Abstract
Cancer sequencing studies have primarily identified cancer-driver genes by the accumulation of protein-altering mutations. An improved method would be annotation-independent, sensitive to unknown distributions of functions within proteins, and inclusive of non-coding drivers. We employed density-based clustering methods in 21 tumor types to detect variably-sized significantly mutated regions (SMRs). SMRs reveal recurrent alterations across a spectrum of coding and non-coding elements, including transcription factor binding sites and untranslated regions mutated in up to ∼15% of specific tumor types. SMRs reveal spatial clustering of mutations at molecular domains and interfaces, often with associated changes in signaling. Mutation frequencies in SMRs demonstrate that distinct protein regions are differentially mutated among tumor types, as exemplified by a linker region of PIK3CA in which biophysical simulations suggest mutations affect regulatory interactions. The functional diversity of SMRs underscores both the varied mechanisms of oncogenic misregulation and the advantage of functionally-agnostic driver identification.
Keywords: Significantly Mutated Regions (SMRs), cancer, exome sequencing, non-coding variation
Introduction
In cancer, driver mutations alter functional elements of diverse nature and size. For example, melanoma drivers include hyper-activating mutations at single amino acid residues (e.g. BRAF V6001), inactivating mutations along tumor suppressor exons (e.g. PTEN1), and regulatory mutations (e.g. TERT promoter2). Cancer genomics projects, such as the The Cancer Genome Atlas (TCGA) and the International Cancer Genome Consortium (ICGC), have substantially expanded our understanding of the landscape of somatic alterations by identifying frequently mutated protein coding genes3–5. However, these studies have focused little attention on systematically analyzing the positional distribution of coding mutations or characterizing non-coding alterations6.
Algorithms to identify cancer-driver genes often examine non-synonymous to synonymous mutation rates across the gene body or recurrently mutated amino acids called mutation hotspots5, as observed in BRAF7, IDH18, and DNA polymerase ε (POLE)9. Yet, these analyses ignore recurrent alterations in the vast intermediate scale of functional coding elements, such as protein subunits or interfaces. Moreover, where mutation clustering within genes has been examined10–12, analyses have employed fixed base-pair windows or identified clusters of non-synonymous mutations, assuming driver mutations exclusively impact protein sequence and ignoring the importance of exon-embedded regulatory elements13–18.
A significant proportion of regulatory elements in the genome occurs proximal to, or even in, exons15,19, suggesting many may be captured by whole-exome sequencing (WES). Efforts to characterize non-coding regulatory variation in cancer genomes have primarily examined either (1) pan-cancer whole-genome sequencing (WGS) data, or (2) predefined regions –such as ETS binding sites, splicing signals, promoters, and untranslated regions (UTRs)– or mutation types20–23. These approaches either presume the relevant targets of disruption, or disregard the established heterogeneity among cancer types at the level of driver genes and pathways5,24,25 as well as in nucleotide-specific mutation probabilities3,4. Yet, systematic analyses of metazoan regulatory activity have revealed substantial tissue and developmental stage specificity26–28, suggesting that mutations in cancer type-specific regulatory features may be significant non-coding drivers of cancer.
To address these diverse limitations, we employed density-based clustering techniques utilizing cancer-, mutation type-, and gene-specific mutation models to identify regions of recurrent mutations in 21 cancer types. This approach permitted the unbiased identification of variably-sized genomic regions recurrently altered by somatic mutations, which we term significantly mutated regions (SMRs). We identified SMRs in numerous well-established cancer-drivers as well as in novel genes and functional elements. Moreover, SMRs were associated with non-coding elements, protein structures, molecular interfaces, and transcriptional and signaling profiles, providing insight into the molecular consequences of accumulating somatic mutations in these regions. Overall, SMRs revealed a rich spectrum of coding and non-coding elements recurrently targeted by somatic alterations that complement gene- and pathway-centric analyses.
Results
Multi-scale detection of significantly mutated regions
We examined ∼3 million previously identified5 somatic, single nucleotide variants (SNVs) from 4,735 tumors of 21 cancer types, recording29 their impact on protein-coding sequences, transcripts, and adjacent regulatory regions (Supplementary Fig. 1). Fully 79.0% (n=2,431,360) of these somatic mutations do not alter protein-coding sequences or their splicing and thus were not previously considered in the analysis of cancer-driver mutations5 (Fig. 1a).
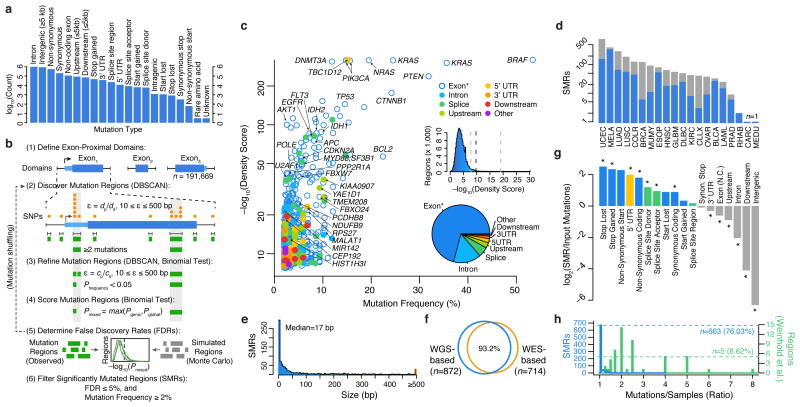
Figure 1.
Identification of significantly mutated regions (SMRs) in 20 cancer types across a broad spectrum of functional elements. (a) Pan-cancer distribution of mutation types in n=3,078,482 somatic single-nucleotide variant (SNV) calls. (b) Exons and exon-proximal domains (±1,000 bp) were scanned for clusters of somatic mutations (orange, DBSCAN). Distance parameter ε is dynamically defined as the average distance of mutated positions (dp) in the domain size (ds). Clusters (green) are divided if sub-clusters with higher mutation densities (P < 0.05, binomial test) are found in a second-pass analysis with ε defined as the average distance of mutated positions (cp) within the cluster of size cs (see Online Methods for density scoring and FDR calculation). (c) Per-cancer mutation frequency and density scores of discovered SMRs (color-coded by type and labelled by associated gene). The distribution of density scores in evaluated regions and SMR region types are shown in insets (middle) and (bottom), respectively. Dashed lines indicate the minimum, median, and maximum density score FDR (5%) thresholds. “Exon*” label refers to coding exons and non-coding genes. (d) Number of SMRs with FDR ≤ 5% and mutation frequency ≥2% per cancer type. Gray bars indicate SMRs with FDR ≤ 5% but mutation frequency <2%. (e) SMR size distribution. (f) Concordance between SMRs discovered by employing background models derived from whole-genome (WGS-based) or whole-exome (WES-based) sequencing. (g) Categories with significant fold change in mutation type representation between SMR-associated and input mutations are denoted (*; P < 0.01). (h) Distribution of the number of mutations per sample in SMRs (blue) and 58 (green) recurrently-altered non-coding regions20.
To discover both coding and non-coding cancer-drivers, we applied an annotation-independent, density-based clustering technique30 to identify 198,247 variably-sized clusters of somatic mutations within exon-proximal domains of the human genome (Fig. 1b; Online Methods). We included synonymous mutations because functionally important non-coding features can be embedded within coding regions13–18.
Mutation density scores within each identified cluster were derived as the Fisher's combined p-value of the individual binomial probabilities of observing k or more mutations for each mutation type within the region in each cancer type (Online Methods). We evaluated mutation density for each cluster using gene-specific and genome-wide models of mutation probability (Supplementary Fig. 2), which were well-correlated (Supplementary Fig. 3a), selecting the more conservative estimate for each cluster as the final density score (Online Methods). Gene-specific mutation probability models accounted for sequence composition (GC-content) as well as differences in local gene expression and replication timing, which have been shown to correlate with somatic mutation rate4. To avoid skewed mutation probability estimates due to selection pressure on exons, we applied a Bayesian framework to derive gene-specific mutation probabilities given intronic mutation probabilities in cancer WGS data3,20 while controlling for differences in sensitivity in WES and WGS (Online Methods).
Although many known cancer genes do not display signals of high mutation density, increasing density scores correlated with stronger enrichments (up to 120×) for somatic SNV-driven cancer genes (n=158) as determined by the Cancer Gene Census (CGC; Supplementary Fig. 3b-c)31,32. Moreover, ∼10% of genes associated with SMRs in the top density score quintile were not found previously in a gene-level analysis5 or in the CGC. Thus, high density scores are enriched for known cancer genes but also nominate novel drivers.
We applied Monte Carlo simulations to select density score thresholds that control the false discovery rate (FDR) to ≤ 5% (Supplementary Fig. 4, Supplementary Table 1). We identified 872 Significantly Mutated Regions (SMRs; Fig. 1c) that were altered in ≥2% of patients in 20 cancer types for further characterization (Fig. 1d). SMRs span 735 genomic regions, which are assigned unique SMR codes (e.g. TP53.1). Note that some SMRs (n=120) appear in more than one cancer type.
