (Figure 1A, B), comprising the addition of myristate, a 14-carbon saturated fatty acid, and palmitate, a 16-carbon saturated fatty acid, onto N-terminal glycines and cysteines of proteins, respectively. Serine and threonine residues can also be modified by different fatty acids, such as O-palmitoleylation and O-octanoylation (Figure 1C, D). In addition, fatty-acylation also occurs on the epsilon-NH2 groups of lysine side chains (Figure 1E). C-phosphatidylethanolaminylation is a less prevalent form of fatty-acylation, which involves conjugation of phosphatidylethanolamine to the C-terminal glycine residue of LC3/Atg8, a key protein in autophagy (Figure 1F). Fatty acids can also be attached to the C-termini of proteins post-translationally through glycosylphosphatidylinositol (GPI) anchors.
Figure 1.
Fatty-acylated proteins in eukaryotes. A) N-myristoylation. B) S-palmitoylation. C) O-palmitoleylation. D) O-octanoylation. E) Nε-fatty-acylation. F) C-phosphatidylethanolaminylation.
Fatty-acylation can not only target proteins to specific membrane compartments, but also broadly influence protein-protein interactions and protein activity [2]. As a result, protein fatty-acylation is now well-recognized to regulate a variety of biological processes in eukaryotes, such as cell division and differentiation, synaptic transmission, immunity, and more [2].
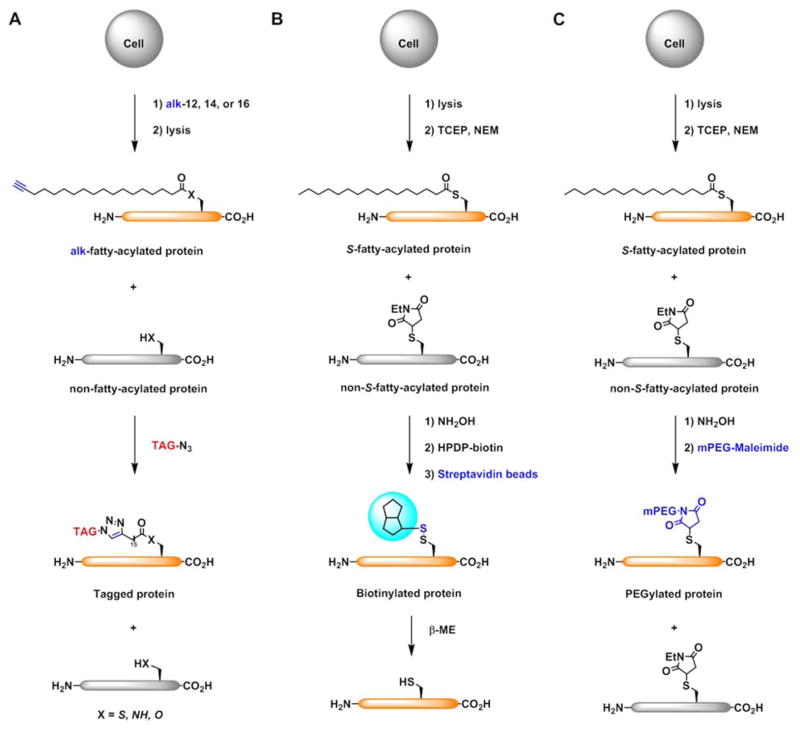
Historically, protein fatty-acylation was difficult to study largely due to lack of specific antibodies and limited detection methods. To overcome these limitations, selective chemical labeling methods have been developed to tag specific forms of protein fatty-acylation [1,3]. For example, fatty acid chemical reporters have provided an efficient approach for non-radioactive detection and large-scale analysis of fatty-acylated proteins when combined with bioorthogonal reactions and mass spectrometry-based proteomics (Figure 2A). Fatty acid chemical reporters contain unique chemical functionality (e.g., alkyne or azide) and can be metabolically incorporated into fatty-acylated proteins [4]. The alkyne or azide tag introduced into fatty-acylated proteins allows bioorthogonal reactions with either fluorophores for rapid and sensitive visualization of fatty-acylated proteins by in-gel fluorescence imaging or affinity tags (e.g., biotin) for selective enrichment and large-scale proteomic identification (Figure 2A). This chemical reporter strategy has been widely employed for the global analysis of N-myristoylated and S-palmitoylated proteins, and can in principle be used for other fatty-acylated proteins [5]. Alternatively, S-fatty-acylated proteins can also be selectively labeled and enriched by the acyl-biotin exchange (ABE) protocol, which exploits the hydroxylamine (NH2OH) sensitivity of thioester bonds in S-palmitoylated proteins (Figure 2B) [6,7]. ABE encompasses reduction of disulfide bonds, capping of free cysteines with N-ethyl maleimide (NEM), cleavage of thioester bonds with NH2OH, capture of newly liberated cysteines with HPDP–biotin, streptavidin pull-down, and elution of S-acylated proteins for western blotting or proteomic analysis. An improved version of ABE that requires fewer steps, i.e., acyl resin-assisted capture (acyl-RAC), has recently been developed [8]. Following capping and NH2OH treatment, newly liberated cysteines are captured on thiosepharose resin via the formation of disulfide bonds, which can subsequently be reduced to elute captured proteins.
Figure 2.
Methods for proteomic and quantitative analysis of fatty-acylated proteins. A) Fatty acid chemical reporter labeling. X = S, NH, O. B) Chemical enrichment of S-palmitoylated proteins. C) NH2OH-mediated acyl-PEGylation exchange (APE) for quantitative analysis of S-palmitoylation.
The selective chemical methods have allowed the large-scale analysis of fatty-acylated proteins, especially N-myristoylated and S-palmitoylated proteins, in different cell-types and animals. Notably, more than 300 fatty-acylated proteins have been identified to date, suggesting broader roles of fatty-acylation in regulating eukaryotic biology than previously appreciated [2]. In this review, we summarize the large-scale N-myristoylome and S-palmitoylome profiling studies, with a focus on those appeared in the past two years, and discuss the current status and unmet challenges for proteome-wide analysis of other fatty-acylated proteins. We then highlight emerging chemical biology and proteomic methods for higher resolution analysis of fatty-acylated proteins and close with an outlook on future developments needed in the fatty-acylation field.
N-myristoylation profiling
Protein N-myristoylation is catalyzed by N-myristoyltransferases (NMTs) that use myristoyl-CoA and typically modify N-terminal glycine residues of proteins co-translationally [9]. Alternatively, post-translational N-myristoylation can occur during apoptosis following caspase cleavage of proteins to expose N-terminal glycine residues. N-myristoylation can control protein subcellular localization and activity by promoting protein-membrane and protein-protein interactions and is involved in a wide variety of cellular processes, ranging from T cell activation, programmed cell death, and microbial infections [9].
