Abstract
Major depressive disorder (MDD) is among the leading causes of worldwide disability. Despite its significant heritability, large-scale genome-wide association studies (GWASs) of MDD have yet to identify robustly associated common variants. Although increased sample sizes are being amassed for the next wave of GWAS, few studies have as yet focused on rare genetic variants in the study of MDD. We sequenced the exons of 1742 synaptic genes previously identified by proteomic experiments. PLINK/SEQ was used to perform single variant, gene burden and gene set analyses. The GeneMANIA interaction database was used to identify protein–protein interaction-based networks. Cases were selected from a familial collection of early-onset, recurrent depression and were compared with screened controls. After extensive quality control, we analyzed 259 cases with familial, early-onset MDD and 334 controls. The distribution of association test statistics for the single variant and gene burden analyses were consistent with the null hypothesis. However, analysis of prioritized gene sets showed a significant association with damaging singleton variants in a Cav2-adaptor gene set (odds ratio = 2.6; P = 0.0008) that survived correction for all gene sets and annotation categories tested (empirical P = 0.049). In addition, we also found statistically significant evidence for an enrichment of rare variants in a protein-based network of 14 genes involved in actin polymerization and dendritic spine formation (nominal P = 0.0031). In conclusion, we have identified a statistically significant gene set and gene network of rare variants that are over-represented in MDD, providing initial evidence that calcium signaling and dendrite regulation may be involved in the etiology of depression.
INTRODUCTION
Major depressive disorder (MDD) is a leading cause of suffering and morbidity worldwide.1 MDD is associated with an increased risk of mortality from both suicide and early morbid sequelae from common medical illnesses.2 Despite this overall burden, little is known about its pathophysiology. Broad risk factors such as family history and early adversity have been well established to increase the risk of MDD in adulthood; yet, how these risk factors give rise to the often dynamic phenotype of MDD remains unknown.3 Twin and molecular-based studies have found consistent evidence for a moderate heritable component, with heritability estimates typically ranging from 30% to 40%.4 However, genome-wide linkage studies have struggled with inconsistent replication3 and meta-analyses of large-scale genome-wide association studies (GWAS) have so far failed to identify findings with genome-wide significance.5 Together, these results suggest that a major variant of large effect is unlikely to exist and are consistent with the presence of prominent genetic heterogeneity.
Current GWAS identify rare variation in the form of large copy-number variants but do not, by design, assay rare single-nucleotide variants (SNVs), which form the bulk of genetic variation in individuals.6 Large-scale surveys of rare variation have recently been made possible by ‘next-generation’ sequencing studies, which have been applied to the study of a number of psychiatric disorders, including intellectual disability,7 autism,8 schizophrenia (SCZ)9,10 and bipolar disorder (BP).11 Although the precise genetic architecture may differ by phenotype, an emerging theme from these early sequencing efforts has been the presence of locus heterogeneity, suggesting the need for initial efforts to focus on gene sets and pathways, as these analyses provide greater power by collating larger numbers of rare variants.
Few studies have so far focused on high-throughput sequencing in MDD. To our knowledge, only one such study of MDD has been performed, but exome sequencing was carried out on only 10 subjects with an ‘extreme’ pharmacological phenotype.12 In the current study, we sought to perform a hypothesis-driven exploration of rare variants in genes found within the synapse, a complex organelle-like structure that has been implicated in the pathophysiology of major mental illness and is the target of almost all currently active psychotropic medications, including antidepressants.13 We leverage the results of recent proteomic studies that have comprehensively catalogued the constituent proteins of the synapse14 to perform high-throughput sequencing on a mutational target with a greater a priori likelihood of being involved in the pathophysiology of major mental illness, such as MDD.
MATERIALS AND METHODS
Sample
Cases were selected from the Genetics of Early-Onset, Recurrent Depression (GenRED) study, which previously ascertained a sample of European-American families with early-onset (age at onset <30 years) depression across six US sites.15 Informed consent was obtained from all cases and original Institutional Review Board's approval was provided by each of the recruiting sites, including Johns Hopkins (IRB approval number: NA_00035775). Cases were interviewed with the Diagnostic Instrument for Genetic Studies and underwent best-estimate diagnostic procedures. For the current study, we selected cases from independent families who had at least one first-degree relative also diagnosed with early-onset depression. These cases had an average age of onset of 18.9 years and had suffered a mean of 7.4 lifetime depressive episodes. Controls of European-American ancestry were selected from the NIMH Genetics Initiative, excluding subjects with a history of psychotic symptoms, BP or sufficient symptoms to meet a Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition, depressive episode criteria.
Synaptic gene target selection
At the onset of this study (2010), we performed an evaluation of all published or publically available proteomic studies focused on the synapse. A comprehensive list of these synaptic proteins was compiled into SynaptomeDB (http://metamoodics.org/SynaptomeDB).16 Our search strategy identified 1886 individual synaptic proteins, consisting of proteins found in synaptic vesicles (n = 107), the presynaptic membrane (n = 336), the presynaptic active zone (n = 209) and the postsynaptic density (n = 1,755).
