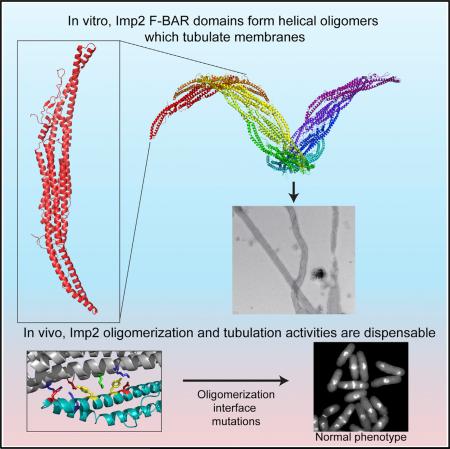
SUMMARY
F-BAR proteins link cellular membranes to the actin cytoskeleton in many biological processes. Here we investigated the function of the Schizosaccharomyces pombe Imp2 F-BAR domain in cytokinesis and find that it is critical for Imp2's role in contractile ring constriction and disassembly. To understand mechanistically how the F-BAR domain functions, we determined its structure, elucidated how it interacts with membranes, and identified an interaction between dimers that allows helical oligomerization and membrane tubulation. Using mutations that block either membrane binding or tubulation, we find that membrane binding is required for Imp2's cytokinetic function but that oligomerization and tubulation, activities often deemed central to F-BAR protein function, are dispensable. Accordingly, F-BARs that do not have the capacity to tubulate membranes functionally substitute for the Imp2 F-BAR, establishing that its major role is as a cell-cycle-regulated bridge between the membrane and Imp2 protein partners, rather than as a driver of membrane curvature.
Graphical Abstract
INTRODUCTION
The Fer/CIP4 Homology-Bin/Amphiphysin/Rvs (F-BAR) protein family is characterized by the presence of an N-terminal membrane-binding F-BAR domain and conserved roles in linking cellular membranes to the actin cytoskeleton (Roberts-Galbraith and Gould, 2010). F-BAR family proteins are found throughout eukaryotes, with 22 members in humans, 4 in Saccharomyces cerevisiae, and 7 in Schizosaccharomyces pombe. They are involved in a diverse array of actin-driven cellular processes including endocytosis, motility, and cytokinesis (Frost et al., 2009).
Even within a single biological process, such as mammalian endocytosis, several different F-BAR proteins collaborate (Qualmann et al., 2011). Studies of the relative timing of recruitment to endocytic sites indicate that multiple F-BARs assemble in a defined order (Taylor et al., 2011). One hypothesis for this observation has been that each F-BAR protein senses or induces a different membrane curvature and/or membrane composition through its F-BAR domain (Mim and Unger, 2012; Qualmann et al., 2011). Structurally, F-BAR domains are shallowly curved, crescent-shaped α-helical dimers with membrane-binding capacity (reviewed in Frost et al., 2009; Mim and Unger, 2012; Qualmann et al., 2011; Suetsugu et al., 2010). Several F-BAR domain proteins, such as those at sites of endocytosis (e.g., human FBP17, CIP4, Pacsin2, and FCHo2), are able to bend membranes into thin tubules when present at high concentration (Henne et al., 2007; Itoh et al., 2005; Tsujita et al., 2006). This process is thought to be accomplished through the formation of oligomeric assemblies of F-BAR domains on membranes that collectively enforce a curvature (Frost et al., 2008; Shimada et al., 2007; Yu and Schulten, 2013). Other F-BAR domains can induce outward curvature in membranes, namely protrusions rather than invaginations (Carlson et al., 2011; Guerrier et al., 2009). Additionally, multiple F-BAR domains lack the ability to deform membranes altogether (McDonald et al., 2015; Moravcevic et al., 2015; Tsujita et al., 2006). It is not clear what properties of an F-BAR domain confer diverse membrane-binding behaviors in vitro, and how these in vitro activities correspond to F-BAR protein physiological functions.
As in mammalian endocytosis, cytokinesis in the fission yeast S. pombe involves multiple F-BAR proteins (three) that arrive in a defined order as a medially placed actomyosin-based contractile ring (CR) is assembled and constricts. To further our understanding of cytokinesis, and of the multiple functions of F-BAR proteins in a single biological process, a clear understanding of the shared and distinct features of these proteins must be obtained. Of the three, Cdc15 (Fankhauser et al., 1995), Imp2 (Demeter and Sazer, 1998), and Rga7 (Arasada and Pollard, 2011; Martín-García et al., 2014), Cdc15 has been most thoroughly characterized. Essential for cytokinesis (Fankhauser et al., 1995; Nurse et al., 1976), Cdc15 is one of the first components detected at the incipient CR (Wu et al., 2003) and is one of the most abundant CR elements (Wu and Pollard, 2005). It acts as a key anchor of the CR (Laporte et al., 2011; Roberts-Galbraith et al., 2009, 2010) and a platform for assembling additional CR components. Specifically, Cdc15 binds membranes and the cytokinetic formin Cdc12 through its F-BAR domain (Carnahan and Gould, 2003; Willet et al., 2015; McDonald et al., 2015) and other proteins involved in cytokinesis through its SH3 domain (Roberts-Galbraith et al., 2009; Ren et al., 2015), and these interactions are modulated by Cdc15's phosphostatus (Roberts-Galbraith et al., 2010). rga7Δ cells have only mild septation defects, and it therefore appears that Rga7 is the least important of the three F-BAR proteins for cytokinesis (Arasada and Pollard, 2015; Martín-García et al., 2014). Cells lacking the Cdc15 paralog Imp2, on the other hand, are strikingly defective in cell division, specifically in the final stages of CR constriction and disassembly (Demeter and Sazer, 1998), consistent with its recruitment to the division site later than Cdc15 (~10 min after spindle pole body separation) (Ren et al., 2015; Wu et al., 2003). Finally, whereas Cdc15 localizes to cell tips during interphase (Arasada and Pollard, 2011; Carnahan and Gould, 2003), Imp2 localizes solely at the CR during cytokinesis (Demeter and Sazer, 1998). Thus, although Imp2 and Cdc15 are paralogs, there are significant differences in their behaviors and functions during the cell cycle.
Because Cdc15's and Imp2's SH3 domains are functionally interchangeable, collaborating to recruit proteins required to stabilize the CR (Ren et al., 2015; Roberts-Galbraith et al., 2009), we tested whether the unique function of Imp2 is dictated, at least in part, by distinct structural and functional properties of its F-BAR domain. We found that the Imp2 F-BAR preferentially bound phosphoinositol-containing membranes and showed no preference for a specific membrane curvature. The Imp2 F-BAR robustly tubulated membranes in vitro and when overexpressed in cultured cells. We determined the crystal structure of the Imp2 F-BAR domain and used it to identify the molecular basis for membrane binding and tubulation. As expected, membrane binding via the F-BAR is strictly required for Imp2's function in cytokinesis but, surprisingly, despite Imp2's prodigious membrane tubulation ability in vitro, tubulation activity is dispensable in vivo. Accordingly, the Imp2 F-BAR can be functionally replaced by the Cdc15 or S. cerevisiae Hof1 F-BAR domains, which do not have the ability to tubulate membranes (McDonald et al., 2015; Moravcevic et al., 2015). These results indicate that membrane tubulation via F-BAR proteins is not required for cytokinesis, raise the possibility that membrane tubulation may not be an essential physiological function of other F-BAR domain proteins that exhibit this activity in vitro, and indicate that the unique function of Imp2 in cytokinesis is dictatedby propertiesof its centraldomain rather thanits F-BAR.