Myristic acid analogs functionalized at the ω-position with an alkyne or azide group, such as alk-11, alk-12, az-11, and az-12, have been developed as chemical reporters to study N-myristoylated proteins [10–12], which have become the method of choice for large-scale proteomic analysis of N-myristoylation. Comparative studies have previously shown that alk-12, in combination with azide-tagged fluorophores or biotin, gives minimal background labeling, and that alk-12 preferentially labels N-myristoylated proteins compared to longer chain fatty acid reporters [13,14]. However, due to highly promiscuous fatty-acylation machinery and fatty acid metabolism, alk-12 has also been shown to label other types of fatty-acylated proteins [14], including Nε-myristoylated proteins [15,16], S-palmitoylated proteins, and GPI-anchor modified proteins [14,17], which can complicate N-myristoylome profiling studies. To differentiate N-myristoylated proteins from other fatty-acylated proteins in proteome-wide studies using alk-12 labeling, several strategies have been applied. The first strategy involves combination of potent NMT inhibitors with alk-12 labeling and quantitative proteomic analysis to quantify the relative abundance of alk-12-labeled proteins in response to NMT inhibitors, which therefore allows reliable identification of NMT substrates (i.e., N-myristoylated proteins). The application of this approach to live malaria parasites identified more than 30 NMT substrates in a proteome-wide analysis, associated with a wide range of functions that are essential for parasite viability [17]. Integrating alk-12-enabled quantitative proteomics with specific chemical inhibition of NMT was further refined and applied to globally profile co-translationally and post-translationally N-myristoylated proteome of human cells [18], which allowed the direct identification of more than 100 NMT substrates in human cells, including 40 proteins that are post-translationally N-myristoylated, following caspase cleavage, during apoptosis. This general strategy has also been applied for global analysis of protein N-myristoylation in parasites such as Leishmania donovani, revealing 30 high confidence NMT substrates with more than half uncharacterized previously [19].
In more complex systems, such as in microbe-infected mammalian cells, NMT inhibition may affect the viability of cells and may not be ideal for proteome-wide profiling of N-myristoylated proteins. In this context, the criteria of possessing an N-terminal glycine can be utilized to filter out other fatty-acylated proteins and simplify the proteomics data for further analysis. This targeted analysis enabled global profiling of host N-myristoylated proteins that are differentially modified in cells infected with herpes simplex virus (HSV) [20] or human immunodeficiency virus 1 (HIV-1) [21]. Recent studies on pathogenic mechanisms of Gram-negative bacteria Shigella revealed that the Shigella type III secretion effector invasion plasmid antigen J (IpaJ) can cleave N-myristoylated glycines of proteins to alter host secretion and trafficking pathways during infection [22]. To determine the protein substrates of IpaJ, we performed N-myristoylome profiling of mammalian protein substrates of IpaJ under different biological contexts using the chemical reporter method coupled with quantitative proteomic analysis [23]. Initial in vitro comparative proteomics of alk-12-labeled cell lysates that were treated with recombinant IpaJ indicated that majority of N-myristoylated proteins are IpaJ substrates in vitro, which suggested N-myristoylated glycine as the minimal element required for IpaJ substrate recognition and proteolysis. Nonetheless, a subsequent whole-cell N-myristoylome profiling on alk-12-labeled cells that were infected with wild-type or IpaJ deletion Shigella strain demonstrated that IpaJ specifically targets Golgi-associated ARF/ARL family GTPases in the context of Shigella infection. Taken together, these large-scale studies greatly advance our understanding of N-myristoylation and also highlight the utility of N-myristoylome profiling for unbiased analysis of drug targets and specific enzymes.
S-palmitoylation profiling
S-palmitoylation (or called S-fatty-acylation) is a reversible modification that is mediated by a family of Asp-His-His-Cys (DHHC)-containing protein acyltransferases (DHHC-PATs) encoded by ZDHHC genes (7 in yeast and ~23 in mammals) and removed by acyl-protein thioesterases (APTs) [24]. The dynamic cycling between S-palmitoylated and non-palmitoylated states is well-recognized as an important mechanism for regulating protein trafficking, activity, stability, and even protein-protein interactions. Moreover, the dysregulation of protein S-palmitoylation and de-palmitoylation cycle is associated with a range of human diseases, including cancer, infectious diseases, and neurodegenerative disorders [25,26].
Over the past decade, tremendous advances have been made towards large-scale S-palmitoylation profiling using ABE method [7] and fatty acid chemical reporters (e.g., 17-octadecynoic acid (17-ODYA), or called alk-16) [5]. These two methods are largely complementary and provide unique coverages of S-palmitoylated proteins. ABE allows selective analysis of S-palmitoylated proteins from cell lysates/tissues, but does not reveal S-palmitoylation dynamics and distinguish S-palmitoylated proteins from other thioester-modified proteins, such as intermediates in ubiquitin conjugation pathway [27]. In contrast, the chemical reporter method enables examination of S-palmitoylation dynamics by classic pulse-chase labeling and selective analysis of fatty-acylated proteins, but requires metabolic labeling of cells with exogenous fatty acid analogs and also labels other fatty-acylated proteins, such as N-myristoylated proteins, Nε-myristoylated proteins, and GPI-anchor modified proteins. Indeed, a recent study using both methods for global analysis of S-palmitoylome in a major human malaria parasite, Plasmodium falciparum, showed that these two methods together provided overlapped lists of S-palmitoylated proteins with high confidence [27].
ABE and chemical reporter strategy, especially when combined with cutting-edge mass spectrometry-based proteomic analysis, has made the large-scale global analysis of S-palmitoylated proteins more robust. Indeed, more than 20 large-scale S-palmitoylome profiling studies have been published from 2006 to 2013 [28], identifying hundreds of new S-palmitoylated candidate proteins and suggesting broader roles of S-palmitoylation in regulating biology across different organisms than previously appreciated. For instance, the first S-palmitoylome profiling study applied the ABE method to the yeast Saccharomyces cerevisiae, identifying 35 new S-palmitoylated proteins and revealing the diverse enzymatic specificities of individual PATs for the first time [7]. Later, a global analysis of rat neural S-palmitoylome using the ABE method identified more than 200 new S-palmitoylated protein candidates with diverse functions, ranging from ion channels to vesicular trafficking factors [29]. Subsequent assessment of S-palmitoylation dynamics in drug-induced neural activity models further highlighted increasing roles of S-palmitoylation in regulating synaptic function. Alternatively, S-palmitoylome profiling studies in dendritic cells (DC2.4) using alk-16 chemical reporter identified more than 150 candidate S-palmitoylated proteins and revealed that S-palmitoylation is crucial for the antiviral activity of interferon-induced transmembrane protein 3 (IFITM3) [30] as well as the activity of Toll-like receptor 2 (TLR2) [31], highlighting an important role for S-palmitoylation in the control of immune responses [32]. In addition, alk-16-mediated S-palmitoylome profiling of fission yeast in different stages of cellular differentiation revealed that precise control of a single palmitoyltransferase (Erf2) expression level can quantitatively regulate the fatty-acylation of specific proteins such as Rho3 GTPase and help drive meiotic entry in Schizosaccharomyces pombe [33]. As noted above, the chemical reporter method is especially useful for examining S-palmitoylation dynamics. Large-scale analysis of global S-palmitoylation dynamics using alk-16 and quantitative SILAC (Stable Isotope Labeling with Amino Acids in Cell Culture) proteomics has been also performed and revealed that majority of proteins are stably S-palmitoylated, while a subset of proteins involved in cell signaling undergoes rapid S-palmitoylation and de-S-palmitoylation cycles [34]. Quantitative proteomics with ABE and SILAM (Stable Isotope Labeling of Mammals) has also enabled protein S-palmitoylation profiling in more complex biological samples, such as in a mouse model of Huntington’s disease [35], which revealed that S-palmitoylation in the supporting glial cells may contribute to the pathogenesis of Huntington’s disease. Overall, these studies highlight the utility of large-scale S-palmitoylation profiling studies to uncover its new functions and regulatory mechanisms in eukaryotes.
