Abstract
Cells alter their gene expression in response to exposure to various environmental changes. Epigenetic mechanisms such as DNA methylation are believed to regulate the alterations in gene expression patterns. In vitro and in vivo studies have documented changes in cellular proliferation, cytoskeletal remodeling, signal transduction, bone mineralization and immune deficiency under the influence of microgravity conditions experienced in space. However microgravity induced changes in the epigenome have not been well characterized. In this study we have used Next-generation Sequencing (NGS) to profile ground-based “simulated” microgravity induced changes on DNA methylation (5-methylcytosine or 5mC), hydroxymethylation (5-hydroxymethylcytosine or 5hmC), and simultaneous gene expression in cultured human lymphoblastoid cells. Our results indicate that simulated microgravity induced alterations in the methylome (~60% of the differentially methylated regions or DMRs are hypomethylated and ~92% of the differentially hydroxymethylated regions or DHMRs are hyperhydroxymethylated). Simulated microgravity also induced differential expression in 370 transcripts that were associated with crucial biological processes such as oxidative stress response, carbohydrate metabolism and regulation of transcription. While we were not able to obtain any global trend correlating the changes of methylation/ hydroxylation with gene expression, we have been able to profile the simulated microgravity induced changes of 5mC over some of the differentially expressed genes that includes five genes undergoing differential methylation over their promoters and twenty five genes undergoing differential methylation over their gene-bodies. To the best of our knowledge, this is the first NGS-based study to profile epigenomic patterns induced by short time exposure of simulated microgravity and we believe that our findings can be a valuable resource for future explorations.
Introduction
During space flight, astronauts are exposed to powerful environmental assaults such as microgravity, cosmic radiation and magnetic fields that have the potential to impinge upon cellular ontogeny through epigenetic modifications [1]. Throughout the evolutionary history, gravity has been a constant factor in defining the architecture and morphology of living beings [2]. Hence a broader understanding of gravity’s influence on biological functions is important for an accurate evaluation of risks associated with the health of astronauts in spaceflights and should be of enormous interest to the scientific community. The effects of microgravity in altering gene expression have been documented in mammalian cells [3, 4] and other model organisms, such as yeast and bacteria [5–7]. Microgravity associated pathological alterations include reduction in bone mass and calcium concentrations [8], alterations in hormonal levels [9], impairment of immunocompetence [10] and apoptosis signaling [11]. Studies of human lymphoblast and lymphoblast cell cultures following periods of simulated microgravity have demonstrated alterations in metabolic processes and DNA repair pathways which could in turn signify an increased susceptibility to malignancy [12, 13]. Collectively, these studies indicate exposure to microgravity during space flight alters gene expression patterns and subsequently cellular physiology.
DNA methylation is regarded as a major epigenetic mechanism and play key roles in regulating cellular processes in living organisms [14, 15]. Biochemically, DNA methylation refers to the addition of a methyl group (CH3) to the 5’ carbon on the pyrimidine ring of cytosine nucleotides (commonly abbreviated as 5mC). Aberrations in genome-wide 5mC patterns are widely prevalent in cancer and other diseases [14, 16–18]. Traditionally DNA methylation marks have been associated with “transcriptionally silent” genes, however the revelations of global methylation studies facilitated by recent advances in Next Generation Sequencing (NGS) tools have established that the role of 5mC in regulating gene expression is complex, varies according to the genomic context and warrants extensive explorations [19–25]. Discovered in 2009, DNA hydroxymethylation (5hmC) is a relatively new epigenetic modification occurring on Cytosine [26, 27] generated by Ten-Eleven Translocation (TET) protein- mediated oxidative catalysis of 5mC [26]. Though, potential roles of 5hmC at promoter and gene bodies are not clearly understood, it is shown to play some role in maintaining and/or promoting gene expression [14, 16–18, 28]. Microgravity induced alteration in DNA methylation patterns have been reported previously [29–31] but the effect of microgravity on 5hmC is virtually unknown. During the time period this study was being conducted there were no reports of a NGS based study documenting the effects of microgravity on the epigenomic landscape.
The goal of our study was to profile genome-wide effects of “simulated” microgravity on 5mC, 5hmC and gene expression patterns employing Next Generation Sequencing (Fig 1). The TK6 lymphoblastoid cell line, derived from T cell blast crisis of a patient with chronic myelogeneous leukemia [32], served as our model organism. TK6 cells are well characterized and have been extensively used as a substitute for peripheral blood lymphocytes for immunological and epidemiological studies [13]. The limited availability of biological specimen subjected to conditions of microgravity in spaceflights makes ground based “simulated” microgravity studies critical in determining thresholds and thorough testing of the model organism before conducting the experiments during space missions [33]. A High Aspect Ratio Vessel (HARV) based rotary cell culture system (initially developed by NASA) was used in our study to “simulate” microgravity in the TK6 cells as has been described previously [34] and compared to a control static cell-culture system under the influence of earth’s gravity. While assessing the merits of ground based “simulation” studies, it has to be appreciated that the effects of gravity cannot be completely negated but reduced to near zero to achieve a state of “functional near weightlessness” [33].
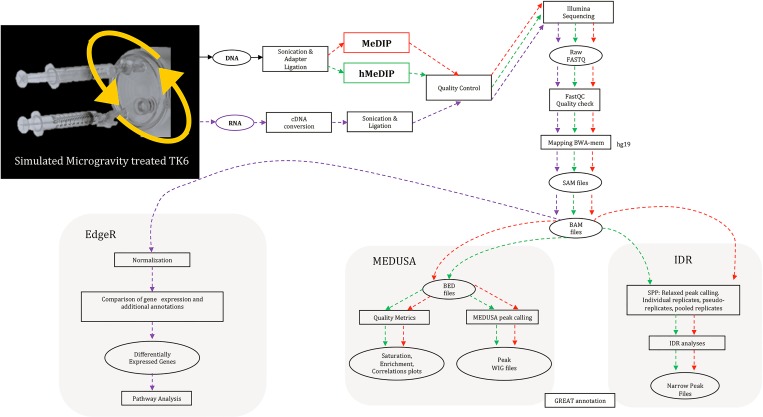
Fig 1. Schematic illustration of the bioinformatics pipeline for MeDIP-seq, hMeDIP-seq and RNA-seq analysis used in our study to understand the DNA methylation and hydroxymethylation and gene expression patterns induced by simulated microgravity.
All steps were done in parallel in TK6 subjected to “simulated” microgravity and static controls under the influence of Earth’s gravitational force.
Materials and Methods
Cell culture
TK6 human lymphoblastoid cells (ATCC, Manassas, VA) were maintained in the log phase of cell growth by culturing in RPMI-1640 (Life Technologies, Grand Island, NY) medium supplemented with 10% Fetal Bovine Serum (Atlanta Biologicals, Flowery Branch, GA) and 1% Penicillin/Streptomycin (Life Technologies, Grand Island, NY) at 37°C in 5% CO2 and 95% air. For ground-based simulation of microgravity, HARV Rotary Cell Culture System (Synthecon, Houston, TX) was used. Actively growing TK6 cells were seeded in the bioreactor at 2 X 105 cells/ml and rotated at 12 rpm/min. In parallel, cells (at the same cellular density i.e. 2 X 105 cells/ml) were maintained in bioreactors in normal gravity (static) condition as controls. The bioreactors were maintained in an incubator at 37°C, with 5% CO2 and 95% air for 48 hours.
DNA isolation, sonication and adapter ligation
Genomic DNA was isolated from the TK6 cells cultured under microgravity and control static conditions using the DNeasy Blood &Tissue kit (Qiagen Inc., Valencia, CA) following manufacturer’s instructions. 2.5 μg of genomic DNA from each sample was sheared using Covaris S2 Device (Covaris Inc., Woburn, MA). Sheared DNA was purified by binding to AmPure beads (Beckman Coulter Inc.) and End-repair performed by incubating sonicated DNA and End repair solution (New England Biolabs Inc., Ipswich, MA) as per manufacturer’s specifications. A-tailing was obtained by incubating the end repaired DNA with dA-tailing mix (New England Biolabs Inc., Ipswich, MA) at 37°C for 30 minutes. At this stage, to facilitate multiplexing each sample was equally divided in two parts (one half for MeDIP and the other half for hMeDIP respectively). Blunt end ligation was performed by incubating the A-tailed DNA samples (1 μg) with unmethylated versions of adapters (IDT Inc., Coralville, IA) based on sequences of the methylated Truseq adapters (Illumina Inc., San Diego, CA) for multiplexing. Thus 8 libraries were prepared as described above. Samples were assayed by qPCR in duplicate and standard curve constructed to establish the molarities of the libraries.
MeDIP-seq /hMeDIP-seq
MeDIP and hMeDIP were performed using the methylated/hydroxymethylated DNA enrichment kits (Diagenode Inc., Denville, NJ) following the manufacturer’s protocol. Briefly, to 1.2 μg of adapter ligated sonicated genomic DNA, three DNA controls (known sequences bearing unmethylated, methylated or hydroxymethylated Cytosines respectively to assess the efficiency of immunoprecipitation reactions) were spiked-in. The concentration of genomic DNA was adjusted to incorporate the addition of the adapter sequences, preserving the appropriate molar ratio between the genomic DNA and anti-5mC/anti-5hmC antibody during MeDIP/hMeDIP as described by Butcher et al. [35]:
Where,
conc.gDNA ➔ adjusted genomic DNA concentration in the adapter ligated libraries,
conc.adapter ligated gDNA ➔ concentration of the adapter ligated gDNA libraries,
bpsonicated gDNA ➔ average size of the pre-ligation sonicated gDNA and
bpadapter DNA ➔ average size of the adapters
After incubation at 95°C to denature the double stranded DNA, immunoprecipitation was performed by incubation with monoclonal antibody directed against 5mC/5hmC (Diagenode Inc., Denville, NJ) and secondary antibody with magnetic bead conjugates (Diagenode Inc., Denville, NJ) overnight at 4°C while being spun continuously at 40 rpm. The captured 5mC/5hmC bearing DNA fragments were separated from the others by magnetic pulldown. After repeated cleanups, the captured DNA was isolated from the magnetic beads bearing antibody using the IPure kit (Diagenode Inc., Denville, NJ). The enrichment of 5mC/5hmC bearing DNA was assessed by performing qPCR on the pre and post immunoprecipitated samples. As a control, an identical immunoprecipitation reaction with mouse IgG instead of monoclonal 5mC/5hmC antibody was performed. The methylated/hydroxymethylated DNA immunoprecipitated libraries were amplified by PCR and submitted to the Purdue Genomics Core Facility for high-throughput sequencing by Hi Seq 2000 (Illumina Inc., San Diego, CA).