RESULTS
The Imp2 F-BAR Is Critical for CR Constriction and Disassembly
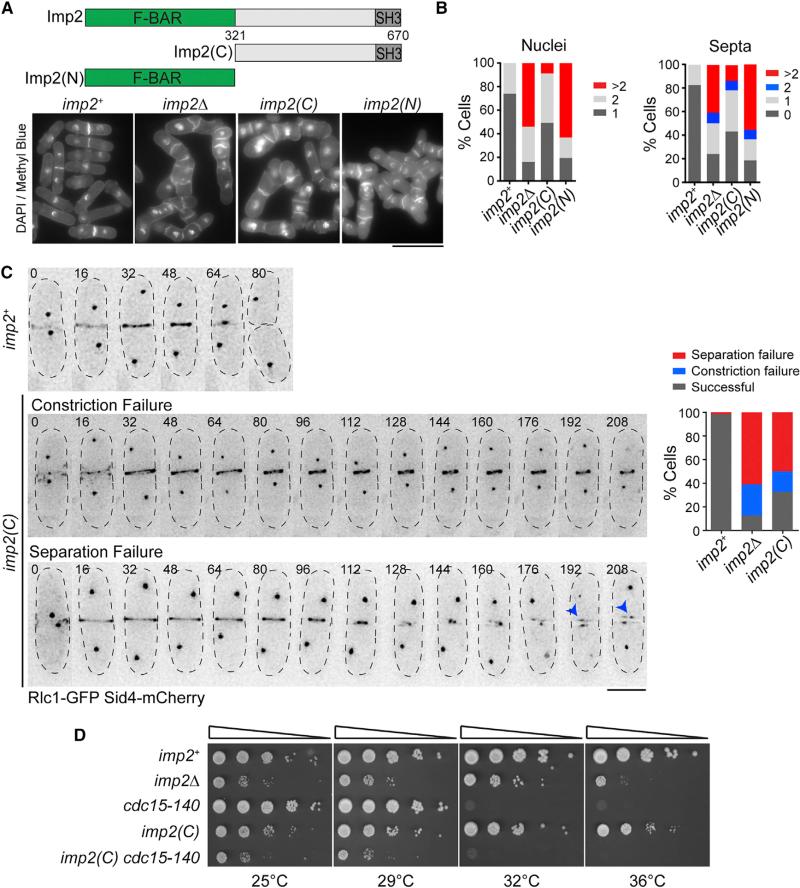
To test the importance of Imp2's F-BAR domain for function, we replaced imp2+ with a truncated version that lacked its F-BAR domain at the endogenous imp2 locus (Figure 1A). Cells producing only Imp2(C) (residues 321–670) were phenotypically similar to imp2Δ, with both alleles displaying a high percentage of multinucleate and multiseptated cells, indicative of failures in cytokinesis (Figures 1A and 1B). Loss of imp2 function via F-BAR truncation was not due to lack of Imp2(C) localization, because GFP-Imp2(C) was detected at the division site in imp2Δ cells (Figure S1A). The incorporation of GFP-Imp2(C) into the CR, where its central and SH3 domains could contribute to CR function (Roberts-Galbraith et al., 2009), is a plausible explanation for why the phenotype of F-BAR domain loss is not quite as severe as that of imp2Δ. Consistent with this interpretation, GFP-Imp2(C) CR localization was abolished with a mutation that prevents the SH3 domain from binding partners, W644S (Figure S1A) (Yu et al., 1994). To identify exactly how imp2(C) cells fail during cytokinesis, we imaged cells producing Sid4-GFP (a spindle pole body component) and Rlc1-GFP (a CR component) to visualize mitotic and cytokinetic progression, respectively. Similar to phenotypes seen previously when imp2 is deleted (Figure S1B) (Demeter and Sazer, 1998), imp2(C) cells often failed to constrict their CRs, disassemble CR remnants, and separate daughter cells (Figure 1C). We quantified the percentage of cells that completed cytokinesis, failed during constriction (defined as cells that build a CR but do not fully constrict it), or failed separation (defined as cells that fully constrict the CR but do not physically separate) (Figure 1C). Because cells lacking the Imp2 F-BAR domain fail cytokinesis >60% of the time, we conclude that the domain is required for Imp2's contribution to recruiting proteins for CR constriction and disassembly such as Rgf3, Fic1, and Pxl1 (Bohnert and Gould, 2012; Cortés et al., 2015; Morrell-Falvey et al., 2005; Ren et al., 2015). Further evidence for the importance of Imp2's F-BAR domain was obtained through genetic analysis. We crossed imp2(C) into a cdc15-140 cytokinesis mutant, which is synthetically lethal with imp2Δ (Demeter and Sazer, 1998). imp2(C) showed a strong negative genetic interaction with cdc15-140 (Figure 1D), confirming genetically a major loss of protein function in the absence of the F-BAR domain. Although essential, the Imp2 F-BAR domain alone is not sufficient for function, because an imp2(N) allele (Figure 1A) displayed identical phenotypes to imp2Δ (Figures 1A and 1B).
Figure 1. The Imp2 F-BAR Domain Is Important for Cytokinesis.
(A) Schematic of Imp2 and truncation alleles Imp2(C) and Imp2(N). Bottom: DAPI- and methyl blue-stained cells of the indicated imp2 genotype. Scale bar, 10 μm.
(B) Cytokinesis phenotype quantifications of cells in (A); n ≥ 300 for each strain.
(C) Representative images (left) and quantification (right) of cytokinesis defects in imp2+, imp2Δ, and imp2(C); n > 30 for each strain. Blue arrowheads indicate persistent contractile ring remnants. Numbers indicate time (min) from spindle pole body duplication. Scale bar, 5 μm.
(D) Serial dilutions of imp2 strains at the indicated temperatures.
See also Figure S1.
Structure of the Imp2 F-BAR Domain
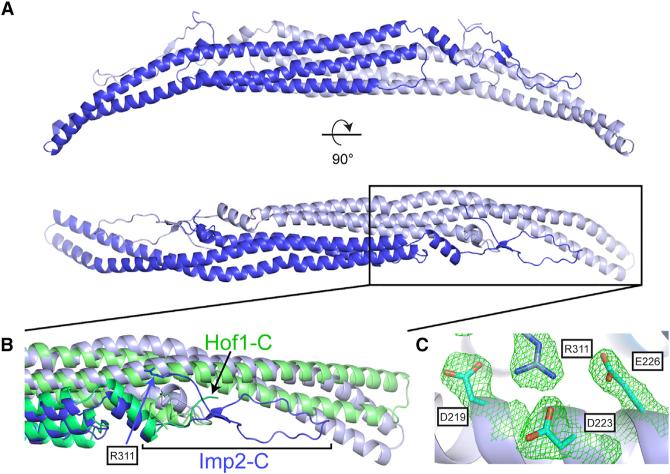
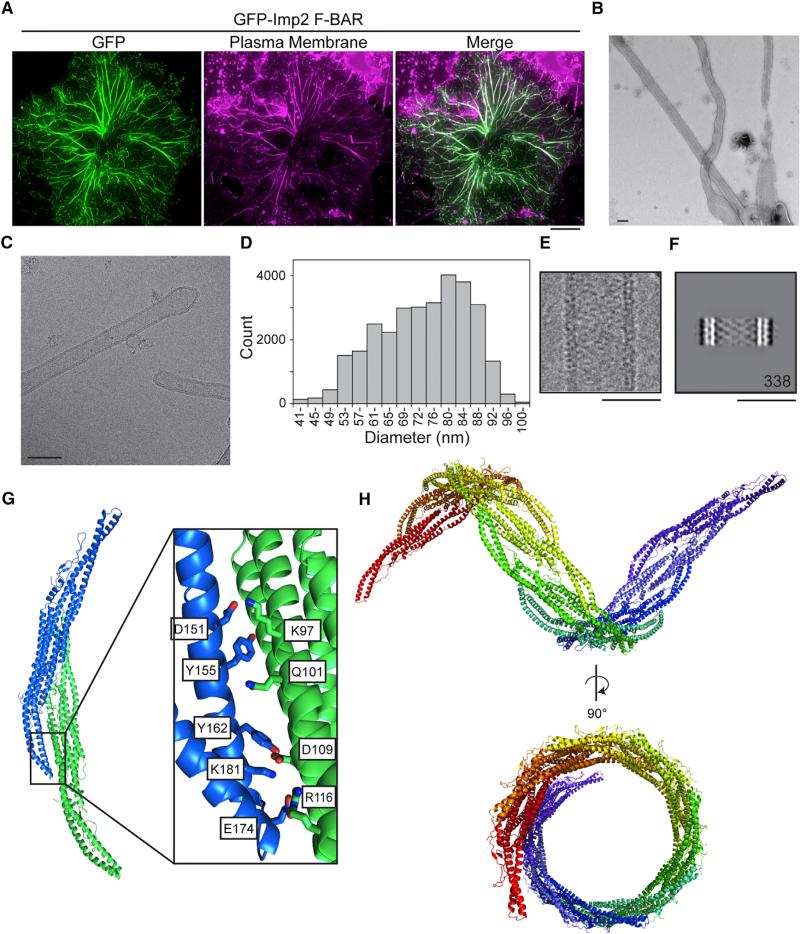
To determine what properties of the Imp2 F-BAR domain drive its cytokinetic function, we took a structural approach. The X-ray crystal structure of the Imp2 F-BAR domain was determined using selenomethionine phasing and fully refined to a resolution of 2.35 Å (Table S1). The Imp2 F-BAR domain adopts a characteristic tightly interwound dimeric BAR domain architecture, with a 5,911-Å2 interface area (Figure 2A). The domain consists of a six-helical dimeric bundle with extended wings that adopt a bent conformation relative to the helical core. Dali server search (Holm and Rosenström, 2010) indicates that the closest structural homolog of Imp2 is the F-BAR domain of S. cerevisiae Hof1 (Moravcevic et al., 2015), with an RMSD (root-mean-square deviation) of 2.7 Å. However, the two structures differ significantly in the overall shape of the F-BAR domain, particularly in wing-tip orientation, with Imp2 exhibiting a significantly higher tip angle (Figure 2B). Additionally, the 30 C-terminal residues of the Imp2 F-BAR domain are well ordered and form an extension of the F-BAR domain compared to Hof1 (Figure 2B). Residues in the extended C terminus make important contacts back to the core of the protein, explaining why a construct encoding a shorter fragment (residues 15–295) was unstable. In particular, a β strand is formed by residues 282–284 and 304–306, and R311 forms intermolecular salt bridges to D219, D223, and E226, located in the core six-helical bundle interface (Figure 2C).