In the past two years, several new S-palmitoylome profiling studies have also been reported. For example, S-palmitoylation chemical reporters have been used to systematically analyze protein S-palmitoylation of host cells during herpes simplex virus (HSV) [20] or human immunodeficiency virus 1 (HIV-1) [21] infection. Interestingly, a fraction of host S-palmitoylated proteins, including regulators of interferon and tetraspanin-family proteins, was selectively suppressed during HSV infection [20]. In addition, a number of HSV proteins, such as glycoproteins and kinases, were shown to be S-palmitoylated during infection, suggesting an important role of S-palmitoylation in viral pathogenesis. In another study, the ABE method has been coupled with 16O/18O-labeling during trypsin digestion for quantitative analysis of the S-palmitoylome of primary human T cells, identifying a set of 280 proteins as robustly palmitoylated [36]. S-palmitoylome profiling of Cryptococcus neoformans with alk-16 labeling identified many fatty-acylated proteins and helped characterize how a specific palmitoyltransferase (i.e., pfa4) contributes to the cell wall morphology and virulence of this fungal pathogen [37], suggesting specific and potent DHHC-PAT inhibitors could be useful for treating fungal infections. Fatty acid reporter profiling with alk-16 has also enabled global analysis of protein S-palmitoylation in Toxoplasma gondii and revealed several new fatty-acylated proteins involved in parasite motility, morphology, and host cell invasion [38].
To integrate S-palmitoylation literature and recent proteomic profiling studies, an online database SwissPalm (http://swisspalm.epfl.ch/) has been established [39], which currently encompasses 5199 S-palmitoylated protein hits from seven species and 535 validated S-palmitoylation sites. Researchers can now readily search for proteins of interest through multiple published S-palmitoylomes in the SwissPalm platform to determine whether specific proteins may be S-palmitoylated, predict their S-palmitoylation sites, as well as identify orthologues and potential functions. In a similar study, the proteomics data from 15 mammalian S-palmitoylome profiling studies were curated into a single compendium of S-palmitoylated proteins, making these large dataset readily accessible to researchers [40]. Notably, bioinformatic analyses of these combined datasets have further highlighted emerging roles of S-palmitoylation in neurological disorders, cancer, and infectious diseases. Together, these combined databases represent extremely valuable resources and will greatly facilitate S-palmitoylation studies.
Proteomic analysis of other fatty-acylated proteins
The proteomic analysis of other fatty-acylated proteins have not been explored so extensively, but are beginning to emerge using metabolic labeling and selective chemical enrichment strategies.
O-fatty-acylation
Limited examples of O-fatty-acylation (Figure 1C, D) have been discovered. Amongst this class of fatty-acylated proteins, Wnt proteins are dually fatty-acylated; for example, Wnt3a is S-palmitoylated on a conserved Cys residue (Cys77) and modified with palmitoleic acid, a monounsaturated C16-fatty acid, on a conserved serine (Ser209) by Porcupine, a membrane-bound O-acyl transferase [41,42]. Other members of the Wnt families, such as xenopus Wnt8 [43] and Wnt1 [44], are also modified by palmitoleic acid on a Ser residue. O-fatty-acylation is essential for the secretion and the function of Wnt proteins [42]. Wnt proteins are conserved signaling molecules essential during embryogenesis and adult tissue homoeostasis. In addition, dysfunctions in Wnt signaling are implicated in many diseases, including cancer [45] and degenerative diseases [46]. Therefore, proper O-fatty-acylation of Wnt may also be important for development and disease progression. Other O-fatty-acylated proteins have also been identified, such as ghrelin, a peptide hormone modified by a C8-fatty acid on Ser3 by GOAT (Ghrelin O-Acyltransferase) [47], as well as histone H4 protein which was shown to be O-palmitoylated on a Ser residue by acyl-CoA:lysophosphatidylcholine acyltransferase (Lpcat1) [48]. Since O-fatty-acylated proteins are readily labeled with chemical reporters, such as az-15 [49] and alk-16 [42], these reagents combined with selective ester cleavage methods and proteomic analysis could reveal novel O-fatty-acylated proteins.
Nε-fatty-acylation
Recently, the discovery of sirtuin-mediated deacylation of ε-N-Lys fatty-acylation (Figure 1E) has suggested more prevalent roles for this modification beyond a few cytokines such as TNF-α and Interleukin-1-α [50,51]. For example, SIRT6, a sirtuin-family deacylase, was shown to hydrolyze specific long chain fatty acids, notably myristate, present on lysine side-chains in mammalian cells using alk-14 metabolic labeling and in vitro biochemical assays [15]. Interestingly, this hydrolysis was found to be critical for facilitating the secretion of TNF-α. Subsequent in vitro studies revealed the specificity of the seven mammalian sirtiun enzymes (SIRT1 through SIRT7) to hydrolyze long-chain fatty acids present on Lys side-chains [16,52,53]. In particular, SIRT2 was shown to also efficiently remove myristoyl group on Lys side-chains using a photocrosslinking chemical probe based on a Lys-myristoylated histone H3 peptide [16], alk-14 labeling, and structural studies [53]. While proteomic studies of ε-N-Lys fatty-acylation have not been reported, the chemical reporter labeling and quantitative proteomics of sirtuin-deficient cells should reveal the diversity and abundance of this modification in different cell types.
C-phosphatidylethanolaminylation
C-phosphatidylethanolaminylation (Figure 1F) of proteins is not a ubiquitous form of fatty-acylation, but is very important in regulating autophagy in eukaryotes. The major modified protein substrates are the ubiquitin-like protein Atg8 in yeast and its orthologues (LC3 isoforms) in mammalian cells [54]. The resulting lipidated Atg8/LC3 facilitates generation of autophagosomal membranes and is essential for the biogenesis of autophagosomes. The abundance and regulatory mechanisms of LC3 isoforms is still under investigation and could also benefit from more sensitive detection and proteomic methods using chemical reporters and selective enrichment strategies.