MeDIP-seq and hMeDIP-seq data processing
FastQC v 0.10.1 [36] was used to assess the quality of the reads and to generate graphical representations of numerous quality metrics (per base sequence quality, GC content and sequence duplication/size distribution levels). The reads were aligned to human reference genome hg19 using BWA v 0.6.2 [37], with default parameters and a maximum insert size of 400 bp. The resulting SAM files were converted to BAM format and sorted using Samtools v0.1.18 [38] as illustrated in (Fig 1). PERL script from the MeDUSA package [39] was used to convert the BAM files to BED format. Since the MEDIPS v1.0 [40] package requires only selective fields as input, the BED format was then reduced to four fields using the UNIX cut option. The MeDUSA pipeline utilizes the Bioconductor package MEDIPS v1.0 and custom R scripts to calculate quality metrics for the MeDIP-seq data were designed. The data was normalized for the size of the sequence libraries by calculating reads per million (RPM) in tiled windows across the genome. Wig files obtained for the normalized read depth following alignment and filtering were presented as RPM. Quality check on the MeDIP-seq data was also performed by calculating CpG enrichment values, saturation plots and coverage plots. Genome-wide correlations between the replicates were performed as a quality check for consistency among the replicates using QCSeqs from the Useq package (v8.40) [41] using a window size of 500 bp, increasing in 250 bp increments and a minimum number of 5 reads in a window.
Identification of DMRs and DHMRs
Peak calling software SPP v1.10 [42], was used to call peaks and rank them based on significance of enrichment (p-values and false discovery rates). IDR (Irreproducible Discovery Rate) framework was used to measure experiment quality in terms of reproducibility [43] and to select the reproducible, consistent peaks (overlapped significant peaks from both replicates) determined based on IDR values. The threshold of 0.05 IDR was used for truncating the peak list as suggested by the developers. The differentially methylated/hydroxymethylated regions identified by IDR analyses were then annotated with their chromosomal locations and feature types for further biological interpretation using custom Perl scripts of MeDUSA package, BEDTools [44] and feature annotation files (GFF files from UCSC) as illustrated in Fig 1. Further annotation (plots for enrichment of 5mC/5hmC) was done using CEAS (v1.02) [45] and proximity of the peaks to the TSS was determined using PeakAnalyzer [46]. The FDR value of 0.05 was used as cut-off for all peak association studies. The complete MeDIP-seq and hmeDIP-seq data was submitted to NCBI GEO (GSE65944) and available in the database http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE65944).
RNA-seq
Total RNA was extracted from cells subjected to simulated microgravity and static control using RNA-STAT-60 (Tel-Test,Inc., Friendswood, TX) using the manufacturer’s instructions. Briefly, 1ml of RNA-STAT solution was added per 106 cells and homogenized for 5 minutes over ice. 1ml of chloroform was added, contents shaken vigorously and centrifuged at 12,000g at 4°C for 15 minutes. The aqueous solution was transferred to corex tube (Corning Inc., Lowell, MA) and 0.8 ml isopropanol added. After incubation of 10 minutes, the contents were centrifuged at 12,000g for another 10 minutes to precipitate the RNA. The RNA pellet was washed with 75% ethanol and centrifuged at 7,500g for 5 minutes at 4°C. The ethanol was aspirated and the RNA pellet dried. The RNA pellet was finally resuspended in DEPC water and submitted to the Purdue Genomic Center for conversion into cDNA, sonication, adapter ligation and sequencing as described previously. The reads (fastq files) were aligned to human reference genome hg19 using Tophat v2.1.0 [47], with default parameters and known transcriptome as illustrated in Fig 1. Alignment results were filtered by Bamutils v0.5.0 [48] to remove reads with multiple mappings. Statistics data of the resulting alignment files were created using Samtools v0.1.18 [49] and Bamutils v0.5.0. The counts of aligned reads mapping to known genes were calculated using bamutils v0.5.0. EdgeR v2.11 [50] was used to compute the differentially expressed genes. Pathway analysis on the set of differentially expressed genes was done using the GeneCodis3 software designed at the Complutense University of Madrid [51].The complete RNA-seq data was submitted to NCBI GEO (GSE65944) and available in the database (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE65944).
Results
Effect of simulated microgravity on cell growth and viability
The effect of simulated microgravity on cell growth and viability 48 hours after the cells were seeded in bioreactors in either the rotating or static condition was determined using Trypan Blue staining method by Automated Cell Counter (Nexcelom Bioscience LLC., Lawrence, MA) in S1 Fig. No significant differences in the percentage of viable cells between the two cell culture conditions after 48 hours was observed. Specifically, 95.1 ± 2.12% of the cells subjected to simulated microgravity were viable, while 91.1 ± 3.54% of the static control cells were determined to be viable. The average cellular diameter (μm) was determined to be 12.6 ± 0.42 and 12.8 ± 0.28 in TK6 cells subjected to microgravity and control respectively. Similar cellular growth rates between the rotating and static culture conditions facilitated ruling out the possibility of cell growth being the major contributor to the changes in the methylation and gene expression patterns.
Changes in 5mC profile following simulated microgravity
We applied MeDIP coupled with high-throughput sequencing to identify the differences in the genome-wide patterns of 5mC upon simulated microgravity on TK6 cells. 2.8x108 and 1.8x108 reads were obtained during MeDIP-seq from TK6 cells subjected to static and simulated microgravity respectively and more than 90% of these reads aligned to the human genome GRCh37/hg19, 2009 Assembly (S1 Table and S2 Fig). Quality assessment generated by FastQC [36] showed satisfactory sequence quality for all measures except for GC content. As GC rich regions of the genome are enriched in MeDIP-seq datasets, this result was not unexpected. The depth of sequencing for MeDIP-seq samples ranged from 2.8X to 6.1X (S1 Table). Cross-correlation analysis was performed as per the ENCODE consortium guidelines [42, 52, 53] and all the samples displayed Normalized Strand Correlation (NSC) and Relative Strand Correlation (RSC) values (S1 Table) characteristic of “high-quality data sets”. The similarity between the replicates was evident as hierarchical dendrogram displayed distinct clustering of biological replicates in two groups (S3 Fig) and sequence coverage analyses displayed that MeDIP-seq reads generated from the samples covered similar number of bases of the reference genome (S4 Fig).
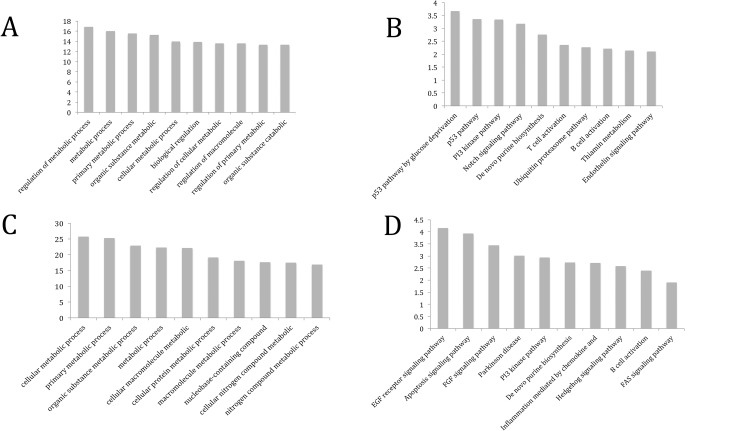
Differentially methylated region (DMRs) were defined as genomic regions in TK6 cells under simulated microgravity that showed alteration in methylation (either increase or decrease) compared to TK6 cells under static conditions. 3204 DMRs (S2 Table & S3 Table) were detected using the IDR pipeline having an IDR cutoff value of 0.05 or less. Of the total DMRs, 1286 (40.14%) were associated with hypermethylation (gain-of-5mC) (S2 Table) and 1918 (59.86%) with hypomethylation (loss-of-5mC) (S3 Table) upon simulated microgravity respectively. The DMRs were further analyzed to determine the overlap of DMR regions with different genomic features by the methylome analysis pipeline described in details by Wilson et al. [39]. Functional genomic distribution analyses indicated that 969 and 1381 genes associated with DMRs have undergone gain-of-5mC and loss-of-5mC respectively (Table 1). Also, 105 hypermethylated and 193 hypomethylated DMRs were observed around -1500 to 1500 bps of Transcription Start Sites (TSS) as demonstrated in Tables 2 & 3. The distribution of the genomic repeat sequences (LINE, SINE and LTR) located within the DMRs has been represented in Table 1. Metadata describing features such as genes, transcripts, Pseudogene, non-coding RNA and other regulatory features present on each DMR has been included in S2 Table & S3 Table. Investigation of annotations from 20 different ontologies from genomic coordinates of DMRs was generated by utilizing Stanford University’s Genomic Regions Enrichment of Annotations Tool (GREAT) version 3.0.0 [54] and included in S4 Table & S5 Table. Gain-of-5mC DMRs induced by simulated microgravity were found to enrich GO Biological Processes like regulation of metabolic process (GO: 0019222), primary metabolic process (GO: 0044238) and cellular metabolic process (GO: 0044237) (Fig 2A). PANTHER Pathway Analysis implicated genes involved in p53 pathway (P00059), PI3 kinase pathway (P00048), T cell activation (P00053) and B cell activation (P00010) to be associated with hypermethylated DMRs (Fig 2B). On the other hand, loss-of-5mC DMRs were observed to enrich GO Biological Processes like cellular metabolic process (GO: 0044237) and primary metabolic process (GO: 0044238) (Fig 2C). PANTHER Pathway Analysis, revealed that these hypomethylated DMRs were associated with genes involved in EGF receptor signaling (P00018), Apoptosis signaling (P00006) and FGF signaling (P00021) pathways among others (Fig 2D).
Table 1. Genome annotation Summary.
The number of genomic features such as CpG islands, CpG shores, ENSEMBL Genes and DNA Repeats (LINE, SINE and LTR) associated with regions undergoing gain-of-5mC/5hmC and loss-of-5mC/5hmC DMRs or DHMRs in TK6 cells cultured under simulated microgravity compared to static condition.
| Features | DMR | DHMR | ||
|---|---|---|---|---|
| Gain-of-5mC | Loss-of-5mC | Gain-of-5hmC | Loss-of-5hmC | |
| CpGI | 23 | 69 | 1 | 0 |
| CpG | 127 | 277 | 5 | 1 |
| Gene | 969 | 1381 | 86 | 7 |
| LINE | 421 | 521 | 47 | 1 |
| SINE | 944 | 1973 | 18 | 11 |
| LTR | 157 | 227 | 28 | 0 |
Table 2. List of hypermethylated DMRs located within +/- 1500 of Transcription Start Sites of genes.
