Abstract
Pacific salmon exposed to sublethal concentrations of organophosphate pesticides (OP) have impaired olfactory function that can lead to loss of behaviors that are essential for survival. These exposures often involve mixtures and can occur at levels below those which inhibit acetylcholinesterase (AChE). In this study, juvenile Coho salmon were exposed for 24 h to either 0.1, 0.5, or 2.5 ppb chlorpyrifos (CPF), 2, 10, or 50 ppb malathion (MAL), or binary mixtures of 0.1 CPF:2 ppb MAL, 0.5 CPF:10 ppb MAL, or 2.5 CPF:10 ppb MAL to mimic single and binary environmental exposures. Microarray analysis of olfactory rosettes from pesticide-exposed salmon revealed differentially expressed genes involved in nervous system function and signaling, aryl hydrocarbon receptor signaling, xenobiotic metabolism, and mitochondrial dysfunction. Coho exposed to OP mixtures exhibited a more pronounced loss in detection of a predatory olfactory cue relative to those exposed to single compounds, whereas respirometry experiments demonstrated that exposure to OPs, individually and in mixtures, reduced maximum respiratory capacity of olfactory rosette mitochondria. The observed molecular, biochemical, and behavioral effects occurred largely in the absence of effects on brain AChE. In summary, our results provide new insights associated with the sublethal neurotoxic effects of OP mixtures relevant to environmental exposures involving molecular and cellular pathways of injury to the salmon olfactory system that underlie neurobehavioral injury.
Keywords: chlorpyrifos, malathion, salmon, olfaction, microarray analysis, mitochondria
Organophosphate pesticides (OPs) are commonly used in public, agricultural, and residential pest control applications. In the United States, OPs accounted for ∼35% (≥33 million pounds) of the total pesticides used in 2007 (EPA, 2011). Of these agents, chlorpyrifos (CPF) and malathion (MAL) remain among the 2 most common active ingredients in OP pesticide formulations. The extensive application of pesticides in agricultural regions has often contaminated surface waters; and CPF and MAL residues have been shown to account for >50% of total OP pesticides detected in Northwest surface waters. Accordingly, salmon inhabiting urban streams in the Pacific Northwest typically contain 2 or more OPs (Anderson and Dugger, 2008; Sargeant et al., 2013), and surface water contamination with OPs has been causatively linked to the decline in Pacific salmon populations (Miller et al., 1989). The aforementioned issue of exposure to pesticide mixtures is a common scenario across aquatic environments nationally (Gilliom, 2007), and complicates the aquatic risk assessment of these agents due to the biochemical and physiological complexities of mixture exposures and the potential for additive, antagonistic, or synergistic interactions.
These interactions among multiple compounds have not been well integrated within regulatory frameworks, with the exception of dose-additivity of anti-cholinesterase agents in assessing risks to humans (EPA, 2002) and to a lesser extent, to aquatic life. Thus, there is the potential that such a framework may not account for sublethal toxicological impacts of pesticide mixture exposures on aquatic ecosystems (Moore and Teed, 2012). Laetz et al. (2009) demonstrated that exposure of juvenile Coho salmon to mixtures of OPs relevant to salmon habitats resulted in additive and synergistic toxic effects, suggesting that single chemical risk assessments may underestimate the impacts of these pesticides in salmon habitats. However, lacking are studies examining the potential mechanisms and biochemical effects of OP mixtures that can underlie loss of salmon fitness.
Foremost among key organ systems of sublethal neurotoxic injuries in salmonids is injury to the olfactory system. The peripheral component of the fish olfactory system consists of rosette-like structures termed olfactory rosettes, which are covered by a sensory epithelium containing olfactory receptor neurons (ORNs) that are highly vulnerable to waterborne toxicants (reviewed in Tierney et al., 2010), including anti-esterase pesticides (Jarrard et al., 2004; Sandahl et al., 2004; Tierney et al., 2006). Along with these cell surface ORNs, a number of signal transduction cascades are critical to olfactory function and behavioral responses (Restrepo et al., 1996). We have shown that exposure to CPF causes a marked inhibition of G-protein-coupled receptor signaling in the olfactory system of zebrafish (Tilton et al., 2011), suggesting that olfactory signaling may be compromised under CPF exposures. However, in the same article, we observed that exposure to the trace metal copper had a more marked disruption of olfactory signal transduction genes than observed with CPF. Maryoung et al. (2015) reported an activation of 4 olfactory signal transduction genes by CPF under hypersaline conditions using genes identified from our zebrafish study. Collectively, these studies suggest that non-acetylcholinesterase (AChE) mechanisms of olfactory neurobehavioral function, such as those involving signaling from the olfactory rosettes, may be important mechanisms of olfactory injury in salmonids receiving pesticide exposures.
In this study, we examined transcriptomic alterations that accompanied olfactory injury by 2 common OPs (CPF and MAL) separately, and as binary mixtures relevant to salmon exposures in the wild. Transcriptome analysis remains a powerful tool to elucidate the fundamental molecular pathways and generate hypotheses regarding mechanisms of sublethal toxicity. Following exposure to the individual OPs and their mixtures, we analyzed genome-wide transcriptional modifications in the olfactory rosettes, odor-driven behaviors, cellular mitochondrial function, and brain AChE to better understand the cellular and molecular events underlying environmentally relevant OP exposures. We also investigated functionally olfactory mitochondrial function as a target for these OPs, as others have proposed mitochondrial dysfunction as a non-cholinergic target of OP exposure (Kaur et al., 2007; Xin et al., 2011). Our hypothesis was that exposure to low environmental levels of binary mixtures of CPF and MAL would cause sublethal neurotoxicity not observed by the single chemicals, and would involve signaling and mitochondrial pathways as AChE-independent modes of olfactory injury.
MATERIALS AND METHODS
Chemicals
MS-222 (Tricaine methanesulfonate) was obtained from Argent Chemical Laboratories (Redmond, Washington). CPF (CAS 2921-88-2; >98% pure) and MAL (CAS No. 121-75-5; >98% pure) were purchased from ChemService Inc (West Chester, Pennsylvania). TRIzol reagent, the SuperScript First-Strand Synthesis System, and the Zero Blunt TOPO PCR Cloning Kit were purchased from Invitrogen (Carlsbad, California). RNeasy mini kit was purchased from Qiagen (Valencia, California). Finnzymes DyNAmo SYBR Green 2-Step qPCR Kit was purchased from Thermo Fisher Scientific (Waltham, Massachusetts). qPCR primers were obtained from Eurofins MWG Operon (Huntsville, Alabama). All other chemicals and solvents were of analytical grade and purchased from standard sources.
Animals
Juvenile Coho salmon (1 year of age, body weight = 15.0 ± 5.7 g) were provided by the Washington Department of Fish and Wildlife (Issaquah, Washington). Fish were fed Bio Vita Fry Feed (Bio-Oregon Inc, Oregon) twice daily using automatic feeders (2% biomass/day) and raised under a natural photoperiod in cylindrical tanks receiving Lake Washington water at the following conditions: 10°C–12°C, 80–120 mg/l total hardness as calcium carbonate, pH 7.4 ± 0.2, and 8.1 mg/l dissolved oxygen content.
Pesticide exposures
All animal welfare and experimental procedures were carried out in accordance with the University of Washington Institutional Animal Care and Use Committee (IACUC) guidelines. Pesticide stock solutions for both single-pesticide and mixture exposures were prepared in 100% ethanol, and nominal concentrations were confirmed by stable-isotope dilution liquid chromatograph tandem mass spectrograph assay (see Supplementary methods and Table S1). Pesticide stock solutions were added to 90-l glass aquaria containing 70-l of lake water and 8 individual fish per treatment group. All aquaria were aerated and their temperature maintained at 11°C via a large, chilled, re-circulating water bath. Exposure concentrations were selected to be below median effective concentrations (EC50) determined for Coho salmon exposed to pesticides by Laetz et al. (2009). Fish were exposed to nominal CPF concentrations of either 0.1, 0.5, or 2.5 ppb (CPF-L, M, and H, respectively), nominal MAL concentrations of 2, 10, or 50 ppb (MAL-L, M, and H, respectively), or 1 of 3 binomial mixture concentrations: mixture-L (0.1 ppb CPF: 2 ppb MAL), mixture-M (0.5 ppb CPF: 10 ppb MAL), mixture-H (2.5 ppb CPF: 10 ppb MAL) in the absence of food for 24 h. Control groups received only carrier and final carrier concentration in each exposure tank was ≤0.0004% of the total volume. Following exposures, fish were euthanized with MS-222 prior to cervical dislocation. The olfactory rosettes were immediately dissected for subsequent RNA isolation and brain tissues were removed, flash frozen in liquid nitrogen, and stored at −80°C until subsequent analysis.