GPI-anchor modification
Glycosylphosphatidylinositols (GPI) are complex glyco-lipids attached to the C-termini of proteins and allow modified proteins to anchor into the cell membrane. GPI-anchored proteins are implicated in many diseases, including prion diseases [55] and cancer [56]. Experimental approaches for profiling of GPI-anchor proteins at a proteome-level are still limited. Recently, a new method was developed to experimentally identify GPI-anchored proteins and their ω-modification sites [57], which involves removal of the GPI-anchor moieties from GPI-anchored peptides using a specific hydrolase enzyme, i.e., phosphatidylinositol-specific phospholipase C (PI-PLC), and aqueous hydrogen fluoride. GPI-anchored peptides can then be readily analyzed by mass spectrometry, therefore allowing identification of GPI-anchor proteins and ω-sites directly. Alternatively, other strategies for proteome-wide scale analysis of GPI-anchor protein took advantage of the chemical structure of GPI-anchors composed of a phosphoethanolamine linker, glycan core, and phospholipid tail, which allowed enrichment of GPI-anchored proteins through metabolic labeling of the lipid tail with alk-12 [17] or the glycan core with GalNAz [58], an azido-tagged analogue of N-azidoacetylgalactosamine. Both chemical reporters led to the identification of known and several novel GPI-anchored proteins in malaria parasite P. falciparum and in HeLa cells, respectively. These strategies could be applied in various organisms to understand the diversity and abundance of GPI-anchored proteins.
Future Directions
Quantitative and specific proteomics
Significant progresses have been made over the past few years in identifying fatty-acylated proteins, thanks to the advances in mass spectrometry analysis and the development of robust tag-enrichment technologies. Large datasets of N-myristoylated and S-palmitoylated proteins have been generated in a wide range of organisms. However, these datasets often include a large number of false-positive hits due to the promiscuousness and metabolism of fatty-acylation reporters as discussed above, and the field now needs to generate more accurate datasets using quantitative chemical proteomics and more specific strategies. Quantitative chemical proteomics allows the identification of fatty-acylated proteins with higher confidence through the assessment of signal over background ratios and is now preferred over spectral counting [59]. In addition, chemical reporters of fatty-acylation can be metabolized in cells and incorporated in a variety of pathways, so more specific strategies, such as the combination of metabolic labeling with acyl transferase/deacetylase enzyme gain/loss of function or other complementary approaches, are often essential to limit the number of false positive hits. For instance, combination of tagging with an inhibitor of the fatty-acylation enzyme for N-myristoylation [17,18] or with a non-selective inhibitor of serine hydrolases that favors fatty acyl-bearing substrates and inhibits de-S-palmitoylation [34,60] has proven particularly powerful. For S-palmitoylation, advantage of the hydroxylamine sensitivity of the thioester linkage can also be used [61], as well as the combination of the two enrichment methods [27], i.e., ABE and metabolic labeling, which should generate higher confidence datasets of S-palmitoylated proteins.
Quantitative analysis of S-palmitoylation
Despite the recent advances in the MS/MS analysis of the fatty-acylated peptides, it remains challenging to identify the number of fatty-acylation sites and the site occupancy. The quantitation of fatty-acylation levels on specific proteins and modification sites is crucial for understanding functional consequences in vivo. To address these challenges, recent mass-shift based labeling methods such as PEG-switch assay [62] or acyl-PEGylation Exchange (APE) (unpublished results of our laboratory) has enabled sensitive detection of endogenous S-palmitoylation levels of specific proteins by western blotting analysis (Figure 2C). Following cell lysis and capping of free cysteines with maleimide, lysates were treated with a 5 kDa PEG-maleimide in the presence of hydroxylamine, which can hydrolyze the palmitate moiety. This method allows the addition of a 5 kDa mass tag for each S-palmitoylation site. SDS-PAGE followed by western blotting analysis of the protein of interest showed several band shifts, corresponding to the number of S-palmitoylation sites. Importantly, the method allows semi-quantitative analysis of the site occupancy by quantification of the several band shifts observed by western blot and should complement large-scale proteomic studies.
Site-identification
While site mutagenesis is often used to identify sites of fatty-acylation, this approach is only straightforward for proteins with a limited number of putative acylated sites such as N-myristoylation. Site-identification can be very difficult for other types of fatty-acylation such as S-palmitoylation and Nε-fatty-acylation that involve multiple Cys or Lys residues for modification. Over the past few years, critical advances have been made for N-myristoylation with the development of new cleavable reagents that can be used in combination with an alkyne-tagged chemical reporter of N-myristoylation, alk-12, and allowed direct identification of the N-myristoylation sites with high confidence [17,18,63]. In this regard, multifunctional affinity reagents comprising an azido group, a rhodamine fluorophore, a biotin moiety, and an arginine or lysine residue have been developed [63]. After metabolic labeling of N-myristoylated proteins with alk-12, bioorthogonal reaction with the multifunctional affinity reagent, streptavidin purification, and trypsin proteolysis, alk-12-tagged N-myristoylated peptides were enriched and then released by trypsin from the affinity matrix for mass spectrometry analysis. In a pioneering study, 69 alk-12-tagged N-myristoylated peptides were directly identified in Hela cells, suggesting the robustness of this approach for high-confidence identification of N-myristoylated proteome [63]. In principle, these reagents could be applicable to detect other amide-linked post-translational modifications, but the ionization method might need to be optimized to detect S-palmitoylation or O-fatty-acylation due to the instability of the thioester and ester bonds. In addition, limited successes have been achieved for S-palmitoylation site characterization using an indirect strategy based on the acyl-RAC [8,20] or ABE strategy [64], sometimes in combination with thiol-reactive cleavable isotope-coded affinity tag (cICAT) reagents or iTRAQ reagents. The main challenges will be to improve the sample preparation to overcome low abundance of the modified peptide and high hydrophobicity of the fatty-acylated peptide. Notably, S-palmitoylation often occurs in or in close proximity to a transmembrane domain, making the tryptic peptide highly hydrophobic and most likely being lost during sample preparation.