Columns display the genomic coordinates of DMRs, Gene Symbol of the corresponding gene, the description of the genome and the exact distance in bp.
| DMR (Chr:Start-End) | Gene Symbol | Description | Distance |
|---|---|---|---|
| 2:74407290–74407690 | MOB1A | MOB kinase activator 1A | -1495 |
| 1:32620788–32621188 | KPNA6 | karyopherin alpha 6 (importin alpha 7) | -1475 |
| 8:48919307–48919707 | UBE2V2 | ubiquitin-conjugating enzyme E2 variant 2 | -1453 |
| 20:10644375–10644775 | JAG1 | jagged 1 | -1421 |
| 19:8526792–8527192 | HNRNPM | heterogeneous nuclear ribonucleoprotein M | -1389 |
| 10:103579850–103580250 | MGEA5 | meningioma expressed antigen 5 (hyaluronidase) | -1354 |
| 1:176177694–176178094 | RFWD2 | ring finger and WD repeat domain 2, E3 ubiquitin protein ligase | -1265 |
| 8:66547493–66547893 | ARMC1 | armadillo repeat containing 1 | -1251 |
| 17:73976545–73976945 | ACOX1 | acyl-CoA oxidase 1, palmitoyl | -1230 |
| 5:137912163–137912563 | HSPA9 | heat shock 70kDa protein 9 (mortalin) | -1230 |
| 1:9710401–9710801 | PIK3CD | phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta | -1189 |
| 17:76820294–76820694 | USP36 | ubiquitin specific peptidase 36 | -1163 |
| 7:144108260–144108660 | NOBOX | NOBOX oogenesis homeobox | -1140 |
| 1:179333515–179333915 | AXDND1 | axonemal dynein light chain domain containing 1 | -1140 |
| 7:150781644–150782044 | AGAP3 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 | -1110 |
| 3:27765104–27765504 | EOMES | eomesodermin | -1098 |
| 1:27096449–27096849 | ARID1A | AT rich interactive domain 1A (SWI-like) | -1071 |
| 17:37608338–37608738 | MED1 | mediator complex subunit 1 | -999 |
| 4:94748859–94749259 | ATOH1 | atonal homolog 1 (Drosophila) | -983 |
| 10:111968848–111969248 | MXI1 | MAX interactor 1, dimerization protein | -941 |
| 20:35488994–35489394 | SOGA1 | suppressor of glucose, autophagy associated 1 | -918 |
| 22:51067323–51067723 | ARSA | arylsulfatase A | -916 |
| 10:112630503–112630903 | PDCD4 | programmed cell death 4 (neoplastic transformation inhibitor) | -862 |
| 16:75468037–75468437 | CFDP1 | craniofacial development protein 1 | -854 |
| 17:4235771–4236171 | UBE2G1 | ubiquitin-conjugating enzyme E2G 1 | -753 |
| 17:47492799–47493199 | PHB | prohibitin | -753 |
| 3:99978894–99979294 | TBC1D23 | TBC1 domain family, member 23 | -750 |
| 17:33469869–33470269 | NLE1 | notchless homolog 1 (Drosophila) | -735 |
| 17:65026584–65026984 | AC005544.1 | Uncharacterized protein | -725 |
| 2:88895888–88896288 | EIF2AK3 | eukaryotic translation initiation factor 2-alpha kinase 3 | -713 |
| 1:150265379–150265779 | MRPS21 | mitochondrial ribosomal protein S21 | -710 |
| 4:37827346–37827746 | PGM2 | phosphoglucomutase 2 | -709 |
| 20:42573450–42573850 | TOX2 | TOX high mobility group box family member 2 | -695 |
| 1:151738242–151738642 | OAZ3 | ornithine decarboxylase antizyme 3 | -689 |
| 11:33277352–33277752 | HIPK3 | homeodomain interacting protein kinase 3 | -666 |
| 15:60691398–60691798 | ANXA2 | annexin A2 | -657 |
| 17:3716988–3717388 | C17orf85 | chromosome 17 open reading frame 85 | -644 |
| 10:8095826–8096226 | GATA3 | GATA binding protein 3 | -630 |
| 9:123295232–123295632 | CDK5RAP2 | CDK5 regulatory subunit associated protein 2 | -599 |
| 7:48019521–48019921 | HUS1 | HUS1 checkpoint homolog (S. pombe) | -571 |
| 12:120109292–120109692 | PRKAB1 | protein kinase, AMP-activated, beta 1 non-catalytic subunit | -557 |
| 15:43637360–43637760 | ADAL | adenosine deaminase-like | -545 |
| 5:122180427–122180827 | SNX24 | sorting nexin 24 | -517 |
| 3:113676612–113677012 | ZDHHC23 | zinc finger, DHHC-type containing 23 | -489 |
| 19:58400668–58401068 | ZNF814 | zinc finger protein 814 | -463 |
| 14:93184061–93184461 | LGMN | legumain | -457 |
| 20:45280352–45280752 | SLC13A3 | solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3 | -454 |
| 2:8977951–8978351 | KIDINS220 | kinase D-interacting substrate, 220kDa | -391 |
| 20:30639472–30639872 | HCK | hemopoietic cell kinase | -319 |
| 5:67521976–67522376 | PIK3R1 | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | -286 |
| 16:25027042–25027442 | ARHGAP17 | Rho GTPase activating protein 17 | -255 |
| 19:11306249–11306649 | KANK2 | KN motif and ankyrin repeat domains 2 | -88 |
| 12:110783800–110784200 | ATP2A2 | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 | -6 |
| 13:78493694–78494094 | EDNRB | endothelin receptor type B | 9 |
| 13:111839015–111839415 | ARHGEF7 | Rho guanine nucleotide exchange factor (GEF) 7 | 14 |
| 3:180633094–180633494 | FXR1 | fragile X mental retardation, autosomal homolog 1 | 46 |
| 12:122457270–122457670 | BCL7A | B-cell CLL/lymphoma 7A | 142 |
| 14:95569392–95569792 | DICER1 | dicer 1, ribonuclease type III | 175 |
| 3:64253200–64253600 | PRICKLE2 | prickle homolog 2 (Drosophila) | 255 |
| 19:45445601–45446001 | APOC4 | apolipoprotein C-IV | 270 |
| 13:114549584–114549984 | GAS6 | growth arrest-specific 6 | 272 |
| 5:68470171–68470571 | CCNB1 | cyclin B1 | 287 |
| 4:89079332–89079732 | ABCG2 | ATP-binding cassette, sub-family G (WHITE), member 2 | 291 |
| 22:41257564–41257964 | DNAJB7 | DnaJ (Hsp40) homolog, subfamily B, member 7 | 366 |
| 15:83676793–83677193 | C15orf40 | chromosome 15 open reading frame 40 | 375 |
| 13:113301551–113301951 | C13orf35 | chromosome 13 open reading frame 35 | 393 |
| 9:130340673–130341073 | FAM129B | family with sequence similarity 129, member B | 395 |
| 21:34804877–34805277 | IFNGR2 | interferon gamma receptor 2 (interferon gamma transducer 1) | 451 |
| 12:70728471–70728871 | CNOT2 | CCR4-NOT transcription complex, subunit 2 | 456 |
| 4:80993052–80993452 | ANTXR2 | anthrax toxin receptor 2 | 465 |
| 19:58741108–58741508 | ZNF544 | zinc finger protein 544 | 474 |
| 11:85565301–85565701 | AP000974.1 | CDNA FLJ26432 fis, clone KDN01418; Uncharacterized protein | 485 |
| 19:40831600–40832000 | C19orf47 | chromosome 19 open reading frame 47 | 530 |
| 5:137070955–137071355 | KLHL3 | kelch-like family member 3 | 549 |
| 19:10120383–10120783 | COL5A3 | collagen, type V, alpha 3 | 564 |
| 17:78389846–78390246 | ENDOV | endonuclease V | 577 |
| 3:101231200–101231600 | SENP7 | SUMO1/sentrin specific peptidase 7 | 628 |
| 13:28673851–28674251 | FLT3 | fms-related tyrosine kinase 3 | 656 |
| 18:32919753–32920153 | ZNF24 | zinc finger protein 24 | 665 |
| 2:202644765–202645165 | ALS2 | amyotrophic lateral sclerosis 2 (juvenile) | 680 |
| 17:17183036–17183436 | COPS3 | COP9 constitutive photomorphogenic homolog subunit 3 (Arabidopsis) | 778 |
| 19:12661227–12661627 | ZNF564 | zinc finger protein 564 | 821 |
| 8:107283104–107283504 | OXR1 | oxidation resistance 1 | 831 |
| 2:25390345–25390745 | POMC | proopiomelanocortin | 895 |
| 3:192959642–192960042 | HRASLS | HRAS-like suppressor | 928 |
| 19:54662449–54662849 | LENG1 | leukocyte receptor cluster (LRC) member 1 | 971 |
| 12:100595414–100595814 | ACTR6 | ARP6 actin-related protein 6 homolog (yeast) | 985 |
| 13:27828691–27829091 | RPL21 | ribosomal protein L21 | 1049 |
| 12:57858360–57858760 | GLI1 | GLI family zinc finger 1 | 1085 |
| 17:74476687–74477087 | RHBDF2 | rhomboid 5 homolog 2 (Drosophila) | 1089 |
| 19:38712475–38712875 | DPF1 | D4, zinc and double PHD fingers family 1 | 1138 |
| 19:53138925–53139325 | ZNF83 | zinc finger protein 83 | 1214 |
| 9:6644198–6644598 | GLDC | glycine dehydrogenase (decarboxylating) | 1252 |
| 3:172362558–172362958 | AC007919.2 | HCG1787166; PRO1163 | 1275 |
| 17:71230451–71230851 | C17orf80 | chromosome 17 open reading frame 80 | 1286 |
| 12:123875689–123876089 | SETD8 | SET domain containing (lysine methyltransferase) 8 | 1300 |
| 7:98479823–98480223 | TRRAP | transformation/transcription domain-associated protein | 1310 |
| 7:72396789–72397189 | POM121 | POM121 transmembrane nucleoporin | 1329 |
| 4:187646331–187646731 | FAT1 | FAT tumor suppressor homolog 1 (Drosophila) | 1345 |
| 2:216256316–216256716 | FN1 | fibronectin 1 | 1354 |
| 17:40654447–40654847 | ATP6V0A1 | ATPase, H+ transporting, lysosomal V0 subunit a1 | 1387 |
| 12:111857341–111857741 | SH2B3 | SH2B adaptor protein 3 | 1397 |
| 12:110925896–110926296 | FAM216A | family with sequence similarity 216, member A | 1400 |
| 2:53996217–53996617 | CHAC2 | ChaC, cation transport regulator homolog 2 (E. coli) | 1488 |
| 2:947917–948317 | SNTG2 | syntrophin, gamma 2 | 1492 |
Table 3. List of hypomethylated DMRs located within +/- 1500 of Transcription Start Sites of genes.