RNA isolation and microarray analysis
Total olfactory rosette RNA was isolated using the RNeasy Mini Kit (Qiagen Inc, Valencia, California) according to the manufacturer’s protocol and stored at −80°C. The quantity (ng/µl) of RNA was determined by measuring the OD260 with a NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, Waltham, Massachusetts) and purity was assessed by measuring OD260/280 and OD260/230 ratios. RNA integrity was characterized using the Agilent RNA 6000 Nano Kit with an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc, Santa Clara, California). Total first strand cDNA was synthesized from DNAse-treated RNA samples using the SuperScript First-Strand Synthesis System (Invitrogen). The cDNA samples were labeled and prepared for hybridization onto an Agilent 4 X 44 K Salmonid cGRASP Gene Expression Microarray (Agilent Technologies, Inc) using the manufacturer’s established protocols. Arrays were hybridized and washed using HS 400 Pro hybridization and wash stations (Tecan Systems, Inc, San Jose, California) and scanned using an Agilent DNA Microarray Scanner (Agilent Technologies, Inc) as previously described (Lefebvre et al., 2012).
Bioinformatics
Data were background-corrected and normalized using the vsn package of Bioconductor. Background values were subtracted local (the median of intensity values in regions around each spot) from foreground values (median of the spot intensity) and normalized using a variance stabilization method (Huber et al., 2002). Data from each treatment were normalized separately and fit using the Bioconductor limma package (Smyth, 2004) to a weighted mixed model that included fixed effects for sample treatment as well as array processing days. Fitting a mixed model accounted for intra-slide correlations (4 arrays per slide), and fitting by-day accounted for inter-day variability. Various comparisons were made using empirical Bayes-adjusted contrasts (Smyth, 2004) after excluding both low-expressing genes and those with a standard deviation <0.5. Gene annotations were based on cGRASP 44 K Salmonid annotation file (preliminary version: 0.2) (http://web.uvic.ca/grasp/microarray/array.html). The microarray data were submitted to Gene Expression Omnibus (GEO).
Examination of the dataset by Ingenuity Pathway Analysis (IPA, http://www.ingenuity.com/) allowed for the identification of biologically relevant canonical pathways, networks, biofunctions, and tox list categories affected by different OP exposures. Only the top ranked (P < .05) biofunctions, ranked using right-tailed Fisher Exact Tests for each OP concentration, were used for further analysis. Venn diagrams showing the counts of overlapping genes using the same sets of data (>1.5-fold up/down-regulated, false-discovery rate-adjusted-P value < .05) were generated to facilitate comparison (see Supplementary methods).
Quantitative real-time PCR (qPCR)
Primers for validation of microarray data were designed using Primer 3 at San Diego Biology Workbench 3.2 (Supplementary Table S2). IPA-identified numerous genes with significant differential expression among treatments, and genes associated with the predominant pathways of impairment were selected for qPCR analysis using the standard curve method (Espinoza et al., 2012) (see Supplementary methods).
Mitochondrial respiration in olfactory rosettes
Olfactory rosettes were harvested from euthanized fish from the CPF-M, MAL-M, and mixture-M groups immediately following behavioral experiments and placed into 20 ml of ice-cold Custodiol HTK Solution (Essential Pharmaceuticals, LLC, Newtown, Pennsylvania) for stable storage (up to 10 h) between mitochondrial respirometry experiments. Olfactory rosettes were transferred to ice-cold respiration buffer (distilled H2O, 210 mM sucrose, 0.5 mM EGTA, 3 mM MgCl2, 10 mM KH2PO4, 20 mM HEPES, 20 mM taurine, 50 mM K-MES, 1 g/l BSA, pH 7.13) prior to initiation of each respirometry experiment. Rates of oxygen consumption were measured in saponin-permeabilized olfactory rosettes by high-resolution respirometry using the Oxygraph-2k (Oroboros Instruments, Innsbruck, Austria). The temperature of the oxygraph chambers matched the temperature of the lake water (11°C), and background oxygen concentration in media was calibrated prior to each experiment (Siegel et al., 2011). Procedures for measurement of mitochondrial electron transport system (ETS) functional parameters were conducted as described (Yeh et al., 2015). Mean oxygen consumption rates are reported as pmoles O2/[sec*mg protein]. The respiratory control ratio (RCR) (ratio of maximum State 3/State 4 respiration) is an indicator of mitochondrial function and coupling of oxidative phosphorylation (Brand and Nicholls, 2011). Similarly, the uncoupled RCR is calculated as the ratio of maximum uncoupled respiration/State 4 respiration.
Effect of OP exposures on neurobehavior and olfactory function
To test olfactory function, independent of brain AChE inhibition, and neuromuscular dysfunction, only Coho salmon from exposure groups: CPF-M, MAL-M, and mixture-M (n = 5 per group) were used for behavioral testing. These exposure groups were selected because they were the highest OP exposures that did not significantly reduce AChE activity or swimming ability (see Supplementary methods). All fish were rinsed for 10 min in a glass flow-through tank receiving lake water prior to behavioral testing to avoid contaminating the Y-maze with OPs. Swimming performance was determined in a laminar flow chamber as previously described by Hawkins and Quinn (1996). In brief, in water conditions similar to rearing, individual fish were placed within the chamber and tested to determine if they could reach 80% (79 cm/s) of the Ucrit (96 cm/s) calculated from similarly sized animals (Jain et al., 1997). This speed, Ucrit, is significant because its achievement is indicative of normal neuromuscular function. Trials consisted of a 10-min acclimation period during which velocity was slowly increased from 0 to 30 cm/s and after which speed was immediately increased by 9 cm/s every 2 min. This procedure was continued until the fish either maintained a speed of ∼79 cm/s for 40 s or failed to match the generated current. Fish were allowed to recover for 5 min prior to Y-maze testing.
The Y-maze used to analyze olfactory-mediated behaviors received lake water at a constant flow rate of 8.3 l/min and was enclosed by a black curtain to minimize the possibility of interference by external visual stimuli. Coho were acclimated for 15 min in the maze prior to the addition of odorant [10−2 M taurocholic acid (TCA) or DI water, control] via peristaltic pump at 10 ml/min. Behavioral responses were recorded for a total of 15 min (5 min prior and 10 min post-odorant addition), and nominal odorant concentrations in the maze were 10−5 M. The quantified behaviors characterized as odorant responses included odorant-induced activity and odor avoidance. Odorant-induced activity was defined as the number of times the fish transitioned between the odorant and non-odorant arm, and odor avoidance was defined as the ratio of time spent the non-odorant arm versus time in the odorant arm. The maze was flushed with clean water and the odorant arm alternated between behavioral trials.
Brain AChE activity
Coho brain AChE activity was determined using a modification of standard methods (Correll and Ehrich, 1991; Ellman et al., 1961). In brief, brains from individual fish were homogenized in 0.1 M sodium phosphate buffer (pH 8.0) prior to centrifugation at 600 × g for 5 min at 4°C. Supernatant was collected and stored at −80°C prior to thawing and incubation at 25°C in sodium phosphate buffer (pH 8.0) containing 1.2 mM DTNB, and addition of acetylthiocholine iodide (4.5 mM). Enzyme activity was measured in triplicate at 25°C after a 5-min pre-incubation using a SpectraMax190 UV–Vis 96-well microplate reader (Molecular Devices, Sunnyvale, California) with absorbance monitored at 412 nm. Samples were assayed in triplicate with appropriate tissue and reagent blanks.
Statistical analysis
Prior to statistical analyses, qPCR and swimming behavior data, and also mitochondrial function data were inspected for homogeneity of variances using the Bartlett’s test and Levene’s test, respectively. qPCR data and swimming behavior test data were assessed for significance using Kruskal-Wallis non-parametric 1-way analysis of variance (ANOVA) followed by a Dunn’s test. Levene’s test determined no significant difference in variance among treatment groups, and thus significance of mitochondrial function data was assessed using parametric ANOVA followed by a Dunnet’s test. All data were considered statistically significant at cutoff of P < .05. All statistical analyses were conducted using GraphPad Prism Ver 5.0 (Graph Pad Software Inc, San Diego, California).
RESULTS
Effect of Pesticides on Toxicity and Brain AChE
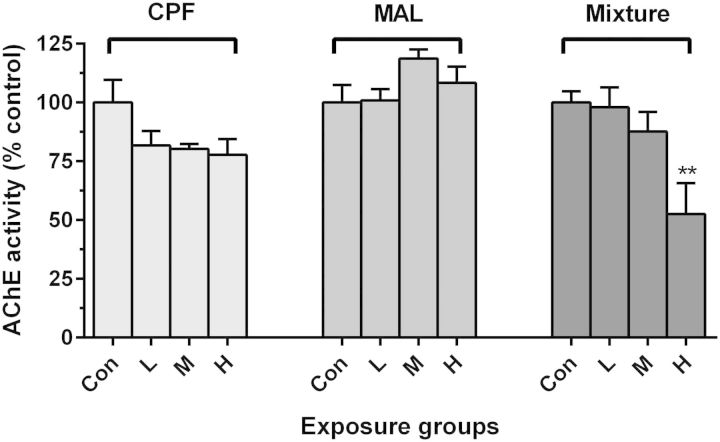
None of the pesticide-exposed fish showed overt physical signs of toxicity (ie, loss of righting reflexes) and no mortalities occurred during exposures. Furthermore, there was no significant inhibition of brain AChE activity following exposure to the pesticides in exposures to single compounds (Figure 1). However, exposure of Coho to the high-dose binary mixture dose elicited a significant reduction in brain AChE activity (47% inhibition; Figure 1). Similarly, no differences were observed in brain AChE activity among fish used in replicate exposures for the swimming behavior analysis and those analyzed for mitochondrial function. (Supplementary Figure S1).