MS/MS analysis of fatty-acylated peptides
Direct MS/MS analysis of the intact endogenous fatty-acylated peptide is important to confirm that the protein is fatty-acylated, to identify or validate the structure of the fatty acid present at the native level, as well as to determine the site occupancy. Analysis of the modified peptides should be one of the priorities of the field in the next few years. While alkyne/azido-tagged fatty acids are known to mimic the natural lipid substrates, they could be metabolized to shorter/longer or unsaturated fatty acid in cells but might still be able to label the protein of interest. This metabolized fatty-acid modification might differ from the fatty-acylation occurring at endogenous level. Furthermore, other modifications could compete with the fatty acid of interest, such as S-nitrosylation for S-palmitoylation, and it is currently difficult to understand the scope and relevance of this competition. Direct and large-scale analysis of fatty-acylated peptides remains challenging due to the high hydrophobicity of the fatty-acylated peptide, poor ionization during MS analysis, and instability of the fatty acid linkage during sample preparation or ionization [65]. Recently, LC-MS analysis of synthetic S-palmitoylated peptides has been reported after careful optimization of sample preparation and LC-MS conditions. It was found that S-palmitoyl groups are largely retained in the presence of TCEP (tris(2-carboxyethyl)phosphine) in neutral Tris buffer, but are significantly lost in the presence of DTT (dithiothreitol) in ammonium bicarbonate buffer. In addition, ETD (electron transfer dissociation) was found to be the best fragmentation method for S-palmitoylated peptide analysis among several ionization methods tested [65]. In another study, a synthetic lipidated peptide standard was also used to optimize LC-MS conditions, which were then adapted to quantify changes in S-palmitoylation site occupancy of purified endogenous pulmonary surfactant protein SP-C [66]. MALDI analysis also allowed to analyze the modified peptides for N-myristoylation and S-palmitoylation but quantification of the site occupancy is limited [67–70].
Fatty acid structure analysis
There is increasing evidence that fatty acids of various chain lengths and structures (unsaturation) can replace the well-characterized fatty acids (e.g., myristate, palmitate, plamitoleic, and octanoate) for protein modification. Several studies have shown that a wide range of dietary and cellular fatty acids could be incorporated at the N-myristoylation site or S-palmitoylation site of Fyn as well as other S-palmitoylated proteins such as GAP [67,68]. Furthermore, recent studies showed that the influenza virus hemagglutinin can be S-acylated with stearate [71] and palmitate. These previously overlooked long-chain fatty-acids might be competitive in regulating protein localization and function. MS/MS analysis of fatty-acylated peptide, together with the development of corresponding chemical reporters, should provide a better understanding of these novel fatty-acylation forms. Alternatively, to overcome the issues of fatty-acylated peptide analysis, the fatty acid can be hydrolyzed from the protein of interest and compared with fatty acid standards or analyzed directly by GC-MS [72,73]. However, this method often requires high purity and large amount (>1 μg) of the protein of interest [73] and determination of the site occupancy is not possible.
Fatty-acylation-dependent protein interactions
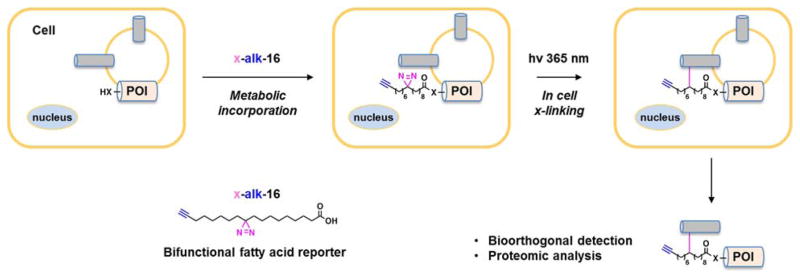
Characterizing the protein complexes and cellular pathways controlled by specific fatty-acylated proteins remains a major frontier. To identify S-palmitoylated membrane protein complexes, a bifunctional 18-carbon palmitic acid reporter containing an internal diazirine and a terminal alkynyl group, x-alk-16, was developed (Figure 3) [74]. This bifunctional 18-carbon palmitic acid reporter metabolically labels S-palmitoylated proteins and can covalently trap protein complexes in cells by UV-crosslinking prior to cell lysis. This bifunctional reporter was used in mammalian cells in combination with quantitative proteomics to study S-palmitoylated IFITM3-interacting proteins and revealed known IFITM3-interacting proteins, such as VAPA, as well as novel interacting proteins, which could be key cellular factors for host resistance to virus infection. A similar method could be applied to identify interacting proteins of other fatty-acylated proteins [75] and study the role of fatty-acylation in regulating protein–protein interactions in cells.
Figure 3.
Photocrosslinking of fatty-acylated protein-protein interactions.
Summary
The development of proteomic strategies for N-myristoylation and S-palmitoylation has significantly expanded the scope and potential functions of these fatty-acylated proteins. The application of specific chemical reporters and selective enrichment methods are poised to make similar breakthroughs for other classes of fatty-acylated proteins. Unravelling the biological significance of these newly discovered candidate fatty-acylated proteins will further require identification of modification sites, robust quantitation in different in vivo settings, and the characterization of key regulatory mechanisms, which are the major directions for future developments in the fatty-acylation field.
Highlights.
Protein fatty-acylation regulates many fundamental biological processes in eukaryotes.
Chemical proteomics has expanded the potential functions of protein fatty-acylation in biology.
New methods are needed for direct identification of fatty-acylation sites and characterization of covalently attached lipids.
Quantitative methods are desired for functional characterization of fatty-acylation regulatory mechanisms.
Acknowledgments
E. T. is a Marie Skłodowska-Curie Fellow. H.C.H. acknowledges support from NIH-NIGMS R01 GM087544 grant and Starr Cancer Consortium I7-A717.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
• of outstanding interest
- 1.Hang HC, Linder ME. Exploring protein lipidation with chemical biology. Chem Rev. 2011;111:6341–6358. doi: 10.1021/cr2001977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Resh MD. Trafficking and signaling by fatty-acylated and prenylated proteins. Nat Chem Biol. 2006;2:584–590. doi: 10.1038/nchembio834. [DOI] [PubMed] [Google Scholar]
- 3.Tate EW, Kalesh KA, Lanyon-Hogg T, Storck EM, Thinon E. Global profiling of protein lipidation using chemical proteomic technologies. Curr Opin Chem Biol. 2015;24:48–57. doi: 10.1016/j.cbpa.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grammel M, Hang HC. Chemical reporters for biological discovery. Nat Chem Biol. 2013;9:475–484. doi: 10.1038/nchembio.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5•.Thinon E, Hang HC. Chemical reporters for exploring protein acylation. Biochem Soc Trans. 2015;43:253–261. doi: 10.1042/BST20150004. This is the most recent review that summarizes almost all chemical reporters in literature for studying protein acylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drisdel RC, Green WN. Labeling and quantifying sites of protein palmitoylation. BioTechniques. 2004;36:276–285. doi: 10.2144/04362RR02. [DOI] [PubMed] [Google Scholar]