Columns display the genomic coordinates of DMRs, Gene Symbol of the corresponding gene, the description of the genome and the exact distance in base pairs.
| DMR (Chr:Start-End) | Gene Symbol | Description | Distance |
|---|---|---|---|
| 1:45958152–45958568 | TESK2 | testis-specific kinase 2 | -1488 |
| 13:21138922–21139338 | IFT88 | intraflagellar transport 88 homolog (Chlamydomonas) | -1455 |
| 12:6831258–6831674 | COPS7A | COP9 constitutive photomorphogenic homolog subunit 7A (Arabidopsis) | -1441 |
| 11:107990630–107991046 | ACAT1 | acetyl-CoA acetyltransferase 1 | -1405 |
| 19:569701–570117 | BSG | basigin (Ok blood group) | -1388 |
| 19:54664797–54665213 | LENG1 | leukocyte receptor cluster (LRC) member 1 | -1385 |
| 14:50232737–50233153 | KLHDC2 | kelch domain containing 2 | -1381 |
| 11:47289128–47289544 | MADD | MAP-kinase activating death domain | -1376 |
| 17:33465350–33465766 | NLE1 | notchless homolog 1 (Drosophila) | -1372 |
| 2:72372691–72373107 | CYP26B1 | cytochrome P450, family 26, subfamily B, polypeptide 1 | -1355 |
| 20:34543689–34544105 | SCAND1 | SCAN domain containing 1 | -1349 |
| 8:76318743–76319159 | HNF4G | hepatocyte nuclear factor 4, gamma | -1320 |
| 17:48946441–48946857 | TOB1 | transducer of ERBB2, 1 | -1310 |
| 16:3931818–3932234 | CREBBP | CREB binding protein | -1299 |
| 2:211306550–211306966 | LANCL1 | LanC lantibiotic synthetase component C-like 1 (bacterial) | -1289 |
| 1:47780891–47781307 | STIL | SCL/TAL1 interrupting locus | -1280 |
| 17:16333898–16334314 | TRPV2 | transient receptor potential cation channel, subfamily V, member 2 | -1263 |
| 3:12706770–12707186 | RAF1 | v-raf-1 murine leukemia viral oncogene homolog 1 | -1253 |
| 19:49577242–49577658 | KCNA7 | potassium voltage-gated channel, shaker-related subfamily, member 7 | -1252 |
| 5:175974933–175975349 | CDHR2 | cadherin-related family member 2 | -1251 |
| 5:156363707–156364123 | TIMD4 | T-cell immunoglobulin and mucin domain containing 4 | -1228 |
| 12:49111699–49112115 | CCNT1 | cyclin T1 | -1226 |
| 17:79607945–79608361 | TSPAN10 | tetraspanin 10 | -1196 |
| 1:25257327–25257743 | RUNX3 | runt-related transcription factor 3 | -1167 |
| 12:110460055–110460471 | ANKRD13A | ankyrin repeat domain 13A | -1161 |
| 22:43011912–43012328 | POLDIP3 | polymerase (DNA-directed), delta interacting protein 3 | -1152 |
| 13:52981570–52981986 | THSD1 | thrombospondin, type I, domain containing 1 | -1149 |
| 9:103203198–103203614 | MSANTD3-TMEFF1 | MSANTD3-TMEFF1 readthrough | -1147 |
| 9:123584680–123585096 | PSMD5 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 5 | -1144 |
| 19:10827435–10827851 | DNM2 | dynamin 2 | -1112 |
| 1:32404887–32405303 | PTP4A2 | protein tyrosine phosphatase type IVA, member 2 | -1107 |
| 2:203129141–203129557 | NOP58 | NOP58 ribonucleoprotein | -1090 |
| 12:53474061–53474477 | SPRYD3 | SPRY domain containing 3 | -1065 |
| 4:87814423–87814839 | C4orf36 | chromosome 4 open reading frame 36 | -1062 |
| 17:29157723–29158139 | ATAD5 | ATPase family, AAA domain containing 5 | -1057 |
| 19:50029617–50030033 | RCN3 | reticulocalbin 3, EF-hand calcium binding domain | -1050 |
| 9:19150115–19150531 | PLIN2 | perilipin 2 | -1047 |
| 7:138666899–138667315 | KIAA1549 | KIAA1549 | -1043 |
| 11:66446181–66446597 | RBM4B | RNA binding motif protein 4B | -997 |
| 16:88783445–88783861 | PIEZO1 | piezo-type mechanosensitive ion channel component 1 | -968 |
| 5:176828391–176828807 | PFN3 | profilin 3 | -962 |
| 6:35309180–35309596 | PPARD | peroxisome proliferator-activated receptor delta | -947 |
| 3:52805678–52806094 | NEK4 | NIMA-related kinase 4 | -921 |
| 14:52291822–52292238 | GNG2 | guanine nucleotide binding protein (G protein), gamma 2 | -883 |
| 1:150292846–150293262 | PRPF3 | PRP3 pre-mRNA processing factor 3 homolog (S. cerevisiae) | -871 |
| 1:28560198–28560614 | DNAJC8 | DnaJ (Hsp40) homolog, subfamily C, member 8 | -870 |
| 17:1626772–1627188 | WDR81 | WD repeat domain 81 | -854 |
| 16:72137332–72137748 | DHX38 | DEAH (Asp-Glu-Ala-His) box polypeptide 38 | -845 |
| 18:77961381–77961797 | PARD6G | par-6 partitioning defective 6 homolog gamma (C. elegans) | -825 |
| 13:42621863–42622279 | DGKH | diacylglycerol kinase, eta | -818 |
| 13:77461149–77461565 | KCTD12 | potassium channel tetramerisation domain containing 12 | -817 |
| 22:43037207–43037623 | ATP5L2 | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G2 | -808 |
| 15:42076825–42077241 | AC073657.1 | -804 | |
| 5:170189356–170189772 | GABRP | gamma-aminobutyric acid (GABA) A receptor, pi | -790 |
| 20:34000467–34000883 | UQCC | ubiquinol-cytochrome c reductase complex chaperone | -731 |
| 9:99802448–99802864 | CTSL2 | cathepsin L2 | -731 |
| 22:50766008–50766424 | DENND6B | DENN/MADD domain containing 6B | -727 |
| 22:19279757–19280173 | CLTCL1 | clathrin, heavy chain-like 1 | -726 |
| 1:154244054–154244470 | HAX1 | HCLS1 associated protein X-1 | -725 |
| 9:125591423–125591839 | PDCL | phosducin-like | -721 |
| 15:40401584–40402000 | BMF | Bcl2 modifying factor | -699 |
| 12:48745657–48746073 | ZNF641 | zinc finger protein 641 | -668 |
| 1:107598393–107598809 | PRMT6 | protein arginine methyltransferase 6 | -666 |
| 5:134073321–134073737 | CAMLG | calcium modulating ligand | -662 |
| 19:2256862–2257278 | JSRP1 | junctional sarcoplasmic reticulum protein 1 | -654 |
| 4:17783579–17783995 | FAM184B | family with sequence similarity 184, member B | -652 |
| 6:25965887–25966303 | TRIM38 | tripartite motif containing 38 | -649 |
| 2:242186700–242187116 | HDLBP | high density lipoprotein binding protein | -629 |
| 7:43909561–43909977 | MRPS24 | mitochondrial ribosomal protein S24 | -613 |
| 20:44440411–44440827 | UBE2C | ubiquitin-conjugating enzyme E2C | -596 |
| 8:99075748–99076164 | C8orf47 | chromosome 8 open reading frame 47 | -583 |
| 8:19680112–19680528 | INTS10 | integrator complex subunit 10 | -579 |
| 15:60885693–60886109 | RORA | RAR-related orphan receptor A | -576 |
| 9:124856243–124856659 | TTLL11 | tubulin tyrosine ligase-like family, member 11 | -566 |
| 12:123948281–123948697 | SNRNP35 | small nuclear ribonucleoprotein 35kDa (U11/U12) | -564 |
| 4:190861171–190861587 | FRG1 | FSHD region gene 1 | -564 |
| 19:11450690–11451106 | RAB3D | RAB3D, member RAS oncogene family | -554 |
| 10:126489606–126490022 | FAM175B | family with sequence similarity 175, member B | -540 |
| 7:129691616–129692032 | ZC3HC1 | zinc finger, C3HC-type containing 1 | -533 |
| 13:103458965–103459381 | RP11-484I6.7 | BIVM-ERCC5 protein | -531 |
| 22:30475426–30475842 | HORMAD2 | HORMA domain containing 2 | -529 |
| 3:155463174–155463590 | PLCH1 | phospholipase C, eta 1 | -526 |
| 19:39970330–39970746 | TIMM50 | translocase of inner mitochondrial membrane 50 homolog (S. cerevisiae) | -514 |
| 19:56347244–56347660 | NLRP4 | NLR family, pyrin domain containing 4 | -492 |
| 1:26871647–26872063 | RPS6KA1 | ribosomal protein S6 kinase, 90kDa, polypeptide 1 | -488 |
| 11:62555950–62556366 | TMEM179B | transmembrane protein 179B | -483 |
| 19:2740422–2740838 | SLC39A3 | solute carrier family 39 (zinc transporter), member 3 | -480 |
| 8:146176515–146176931 | ZNF16 | zinc finger protein 16 | -449 |
| 2:175202383–175202799 | AC018470.1 | Uncharacterized protein FLJ46347 | -440 |
| 3:141120576–141120992 | ZBTB38 | zinc finger and BTB domain containing 38 | -407 |
| 1:27213196–27213612 | GPN2 | GPN-loop GTPase 2 | -388 |
| 19:12946378–12946794 | RTBDN | retbindin | -344 |
| 10:28032598–28033014 | MKX | mohawk homeobox | -332 |
| 6:44923360–44923776 | SUPT3H | suppressor of Ty 3 homolog (S. cerevisiae) | -321 |
| 21:34926838–34927254 | SON | SON DNA binding protein | -309 |
| 20:4880379–4880795 | SLC23A2 | solute carrier family 23 (nucleobase transporters), member 2 | -294 |
| 21:45078094–45078510 | HSF2BP | heat shock transcription factor 2 binding protein | -277 |