Figure 2. Structure of the Imp2 F-BAR Domain.
(A) Crystal structure of the Imp2 F-BAR domain dimer with subunits colored in dark and light blue.
(B) Close-up view of the wing-tip orientation and C-terminal residues of Imp2 (blue), which diverge significantly from Hof1 (green).
(C) Close-up view of the intermolecular salt-bridge network formed by the F-BAR C terminus (2Fo – Fc contoured at 1.1σ).
Structural Basis for Imp2 Membrane Binding
Recombinantly produced and purified Imp2 F-BAR domain (Figure S2A) binds biological membranes rich in phosphorylated phosphatidylinositols (PIPs) in a salt-dependent manner (Figure S2B). The domain bound maximally to synthetic liposomes containing at least 20% phosphatidylserine (PS) (Figure S2C) and showed no preference for membrane curvature, binding liposomes between 100 and 800 nm in diameter equally well (Figure S2D). The domain bound liposomes of various compositions in a cooperative manner, with a preference for liposomes containing negatively charged PIPs and PS (Figures S2E and S2F). Interestingly, this F-BAR domain bound most strongly to liposomes containing PI(4)P compared with PI(3,4,5)P3, indicating a mild preference for the PI(4)P lipid head group.
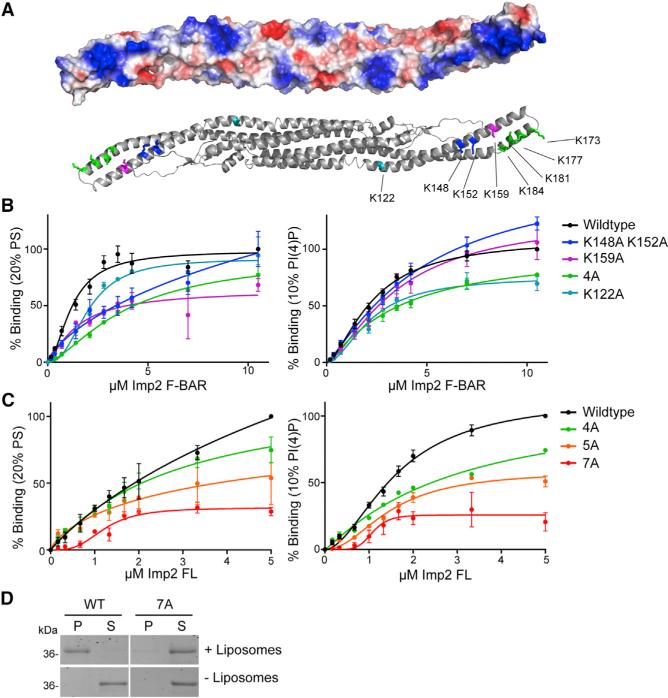
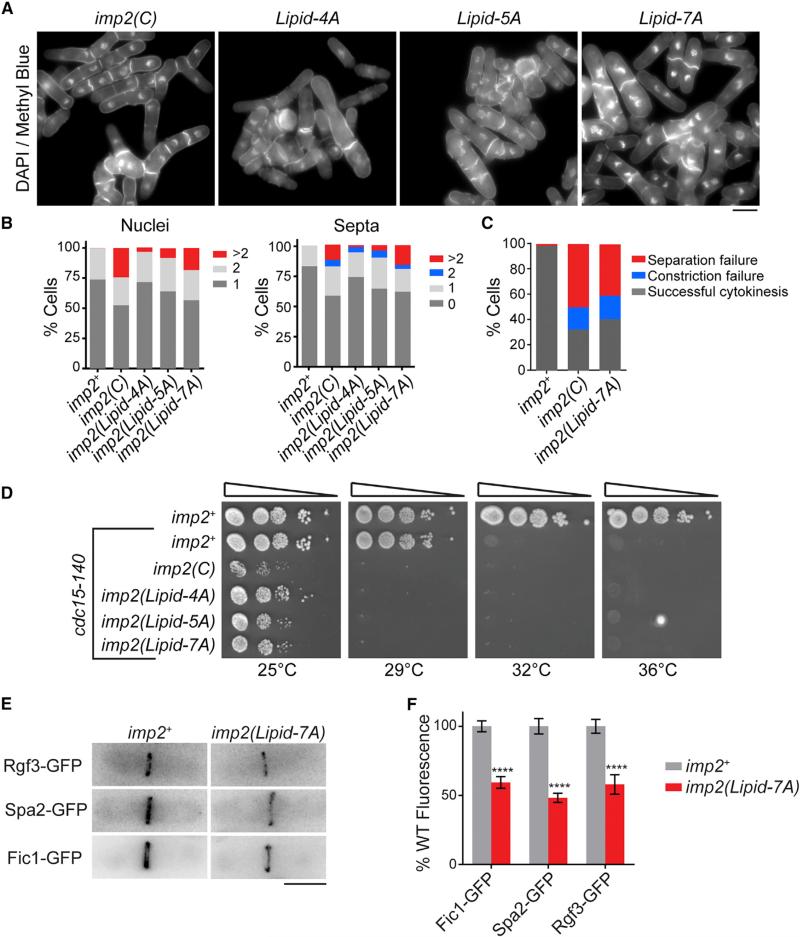
To identify how the Imp2 F-BAR binds to negatively charged phospholipids, the electrostatic surface of the dimer was examined. The concave surface of the dimer contains extensive positive charge, particularly in the domain's bent wings, and an additional positively charged patch was present near the base of each wing in the dimeric core (Figure 3A). These basic regions are composed of clusters of lysine residues, similar in position to those used by the FBP17 and FCHo2 F-BARs to bind membranes (Frost et al., 2008; Henne et al., 2007). To determine the role of these lysine residues in Imp2 F-BAR function, several mutations were generated: K159A, K148A/K152A, and K173A/K177A/K181A/K184A (Lipid-4A) (Figure 3A). We also generated a mutation that disrupted the basic patch at the base of the wing, K122A (Figure 3A). Mutations of these basic residues, particularly the Lipid-4A combination, K159A, and K122A, resulted in decreased affinity of the F-BAR domain for liposomes, confirming the important role of these basic surfaces in Imp2 F-BAR domain membrane binding (Figure 3B). Interestingly, the K122 patch appears to be responsible for the PI(4)P preference, because binding to PI(4)P-containing liposomes decreased more than binding to PS-containing liposomes in the K122A mutant.
Figure 3. Mechanism of Imp2 F-BAR Domain Membrane Binding.
(A) Electrostatic potential map of the concave surface of the Imp2 F-BAR domain. Blue, positive potential; red, negative potential; white, near neutral. Bottom: ribbon model depicting predicted membrane-binding residues.
(B) Binding assays of wild-type (WT) or membrane-binding mutant Imp2 F-BAR domains and liposomes composed of 60% PC/20% PE/20% PS or 70% PC/20% PE/10% PI(4)P. 4A consists of K173A, K177A, K181A, and K184A.
(C) Binding assays of wild-type or multisite membrane-binding mutations in full-length (FL) purified Imp2. 4A: as in (B); 5A: 4A + K159A; 7A: 5A + K148A and K152A.
Error bars in all panels indicate SEM from at least three experiments.
(D) Representative liposome copelleting assay between 1 μM full-length Imp2(7A) and Folch fraction liposomes. P, pellet; S, supernatant.
To further reduce membrane binding, we designed a Lipid-5A mutant consisting of Lipid-4A and K159A, and a Lipid-7A mutant composed of all seven basic wing residues converted to alanine. F-BAR domains containing these mutations proved difficult to purify; however, full-length Imp2-Lipid-4A, -5A, and -7A were readily purified. Testing the full-length proteins in liposome binding assays showed that Imp2 bound membranes whereas Imp2-Lipid-4A, -5A, and -7A had reduced binding (Figure 3C). Despite retaining a low level of binding to synthetic liposomes at saturating concentrations, the 7A combination mutant completely abolished copelleting with Folch fraction liposomes (Figure 3D). We conclude that the basic residues within the wing of the Imp2 F-BAR domain are necessary for Imp2 to associate with membranes.
Having identified how the Imp2 F-BAR domain binds membranes, we next tested the importance of membrane binding for Imp2 function in vivo by replacing the wild-type gene with lipid-binding mutants at the imp2 endogenous locus. imp2-Lipid-4A, -5A, and -7A mutants displayed progressively worsening phenotypes similar to imp2(C) (Figure 4A), with cells often failing cytokinesis to become multinucleated and multiseptated (Figures 4A–4C). Also like imp2(C), the imp2 membrane-binding mutants showed severe negative genetic interactions with cdc15-140 (Figure 4D). When Imp2 cannot bind membranes, Imp2 levels at the ring were decreased, although total cellular amounts remained near wild-type levels (Figures S3A and S3B). Recruitment of important Imp2 SH3 domain interactors to the CR was impaired according to Imp2 reduction (Figures 4E and 4F). We conclude that membrane binding is a necessary function of the Imp2 F-BAR domain in cytokinesis to form an efficient scaffold for its partners.
