Abstract
Follicular lymphoma is an incurable B-cell malignancy1 characterized by the t(14;18) and mutations in one or more components of the epigenome2,3. Whilst frequent gene mutations in signaling pathways, including JAK-STAT, NOTCH and NF-κB, have also been defined2–7, the spectrum of these mutations typically overlap with the closely-related diffuse large B cell lymphoma (DLBCL)6–13. A combination of discovery exome and extended targeted sequencing revealed recurrent somatic mutations in RRAGC uniquely enriched in FL patients (17%). More than half of the mutations preferentially co-occurred with ATP6V1B2 and ATP6AP1 mutations, components of the vacuolar H+-adenosine triphosphate ATPase (v-ATPase) known to be necessary for amino acid-induced mTORC1 activation. The RagC mutants increased raptor binding whilst rendering mTORC1 signaling resistant to amino acid deprivation. Collectively, the activating nature of the RRAGC mutations, their existence within the dominant clone and stability during disease progression supports their potential as an excellent candidate to be therapeutically exploited.
Follicular lymphoma (FL) is one of the commonest non-Hodgkin’s lymphomas (NHLs). Whilst the majority of affected individuals exhibit a characteristic protracted disease course with multiple relapses, others develop aggressive disease and histological transformation with shortened overall survival. Genome-wide profiling studies have primarily focused on single time-point analyses or the subset of patients that have undergone histological transformation in order to determine the genetic mediators of progression2,3. To gain further insight into the genetic diversity of FL, we undertook temporal analyses on individuals diagnosed with FL that underwent several relapse episodes without transformation. These data uncovered recurrent mutations in components of the mTORC1 signaling pathway, specific to FL.
Exome sequencing was performed on 24 tumors (from 5 patients) and matched constitutional DNA, with an average sequencing depth of 140x and 97.5% of the targeted bases covered by >10-fold (Online Methods and Supplementary Table 1). The clinical course from diagnoses to last follow-up ranged from 12.5 to 25 years (Supplementary Table 2 and Supplementary Fig. 1). A median of 94 non-synonymous mutations per tumor were identified and validated mutations of interest by a combination of Sanger and tagged-amplicon sequencing (Supplementary Tables 3 and 4). Consistent with our earlier longitudinal study of paired FL and transformed FL2, tumors from the same individual confirmed a branched evolutionary pattern and demonstrated that all tumors evolve from a dominant ancestral clone (Supplementary Fig. 2). Moreover, mutations in KMT2D, CREBBP and MEF2B were present on the trunks of the phylogenetic trees in all five individuals, consistent with the role of epigenetic deregulation as critical early events in the majority of FLs2,3,14,15.
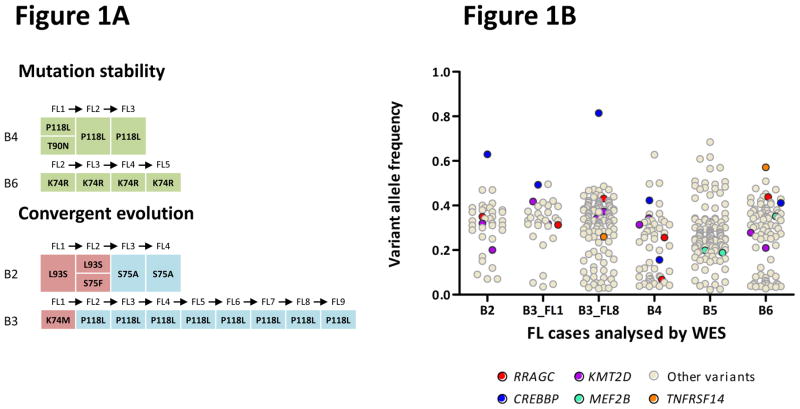
Remarkably, our data disclosed a novel finding of somatic non-silent mutations in the gene RRAGC, which encodes a Ras-related GTP-binding protein (RagC), occurring in four of the five cases. Notably, in cases B4 and B6, the RRAGC mutations (p.Pro118Leu and p.Lys74Arg) were conserved during disease progression whereas in cases B2 and B3, a convergent pattern of clonal selection was seen with different mutations occurring at different time points in the disease evolution (Fig. 1a). Copy number variation were rarely observed at the RRAGC locus, 1p34.3, in both our current data and previous single-nucleotide polymorphism (SNP) array datasets2 (Supplementary Fig. 3). These together with the RRAGC variant allele frequencies (VAF) were consistent with heterozygous mutations (VAF range: 0.17–0.5), whilst clonality plots verified that the VAFs were comparable to those of early driver mutations demonstrating that the RRAGC mutations reside within the dominant clone of the tumor biopsies (Fig. 1b).
Figure 1.
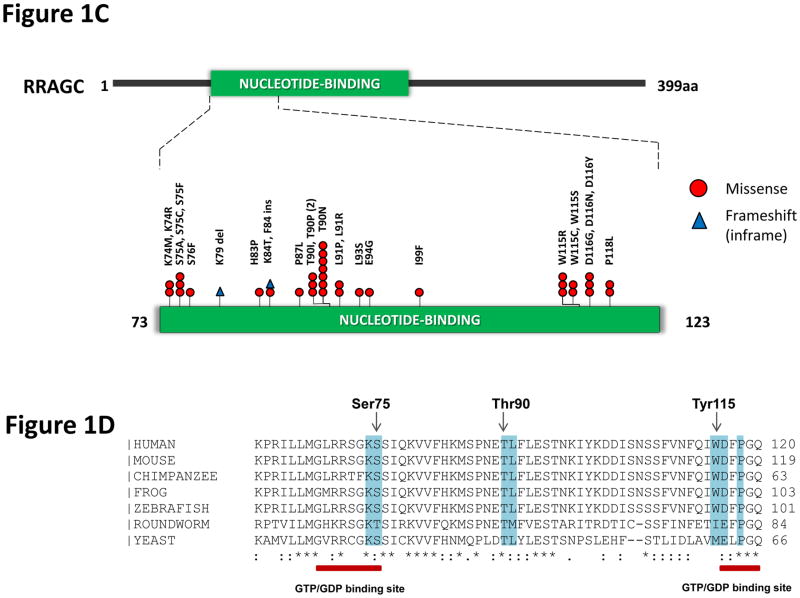
Identification of frequent RRAGC mutations in FL. (a) RRAGC mutations show two different patterns of conservation in successive tumor biopsies during FL progression in the discovery WES cases: mutation stability and convergent evolution. (b) Variant allele frequency (VAF) distribution and density for all the non-synonymous mutations identified in the 5 WES cases. In each case, the first available biopsy is depicted, with the exception of B3 where two time points are illustrated (B3_FL1 and B3_FL8). (c) Schema of the protein domain and locations of the RRAGC mutations identified in this study (NCBI protein reference sequence: NP_071440.1). Thirty-seven mutations affecting 32 cases. ‘^’ denotes a second RRAGC mutation occurring in a different disease event from the same patient. ‘*’ represents a second RRAGC mutation within the same biopsy of a particular patient. Multiple circles for the same amino acid represent multiple cases with mutations affecting the same residue. (d) Sequence alignment of a section of the RRAGC nucleotide binding domain. Conserved residues across all the listed species is indicated by an asterisk (*). The location of the GTP/GDP binding sites are indicated by the red horizontal bar (locations: aa 68–75; and aa 116–120) whilst the recurrent hotspot residues are highlighted by the light blue vertical panel.