| 13:50070769–50071185 | PHF11 | PHD finger protein 11 | -272 |
| 16:22018485–22018901 | C16orf52 | chromosome 16 open reading frame 52 | -266 |
| 11:17372825–17373241 | NCR3LG1 | natural killer cell cytotoxicity receptor 3 ligand 1 | -240 |
| 20:36149179–36149595 | NNAT | neuronatin | -230 |
| 14:96670598–96671014 | BDKRB2 | bradykinin receptor B2 | -210 |
| 6:84419386–84419802 | SNAP91 | synaptosomal-associated protein, 91kDa | -184 |
| 17:57983959–57984375 | RPS6KB1 | ribosomal protein S6 kinase, 70kDa, polypeptide 1 | -182 |
| 9:130660260–130660676 | ST6GALNAC6 | ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 | -177 |
| 1:107683060–107683476 | NTNG1 | netrin G1 | -174 |
| 18:2846660–2847076 | EMILIN2 | elastin microfibril interfacer 2 | -160 |
| 17:73528230–73528646 | LLGL2 | lethal giant larvae homolog 2 (Drosophila) | -136 |
| 3:55521255–55521671 | WNT5A | wingless-type MMTV integration site family, member 5A | -132 |
| 4:99578749–99579165 | TSPAN5 | tetraspanin 5 | -114 |
| 17:72580766–72581182 | C17orf77 | chromosome 17 open reading frame 77 | -83 |
| 17:73085977–73086393 | SLC16A5 | solute carrier family 16, member 5 (monocarboxylic acid transporter 6) | -72 |
| 11:62445317–62445733 | UBXN1 | UBX domain protein 1 | -70 |
| 2:219906078–219906494 | CCDC108 | coiled-coil domain containing 108 | -41 |
| 1:47697216–47697632 | TAL1 | T-cell acute lymphocytic leukemia 1 | -37 |
| 17:73230571–73230987 | NUP85 | nucleoporin 85kDa | -20 |
| 4:111397002–111397418 | ENPEP | glutamyl aminopeptidase (aminopeptidase A) | -19 |
| 19:11658471–11658887 | CNN1 | calponin 1, basic, smooth muscle | 24 |
| 4:47839834–47840250 | CORIN | corin, serine peptidase | 47 |
| 15:55790255–55790671 | DYX1C1 | dyslexia susceptibility 1 candidate 1 | 83 |
| 21:34924435–34924851 | SON | SON DNA binding protein | 89 |
| 15:57025972–57026388 | ZNF280D | zinc finger protein 280D | 104 |
| 6:137815204–137815620 | OLIG3 | oligodendrocyte transcription factor 3 | 119 |
| 14:23652505–23652921 | SLC7A8 | solute carrier family 7 (amino acid transporter light chain, L system), member 8 | 136 |
| 12:31812613–31813029 | METTL20 | methyltransferase like 20 | 176 |
| 14:72400014–72400430 | RGS6 | regulator of G-protein signaling 6 | 273 |
| 2:98703769–98704185 | VWA3B | von Willebrand factor A domain containing 3B | 278 |
| 3:186281629–186282045 | TBCCD1 | TBCC domain containing 1 | 297 |
| 6:43112135–43112551 | PTK7 | PTK7 protein tyrosine kinase 7 | 306 |
| 14:64956734–64957150 | ZBTB25 | zinc finger and BTB domain containing 25 | 309 |
| 2:216239872–216240288 | FN1 | fibronectin 1 | 332 |
| 19:14217131–14217547 | PRKACA | protein kinase, cAMP-dependent, catalytic, alpha | 333 |
| 20:62903727–62904143 | PCMTD2 | protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 | 385 |
| 14:32545502–32545918 | ARHGAP5 | Rho GTPase activating protein 5 | 390 |
| 2:24271847–24272263 | C2orf44 | chromosome 2 open reading frame 44 | 390 |
| 11:63754509–63754925 | OTUB1 | OTU domain, ubiquitin aldehyde binding 1 | 403 |
| 3:188665201–188665617 | TPRG1 | tumor protein p63 regulated 1 | 406 |
| 5:96294368–96294784 | LNPEP | leucyl/cystinyl aminopeptidase | 421 |
| 1:151694242–151694658 | RIIAD1 | regulatory subunit of type II PKA R-subunit (RIIa) domain containing 1 | 437 |
| 19:16204630–16205046 | TPM4 | tropomyosin 4 | 456 |
| 11:71163246–71163662 | DHCR7 | 7-dehydrocholesterol reductase | 460 |
| 12:3120506–3120922 | TEAD4 | TEA domain family member 4 | 504 |
| 1:200012019–200012435 | NR5A2 | nuclear receptor subfamily 5, group A, member 2 | 510 |
| 19:6007516–6007932 | RFX2 | regulatory factor X, 2 (influences HLA class II expression) | 513 |
| 2:216300012–216300428 | FN1 | fibronectin 1 | 570 |
| 16:57474089–57474505 | CIAPIN1 | cytokine induced apoptosis inhibitor 1 | 590 |
| 3:42002666–42003082 | ULK4 | unc-51-like kinase 4 (C. elegans) | 612 |
| 1:228644735–228645151 | HIST3H2A | histone cluster 3, H2a | 617 |
| 19:47735191–47735607 | BBC3 | BCL2 binding component 3 | 624 |
| 13:36919583–36919999 | SPG20 | spastic paraplegia 20 (Troyer syndrome) | 629 |
| 22:30477422–30477838 | HORMAD2 | HORMA domain containing 2 | 630 |
| 2:235404366–235404782 | ARL4C | ADP-ribosylation factor-like 4C | 670 |
| 10:7860937–7861353 | TAF3 | TAF3 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 140kDa | 678 |
| 12:58160138–58160554 | CYP27B1 | cytochrome P450, family 27, subfamily B, polypeptide 1 | 688 |
| 6:35420636–35421052 | FANCE | Fanconi anemia, complementation group E | 706 |
| 19:13988601–13989017 | NANOS3 | nanos homolog 3 (Drosophila) | 746 |
| 10:74870845–74871261 | NUDT13 | nudix (nucleoside diphosphate linked moiety X)-type motif 13 | 765 |
| 6:27115481–27115897 | HIST1H2AH | histone cluster 1, H2ah | 828 |
| 1:247266524–247266940 | ZNF669 | zinc finger protein 669 | 848 |
| 19:47216200–47216616 | PRKD2 | protein kinase D2 | 872 |
| 14:21946061–21946477 | TOX4 | TOX high mobility group box family member 4 | 886 |
| 19:11668857–11669273 | ELOF1 | elongation factor 1 homolog (S. cerevisiae) | 895 |
| 17:36996603–36997019 | C17orf98 | chromosome 17 open reading frame 98 | 897 |
| 20:31123045–31123461 | C20orf112 | chromosome 20 open reading frame 112 | 947 |
| 12:50786921–50787337 | LARP4 | La ribonucleoprotein domain family, member 4 | 963 |
| 20:62340208–62340624 | ZGPAT | zinc finger, CCCH-type with G patch domain | 974 |
| 6:44188171–44188587 | SLC29A1 | solute carrier family 29 (nucleoside transporters), member 1 | 986 |
| 3:185001603–185002019 | MAP3K13 | mitogen-activated protein kinase kinase kinase 13 | 997 |
| 12:65089089–65089505 | AC025262.1 | Mesenchymal stem cell protein DSC96 | 1032 |
| 9:19050250–19050666 | RRAGA | Ras-related GTP binding A | 1086 |
| 16:1878116–1878532 | FAHD1 | fumarylacetoacetate hydrolase domain containing 1 | 1099 |
| 1:35323305–35323721 | SMIM12 | small integral membrane protein 12 | 1133 |
| 20:62461205–62461621 | ZBTB46 | zinc finger and BTB domain containing 46 | 1154 |
| 19:17531871–17532287 | MVB12A | multivesicular body subunit 12A | 1159 |
| 11:638254–638670 | DRD4 | dopamine receptor D4 | 1169 |
| 15:79576111–79576527 | ANKRD34C | ankyrin repeat domain 34C | 1173 |
| 3:55519903–55520319 | WNT5A | wingless-type MMTV integration site family, member 5A | 1220 |
| 7:148786349–148786765 | ZNF786 | zinc finger protein 786 | 1240 |
| 19:39437782–39438198 | FBXO17 | F-box protein 17 | 1253 |
| 19:47352074–47352490 | AP2S1 | adaptor-related protein complex 2, sigma 1 subunit | 1291 |
| 2:201751795–201752211 | PPIL3 | peptidylprolyl isomerase (cyclophilin)-like 3 | 1299 |
| 12:92529288–92529704 | RP11-24B21.1 | uncharacterized protein LOC256021 isoform 1 | 1301 |
| 1:152007979–152008395 | S100A11 | S100 calcium binding protein A11 | 1324 |
| 13:103424546–103424962 | TEX30 | testis expressed 30 | 1351 |
| 6:107779175–107779591 | PDSS2 | prenyl (decaprenyl) diphosphate synthase, subunit 2 | 1377 |
| 1:31380013–31380429 | SDC3 | syndecan 3 | 1387 |
| 22:31004379–31004795 | TCN2 | transcobalamin II | 1396 |
| 20:44423810–44424226 | DNTTIP1 | deoxynucleotidyltransferase, terminal, interacting protein 1 | 1414 |
| 19:46086441–46086857 | OPA3 | optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) | 1428 |
| 3:160115349–160115765 | IFT80 | intraflagellar transport 80 homolog (Chlamydomonas) | 1438 |
| 14:23282743–23283159 | SLC7A7 | solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 | 1440 |
| 18:686337–686753 | ENOSF1 | enolase superfamily member 1 | 1459 |
| 8:135650448–135650864 | ZFAT | zinc finger and AT hook domain containing | 1467 |
Fig 2. Pathways illustrating the network of genomic loci involved with (A & B) Regions undergoing increase in 5mC content and (C & D) decrease in 5mC contents, upon simulated microgravity.