Figure 4. Membrane Binding Is an Essential Function of the Imp2 F-BAR Domain.
(A) DAPI- and methyl blue-stained cells of the indicated imp2 genotype. Lipid mutations are described in Figure 3C. Scale bar, 4 μm.
(B) Quantification of cytokinesis phenotypes of cells in (A); n ≥ 300 for each strain.
(C) Cytokinesis failure phenotype quantifications from time-lapse imaging of Rlc1-GFP and Sid4-GFP in the indicated imp2 strains; n ≥ 30 for each strain. Data for imp2+ and imp2(C) are from Figure 1C.
(D) Serial dilution assay of imp2 membrane-binding mutants at the indicated temperatures.
(E) Representative images of GFP-tagged Imp2 SH3 interactors in the indicated imp2 strains. Scale bar, 5 μm.
(F) Quantification of Imp2 SH3-binding partners in the contractile ring from (E). Error bars indicate SEM; n > 60 for each condition. ****p < 0.0001.
See also Figure S3.
The Imp2 F-BAR Tubulates Membranes by Forming Higher-Order Structures
Several F-BAR domains not only bind membranes but bend and tubulate them when present at high concentration (Frost et al., 2008; Itoh et al., 2005; Tsujita et al., 2006). Indeed, F-BAR domains have been hypothesized to induce or stabilize the curvature present at the division site during S. pombe cytokinesis (Frost et al., 2009). Because Cdc15 does not tubulate membranes (McDonald et al., 2015), we tested whether tubulation was an activity of the Imp2 F-BAR and whether this activity is in fact important during cytokinesis. We found that the Imp2 F-BAR tubulated the plasma membrane of COS-7 cells when overexpressed (Figure 5A). It also tubulated synthetic liposomes in vitro, as detected by negative-stain electron microscopy (EM) (Figure 5B) and live imaging of giant unilamellar vesicles (GUVs) (Movie S1).
Figure 5. The Imp2 F-BAR Domain Tubulates Membranes through Helical Oligomerization.
(A) GFP-Imp2 F-BAR domain expressed in COS-7 cells and costained with CellMask Orange plasma membrane dye. Scale bar, 10 μm.
(B) Imp2 F-BAR domain incubated with liposomes composed of 50% PS/35% PC/10% PE/5% PI and visualized with negative-stain EM. Scale bar, 100 nm.
(C) Cryo-EM image of an Imp2 F-BAR domain-coated membrane tubule in vitrified ice. Scale bar, 100 nm.
(D) Size distribution of Imp2 F-BAR domain-induced liposome tubules measured from cryo-EM images.
(E) Representative Imp2 liposome tubule as used to generate the class average in (F). Scale bar, 50 nm.
(F) Class average of Imp2 F-BAR domain-coated tubules with a diameter of 50 nm; 338 images were included in the average. Scale bar, 50 nm.
(G) Packing of the dimer of dimers present in Imp2 F-BAR domain crystals. Individual dimers are represented as blue and green. The inset shows key interface residues.
(H) Extending the dimer-dimer contacts between Imp2 F-BARs results in a helical-filament structure. Each dimer is depicted in a different color.
To investigate how the Imp2 F-BAR domain tubulates membranes in detail, the organization of Imp2 F-BAR coats on membranes was more closely examined using cryo-EM (Figure 5C). Images of tubules in vitrified ice showed that, similar to the CIP4 F-BAR (Frost et al., 2008), the Imp2 F-BAR forms tubules spanning a range of diameters (Figure 5D). The Imp2 F-BAR domain could be seen coating the membrane of individual tubules (Figure 5E), and class averages of Imp2-coated tubules of similar widths revealed an ordered pattern on the membrane (Figure 5F). Despite this ordered pattern, a high degree of heterogeneity of the Imp2 F-BAR tubules precluded determination of a high-resolution structure. This heterogeneity, which included differences in diameter and F-BAR lattice packing even within individual tubules, was not improved using low-temperature annealing protocols that have been used previously to determine an F-BAR tubule structure (Frost et al., 2008).
Membrane tubulation by F-BAR domains is expected to rely upon membrane binding. Therefore, we tested whether mutations of the membrane-binding residues on the wing and core interfered with membrane tubulation by Imp2 F-BAR domains. The K122A core patch mutant retained tubulation activity, although at a much-reduced level, whereas the wing patch mutants lacked tubulation activity (Figure S3). Therefore, membrane binding is required for the Imp2 F-BAR domain to tubulate membranes.
Imp2 F-BAR Helical Oligomerization Supports Membrane Tubulation
Membrane tubulation by F-BARs is proposed to occur through a scaffolding mechanism where F-BAR domains organize into defined oligomers that collectively deform the membrane (Frost et al., 2008; Shimada et al., 2007; Yu and Schulten, 2013). To determine the mechanism of Imp2 F-BAR oligomerization, we analyzed the packing of the Imp2 F-BAR domain in crystals. The domain organized into a higher-order dimer of dimers in the crystal (Figure 5G), formed by reciprocal packing of the wing of one dimer with the core of the next. The interaction surface was significant, with 804 Å2 of interface area. The dimer of dimers showed a systematic shift in register with the addition of each dimer subunit that could support the formation of an oligomeric filament. We computationally extended the dimer of dimers, assuming symmetric wing-to-core interactions, to produce a model of an Imp2 F-BAR filament (Figure 5H; Movie S2). Strikingly, the filament is helical, suggesting that this conformation of Imp2 F-BAR oligomers on a membrane may be responsible for its tubulation activity.
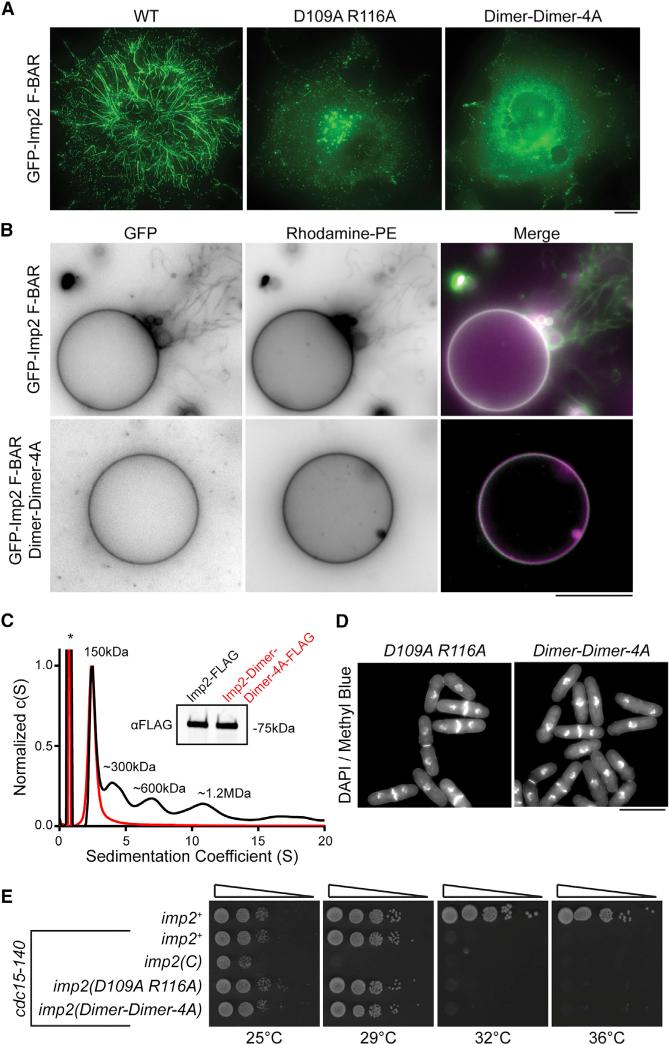
A detailed examination of the dimer-of-dimers crystal packing showed that the interface is formed by a series of salt bridges and hydrogen bonds between wing tip residues (D151, Y155, Y162, E174, K181) and dimer core residues (K97, Q101, D109, R116) (Figure 5G). To test this helical oligomerization model and the role of tip-to-core dimer-dimer interactions in Imp2 membrane tubulation, alanine point mutations in the dimer core residues (D109A, R166A and K97A, Q101A, D109A, R116A [Dimer-Dimer-4A]) were produced. Dimer-dimer mutants were completely deficient in their ability to tubulate membranes when overexpressed in COS-7 cells (Figure 6A), supporting our model of tubulation. Additionally, these mutations abolished the ability of the Imp2 F-BAR to tubulate GUVs in vitro (Figure 6B). Mutation of these residues had no significant effect on the affinity of the Imp2 F-BAR for membranes, indicating that constructs containing these point mutants (K97A, Q101A, D109A, and R116A) were properly folded and were not deficient in membrane binding (Figure S4A).
Figure 6. Imp2 Oligomerization Is Essential for Tubulation but Dispensable for Cytokinesis.