Capture design
We used an Agilent Sure Select Custom Panel to design baits targeting the synaptome genes (n = 50 079 baits) based on the bait sequences of the Agilent Sure Select All Exome Version 2 Capture Kit (Agilent, Santa Clara, CA, USA). We added additional baits to provide further coverage of GC-rich genes (n = 5,251) and miRNA-binding sites (n = 973). In all, the capture targeted 6.7 Mb of genomic sequence.
High-throughput sequencing
We used the Illumina TruSeq DNA Sample Preparation Kit (Illumina, CA, USA) with 12 multiplexed adapters. Multiplexed DNA libraries were pooled at equal molar ratio before hybridization with the custom Agilent Sure Select probes. Following elution of captured sequence, we performed 6–8 cycles of PCR-based amplification. Samples passing standard quality-control (QC) metrics were sequenced on the Illumina HiSeq2000 (Illumina, CA, USA). Twelve multiplexed samples were sequenced per lane using 75 base pair-end read module.
Data processing and QC
The various QC steps are highlighted in Supplementary Figure S1. After separating the reads into individual samples based on the index barcode, reads were aligned to the human reference genome (UCSC hg19) using the BWA aligner,17 allowing for two mismatches in the 30-base seed. Picard (http://picard.sourceforge.net/) was used to fix mate pair mismatches and to remove reads with identical outer mapping coordinates, which represent likely PCR artifacts. Target coverage for the Agilent Sure Select capture was assessed using Picard’s HSmetrics utility. Genome Analysis Toolkit (GATK) was used to generate SNV and small indel calls within the targeted regions.18 We combined all cases and controls and performed multisample variant calling using GATK’s Unified Genotyper with variant recalibration. SNV clusters (defined as >3 SNVs per 10 bases) and SNVs falling within a called indel region were masked. We used variant quality score recalibration to generate a final list of high-confidence SNVs and indels as recommended by GATK documentation.19,20 We also added three additional QC steps to reduce the rate of false-positive calls: genotype quality >20; a depth-of-coverage of at least 10; and an average allele balance for heterozygous genotypes between 0.2 and 0.8.
Variant Call Format (VCF) files were converted to PLINK file format using VCF tools and custom scripts. PLINK was subsequently used to remove individuals with >50% missing genotype, followed by the removal of variants with a missing rate >10% and variants in Hardy–Weinberg Disequilibrium (P < 1 × 10−6). After these QC steps, the study-wide missing rate was 97.6% (Supplementary Table S1). Principal component analysis (PCA) of the case–control sample was performed using Eigenstrat21 to assess for potential population stratification using common sequenced variants (minor allele frequency (MAF) >5%) in approximate linkage equilibrium (Supplementary Figure S2). We inspected the top axes of variation in each PCA component and removed three outlier individuals, with the remaining samples showing appropriate clustering consistent with a European-American sample. After applying all the above QC steps to an initial sample of 308 cases and 353 controls, the final sample size was 259 cases and 334 controls.
Annotation
Identified variants were annotated with ANNOVAR22 using Ensembl version 63 as the reference database. We considered two functional categories of variants in the analyses described below: loss of function and damaging. We categorized loss-of-function variants as those that were stopgain, stoploss, canonical splicing or indel frameshift variants. Damaging variants included loss-of-function variants, in addition to the broader class of missense variants that were assessed as potentially damaging by either SIFT or Polyphen-2 using default thresholds (SIFT >0.95 or PolyPhen >0.85). We also considered three different frequency cutoff categories: singletons (or mac1), MAF <1% (maf01), and MAF <5% (maf05). We defined allelic thresholds based on estimates from the European-American and ALL populations of the NHLBI-ESP and the European-American and ALL populations of the 1000 Genomes April 2012 release.
Statistical analysis
We carried out a coordinated series of genetic association tests at the level of the individual variant(s), individual gene(s) and previously defined gene sets. For variant-level tests, we used logistic regression with Firth's penalized likelihood method,23 controlling for the top five ancestry-based principal components. Firth’s method provides unbiased estimates of the logistic regression model when there is sparse data,24,25 as is often the case with rare variants. For the gene-level tests, we used two complementary approaches. We used PLINK/SEQ to calculate gene burden with the SMP (statistic/matrix/permutation) utility as described in Purcell et al.,9 to control for any study-wide differences in the rates of variation across cases and controls. Significance of the gene burden results is evaluated with 10 000 permutations. We also used EPACTS (http://genome.sph.umich.edu/wiki/EPACTS) to calculate SKAT-O tests.26 The PLINK/SEQ burden test is a one-sided that assumes rare functional alleles increase risk for disease, while SKAT-O allows for both risk and protective alleles. We carried out these gene-level tests using the predefined functional and allele frequency categories.
Finally, we used two complementary approaches for gene set-level tests. First, we tested for a burden of risk variants in sets of genes that were based on prior evidence. We followed the approach of a recent SCZ exome sequencing study of SCZ,9 which defined a number of gene sets from the literature, including prior studies of GWAS, copy-number and de novo variants, as well as proteome-based studies of the postsynaptic density and the Cav2 calcium channel. Although the prior evidence implicating any specific type of genetic variation to MDD is much more limited, evidence of shared variation across multiple psychiatric phenotypes, including MDD and SCZ,27 led us to consider the same primary gene sets used in the SCZ exome sequencing study,9 with the added inclusion of two gene sets that include the top published findings from Psychiatric Genomics Consortium GWAS of MDD5 and BP28 (genes within 300 kb of associated loci with meta-analysis association P < 10−4). Further details, including gene set membership, are available in Supplementary Table S4. We tested only predefined gene sets with at least five genes sequenced in our study. To obtain a study-wide level of significance accounting for all gene sets tested and the different analyses carried out with these gene sets, we repeated the entire gene set testing procedure after randomly permuting case–control labels 1000 times.