We classified SMRs into high-, medium-, and low-confidence sets on the basis of their density scores and contribution from mutator samples (Supplementary Table 2, Online Methods). We observed correspondingly high (63.3×, P = 2.5 × 10–46), medium (6.2×, P = 2.6 × 10–10), and low (5.0×, P = 5.0 × 10–4) enrichments for somatic SNV-driven cancer genes in these sets. To control for unaccounted processes that could result in clusters of mutations with no selective advantage in cancer, we leveraged single-nucleotide and tri-nucleotide density scores from intronic mutation clusters under the assumption that these are non-functional (Online Methods). This procedure identified 205 ‘robust’ SMRs that passed a false discovery threshold (FDR ≤ 5%) in these secondary tests, or were found in multiple cancer types. Fully 95.0% of high-confidence SMRs in cancer types where these tests could be applied satisfied these stringent alternate criteria (Supplementary Fig. 5). Over 87% of SMRs were contained within mappable (100 bp) regions of the genome, and an analysis of 6,179 recently-published breakpoints33 yielded a single SMR (in PTEN) within 50 bp of a resolved breakpoint, suggesting that the observed mutation density in SMRs is not attributable to mapping artifacts.
SMRs display a wide range of sizes (Fig. 1e, median = 17 bp, range 1-2,041 bp), are robust to distinct mutation background models (Fig. 1f, Online Methods), are not driven by unaccounted mutation contexts (Supplementary Fig. 6), and are enriched in protein-coding, 5′ UTR and splice-site mutations (Fig. 1g, P < 0.01). Importantly, SMRs are not driven by samples that contribute large numbers of mutations per region (Fig. 1h). This is in contrast to recently proposed regions of recurrent alteration20 where as few as five were driven exclusively by distinct samples (P = 6.0 × 10–45, Wilcoxon rank sum test). Thus, we have identified a diverse set of variably-sized SMRs targeted by recurrent somatic alterations and sought to characterize their relevance to functional elements and cancer genes.
SMRs enrich known and implicate novel cancer genes
SMRs are predicted to have diverse impacts on 610 genes and are 8.35×-fold enriched in known somatic cancer genes (Lawrence et al.5 or CGC, P = 8.1 × 10–49, hypergeometric test), affecting a total of 91 known drivers, including canonical oncogenes (e.g. BRAF, KRAS, NRAS, PIK3CA, and CTNNB1) and tumor suppressors (e.g. PTEN, TP53, and APC). SMR-associated genes also include 17 CGC genes previously undetected in a gene-level analysis5, such as established oncogenes like BCL2 and PIM1 and the cancer-associated non-coding gene, MALAT1. Most coding SMRs are driven by non-synonymous mutations (Supplementary Fig. 7), demonstrating that SMRs capture positive selection primarily acting on protein alterations. In total, SMRs implicate 26 known cancer genes to an additional 31 gene-to-cancer-type associations not uncovered by a gene-level analysis5 (Supplementary Table 3). We note, however, that most known cancer genes do not harbor regions of dense mutation recurrence within these data (see Supplementary Discussion, Supplementary Fig. 8), suggesting that SMR identification complements gene-level approaches.
We discovered SMRs in multiple novel cancer-driver genes, including the breast cancer-associated antigen and putative transcription factor ANKRD30A34, in which ∼21% of melanomas harbor mutations within one or more of three SMRs. Mutations in these SMRs were validated in WGS data from 6 of 17 cutaneous melanomas3,20. Within the entire gene-body, 27 of 118 WES and 10 of 17 WGS datasets from melanoma patients harbor somatic protein-altering mutations in ANKRD30A. Overall, of the 185 high-confidence SMRs, 16 were associated with novel cancer-driver genes (Supplementary Table 4). As expected on the basis of methodological differences, these putative novel cancer-drivers are primarily (∼81%) driven by non-coding alterations, as discussed in the next section.
SMRs implicate diverse non-coding regulatory features
A significant proportion (31.2%; P < 2.2 × 10–16, proportions test) of SMRs are not predicted to affect protein sequences, highlighting the potential to discover pathological non-coding variation in WES data. In total, 130 SMRs lay within open chromatin28 and are enriched in promoter (Q = 4.0 × 10–9, 4.9×) and 5′ UTR features (Q = 4.4 × 10–10, 6.0×; Supplementary Table 5). Three promoter SMRs (n=26) coincide with regions deemed significantly mutated in a pan-cancer analysis of WGS data20. Across all cancer types, small (≤25 bp) non-coding SMRs were enriched in binding sequences for ETS oncogene family (Q = 2.6 × 10–6, 7.4×) and winged-helix repressor (Q = 2.0 × 10–4, 3.2×) TFs (Fig. 2a, Supplementary Table 6). We also detected cancer-specific TF motif enrichments within SMRs from diffuse large B-cell lymphoma, melanoma, and rhabdosarcoma (Fig. 2b, Supplementary Table 7).
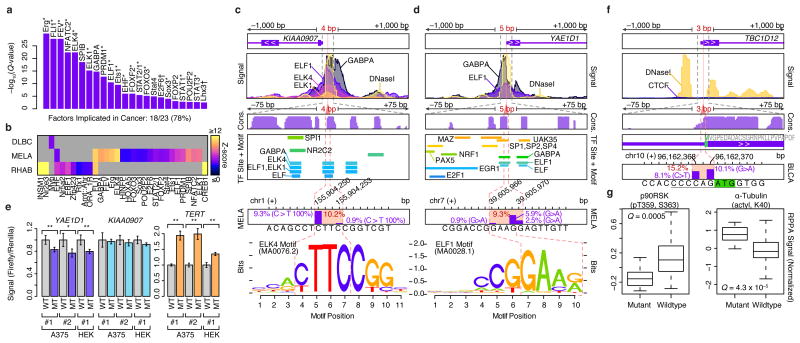
Figure 2.
Non-coding SMRs recurrently alter promoters and 5′ UTRs. (a) Transcription factors (TFs) with enriched (Q < 0.01) motifs in small SMRs (≤25bp) across all cancer types are shown. 18 of the 23 TFs are known cancer-associated TFs (*) or associated with cell-cycle control or developmental roles (†). (b) Cancer-specific motif enrichment analysis. (c) Gene structure, ENCODE ChIP-seq and DNaseI signals, vertebrate conservation (phastCons 100way), Factorbook TF binding sites and motif occurrences, and somatic mutation frequencies at melanoma SMRs in KIAA0907 and (d) YAE1D1 promoter regions are shown at multiple scales (±1,000, ±75, and ±7 bp). Mutation frequency within each SMR (red) and at each position (purple bars) are shown. Motifs of ETS-family binding sites that overlap the SMRs are highlighted. (e) Luciferase reporter signal from wildtype (WT) and mutant (MT) promoters in three experiments performed in melanoma (A375) and HEK 293T cells with independent plasmid DNA preps (#1-2). For each experiment, three replicates were performed. Luciferase/renilla signals are shown, and are normalized by the mean WT signal per experiment. Two asterisks denotes P < 0.05 in two-sided t-tests; one asterisk denotes P < 0.1. Error bars indicate s.d. (f) Gene-structure, ENCODE CTCF and DNaseI signals, vertebrate conservation (phastCons 100way) at the 5′ UTR TBC1D12 bladder cancer SMR are shown at multiple scales. Start codon position is highlighted in green and Kozak sequence is underlined. (g) Relative protein and post-translational modification signals of wildtype (n=78) and mutant (TBC1D12.1 SMR-altered, n=14) bladder tumors. Central band, box boundaries, and whiskers correspond to the median, the interquartile range, and the highest/lowest points within 1.5× the interquartile range, respectively.
We discovered (4 and 5 bp) SMRs within open chromatin sites of the KIAA0907 and YAE1D1 promoters that were altered in 10.2% and 9.3% of WES melanomas (Fig. 2c,d), respectively. Somatic mutations in these SMRs were confirmed in WGS data of melanomas (n=1 for KIAA0907 and n=2 for YAE1D1 of n=17, respectively)3,20. Yet, these regions did not reach significance in a pan-cancer analysis20, highlighting cancer-specificity in non-coding alterations. In both SMRs, mutations alter core-recognition sequences within in vivo ETS factor binding sites (ENCODE), with varying effects on ETS primary sequence preferences. KIAA0907 encodes a putative RNA-binding protein. However, intronic sequences in this gene harbor SNORA42, an H/ACA class snoRNA with increased expression in lung and colorectal cancer35,36, suggesting promoter SMR alterations may enhance transcription at this locus. However, we observed no detectable changes in mutant KIAA0907 reporter gene expression (Fig. 2e). Whereas YAE1D1 promoter mutations reduce reporter gene expression (Fig. 2e), RNA-level overexpression of YAE1D1 has previously been observed in lower crypt-like colorectal cancer37, and a small cohort of melanoma samples showed increased YAE1D1 protein levels compared to untransformed melanocytes38.