(A) GFP-Imp2 F-BAR domains with the indicated mutations were expressed in COS-7 cells. Scale bar, 10 μm.
(B) Dimer-dimer contact mutations abolish tubulation activity of the GFP-Imp2 F-BAR domain on giant unilamellar vesicles composed of 64% PC/20% PE/10% PS/5% PI(4)P/1% Rhodamine-PE. Scale bar, 10 μm.
(C) Sedimentation velocity analytical ultracentrifugation trace of full-length Imp2 and Imp2(Dimer-Dimer-4A) purified from S. pombe. The asterisk indicates a 3× FLAG peptide contaminant. Inset: αFLAG western blot of Imp2 and Imp2(Dimer-Dimer-4A) samples.
(D) DAPI- and methyl blue-stained cells of the indicated imp2 genotype. Scale bar, 10 μm. See Figure S4B for quantification.
(E) Serial dilutions of imp2 mutant strains at the indicated temperatures. Dimer-Dimer-4A: K97A, Q101A, D109A, R116A.
See also Figure S4.
The Imp2 F-BAR Domain's Cytokinetic Function Does Not Require Tubulation or Oligomerization
Despite multiple F-BAR proteins’ ability to tubulate membranes in vitro, the importance of tubulation activity for the physiological function of any F-BAR is unknown. We directly tested the importance of membrane tubulation in Imp2's physiological function in cytokinesis by integrating both tubulation-deficient dimer-dimer mutants (D109A, R116A and Dimer-Dimer-4A) into the imp2 endogenous locus. Although we determined that these mutants can no longer tubulate membranes in vivo or in vitro (Figures 6A and 6B), we first tested whether the mutants retained any oligomerization activity in S. pombe. To this end, Imp2 and the Imp2-Dimer-Dimer-4A mutant were purified from S. pombe cell lysates and assayed by analytical ultracentrifugation. Whereas wild-type Imp2 was present in multiple oligomeric species, the Dimer-Dimer-4A mutant was strictly dimeric (Figure 6C), confirming complete loss of oligomerization in vivo along with loss of tubulation activity. Surprisingly, these mutant cells had wild-type morphologies with no evidence of cytokinetic defects (Figure 6D; Figure S4B). The absence of compromised cytokinetic function was confirmed genetically, as dimer-dimer mutants displayed no synthetic genetic interactions with cdc15-140 (Figure 6E). We observed a minor decrease in the levels of Imp2-Dimer-Dimer-4A at the contractile ring (Figure S4C), but this did not result in observable cytokinetic phenotypes. Therefore, despite a strong ability to tubulate membranes in vitro or when overexpressed in vivo, tubulation and oligomerization are not necessary for Imp2's function in cytokinesis.
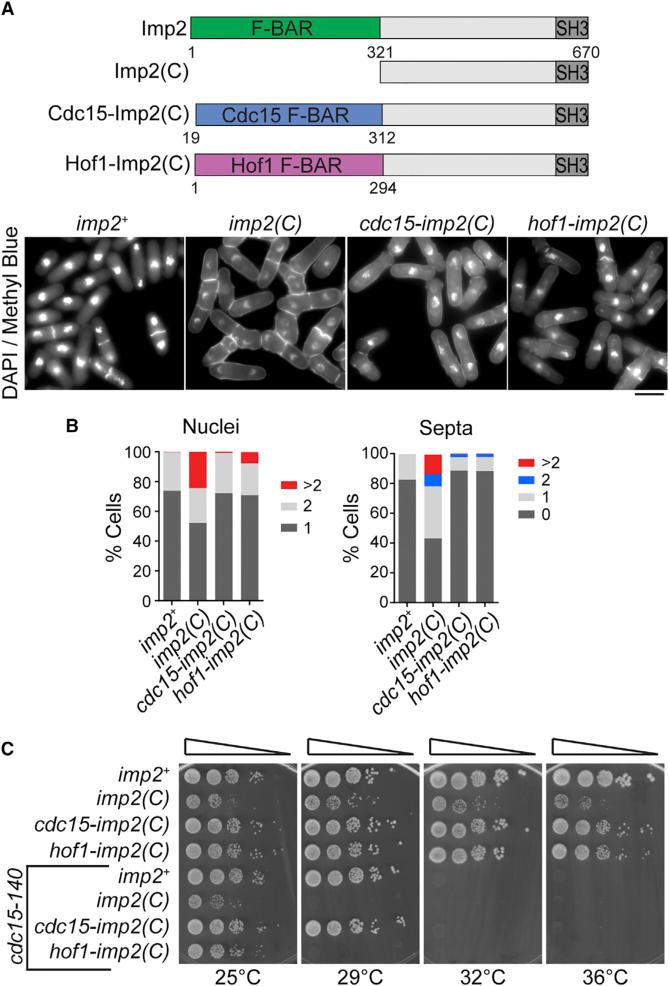
To rigorously test this conclusion, we created synthetic fusion proteins that replaced Imp2's F-BAR domain with the F-BAR domains from Imp2's paralog Cdc15 or homolog S. cerevisiae Hof1 (Figure 7A, top), which, unlike Imp2's F-BAR domain, do not tubulate membranes (McDonald et al., 2015; Moravcevic et al., 2015). Both Cdc15 and Hof1 F-BAR domains successfully replaced Imp2 F-BAR domain function in cytokinesis (Figures 7A and 7B) and in growth (Figure 7C), and the fusion proteins localized to the division site normally (Figure S5). We conclude that Imp2's F-BAR domain does not need to bend the membrane but instead operates as a temporally restricted bridge between the membrane and other proteins that bind Imp2.
Figure 7. Nontubulating F-BAR Domains Can Complement Imp2 F-BAR Domain Function.
(A) Schematic of Imp2 F-BAR domain replacement constructs. Bottom: DAPI- and methyl blue-stained cells of the indicated imp2 genotype. Scale bar, 5 μm.
(B) Cytokinesis phenotype quantifications of cells in (A); n ≥ 300 for each strain.
(C) Serial dilutions of imp2 strains at the indicated temperatures.
See also Figure S5.
DISCUSSION
We have determined that the Imp2 F-BAR domain is critical for proper CR constriction, septation, and disassembly during cytokinesis. We determined the structure of the domain, as well as the domain's mechanism of membrane binding and helical oligomerization-based tubulation. Using separate sets of mutations that affected either Imp2 membrane-binding or tubulation activities, we surprisingly found that Imp2's cytokinetic functions rely only on F-BAR membrane binding, rather than tubulation activity, and can be replaced by other nontubulating cytokinetic F-BAR domains. Our data suggest that the Imp2 F-BAR domain acts only as a membrane-bound tether to properly orient and scaffold cytokinetic partners that interact with Imp2's other domains.
Diverse Modes of F-BAR Oligomerization Drive Tubulation
Our model of Imp2 F-BAR supermolecular assembly derived from the Imp2 crystal structure predicts that the Imp2 F-BAR domain can oligomerize to form a helical structure with a diameter of ~130 Å. The diameters of tubules formed by the Imp2 F-BAR range from ~50–100 nm in vitro to ~100–300 nm in cultured cells. This variability in tubule diameter induced by the Imp2 F-BAR domain suggests that the dimer-dimer contact interfaces have significant flexibility. The wing angles of individual Imp2 F-BAR dimers may also be flexible, introducing another degree of freedom to helical filaments. Flexibility at the Imp2 F-BAR-to-F-BAR interfaces was also suggested by our EM data of Imp2 F-BAR domain-coated tubules. Although striations of a helical Imp2 F-BAR domain coat were visible on individual tubules, this membrane coat was irregular and varied down a tubule's length. Whether structural flexibility is important for Imp2 F-BAR function outside the context of an oligomeric structure awaits further analysis.
We defined mutations that disrupt the observed superhelical oligomeric interface of Imp2 and demonstrated that these mutant proteins completely lack tubulation activity. Our model, therefore, provides additional evidence for a “scaffolding” mechanism of F-BAR domain membrane tubulation (Frost et al., 2008; Shimada et al., 2007; Yu and Schulten, 2013) that could be relevant to other F-BAR domains. Indeed, our model is similar in principle to EM reconstructions of CIP4 membrane tubules (Frost et al., 2008) but diverges in the physical mechanism of F-BAR-to-F-BAR interactions. Multiple interactions were present in CIP4 tubule reconstructions between F-BARs, including lateral interactions between adjacent dimers and tip-to-tip interactions between F-BAR domain wings. Imp2 F-BAR-to-F-BAR interactions, on the other hand, occur between the tip of one dimer and the core of an adjacent dimer. Mutation of this single interface is sufficient to disrupt all oligomerization and subsequent tubule formation. Unsurprisingly, the residues involved in CIP4 oligomerization are distinct from those identified in Imp2. Therefore, this common in vitro ability of F-BARs to tubulate membranes using a scaffolding mechanism can be mediated by diverse F-BAR-to-F-BAR interactions.