FIG. 1.
Effect of CPF, MAL, and mixture exposures on Coho brain AChE activity. Asterisks (*) indicate statistically significant difference between treatment groups and its specific control samples (ANOVA, **P < .01, mean ± SEM of n = 8 individuals).
Microarray Analysis of Differentially Expressed Genes by CPF, MAL and Mixture Exposures
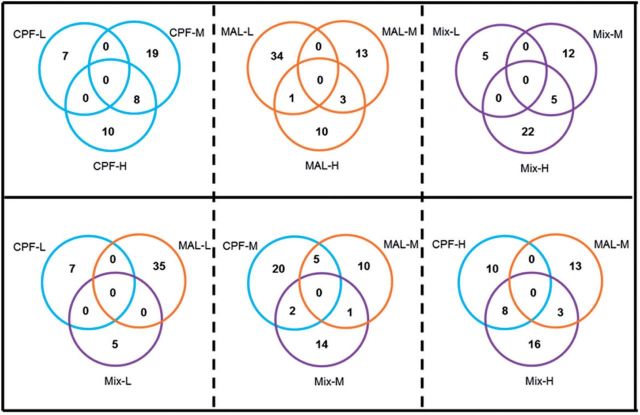
Microarray analysis of Coho olfactory rosettes revealed 1575 differentially expressed genes in at least 1 exposure group at cutoffs of >1.5-fold up/down-regulation and adjusted-P values of < .05 (GEO accession GSE47984 for the microarray dataset). There were 8, 28, and 138 differentially expressed genes in CPF-L, M, and H groups, respectively, and 56, 636, and 443 differentially expressed genes in MAL-L, M, and H groups, respectively. Binary mixture exposures resulted in the modulation of the expression of 9, 624, and 311 genes in the low, medium, and high concentration groups, respectively (Supplementary Table S3). There was no overlap of altered genes within the 3 CPF concentrations and little overlap within the MAL (15 genes) or mixture groups (6 genes) (Figure 2). Most of the commonly altered genes were downregulated relative to controls. Table 1 shows dynamin 1-like (DNM1L) was commonly changed among the medium doses (CPF-M, MAL-M, and Mix-M) whereas genes such as polymerase (RNA) II (POLR2H), thiopurine S-methyltransferase (TPMT), polyadenylate-binding protein-interacting protein 2 (PAIP2), cell growth regulator with ring finger domain 1 (CGRRF1), adenylate kinase 6 (AK6), and prostaglandin E synthase 3 (cytosolic) (PTGES3) which are involved in various biological processes such as metabolic process, translational initiation regulation, and DNA repair, were commonly altered among the CPF-H, MAL-M, and mixture-H groups (Table 1). With the exception of CGRRF1, genes expression within the CPF-H and MAL-M groups was opposite to those seen in the mixture-H groups. Microarray validation by qPCR analysis of 6 genes identified by IPA, including CYP1A1, SOD2, NFKB2, HSP90AA1, HES1, and EIF4A3 revealed similar trends between the 2 methodologies (Figure 3).
FIG. 2.
Venn diagram analysis of the differentially expressed genes following OP exposures. The number of transcripts meeting the cutoff (>1.5-fold up/down-regulated, adjusted-P value < .05) for each OP treatment and concentration are contained within each section of the labeled circle. The number of genes in the Venn diagram is based on unique probeset IDs, not gene symbols.
TABLE 1.
Salmon Olfactory Genes That Were Commonly Altered Among 3 Different Exposure Treatmentsa
| Genes differentially expressed in common among MAL-L, MAL-M, MAL-H dosing groups | ||||||
|---|---|---|---|---|---|---|
| Symbol | Description | Fold-change |
Ensembl | |||
| MAL-L | MAL-M | MAL-H | ||||
| FRAP1 | Mechanistic target of rapamycin (serine/threonine kinase) | 0.49 | 0.55 | 0.53 | ENSG00000198793 | |
| MYST2 | K(lysine) acetyltransferase 7 | 0.46 | 0.42 | 0.44 | ENSG00000136504 | |
| JUN | Jun oncogene | 0.41 | 0.41 | 0.45 | ENSMUSG00000052684 | |
| ZIC5 | Zic family member 5 | 0.35 | 0.38 | 0.36 | ENSG00000139800 | |
| TAP2 | Transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) | 0.42 | 0.45 | 0.42 | ENSRNOG00000000455 | |
| XIRP1 | Xin actin-binding repeat containing 1 | 0.40 | 0.28 | 0.36 | ENSDARG00000030722 | |
| ITGA4 | Integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) | 0.47 | 0.50 | 0.46 | ENSG00000115232 | |
| ENPEP | Glutamyl aminopeptidase | 0.49 | 0.44 | 0.44 | ||
| ZCWPW2 | Zinc finger, CW type with PWWP domain 2 | 0.46 | 0.49 | 0.51 | ENSG00000206559 | |
| CALCA | Calcitonin | 0.45 | 0.42 | 0.47 | ENSGALG00000006054 | |
| QTRTD1 | Queuine tRNA-ribosyltransferase domain containing 1 | 0.44 | 0.46 | 0.40 | ENSG00000151576 | |
| LBR | Lamin B receptor | 0.51 | 0.55 | 0.51 | ENSRNOG00000003103 | |
| Genes differentially expressed in common among Mix-L, Mix-M, Mix-H dosing groups | ||||||
| Symbol | Description | Fold-change | Ensembl | |||
| Mix-L | Mix-M | Mix-H | ||||
| PRSS27 | Protease, serine 27 | 0.52 | 0.53 | 0.39 | ENSG00000172382 | |
| SSR4 | Signal sequence receptor, delta | 0.35 | 0.36 | 0.36 | ENSMUSG00000002014 | |
| MXRA8 | Matrix-remodeling-associated protein 8 | 0.34 | 0.34 | 0.25 | ENSGALG00000001561 | |
| PYGM | Phosphorylase, glycogen, muscle | 0.61 | 0.60 | 0.53 | ENSBTAG00000024240 | |
| Genes differentially expressed in common among CPF-M, MAL-M, and Mix-M dosing groups | ||||||
| Symbol | Description | Fold-change | Ensembl | |||
| CPF-M | MAL-M | Mix-M | ||||
| DNM1L | Dynamin 1-like | 0.61 | 0.65 | 0.57 | ENSDARG00000015006 | |
| Genes differentially expressed in common among CPF-H, MAL-M, and Mix-H dosing groups | ||||||
| Symbol | Description | Fold-change | Ensembl | |||
| CPF-H | MAL-M | Mix-H | ||||
| POLR2H | Polymerase (RNA) II (DNA directed) polypeptide H | 0.64 | 0.50 | 1.85 | ENSG00000163882 | |
| TPMT | Thiopurine S-methyltransferase | 0.66 | 0.62 | 1.79 | ENSDARG00000055974 | |
| PAIP2 | Polyadenylate-binding protein-interacting protein 2 | 0.67 | 0.48 | 1.74 | ||
| CGRRF1 | Cell growth regulator with ring finger domain 1 | 0.50 | 0.59 | 0.42 | ENSG00000100532 | |
| AK6 | Adenylate kinase 6 | 0.65 | 0.52 | 1.74 | ||
| PTGES3 | Prostaglandin E synthase 3 (cytosolic) | 0.66 | 0.48 | 1.98 | ENSMUSG00000071072 | |
aGenes without any reference ID (e.g., entrezgene, ensemble, unigene, refseq) are not listed here.
FIG. 3.