To determine the prevalence of RRAGC mutations, targeted sequencing was performed in an extension cohort of 141 FL samples (including the original 5 cases) and 32 cases with paired transformed FL. RRAGC mutations were present in 17% of cases (Table 1). The mutations were predominantly missense, with exception of two in-frame frameshift mutations, restricted to exons 1 and 2 (Fig. 1c and Supplementary Table 5). The clustering of mutations corresponded to the nucleotide-binding domain with hotspots centering on amino acids p.Ser75, p.Thr90, p.Try115, p.Asp116 and p.Pro118, residues highly conserved between species (Fig. 1c and Fig. 1d). In 10 patients with constitutional DNA, the somatic nature of the mutations was confirmed. To investigate the full complement of RRAGC mutations in other malignancies, we performed Sanger sequencing, restricting our analyses to exon 1 and 2, in a further 329 related mature B-cell NHLs and 51 B-cell lymphoma cell lines alongside an analysis of publically available sequencing datasets. RRAGC mutations were absent in other hematological malignancies, including myeloid and other mature B-cell NHL entities (Table 1) with the exception of infrequent mutations in the closely-related DLBCL. We found that RRAGC was rarely mutated in non-hematological neoplasms (0.3% of nearly 10,000 samples) included in the Cancer Genomics database (cBioportal)21, with the majority of mutations arising in residues beyond p.Pro118 (Supplementary Table 6). RRAGC mutations are therefore highly enriched in FL with their nature and frequency suggesting that the changes are likely to be functionally relevant in this lymphoma.
Table 1.
Frequency of RRAGC mutations in lymphoma and other hematological malignancies
| Tumor type | Occurrence No./total No. | Frequency |
|---|---|---|
| FL | 25/141 | 17.7% |
| Diagnostic | 13/94 | 13.8% |
| Relapse | 12/47 | 25.5% |
| FL and tFL pairs | 6/32# | 18.8% |
| DLBCL^ | 3/174 | 1.7% |
| GCB | 1/67 | 1.5% |
| ABC | 1/43 | 2.3% |
| PMBL | 1/29 | 3.4% |
| U | 0/35 | 0% |
| DLBCL* | 1/185 | 0.5% |
| B cell lymphomas | ||
| Burkitt lymphoma* | 0/42 | 0% |
| CLL/SLL^ | 0/96 | 0% |
| CLL* | 0/258 | 0% |
| MCL* | 0/29 | 0% |
| SMZL^ | 0/48 | 0% |
| Other B-cell lymphomas | 0/48 | 0% |
| Cell lines (B-NHL) | 2/51 | 3.9% |
| Other hematological malignancies | ||
| AML* | 0/200 | 0% |
| CML* | 0/129 | 0% |
| MM* | 0/203 | 0% |
| Benign reactive lymph nodes | 0/10 | 0% |
tFL, transformed follicular lymphoma; DLBCL, diffuse large B cell lymphoma; GCB, germinal center B-cell subtype DLBCL defined by gene-expression profiling (GEP); ABC, activated B-cell subtype of DLBCL defined by GEP; PMBL, primary mediastinal B-cell lymphoma defined by GEP; U, unclassifiable by GEP; CLL, chronic lymphocytic leukemia; MCL, mantle cell lymphoma; SMZL, splenic marginal zone lymphoma; AML, acute myeloid leukemia; CML, chronic myeloid leukemia; MM, multiple myeloma.
comprising 5 cases with mutations in both FL and tFL and 1 case with only the tFL sample
Sanger sequencing restricted to RRAGC exon 1 and 2
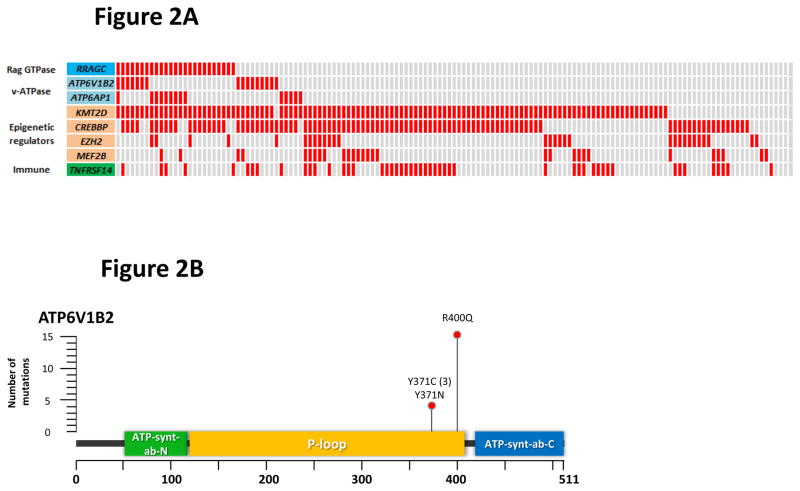
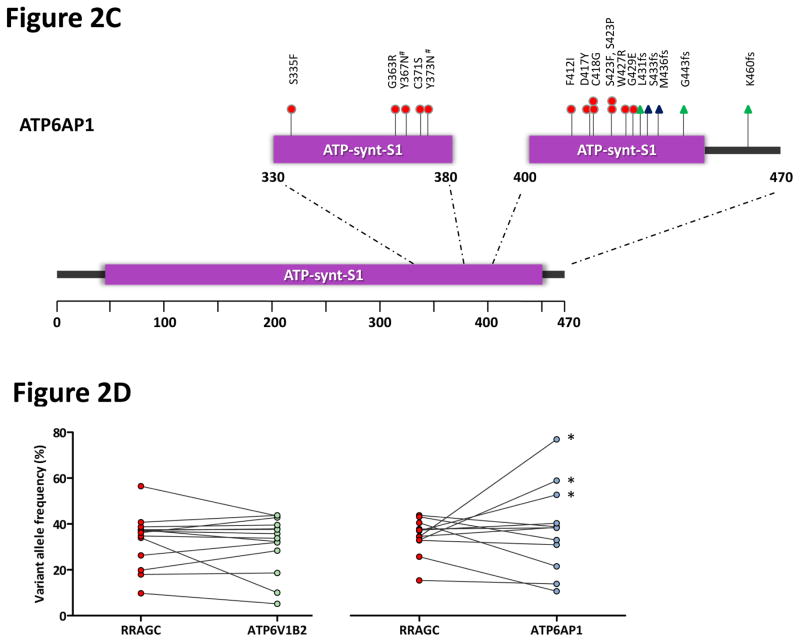