Changes in 5hmC profile upon simulated-microgravity
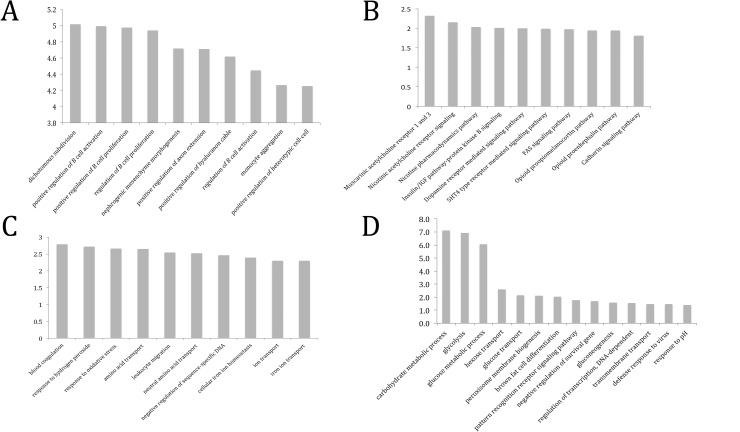
We applied hMeDIP analyses coupled with high-throughput sequencing to identify the differences in the genome-wide patterns of 5hmC upon simulated microgravity on TK6 cells. 2.7x108 and 1.4x108 reads were obtained during hMeDIP-seq from TK6 cells under static and simulated microgravity respectively and more than 90% of these read uniquely aligned to the human genome GRCh37/hg19, 2009 Assembly (S1 Table and S2 Fig). The depth of sequencing for the hmeDIP-seq samples ranged from 1.8X to 4.6X depending on the sample (S1 Table). Cross-correlation analysis was performed as per the ENCODE consortium guidelines [42, 52, 53] and all the samples displayed Normalized Strand Correlation (NSC) and Relative Strand Correlation (RSC) values greater than the minimum threshold (S1 Table). The consistency of reads in the biological replicates were observed through the cluster analysis (S3 Fig) and coverage analysis (S5 Fig). Of the 167 Differentially Hydroxymethylated Regions (DHMRs) (S6 Table & S7 Table) generated at IDR < 0.05, 154 (92.2%) were associated with hyper-hydroxymethylation (gain-of-5hmC) (S6 Table) and 13 (7.8%) with hypo-hydroxymethylation (loss-of-5hmC) (S7 Table) upon simulated microgravity respectively. The overlap of DHMRs with different genomic features indicated that 86 and 7 genes were associated with gain-of-5hmC and loss-of-5hmC DHMRs respectively (Table 1). Also, 5 gain-of-5hmC (Table 4) and2 loss-of-5hmC (Table 5) DHMRs were observed around -1500 to 1500 bps of Transcription Start Sites (TSS). The distribution of DNA repeat regions present within the DHMRs was represented in Table 1. Metadata describing each DHMR was included in S6 Table & S7 Table. Investigation of GREAT version 3.0.0 ontology annotation [54] was included in S8 Table. Gain-of-5hmC DHMRs induced by simulated microgravity were found to be associated with genes that enriched in GO Biological Processes like positive regulation of B cell activation (GO: 0050871), positive regulation of B cell proliferation (GO: 0030890) and positive regulation of cell-cell adhesion (GO: 0034116) among others (Fig 3A). Panther Pathway Analysis of these gan-of-5hmC DHMRs implicated the muscarinic acetylcholine receptor signaling (P00042), insulin/IGF pathway-protein kinase B signaling cascade (P00033) and Fas signaling (P00020) among others (Fig 3B), Due to an extremely small gene set associated with loss-of-5hmC DHMRs, significant p-values of pathway associations were not obtained and have not been reported here.
Table 4. List of hyper-hydroxymethylated DHMRs located within +/- 1500 of Transcription Start Sites of genes.
Columns display the genomic coordinates of DHMRs, Gene Symbol of the corresponding gene, the description of the genome and the exact distance in base pairs.
| DHMR(Chr:Start-End) | Gene Symbol | Description | Distance |
|---|---|---|---|
| 7:73103920–73104390 | WBSCR22 | Williams Beuren syndrome chromosome region 22 | -1091 |
| 19:11319910–11320380 | DOCK6 | dedicator of cytokinesis 6 | -525 |
| 19:6678748–6679218 | C3 | complement component 3 | 229 |
| 20:29977639–29978109 | DEFB119 | defensin, beta 119 | 412 |
| 11:117745746–117746216 | FXYD6 | FXYD domain containing ion transport regulator 6 | 1359 |
Table 5. List of hypo-hydroxymethylated DHMRs located within +/- 1500 of Transcription Start Sites of genes.
Columns display the genomic coordinates of DHMRs, Gene Symbol of the corresponding gene, the description of the genome and the exact distance in base pairs
| DHMR(Chr:Start-End) | Gene Symbol | Description | Distance |
|---|---|---|---|
| 3:96336458–96336934 | MTRNR2L2 | MT-RNR2-like 2 | -579 |
| 3:96336159–96336635 | MTRNR2L2 | MT-RNR2-like 2 | -280 |
Fig 3. Pathways illustrating the network of genomic loci involved with (A & B) Regions undergoing increase in 5hmC content and (C & D) differential gene expression.
Changes in the transcriptome upon simulated-microgravity
In TK6 cells, simulated microgravity induced differential expression of 370 transcripts out of 22,376 transcripts analyzed (FDR<0.1) compared to static control (S9 Table). 271 (73.24%) differentially expressed transcripts were associated with a decrease in expression, while 99 (26.76%) differentially expressed transcripts were associated with an increase in gene expression. 17 (4.59%) genes were associated with a drastic change of differentially expression (greater than 2 fold increase or decrease), while the vast majority were associated with intermediate (0–2 fold) change in differential expression. Furthermore, the pathway analysis (S10 Table) of transcriptionally upregulated genes showed enrichment of GO Biological Processes such as response to oxidative stress (GO:0006979) and ion transport (GO:0006811) (Fig 3C), while the downregulated genes could be linked to regulation of DNA-dependent transcription (GO:0006355) and carbohydrate metabolic processes (GO:0005975) (Fig 3D). Some of the top upregulated genes include CHAC1, TRPA1, ATAD3C, INHBE, CTH, HMOX, HBD, SPG20, CACNA1D and PTGER4, while the top downregulated genes were GOLGA6L9, PFKFB4, FBXO17, ITGA6, PIK3R6, SLC2A5, INSIG2, AKAP6, HILPDA and POU2F3 (S9 Table).
Correlation between simulated microgravity induced DMRS/DHMRs and gene expression
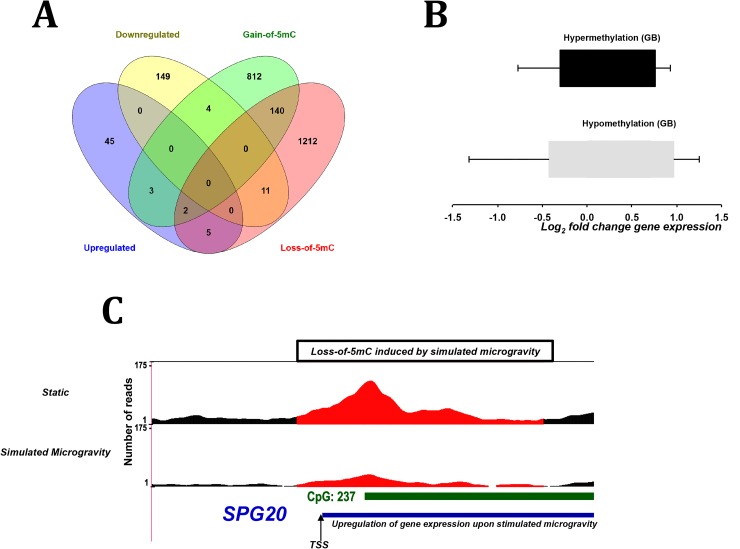
A comparison of the simulated microgravity induced differentially expressed genes (S9 Table) with DMRs located at gene promoters (Tables 2 & 3) revealed that two transcriptionally upregulated genes (TSPAN5 and SPG20) were associated with loss-of-5mC at their promoter and three transcriptionally downregulated genes (PLIN2, MAP3K13 and FBXO1) were associated with loss-of-5mC at their promoter. None of the gene promoters linked to DHMRs (Tables 4 & 5) were found to be differentially expressed (S9 Table). Similarly, the comparison of simulated microgravity induced differentially expressed genes with DMRs/DHMRs located at gene bodies revealed that 25 differentially expressed associated with DMRs at their gene bodies and none of the differentially expressed genes associated with DHMRs at their gene bodies. The relationship between methylation status at gene bodies and their respective transcriptional activity of these 25 differentially expressed genes did not show any significant correlation by Fisher’s Exact Test (Fig 4A) and could be divided into five distinct groups, (i) five transcriptionally upregulated genes with loss-of-5mC DMRs at their gene bodies (CTH, CACNA1D, SPG20, PLS1 and SLC39A14), (ii) eleven transcriptionally downregulated genes with loss-of-5mC DMRs at their gene bodies (FBXO17, AKAP6, RIT1, GTF2IRD2P1, MSTO1, PMS2CL, MAP3K13, ST3GAL1, NCKIPSD, MAST1 and MSTO2P), (iii) three transcriptionally downregulated genes associated with gain-of-5mC DMRs at their gene bodies (CACNB2, WDR45B and CABLES1), (iv) three transcriptionally upregulated genes with gain-of-5mC DMRs at their gene bodies (CASZ1, VCL and ATF3) and (v) two transcriptionally upregulated genes with gain-of-5mC as well as loss-of-5mC DMRs at their gene bodies (ARID5B and TSPAN5) (Fig 4B). The comparison of DMRs and DHMRs located at gene bodies (S6 Fig) yielded six overlapping groups namely (i) 140 genes were associated with gain-of-5mC and loss-of-5mC DMRs at their gene bodies, (ii) eight were genes were associated with loss-of-5mC DMRs and gain-of-5hmC DHMRs, (iii) five gene were associated with gain-of-5mC and loss-of-5mC DMRs as well as gain-of-5hmC DHMRs at their gene bodies, (iv) seven genes were associated with gain-of-5mC DMRs and gain-of-5hmC DHMRs at their gene bodies, (v) one gene was associated with gain-of-5mC DMR and loss-of-5hmC DHMR at its gene body and (vi) two genes were associated with gain-of-5hmC DHMRs and loss-of-5hmC DHMRs at their gene bodies.
Fig 4. Overlap of simulated microgravity induced differentially expressed genes and genes undergoing differential methylation over gene-bodies (A) Fisher’s test showing no significant correlation between direction of methylation changes over gene bodies and their relative expression, (B)Venn diagram showing the overlap of differentially expressed genes with DMRs associated with gene bodies, (C) Statistical Representation of a gene SPG20, which underwent upregulation at the transcript level and a simultaneous decrease in 5mC levels at its promoter upon simulated microgravity.
Discussion
The objective of this ground-based study was to map the genome-wide effects of simulated microgravity on DNA methylation, hydroxymethylation; and gene expression patterns in TK6 lymphoblastoid cells by a powerful Next Generation Sequencing pipeline. Although on the basis of numerous studies reporting microgravity-induced physiological changes in living organisms ranging from prokaryotes to humans, it has been speculated that microgravity-induced changes may occur in the methylome, very little is known about the effects of microgravity on DNA methylation. In 2009, Ou et al reported hypermethylation of a set of genes and transposable elements in rice (Oryza sativa L.) plants germinating from space-flown seeds [29]. Ou et al also reported that the spaceflight-induced hypermethylated genes did not generally correlate with alterations in their gene expression status [29]. In 2010, Singh et al reported that human T-lymphocytes subjected to simulated microgravity underwent global DNA hypomethylation on the basis of Methylation Sensitive-Random Amplified Polymorphic DNA (MS-RAPD)-PCR analysis [30]. However, since MS-RAPD-PCR is unable to identify specific methylated sites, the study by Singh et al could not report the target genes associated with the simulated-microgravity induced DNA hypomethylation. In 2011, Ou et al validated their previous finding that spaceflight induced hypermethylation of DNA (the frequency of spaceflight-induced hypermethylation was demonstrated to be nearly double of spaceflight-induced hypomethylation events) by assessing a larger genomic subset comprising 467 loci [31], though it was not evident if any study to correlate changes in DNA methylation with gene expression were further conducted.