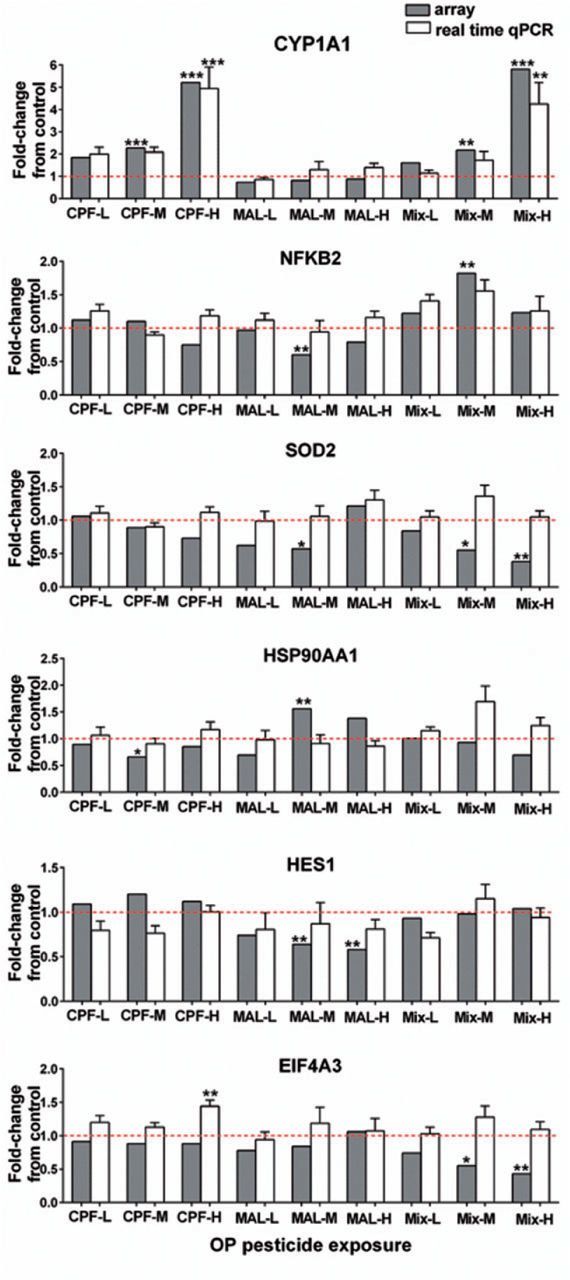
Confirmation of the microarray data by quantitative real-time PCR. Six genes were analyzed for fold-change from the controls as both by microarray analysis (gray bars) and by qPCR (white bars). The PCR results were normalized using GAPDH as internal. qPCR data represent the mean ± SEM of n = 8 individuals. Asterisk indicates statistically significant differences in expression relative to control values (*P < .05, **P < .01, ***P < .001). The dash line indicates 1-fold change, above the line suggests induction, and below the line suggests a reduction in gene expression.
Identification of Gene Pathways Altered by Single Compounds and Their Mixtures
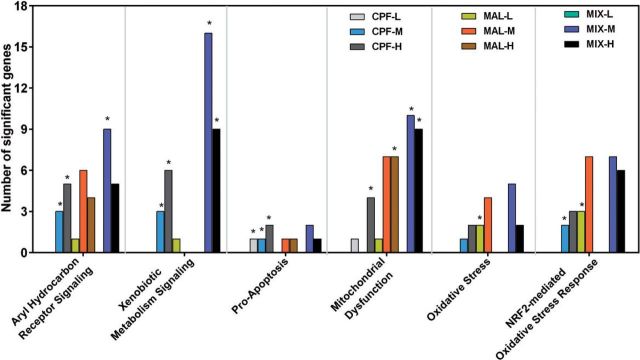
IPA identified the biological functions, canonical pathways, and toxicity lists that were significantly affected by individual and mixed OPs relative to controls. A total of 1329 differentially expressed genes (>1.5-fold up/down-regulated, adjusted-P value < .05) were mapped to their corresponding mammalian orthologous gene object in the IPA database using the gene symbol as an identifier. We evaluated the biofunction of significantly differentially expressed target genes based upon functional similarity using the “Molecular and Cellular Function” (MCF) (Supplementary Table S4) and “Physiological System Development and Function” (PSDF) categories (Supplementary Table S5). Cell Morphology, Cellular Development, Cellular Growth, and Proliferation were shared commonly among CPF, MAL, and mixture groups that ranked the top 5 significant MCFs (P < .05) (Figure 4; Supplementary Table S4).
FIG. 4.
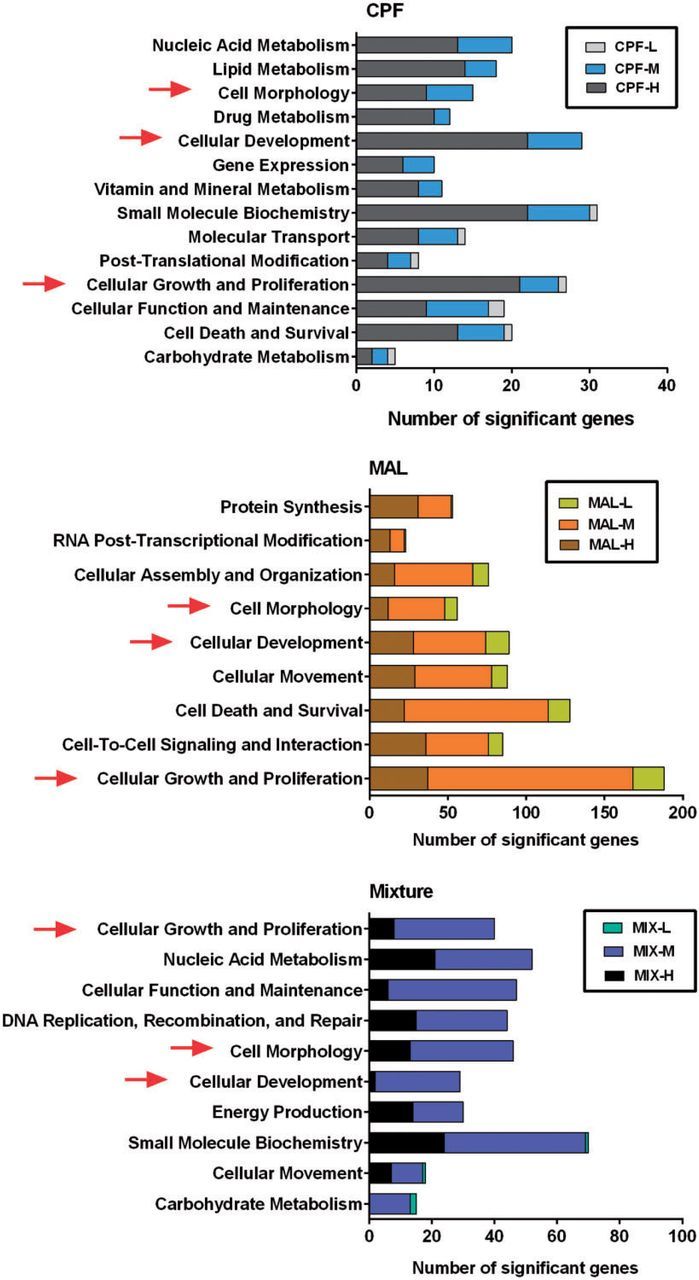
Comparison of number genes related to top 5 altered “Molecular and Cellular Function” in IPA analysis (P < .05; details see Supplementary Table S4). Arrows indicate the commonly altered MCF categories in both single compound exposure and binary mixture exposure. Comparison of number genes related to altered “Tox List” in IPA analysis (*P < .05; for details see Supplementary Table S7).
We compared the significantly altered canonical pathways in single compound versus mixture exposures and observed the greatest overlap between medium and high doses for each exposure group. More importantly, some cellular signaling pathways were commonly altered among the single compound and mixture groups. For example, AhR Signaling and Xenobiotic Metabolism Signaling pathways were altered in both CPF-M and mixture-M groups; Actin Cytoskeleton Signaling was affected in both MAL-M and mixture-M groups. In total, 8 pathways were overlapping between CPF-H and mixture-H, including Xenobiotic Metabolism Signaling and Mitochondrial Dysfunction, whereas 3 pathways were commonly altered between MAL-H and mixture-H groups (Supplementary Table S6). Among all the altered signaling pathways (P < .05), Protein Kinase A Signaling (P = .020, 18 genes), Axonal Guidance Signaling (P = .014, 17 genes), and Xenobiotic Metabolism Signaling (P = .0005, 16 genes) had the largest number of differentially expressed genes in the MAL-M and mixture-M groups (Supplementary Table S6). Some of these metabolic and signaling pathways represent the significant toxicological impacts of OP pesticides on the olfactory system (Figure 4; Supplementary Table S7).
Genes involved in “neurotransmitter and other nervous system signaling”, “growth factor and organ development”, and “second messenger signaling” that relates to olfactory function were persistently down-regulated among most treatments (Supplementary Figure S4 and Supplementary Table S8). However, some genes, such as ARPCIA, GSN, MYH6, PRKAG1, and PAK6, which are primarily involved in actin binding and protein kinase activity, were inversely expressed in mixed, versus single compound, exposures.
Effect of OP Pesticides on Mitochondrial Function
Exposure to the medium dose of CPF, MAL, and the mixture did not modulate State 3 respiration, or flux through complexes II and IV (Supplementary Table S9). Exposure to OPs individually and in mixtures also did not modulate State 4 respiration in olfactory rosette mitochondria (Figure 5A). However, ETS respiratory capacity (ie, uncoupled respiration) was reduced by 38% and 36% by exposure to MAL-M and the mixture-M, respectively (Figure 5B). As a result, the uncoupled RCRs (uncoupled RCR) of Coho olfactory rosette mitochondria exposed to MAL-M were reduced by 46% relative to controls (Figure 5C). A non-significant 18% reduction in uncoupled RCR resulted from exposure to mixture-M. Rates of maximum uncoupled respiration, and uncoupled RCRs were similar in the CPF-M and control groups (Figs. 5B and C).