RagC is one of four members of the Rag GTPase family in mammals, which form obligate heterodimers between RagA/B and RagC/D22,23. The Rag GTPases form a supercomplex on the lysosomal surface with Ragulator, the v-ATPase and SLC38A9 (Supplementary Fig. 4), essential for inducing mechanistic target of rapamycin complex 1 (mTORC1) activation in response to amino acid sufficiency24-31. Other members of the Rag GTPase family and mTORC1 components (RRAGA, RRAGB, RRAGD, MTOR, RPTOR and MLST8) were infrequently mutated (Supplementary Table 5). To expand this search, we examined our FL datasets to ascertain if other regulatory complexes upstream and downstream of Rag GTPases were subject to genetic aberrations. This approach uncovered mutations in two subunits of the v-ATPase complex, ATP6V1B2 and ATP6AP1. The v-ATPase complex resides within intracellular compartments such as the lysosome and is composed of two domains, a cytosolic V1 domain responsible for ATP hydrolysis and a transmembrane V0 that enables proton translocation32. ATP6V1B2 is a non-catalytic subunit within the V1 domain, and ATP6AP1 is thought to be an accessory subunit that regulates the function of the v-ATPase complex33. To assess the relationship between these v-ATPase subunit mutations, RRAGC and FL-associated genes, we resequenced our extension cohort of 141 FL cases identifying 11.3% and 9.9% of cases with ATP6V1B2 and ATP6AP1 mutations, respectively (Fig. 2a and Supplementary Table 7, 8). Interestingly, mutations in RRAGC, ATP6V1B2 and ATP6AP1 showed strong correlations with more than half of the RRAGC mutations co-occurring with either ATP6V1B2 or ATP6AP1 (Fisher’s exact test, P < 0.0001), whereas ATP6V1B2 and ATP6AP1 mutations were mutually exclusive. Mutations in ATP6V1B2 were all missense, with a hotspot at c.1199G>A; p.Arg400Gln representing 80% of all mutations detected (Fig. 2b). In comparison, mutations in ATP6AP1 included both missense and frameshift mutations most localizing to the C-terminal end of the ATP-synthase domain (Fig. 2c). Deep targeted sequencing (mean coverage 4655x) of 15 co-mutated cases demonstrated no definitive hierarchy in mutation order of RRAGC and ATP6V1B2 or ATP6AP1 suggesting that both alterations are acquired concomitantly during similar clonal selective sweeps (Fig. 2d). Together, over a quarter of patients (27.4%; 39 of 141 cases) had mutations in one or more of the three genes.
Figure 2.
Frequent and co-occurring mutations in ATP6V1B2 and ATP6AP1. (a) The heatmap shows the distribution of mutations in RRAGC, ATP6V1B2, ATP6AP1 and other known FL-associated genes in 141 FL cases. Each column represents an individual case and each row denotes a specific gene. Red indicates the presence of mutations, and light grey indicates the absence. (b) Schema of the protein domain and locations of the identified ATP6V1B2 mutations (NCBI protein reference sequence: NP_001684.2). (c) Schema of the protein domain and locations of the identified ATP6AP1 mutations (NCBI protein reference sequence: NP_001174.2). Red circles represent missense mutations, blue triangles are in-frame indels and green triangles are out-of-frame indels. The ‘#’ indicates mutations occurring in the same case. (d) Comparison of the allele frequencies in 26 co-mutated (RRAGC vs. ATP6V1B2 and RRAGC vs. ATP6AP1) samples (comprising 15 cases, some with multiple biopsies). Male cases are marked with an asterisk (*) and demonstrate expected increases in allelic frequencies of ATP6AP1 mutations as the gene locus resides on the X chromosome.
To begin understanding the pathogenic role of RRAGC mutations, we examined RNA-seq data in 13 FL cases (Supplementary Table 9) and identified 257 differentially expressed genes between mutated and wild-type cases (Supplementary Fig. 5a). There was no difference in RRAGC expression between mutated and wild-type cases (Supplementary Fig. 5b). Gene set enrichment analyses (GSEA) showed that RRAGC-mutated cases were characterized by up-regulated expression for gene sets involved in translation regulation, well-known downstream mTOR targets34,35 (Supplementary Fig. 6 and Supplementary Table 10), implicating altered signaling as a consequence of these mutations.