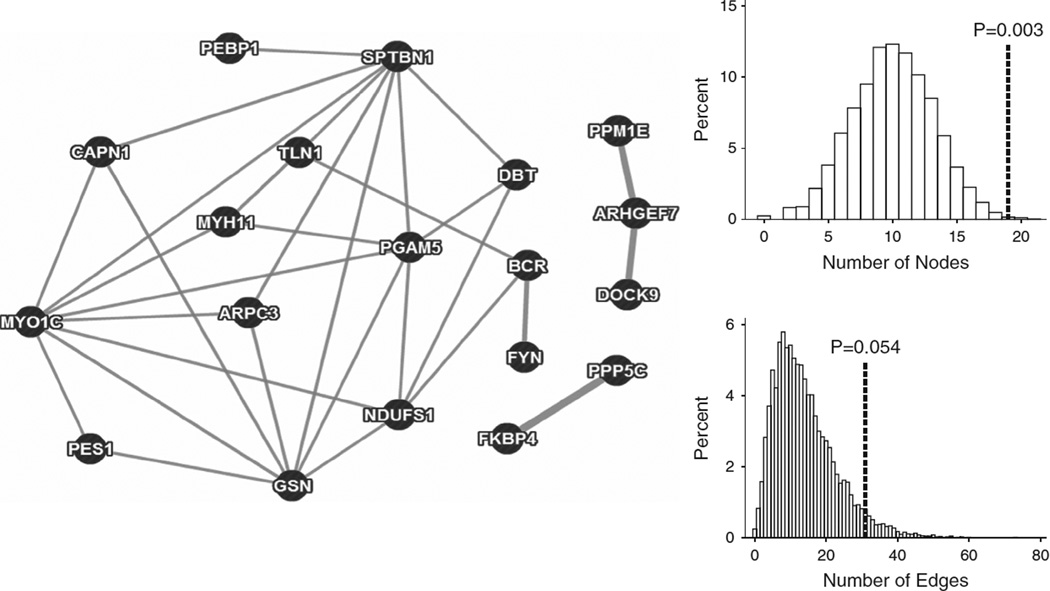
We also used a more agnostic gene set strategy by adapting an approach used in a previous study29 to examine whether genes with nominal evidence of association from our PLINK/SEQ-based gene-level test were more likely to interact with each other than expected by chance, thereby suggesting a common molecular pathway. We downloaded the direct protein–protein interaction data set from GeneMANIA30 and calculated the number of nodes and edges between the top associated genes in our observed data, where nodes represents proteins and edges represent interactions between any two proteins. To evaluate the significance of the observed network, we randomly selected an equal number of genes from the GeneMANIA database that were also sequenced in our study and were matched by gene size (±10 kb) with our observed genes. We repeated this 10 000 times to calculate the number of times that the number of nodes or edges in the randomly drawn genes was greater than what was found in the observed data.
RESULTS
After extensive QC (summarized in Supplementary Figure S1), the final cleaned data set included 259 cases with early-onset, recurrent major depression and 334 controls sequenced to an average depth of 50 ×, with 88.1% of bases being covered with at least 6 × reads. In total, there were 1742 genes with ≥70% of bases covered at 6 ×, which was used as a minimum coverage threshold for downstream analyses. Basic QC metrics are shown in Supplementary Table S1. Controls were sequenced to slightly higher depth, but there were otherwise minimal differences across QC metrics between cases and controls, including a comparable distribution of variants across annotation classes in cases and controls.
Single variant and gene-based analyses
Our results for the single-variant analyses were consistent with the null hypothesis (Supplementary Table S2 and Supplementary Figure S3). We subsequently performed gene-based tests using three frequency cutoffs (maf05, maf01 and singletons or mac1) and two functional categories representing a ‘damaging’ (defined by SIFT or Polyphen2) or ‘loss-of function’ effect. We implemented gene-based testing using PLINK/SEQ’s one-sided burden test or SKAT-O, a kernel-based test that is more sensitive to the presence of both protective and risk variants. The results, shown in Supplementary Table S3 and Supplementary Figure S4, show a number of genes with nominally significant enrichment for rare damaging variants, although no single gene-based association result survived correction for the full number of genes tested (best gene burden association P = 0.002).