FIG. 5.
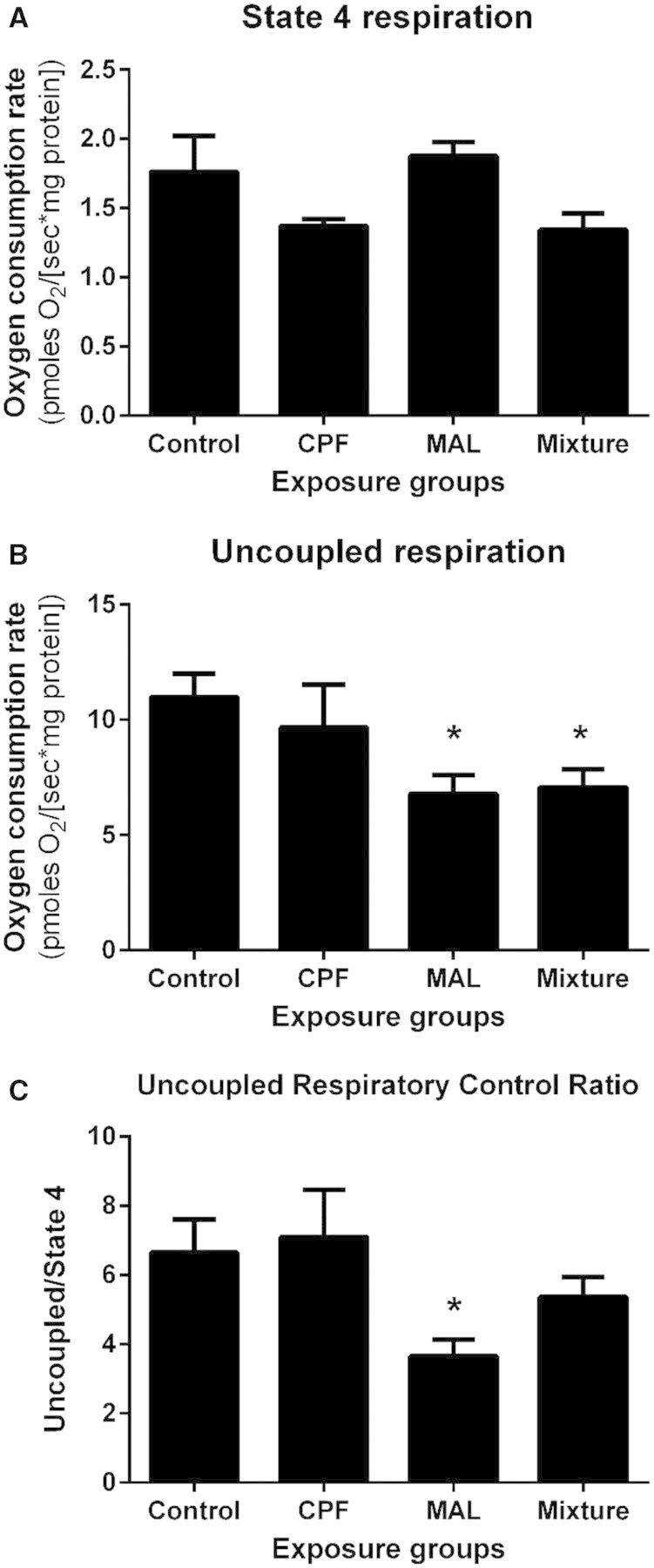
Effect of pesticides on uncoupled RCR in Coho olfactory rosettes. Uncoupled RCRs for each exposure group were calculated by the ratio of uncoupled respiration/State 4 respiration. Asterisk indicates statistically significant differences in treatment groups relative to controls at P < .05. Data represent mean ± SEM of n = 4–6 individuals.
Swimming Performance and Odor-Driven Behaviors
As observed in Figure 6A, there were no treatment-related differences observed in overall swimming behavior associated with muscular function. In contrast, Coho exposed to the mixture-M exhibited an 88% reduction in odorant avoidance compared with unexposed controls. This effect occurred in the absence of an effect on brain AChE. Both CPF-M and MAL-M exposed fish exhibited an extensive, but non-statistically significant, 52% reduction in odorant avoidance compared with controls (Figure 6B). Fish in the mixture-M group showed a loss of odorant-induced activity (reduced by 97%) relative to controls (Figure 6C), whereas single compound exposed fish exhibited a less pronounced effect (71% and 65% reduction in CPF-M and MAL-M groups, respectively). These aforementioned losses in odorant-induced behavioral activity occurred in the absence of effects on brain AChE. Olfactory behavior did not differ among treatment groups prior to the onset of odorant administration, and did not differ among exposure replicates groups.
FIG. 6.
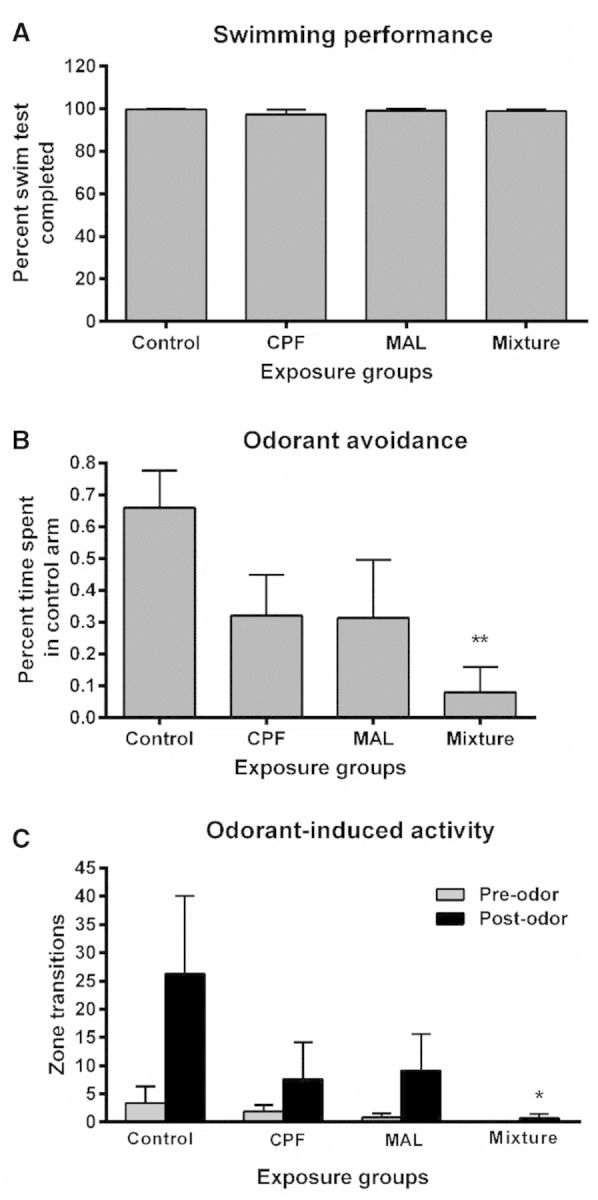
Behavioral responses to TCA or DI water. A, Swimming performance. B, Odorant avoidance. C, Odorant induced activity. Pre-odor activity datasets are represented as grey bars and post-odor activity datasets are represented as black bars. All data represent the mean ± SEM of n = 10 individuals. Asterisk indicates statistically significant differences in treatment groups relative to controls (*P < .05, **P < .01).
DISCUSSION
Many of the studies reported in the aquatic toxicology literature involving environmental pesticide exposures to fish have had a primary focus on the inhibition of AChE, a common mechanism of neurotoxicity of OPs (Annau, 1992; Fulton and Key, 2001; Mileson et al., 1998). In the case of OP mixtures, certain organophosphate and carbamate insecticides inhibit AChE in Coho salmon olfactory rosettes (Jarrard et al., 2004) and brain AChE activity (Laetz et al., 2009), which were linked to loss of swimming behavior (Laetz et al., 2013). However, there are no known cholinergic synapses in the olfactory epithelium (Scholz et al., 2006), suggesting the causal mechanisms of injury to the fish olfactory system are complex and can involve non-AChE-dependent mechanisms, such as those that affect olfactory-driven neurobehaviors (ie, olfactory signal transduction pathways). Using zebrafish, a well-defined genetic laboratory fish model, we previously demonstrated that exposure to CPF disrupts olfactory function via inhibition of G-protein-coupled receptor signaling, leading to an olfactory system that is less responsive to odorants (Tilton et al., 2011). In the case of salmon, exposure to OPs can inhibit olfactory-dependent behavioral processes such as prey capture, predator detection, and migration, all of which are critical for survival (Morgan and Kiceniuk, 1990; Scholz et al., 2000; Tierney et al., 2010). CPF is among the more potent OP olfactory toxicants, and CPF exposures below 3 ppb have been shown to diminish odorant-evoked potentials in sensory epithelia by 25%–70%, inhibiting swimming and feeding behaviors (Sandahl et al., 2004, 2005).