Of the 13 F-BAR domain structures available (Edeling et al., 2009; Henne et al., 2007; Moravcevic et al., 2015; Rao et al., 2010; Reider et al., 2009; Shimada et al., 2007; Wang et al., 2009), the Imp2 crystal structure is most similar to that of its homolog in S. cerevisiae, Hof1, with an RMSD of 2.7 Å. Despite this considerable similarity, the Hof1 F-BAR domain was not observed to tubulate membranes when expressed in cultured cells (Moravcevic et al., 2015). Neither the interaction interface we identified in the Imp2 F-BAR nor the one identified in CIP4 (Frost et al., 2008) is conserved in Hof1, and this may explain why Hof1 does not tubulate membranes when over-expressed (Moravcevic et al., 2015). As the sole cytokinetic F-BAR in S. cerevisiae, Hof1's inability to tubulate membranes is consistent with our conclusion that membrane tubulation by F-BAR proteins is not required during cytokinesis in yeast.
Are F-BAR Domain Oligomerization and Tubulation Important In Vivo?
Our results show that although the Imp2 F-BAR domain can tubulate membranes in vitro, it does not require this activity for its function in vivo. In contrast to prevailing models derived from in vitro analyses (Frost et al., 2009; Mim and Unger, 2012; Qualmann et al., 2011; Suetsugu et al., 2010), these results combined with the findings that the Drosophila Nwk1 F-BAR, the Cdc15 F-BAR, and six other human F-BAR domains also do not tubulate membranes (Becalska et al., 2013; Kelley et al., 2015; McDonald et al., 2015) indicate that membrane tubulation is not an essential physiological function of all F-BAR domains. It is possible that the observed membrane tubulation by some F-BAR domains is due to their overexpression, with membrane deformation unnecessary for their biological function, as in the case of Imp2. High levels of molecular crowding can lead to membrane tubulation by certain proteins when locally concentrated at membranes (Stachowiak et al., 2012). On the other hand, it is not yet clear what function, if any, lateral F-BAR-to-F-BAR contacts perform in vivo, if not membrane tubulation. It is possible that less extensive oligomeric membrane-bound structures exist, such as the clusters of F-BAR proteins at dendritic spines (Schneider et al., 2014), that could reinforce certain structures or cluster-specific phosphoinositides (Zhao et al., 2013). Our data indicate that Imp2 is present as an oligomeric species in solution that may play a minor role in recruitment to the CR, because oligomerization mutants localize less robustly. However, the decrease in CR localization when oligomerization was inhibited did not lead to any observable phenotype. It will be important to clarify the extent to which other F-BAR domains utilize their lateral contacts in normal physiological function.
Function of F-BAR Proteins in Cytokinesis
Combining our results with those from other studies (Moravcevic et al., 2015; Nishihama et al., 2009; Oh et al., 2013; Ren et al., 2015; Roberts-Galbraith et al., 2009, 2010) leads us to suggest that the F-BAR proteins participating in cytokinesis serve as regulatable linkers between the membrane and additional division machinery rather than as direct engines of membrane deformation. The results of our F-BAR domain-swapping experiments further indicate that considerable plasticity is allowed in the membrane-binding module offered by different F-BAR proteins. Given that Cdc15's and Imp2's SH3 domains are functionally interchangeable (Ren et al., 2015) and Cdc15's F-BAR can substitute for Imp2's, the central region of these two proteins must specify important functional distinctions. Cdc15's unstructured central region controls the conformation of the protein through extensive regulatory phosphorylation (Roberts-Galbraith et al., 2010; Ullal et al., 2015), and is also proposed to mediate an undefined interaction with the Golgi network (Arasada and Pollard, 2014). The central region of S. cerevisiae Hof1 binds septins and myosin II (Meitinger et al., 2011; Oh et al., 2013), and tunes these interactions with cell-cycle-regulated phosphorylation (Meitinger et al., 2011, 2013). Imp2's central domain is also differentially phosphorylated throughout the cell cycle (unpublished data), hinting that Imp2 function may also be regulated via phosphorylation. Future work will be directed at understanding whether differential phosphoregulation underlies the distinct temporal requirements of Cdc15 and Imp2 in cytokinesis, and the contribution of Imp2's central region to regulation of its membrane and protein-binding domains.
EXPERIMENTAL PROCEDURES
Yeast Strains, Media, and Genetic Methods
S. pombe strains used in this study (Table S2) were grown in yeast extract or minimal medium with appropriate supplements (Moreno et al., 1991). S. pombe transformations were performed using a lithium acetate method (Keeney and Boeke, 1994). Regulated expression of genes from the nmt1 promoter (Basi et al., 1993; Maundrell, 1993) was achieved by growth in the presence of thiamine (promoter repressed) and then washing into medium lacking thiamine (promoter nonrepressed). Nuclei and septa were visualized by ethanol fixation and 4′,6-diamidino-2-phenylindole (DAPI) and methyl blue staining. Cassettes encoding tags and selectable markers were inserted into the S. pombe genome at the 3′ end of open reading frames as previously described (Bähler et al., 1998). All transformants were confirmed by PCR and sequencing.
Protein Purification and Crystallization
Recombinant Imp2 F-BAR domain (15–320), full-length Imp2, and GFP-Imp2 F-BAR domain (15–320) constructs were produced in Escherichia coli Rosetta 2 cells (Novagen) grown in Terrific broth or T7 Express Crystal (NEB) in M9 salts supplemented with selenomethionine (SeMet). Protein was purified over His-Bind resin (Novagen) or HIS-Select HF resin (Sigma) in the presence of 1% NP-40 and 5 mM β-mercaptoethanol according to the manufacturer's protocol. Protein for crystallization was incubated overnight with thrombin to remove the His tag. Protein was further purified with a cation-exchange chromatography column (GE Healthcare HiTrap SP) and concentrated with an Amicon Ultra centrifugal filter (Millipore) to 4–10 mg/ml. Protein crystals were produced by hanging drop vapor diffusion using a mosquito crystallization robot (TTP Labtech). A high-quality single crystal grew within 2–3 weeks from a 1:1 mixture of 5.4 mg/ml Imp2:18% PEG 3350, 0.2 M ammonium formate.
Native and SeMet single-wavelength anomalous diffraction (SAD) data were collected on the 22-ID beamline of the Southeast Regional Collaborative Access Team at the Advanced Photon Source at Argonne National Laboratory to 2.35 and 2.7 Å, respectively (Table S1). Data were processed using HKL2000 (Otwinowski and Minor, 1997). Initial attempts to determine the structure by molecular replacement were not successful. Instead, PHENIX (Adams et al., 2010), employing HYSS (Grosse-Kunstleve and Adams, 2003) and Phaser (McCoy et al., 2007), was used for SeMet SAD phasing. RESOLVE (Terwilliger et al., 2008) was used to extend the resolution to 2.35 Å and generate an initial structural model. The structure was then fully built and refined via iterative model building and refinement using Coot (Emsley et al., 2010) and REFMAC5 (Murshudov et al., 1997) or PHENIX (Adams et al., 2010), respectively. Diffraction data and final refinement statistics are summarized in Table S1. Molecular graphics were prepared using PyMOL (Schrödinger).
Liposome Assays
All lipids were obtained from Avanti Polar Lipids except for Folch fraction samples (Sigma). Liposomes were prepared as previously described (Henne et al., 2007). Briefly, CHCl3 lipid stocks were mixed at the desired ratios and dried under N2 gas, and chloroform was removed under high vacuum. Liposomes were hydrated in 10 mM HEPES (pH 7.4), 100 mM KCl, 1 mM DTT to a concentration of 1 mg/ml, subjected to ten freeze-thaw cycles, and extruded through polycarbonate filters of different sizes (Whatman) with a Mini-Extruder (Avanti Polar Lipids). Giant unilamellar vesicles were formed by drying 10 μl of a 10 mg/mL lipid stock (69% 1,2-dioleoyl-sn-glycero-3-phosphocholine [DOPC], 15% 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine [DOPE], 10% 1,2-dioleoyl-sn-glycero-3-phospho-L-serine [DOPS], 5% PI(4)P, 1% Rhodamine-PE) on indium-tin-oxide-coated glass coverslips (Sigma-Aldrich) under vacuum. A 2-mm chamber was assembled between the lipid-coated coverslips and filled with a 20 mM HEPES (pH 7.4), 500 mM sucrose buffer through which a 10-Hz, 2.5-V sinusoidal current was passed for 2 hr to form the GUVs. After formation, a glucose solution was added to a final concentration of 500 mM and NaCl to 100 mM. Recombinant GFP-F-BAR solutions were mixed with GUVs at a final concentration of 10 μM before imaging in a 0.5-mm chamber.
For liposome copelleting binding assays, the Imp2 F-BAR was added to 0.5 mg/ml (final concentration) liposomes for 15 min at room temperature before centrifugation at 150,000 × g in an Optima TL ultracentrifuge for 15 min at 25°C. Pellet and supernatant fractions were resuspended in equal volumes and analyzed by SDS-PAGE. Protein bands were visualized by Coomassie staining and quantified with LI-COR Odyssey software.