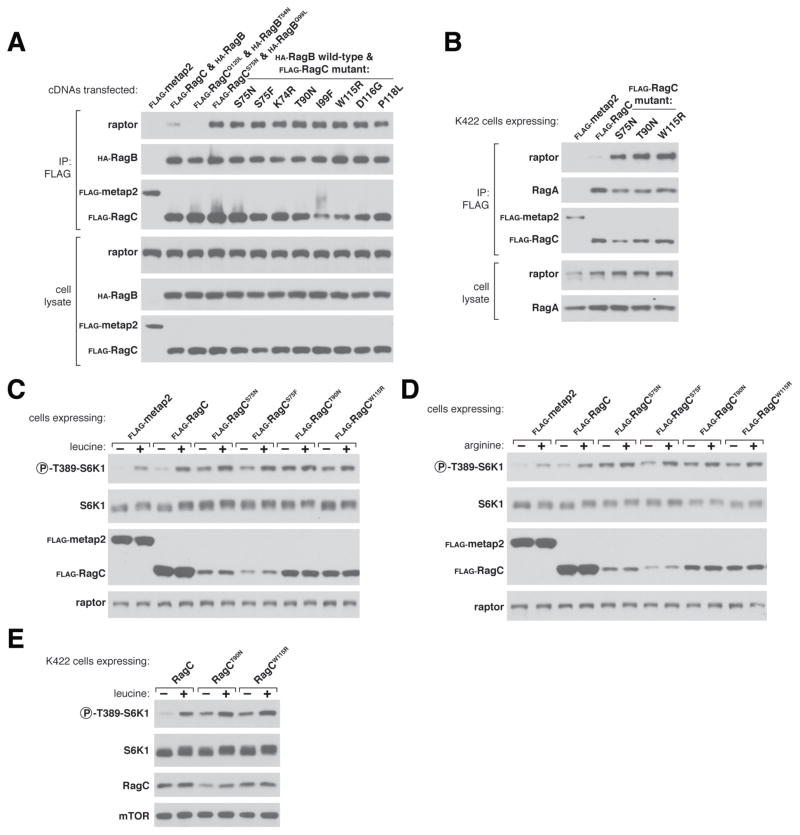
The direct binding of Rag GTPase heterodimers to mTORC1 is a key event in the activation of mTORC1 by amino acids23–29. Under these conditions, the active Rag heterodimer, composed of GTP-loaded RagA/B bound to GDP-loaded RagC/D, directly interacts with raptor (Supplementary Fig. 4), a component of mTORC125. We first assessed the effects of 8 RagC mutants detected in our FL series (p.Lys74Arg, p.Ser75Asn, p.Ser75Phe, p.Thr90Asn, p.Ile99Phe, p.Tyr115Arg, p.Asp116Gly and p.Pro118Leu) on their raptor binding capacity by co-expressing each RagC mutant (RagCmut) together with wild-type RagB in HEK-293T cells. These RagB-RagCmut heterodimers co-immunoprecipitated substantially more endogenous raptor than a fully wild-type RagB-RagC heterodimer (Fig. 3a). Importantly, the increased raptor binding was specific to the identified FL RRAGC mutations, as RagC mutations in other cancer types did not demonstrate the same capacity (Supplementary Fig. 7a). Interestingly, the increased raptor binding observed with the RagCmut was similar to that seen with RagC p.Ser75Asn, a previously characterized mutant with decreased affinity for GTP that therefore functions like a ‘GDP-bound’ mutant, mimicking the RagC conformation that is necessary for mTORC1 activation by amino acids25, 36–39 (Fig. 3a). The similar increase in raptor binding caused by the RagC mutants and RagC p.Ser75Asn provided the first indication that the RRAGC mutations observed in FL are likely activating. To examine the impact of these mutations in B-cell lymphomas, we stably expressed three recurring FL mutants (p.Ser75Phe, p.Thr90Asn and p.Trp115Arg) and the ‘GDP-binding’ mutant, RagC p.Ser75Asn, in four germinal center NHL cell lines (Karpas-422, Raji, OCI-Ly7 and OCI-Ly8) and these reaffirmed our findings of increased raptor binding over wild-type RagC (Fig. 3b and Supplementary Fig. 7b–d). To test the effects of the RagC mutants on mTORC1 signaling, we stably expressed these four mutants in HEK-293T cells. Overexpression of all four mutants increased mTORC1 activity, even under complete amino acid deprivation as detected by the phosphorylation of S6 kinase 1 (S6K1), an established mTORC1 substrate (Supplementary Fig. 7e). Furthermore, all RagCmuts tested rendered mTORC1 signaling partially or fully insensitive to leucine or arginine deprivation, amino acids of particular importance for mTORC1 pathway activation 24, 40, 41 (Figs. 3c, 3d and Supplementary Fig. 7f, 7g). Similarly, in Karpas-422 cells, overexpression of two mutants, p.Thr90Asn and p.Trp115Arg, but not wild-type RagC, led to increased mTORC1 signaling in the absence of leucine, validating the mTORC1 activating properties of the RagC mutants (Fig. 3e and Supplementary Fig. 7h).
Figure 3.
Effects of RagC mutants on mTORC1 signaling. (a) Rag heterodimers containing the RagC mutants co-immunoprecipitate the largest amounts of raptor, an mTORC1 component, similar to the previously characterized RagCS75N mutant. Anti-FLAG immunoprecipitates were collected from HEK-293T cells transiently expressing the indicated cDNAs, and cell lysates and immunoprecipitates were analyzed by immunoblotting. RagBQ99L and RagCQ120L are ‘GTP-locked’ mutants36, 47, while RagCS75N and RagBT54N function as ‘GDP-binding’ mutants25, 36–39. (b) Two recurrent mutants from FL, RagCT90N and RagCW115R, and the previously characterized RagCS75N mutant co-immunoprecipitate more endogenous raptor than wild-type RagC in Karpas-422 cells, a B cell lymphoma line of germinal center origin. Anti-FLAG immunoprecipitates from Karpas-422 cells stably expressing the indicated proteins were collected and analyzed as in (a). (c) Stable overexpression of RagCS75N, RagCS75F, RagCT90N, and RagCW115R render cells partially or fully insensitive to leucine deprivation. HEK-293T cells stably expressing the indicated proteins were starved of leucine for 50 minutes and re-stimulated with leucine for 10 minutes. Cell lysates were analyzed by immunoblotting for the indicated proteins. (d) Stable overexpression of the indicated RagC mutants leads to an increase in mTORC1 signaling in the absence of arginine. HEK-293T cells stably expressing the indicated proteins were starved of arginine for 50 minutes, restimulated with arginine for 10 minutes, and analyzed as in (b). (e) Stable overexpression of the indicated RagC mutants, but not wild-type RagC, leads to increased mTORC1 signaling in the absence of leucine in a B cell lymphoma line. Karpas-422 cells stably expressing the indicated proteins were treated and analyzed as in (b).