In addition to SMRs that impact promoter regions, we observed 32 SMRs in 5′ and 3′ UTRs, including putative miRNA target sites39 Most strikingly, we discovered a 3 bp SMR in the 5′ UTR of TBC1D12 that is mutated in ∼15% of bladder cancers (Fig. 2f). Recurrent mutations were positioned near the start codon (Kozak region positions –1 and –3), suggesting a role in translational control. Mutations in this SMR were validated in whole-genome sequences of 7 cancer types, including 2 of 20 bladder cancers, 2 of 40 lung adenomas, and 3 of 172 breast cancers3,20. Bladder tumors with mutations in this SMR display altered RPS6KA1 (p90RSK) phosphorylation (P = 0.0005, t-test, Benjamini-Hochberg), a signal of increased cell-cycle proliferation40, and α-Tubulin (P = 4.3 × 10–5, t-test, Benjamini-Hochberg) levels, as determined by reverse-phase protein array (RPPA) assays41 (Fig. 2g, Online Methods). These results establish the utility of WES data for identifying recurrently mutated non-coding regions and our SMR identification method in pinpointing potentially functional non-coding alterations in cancer.
SMRs permit high-resolution analysis of coding alterations
As expected, most exome-derived SMRs lay within protein-coding regions. The identification of SMRs across multiple cancer types permitted a systematic analysis of differential mutation frequencies with sub-genic and cancer type resolution. Although many protein domains show high burdens of somatic mutation in multiple cancers, protein domains can show remarkable cancer type-specific burdens of mutation as exemplified by VHL in kidney clear-cell carcinoma and SET in diffuse large B-cell lymphoma (Fig. 3a).
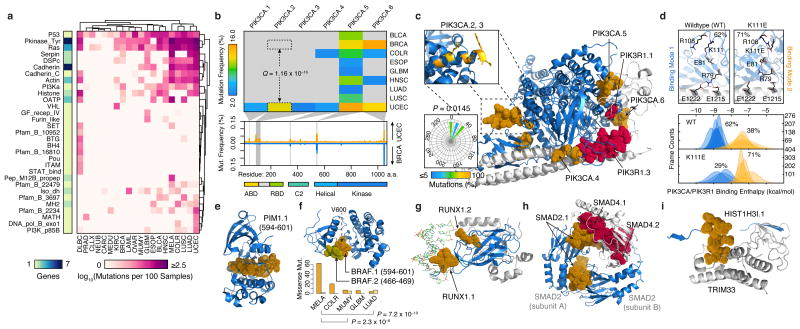
Figure 3.
Structural mapping of SMRs onto proteins and complexes reveals differentially-altered regions among cancers and molecular interfaces targeted by recurrent alterations. (a) Non-synonymous mutation frequency per PFAM protein domain, per cancer, per residue. Number of genes per domain is shown (left). (b) Mutation frequency matrix of PIK3CA SMRs across cancer types, and comparison of per residue mutation frequency of PIK3CA domains46 in endometrial (UCEC; orange) and breast cancer (BRCA; blue) samples. Gray bars indicate SMRs within PIK3CA. (c) Co-crystal structure of the PIK3CA (p110α; blue) and PIK3R1 (p85α; gray) interaction (PDB: 2RDO, 2IUG, 3HIZ). Residues within endometrial cancer SMRs on PIK3CA (orange) and PIK3R1 (red) are rendered as solvent-accessible surfaces. Insets display mutated residues within the PIK3CA.2, PIK3CA.3 SMR α-helix (yellow, top) and their corresponding side-chain dihedral angles (bottom). (d) Molecular dynamics simulations suggest PIK3CA–PIK3R1 binding is bimodal (bottom). Mutations within the PIK3CA.2, PIK3CA.3 SMR α-helix interfere with R79 binding contacts at the PIK3R1 interface, as shown in the wildtype and K111E mutant. Molecular structures of spatially-clustered (e) mutations (diffuse large B-cell lymphoma) and (f) SMRs (multiple myeloma), (g) a DNA (green) interface SMR, (h) reciprocal protein interface SMRs, and (i) a histone H3.1 SMR in the TRIM33 interface. Structural alignments and molecular visualizations prepared with PyMOL (Schrödinger). The relative proportions of BRAF.1 and BRAF.2 missense mutations per cancer type are shown in (f). PDB codes for (e-i) are 3CXW, 1UWH, 1H9D, 1U7V, and 3U5N, respectively.
Among genes (n=94) with multiple SMRs, we detected 48 SMRs that are differentially mutated between cancer types (Supplementary Table 8). A striking example of this differential targeting occurs within the catalytic subunit of the phosphoinositide 3-kinase, PIK3CA (p110α), a key oncogene implicated in a range of human cancers42,43. We detected six SMRs in PIK3CA across eight cancer types (Fig. 3b), with multiple cancer types displaying SMRs in the helical (PIK3CA.5) and kinase (PIK3CA.6) domains. In contrast, we observed cancer-specific SMRs (PIK3CA.2, PIK3CA.3) affecting an α-helical region between the adaptor binding domain (ABD) and linker domains of PIK3CA. Up to 14% of uterine corpus endometrial carcinomas harbor alterations in these intron-separated SMRs although these regions are not highly recurrently altered in other cancers. For example, we observed significant (Q = 1.2 × 10–16, proportions test) differences in PIK3CA.2 alteration frequencies in endometrial and breast cancers (Fig. 3b) and further validated these differences (P = 0.02, proportions test) in whole-genome sequences3,20. These findings indicate that previously described differences44 in total PIK3CA mutation frequencies between endometrial and breast cancers could in part be localized to this region.
Although the oncogenic effects of recurrent mutations in the ABD (PIK3CA.1), C2 (PIK3CA.4), helical (PIK3CA.5) and kinase (PIK3CA.6) domains of PIK3CA have been previously described, mutations in this ABD–RBD linker region are poorly understood45–48. Interestingly, missense mutations within this region are directionally orientated to one side of the α-helix (P = 0.0145, Rayleigh test), suggesting alterations to a molecular interface (Fig. 3c). Large-scale molecular dynamics simulations of PIK3CA–PIK3R1 indicate that PIK3CA.2 (K111E) and PIK3CA.3 (G118D) mutations can alter intermolecular salt bridge patterns at R79, which may result in a 1.8 kcal/mol loss of binding interactions compared to wildtype PIK3CA (Fig. 3d, Supplementary Fig. 9; Online Methods). Taken together, these results suggest a previously unrecognized mechanism of oncogenic alteration in PIK3CA.
To systematically characterize the location of alterations with respect to three-dimensional protein structures, we leveraged structural information from 428 SMR-associated and known cancer genes. We detected n=46 proteins with three-dimensional clustering of missense mutations (Supplementary Table 9), as exemplified by PIM1, an SMR-associated serine/threonine kinase proto-oncogene (Fig. 3e; Online Methods). This approach also identified three-dimensional clustering between BRAFV600 and BRAFP-loop SMRs (Fig. 3f), regions where mutations have been shown to function through distinct mechanisms49. Moreover, we found that BRAFV600 mutations are more frequent in melanoma and colorectal cancers, whereas BRAFP-loop mutations are more common in multiple myeloma and lung adenomas (P < 0.01, proportions test). In total, seven of 16 proteins with multiple SMRs displayed significant SMR three-dimensional clustering (Supplementary Table 10), which is consistent with frequent spatial coherence in pathogenic alterations.
We next sought to identify SMRs that might affect the molecular interfaces of protein-protein and DNA-protein interactions, a recognized yet understudied mechanism of cancer-driver mutations50–52. We examined intermolecular distances between SMR residues and interacting proteins or DNA and identified 17 SMRs that likely alter molecular interfaces (Table 1; Online Methods). These include 15 molecular interfaces of protein-protein and DNA-protein interactions with established cancer associations, such as the substrate-binding cleft of SPOP53 and DNA-binding interfaces on RUNX1 (Fig. 3g). We detected reciprocal SMRs at all electrostatic interfaces of the SMAD2–SMAD4 heterotrimer in colorectal cancer (Fig. 3h), as have been recently described54, and reciprocal SMRs at the regulatory PIK3CA–PIK3R1 interface in endometrial cancer (Fig. 3b). Together, these results highlight the robustness of SMRs in detecting validated driver alterations in molecular interfaces (Supplementary Fig. 10). In addition, SMRs pinpoint recurrent alterations at the interface between histone H3.1 (Fig. 3i) and TRIM33, an E3 ubiquitin ligase, and at the DNA-protein interface of histone H2B (Supplementary Fig. 11). These findings underscore and extend recent associations between altered epigenetic regulation and histone alterations in tumorigenesis55.