In this study, we demonstrated that in vivo exposures to binary mixtures of OPs at environmentally relevant concentrations disrupt molecular biochemical pathways involved in ORN energetics, metabolism, and oxidative damage, which were associated with partial inhibition of olfactory mitochondrial function, and also the olfactory neurobehavioral injury. The loss of detection of the ability of Coho to detect a predatory cue, as observed in this study, has been linked to loss of feeding and predator avoidance behaviors, and increased mortality risk (McIntyre et al., 2008, 2012). Although it was not the intent of this study to establish OP benchmark doses or evaluate in a rigorous statistical approach the potential for additive or synergistic interactions on these sublethal toxic effects, our data suggest a generally additive effect of OP exposures on sublethal endpoints of toxicity. Our results are consistent with other studies suggesting that exposures to mixtures of pesticides commonly encountered in fish habitats pose a more severe threat than can be assessed by a single chemical risk assessment (Laetz et al., 2009, 2013).
Proper olfactory function relies on the complex coordination of many biochemical signal transduction cascades (Restrepo et al., 1996; Schild and Restrepo, 1998). Odorants bind to G-protein-coupled receptors which subsequently trigger cAMP-mediated cyclic nucleotide-gated (CNG) channels (Nakamura and Gold, 1987), propagating the signal from the neuronal body down the axon to the olfactory bulb and telencephalon. As we have shown, CPF, MAL, and their mixtures can negatively impact many important signal transduction pathways and mitochondrial function, both of which are central to ORN health and function. Inhibition of action potentials generated from ORNs in response to an olfactory stimulus, as measured by electro-olfactogram (EOG) analysis has been shown to block subsequent olfactory-driven behaviors (Sandahl et al., 2004). However, EOG analysis cannot discriminate the signal transduction pathway components which may be dysregulated by pesticides. In contrast, our study shows that OP exposures inhibit ORN molecular signaling pathways and mitochondrial function, significantly impaired olfactory-driven behaviors. More importantly, these neurobehavioral impairments were independent of neuromuscular dysfunction and brain AChE activity, emphasizing olfactory specific toxicity.
Other studies in rodents have suggested non-cholinergic mechanisms of CPF neurotoxicity, including those potentially interfere with signaling cascades (Schuh et al., 2002; Song et al., 1997); alter the expression of nuclear transcription factors which regulate cell replication and differentiation (Crumpton et al., 2000); and induce oxidative stress, all of which can lead to cellular damage and apoptosis (Saulsbury et al., 2009). Similarly, we observed expression changes in genes from numerous pathways involved in critical cellular functions in salmon olfactory rosettes following CPF exposures, in particular, calcium signaling genes (CALR precursor), and genes involved in tight junction signaling, including adhesion-related genes TJR1, tumor necrosis factor (TNF), and MYH9. Down-regulation of these genes could disrupt signal transduction cascades and interfere with neuronal growth and cell-cell connection. Similarly, others have reported a number of dysregulated olfactory receptor genes following CPF exposure (2 mg/kg) in the forebrains of rats (Ray et al., 2010). Although we did not detect extensive gene expression changes in Coho olfactory receptor genes in this study, this could be a result of incomplete coverage of the Agilent array platform with regard to salmon-specific odorant receptors. We are addressing this shortcoming by developing a salmon-specific olfactory sensory neuron array for follow-up studies. In general, however, our results are consistent with the findings of our previous zebrafish study which suggested that CPF may impair olfactory function via signal transduction pathways, despite having less of an overall impact on olfactory genes when compared with metals such as copper (Tilton et al., 2011).
The non-cholinergic mechanisms of MAL-induced neurotoxicity may be less clear than for CPF. We observed more differentially expressed genes and disruption of molecular pathways in MAL exposures relative to CPF, even in the lower concentration groups. For example, the MAL-M exposure significantly altered genes involved in the protein kinase A signaling pathway, which is essential in maintaining olfactory signal transduction, including odor-induced response (Vielma et al., 2008) and axon pathfinding in fish ORNs (Yoshida et al., 2002). In vitro studies have suggested involvement of calcium signaling in MAL-induced neurotoxicity, likely via inhibiting calmodulin (CaM) activity (Pala et al., 1991). We observed a down-regulation of CAMK2A, which may also suggest an interference of calcium signaling by MAL exposure. Other nervous system signaling pathways, such as Notch and Actin Cytoskeleton Signaling, could also be targets of MAL exposure. For example, MAL-induced disruption of actin microfilaments was related to a loss of cellular adhesion and changes in cellular morphology in mammalian cells (Cabello et al., 2003). In our study, we observed significant alteration in genes related to actin cytoskeleton in the MAL-M group, which may affect cellular movement and morphology. Several genes of the basic helix-loop-helix class (HES1, HES5, and HES6), which were down-regulated in the MAL (medium and high dose) groups, are components of Notch signaling and regulate critical genes in the neurogenesis process (Cau et al., 2000). The fact that we observed little overlap between CPF and MAL with respect to significantly altered genes and pathways suggests some different non-AChE mechanisms of action by these 2 chemicals.
The observed changes in protein synthesis-related genes, such as eukaryotic translation elongation factors (EEF2, EIF3C, and EIF4EBP2), STAT3, and TNF, suggest an influence of CPF on cell growth and proliferation. Also, CYP1A, which has been widely used as a biomarker for the assessment of environmental pollutants in aquatic animals and is transcriptionally induced by the AhR, was up-regulated in the CPF-M and CPF-H groups (2.3- and 4.3-fold, respectively). This observation is similar to an earlier study that showed increased CYP1A family mRNA and enzyme activity in common carp liver after CPF exposure (11.6 and 116 μg/l) (Xing et al., 2014), suggesting a role for CYP1 family members in the bioactivation of CPF in vivo.
CPF and MAL have been observed to have inhibitory effects on mitochondrial function in rodents (Karami-Mohajeri and Abdollahi, 2013), and results from our microarray study, namely the dysregulation of NADH dehydrogenase (ubiquinone) 1 beta subcomplex subunit 11, mitochondrial precursor NDUFB11, NADH dehydrogenase subunits MT-ND4 and MT-ND5, complex III cytochrome b5 type A, complex IV cytochrome c oxidase subunit I (MT-COX1), and cytochrome c oxidase subunit III (MT-COX3), supported the hypothesis that olfactory mitochondria may be targets of these agents. Among the exposure groups, the mixture-M group had the largest number of altered genes in mitochondrial function-related pathways, followed by the MAL-M group, and the CPF-M group. Interestingly, while these genes (ie, MT-COX1, MT-COX3, and MT-ND4) were largely down-regulated in the mixture-M group, the MAL-M group were up-regulated, and none of the above genes significantly changed in the CPF-M group. These observations suggest a modulatory effect on mitochondrial gene expression in the presence of mixtures, or differential effects of the two compounds. It is also important to point out that cellular gene expression changes following chemical exposures are dynamic, and we analyzed our transcriptomic data at only one time point (24 h) following OP exposures. Thus, this study does not address in detail the sequence of dynamic changes in gene expression and compensatory mechanisms in the olfactory mitochondria that may occur following chemical exposures.
In addition to cellular energetics, mitochondria also play a critical role in the production of reactive oxygen species (ROS), which can lead to oxidative damage (Dikalov, 2011; Marchi et al., 2012). The major sites for the generation of ROS are believed to be complexes I and III, where superoxide () is generated and then converted to H2O2 by superoxide dismutase (SOD) (Brookes, 2005; Karami-Mohajeri and Abdollahi, 2013). SOD2 is a key scavenger of in the mitochondrial matrix. Therefore, the decreased expression of SOD2 (45%–62% decrease) in the pesticide mixture group is suggestive of increased oxidative stress in the olfactory system of our animals. Consistent with these observations was our observations of down-regulated glutathione S-transferase omega 1 (GSTO1) in CPF-H, MAL-M, and MAL-H groups, whereas there was induction of GSTO1 by OP mixtures. Although we did not measure the oxidative stress levels in this study using biochemical techniques, the dysregulation of antioxidant genes such as SOD2 and GSTO1 suggests the disruption of redox status in the ORNs. These observations may be linked to the loss of olfactory function, especially when considered with the concomitant partial inhibition of mitochondrial function observed in olfactory rosettes on exposure to the mixtures. The aforementioned hypothesis is consistent with another report showing an elevation in glutathione S-transferase enzymatic activity, presumably has an adaptive response, in rainbow trout receiving pesticide mixture exposures (Tierney et al., 2008).