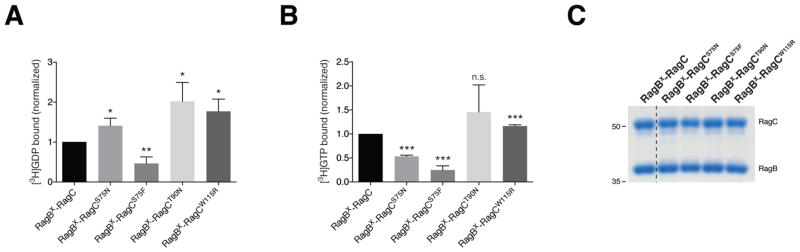
To determine if RagC mutations affect their capacity to bind guanine nucleotides, we employed a specific in vitro assay in which nucleotide binding to purified Rag heterodimers could be assessed by purifying wild-type and mutant RagC in complex with a RagB mutant (p.Asp163Asn) that preferentially binds to xanthosine nucleotides. Employing this RagB mutant allowed us to measure guanine nucleotide binding to the RagC mutant only, even in the presence of RagB42,43. Two classes of RagC mutants emerged from this analysis. One class, including RagC Ser75Asn and Ser75Phe, had significantly decreased affinity for GTP in comparison to wild-type RagC, and a preference for binding GDP over GTP (Figs. 4a–b). These mutants are analogous to the Ras Ser17Asn mutant, which disrupts coordination of the magnesium cofactor leading to decreased affinity for all nucleotides37,38. Interestingly, this Ras mutant suppresses signaling, not through decreased GTP binding, but rather through its high affinity for ras guanine nucleotide exchange factors (GEFs), thus preventing guanine nucleotide exchange on wild-type Ras38, 44, 45. Further studies will be needed to uncover if the same mechanism is true for the RagC mutants, as a GEF for RagC has yet to be identified. The second class of mutants, Thr90Asn and Trp115Arg, display a slight preference for GDP binding over GTP, without an overall decrease in GTP binding in comparison to wild-type RagC (Fig. 4a, b). While the relative nucleotide affinity of these mutants is biased towards GDP, this may not account for their signaling effects in cells, as intracellular GTP concentrations are 10–20 times higher than those of GDP46. Importantly, the levels of RagC were consistent in our assays indicating that the variation in nucleotide binding cannot be accounted for by differences in protein levels (Fig. 4c). While this second class of mutants may activate the mTORC1 pathway through mechanisms not involving changes in nucleotide loading, further work is needed to uncover the exact mechanism through which they lead to increased raptor binding.
Figure 4.
RagC mutants alter the affinity of nucleotide binding. (a) Three RagC mutants exhibit increased GDP binding, while RagCS75F decreased GDP binding. Nucleotide binding assays were performed with the indicated RagC heterodimer incubated with [3H]GDP and binding assessed using a filter-binding assay. Each value represents the normalized mean +/− SD for n = 3. Statistical differences are assessed comparing each sample to the binding observed with the RagB-RagC wild-type heterodimer. (b) RagCS75N and RagCS75F significantly decrease GTP binding, while RagCT90N does not affect GTP binding with RagCW115R slightly increasing this activity. Nucleotide binding assays were performed as in (B) but incubated with [3H]GTP. Each value represents the normalized mean +/− SD for n = 3. Statistical differences are assessed comparing each sample to the binding observed with the RagB-RagC wild-type heterodimer. (c) RagB and RagC protein levels in the nucleotide binding assays are consistent. Aliquots of the purified Rag heterodimers used in the nucleotide binding assays were resolved on an SDS-PAGE gel and stained with Coomassie. *P<0.05; **P<0.01; ***P<0.005.
As the v-ATPase complex is functionally linked with the Rag GTPases and Ragulator in sensing amino acids and activating mTORC1 signaling29, the co-existence of RRAGC with either ATP6V1B2 or ATP6AP1 mutations raises the question of a functional epistasis. ATP-hydrolysis and the v-ATPase rotator conformation are crucial for relaying the amino acid signal from the lysosomal lumen to the Rag GTPases29 and therefore, our working hypothesis is that mutations in these v-ATPase subunits help convey a ‘false’ amino acid sufficiency signal or alter interactions between v-ATPase, Ragulator and the Rag GTPases, which requires experimental clarification.
In conclusion, our study identifies frequent mutations in components of the lysosome-centric mTORC1 signaling cascade in FL. We demonstrate that RRAGC mutants confer a gain-of-function mechanism by bypassing the amino acid deprivation state to activate mTORC1 signaling. Together, the emergence of frequent activating RRAGC mutations that are clonally represented and maintained during progression is particularly valuable and might be exploited as a therapeutic target, however, its utility as a predictive biomarker of mTOR inhibitor sensitivity warrants further investigation.
ONLINE METHODS
Patients and samples
Samples were obtained from individuals with FL and non-FL tumors following approval from the Institutional Review Board and local ethics committee of all participating centers. Informed written consent was obtained from all individuals. The discovery cohort (Supplementary Table 2 and Supplementary Fig. 1) comprised five patients who had not undergone histologic transformation and selected on the basis of available fresh frozen tumor lymph node biopsies, matched constitutional DNA and samples from multiple disease episodes. The clonality between tumor biopsies obtained from multiple disease episodes from an individual patient were confirmed by BCL2-IGH breakpoint analysis as previously described2. The FL validation cohort (Supplementary Table 11) comprised either diagnostic or relapse FL (n = 141 cases) or paired FL-tFL tumor biopsies (n=32 cases) obtained from two centers (Barts Cancer Institute and University of Southampton). The clinical characteristics of the cohort are shown in Supplementary Table 12. Non-FL tumors for validation included DNA from 174 DLBCL, 96 CLL and 48 SMZL tumors. Histology of all tumors was confirmed by pathological review.
Whole exome sequencing and analysis
Whole exome capture libraries were constructed from 2–3ug of tumor or constitutional DNA after shearing, end repair, phosphorylation and ligation to barcoded sequencing adaptor, using the Human All Exon V5 SureSelect XT (Agilent Technologies). Enriched exome libraries were multiplexed and sequenced on the HiSeq 2500 (Illumina) to generate 100bp paired-end reads. Sequencing metrics are provided in Supplementary Table 1. The processing and analysis of whole exome sequencing data were performed using our previous pipeline2. Briefly, sequencing reads were aligned to the reference genome hg19, using Burrows-Wheeler Aligner (BWA)48. Local alignments and base quality scores were adjusted using the Genome Analysis Toolkit (GATK)49 version 2.5.2.
Variant detection and mutation annotation
Somatic SNVs and indels were identified using the Strelka pipeline as previously described2. For each sample, the number of reads supporting the reference and variant alleles at each position was extracted. VAFs were calculated by dividing the number of supporting variant reads to the total reads obtained. To improve the variant calls across all the tumors from the same patient, identified variants were further genotyped and verified across all tumors and matched normal using VarScan2’s multi-sample calling method ‘mpileup2cns’50, based on reads with mapping quality > 30 and minimum base quality of 20 at the targeted site. Annotation of variants was attained using the SNPnexus tool51.