Gene set and network analyses
Given the limited power of our sample to detect effects at the variant or gene level31, our primary focus was on the analysis of gene sets and gene networks, which could aggregate a sufficient number of rare variants for an enrichment analysis to reach statistical significance. Based on evidence suggesting that the genetic etiology of MDD partially overlaps with that of BP and SCZ,27 we used prior findings from these related psychiatric phenotypes, as well as two gene sets comprised of the top loci from the published Psychiatric Genomics Consortium GWAS meta-analysis of BP and MDD (genes within 300 kb of a locus with a meta-analysis association P < 10−4). The 41 prioritized gene sets (described in Supplementary Table S4) generally fall into four main classes: (1) statistically defined associations from prior studies of common and rare variation in SCZ, BP and MDD; (2) proteome-based gene sets of the postsynaptic density and its substructures; (3) proteome-based gene sets of the Cav2 calcium channel; and (4) annotation-based gene sets of the major ion channels. We tested for the enrichment of risk variants in gene sets that contained at least five genes in our post-QC sequenced sample, using the same three allele frequency (maf05, maf01 and mac1) and two functional annotation (damaging and loss of function) categories from the gene-level analyses. The top results are shown in Table 1, with the full results shown in Supplementary Table S5. The strongest association was in the calcium channel Cav2-channel ‘adaptor’ gene set, which showed an odds ratio (OR) = 2.6 (P = 0.0008) in the damaging singleton category and showed at least nominal evidence of association across the various annotation classes. This proteomically defined gene set consisted of 20 genes, including 11 that were sequenced in our study, of which 7 had over-representation of rare variants in cases compared with controls (STXBP5, RIMS1, CTNNB1, DMXL2, SYN1, YWHAB, YWHAH). This gene set remained significant after accounting for the multiple testing of the different frequency and annotation categories via permutation (empirical P = 0.049).
Table 1.
Top results from the prespecified gene set analysis showing all associations with a nominal P < 0.05
| Gene/set | Annotation/frequency category | Variants in affected/unaffected | OR | P-value | P-value (adjusted) |
|---|---|---|---|---|---|
| Cav2 adaptors | Damaging/mac1 | 22/11 | 2.58 | 0.0008 | 0.049 |
| Neuronal synaptic vesicle | Lof/maf05 | 10/3 | 4.30 | 0.0171 | 0.780 |
| Cav2 adaptors | Damaging/maf05 | 41/35 | 1.51 | 0.0174 | 0.785 |
| Neuronal synaptic vesicle | Lof/maf01 | 10/3 | 4.30 | 0.0187 | 0.801 |
| Neuronal PSD | Lof/mac1 | 32/32 | 1.29 | 0.0211 | 0.834 |
| Cav2 adaptors | Damaging/maf01 | 32/25 | 1.65 | 0.0221 | 0.843 |
| Neuronal synaptic vesicle | Lof/mac1 | 8/3 | 3.44 | 0.0224 | 0.848 |
| Neuronal early endosomes | Damaging/mac1 | 4/1 | 5.16 | 0.0240 | 0.866 |
| Neuronal plasma membrane | Damaging/mac1 | 20/11 | 2.34 | 0.0244 | 0.875 |
| Neuronal plasma membrane | Damaging/maf01 | 29/23 | 1.63 | 0.0279 | 0.907 |
| Neuronal plasma membrane | Damaging/maf05 | 38/34 | 1.44 | 0.0390 | 0.954 |
| Neuronal Golgi | Damaging/mac1 | 11/9 | 1.58 | 0.0421 | 0.966 |
| MDD GWAS (P < 1 × 10−4) | Lof/mac1 | 8/4 | 2.58 | 0.0495 | 0.979 |
| Neuronal ARC | Damaging/maf05 | 46/40 | 1.48 | 0.0499 | 0.981 |
Abbreviations: GWAS, genome-wide association studies; MDD, major depressive disorder; OR, odds ratio; PSD, postsynaptic density.
In an alternative network-based approach, we selected all genes with a PLINK/SEQ gene burden association P-value < 0.05 (n = 28 for maf05 and n = 16 for maf01) and tested whether these genes were more interconnected than expected by chance. We evaluated how many times these nominally significant genes were observed to interact in the curated protein–protein interaction database from GeneMANIA and compared this with 10 000 randomly drawn sets of an equal number of genes, which were sequenced in our study and matched by gene size (±10 kb). This analysis found an interconnected network of proteins under in maf05 frequency threshold (shown in Figure 1), which was statistically significant for the number of proteins found to interact with each other (nodes; P = 0.0031) and near significant for the number of pairwise interactions between these proteins (edges; P = 0.054). The analysis of the smaller list of nominally associated genes with maf01 also showed an excess number of nodes and edges, but these were not statistically significant, with empirical P-values of 0.053 and 0.11, respectively.
Figure 1.
Protein–protein interaction network identified using GeneMANIA (direct interaction database). The histograms showed the null distribution, with the line showing the analysis’ observed results and its empirical P-value.
DISCUSSION
MDD remains one of the few prominently heritable phenotypes that have yet to yield reliable genetic findings, suggesting the presence of extensive etiological and phenotypic heterogeneity. In this study, we have performed an initial study of rare variation in a large subset of genes with a priori relevance to MDD and have attempted to improve our chances of detecting a meaningful association by focusing on subjects with early-onset, recurrent MDD, which may be a more heritable form of the illness.15 Although our sample size was insufficient to identify a statistically significant rare variant burden at the single gene level, we were able to identify a significantly associated gene set and a gene network with enriched protein–protein interactions. Together, these findings provide initial evidence that calcium channels and dendrite regulation may be involved in the pathophysiology of depression.