- 7.Roth AF, Wan J, Bailey AO, Sun B, Kuchar JA, Green WN, Phinney BS, Yates JR, Iii, Davis NG. Global Analysis of Protein Palmitoylation in Yeast. Cell. 2006;125:1003–1013. doi: 10.1016/j.cell.2006.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forrester MT, Hess DT, Thompson JW, Hultman R, Moseley MA, Stamler JS, Casey PJ. Site-specific analysis of protein S-acylation by resin-assisted capture. J Lipid Res. 2011;52:393–398. doi: 10.1194/jlr.D011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wright M, Heal W, Mann D, Tate E. Protein myristoylation in health and disease. J Chem Biol. 2010;3:19–35. doi: 10.1007/s12154-009-0032-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charron G, Wilson J, Hang HC. Chemical tools for understanding protein lipidation in eukaryotes. Curr Opin Chem Biol. 2009;13:382–391. doi: 10.1016/j.cbpa.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 11.Hannoush RN, Sun J. The chemical toolbox for monitoring protein fatty acylation and prenylation. Nat Chem Biol. 2010;6:498–506. doi: 10.1038/nchembio.388. [DOI] [PubMed] [Google Scholar]
- 12.Hang HC, Wilson JP, Charron G. Bioorthogonal Chemical Reporters for Analyzing Protein Lipidation and Lipid Trafficking. Acc Chem Res. 2011;44:699–708. doi: 10.1021/ar200063v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charron G, Zhang MM, Yount JS, Wilson J, Raghavan AS, Shamir E, Hang HC. Robust Fluorescent Detection of Protein Fatty-Acylation with Chemical Reporters. J Am Chem Soc. 2009;131:4967–4975. doi: 10.1021/ja810122f. [DOI] [PubMed] [Google Scholar]
- 14.Wilson JP, Raghavan AS, Yang Y-Y, Charron G, Hang HC. Proteomic Analysis of Fatty-acylated Proteins in Mammalian Cells with Chemical Reporters Reveals S-Acylation of Histone H3 Variants. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15•.Jiang H, Khan S, Wang Y, Charron G, He B, Sebastian C, Du J, Kim R, Ge E, Mostoslavsky R, et al. SIRT6 regulates TNF-[agr] secretion through hydrolysis of long-chain fatty acyl lysine. Nature. 2013;496:110–113. doi: 10.1038/nature12038. This paper reports SIRT6 can hydrolyze long chain fatty acids present on lysine side-chains in cells and suggests more prevalent roles for Nε-fatty-acylation in biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Z, Yang T, Li X, Peng T, Hang HC, Li XD. Integrative chemical biology approaches for identification and characterization of “erasers” for fatty-acid-acylated lysine residues within proteins. Angew Chem Int Ed Engl. 2015;54:1149–1152. doi: 10.1002/anie.201408763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17••.Wright MH, Clough B, Rackham MD, Rangachari K, Brannigan JA, Grainger M, Moss DK, Bottrill AR, Heal WP, Broncel M, et al. Validation of N-myristoyltransferase as an antimalarial drug target using an integrated chemical biology approach. Nat Chem. 2014;6:112–121. doi: 10.1038/nchem.1830. This study integrates N-myristoylation chemical reporter, selective NMT inhibitors, and quantitative proteomics to globally profile N-myristoylation in malaria parasites and validate NMT as a drug target. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18••.Thinon E, Serwa RA, Broncel M, Brannigan JA, Brassat U, Wright MH, Heal WP, Wilkinson AJ, Mann DJ, Tate EW. Global profiling of co- and post-translationally N-myristoylated proteomes in human cells. Nat Commun. 2014;5:4919. doi: 10.1038/ncomms5919. This paper reports the global profiling of N-myristoylation in human cells using N-myristoylation chemical reporter, selective NMT inhibitors, and quantitative proteomics, along with the crystal structures of human NMTs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright Megan H, Paape D, Storck Elisabeth M, Serwa Remigiusz A, Smith Deborah F, Tate Edward W. Global Analysis of Protein N-Myristoylation and Exploration of N-Myristoyltransferase as a Drug Target in the Neglected Human Pathogen Leishmania donovani. Chem Biol. 2015;22:342–354. doi: 10.1016/j.chembiol.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20•.Serwa Remigiusz A, Abaitua F, Krause E, Tate Edward W, O’Hare P. Systems Analysis of Protein Fatty Acylation in Herpes Simplex Virus-Infected Cells Using Chemical Proteomics. Chem Biol. 2015;22:1008–1017. doi: 10.1016/j.chembiol.2015.06.024. This paper utilizes both the chemical reporter strategy and ABE method to globally analyze host and virus protein fatty-acylation during infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colquhoun DR, Lyashkov AE, Mohien CU, Aquino VN, Bullock BT, Dinglasan RR, Agnew BJ, Graham DRM. Bioorthogonal mimetics of palmitoyl-CoA and myristoyl-CoA and their subsequent isolation by click chemistry and characterization by mass spectrometry reveal novel acylated host-proteins modified by HIV-1 infection. Proteomics. 2015;15:2066–2077. doi: 10.1002/pmic.201500063. [DOI] [PubMed] [Google Scholar]
- 22•.Burnaevskiy N, Fox TG, Plymire DA, Ertelt JM, Weigele BA, Selyunin AS, Way SS, Patrie SM, Alto NM. Proteolytic elimination of N-myristoyl modifications by the Shigella virulence factor IpaJ. Nature. 2013;496:106–109. doi: 10.1038/nature12004. This paper reports the first example of elimination of N-myristoylation by a secreted bacterial effector protein IpaJ from Shigella. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23••.Burnaevskiy N, Peng T, Reddick LE, Hang Howard C, Alto Neal M. Myristoylome Profiling Reveals a Concerted Mechanism of ARF GTPase Deacylation by the Bacterial Protease IpaJ. Mol Cell. 2015;58:110–122. doi: 10.1016/j.molcel.2015.01.040. In this study, N-myristoylation chemical reporter and quantitative proteomics were utilized to globally profile the protein substrates of IpaJ under different physiological conditions, which help determine the substrate scope of IpaJ and its mechanisms for substrate selection and cleavage. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linder ME, Deschenes RJ. Palmitoylation: policing protein stability and traffic. Nat Rev Mol Cell Biol. 2007;8:74–84. doi: 10.1038/nrm2084. [DOI] [PubMed] [Google Scholar]
- 25.Chavda B, Arnott JA, Planey SL. Targeting protein palmitoylation: selective inhibitors and implications in disease. Expert Opin Drug Discov. 2014;9:1005–1019. doi: 10.1517/17460441.2014.933802. [DOI] [PubMed] [Google Scholar]