Table 1. Recurrently altered protein interfaces uncovered by SMRs.
| Protein (i) | Partner (j) | PDB | Chain (i) | Chain (j) | Region | Avg. Distance (Å) | Distance Ratio | Q-value | Status |
|---|---|---|---|---|---|---|---|---|---|
| VHL | TCEB1 | 3ZUN | I | H | chr3:10191469:10191513 | 7.259 | 0.395 | 7.62 × 10-10 | Known |
| VHL | TCEB2 | 1LQB | C | A | chr3:10191469:10191513 | 9.867 | 0.367 | 7.62 × 10-10 | Known |
| SPOP | H2AFY | 3HQH | A | M | chr17:47696421:47696467 | 7.962 | 0.462 | 3.72 × 10-8 | Known |
| SMAD2 | SMAD4 | 1U7V | A | C | chr18:45374881:45374945 | 9.231 | 0.460 | 5.61 × 10-8 | Known |
| HIST1H2BK | DNA | 2CV5 | D | J | chr6:27114446:27114519 | 9.730 | 0.520 | 3.27 × 10-7 | Novel |
| TP53 | TP53BP1 | 1KZY | B | D | chr17:7578369:7578556 | 13.253 | 0.556 | 5.13 × 10-7 | Known |
| SMAD4 | SMAD2 | 1U7V | B | C | chr18:48604665:48604797 | 11.878 | 0.694 | 5.13 × 10-7 | Known |
| DNMT3A | DNMT3L | 2QRV | E | F | chr2:25463271:25463308 | 10.112 | 0.380 | 5.13 × 10-7 | Known |
| SMAD4 | SMAD3 | 1U7F | B | C | chr18:48604665:48604797 | 11.883 | 0.700 | 1.94 × 10-6 | Known |
| PIK3CA | PIK3R1 | 3HHM | A | B | chr3:178936070:178936099 | 9.028 | 0.335 | 2.56 × 10-6 | Known |
| RUNX1 | DNA | 1H9D | C | H | chr21:36231782:36231792 | 8.957 | 0.351 | 0.001 | Known |
| HIST1H3I | TRIM33 | 3U5N | D | A | chr6:27839651:27840062 | 11.480 | 0.610 | 0.001 | Novel |
| HIST1H2BK | HIST1H4* | 2CV5 | D | F | chr6:27114446:27114519 | 13.680 | 0.664 | 0.002 | Novel |
| PPP2R1A | PPP2R5C | 2NPP | D | E | chr19:52716323:52716329 | 7.313 | 0.247 | 0.007 | Known |
| HRAS | RASA1 | 1WQ1 | R | G | chr11:534283:534291 | 5.302 | 0.350 | 0.007 | Known |
| PIK3R1 | PIK3CA | 3HIZ | B | A | chr5:67589138:67589149 | 6.713 | 0.567 | 0.008 | Known |
| NFE2L2 | KEAP1 | 2FLU | P | X | chr2:178098799:178098815 | 6.157 | 0.566 | 0.009 | Known |
| EGFR | EGF | 3NJP | B | A | chr7:55233035:55233043 | 8.763 | 0.386 | 0.019 | Known |
| FGFR2 | FGF8 | 2FDB | R | M | chr10:123279674:123279677 | 10.288 | 0.413 | 0.036 | Known |
| FBXW7 | SKP1 | 2OVR | B | C | chr4:153249384:153249385 | 9.352 | 0.346 | 0.036 | Known |
| FGFR2 | FGF2 | 1EV2 | H | A | chr10:123279674:123279677 | 11.685 | 0.406 | 0.037 | Known |
Indicates multiple components partner proteins identified. “Status” indicates whether the SMR-harboring protein (i) is a known or novel cancer-driver gene.
Molecular signatures highlight impact of SMR alterations
We sought to determine the potential functional impact of SMR alterations by their association with molecular signatures. We leveraged RNA-seq, reverse-phase protein array (RPPA), and clinical data to ask whether: (1) SMRs alterations associate with distinct molecular signatures or survival outcomes, (2) SMR alterations correlate with similar molecular profiles in distinct cancers, (3) same-gene SMR alterations associate with similar or different molecular signatures.
We found that mutations in SMRs were associated with diverse changes in RNA expression, signaling pathways, and patient survival (Fig. 4a, Supplementary Tables 11–14; Online Methods)56. These analyses revealed previously unappreciated connections between recurrent somatic mutations and molecular signatures, which highlight recurrent GSK3 pathway alterations in endometrial cancer and mTOR, EIF4 and EGF pathway alterations in glioblastoma (Supplementary Table 15). For example, synonymous point mutations in a bladder cancer SMR in sorting nexin 19 (SNX19) were associated with significant increases in protein expression levels of RAB25 (P = 2.5 × 10–27, t-test; Fig. 4b; Supplementary Table 12), a RAS family GTPase that promotes ovarian and breast cancer progression (57,58. These increases are consistent with RNA expression differences of RAB25 (P = 0.02; Wilcoxon rank sum test; Fig. 4c). Intriguingly, both SNX19 and RAB25 are implicated in intracellular trafficking, but the mechanism by which synonymous mutations in SNX19 correlate with RAB25 expression remains to be determined. In both SNX19 and NDUFA13, SMRs with clusters of synonymous mutation overlap open chromatin sites28 , suggesting potential regulatory impacts.
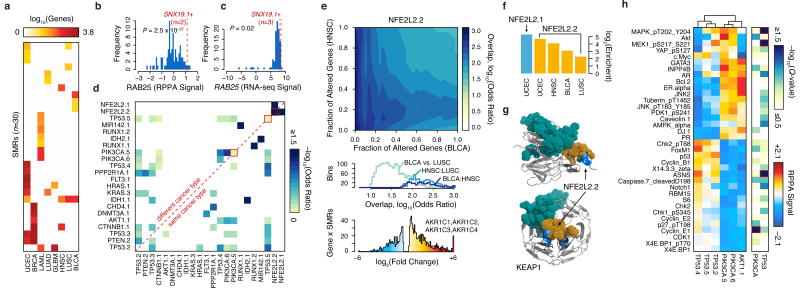
Figure 4.
SMRs are associated with distinct molecular signatures. (a) Matched RNA-seq data for nine cancers revealed that mutations in 30 distinct SMRs associated with ≥10 differentially expressed genes (FDR < 5%). (b) Normalized reverse phase protein array (RPPA) and (c) RNA-seq signals for RAB25 are plotted. Red lines indicate signals for samples with mutated SNX19 SMR. (d) Similarity between differentially expressed gene sets associated with mutations in each SMR pair. (e) Overlap between differentially expressed genes associated with altered NFE2L2.2 in bladder cancer (BLCA) and head and neck carcinoma (HNSC) is shown (top). Differentially expressed genes are sorted by p-value and similarity is quantified by Fisher's exact test odds ratio. The distribution of odds ratios of similarity is summarized for three comparisons (middle). Samples with NFE2L2.2 mutations exhibit highly increased expression of aldo-keto reductase enzymes (bottom). (f) The relative enrichment for oxidoreductase activity (GO:0016616) for specific cancer types (Supplementary Table 13). (g) Structure of SMR NFE2L2.2 (orange) in the KEAP1-binding domain (PDB: 3WN7). A sector of recurrent alterations on KEAP1 (teal) did not pass our 2% frequency cutoff. (h) Breast cancer patients were grouped by mutations in six SMRs in PIK3CA, AKT1, and TP53. Normalized RPPA-based expression was obtained from The Cancer Proteome Atlas (TCPA)41. The median RPPA signal for 36 markers and q-value (Kruskal-Wallis test) of differential expression between SMRs of TP53 or of PIK3CA are plotted (red highlights markers with significant intragenic differences, Q < 0.05).
We identified concordant changes in gene expression between SMR pairs, revealing potential functional relationships among 23 SMRs from 17 genes (Fig. 4d). These included multiple well-established mechanistic relationships, many of which were supported by RPPA measurements41, such as between PIK3CA and AKT1. Furthermore, this analysis revealed that mutations in the same SMR in different cancers can elicit similar molecular profiles in distinct cancers. For instance, we found that SMR alterations in the oncogenic transcription factor NFE2L259 were associated with large, concordant transcriptomic changes in four distinct cancer types (bladder, endometrial, lung squamous cell carcinoma, and head and neck cancer; Fig. 4e). The four genes with the highest increases in gene expression among endometrial cancer samples with alterations in NFE2L2.1 were the aldo-keto reductases AKR1C1-4 (Fig. 4e), which contribute to altered androgen metabolism and have been implicated in multiple cancer types60–62. Across all four cancer types, transcriptomic changes associated with NFE2L2 SMR alterations were highly enriched for oxidoreductases acting on the CH-OH group of donors, NAD or NADP as acceptors (4.9-39.0×, P ≤ 0.001, Benjamini-Hochberg, Fig. 4f). Mutations in KEAP1, a NFE2L2 binding partner, recapitulated the expression changes observed in patients with mutations in NFE2L2 SMRs (Fig. 4g; Supplementary Fig. 12; P < 0.01, Benjamini-Hochberg).
The identified SMRs also permitted interrogation of mutations in different regions of a given gene with respect to associated molecular signatures. For example in breast cancer, alterations in distinct SMRs within TP53 were associated with highly similar changes in protein-levels. Yet, we observed SMR-specific differences in ASNS levels and MAPK, MEK1 phosphorylation among TP53 SMR-altered samples (Fig. 4h, Q < 0.01). These results establish differences in the molecular signatures associated with same-gene SMR alterations and are consistent with pleiotropy in established oncogenes and tumor suppressors63,64.