Electron Microscopy
Folch fraction liposomes or synthetic liposomes composed of 10% phosphatidylethanolamine (PE), 5% PI, 50% PS, and 35% phosphatidylcholine (PC) (Frost et al., 2008) at 1 mg/ml were preincubated with the Imp2 F-BAR domain at a lipid-to-protein ratio of 2:1 for 15 min at room temperature. Negative-stain grids were prepared by applying 2.5 μl of F-BAR-bound liposome sample to a glow-discharged, carbon-coated 400-Cu mesh grid and stained with 0.75% uranyl formate (Ohi et al., 2004). Negative-stain samples were imaged on an FEI 100-kV Morgagni electron microscope with a 1K × 1K AMT CCD camera. For preparation of samples in vitrified ice, 2 μl of sample was applied to a Quantifoil R2/1 holey carbon grid (Quantifoil Micro Tools), blotted for 2 s, and plunged into liquid ethane using a Vitrobot Mark III (FEI). Cryo samples were examined at liquid-nitrogen temperatures using an FEI Polara at 200 kV equipped with a field emission electron gun. Samples were imaged under low-dose conditions (total electron dose of 20 e−/Å2), at a nominal magnification of 59,0003× (1.97 Å per pixel), and using defocus values ranging from −3.0 to −5.0 μm on a 4K × 4K Gatan UltraScan CCD camera.
Cell Culture
COS-7 cells were cultured in DMEM containing 10% FBS on fibronectin (Sigma)-coated glass coverslips. Transfections were performed using Lipofectamine LTX reagent (Invitrogen) according to the manufacturer's protocol and imaged after 24 hr. Cells in Figure 5A were costained with CellMask Orange plasma membrane dye immediately before imaging (Invitrogen).
Microscopy
GUV, COS-7 cell, and S. pombe imaging was performed on a Personal DeltaVision microscope system (Applied Precision) that includes an Olympus IX71 microscope, a 603 NA 1.42 planApo objective, and a Photometrics CoolSNAP HQ2 camera. Time-lapse imaging was performed on log-phase cells using an ONIX microfluidics perfusion system, flowing 25°C yeast extract media through the chamber at 5 psi throughout imaging. Image stacks were deconvolved and projected using softWoRx imaging software (GE Healthcare Life Sciences).
Supplementary Material
Highlights.
The S. pombe Imp2 F-BAR domain binds and tubulates membranes in vitro
Helical oligomers formed by the F-BAR underlie its ability to tubulate membranes
Canonical F-BAR tubulation activity is dispensable for Imp2 cytokinetic function
Yeast cytokinesis does not require membrane tubulation by F-BAR domain proteins
ACKNOWLEDGMENTS
The authors thank Dr. Rachel Roberts-Galbraith and members of the K.L.G. laboratory, especially Dr. Janel Beckley, for critical review of the manuscript; the Vanderbilt Center for Structural Biology for Analytical Ultracentrifuge and Electron Microscopy Facility support; and NIH grant P20GM103486 for University of Kentucky core support. N.A.M. was supported by AHA fellowship 15PRE21780003. This work was supported by NIH grant GM101035 to K.L.G.
Footnotes
AUTHOR CONTRIBUTIONS
Conceptualization, N.A.M., M.D.O., C.W.V.K., and K.L.G.; Investigation, N.A.M., Y.T., A.F., and P.X.; Writing, N.A.M., M.D.O., C.W.V.K., and K.L.G.
ACCESSION NUMBERS
The accession number for the atomic coordinates and structure factors reported in this paper is PDB: 5C1F.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, five figures, two tables, and two movies and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2015.12.062.
REFERENCES
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arasada R, Pollard TD. Distinct roles for F-BAR proteins Cdc15p and Bzz1p in actin polymerization at sites of endocytosis in fission yeast. Curr. Biol. 2011;21:1450–1459. doi: 10.1016/j.cub.2011.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arasada R, Pollard TD. Contractile ring stability in S. pombe depends on F-BAR protein Cdc15p and Bgs1p transport from the Golgi complex. Cell Rep. 2014;8:1533–1544. doi: 10.1016/j.celrep.2014.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arasada R, Pollard TD. A role for F-BAR protein Rga7p during cytokinesis in S. pombe. J. Cell Sci. 2015;128:2259–2268. doi: 10.1242/jcs.162974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, III, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Basi G, Schmid E, Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 1993;123:131–136. doi: 10.1016/0378-1119(93)90552-e. [DOI] [PubMed] [Google Scholar]
- Becalska AN, Kelley CF, Berciu C, Stanishneva-Konovalova TB, Fu X, Wang S, Sokolova OS, Nicastro D, Rodal AA. Formation of membrane ridges and scallops by the F-BAR protein Nervous Wreck. Mol. Biol. Cell. 2013;24:2406–2418. doi: 10.1091/mbc.E13-05-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert KA, Gould KL. Cytokinesis-based constraints on polarized cell growth in fission yeast. PLoS Genet. 2012;8:e1003004. doi: 10.1371/journal.pgen.1003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson BR, Lloyd KE, Kruszewski A, Kim I-H, Rodriguiz RM, Heindel C, Faytell M, Dudek SM, Wetsel WC, Soderling SH. WRP/srGAP3 facilitates the initiation of spine development by an inverse F-BAR domain, and its loss impairs long-term memory. J. Neurosci. 2011;31:2447–2460. doi: 10.1523/JNEUROSCI.4433-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnahan RH, Gould KL. The PCH family protein, Cdc15p, recruits two F-actin nucleation pathways to coordinate cytokinetic actin ring formation in Schizosaccharomyces pombe. J. Cell Biol. 2003;162:851–862. doi: 10.1083/jcb.200305012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortés JC, Pujol N, Sato M, Pinar M, Ramos M, Moreno B, Osumi M, Ribas JC, Pérez P. Cooperation between paxillin-like protein Pxl1 and glucan synthase Bgs1 is essential for actomyosin ring stability and septum formation in fission yeast. PLoS Genet. 2015;11:e1005358. doi: 10.1371/journal.pgen.1005358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeter J, Sazer S. imp2, a new component of the actin ring in the fission yeast Schizosaccharomyces pombe. J. Cell Biol. 1998;143:415–427. doi: 10.1083/jcb.143.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edeling MA, Sanker S, Shima T, Umasankar PK, Höning S, Kim HY, Davidson LA, Watkins SC, Tsang M, Owen DJ, Traub LM. Structural requirements for PACSIN/Syndapin operation during zebrafish embryonic notochord development. PLoS One. 2009;4:e8150. doi: 10.1371/journal.pone.0008150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Reymond A, Cerutti L, Utzig S, Hofmann K, Simanis V. The S. pombe cdc15 gene is a key element in the reorganization of F-actin at mitosis. Cell. 1995;82:435–444. doi: 10.1016/0092-8674(95)90432-8. [DOI] [PubMed] [Google Scholar]
- Frost A, Perera R, Roux A, Spasov K, Destaing O, Egelman EH, De Camilli P, Unger VM. Structural basis of membrane invagination by F-BAR domains. Cell. 2008;132:807–817. doi: 10.1016/j.cell.2007.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost A, Unger VM, De Camilli P. The BAR domain superfamily: membrane-molding macromolecules. Cell. 2009;137:191–196. doi: 10.1016/j.cell.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse-Kunstleve RW, Adams PD. Substructure search procedures for macromolecular structures. Acta Crystallogr. D Biol. Crystallogr. 2003;59:1966–1973. doi: 10.1107/s0907444903018043. [DOI] [PubMed] [Google Scholar]
- Guerrier S, Coutinho-Budd J, Sassa T, Gresset A, Jordan NV, Chen K, Jin W-L, Frost A, Polleux F. The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell. 2009;138:990–1004. doi: 10.1016/j.cell.2009.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne WM, Kent HM, Ford MGJ, Hegde BG, Daumke O, Butler PJG, Mittal R, Langen R, Evans PR, McMahon HT. Structure and analysis of FCHo2 F-BAR domain: a dimerizing and membrane recruitment module that effects membrane curvature. Structure. 2007;15:839–852. doi: 10.1016/j.str.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Erdmann KS, Roux A, Habermann B, Werner H, De Camilli P. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev. Cell. 2005;9:791–804. doi: 10.1016/j.devcel.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Keeney JB, Boeke JD. Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics. 1994;136:849–856. doi: 10.1093/genetics/136.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley CF, Becalska AN, Berciu C, Nicastro D, Rodal AA. Assembly of actin filaments and microtubules in Nwk F-BAR-induced membrane deformations. Commun. Integr. Biol. 2015;8:e1000703. doi: 10.1080/19420889.2014.1000703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D, Coffman VC, Lee I-J, Wu J-Q. Assembly and architecture of precursor nodes during fission yeast cytokinesis. J. Cell Biol. 2011;192:1005–1021. doi: 10.1083/jcb.201008171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-García R, Coll PM, Pérez P. F-BAR domain protein Rga7 collaborates with Cdc15 and Imp2 to ensure proper cytokinesis in fission yeast. J. Cell Sci. 2014;127:4146–4158. doi: 10.1242/jcs.146233. [DOI] [PubMed] [Google Scholar]
- Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald NA, Vander Kooi CW, Ohi MD, Gould KL. Oligomerization but not membrane bending underlies the function of certain F-BAR proteins in cell motility and cytokinesis. Dev. Cell. 2015;35:725–736. doi: 10.1016/j.devcel.2015.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger F, Boehm ME, Hofmann A, Hub B, Zentgraf H, Lehmann WD, Pereira G. Phosphorylation-dependent regulation of the F-BAR protein Hof1 during cytokinesis. Genes Dev. 2011;25:875–888. doi: 10.1101/gad.622411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger F, Palani S, Hub B, Pereira G. Dual function of the NDR-kinase Dbf2 in the regulation of the F-BAR protein Hof1 during cytokinesis. Mol. Biol. Cell. 2013;24:1290–1304. doi: 10.1091/mbc.E12-08-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mim C, Unger VM. Membrane curvature and its generation by BAR proteins. Trends Biochem. Sci. 2012;37:526–533. doi: 10.1016/j.tibs.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moravcevic K, Alvarado D, Schmitz KR, Kenniston JA, Mendrola JM, Ferguson KM, Lemmon MA. Comparison of Saccharomyces cerevisiae F-BAR domain structures reveals a conserved inositol phosphate binding site. Structure. 2015;23:352–363. doi: 10.1016/j.str.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Morrell-Falvey JL, Ren L, Feoktistova A, Haese GD, Gould KL. Cell wall remodeling at the fission yeast cell division site requires the Rho-GEF Rgf3p. J. Cell Sci. 2005;118:5563–5573. doi: 10.1242/jcs.02664. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macro-molecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nishihama R, Schreiter JH, Onishi M, Vallen EA, Hanna J, Moravcevic K, Lippincott MF, Han H, Lemmon MA, Pringle JR, Bi E. Role of Inn1 and its interactions with Hof1 and Cyk3 in promoting cleavage furrow and septum formation in S. cerevisiae. J. Cell Biol. 2009;185:995–1012. doi: 10.1083/jcb.200903125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P, Thuriaux P, Nasmyth K. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 1976;146:167–178. doi: 10.1007/BF00268085. [DOI] [PubMed] [Google Scholar]
- Oh Y, Schreiter J, Nishihama R, Wloka C, Bi E. Targeting and functional mechanisms of the cytokinesis-related F-BAR protein Hof1 during the cell cycle. Mol. Biol. Cell. 2013;24:1305–1320. doi: 10.1091/mbc.E12-11-0804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi M, Li Y, Cheng Y, Walz T. Negative staining and image classification—powerful tools in modern electron microscopy. Biol. Proced. Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Qualmann B, Koch D, Kessels MM. Let's go bananas: revisiting the endocytic BAR code. EMBO J. 2011;30:3501–3515. doi: 10.1038/emboj.2011.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao Y, Ma Q, Vahedi-Faridi A, Sundborger A, Pechstein A, Puchkov D, Luo L, Shupliakov O, Saenger W, Haucke V. Molecular basis for SH3 domain regulation of F-BAR-mediated membrane deformation. Proc. Natl. Acad. Sci. USA. 2010;107:8213–8218. doi: 10.1073/pnas.1003478107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reider A, Barker SL, Mishra SK, Im YJ, Maldonado-Báez L, Hurley JH, Traub LM, Wendland B. Syp1 is a conserved endocytic adaptor that contains domains involved in cargo selection and membrane tubulation. EMBO J. 2009;28:3103–3116. doi: 10.1038/emboj.2009.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren L, Willet AH, Roberts-Galbraith RH, McDonald NA, Feoktistova A, Chen J-S, Huang H, Guillen R, Boone C, Sidhu SS, et al. The Cdc15 and Imp2 SH3 domains cooperatively scaffold a network of proteins that redundantly ensure efficient cell division in fission yeast. Mol. Biol. Cell. 2015;26:256–269. doi: 10.1091/mbc.E14-10-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts-Galbraith RH, Gould KL. Setting the F-BAR: functions and regulation of the F-BAR protein family. Cell Cycle. 2010;9:4091–4097. doi: 10.4161/cc.9.20.13587. [DOI] [PubMed] [Google Scholar]
- Roberts-Galbraith RH, Chen J-S, Wang J, Gould KL. The SH3 domains of two PCH family members cooperate in assembly of the Schizosaccharomyces pombe contractile ring. J. Cell Biol. 2009;184:113–127. doi: 10.1083/jcb.200806044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts-Galbraith RH, Ohi MD, Ballif BA, Chen J-S, McLeod I, McDonald WH, Gygi SP, Yates JR, III, Gould KL. Dephosphorylation of F-BAR protein Cdc15 modulates its conformation and stimulates its scaffolding activity at the cell division site. Mol. Cell. 2010;39:86–99. doi: 10.1016/j.molcel.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider K, Seemann E, Liebmann L, Ahuja R, Koch D, Westermann M, Hübner CA, Kessels MM, Qualmann B. ProSAP1 and membrane nanodomain-associated syndapin I promote postsynapse formation and function. J. Cell Biol. 2014;205:197–215. doi: 10.1083/jcb.201307088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada A, Niwa H, Tsujita K, Suetsugu S, Nitta K, Hanawa-Suetsugu K, Akasaka R, Nishino Y, Toyama M, Chen L, et al. Curved EFC/F-BAR-domain dimers are joined end to end into a filament for membrane invagination in endocytosis. Cell. 2007;129:761–772. doi: 10.1016/j.cell.2007.03.040. [DOI] [PubMed] [Google Scholar]
- Stachowiak JC, Schmid EM, Ryan CJ, Ann HS, Sasaki DY, Sherman MB, Geissler PL, Fletcher DA, Hayden CC. Membrane bending by protein-protein crowding. Nat. Cell Biol. 2012;14:944–949. doi: 10.1038/ncb2561. [DOI] [PubMed] [Google Scholar]
- Suetsugu S, Toyooka K, Senju Y. Subcellular membrane curvature mediated by the BAR domain superfamily proteins. Semin. Cell Dev. Biol. 2010;21:340–349. doi: 10.1016/j.semcdb.2009.12.002. [DOI] [PubMed] [Google Scholar]
- Taylor MJ, Perrais D, Merrifield CJ. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 2011;9:e1000604. doi: 10.1371/journal.pbio.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Adams PD, Read RJ, Zwart PH, Hung LW. Iterative-build OMIT maps: map improvement by iterative model building and refinement without model bias. Acta Crystallogr. D Biol. Crystallogr. 2008;64:515–524. doi: 10.1107/S0907444908004319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujita K, Suetsugu S, Sasaki N, Furutani M, Oikawa T, Takenawa T. Coordination between the actin cytoskeleton and membrane deformation by a novel membrane tubulation domain of PCH proteins is involved in endocytosis. J. Cell Biol. 2006;172:269–279. doi: 10.1083/jcb.200508091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullal P, McDonald NA, Chen J-S, Lo Presti L, Roberts-Galbraith RH, Gould KL, Martin SG. The DYRK-family kinase Pom1 phosphor-ylates the F-BAR protein Cdc15 to prevent division at cell poles. J. Cell Biol. 2015;211:653–668. doi: 10.1083/jcb.201504073. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang Q, Navarro MVAS, Peng G, Molinelli E, Goh SL, Judson BL, Rajashankar KR, Sondermann H. Molecular mechanism of membrane constriction and tubulation mediated by the F-BAR protein Pacsin/Syndapin. Proc. Natl. Acad. Sci. USA. 2009;106:12700–12705. doi: 10.1073/pnas.0902974106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willet AH, McDonald NA, Bohnert KA, Baird MA, Allen JR, Davidson MW, Gould KL. The F-BAR Cdc15 promotes contractile ring formation through the direct recruitment of the formin Cdc12. J. Cell Biol. 2015;208:391–399. doi: 10.1083/jcb.201411097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J-Q, Pollard TD. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- Wu J-Q, Kuhn JR, Kovar DR, Pollard TD. Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev. Cell. 2003;5:723–734. doi: 10.1016/s1534-5807(03)00324-1. [DOI] [PubMed] [Google Scholar]
- Yu H, Schulten K. Membrane sculpting by F-BAR domains studied by molecular dynamics simulations. PLoS Comput. Biol. 2013;9:e1002892. doi: 10.1371/journal.pcbi.1002892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Chen JK, Feng S, Dalgarno DC, Brauer AW, Schreiber SL. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell. 1994;76:933–945. doi: 10.1016/0092-8674(94)90367-0. [DOI] [PubMed] [Google Scholar]
- Zhao H, Michelot A, Koskela EV, Tkach V, Stamou D, Drubin DG, Lappalainen P. Membrane-sculpting BAR domains generate stable lipid microdomains. Cell Rep. 2013;4:1213–1223. doi: 10.1016/j.celrep.2013.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.