Our respirometry experiments provided a linkage among biochemical outcomes associated with the disruption of mitochondrial gene expression and indicated that exposure to OPs, individually and in mixtures, reduced functionality of olfactory rosette mitochondria. The maximum achievable rates of mitochondrial respiration (ie, uncoupled respiration) were significantly lower in the MAL-M and mixture-M groups relative to controls, indicating inhibition of mitochondrial respiratory capacity. Furthermore, the significant reduction of uncoupled RCR caused by MAL-M exposure indicated a reduced efficiency of mitochondrial oxidative phosphorylation. Collectively, these results suggest exposure to MAL-M reduces the ability of Coho salmon olfactory rosette mitochondria to generate ATP. Although the mixture-M significantly lowered uncoupled mitochondrial respiration, the mean rate of State 4 respiration was lower than that of the control group, and thus the uncoupled RCRs were not significantly different from controls. A previous report indicated that CPF significantly inhibits activity of complex I of the mitochondrial ETS in a rat neuronal cell line (Lee et al., 2012). Similarly, MAL-M was reported to decrease State 3 respiration in rat liver mitochondria (Spetale et al., 1977). These aforementioned studies involved exposures to CPF and MAL in the micromolar range, far exceeding the environmentally relevant part per billion doses used in this study, and may explain why we did not observe a significant decrease in State 3 respiration from the OP exposures. The effect of OP exposure on cellular mitochondrial copy number within olfactory rosettes was not determined in this study, but could provide insight into the mechanism underlying the significant reduction of maximum respiratory capacity by MAL and the mixture groups.
In summary, we have observed olfactory neurobehavioral dysfunction in salmon exposed to levels of OP mixture concentrations relevant to environmental exposures that were linked at the molecular level to disruption of global olfactory gene expression and to changes in ORN function and impairment of olfactory mitochondrial function at the cellular level. Moreover, the observed inhibition of olfactory-driven behaviors occurred in the absence of pesticide effects on brain AChE. Our findings add mechanistic data in support of other studies that showed OPs can have additive or synergistic toxicity in fish (Laetz et al., 2009, 2013), thus highlighting the need to incorporate mixture models into the current pesticide-risk assessment framework that target low-level sublethal chemical mixtures on ecologically important aquatic species.
FUNDING
National Institutes of Health Superfund Research Program [P42-ES004696], the UW Center for Ecogenetics & Environmental Health [P30ES07033], and the University of Washington Sea Grant Program, pursuant to National Oceanic and Atmospheric Administration Project R/OCEH-8 [NA10OAR4170057].
Supplementary Material
ACKNOWLEDGMENTS
The authors appreciate the assistance of Dr Brian Beckman and Abby Tillotson at NOAA fisheries, Seattle, Washington, who provided the juvenile Coho salmon for these experiments. We also thank Dr Thomas P. Quinn at University of Washington for providing the salmon swimming chamber. The authors declare that they have no competing financial interests.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
REFERENCES
- Anderson P. D., Dugger D. (2008). Surface Water Monitoring Program for Pesticides in Salmonid-Bearing Streams, 2007 Data Summary. Washington State of Department of Ecology, Olympia, WA. [Google Scholar]
- Annau Z. (1992). Neurobehavioral effects of organophosphorus compounds. In Organophosphates—Chemistry, Fate, and Effects (Chambers J. E., Levi P. E., Eds.), pp. 419–432. Academic Press, San Diego. [Google Scholar]
- Brand M. D., Nicholls D. G. (2011). Assessing mitochondrial dysfunction in cells. Biochem. J. 435, 297–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes P. S. (2005). Mitochondrial H(+) leak and ROS generation: an odd couple. Free Radic. Biol. Med. 38, 12–23. [DOI] [PubMed] [Google Scholar]
- Cabello G., Galaz S., Botella L., Calaf G., Pacheco M., Stockert J. C., Villanueva A., Canete M., Juarranz A. (2003). The pesticide malathion induces alterations in actin cytoskeleton and in cell adhesion of cultured breast carcinoma cells. Int. J. Oncol. 23, 697–704. [DOI] [PubMed] [Google Scholar]
- Cau E., Gradwohl G., Casarosa S., Kageyama R., Guillemot F. (2000). Hes genes regulate sequential stages of neurogenesis in the olfactory epithelium. Development 127, 2323–2332. [DOI] [PubMed] [Google Scholar]
- Correll L., Ehrich M. (1991). A microassay method for neurotoxic esterase determinations. Fund. Appl. Toxicol. 16, 110–116. [DOI] [PubMed] [Google Scholar]
- Crumpton T. L., Seidler F. J., Slotkin T. A. (2000). Developmental neurotoxicity of chlorpyrifos in vivo and in vitro: effects on nuclear transcription factors involved in cell replication and differentiation. Brain Res. 857, 87–98. [DOI] [PubMed] [Google Scholar]
- Dikalov S. (2011). Cross talk between mitochondria and NADPH oxidases. Free Radic. Biol. Med. 51, 1289–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellman G. L., Courtney K. D., Andres V., Jr, Feather-Stone R. M. (1961). A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem. Pharmacol. 7, 88–95. [DOI] [PubMed] [Google Scholar]
- EPA (2002). Guidance on Cumulative Risk Assessment of Pesticide Chemicals That Have a Common Mechanism of Toxicity. EPA, Washington, DC. [Google Scholar]
- EPA (2011). Pesticides Industry Sales and Usage: 2006 and 2007 Market Estimates. EPA, Washington, DC. [Google Scholar]
- Espinoza H. M., Williams C. R., Gallagher E. P. (2012). Effect of cadmium on glutathione S-transferase and metallothionein gene expression in coho salmon liver, gill and olfactory tissues. Aquat. Toxicol. 110–111, 37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton M. H., Key P. B. (2001). Acetylcholinesterase inhibition in estuarine fish and invertebrates as an indicator of organophosphorus insecticide exposure and effects. Environ. Toxicol. Chem./SETAC 20, 37–45. [DOI] [PubMed] [Google Scholar]
- Gilliom R. J. (2007). Pesticides in U.S. streams and groundwater. Environ. Sci. Technol. 41, 3408–3414. [DOI] [PubMed] [Google Scholar]
- Hawkins D. K., Quinn T. P. (1996). Critical swimming velocity and associated morphology of juvenile coastal cutthroat trout (Oncorhynchus clarki clarki), steelhead trout (Oncorhynchus mykiss), and their hybrids. Can. J. Fish. Aquat. Sci. 53, 1487–1496. [Google Scholar]
- Huber W., von Heydebreck A., Sultmann H., Poustka A., Vingron M. (2002). Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18(Suppl 1), S96–S104. [DOI] [PubMed] [Google Scholar]
- Jain K. E., Hamilton J. C., Farrell A. P. (1997). Use of a ramp velocity test to measure critical swimming speed in rainbow trout (Onchorhynchus mykiss). Comp. Biochem. Physiol. A Physio. 117, 441–444. [Google Scholar]
- Jarrard H. E., Delaney K. R., Kennedy C. J. (2004). Impacts of carbamate pesticides on olfactory neurophysiology and cholinesterase activity in coho salmon (Oncorhynchus kisutch). Aquat. Toxicol. 69, 133–148. [DOI] [PubMed] [Google Scholar]
- Karami-Mohajeri S., Abdollahi M. (2013). Mitochondrial dysfunction and organophosphorus compounds. Toxicol. Appl. Pharmacol. 270, 39–44. [DOI] [PubMed] [Google Scholar]
- Kaur P., Radotra B., Minz R. W., Gill K. D. (2007). Impaired mitochondrial energy metabolism and neuronal apoptotic cell death after chronic dichlorvos (OP) exposure in rat brain. Neurotoxicology 28, 1208–1219. [DOI] [PubMed] [Google Scholar]
- Laetz C. A., Baldwin D. H., Collier T. K., Hebert V., Stark J. D., Scholz N. L. (2009). The synergistic toxicity of pesticide mixtures: implications for risk assessment and the conservation of endangered Pacific salmon. Environ. Health Perspect. 117, 348–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laetz C. A., Baldwin D. H., Hebert V. R., Stark J. D., Scholz N. (2013). The interactive neurobehavioral toxicity of diazinon, malathion, and ethoprop to juvenile coho salmon. Environ. Sci. Technol. 47, 2925–2931. [DOI] [PubMed] [Google Scholar]
- Lee J. E., Park J. H., Shin I. C., Koh H. C. (2012). Reactive oxygen species regulated mitochondria-mediated apoptosis in PC12 cells exposed to chlorpyrifos. Toxicol. Appl. Pharmacol. 263, 148–162. [DOI] [PubMed] [Google Scholar]