We tested a number of prespecified gene sets with prior evidence for involvement in mental disorders such as SCZ and BP, reasoning that shared genetic risk across common variants between these disorders and MDD27 would also be seen for rare variants. Indeed, out of the 41 prioritized gene sets, only one gene set (damaging, singleton variants in the Cav2-adapter) showed sufficient evidence for association (OR = 2.6; P = 0.0008) to remain significant after a permutation procedure accounting for all the annotation and frequency classes tested (empirical P = 0.049). This gene set showed nominal evidence for association in both the damaging maf5 (OR = 1.51, P = 0.0174) and the damaging maf1 (OR = 1.65, P = 0.0221) categories, but its stronger evidence for association with the singleton category is consistent with findings from theoretical models32 and from studies of other neuropsychiatric disorders9,10 that point to rarer mutations being potentially more penetrant. The Cav2-adaptor gene set, as part of a broader set of Cav2-related genes, was initially reported in a proteomic analysis of proteins associated with the central Cav2 α1 subunit and its four β accessory subunits.33 The Cav2-adaptor gene set consists of a diverse group of genes encoding proteins involved in scaffolding, exocytosis and intracellular signaling. Among the genes contributing to the gene set association signal, there is a notable convergence on the process of synaptic vesicle exocytosis (RIMS1, STXBP5, CTNNB1, DMXL2, SYN1).34–38 Not unexpectedly, a number of rare variants within these genes have also been found in brain disorders, such as autism39,40 and epilepsy.41 RIMS1 is also notable for being one of the 108 recently identified genome-wide significant loci in SCZ.42
The complimentary protein–protein interaction analysis was driven by a network of 14 genes that included several of actin-based cytoskeleton genes (GSN, ARPC3, SPTBN1, CALP1, MYO1C). Gelsolin (GSN) is an actin-binding protein, which regulates actin polymerization by ‘severing’ actin polymers when activated by calcium signaling.43,44 ARPC3 encodes a critical subunit of the actin-related protein 2/3 complex, which is a regulator of actin polymerization and is involved in the formation of dendritic spines.45 SPTBN1, also known as βII-spectrin, is a scaffolding protein that binds the actin cytoskeleton to stabilize axonal projections46 and induce neurite formation.47 Intriguingly, βIIspectrin is also a substrate of calpain 1 (CALP1), which was also identified in the network analysis, and is a calcium-activated protease and potential mediator of corticotropin-releasing hormone-induced loss of dendritic spines.48 Finally, MYO1C, is a widely expressed myosin molecule with binding affinity for actin and the cell-signaling membrane inositide PIP2.49 Its role in the synapse remains unknown, although it has been found to have a role in axonal guidance50 and in mechanical amplification of electrical stimuli in vestibular hair cells.51
The genes identified in this network analysis converge on the role of actin polymerization and regulation of dendritic spines, which may underlie the key mechanisms responsible for synaptic plasticity.52 More broadly, abnormalities of size, density and shape of dendritic spines have been hypothesized to have a role in various brain disorders, including intellectual disability,53 autism,54 Alzheimer’s disease,55 SCZ and BP,56 as well as MDD.57 There is also increasing evidence to suggest that they may additionally be involved in the regulation of mood: lowered spine densities have been found in postmortem brains with two prototypical mood disorders (BP and MDD)56,57; in addition, chronic stress, long considered one of the most robust risk factors for mood disorders, has been associated with excessive pruning of dendritic spines;58 and finally, the antidepressant ‘effect’ of both traditional and novel pharmacological agents has been proposed to be mediated by increased formation of dendritic spines in the prefrontal cortex.58,59
As one of the first attempts to investigate the role of rare variants in MDD, our study should be seen in the context of a number of limitations. First, the sample size is modest for genetic studies of rare variation, leading us to focus the primary analyses on gene sets and pathways, as associations in these aggregate ‘analysis units’ are more likely to be detected in smaller sample sizes.9 Power analyses (Supplementary Figure S5a) based on this study’s sample size revealed sufficient power (>80%) to detect gene burden results with ORs > 2.5 under cumulative burden frequencies of approximately 10%. Yet, in scenarios where the burden frequencies were <5%, this study only had sufficient power to detect gene burden results with relatively high effect sizes (OR > 3). Power calculation of the gene set analyses, however, showed significantly higher power to detect associations with gene sets with OR > 2.0 given the gene set cumulative frequencies that were seen in our study (Supplementary Figure S5b). Nevertheless, even in the gene set analyses, our study sample was significantly underpowered to detect associations with ORs < 2.0. A second important limitation is that, although we surveyed 1742 genes hypothesized to have greater a priori likelihood of being associated with MDD, an exome or whole genome approach would have offered a more comprehensive overview of genes potentially associated with MDD, albeit one with a greater burden of multiple testing and, therefore, a need for larger sample sizes. Third, our sequencing yielded a ‘medium’ level of coverage, which may have missed or miscalled particularly rare variants, such as singletons. To allay these concerns, we applied stringent QC procedures and used the Integrative Genomics Viewer60 to individually visualize all the variants in the significant gene set and in protein–protein interaction network. In addition, we performed Sanger sequencing and successfully validated all variants found in cases within the ‘singleton Cav2-adaptor’ gene set category for which we had additional DNA (20 out of 22 variants). Fourth, our sequencing study was likely limited by what has been termed the ‘annotation gap,’ where the known and unknown imprecision of bioinformatics annotation may have led to the inclusion of false positives and false negatives in our analysis of ‘damaging’ variants.61 As sequencing becomes more mainstream, functional annotation will likely become increasingly accurate; however, for the time being, we have, by necessity, resorted to the most widely used tools. Finally, our sequencing strategy has focused only on the more ‘interpretable’ exonic variations, with no systematic sequencing of non-coding regions, which represent an even more significant annotation challenge.