Sanger sequencing of genomic DNA
Direct bidirectional Sanger sequencing of RRAGC exon 1 and 2, ATP6V1B2 (all exons), ATP6AP1 (all exons) and ATP6AP2 (all exons) from genomic DNA from tumors and matched normal samples was performed following direct PCR amplification, using specific primers, and enzymatic clean up using Exo-Sap (USB Corporation).
Phylogenetic analyses
Evolutionary trees were reconstructed for each individual based on the distance matrix between GL, FL and relapse FL samples derived from the numbers of somatic non-synonymous variants from each biopsy, using the Neighbor-Joining algorithm52 implemented in the PHYLIP package as previously reported2. Once the consensus phylogenetic tree was determined, it was redrawn starting from GL leading to the putative CPC, then to FL and subsequent relapse FL samples, with the branch length proportional to the number of somatic changes i.e. genetic distance between the samples.
Copy number variation of the RRAGC locus
Copy number variation and copy neutral loss of heterozygosity for ATP6AP1, ATP6V1B2, RRAGC and TNFRSF14 gene loci were extracted from our previous SNP array analyses using the methodology previously described2. To detect copy number imbalances from our discovery WES data, VarScan2 “copynumber” module was first employed, using the minimum read coverage as 20, and both mapping and base qualities as ≥ 20 for usable reads, to generate raw copy number calls. Raw calls were adjusted for GC content and re-centered to 0 based on the modal LogR value determined by kernel density estimates, using VarScan2 “copyCaller” module. Outliers were identified and modified using the data winsorizing procedure. The DNAcopy R Bioconductor package (Seshan VE and Olshen A. DNAcopy: DNA copy number data analysis. R package version 1.40.0) was employed to identify joint segments of LogR values using the circular binary segmentation (CBS) algorithm. To identify regions of LOH, variants (including SNPs and short indels) against the reference genome were first identified for paired normal and tumour samples using VarScan2. Next, B-allele frequency (BAF) files were created, allowing the minimum read depth of 10 for both tumour and normal. The ASCAT R package53 was then used to assess CNAs and LOH regions, using the logR and BAF files derived from VarScan variant calls, with the depth information normalized by dividing the depth of each variant by the median depth across all variants.
Targeted sequencing of Rag GTPases and mTORC1-associated genes
Target-specific primers for FL-associated genes2 and 7 mTORC1-associated genes (RRAGA, RRAGB, RRAGC, RRAGD, MTOR, RPTOR and MLST8) were custom designed using Fluidigm’s D3 Assay design service. Targeted enrichment was performed by Access Array (Fluidigm) in a multiplex format using genomic DNA (50ng) according to the manufacturer’s Multiplex Amplicon Tagging Protocol. The multiplexed library pools were sequenced on the Illumina Miseq platform. All samples were screened in duplicate with the inclusion of normal tonsil DNA controls in each run. Variants were called and annotated as previously described2. In brief, reads were aligned to hg19 using BOWTIE254. SAMtools55 were used to generate sorted BAM files and the VarScan2 tool was used to examine the pileup file to call variants.
Deep tagged-amplicon sequencing for RRAGC, ATP6V1B2 and ATP6AP1 genes
Universal adapter sequences were tagged to the 5′ and 3′ end of target-specific primers of approximately 200±20bp in length. Based on our initial experiments that showed clustering of variants within specific exons of the 3 genes, subsequent analyses were restricted to RRAGC (exons 1 and 2), ATP6V1B2 (exons 11 and 12) and ATP6AP1 (exons 9 and 10). Primer sequences are shown in Supplementary Table 13. 100ng of genomic DNA were amplified in 2 to 4-plex PCR reactions using non-overlapping tagged-primers with the HotStar Taq Plus kit (Qiagen) under limited cycling conditions. Amplified PCR fragments were subsequently pooled in equimolar ratios by sample and prepared for sequencing with the attachment sample specific indexes and Illumina adaptor sequences. Indexed libraries were pooled and sequenced on a single lane of an Illumina MiSeq instrument using the V2 300-cycles Miseq reagent kit (Illumina) generating 150-bp paired end reads. Each sample was screened in duplicate. Variant calling and annotation are as described above.
RNA-sequencing analysis
RNA-seq data for all 13 FL samples (5 mutants and 8 wild-type) were downloaded from the International Cancer Genome Consortium (ICGC) data repository (see URLs). Details of the samples are summarized in Supplementary Table 9. Raw read counts for all annotated ENSEMBL genes across the 13 samples were extracted from the “exp_seq.MALY-DE.tsv” file in the ICGC data repository. Only genes that achieved at least one read count per million reads (CPM) in at least five samples were selected, with these criteria producing 22,126 filtered genes in total. After applying scale normalization, read counts were converted to log2-cpm using the voom function56 with associated weights ready for linear modeling. Differential gene expression (DGE) analyses between mutant and wild-type groups were further performed using the limma R package, which powers DGE analyses for RNA-seq and microarray data 57. A double threshold of raw p value <0.01 and an absolute fold change > 2 were used to define significantly differentially expressed (DE) genes (Supplementary Figure 5a). Based on the t-statistic of filtered genes from the Iimma test, GSEA was performed against the predefined curated gene sets (c2) acquired from the MSigDB collection58, including KEGG and Reactome gene sets. Top significantly enriched gene sets were selected based on FDR q-value <0.05 (Supplementary Table 10).
Materials
Reagents were obtained from the following sources: HRP-labeled anti-mouse and anti-rabbit secondary antibody from Santa Cruz Biotechnology; antibodies to phospho-T389 S6K1, S6K1, mTOR, and FLAG epitope from Cell Signaling Technology; antibody to the HA epitope from Bethyl laboratories; antibody to raptor from Millipore. RPMI, FLAG M2 affinity gel, GTP, GDP, and amino acids from Sigma Aldrich; XDP and XTP from Jena Biosciences; [3H]-labeled GTP and GDP from Perkin Elmer; DMEM from SAFC Biosciences; Complete Protease Cocktail from Roche; Inactivated Fetal Calf Serum (IFS) and simply blue stain from Invitrogen; amino acid-free RPMI from US Biologicals.
Cell lines and tissue culture
HEK-293T cells were cultured in DMEM 10% IFS supplemented with 2 mM glutamine, penicillin (100 IU/mL), and streptomycin (100 μg/mL). Karpas-422, Raji, OCI-Ly7, and OCI-Ly8 cells were cultured in RPMI 10% IFS supplemented with 2 mM glutamine, penicillin (100 IU/mL), and streptomycin (100 μg/mL). All cell lines were maintained at 37°C and 5% CO2.