The disparity between the conclusions of the studies conducted by Ou et al and Singh et al could be attributed to several factors, but we think that the following might be important to consider: (i) differences in mechanisms establishing and maintaining DNA methylation patterns in plants and animals (for a comprehensive review refer to [55]), (ii) while Ou et al’s investigation was based on spaceflight-induced “epigenetic memory” being transmitted from the seeds to the sapling, Singh et al had investigated the simulated microgravity-influenced changes in DNA methylation in immortalized T-lymphocyte cell cultures that might not be inheritable and (iii) while Singh et al had investigated the effects of only simulated microgravity on DNA methylation, Ou et al was investigating the effects of numerous factors like cosmic radiation, microgravity and space magnetic fields encountered during spaceflight. These reports therefore provided a strong basis for us to perform this study with advanced methods such as MeDIP-seq, hMeDIP-seq and RNA-seq to explore the relationship between the methylome and the transcriptome in microgravity exposed cells. To the best of our knowledge, this is the first report profiling the effects of simulated microgravity on the epigenomic landscape of human cells. 3204 DMRs and 2116 DHMRs distributed throughout the genome were identified in TK6 cells subjected to simulated microgravity. The majority of the DMRs (59.86%) were identified to undergo hypomethylation, which was consistent with the findings of Singh et al [30]. On the other hand the majority of DHMRs (92.2%) were associated with hyper hydroxymethylation.
Additionally, we have been able to perform ontology based annotations to obtain information about the biological processes that might be affected by genes associated with simulated microgravity induced changes occurring in the methylome. In particular, genes involved in primary metabolic processes, immune functions and the p53 pathway seems to be undergoing changes in their methylation/hydroxymethylation status under the influence of simulated microgravity. An early study on lymphoblastoid cells subjected to 48 hours of simulated microgravity by Degan et al. reported decrease of cellular ATP content, suggesting a simulated microgravity induced alteration in cellular metabolism [56]. It remains to be seen how simulated microgravity induced changes over the methylation levels of p53 effector genes play in TK6 which expresses the wild-type p53 [57], a tumor suppressor functioning extensively in the DNA repair pathway. Reduction of global methylation has been proposed to be a hallmark of genomic instability [14, 58] and it remains to be seen if the extensive loss-of-5mC induced by simulated microgravity reported in this study has any functional implications. Another finding in TK6 cell line, which was originally derived from a patient with T- blast crisis [32, 59, 60] and could potentially harbor progenitor forms of lymphocytes, pertains to changes in methylation/hydroxymethylation patterns over genes involved in lymphocyte development and activation cultured under simulated microgravity conditions. Interestingly whole-exome sequencing has revealed similarities in the genomic content of lymphocytes and lymphoblastoid cells [61], and thus in light of our findings TK6 lymphoblastoid cells may emerge as a good model to study B and T- lymphocyte development and activation in in vitro genomic studies.
Our study also revealed that simulated microgravity could alter the expression of 370 transcripts, however only 17 of these underwent greater than 2-fold change of up/downregulation. The transcriptionally upregulated genes showed enrichment of pathways involving response to oxidative stress and negative regulation of gene expression, while the downregulated genes could be linked to pathways responsible for glucose metabolism and transcription regulation. While our study illustrated that there was no direct relationship between differentially expressed genes and changes in 5mC/5hmC over its promoters/gene bodies, we have been able to determine the methylation status of individual genes implicated in earlier studies to be affected in transcriptional or translational activities on exposure to simulated microgravity in ground based studies or in spaceflights. For instance, the voltage-dependent calcium channel L-type, alpha 1D (CACNA1D) gene transcript was observed to be differentially expressed in human T-lymphocytes subjected to microgravity conditions during spaceflight compared to ground static controls [49]. While, we observed a nominal increase at the transcript level for CACNA1D, we observed a decrease in 5mC levels over its gene body under simulated microgravity.
In another study, the Activating Transcription Factor-3 (ATF3) has been implicated to be differentially expressed upon being subjected to microgravity during spaceflight in cultured HUVEC cells [62]. Our RNA-seq data illustrated an increase of 1.3 fold in the transcript level of ATF3 and a decrease in the 5mC levels over its gene body under the influence of simulated microgravity. Interestingly, ATF3, a member of the ATF/CREB family of transcription factors, has been observed to be upregulated when cells are exposed to stress conditions [50]. On the other hand, Integrin alpha-6 (ITGA6) which is an integral cell surface protein has been observed to be down-regulated at the transcriptional scale during short-term weightlessness produced by parabolic maneuvers in human cells [51]. While RNA-seq revealed a decrease of ITGA6 transcript by more than 2-fold, we were not able to observe changes in the 5mC and 5hmC profile over its gene body or promoter, implying that possibly mechanisms other than DNA methylation might be involved in its regulation.
Some of the novel gene functions that we have linked with DNA methylation status include the F-Box Protein 17 (FBXO17), which constitutes one of the four subunits of the ubiquitin-protein-ligase complex called SKP1-cullin-F-box (SCFs) and mediates substrate specificity [63, 64]. While the transcript level of FBXO17 was observed to be downregulated by 2.47 fold, the 5mC levels over the gene body of FBXO17 (chr19:39437782–39438198) decreased in TK6 cells subjected to simulated microgravity. Recently it has been demonstrated that the recruitment of F-box motif bearing homologous protein in yeast Met30 is regulated by a complex mechanism and has been implicated in stress response [65, 66]. In sync with these observations, reduction of 5mC levels over gene bodies of other F-Box motif containing proteins such as FBXO31 (chr16:87421262–87421678) and FBXO42 (chr1:16674945–16675361) and promoter of FBXO5 (2024 bps upstream of TSS; chr6:153306530–153306946), was also observed though their transcripts were not differentially expressed. Interestingly, genes which function as molecular mechano-sensors like Vinculin (VCL) or mediate stress-signal transduction events like Tetraspan-5 (TSPAN5) were also seen to undergo changes in its gene methylation levels and expression. Similarly, other genes involved in the Metabolic process (GO:0008152) like Cystathionine gamma-lyase (CTH), Phospholipid scramblase-1 (PLS1) Microtubule-associated serine/threonine-protein kinase-1 (MAST1), Zinc finger protein castor homolog-1 (CASZ1), CMP-N-acetylneuraminate-beta-galactosamide-alpha-2,3-sialyltransferase-1 (ST3GAL1), AT-rich interactive domain-containing protein-5B (ARID5B), Mitogen-activated protein kinase-13 (MAP3K13) and Perilipin-2 (PLIN2) were implicated in this study contributing to mechano-stress response. Though our study does not show a global correlation between methylation status and transcriptional activity, the simulated microgravity induced changes over SPG20 (a gene implicated in endosomal trafficking and mitochondrial functions) recapitulates the conventional theory of decrease in promoter methylation corresponding to elevated gene activity. This novel finding suggests that methylation-dependent transcriptional activity is not a genome-wide phenomenon, instead it may be applicable for specific genes.
Thus, in conclusion we believe that 48 hours of treatment with simulated microgravity triggered changes in the transcriptome particularly involving biological processes such as negative regulation of transcription, response to stress and reduction in carbohydrate metabolic processes. This study revealed that simulated microgravity influenced alteration of genome-wide 5mC and 5hmC patterns, however no correlation was found between DMRs/DHMRs situated at gene bodies and promoters and their transcriptional status. While it has been long held that genes with methylated promoters are transcriptionally silent, recent studies have uncovered the association of methylated gene promoters with both transcriptionally active and inactive genes [20, 21, 67–70]. On the other hand, gene body methylation has been observed to be positively correlated with gene expression in some studies [71, 72] and no such correlation has been found in others [22, 73–75]. Recent deep-sequencing based explorations have challenged the traditional paradigm and illustrated complexities of the nature of relationship between DNA methylation and gene expression [19–25]. It is also conceivable that pronounced alterations in epigenetic patterns may take place in cells subjected to prolonged microgravity environments. The ground-based microgravity simulators like the one used in our study have undoubtedly enhanced our understanding of microgravity but it has to be pointed out that the principle of “simulating” microgravity involves changing the direction of Earth’s gravity subjected to the samples through continuous rotation and represent “functional near weightlessness”. While this is the first study to profile the simulated microgravity induced changes in 5mC/5hmC patterns and gene expression simultaneously providing a perspective of epigenetic alterations we could expect during short-term exposures, our understanding is far from complete. We believe that genes involved in altered biological processes identified in this study will be of considerable interest and provide a valuable resource for future investigations. Finally, in the interest of astronauts who are exposed to microgravity for prolonged periods of time, future studies should focus on performing time course experiments monitoring the influence of “real” and “simulated” microgravity exposure on a variety of models to determine the precise effects of microgravity on the epigenome
Supporting Information
(TIF)
The total number of reads (white) and the total number of unique reads aligned to the human genome (blue) obtained by performing hmeDIP-seq and meDIP-seq on TK6 cells cultured under static (control) and simulated microgravity (12 rpm) conditions for 48 hours. S1 Table demonstrates the exact numbers and percentage of mapped reads.
(TIF)
(TIF)
Color of these lines represent the fold coverage of the CpGs as shown in the legend.
(TIF)
Color of these lines represent the fold coverage of the CpGs as shown in the legend.
(TIF)
(TIF)
(XLSX)
For every DMR identified, a description of the genomic features found in this region has been provided. The columns represent the following information: (A) Genomic coordinates of the region defined as a DMR and (B) ENCODE IDs of features (such as gene, transcript, pseudogene, non-coding RNA or other regulatory feature) present in the region.
(XLSX)
For every DMR identified, a description of the genomic features found in this region has been provided. The columns represent the following information: (A) Genomic coordinates of the region defined as a DMR and (B) ENCODE IDs of features (such as gene, transcript, pseudogene, non-coding RNA or other regulatory feature) present in the region.
(XLSX)
The columns represents the respective ontology name, term name / identifier, term description, binomial rank, binomial p-value (uncorrected), binomial Bonferroni corrected p-value, binomial FDR q-value and names of gene hits generated by GREAT version 3.0.0; Species assembly: hg19 and association rule: Basal+extension: 5000 bp upstream, 1000 bp downstream, 1000000 bp max extension, curated regulatory domains included.
(XLSX)
The columns represents the respective ontology name, term name / identifier, term description, binomial rank, binomial p-value (uncorrected), binomial Bonferroni corrected p-value, binomial FDR q-value and names of gene hits generated by GREAT version 3.0.0; Species assembly: hg19 and association rule: Basal+extension: 5000 bp upstream, 1000 bp downstream, 1000000 bp max extension, curated regulatory domains included.
(XLSX)
For every DHMR identified, a description of the genomic features found in this region has been provided. The columns represent the following information: (A) Genomic coordinates of the region defined as a DHMR and (B) ENCODE IDs of features (such as gene, transcript, pseudogene, non-coding RNA or other regulatory feature) present in the region.