In conclusion, this study suggests that the evaluation of rare variants may be a promising endeavor even for phenotypes as heterogeneous as depression. Limited success in GWAS may not imply lack of success in rare variant studies, as highlighted by the successful identification of rare, but not yet common, variants in autism.62 Although our findings need to be replicated in independent samples, the presence of a statistically significant gene set and network analysis suggests that focusing on rare variants may be a tractable way to elucidate at least part of the genetic underpinnings of MDD.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the Johns Hopkins University Brain Science Institute (TW, JBP, FSG). Data and biomaterials were collected in six projects that participated in the NIMH Genetics of Recurrent Early-Onset Depression (GenRED) project. From 1999 to 2003, the principal investigators and co-investigators were New York State Psychiatric Institute, New York, R01-MH060912 (Myrna Weissman, PhD and James K Knowles, MD, PhD); University of Pittsburgh, R01-MH060866 (George S Zubenko, MD, PhD, and Wendy N Zubenko, EdD, RN, CS); Johns Hopkins University, Baltimore, R01-MH059552 (J Raymond DePaulo, MD, Melvin McInnis, MD and Dean MacKinnon, MD); University of Pennsylvania, Philadelphia, R01-MH61686 (Doug Levinson, MD [GenRED coordinator], Madeleine M Gladis, PhD, Kathleen Murphy-Eberenz, PhD and Peter Holmans, PhD [University of Wales College of Medicine]); University of Iowa, Iowa City, R01-MH059542 (Raymond Crowe, MD and William H Coryell, MD); Rush University Medical Center, Chicago, R01-MH059541-05 (William Scheftner, MD, Rush-Presbyterian).
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
REFERENCES
- 1.Whiteford HA, Degenhardt L, Rehm J, Baxter AJ, Ferrari AJ, Erskine HE, et al. Global burden of disease attributable to mental and substance use disorders: Findings from the global burden of disease study 2010. Lancet. 2013;382:1575–1586. doi: 10.1016/S0140-6736(13)61611-6. [DOI] [PubMed] [Google Scholar]
- 2.Ferrari AJ, Charlson FJ, Norman RE, Patten SB, Freedman G, Murray CJ, et al. Burden of depressive disorders by country, sex, age, and year: Findings from the global burden of disease study 2010. PLoS Med. 2013;10:e1001547. doi: 10.1371/journal.pmed.1001547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flint J, Kendler KS. The genetics of major depression. Neuron. 2014;81:484–503. doi: 10.1016/j.neuron.2014.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sullivan PF, Neale MC, Kendler KS. Genetic epidemiology of major depression: review and meta-analysis. Am J Psychiatry. 2000;157:1552–1562. doi: 10.1176/appi.ajp.157.10.1552. [DOI] [PubMed] [Google Scholar]
- 5.Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium. Ripke S, Wray NR, Lewis CM, Hamilton SP, Weissman MM, et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry. 2013;18:497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pritchard JK. Are rare variants responsible for susceptibility to complex diseases? Am J Hum Genet. 2001;69:124–137. doi: 10.1086/321272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilissen C, Hehir-Kwa JY, Thung DT, van de Vorst M, van Bon BW, Willemsen MH, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–347. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
- 8.Krumm N, O'Roak BJ, Shendure J, Eichler EE. A de novo convergence of autism genetics and molecular neuroscience. Trends Neurosci. 2014;37:95–105. doi: 10.1016/j.tins.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Georgi B, Craig D, Kember RL, Liu W, Lindquist I, Nasser S, et al. Genomic view of bipolar disorder revealed by whole genome sequencing in a genetic isolate. PLoS Genet. 2014;10:e1004229. doi: 10.1371/journal.pgen.1004229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tammiste A, Jiang T, Fischer K, Magi R, Krjutskov K, Pettai K, et al. Whole-exome sequencing identifies a polymorphism in the BMP5 gene associated with SSRI treatment response in major depression. J Psychopharmacol. 2013;27:915–920. doi: 10.1177/0269881113499829. [DOI] [PubMed] [Google Scholar]
- 13.Grant SG. Synaptopathies: diseases of the synaptome. Curr Opin Neurobiol. 2012;22:522–529. doi: 10.1016/j.conb.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 14.Nithianantharajah J, Komiyama NH, McKechanie A, Johnstone M, Blackwood DH, St Clair D, et al. Synaptic scaffold evolution generated components of vertebrate cognitive complexity. Nat Neurosci. 2013;16:16–24. doi: 10.1038/nn.3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levinson DF, Zubenko GS, Crowe RR, DePaulo RJ, Scheftner WS, Weissman MM, et al. Genetics of recurrent early-onset depression (GenRED): Design and preliminary clinical characteristics of a repository sample for genetic linkage studies. Am J Med Genet B Neuropsychiatr Genet. 2003;119:118–130. doi: 10.1002/ajmg.b.20009. [DOI] [PubMed] [Google Scholar]