- Lefebvre K. A., Frame E. R., Gulland F., Hansen J. D., Kendrick P. S., Beyer R. P., Bammler T. K., Farin F. M., Hiolski E. M., Smith D. R., et al. (2012). A novel antibody-based biomarker for chronic algal toxin exposure and sub-acute neurotoxicity. PLoS One 7, e36213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchi S., Giorgi C., Suski J. M., Agnoletto C., Bononi A., Bonora M., De Marchi E., Missiroli S., Patergnani S., Poletti F., et al. (2012). Mitochondria-ros crosstalk in the control of cell death and aging. J. Signal Transduct. 2012, 329635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maryoung L. A., Blunt B., Tierney K. B., Schlenk D. (2015). Sublethal toxicity of chlorpyrifos to salmonid olfaction after hypersaline acclimation. Aquat. Toxicol. 161, 94–101. [DOI] [PubMed] [Google Scholar]
- McIntyre J. K., Baldwin D. H., Beauchamp D. A., Scholz N. L. (2012). Low-level copper exposures increase visibility and vulnerability of juvenile coho salmon to cutthroat trout predators. Ecol. Appl. 22, 1460–1471. [DOI] [PubMed] [Google Scholar]
- McIntyre J. K., Baldwin D. H., Meador J. P., Scholz N. L. (2008). Chemosensory deprivation in juvenile coho salmon exposed to dissolved copper under varying water chemistry conditions. Environ. Sci. Technol. 42, 1352–1358. [DOI] [PubMed] [Google Scholar]
- Mileson B. E., Chambers J. E., Chen W. L., Dettbarn W., Ehrich M., Eldefrawi A. T., Gaylor D. W., Hamernik K., Hodgson E., Karczmar A. G., et al. (1998). Common mechanism of toxicity: a case study of organophosphorus pesticides. Toxicol. Sci. 41, 8–20. [DOI] [PubMed] [Google Scholar]
- Miller R. R., Williams J. D., Williams J. E. (1989). Extinctions of North American fishes during the past century. Fisheries 14, 22–38. [Google Scholar]
- Moore D. R., Teed R. S. (2012). Risks of carbamate and organophosphate pesticide mixtures to salmon in the Pacific Northwest. Integr. Environ. Assess. Manag. 9, 70–78. [DOI] [PubMed] [Google Scholar]
- Morgan M. J., Kiceniuk J. W. (1990). Effect of fenitrothion on the foraging behavior of juvenile Atlantic salmon. Environ. Toxicol. Chem. 9, 489–495. [Google Scholar]
- Nakamura T., Gold G. H. (1987). A cyclic nucleotide-gated conductance in olfactory receptor cilia. Nature 325, 442–444. [DOI] [PubMed] [Google Scholar]
- Pala I., Vig P. J., Desaiah D., Srinivasan A. (1991). In vitro effects of organophosphorus compounds on calmodulin activity. J. Appl. Toxicol. JAT 11, 391–395. [DOI] [PubMed] [Google Scholar]
- Ray A., Liu J., Ayoubi P., Pope C. (2010). Dose-related gene expression changes in forebrain following acute, low-level chlorpyrifos exposure in neonatal rats. Toxicol. Appl. Pharmacol. 248, 144–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo D., Teeter J. H., Schild D. (1996). Second messenger signaling in olfactory transduction. J. Neurobiol. 30, 37–48. [DOI] [PubMed] [Google Scholar]
- Sandahl J. F., Baldwin D. H., Jenkins J. J., Scholz N. L. (2004). Odor-evoked field potentials as indicators of sublethal neurotoxicity in juvenile coho salmon (Oncorhynchus kisutch) exposed to copper, chlorpyrifos, or esfenvalerate. Can. J. Fish. Aquat. 61, 404–413. [Google Scholar]
- Sandahl J. F., Baldwin D. H., Jenkins J. J., Scholz N. L. (2005). Comparative thresholds for acetylcholinesterase inhibition and behavioral impairment in coho salmon exposed to chlorpyrifos. Environ. Toxicol. Chem./SETAC 24, 136–145. [DOI] [PubMed] [Google Scholar]
- Sargeant D., Newell E., Anderson P., Cook A. (2013). Surface Water Monitoring Program for Pesticides in Salmon – Bearing Streams, 2009 – 2011 Triennial Report. A Cooperative Study by the Washington State Departments of Ecology and Agriculture. Washington State Department of Ecology Publication 13-03-002. Department of Agriculture Publication AGR PUB 102 – 377 Washington State of Departments of Ecology, Olympia, WA. [Google Scholar]
- Saulsbury M. D., Heyliger S. O., Wang K., Johnson D. J. (2009). Chlorpyrifos induces oxidative stress in oligodendrocyte progenitor cells. Toxicology 259, 1–9. [DOI] [PubMed] [Google Scholar]
- Schild D., Restrepo D. (1998). Transduction mechanisms in vertebrate olfactory receptor cells. Physiol. Rev. 78, 429–466. [DOI] [PubMed] [Google Scholar]
- Scholz N. L., Truelove N. K., French B. L., Berejikian B. A., Quinn T. P., Casillas E., Collier T. K. (2000). Diazinon disrupts antipredator and homing behaviors in chinook salmon (Oncorhynchus tshawytscha). Can. J. Fish. Aquat. Sci. 57, 1911–1918. [Google Scholar]
- Scholz N. L., Truelove N. K., Labenia J. S., Baldwin D. H., Collier T. K. (2006). Dose-additive inhibition of chinook salmon acetylcholinesterase activity by mixtures of organophosphate and carbamate insecticides. Environ. Toxicol. Chem./SETAC 25, 1200–1207. [DOI] [PubMed] [Google Scholar]
- Schuh R. A., Lein P. J., Beckles R. A., Jett D. A. (2002). Noncholinesterase mechanisms of chlorpyrifos neurotoxicity: altered phosphorylation of Ca2+/cAMP response element binding protein in cultured neurons. Toxicol. Appl. Pharmacol. 182, 176–185. [DOI] [PubMed] [Google Scholar]
- Siegel M. P., Kruse S. E., Knowels G., Salmon A., Beyer R., Xie H., Van Remmen H., Smith S. R., Marcinek D. J. (2011). Reduced coupling of oxidative phosphorylation in vivo precedes electron transport chain defects due to mild oxidative stress in mice. PLoS One 6, e26963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G. K. (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, Article3. [DOI] [PubMed] [Google Scholar]
- Song X., Seidler F. J., Saleh J. L., Zhang J., Padilla S., Slotkin T. A. (1997). Cellular mechanisms for developmental toxicity of chlorpyrifos: targeting the adenylyl cyclase signaling cascade. Toxicol. Appl. Pharmacol. 145, 158–174. [DOI] [PubMed] [Google Scholar]
- Spetale M. R., Morisoli L. S., Rodriguez Garay E. A. (1977). The effect of organophosphorus compounds on respiration by rat liver mitochondria. Il Farmaco; Edizione Scientifica 32, 116–122. [PubMed] [Google Scholar]
- Tierney K. B., Baldwin D. H., Hara T. J., Ross P. S., Scholz N. L., Kennedy C. J. (2010). Olfactory toxicity in fishes. Aquat. Toxicol. 96, 2–26. [DOI] [PubMed] [Google Scholar]
- Tierney K. B., Ross P. S., Jarrard H. E., Delaney K. R., Kennedy C. J. (2006). Changes in juvenile coho salmon electro-olfactogram during and after short-term exposure to current-use pesticides. Environ. Toxicol. Chem./SETAC 25, 2809–2817. [DOI] [PubMed] [Google Scholar]
- Tierney K. B., Sampson J. L., Ross P. S., Sekela M. A., Kennedy C. J. (2008). Salmon olfaction is impaired by an environmentally realistic pesticide mixture. Environ. Sci. Technol. 42, 4996–5001. [DOI] [PubMed] [Google Scholar]
- Tilton F. A., Tilton S. C., Bammler T. K., Beyer R. P., Stapleton P. L., Scholz N. L., Gallagher E. P. (2011). Transcriptional impact of organophosphate and metal mixtures on olfaction: copper dominates the chlorpyrifos-induced response in adult zebrafish. Aquat. Toxicol. 102, 205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielma A., Ardiles A., Delgado L., Schmachtenberg O. (2008). The elusive crypt olfactory receptor neuron: evidence for its stimulation by amino acids and cAMP pathway agonists. J. Exp. Biol. 211, 2417–2422. [DOI] [PubMed] [Google Scholar]
- Xin X., Zeng T., Dou D. D., Zhao S., Du J. Y., Pei J. J., Xie K. Q., Zhao X. L. (2011). Changes of mitochondrial ultrastructures and function in central nervous tissue of hens treated with tri-ortho-cresyl phosphate (TOCP). Hum. Exp. Toxicol. 30, 1062–1072. [DOI] [PubMed] [Google Scholar]
- Xing H., Zhang Z., Yao H., Liu T., Wang L., Xu S., Li S. (2014). Effects of atrazine and chlorpyrifos on cytochrome P450 in common carp liver. Chemosphere 104, 244–250. [DOI] [PubMed] [Google Scholar]
- Yeh A., Kruse S. E., Marcinek D. J., Gallagher E. P. (2015). Effect of omega-3 fatty acid oxidation products on the cellular and mitochondrial toxicity of BDE 47. Toxicol. In Vitro 29, 672–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Ito A., Matsuda N., Mishina M. (2002). Regulation by protein kinase A switching of axonal pathfinding of zebrafish olfactory sensory neurons through the olfactory placode-olfactory bulb boundary. J. Neurosci. 22, 4964–4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.