The structure of cancer mutations remains largely unseen
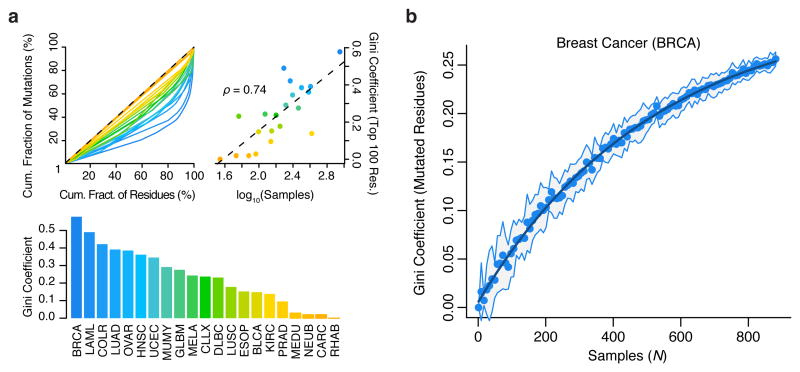
SMR analysis leverages structure in the distribution of somatic driver mutations to identify cancer-associated regions. We sought an alternative metric to assess the structure in the distribution of somatic coding mutations analyzed here by measuring the Gini coefficient of amino acid substitutions per residue in each cancer (Fig. 5a). Gini coefficients of dispersion were well-correlated with sample numbers (Spearman's ρ = 0.74). Subsampling demonstrates that even with sample numbers >850, a large proportion of the structure of protein-altering mutations in breast cancer remains unseen (Fig. 5b). These findings highlight the value of increasing cancer sample sizes in assessing the landscape of driver mutations.
Figure 5.
Structure in the distribution of cancer mutations remains largely uncharacterized. Gini coefficients of dispersion were calculated as the fraction of non-synonymous mutations contained per residue, across ∼19,000 proteins. (a) Lorenz curves (top-left), Gini-coefficients (top-right), and their correlation with tumor sample numbers (bottom) are shown. (b) Gini coefficients of non-synonymous mutation frequency in breast cancer as a function of (bootstrapped) sample size. Line of exponential fit is shown in dark blue. For comparisons between cancer types (a), the Gini coefficients were computed exclusively on the 100 most mutated residues per cancer.
Discussion
With few exceptions, studies of disease-associated variation have focused on identifying predefined functional units with recurrent alterations. This approach not only assumes accurate annotations but ignores the largely uncharacterized spectrum of functional elements that may be the targets of pathologic variants. Our approach avoids these limitations and complements existing gene-level and pathway-based strategies for discovering cancer-drivers by identifying variably-sized SMRs across 20 cancer types (Supplementary Table 16). SMR-associated genes include known cancer genes, such as PIM1 and MIR142 that were missed by gene-level analyses, as well as multiple novel genes with potential roles in cancer development.
Cancer SMRs target a diverse spectrum of functional elements in the genome, including single amino acids, complete coding exons and protein domains, miRNAs, 5′ UTRs, splice sites, and TF binding sites among others. This functional diversity underscores both the varied mechanisms of oncogenic misregulation and the advantage of functionally-agnostic detection approaches. Notably, several of the most frequently altered SMRs lay within non-coding regions. Strikingly, 17 out of 39 promoter and 5′ UTR melanoma SMRs overlap the core recognition sequences of in vivo ETS-family binding sites (Odds Ratio = 15.2, P = 1.5 × 10–11, Fisher's exact test). In addition, ∼15% of bladder cancer patients harbor 5′ UTR alterations in TBC1D12. Together, these results extend the support for non-coding drivers in cancer20,23,65 and establish the potential for discovering non-coding variation in WES.
The identification of SMRs provides a sub-genic, cancer-specific analysis of somatic mutations and associated molecular signatures. Cancer type differences in SMR mutation frequencies within BRAF, EGFR, and a mechanistically uncharacterized α-helix in PIK3CA demonstrate substructure in the distribution of somatic mutations between cancers, a property that may arise from pleiotropic functions. The close geometric proximity and directional uniformity of mutations in this helix suggest that PIK3CA.2 and PIK3CA.3 mutations function through similar mechanisms. Moreover, biophysical simulations indicate that mutations in both SMRs result in an elevated basal signaling activity of catalytic PIK3CA by way of weakened interactions with the regulatory PIK3R1 protein. These findings are concordant with recent biochemical evidence48. Consistent with pleiotropic dependencies, alterations to SMRs within a single gene can be associated with distinct molecular signatures, as exemplified by TP53 SMRs in breast cancers. Together, these results provide robust support for sub-genic functional targeting in distinct cancers and genes, and future efforts to examine SMR mutations in conjunction with clinical data in significantly larger patient cohorts may permit assessment of the prognostic value of SMRs.
SMR detection would benefit from further improvements of somatic mutation models. Here, we have applied cancer-specific models that take into account variation in somatic mutation rates throughout the genome. We controlled for mutational effects stemming from differences in replication timing and gene expression4,66. In addition, our models capture nucleotide-specific mutation probabilities3, account for strand-specificity67, leverage WGS mutation frequencies to limit effects from purifying selection on exons, and control mutation processes that may result in mutation clustering and tri-nucleotide mutation biases3. However, tumor-specific DNA repair defects3,66,68,69 and cell-type specific chromatin context70 also contribute to somatic mutation rates. Mutation models that account for cell-type specific expression and chromatin context at refined scales may require sequencing cohorts of matched normal tissue and increased sample sizes.
Although the sequencing of additional cancer genomes will further identification of novel cancer-driver genes5, characterizing the biochemical and cellular consequences of individual mutations is critical. We demonstrate that identifying the spatial distribution of mutation recurrence in the genome, when combined with additional genomic, biophysical, structural, or phenotypic information, often enhances mechanistic insights. Applying recently-developed high-throughput approaches71–73 to directly interrogate variation within SMRs may allow further understanding of the molecular mechanisms driving cancer and facilitate diagnostics and therapeutics development.
URLs
Data from Lawrence et al.5 was obtained from the TumorPortal through: http://cancergenome.broadinstitute.org/data/per_ttype_mafs/PanCan.maf; TCGA Data Portal, https://tcga-data.nci.nih.gov/tcga; UCSC cancer browser, http://genome-cancer.ucsc.edu.
Methods
Methods and any associated references are available in the online version of the paper.
Online Methods
Scientific computing was performed within Python74,75 and R environments. Data structure and genomic interval operations were performed with PANDAS76 and Pybedtools77, respectively. Statistical computing was performed with SciPy and NumPy78, and machine learning methods were implemented with SciKit Learn79. Structural and sequence alignment analyses were performed with BioPython80, PyMOL (Schrödinger) modules, and custom scripts. Reverse-Phase Protein Array (RPPA), RNA-seq, and survival analyses were performed in R and open-source packages (as described below).
Uniform Variant Annotation
3,185,590 uniformly-processed5, whole-exome sequencing (WES) somatic variant calls from 21 cancer types were downloaded from indicated URL. We applied snpEff29 to uniformly annotate n=3,078,482 (96.6%) single-nucleotide variant (SNV) calls from 4,735 tumors recording (GRCh37.66) mutation impact in protein-coding regions, transcribed regions (coding plus non-coding exons, introns, 5′ UTR, and 3′ UTR), and gene-associated regions (transcribed 5 kb upstream and 5 kb downstream) and standardize gene-name assignments. These procedures standardized gene-name assignments at multiple scales and removed gene assignments to “?” (n=64), “---” (n=130,728) in the original file. In addition, this procedure reduced variant calls unassigned to any genes (“Unknown”, n=1,239,475) to n=899,731 intergenic calls (>5 kb from annotated exons). This procedure was also applied to annotate n=11,461,951 whole-genome sequencing (WGS) somatic SNV calls from 23 cancer types3,20.
Mutation Probability Models
For each tumor type and gene, we calculated multiple distinct mutation probabilities. First, we calculated the frequency of transitions and transversions within the mappable, exonic regions of each gene to derive ‘Exonic’ mutation probabilities for each gene in the hg19 human genome assembly using WES data. Specifically, these probabilities indicate the fraction of mappable (100 bp), exonic reference bases (e.g. adenines) in each gene that were somatically mutated to a specific base (e.g. cytosine) per sample, in the cohort of tumor-specific WES data.
Because expression levels and replication timing have been shown to be major co-variates of somatic mutation probability in the genome, we sought to refine our mutation probability models for each gene using this information. For each gene, and in each tumor type, we identified the set of genes most similar in the expression, replication time, and GC-content (gene-level features). We used previously compiled4 expression and replication timing data and derived feature-specific weights defined as the rank correlation between gene features and the observed exonic mutation probabilities in each tumor type. We then converted gene features into their percentile ranks. Genes were sorted sequentially based on the gene feature weights and the neighborhood of the 500 closest genes were selected for each query gene. We then measured the sum of correlation-weighted, absolute feature distances between gene pairs within the 500 gene rank neighborhood. For each gene, we selected the ≤200 most similar genes with a normalized distance score ≤ 1. Lastly, we averaged the ‘Exonic’ mutation probability per transition/transversion to derive a set of ‘Matched’ mutation probabilities.
To avoid skewed mutation probabilities due to increased selection pressure on exons, we utilized a pan-cancer whole-genome sequencing (WGS)3,20 data in conjunction with cancer-specific WES data. We employed a Bayesian framework to derive posterior mutation probabilities for each transition and transversion per gene in each of the analyzed cancer types. Specifically, we modeled the likelihood of observing a mutation as a binomial distribution. We placed a prior Beta distribution on the mutation probability for each mutation type. The prior distribution was parameterized with parameters α = μ * ν and β = (1 – μ) * ν, where μ is the per base mutation probability in the WES data and ν is the number of exome sequencing samples in each cancer type. This parameterization enables the variance of the prior distribution to scale inversely with the sample size. We utilized the set of genes (≤200) that are matched to the analyzed gene as described above. We used all observed intronic WGS mutations in this cancer-specific matched set to calculate the posterior mutation probability for the analyzed gene. In this framework, the posterior distribution is also another Beta distribution. We then assigned the expected value of the posterior probability distribution as the estimate of the mutation probability for each transition/transversion (n=12). Finally, we calibrated the posterior mutation probabilities by the cancer-specific transition/transversion rates such that the median ‘Bayesian’ mutation probability is equal to the mean cancer-specific ‘Exonic’ mutation rate.