- 16.Pirooznia M, Wang T, Avramopoulos D, Valle D, Thomas G, Huganir RL, et al. SynaptomeDB: an ontology-based knowledgebase for synaptic genes. Bioinformatics. 2012;28:897–899. doi: 10.1093/bioinformatics/bts040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pirooznia M, Kramer M, Parla J, Goes FS, Potash JB, McCombie WR, et al. Validation and assessment of variant calling pipelines for next-generation sequencing. Hum Genomics. 2014;8 doi: 10.1186/1479-7364-8-14. 7364–8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 22.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bull SB, Lewinger JP, Lee SS. Confidence intervals for multinomial logistic regression in sparse data. Stat Med. 2007;26:903–918. doi: 10.1002/sim.2518. [DOI] [PubMed] [Google Scholar]
- 24.Heinze G, Schemper M. A solution to the problem of separation in logistic regression. Stat Med. 2002;21:2409–2419. doi: 10.1002/sim.1047. [DOI] [PubMed] [Google Scholar]
- 25.Heinze G. A comparative investigation of methods for logistic regression with separated or nearly separated data. Stat Med. 2006;25:4216–4226. doi: 10.1002/sim.2687. [DOI] [PubMed] [Google Scholar]
- 26.Lee S, Wu MC, Lin X. Optimal tests for rare variant effects in sequencing association studies. Biostatistics. 2012;13:762–775. doi: 10.1093/biostatistics/kxs014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cross-Disorder Group of the Psychiatric Genomics Consortium. Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Psychiatric GWAS Consortium Bipolar Disorder Working Group. Sklar P, Ripke S, Scott LJ, Andreassen OA, Cichon S, et al. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43:977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gulsuner S, Walsh T, Watts AC, Lee MK, Thornton AM, Casadei S, et al. Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell. 2013;154:518–529. doi: 10.1016/j.cell.2013.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zuberi K, Franz M, Rodriguez H, Montojo J, Lopes CT, Bader GD, et al. GeneMANIA prediction server 2013 update. Nucleic Acids Res. 2013;41:W115–W122. doi: 10.1093/nar/gkt533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zuk O, Schaffner SF, Samocha K, Do R, Hechter E, Kathiresan S, et al. Searching for missing heritability: designing rare variant association studies. Proc Natl Acad Sci USA. 2014;111:E455–E464. doi: 10.1073/pnas.1322563111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agarwala V, Flannick J, Sunyaev S, GoT2D Consortium. Altshuler D. Evaluating empirical bounds on complex disease genetic architecture. Nat Genet. 2013;45:1418–1427. doi: 10.1038/ng.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muller CS, Haupt A, Bildl W, Schindler J, Knaus HG, Meissner M, et al. Quantitative proteomics of the Cav2 channel nano-environments in the mammalian brain. Proc Natl Acad Sci USA. 2010;107:14950–14957. doi: 10.1073/pnas.1005940107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaeser PS, Deng L, Fan M, Sudhof TC. RIM genes differentially contribute to organizing presynaptic release sites. Proc Natl Acad Sci USA. 2012;109:11830–11835. doi: 10.1073/pnas.1209318109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fujita Y, Shirataki H, Sakisaka T, Asakura T, Ohya T, Kotani H, et al. Tomosyn: a syntaxin-1-binding protein that forms a novel complex in the neurotransmitter release process. Neuron. 1998;20:905–915. doi: 10.1016/s0896-6273(00)80472-9. [DOI] [PubMed] [Google Scholar]
- 36.Taylor AM, Wu J, Tai HC, Schuman EM. Axonal translation of beta-catenin regulates synaptic vesicle dynamics. J Neurosci. 2013;33:5584–5589. doi: 10.1523/JNEUROSCI.2944-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagano F, Kawabe H, Nakanishi H, Shinohara M, Deguchi-Tawarada M, Takeuchi M, et al. Rabconnectin-3, a novel protein that binds both GDP/GTP exchange protein and GTPase-activating protein for Rab3 small G protein family. J Biol Chem. 2002;277:9629–9632. doi: 10.1074/jbc.C100730200. [DOI] [PubMed] [Google Scholar]
- 38.Sudhof TC, Czernik AJ, Kao HT, Takei K, Johnston PA, Horiuchi A, et al. Synapsins: mosaics of shared and individual domains in a family of synaptic vesicle phosphoproteins. Science. 1989;245:1474–1480. doi: 10.1126/science.2506642. [DOI] [PubMed] [Google Scholar]
- 39.Cukier HN, Dueker ND, Slifer SH, Lee JM, Whitehead PL, Lalanne E, et al. Exome sequencing of extended families with autism reveals genes shared across neurodevelopmental and neuropsychiatric disorders. Mol Autism. 2014;5 doi: 10.1186/2040-2392-5-1. 2392–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dong S, Walker MF, Carriero NJ, DiCola M, Willsey AJ, Ye AY, et al. De novo insertions and deletions of predominantly paternal origin are associated with autism spectrum disorder. Cell Rep. 2014;9:16–23. doi: 10.1016/j.celrep.2014.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lignani G, Raimondi A, Ferrea E, Rocchi A, Paonessa F, Cesca F, et al. Epileptogenic Q555X SYN1 mutant triggers imbalances in release dynamics and short-term plasticity. Hum Mol Genet. 2013;22:2186–2199. doi: 10.1093/hmg/ddt071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Janmey PA, Stossel TP. Modulation of gelsolin function by phosphatidylinositol 4,5-bisphosphate. Nature. 1987;325:362–364. doi: 10.1038/325362a0. [DOI] [PubMed] [Google Scholar]