- 26.Yeste-Velasco M, Linder ME, Lu Y-J. Protein S-palmitoylation and cancer. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer. 2015;1856:107–120. doi: 10.1016/j.bbcan.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 27••.Jones Matthew L, Collins Mark O, Goulding D, Choudhary Jyoti S, Rayner Julian C. Analysis of Protein Palmitoylation Reveals a Pervasive Role in Plasmodium Development and Pathogenesis. Cell Host Microbe. 2012;12:246–258. doi: 10.1016/j.chom.2012.06.005. In this paper, the authors compared the chemical reporter strategy and ABE method in parallel for globally profiling protein S-palmitoylation in malaria parasite Plasmodium, and demonstrated that these two methods are largely complementary and provide unique coverages of S-palmitoylated proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou B, An M, Freeman MR, Yang W. Technologies and Challenges in Proteomic Analysis of Protein S-acylation. J Proteomics Bioinform. 2014;7:256–263. doi: 10.4172/jpb.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang R, Wan J, Arstikaitis P, Takahashi H, Huang K, Bailey AO, Thompson JX, Roth AF, Drisdel RC, Mastro R, et al. Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature. 2008;456:904–909. doi: 10.1038/nature07605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yount JS, Moltedo B, Yang YY, Charron G, Moran TM, Lopez CB, Hang HC. Palmitoylome profiling reveals S-palmitoylation-dependent antiviral activity of IFITM3. Nat Chem Biol. 2010;6:610–614. doi: 10.1038/nchembio.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chesarino NM, Hach JC, Chen JL, Zaro BW, Rajaram MV, Turner J, Schlesinger LS, Pratt MR, Hang HC, Yount JS. Chemoproteomics reveals Toll-like receptor fatty acylation. BMC Biol. 2014;12:91. doi: 10.1186/s12915-014-0091-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yount JS, Zhang MM, Hang HC. Emerging roles for protein S-palmitoylation in immunity from chemical proteomics. Curr Opin Chem Biol. 2013;17:27–33. doi: 10.1016/j.cbpa.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang MM, Wu P-YJ, Kelly FD, Nurse P, Hang HC. Quantitative Control of Protein S-Palmitoylation Regulates Meiotic Entry in Fission Yeast. PLoS Biol. 2013;11:e1001597. doi: 10.1371/journal.pbio.1001597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin BR, Wang C, Adibekian A, Tully SE, Cravatt BF. Global profiling of dynamic protein palmitoylation. Nat Methods. 2012;9:84–89. doi: 10.1038/nmeth.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wan J, Savas JN, Roth AF, Sanders SS, Singaraja RR, Hayden MR, Yates JR, 3rd, Davis NG. Tracking brain palmitoylation change: predominance of glial change in a mouse model of Huntington’s disease. Chem Biol. 2013;20:1421–1434. doi: 10.1016/j.chembiol.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morrison E, Kuropka B, Kliche S, Brügger B, Krause E, Freund C. Quantitative analysis of the human T cell palmitome. Sci Rep. 2015;5:11598. doi: 10.1038/srep11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37•.Santiago-Tirado FH, Peng T, Yang M, Hang HC, Doering TL. A Single Protein S-acyl Transferase Acts through Diverse Substrates to Determine Cryptococcal Morphology, Stress Tolerance, and Pathogenic Outcome. PLoS Pathog. 2015;11:e1004908. doi: 10.1371/journal.ppat.1004908. Bioorthogonal S-palmitoylome profiling was performed to determine the specific substrates of PFA4, a putative protein S-acyltransferase that regulates the morphology, stress tolerance, and virulence of Cryptococcus neoformans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Foe Ian T, Child Matthew A, Majmudar Jaimeen D, Krishnamurthy S, van der Linden Wouter A, Ward Gary E, Martin Brent R, Bogyo M. Global Analysis of Palmitoylated Proteins in Toxoplasma gondii. Cell Host Microbe. 18:501–511. doi: 10.1016/j.chom.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39••.Blanc M, David F, Abrami L, Migliozzi D, Armand F, Bürgi J, van der Goot F. SwissPalm: Protein Palmitoylation database [v1; ref status: indexed, http://f1000res/5co] F1000Research. 2015;4:261. doi: 10.12688/f1000research.6464.1. In this study, proteomics data from 19 large-scale S-palmitoylome profiling studies were combined into a single database SwissPalm that is readily accessible for researchers to search for proteins of interest. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40••.Sanders SS, Martin DDO, Butland SL, Lavallée-Adam M, Calzolari D, Kay C, Yates JR, III, Hayden MR. Curation of the Mammalian Palmitoylome Indicates a Pivotal Role for Palmitoylation in Diseases and Disorders of the Nervous System and Cancers. PLoS Comput Biol. 2015;11:e1004405. doi: 10.1371/journal.pcbi.1004405. In this study, proteomics data from 15 mammalian S-palmitoylome profiling studies were curated into a single compendium of S-palmitoylated proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takada R, Satomi Y, Kurata T, Ueno N, Norioka S, Kondoh H, Takao T, Takada S. Monounsaturated fatty acid modification of Wnt protein: its role in Wnt secretion. Dev Cell. 2006;11:791–801. doi: 10.1016/j.devcel.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 42.Gao X, Hannoush RN. Single-cell imaging of Wnt palmitoylation by the acyltransferase porcupine. Nat Chem Biol. 2014;10:61–68. doi: 10.1038/nchembio.1392. [DOI] [PubMed] [Google Scholar]
- 43.Janda CY, Waghray D, Levin AM, Thomas C, Garcia KC. Structural basis of Wnt recognition by Frizzled. Science. 2012;337:59–64. doi: 10.1126/science.1222879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miranda M, Galli LM, Enriquez M, Szabo LA, Gao X, Hannoush RN, Burrus LW. Identification of the WNT1 residues required for palmitoylation by Porcupine. FEBS Lett. 2014;588:4815–4824. doi: 10.1016/j.febslet.2014.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu J, Pan S, Hsieh MH, Ng N, Sun F, Wang T, Kasibhatla S, Schuller AG, Li AG, Cheng D, et al. Targeting Wnt-driven cancer through the inhibition of Porcupine by LGK974. Proc Natl Acad Sci U S A. 2013;110:20224–20229. doi: 10.1073/pnas.1314239110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Purro SA, Galli S, Salinas PC. Dysfunction of Wnt signaling and synaptic disassembly in neurodegenerative diseases. J Mol Cell Biol. 2014;6:75–80. doi: 10.1093/jmcb/mjt049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang J, Brown MS, Liang G, Grishin NV, Goldstein JL. Identification of the acyltransferase that octanoylates ghrelin, an appetite-stimulating peptide hormone. Cell. 2008;132:387–396. doi: 10.1016/j.cell.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 48.Zou C, Ellis BM, Smith RM, Chen BB, Zhao Y, Mallampalli RK. Acyl-CoA:lysophosphatidylcholine acyltransferase I (Lpcat1) catalyzes histone protein O-palmitoylation to regulate mRNA synthesis. J Biol Chem. 2011;286:28019–28025. doi: 10.1074/jbc.M111.253385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ching W, Hang HC, Nusse R. Lipid-independent secretion of a Drosophila Wnt protein. J Biol Chem. 2008;283:17092–17098. doi: 10.1074/jbc.M802059200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stevenson FT, Bursten SL, Locksley RM, Lovett DH. Myristyl acylation of the tumor necrosis factor alpha precursor on specific lysine residues. J Exp Med. 1992;176:1053–1062. doi: 10.1084/jem.176.