Virus production and viral transduction
The production of lentiviruses was achieved by transfection of viral HEK-293T cells with pLJM60-FLAG-metap2 or pLJM60-FLAG-RagC (wild-type or mutant) constructs, with the VSV-G envelope and CMV ΔVPR packaging plasmids. Similarly, the production of retroviruses for infection of the Karpas-422 cells was achieved by transfection of viral HEK-293T cells with pMXs-RagC (wild-type or mutant) constructs, with the VSV-G envelope and gag/pol packaging plasmids. Twenty-four hours after transfection, the media was changed to DME with 30% IFS. After another 24 hours, the virus-containing supernatant was collected from the cells and passed through a 0.45 μm filter. Target cells were plated in 6-well plates with virus containing media and 8 μg/mL polybrene. Spin infections were performed by centrifugation at 2,200 rpm for 1 hour. Twenty-four hours later, the media was changed to fresh media containing either puromycin (when infected with the lentivirus) or blasticidin (when infected with the retrovirus) for selection.
Cell lysis and immunoprecipitation
Cells were rinsed once with ice-cold PBS and immediately lysed with Triton lysis buffer (1% Triton, 10 mM β-glycerol phosphate, 10 mM pyrophosphate, 40 mM Hepes pH 7.4, 2.5 mM MgCl2 and 1 tablet of EDTA-free protease inhibitor [Roche] (per 25 ml buffer). The cell lysates were centrifuged at 13,000 rpm in a microcentrifuge at 4°C for 10 minutes. For anti-FLAG-immunoprecipitations, the FLAG-M2 affinity gel was washed three times with lysis buffer. 30 μl of a 50% slurry of the affinity gel in lysis buffer was then added to cleared cell lysates and rotated for 2 hours at 4°C. The beads were washed once with lysis buffer and 3 times with lysis buffer containing 500 mM NaCl after the incubation. Immunoprecipitated proteins were denatured by the addition of 50 μl of sample buffer and boiled for 5 minutes as described24, resolved by 8%–16% SDS-PAGE, and analyzed by immunoblotting.
For co-transfection experiments in HEK-293T cells, 2 million cells were plated in 10 cm culture dishes. Twenty-four hours later, cells were transfected via the polyethylenimine method59 with the pRK5-based cDNA expression plasmids indicated in the figures in the following amounts: 800 ng Flag-Metap2 or 400 ng Flag-RagC (wild-type and mutants); 400 ng of RagB (wild-type and mutants). The total amount of plasmid DNA in each transfection was normalized to 5 μg with empty pRK5. Thirty-six hours after transfection, cells were lysed as described above.
For experiments that required amino acid, leucine, or arginine starvation or restimulation, cells were treated as previously described35. Briefly, cells were incubated in amino acid, leucine, or arginine free RPMI for 50 minutes and then stimulated with amino acids, leucine, or arginine for 10 minutes.
Nucleotide binding assays
40 pmols of FLAG-RagC (wild-type or mutant)-HA-RagBD163N were loaded with either 8 μCi of [3H]GDP or 8 μCi of [3H]GTP (5–20 Ci/mmol) and co-loaded with either 62.5 nM XDP or 62.5 nM XTP in a total volume of 80ul of CHAPS buffer, supplemented with 2.5 mM DTT, 10 μg BSA, and 6.25 mM EDTA. The CHAPS buffer contained 0.3% CHAPS, 40 mM HEPES [pH 7.4], and 30 mM NaCl. The complexes were rotated for 10 minutes at room temperature and then stabilized with 25 mM MgCl2, rotated for another 10 minutes at room temperature, and then incubated on ice for 1 hour to allow the binding reaction to occur. 10 μl samples were taken, in triplicate, and spotted on nitrocellulose filters, which were washed three times with 1 ml of wash buffer (1.5% CHAPS, 40 mM HEPES [pH 7.4], 150 mM NaCl, and 5 mM MgCl2). Filter-associated radioactivity was quantified using a TriCarb scintillation counter (PerkinElmer).
Statistical analysis
Fisher’s exact tests were used for comparison between two groups. For analysis of the nucleotide binding assay groups, two-tailed t tests were used. P values of less than 0.05 were considered to indicate statistical significance.
Supplementary Material
Acknowledgments
We are indebted to the patients for donating tumor specimens as part of this study. We would like to thank G. Clark at the Francis Crick Institute for automated DNA sequencing and the Genome Centre for Illumina Miseq sequencing. We acknowledge the support of Barts, Cambridge, Leeds and Southampton’s Experimental Cancer Medicine and Cancer Research UK Centers. This work was supported by grants from Kay Kendall Leukaemia Fund and Cancer Research UK (awarded to J.F.) J.O. is a recipient of the Kay Kendall Leukaemia Fund (KKLF) Junior Clinical Research Fellowship (KKL 557).
Footnotes
URLs. cBioportal for Cancer Genomics, http://www.cbioportal.org/; ICGC data portal, https://dcc.icgc.org/
Accession codes. Exome sequencing data have been deposited at the European Genome-phenome Archive (EGA) under accession number EGAS00001001190.
AUTHOR CONTRIBUTIONS
J.O. and J.F. conceived the study. J.O., D.M.S. and J.F. directed the study. C.M., G.P., P.J., A.D., J.C.S., M.D., S.B., A.J., T.A.L., R.A., S.M. and J.G.G. provided patient samples and clinical data. M.C., A.J. and M.D. conducted pathological review of specimens. J.M. collated clinical information. S.I. prepared and processed samples. H.Q. provided cell lines DNA. J.O., R.L.W., S.A., L.W., B.M.C., L.E-I., A.F.A-S., A.C., C.B. and R.Z. performed experiments. J.W., J.A.G-A., S.H.B. and C.C. performed the bioinformatic analysis. J.R. and R.S. coordinated and verified the ICGC dataset. J.O., R.L.W., J.W., D.M.S and J.F. analyzed and interpreted the data. J.O., R.L.W. and J.F. wrote the manuscript. All authors read and approved the final manuscript.
COMPETING FINANCIAL INTERESTS
We declare no competing financial interests.