- 44.Lin KM, Wenegieme E, Lu PJ, Chen CS, Yin HL. Gelsolin binding to phosphatidylinositol 4,5-bisphosphate is modulated by calcium and pH. J Biol Chem. 1997;272:20443–20450. doi: 10.1074/jbc.272.33.20443. [DOI] [PubMed] [Google Scholar]
- 45.Kim IH, Racz B, Wang H, Burianek L, Weinberg R, Yasuda R, et al. Disruption of Arp2/3 results in asymmetric structural plasticity of dendritic spines and progressive synaptic and behavioral abnormalities. J Neurosci. 2013;33:6081–6092. doi: 10.1523/JNEUROSCI.0035-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu K, Zhong G, Zhuang X. Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science. 2013;339:452–456. doi: 10.1126/science.1232251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee HJ, Lee K, Im H. Alpha-synuclein modulates neurite outgrowth by interacting with SPTBN1. Biochem Biophys Res Commun. 2012;424:497–502. doi: 10.1016/j.bbrc.2012.06.143. [DOI] [PubMed] [Google Scholar]
- 48.Andres AL, Regev L, Phi L, Seese RR, Chen Y, Gall CM, et al. NMDA receptor activation and calpain contribute to disruption of dendritic spines by the stress neuropeptide CRH. J Neurosci. 2013;33:16945–16960. doi: 10.1523/JNEUROSCI.1445-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hokanson DE, Laakso JM, Lin T, Sept D, Ostap EM. Myo1c binds phosphoinositides through a putative pleckstrin homology domain. Mol Biol Cell. 2006;17:4856–4865. doi: 10.1091/mbc.E06-05-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang FS, Liu CW, Diefenbach TJ, Jay DG. Modeling the role of myosin 1c in neuronal growth cone turning. Biophys J. 2003;85:3319–3328. doi: 10.1016/S0006-3495(03)74751-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stauffer EA, Scarborough JD, Hirono M, Miller ED, Shah K, Mercer JA, et al. Fast adaptation in vestibular hair cells requires myosin-1c activity. Neuron. 2005;47:541–553. doi: 10.1016/j.neuron.2005.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bellot A, Guivernau B, Tajes M, Bosch-Morato M, Valls-Comamala V, Munoz FJ. The structure and function of actin cytoskeleton in mature glutamatergic dendritic spines. Brain Res. 2014;1573:1–16. doi: 10.1016/j.brainres.2014.05.024. [DOI] [PubMed] [Google Scholar]
- 53.Clement JP, Aceti M, Creson TK, Ozkan ED, Shi Y, Reish NJ, et al. Pathogenic SYNGAP1 mutations impair cognitive development by disrupting maturation of dendritic spine synapses. Cell. 2012;151:709–723. doi: 10.1016/j.cell.2012.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang G, Gudsnuk K, Kuo SH, Cotrina ML, Rosoklija G, Sosunov A, et al. Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron. 2014;83:1131–1143. doi: 10.1016/j.neuron.2014.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wei W, Nguyen LN, Kessels HW, Hagiwara H, Sisodia S, Malinow R. Amyloid beta from axons and dendrites reduces local spine number and plasticity. Nat Neurosci. 2010;13:190–196. doi: 10.1038/nn.2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Konopaske GT, Lange N, Coyle JT, Benes FM. Prefrontal cortical dendritic spine pathology in schizophrenia and bipolar disorder. JAMA Psychiatry. 2014;71:1323–1331. doi: 10.1001/jamapsychiatry.2014.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kang HJ, Voleti B, Hajszan T, Rajkowska G, Stockmeier CA, Licznerski P, et al. Decreased expression of synapse-related genes and loss of synapses in major depressive disorder. Nat Med. 2012;18:1413–1417. doi: 10.1038/nm.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Licznerski P, Duman RS. Remodeling of axo-spinous synapses in the pathophysiology and treatment of depression. Neuroscience. 2013;251:33–50. doi: 10.1016/j.neuroscience.2012.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li N, Lee B, Liu RJ, Banasr M, Dwyer JM, Iwata M, et al. mTOR-dependent synapse formation underlies the rapid antidepressant effects of NMDA antagonists. Science. 2010;329:959–964. doi: 10.1126/science.1190287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McCarthy DJ, Humburg P, Kanapin A, Rivas MA, Gaulton K, Cazier JB, et al. Choice of transcripts and software has a large effect on variant annotation. Genome Med. 2014;6:26. doi: 10.1186/gm543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Berg JM, Geschwind DH. Autism genetics: searching for specificity and convergence. Genome Biol. 2012;13:247. doi: 10.1186/gb-2012-13-7-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.