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stevenson FT, Bursten SL, Fanton C, Locksley RM, Lovett DH. The 31-kDa precursor of interleukin 1 alpha is myristoylated on specific lysines within the 16-kDa N-terminal propiece. Proc Natl Acad Sci U S A. 1993;90:7245–7249. doi: 10.1073/pnas.90.15.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feldman JL, Baeza J, Denu JM. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J Biol Chem. 2013;288:31350–31356. doi: 10.1074/jbc.C113.511261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Teng YB, Jing H, Aramsangtienchai P, He B, Khan S, Hu J, Lin H, Hao Q. Efficient demyristoylase activity of SIRT2 revealed by kinetic and structural studies. Sci Rep. 2015;5:8529. doi: 10.1038/srep08529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 55.Godsave SF, Peters PJ, Wille H. Subcellular distribution of the prion protein in sickness and in health. Virus Res. 2015;207:136–145. doi: 10.1016/j.virusres.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 56.Gamage DG, Hendrickson TL. GPI transamidase and GPI anchored proteins: oncogenes and biomarkers for cancer. Crit Rev Biochem Mol Biol. 2013;48:446–464. doi: 10.3109/10409238.2013.831024. [DOI] [PubMed] [Google Scholar]
- 57.Masuishi Y, Nomura A, Okayama A, Kimura Y, Arakawa N, Hirano H. Mass Spectrometric Identification of Glycosylphosphatidylinositol-Anchored Peptides. J Proteome Res. 2013;12:4617–4626. doi: 10.1021/pr4004807. [DOI] [PubMed] [Google Scholar]
- 58.Cortes LK, Vainauskas S, Dai N, McClung CM, Shah M, Benner JS, Correa IR, Jr, VerBerkmoes NC, Taron CH. Proteomic identification of mammalian cell surface derived glycosylphosphatidylinositol-anchored proteins through selective glycan enrichment. Proteomics. 2014;14:2471–2484. doi: 10.1002/pmic.201400148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Z, Adams RM, Chourey K, Hurst GB, Hettich RL, Pan C. Systematic Comparison of Label-Free, Metabolic Labeling, and Isobaric Chemical Labeling for Quantitative Proteomics on LTQ Orbitrap Velos. J Proteome Res. 2012;11:1582–1590. doi: 10.1021/pr200748h. [DOI] [PubMed] [Google Scholar]
- 60.Tom CT, Martin BR. Fat Chance! Getting a Grip on a Slippery Modification. ACS Chem Biol. 2012 doi: 10.1021/cb300607e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martin BR, Cravatt BF. Large-scale profiling of protein palmitoylation in mammalian cells. Nat Methods. 2009;6:135–138. doi: 10.1038/nmeth.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Howie J, Reilly L, Fraser NJ, Vlachaki Walker JM, Wypijewski KJ, Ashford MLJ, Calaghan SC, McClafferty H, Tian L, Shipston MJ, et al. Substrate recognition by the cell surface palmitoyl transferase DHHC5. Proc Natl Acad Sci U S A. 2014;111:17534–17539. doi: 10.1073/pnas.1413627111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63••.Broncel M, Serwa RA, Ciepla P, Krause E, Dallman MJ, Magee AI, Tate EW. Multifunctional Reagents for Quantitative Proteome-Wide Analysis of Protein Modification in Human Cells and Dynamic Profiling of Protein Lipidation During Vertebrate Development. Angew Chem, Int Ed. 2015;54:5948–5951. doi: 10.1002/anie.201500342. In this study, multifunctional affinity reagents were developed and applied to directly detect alkyne-tagged N-myristoylated peptides by mass spectrometry with high-confidence for identification of N-myristoylated proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang W, Di Vizio D, Kirchner M, Steen H, Freeman MR. Proteome scale characterization of human S-acylated proteins in lipid raft-enriched and non-raft membranes. Mol Cell Proteomics. 2010;9:54–70. doi: 10.1074/mcp.M800448-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ji Y, Leymarie N, Haeussler DJ, Bachschmid MM, Costello CE, Lin C. Direct detection of S-palmitoylation by mass spectrometry. Anal Chem. 2013;85:11952–11959. doi: 10.1021/ac402850s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harayama T, Shindou H, Kita Y, Otsubo E, Ikeda K, Chida S, Weaver TE, Shimizu T. Establishment of LC-MS methods for the analysis of palmitoylated surfactant proteins. J Lipid Res. 2015;56:1370–1379. doi: 10.1194/jlr.D060236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liang X, Nazarian A, Erdjument-Bromage H, Bornmann W, Tempst P, Resh MD. Heterogeneous fatty acylation of Src family kinases with polyunsaturated fatty acids regulates raft localization and signal transduction. J Biol Chem. 2001;276:30987–30994. doi: 10.1074/jbc.M104018200. [DOI] [PubMed] [Google Scholar]
- 68.Liang X, Lu Y, Neubert TA, Resh MD. Mass spectrometric analysis of GAP-43/neuromodulin reveals the presence of a variety of fatty acylated species. J Biol Chem. 2002;277:33032–33040. doi: 10.1074/jbc.M204607200. [DOI] [PubMed] [Google Scholar]
- 69.Xue L, Gollapalli DR, Maiti P, Jahng WJ, Rando RR. A palmitoylation switch mechanism in the regulation of the visual cycle. Cell. 2004;117:761–771. doi: 10.1016/j.cell.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 70.Moriya K, Nagatoshi K, Noriyasu Y, Okamura T, Takamitsu E, Suzuki T, Utsumi T. Protein N-myristoylation plays a critical role in the endoplasmic reticulum morphological change induced by overexpression of protein Lunapark, an integral membrane protein of the endoplasmic reticulum. PLoS ONE. 2013;8:e78235. doi: 10.1371/journal.pone.0078235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brett K, Kordyukova LV, Serebryakova MV, Mintaev RR, Alexeevski AV, Veit M. Site-specific S-acylation of influenza virus hemagglutinin: the location of the acylation site relative to the membrane border is the decisive factor for attachment of stearate. J Biol Chem. 2014;289:34978–34989. doi: 10.1074/jbc.M114.586180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sorek N, Akerman A, Yalovsky S. Analysis of protein prenylation and S-acylation using gas chromatography-coupled mass spectrometry. Methods Mol Biol. 2013;1043:121–134. doi: 10.1007/978-1-62703-532-3_13. [DOI] [PubMed] [Google Scholar]
- 73.Sorek N, Yalovsky S. Analysis of protein S-acylation by gas chromatography-coupled mass spectrometry using purified proteins. Nat Protoc. 2010;5:834–840. doi: 10.1038/nprot.2010.33. [DOI] [PubMed] [Google Scholar]
- 74••.Peng T, Hang HC. Bifunctional fatty acid chemical reporter for analyzing S-palmitoylated membrane protein-protein interactions in mammalian cells. J Am Chem Soc. 2015;137:556–559. doi: 10.1021/ja502109n. This paper demonstrates that a bifunctional fatty acid with an internal diazirine and a terminal alkyne is readily incorporated into S-palmitoylated proteins and induces photocrosslinking of S-palmitoylated proteins with their interacting partners in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Peng T, Yuan X, Hang HC. Turning the spotlight on protein-lipid interactions in cells. Curr Opin Chem Biol. 2014;21:144–153. doi: 10.1016/j.cbpa.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]