We computed a ‘Global’ mutation probability per tumor type as the average probability of transitions and transversions across all genes as observed in ‘Exonic’ mutation probabilities in each tumor type. The distributions of WES-derived (‘Exonic’, ‘Matched’, and ‘Global’) as well as WGS-derived (‘Bayesian’) mutation probabilities varied strongly between cancer types (Supplementary Fig. 2a) and among genes within individual cancer types, highlighting the importance of such cancer- and gene-specific treatment of background mutation probabilities3,4. Complementary mutation probabilities are well-correlated (Supplementary Fig. 2b). The ‘Bayesian’ and ‘Matched’ mutation probabilities are well-correlated among genes (Supplementary Fig. 2c), though ‘Bayesian’ mutation probabilities are better-correlated (Supplementary Fig. 2d) with the observed WGS intronic mutation densities. These ‘Bayesian’ (WGS-based) and ‘Matched’ (WES-based) mutation probabilities were used for the comparison presented in Fig. 1f.
Lastly, to account for tri-nucleotide biases3,4 in diverse mutation processes and cancer types, we computed ‘Trinucleotide’ mutation probability models for each tumor type. Specifically, ‘Trinucleotide’ mutation probabilities were calculated as the fraction of mappable (100 bp), exonic reference bases (e.g. adenines, A) within specific tri-nucleotide contexts (e.g. CAG) that were somatically mutated to a specific base (e.g. cytosine, CAG>CCG) per sample, in the cohort of tumor-specific WES data.
Mutation Domain Definition
We extended Ensembl (75) exonic regions by 0 bp and 1,000 bp and merged regions to define n=305,145 ‘concise’ (C) and n=191,669 ‘expanded’ (E) genomic domains in which mutation clusters were evaluated (see below). We identified the n=279,979 ‘concise’ and n=175,228 ‘expanded’ domains in which over ≥90% of positions are fully mappable with single-end 100 bp reads (ENCODE, UCSC Genome Browser). For each set of domains, we computed the number of possible genomic ranges (start, stop), which for the ‘expanded’ set amounts to 1,005,774,400,023 ranges (1012.0025). In addition, we removed ‘blacklisted’ regions of the human genome previously defined by the ENCODE project81.
Mutator Sample Identification
Samples harboring aberrantly high burdens of mutations in each tumor type were detected using median absolute deviation (MAD) outlier detection on the distribution of mutations (logn) per sample. As a threshold for consistency, mutator (outlier) samples were selected as those exceeding two standard deviations (s.d.).
Mutation Cluster Identification
We deployed density-based spatial clustering of applications with noise (DBSCAN) to detect clusters of ≥2 SNVs within exonic domains (above), evaluating density-reachability within ε base-pairs in each cancer type. The reachability parameter, ε, was dynamically defined with ε=dp/ds where dp and ds refer to the number of mutated positions (base-pairs) and the base-pair size of the domain d, thresholded to 10 ≤ ε ≤ 500 bp. In contrast to sliding window approaches or k-means spatial clustering, DBSCAN is not confined to evaluating predefined cluster sizes or numbers and tolerates noise in spatial density, whereby distal mutations are not assigned to clusters. Detected mutation clusters were refined where subclusters of ≥2 SNVs with significantly higher (P < 0.05, binomial test) mutation densities (mutated tumor samples per kb) existed.
Mutation Cluster Scoring
The significance of the observed mutation densities in each cluster was determined as Fisher's combined binomial probability of sampling the observed (k) or more mutations for each mutation type within the region. For each region, we computed the above density scores with the previously described ‘Exonic’, ‘Matched’, ‘Bayesian’, and ‘Global’ somatic mutation probabilities. As the primary density score (PDensity), we selected the most conservative of the ‘Bayesian’ and ‘Global’ density scores, max(PBayesian, PGlobal). Finally, we computed a tri-nucleotide mutation density (PTrinucleotide) score for each region using the ‘Trinucleotide’ somatic mutation probabilities.
Mutation Cluster Thresholding
We applied the procedures above to detect and evaluate mutation clusters in two sets of ‘concise’ (C) and ‘expanded’ (E) query domains (described in Mutation Domain Definition). 117,148/198,718 of the mutation clusters identified in E fall within the C query domains, respectively, indicating a 1.7× increase in clusters within the 1,000 bp-expanded domains.
Empirical false discovery rates (FDRs) were calculated from ten simulations performed by randomizing mutations within C domains in each tumor type, simulating a total of 30,784,820 mutations across cancer types. In each simulation, the positions of the observed mutations in each domain and tumor type, were randomized while maintaining reference base identity to retain the observed ‘Global’ mutation probabilities per transition and transversion (n=12). In each iteration, mutation cluster detection, refinement, and scoring procedures were repeated as above. For each simulation, we computed the density score (PDensity) threshold that guarantees a FDR ≤ 5%, whereby false and true discoveries are computed as the number clusters from simulated (randomized) and observed domain mutations, respectively. We excluded clusters with outlier density scores from the false discovery set if the clusters were associated with Cancer Gene Census (CGC) genes (n=522)31,32, as these regions would not represent false discoveries. For each tumor type, the expectation value (i.e. average) of FDR ≤ 5% simulation thresholds was defined as the final tumor-specific FDR threshold. To control FDRs to ≤5% in the E domains, where mutations cannot be randomized owing to the decreased certainty of WES coverage, we adjusted FDRs from C domains by the 1.7× increase in E/C clusters in each tumor type. ‘Expanded’ (E) domain 5% FDR thresholds per tumor type are provided in Supplementary Table 1.
To assess the robustness of the FDR cutoffs, we expanded the number of simulations to 90× and confirmed a 99.2% overlap (Jaccard index) in the 5% FDR-thresholded clusters (Supplementary Fig. 4e-g).
We reiterated mutation cluster FDR estimation and filtering using an alternate, conservative density score, PAlternate = max(PMatched, PGlobal), resulting in 714 regions. Fully 93.2% of these regions were identified as SMRs on the basis of the primary density scores (PDensity).
Mutation Cluster Filtering
As a final step in calling significantly mutated regions (SMRs), we selected clusters with density scores (PDensity) at the 5% FDR threshold and that were mutated in ≥2% of samples in each cancer type. Lastly, clusters associated with pseudogenes, olfactory receptors, and other repetitive gene-classes, were removed. This procedure resulted in 872 significantly mutated regions (SMRs), from 735 unique genomic regions, in 20 distinct cancer types.
Mutation Cluster Annotation
SMRs were annotated on the basis of mutation impacts on coding, transcribed, and gene-associated regions (see Uniform Variant Annotation). For SMRs associated with multiple genes (i.e. overlapping annotations), we preferentially assigned SMRs to (1) previously known cancer-driver genes (as defined by Lawrence et al. or the CGC), or (2) the gene impacted by the most severe type of mutation. Where mutation impact was insufficient to resolve multiple gene assignments, we selected the gene impacted by the largest number of mutations within the SMR. On this basis, we assign each SMR to a single gene, recording the types of mutation impacts on the gene, and the class of region affected. Region classes include: exon (coding region and non-coding gene), intron, splice, upstream, 5′ UTR, 3′ UTR, downstream, and other (intergenic). Mutation impacts (from snpEff) include in order of severity: rare amino acid, splice-site acceptor, splice-site donor, start lost, stop lost, stop gained, non-synonymous coding, splice-site branch U12, non-synonymous start, non-synonymous stop, splice-site region, splice-site branch, start gained, synonymous coding, synonymous start, synonymous stop,non-coding gene (“exon”), 3′ UTR, 5′ UTR, miRNA, intron, upstream, downstream, and intergenic.
Mutation Cluster Classification
SMRs were classified into ‘high-’, ‘medium-’, and ‘low-confidence’ sets as follows. First, SMRs in which alterations fall below the 2% mutation frequency threshold following mutator sample (as defined above) removal were labeled as mutator-driven SMRs. Among SMRs robust to mutator removal, those with FDR-corrected density scores significant at adjusted P < 0.05 following Bonferroni correction (PDensity ≤ 5.2 × 10–17) were classified as high-confidence. Mutator-driven SMRs were classified as low-confidence. SMRs that did not meet the high-confidence or low-confidence criteria were deemed medium-confidence.