References
- 1.Swenson WT, et al. Improved survival of follicular lymphoma patients in the United States. J Clin Oncol. 2005;23:5019–26. doi: 10.1200/JCO.2005.04.503. [DOI] [PubMed] [Google Scholar]
- 2.Okosun J, et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat Genet. 2014;46:176–81. doi: 10.1038/ng.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pasqualucci L, et al. Genetics of follicular lymphoma transformation. Cell Rep. 2014;6:130–40. doi: 10.1016/j.celrep.2013.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karube K, et al. Recurrent mutations of NOTCH genes in follicular lymphoma identify a distinctive subset of tumours. J Pathol. 2014;234:423–30. doi: 10.1002/path.4428. [DOI] [PubMed] [Google Scholar]
- 5.Yildiz M, et al. Activating STAT6 mutations in follicular lymphoma. Blood. 2014 doi: 10.1182/blood-2014-06-582650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morin RD, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42:181–5. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pasqualucci L, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471:189–95. doi: 10.1038/nature09730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Compagno M, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459:717–21. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lenz G, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319:1676–9. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- 10.Lohr JG, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A. 2012;109:3879–84. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ngo VN, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470:115–9. doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morin RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pasqualucci L, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43:830–7. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bödör C, et al. EZH2 mutations are frequent and represent an early event in follicular lymphoma. Blood. 2013;122:3165–8. doi: 10.1182/blood-2013-04-496893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green MR, et al. Hierarchy in somatic mutations arising during genomic evolution and progression of follicular lymphoma. Blood. 2013;121:1604–11. doi: 10.1182/blood-2012-09-457283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beà S, et al. Landscape of somatic mutations and clonal evolution in mantle cell lymphoma. Proc Natl Acad Sci U S A. 2013;110:18250–5. doi: 10.1073/pnas.1314608110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landau DA, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–26. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lohr JG, et al. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell. 2014;25:91–101. doi: 10.1016/j.ccr.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morin RD, et al. Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing. Blood. 2013;122:1256–65. doi: 10.1182/blood-2013-02-483727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmitz R, et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012;490:116–20. doi: 10.1038/nature11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakashima N, Noguchi E, Nishimoto T. Saccharomyces cerevisiae putative G protein, Gtr1p, which forms complexes with itself and a novel protein designated as Gtr2p, negatively regulates the Ran/Gsp1p G protein cycle through Gtr2p. Genetics. 1999;152:853–67. doi: 10.1093/genetics/152.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sekiguchi T, Hirose E, Nakashima N, Ii M, Nishimoto T. Novel G proteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B. J Biol Chem. 2001;276:7246–57. doi: 10.1074/jbc.M004389200. [DOI] [PubMed] [Google Scholar]
- 24.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–45. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sancak Y, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bar-Peled L, et al. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340:1100–6. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150:1196–208. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sancak Y, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zoncu R, et al. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–83. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rebsamen M, et al. SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature. 2015 doi: 10.1038/nature14107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang S, et al. Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science. 2015;347:188–94. doi: 10.1126/science.1257132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol. 2007;8:917–29. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- 33.Jansen EJ, Martens GJ. Novel insights into V-ATPase functioning: distinct roles for its accessory subunits ATP6AP1/Ac45 and ATP6AP2/(pro) renin receptor. Curr Protein Pept Sci. 2012;13:124–33. doi: 10.2174/138920312800493160. [DOI] [PubMed] [Google Scholar]
- 34.Iadevaia V, Huo Y, Zhang Z, Foster LJ, Proud CG. Roles of the mammalian target of rapamycin, mTOR, in controlling ribosome biogenesis and protein synthesis. Biochem Soc Trans. 2012;40:168–72. doi: 10.1042/BST20110682. [DOI] [PubMed] [Google Scholar]
- 35.Ben-Sahra I, Howell JJ, Asara JM, Manning BD. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science. 2013;339:1323–8. doi: 10.1126/science.1228792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsun ZY, et al. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell. 2013;52:495–505. doi: 10.1016/j.molcel.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feig LA, Cooper GM. Relationship among guanine nucleotide exchange, GTP hydrolysis, and transforming potential of mutated ras proteins. Mol Cell Biol. 1988;8:2472–8. doi: 10.1128/mcb.8.6.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feig LA. Tools of the trade: use of dominant-inhibitory mutants of Ras-family GTPases. Nat Cell Biol. 1999;1:E25–7. doi: 10.1038/10018. [DOI] [PubMed] [Google Scholar]
- 39.John J, et al. Kinetic and structural analysis of the Mg(2+)-binding site of the guanine nucleotide-binding protein p21H-ras. J Biol Chem. 1993;268:923–9. [PubMed] [Google Scholar]
- 40.Hara K, et al. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273:14484–94. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- 41.Nicklin P, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–34. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoffenberg S, et al. Functional and structural interactions of the Rab5 D136N mutant with xanthine nucleotides. Biochem Biophys Res Commun. 1995;215:241–9. doi: 10.1006/bbrc.1995.2459. [DOI] [PubMed] [Google Scholar]
- 43.Schmidt G, et al. Biochemical and biological consequences of changing the specificity of p21ras from guanosine to xanthosine nucleotides. Oncogene. 1996;12:87–96. [PubMed] [Google Scholar]
- 44.Farnsworth CL, Feig LA. Dominant inhibitory mutations in the Mg(2+)-binding site of RasH prevent its activation by GTP. Mol Cell Biol. 1991;11:4822–9. doi: 10.1128/mcb.11.10.4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lai CC, Boguski M, Broek D, Powers S. Influence of guanine nucleotides on complex formation between Ras and CDC25 proteins. Mol Cell Biol. 1993;13:1345–52. doi: 10.1128/mcb.13.3.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Proud CS. Guanine nucleotides, protein phosphorylation and the control of translation. Trends Biochem Sci. 1986;11:73–77. [Google Scholar]
- 47.Krengel U, et al. Three-dimensional structures of H-ras p21 mutants: molecular basis for their inability to function as signal switch molecules. Cell. 1990;62:539–48. doi: 10.1016/0092-8674(90)90018-a. [DOI] [PubMed] [Google Scholar]
- 48.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler etc. Bioinformatics. 2009 doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koboldt DC, et al. VarScan2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–76. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dayem Ullah AZ, Lemoine NR, Chelala C. SNPnexus: a web server for functional annotation of novel and publicly known genetic variants (2012 update) Nucleic Acids Res. 2012;40:W65–70. doi: 10.1093/nar/gks364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 53.Van Loo P, et al. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci U S A. 2010;107:16910–5. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boussif O, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci U S A. 1995;92:7297–301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.