To control for unaccounted mutation processes that could result in clusters of mutations with no selective advantage in cancer, we introduced the assumption that intronic mutations are primarily composed of passenger mutations and treated intronic clusters as false discoveries. For each cancer type, the distribution of density scores from intronic mutation clusters was modeled with Gaussian Kernel-Density Estimation (KDE) to derive p-value and q-value (FDR) estimates that limit the false discovery rate to ≤5%. This approach is limited to the ten cancer types with sufficient intronic mutation clusters to permit KD-estimates of their distribution of mutation density scores (Supplementary Fig. 5). A threshold of n ≥ 100 intronic mutation clusters was determined on the basis of the stability of FDR thresholds as determined by subsampling intronic mutation clusters in melanoma (data not shown). We applied this approach to control false discovery rates on two metrics: First, to account for unaccounted mutation clustering, we apply this approach on our expression-, replication timing-, and sequence (GC%) composition-controlled single-nucleotide probabilities (PDensity). Second, to account for biases in tri-nucleotide mutation frequencies in each cancer type, we apply this approach on tri-nucleotide density scores (PTrinucleotide). SMRs discovered in multiple cancer types and non-mutator-driven SMRs compliant with intron-based FDR ≤ 5% thresholds (PDensity, PTrinucleotide both) were classified as ‘robust’.
Mutation Cluster Labeling
SMRs with higher than expected APOBEC mutation signatures69 were labeled (Supplementary Fig. 6d, see Mutation Trinucleotide Analysis below). Finally, we annotated SMRs with respect to their 35 bp uniqueness and alignability with 50, 75, and 100 bp single-end reads. SMRs coordinates and corresponding annotations are provided in Supplementary Table 2.
Mutation Trinucleotide Analysis
We evaluated the frequency of trinucleotide sequence contexts as a subset of these (TCW) have been previously shown to differ significantly in mutation frequencies from other single-nucleotide contexts owing to APOBEC mutational processes69. Although APOBEC mutation signatures are identifiable in the data, our SMRs are depleted for such signatures (Supplementary Fig. 6a), suggesting the background models conservatively control for this mutation signature. Moreover, we extended these analyses to examine two important metrics:
unaccounted trinucleotide biases measured as the deviation in the observed trinucleotide mutation frequencies on the basis of single-nucleotide frequencies, and
fold change in frequencies of trinucleotide contexts in the SMR mutations compared to the input mutations
We observed a low correlation between the unaccounted-for trinucleotide biases and the fold change in trinucleotide contexts in diverse cancer types (Supplementary Fig. 6b), further supporting the conclusion that SMRs are not driven by unaccounted-for trinucleotide mutation signatures. These analyses are restricted to cancer types (n=6) that have ≥250 SMR mutation sites to prevent noise from cancer types with low numbers of SMR mutations. These cancer types encompass 79% of SMRs. Across cancer types, unaccounted-for trinucleotide frequencies account for only ∼7.9% of SMR sequences. For completeness, we have calculated within each SMR the fraction of mutations that are consistent with APOBEC signatures (Supplementary Fig. 6c). As shown in Supplementary Fig. 6d, only 4% of SMRs show higher than expected APOBEC mutation signatures following Holmes-Bonferroni corrected. Raw (uncorrected) p-values would indicate that 12% of SMRs have higher than expected APOBEC mutation signatures.
For additional methods describing (1) Transcription Factor Motif Enrichments, (2) Protein Structure Mapping, (3) Mutation Spatial Clustering, (4) Mutation Dihedral Angles, (5) Molecular Dynamics of PIK3CA/PIK3R1 Binding, (6) RNA-seq Analysis, (7) Reverse-Phase Protein Array (RPPA) Analysis, (8) Functional Enrichment Analysis, (9) Survival Analysis, (10) miRNA Target Site Analysis, and (11) Luciferase Assays, please see Supplementary Note.
Code Availability
The Python and R scripts to process the data and conduct the analyses described herein are available from the authors by request.
Supplementary Material
Acknowledgments
We thank the TCGA, ICGC, and TCPA for making these large-scale cancer datasets available to the scientific community. C.L.A. was supported by NIH grants 3U54DK10255602 and 1P50HG00773501. C.C. was supported by the Child Health Research Institute, the Lucile Packard Foundation for Children's Health and Stanford CTSA grant number UL1TR000093. J.A.R. was supported the Damon Runyon Cancer Research Foundation and NIH award 1U01HG007919-01. G.K. acknowledges support from the Lawrence Scholars Program, the NIH Simbios Program (U54 GM072970), and the Center for Molecular Analysis and Design at Stanford University. Biophysical simulations were supported by the Blue Waters project via National Science Foundation awards OCI-0725070 and ACI-1238993 and the state of Illinois. Further support was provided by the National Center for Multiscale Modeling of Biological Systems (P41GM103712-S1) through Anton-1 resources provided by the Pittsburgh Supercomputing Center under grant number PSCA13072P. This work was supported by the Rita Allen Foundation and NIH grant P50HG007735. We thank H. Tang for discussions regarding statistical analyses. We thank M. M. Winslow, D. M. Fowler, S. Fields, and D. E. Webster for critical reading and suggestions to the manuscript.
Footnotes
Author Contributions: C.L.A. and W.J.G. conceived of the project, and all authors designed experiments and methods. C.L.A. and C.C. developed methods for detection and analysis of significantly mutated regions (SMRs). C.L.A. constructed uniform annotations and non-Bayesian mutation probability models, and performed density-based clustering, scoring and empirical false discovery estimation (simulations), as well as regulatory (non-coding), structural (coding), frequency, and WGS recurrence analyses. C.C. constructed Bayesian mutation probability models, and performed RNA-seq, RPPA, and survival outcome analyses. J.A.R. designed and performed luciferase assays. G.K. carried out biophysical simulations, performed Hidden Markov Model based state decompositions, and computed binding enthalpies. C.L.A., C.C., J.A.R., G.K., M.P.S., and W.J.G. wrote the manuscript.
Competing Financial Interests: M.P.S. is co-founder and a member of the scientific advisory board (SAB) of Personalis, and member of the SAB of Genapsys and Axiomx. W.J.G. is a co-founder of Epinomics. A patent application has been filed by Stanford University with C.L.A., C.C., J.A.R., M.P.S., and W.J.G. named as inventors.
References
- 1.Hodis E, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang FW, et al. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339:957–959. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lawrence MS, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding L, Wendl MC, McMichael JF, Raphael BJ. Expanding the computational toolbox for mining cancer genomes. Nat Rev Genet. 2014;15:556–570. doi: 10.1038/nrg3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 8.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kane DP, Shcherbakova PV. A common cancer-associated DNA polymerase ε mutation causes an exceptionally strong mutator phenotype, indicating fidelity defects distinct from loss of proofreading. Cancer Res. 2014;74:1895–1901. doi: 10.1158/0008-5472.CAN-13-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dees ND, et al. MuSiC: identifying mutational significance in cancer genomes. Genome Res. 2012;22:1589–1598. doi: 10.1101/gr.134635.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamborero D, Gonzalez-Perez A, Lopez-Bigas N. OncodriveCLUST: exploiting the positional clustering of somatic mutations to identify cancer genes. Bioinformatics. 2013;29:2238–2244. doi: 10.1093/bioinformatics/btt395. [DOI] [PubMed] [Google Scholar]
- 12.Porta-Pardo E, Godzik A. e-Driver: a novel method to identify protein regions driving cancer Bioinformatics. 2014;30:3109–3114. doi: 10.1093/bioinformatics/btu499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schnall-Levin M, Zhao Y, Perrimon N, Berger B. Conserved microRNA targeting in Drosophila is as widespread in coding regions as in 3′UTRs. Proc Natl Acad Sci USA. 2010;107:15751–15756. doi: 10.1073/pnas.1006172107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cenik C, et al. Genome analysis reveals interplay between 5′UTR introns and nuclear mRNA export for secretory and mitochondrial genes. PLoS Genet. 2011;7:e1001366. doi: 10.1371/journal.pgen.1001366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stergachis AB, et al. Exonic transcription factor binding directs codon choice and affects protein evolution. Science. 2013;342:1367–1372. doi: 10.1126/science.1243490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolfe AL, et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature. 2014;513:65–70. doi: 10.1038/nature13485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiong HY, et al. RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease Science. 2015;347:1254806. doi: 10.1126/science.1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins. Nat Rev Genet. 2014;15:829–845. doi: 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.ENCODE Project Consortium et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weinhold N, Jacobsen A, Schultz N, Sander C, Lee W. Genome-wide analysis of noncoding regulatory mutations in cancer. Nat Genet. 2014;46:1160–1165. doi: 10.1038/ng.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fredriksson NJ, Ny L, Nilsson JA, Larsson E. Systematic analysis of noncoding somatic mutations and gene expression alterations across 14 tumor types. Nat Genet. 2014;46:1258–1263. doi: 10.1038/ng.3141. [DOI] [PubMed] [Google Scholar]
- 22.Supek F, Miñana B, Valcárcel J, Gabaldón T, Lehner B. Synonymous mutations frequently act as driver mutations in human cancers. Cell. 2014;156:1324–1335. doi: 10.1016/j.cell.2014.01.051. [DOI] [PubMed] [Google Scholar]
- 23.Melton C, Reuter JA, Spacek DV, Snyder M. Recurrent somatic mutations in regulatory regions of human cancer genomes. Nat Genet. 2015;47:710–716. doi: 10.1038/ng.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofree M, Shen JP, Carter H, Gross A, Ideker T. Network-based stratification of tumor mutations. Nat Methods. 2013;10:1108–1115. doi: 10.1038/nmeth.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leiserson MDM, et al. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat Genet. 2015;47:106–114. doi: 10.1038/ng.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Araya CL, et al. Regulatory analysis of the C. elegans genome with spatiotemporal resolution Nature. 2014;512:400–405. doi: 10.1038/nature13497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stergachis AB, et al. Conservation of trans-acting circuitry during mammalian regulatory evolution. Nature. 2014;515:365–370. doi: 10.1038/nature13972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin E, Kriegel HP, Jörg S, Xiaowei X. A density-based algorithm for discovering clusters in large spatial databases with noise. KDD. 1996 doi:10.1.1.71.1980. [Google Scholar]
- 31.Futreal AP, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santarius T, Shipley J, Brewer D, Stratton MR, Cooper CS. A census of amplified and overexpressed human cancer genes. Nat Rev Cancer. 2010;10:59–64. doi: 10.1038/nrc2771. [DOI] [PubMed] [Google Scholar]
- 33.Malhotra A, et al. Breakpoint profiling of 64 cancer genomes reveals numerous complex rearrangements spawned by homology-independent mechanisms. Genome Res. 2013;23:762–776. doi: 10.1101/gr.143677.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jäger D, et al. Identification of a tissue-specific putative transcription factor in breast tissue by serological screening of a breast cancer library. Cancer Res. 2001;61:2055–2061. [PubMed] [Google Scholar]
- 35.Mei YP, et al. Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis. Oncogene. 2012;31:2794–2804. doi: 10.1038/onc.2011.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okugawa Y, et al. Clinical significance of SNORA42 as an oncogene and a prognostic biomarker in colorectal cancer. Gut. 2015 doi: 10.1136/gutjnl-2015-309359. gutjnl–2015–309359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Budinska E, et al. Gene expression patterns unveil a new level of molecular heterogeneity in colorectal cancer. J Pathol. 2013;231:63–76. doi: 10.1002/path.4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uhlén M, et al. Proteomics. Tissue-based map of the human proteome Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 39.Vejnar CE, Zdobnov EM. MiRmap: comprehensive prediction of microRNA target repression strength. Nucleic Acids Res. 2012;40:11673–11683. doi: 10.1093/nar/gks901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lara R, Seckl MJ, Pardo OE. The p90 RSK family members: common functions and isoform specificity. Cancer Res. 2013;73:5301–5308. doi: 10.1158/0008-5472.CAN-12-4448. [DOI] [PubMed] [Google Scholar]
- 41.Li J, et al. TCPA: a resource for cancer functional proteomics data. Nat Methods. 2013;10:1046–1047. doi: 10.1038/nmeth.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Samuels Y, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 43.Thorpe LM, Yuzugullu H, Zhao JJ. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat Rev Cancer. 2014;15:7–24. doi: 10.1038/nrc3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cancer Genome Atlas Research Network et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miled N, et al. Mechanism of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science. 2007;317:239–242. doi: 10.1126/science.1135394. [DOI] [PubMed] [Google Scholar]
- 46.Huang CH, et al. The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations. Science. 2007;318:1744–1748. doi: 10.1126/science.1150799. [DOI] [PubMed] [Google Scholar]
- 47.Gkeka P, et al. Investigating the Structure and Dynamics of the PIK3CA Wild-Type and H1047R Oncogenic Mutant. PLoS Comput Biol. 2014;10:e1003895. doi: 10.1371/journal.pcbi.1003895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burke JE, Perisic O, Masson GR, Vadas O, Williams RL. Oncogenic mutations mimic and enhance dynamic events in the natural activation of phosphoinositide 3-kinase p110α (PIK3CA) Proc Natl Acad Sci USA. 2012;109:15259–15264. doi: 10.1073/pnas.1205508109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haling JR, et al. Structure of the BRAF-MEK complex reveals a kinase activity independent role for BRAF in MAPK signaling. Cancer Cell. 2014;26:402–413. doi: 10.1016/j.ccr.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 50.Kar G, Gursoy A, Keskin O. Human cancer protein-protein interaction network: a structural perspective. PLoS Comput Biol. 2009;5:e1000601. doi: 10.1371/journal.pcbi.1000601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ghersi D, Singh M. Interaction-based discovery of functionally important genes in cancers. Nucleic Acids Res. 2014;42:e18. doi: 10.1093/nar/gkt1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng F, et al. Studying tumorigenesis through network evolution and somatic mutational perturbations in the cancer interactome. Mol Biol Evol. 2014;31:2156–2169. doi: 10.1093/molbev/msu167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barbieri CE, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fleming NI, et al. SMAD2, SMAD3 and SMAD4 mutations in colorectal cancer. Cancer Res. 2013;73:725–735. doi: 10.1158/0008-5472.CAN-12-2706. [DOI] [PubMed] [Google Scholar]
- 55.Yuen BTK, Knoepfler PS. Histone H3.3 mutations: a variant path to cancer. Cancer Cell. 2013;24:567–574. doi: 10.1016/j.ccr.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hornbeck PV, et al. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–70. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cheng KW, et al. The RAB25 small GTPase determines aggressiveness of ovarian and breast cancers. Nat Med. 2004;10:1251–1256. doi: 10.1038/nm1125. [DOI] [PubMed] [Google Scholar]
- 58.Zhang J, et al. Overexpression of Rab25 contributes to metastasis of bladder cancer through induction of epithelial-mesenchymal transition and activation of Akt/GSK-3β/Snail signaling. Carcinogenesis. 2013;34:2401–2408. doi: 10.1093/carcin/bgt187. [DOI] [PubMed] [Google Scholar]
- 59.DeNicola GM, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ji Q, et al. Selective loss of AKR1C1 and AKR1C2 in breast cancer and their potential effect on progesterone signaling. Cancer Res. 2004;64:7610–7617. doi: 10.1158/0008-5472.CAN-04-1608. [DOI] [PubMed] [Google Scholar]
- 61.Stanbrough M, et al. Increased expression of genes converting adrenal androgens to testosterone in androgen-independent prostate cancer. Cancer Res. 2006;66:2815–2825. doi: 10.1158/0008-5472.CAN-05-4000. [DOI] [PubMed] [Google Scholar]
- 62.Riᜎner TL, Šmuc T, Rupreht R, Šinkovec J, Penning TM. AKR1C1 and AKR1C3 may determine progesterone and estrogen ratios in endometrial cancer. Mol Cell Endocrinol. 2006;248:126–135. doi: 10.1016/j.mce.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 63.Zhao L, Vogt PK. Helical domain and kinase domain mutations in p110α of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proceedings of the National Academy of Sciences. 2008;105:2652–2657. doi: 10.1073/pnas.0712169105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu X, et al. Activation of diverse signalling pathways by oncogenic PIK3CA mutations. Nat Commun. 2014;5:4961. doi: 10.1038/ncomms5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Puente XS, et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015;526:519–524. doi: 10.1038/nature14666. [DOI] [PubMed] [Google Scholar]
- 66.Supek F, Lehner B. Differential DNA mismatch repair underlies mutation rate variation across the human genome. Nature. 2015;521:81–84. doi: 10.1038/nature14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reijns MAM, et al. Lagging-strand replication shapes the mutational landscape of the genome. Nature. 2015;518:502–506. doi: 10.1038/nature14183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481:287–294. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]
- 69.Roberts SA, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Polak P, et al. Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature. 2015;518:360–364. doi: 10.1038/nature14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Araya CL, et al. A fundamental protein property, thermodynamic stability, revealed solely from large-scale measurements of protein function. Proceedings of the National Academy of Sciences. 2012;109:16858–16863. doi: 10.1073/pnas.1209751109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Buenrostro JD, et al. Quantitative analysis of RNA-protein interactions on a massively parallel array reveals biophysical and evolutionary landscapes. Nat Biotechnol. 2014;32:562–568. doi: 10.1038/nbt.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guenther UP, et al. Hidden specificity in an apparently nonspecific RNA-binding protein. Nature. 2013;502:385–388. doi: 10.1038/nature12543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Oliphant TE. Python for Scientific Computing. Computing in Science Engineering. 2007;9:10–20. [Google Scholar]
- 75.Millman KJ, Aivazis M. Python for Scientists and Engineers. Computing in Science Engineering. 2011;13:9–12. [Google Scholar]
- 76.McKinney W. Data Structures for Statistical Computing in Python. In: van der Walt S, Millman J, editors. Proceedings of the 9th Python in Science Conference. 2010. pp. 51–56. [Google Scholar]
- 77.Dale RK, Pedersen BS, Quinlan AR. Pybedtools: a flexible Python library for manipulating genomic datasets and annotations. Bioinformatics. 2011;27:3423–3424. doi: 10.1093/bioinformatics/btr539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Van der Walt S, Colbert SC, Varoquaux G. The NumPy Array: A Structure for Efficient Numerical Computation. Computing in Science Engineering. 2011;13:22–30. [Google Scholar]
- 79.Pedregosa F, et al. Scikit-learn: Machine Learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 80.Cock PJA, et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Boyle AP, et al. Comparative analysis of regulatory information and circuits across distant species. Nature. 2014;512:453–456. doi: 10.1038/nature13668. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.