(XLSX)
The columns represent the following information for each identified DHMR: (A) Genomic coordinates of the region defined as a DHMR and (B) ENCODE IDs of features (such as gene, transcript, pseudogene, non-coding RNA or other regulatory feature) present in the region.
(XLSX)
The columns represents the respective ontology name, term name / identifier, term description, binomial rank, binomial p-value (uncorrected), binomial Bonferroni corrected p-value, binomial FDR q-value and names of gene hits generated by GREAT version 3.0.0; Species assembly: hg19 and association rule: Basal+extension: 5000 bp upstream, 1000 bp downstream, 1000000 bp max extension, curated regulatory domains included.
(XLSX)
The columns represent geneID, name of gene from UCSC Genome Browser (duplicates exist because multiple geneID can map to same gene), chromosome location, strand: + or–, transcription start position, transcription end position, the log2-fold-change of gene expression, the average log2-counts-per-million of comparison, p-value of comparison, false discovery rate (corrected p-value) of comparison.
(XLSX)
(XLSX)
Acknowledgments
We would like to thank the National Aeronautics and Space Administration (NASA) task “Defining Epigenetic Programming During Flight Expeditions in Differentiating Embryonic Stem Cells” (NNX12AN09G) for the funding. Partial support provided by the W.M. Keck Foundation; National Institute of Health (NIH), National Cancer Institute (NCI) R25CA128770 Cancer Prevention Internship Program; Indiana Clinical and Translational Sciences Institute (CTSI) (TR000006) and Purdue Center for Cancer Research (P30CA023168) is appreciated. We thank Dr. Phillip Miguel for generating the NGS data and Dr. Hongyu Gao for submitting the Sequencing data in NCBI GEO.
Data Availability
All relevant data are within the paper and its Supporting Information files. Raw sequencing data is deposited at GEO (Accession Number: GSE65944) and available in the database (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE65944).
Funding Statement
The authors would like to acknowledge the following agencies for their support in funding this study: a) NASA (NNX12AN09G); b) NIH, NCI R25CA128770 Cancer Prevention Internship Program; c) Indiana CTSI (TR000006); d) W.M. Keck Foundation; e) Purdue Center for Cancer Research (P30CA023168). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Aponte VM, Finch DS, Klaus DM. Considerations for non-invasive in-flight monitoring of astronaut immune status with potential use of MEMS and NEMS devices. Life Sciences. 2006;79(14):1317–33. 10.1016/j.lfs.2006.04.007 . [DOI] [PubMed] [Google Scholar]
- 2.Morey-Holton ER, Arnaud S. Skeletal Responses to Spaceflight. Advances in Space Biology and Medicine. 1991;1:37–69. 10.1016/S1569-2574(08)60120-3 [DOI] [PubMed] [Google Scholar]
- 3.Hammond TG, Lewis FC, Goodwin TJ, Linnehan RM, Wolf DA, Hire KP, et al. Gene expression in space. Nature Medicine. 1999;5(4):359–. 10.1038/7331 . [DOI] [PubMed] [Google Scholar]
- 4.Hammond TG, Benes E, O'Reilly KC, Wolf DA, Linnehan RM, Taher A, et al. Mechanical culture conditions effect gene expression: gravity-induced changes on the space shuttle. Physiological Genomics. 2000;3(3):163–73. . [DOI] [PubMed] [Google Scholar]
- 5.Shimada N, Sokunbi G, Moorman SJ. Changes in gravitational force affect gene expression in developing organ systems at different developmental times. Bmc Developmental Biology. 2005;5 10.1186/1471-213x-5-10 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson JW, Ott CM, Ramamurthy R, Porwollik S, McClelland M, Pierson DL, et al. Low-shear modeled microgravity alters the Salmonella enterica serovar typhimurium stress response in an RpoS-independent manner. Applied and Environmental Microbiology. 2002;68(11):5408–16. 10.1128/aem.68.11.5408-5416.2002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson JW, Ott CM, Bentrup KHz, Ramamurthy R, Quick L, Porwollik S, et al. Space flight alters bacterial gene expression and virulence and reveals a role for global regulator Hfq. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(41):16299–304. 10.1073/pnas.0707155104 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carmeliet G, Vico L, Bouillon R. Space flight: A challenge for normal bone homeostasis. Critical Reviews in Eukaryotic Gene Expression. 2001;11(1–3):131–44. . [PubMed] [Google Scholar]
- 9.Strollo F, Norsk P, Roecker L, Strollo G, More M, Bollanti L, et al. Indirect evidence of CNS adrenergic pathways activation during spaceflight. Aviation Space and Environmental Medicine. 1998;69(8):777–80. . [PubMed] [Google Scholar]
- 10.Sonnenfeld G, Butel JS, Shearer WT. Effects of the space flight environment on the immune system. Reviews on environmental health. 2003;18(1):1–17. . [DOI] [PubMed] [Google Scholar]
- 11.Dang B, Yang Y, Zhang E, Li W, Mi X, Meng Y, et al. Simulated microgravity increases heavy ion radiation-induced apoptosis in human B lymphoblasts. Life Sciences. 2014;97(2):123–8. 10.1016/j.lfs.2013.12.008 . [DOI] [PubMed] [Google Scholar]
- 12.Degan P, Sancandi M, Zunino A, Ottaggio L, Viaggi S, Cesarone F, et al. Exposure of human lymphocytes and lymphoblastoid cells to simulated microgravity strongly affects energy metabolism and DNA repair. Journal of Cellular Biochemistry. 2005;94(3):460–9. 10.1002/jcb.20302 . [DOI] [PubMed] [Google Scholar]
- 13.Hussain T, Mulherkar R. Lymphoblastoid Cell lines: a Continuous in Vitro Source of Cells to Study Carcinogen Sensitivity and DNA Repair. International journal of molecular and cellular medicine. 2012;1(2):75–87. . [PMC free article] [PubMed] [Google Scholar]
- 14.Baylin SB, Jones PA. A decade of exploring the cancer epigenome—biological and translational implications. Nature Reviews Cancer. 2011;11(10):726–34. 10.1038/nrc3130 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laird PW. Principles and challenges of genome-wide DNA methylation analysis. Nat Rev Genet. 2010;11(3):191–203. 10.1038/nrg2732 [DOI] [PubMed] [Google Scholar]
- 16.Haffner MC, Chaux A, Meeker AK, Esopi DM, Gerber J, Pellakuru LG, et al. Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget. 2011;2(8):627–37. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li W, Liu M. Distribution of 5-hydroxymethylcytosine in different human tissues. Journal of nucleic acids. 2011;2011:870726 10.4061/2011/870726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20(1):11–24. 10.1016/j.ccr.2011.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wagner JR. The relationship between DNA methylation, genetic and expression inter- individual variation in untransformed human fibroblasts. Genome Biology. 2014;15(2):R37 10.1186/gb-2014-15-2-r37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lou S. Whole- genome bisulfite sequencing of multiple individuals reveals complementary roles of promoter and gene body methylation in transcriptional regulation. Genome Biology. 2014;15(7):408 10.1186/s13059-014-0408-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kulis M. Epigenomic analysis detects widespread gene- body DNA hypomethylation in chronic lymphocytic leukemia. Nature Genetics. 2012;44(11):1236–43. 10.1038/ng.2443 [DOI] [PubMed] [Google Scholar]
- 22.Maunakea AK. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466(7303):253–8. 10.1038/nature09165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jun W. Characterization of tissue- specific differential DNA methylation suggests distinct modes of positive and negative gene expression regulation. BMC Genomics. 2015;16(1):1–12. 10.1186/s12864-015-1271-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rafael AI, Christine L-A, Bo W, Zhijin W, Carolina M, Patrick O, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue- specific CpG island shores. Nature Genetics. 2009;41(2):178 10.1038/ng.298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peter AJ. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nature Reviews Genetics. 2012;13(7):484 10.1038/nrg3230 [DOI] [PubMed] [Google Scholar]
- 26.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET1. Science. 2009;324(5929):930–5. 10.1126/science.1170116 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kriaucionis S, Heintz N. The Nuclear DNA Base 5-Hydroxymethylcytosine Is Present in Purkinje Neurons and the Brain. Science. 2009;324(5929):929–30. 10.1126/science.1169786 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kellinger MW, Song CX, Chong J, Lu XY, He C, Wang D. 5-formylcytosine and 5-carboxylcytosine reduce the rate and substrate specificity of RNA polymerase II transcription. Nature Structural & Molecular Biology. 2012;19(8):831–3. 10.1038/nsmb.2346 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ou X, Long L, Zhang Y, Xue Y, Liu J, Lin X, et al. Spaceflight induces both transient and heritable alterations in DNA methylation and gene expression in rice (Oryza sativa L.). Mutation Research-Fundamental and Molecular Mechanisms of Mutagenesis. 2009;662(1–2):44–53. 10.1016/j.mrfmmm.2008.12.004 . [DOI] [PubMed] [Google Scholar]
- 30.Singh KP, Kumari R, DuMond JW. Simulated Microgravity-Induced Epigenetic Changes in Human Lymphocytes. Journal of Cellular Biochemistry. 2010;111(1):123–9. 10.1002/jcb.22674 . [DOI] [PubMed] [Google Scholar]
- 31.Ou X, Long L, Wu Y, Yu Y, Lin X, Qi X, et al. Spaceflight-induced genetic and epigenetic changes in the rice (Oryza sativa L.) genome are independent of each other. Genome. 2010;53(7):524–32. 10.1139/g10-030 . [DOI] [PubMed] [Google Scholar]
- 32.Watanabe T, Kataoka T, Mizuta S, Kobayashi M, Uchida T, Imai K, et al. ESTABLISHMENT AND CHARACTERIZATION OF A NOVEL CELL-LINE, TK-6, DERIVED FROM T-CELL BLAST CRISIS OF CHRONIC MYELOGENOUS LEUKEMIA, WITH THE SECRETION OF PARATHYROID HORMONE-RELATED PROTEIN. Leukemia. 1995;9(11):1926–34. . [PubMed] [Google Scholar]
- 33.Herranz R. Ground- based facilities for simulation of microgravity: organism- specific recommendations for their use, and recommended terminology. Astrobiology. 2013;13(1):1 10.1089/ast.2012.0876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mangala LS, Zhang Y, He Z, Emami K, Ramesh GT, Story M, et al. Effects of Simulated Microgravity on Expression Profile of MicroRNA in Human Lymphoblastoid Cells. Journal of Biological Chemistry. 2011;286(37):32483–90. 10.1074/jbc.M111.267765 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Butcher LM, Beck S. AutoMeDIP-seq: A high-throughput, whole genome, DNA methylation assay. Methods. 2010;52(3):223–31. 10.1016/j.ymeth.2010.04.003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andrews S. FastQC: A quality control tool for high throughput sequence data. 2009. Available: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 37.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. Epub 2009/05/20. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson G, Dhami P, Feber A, Cortazar D, Suzuki Y, Schulz R, et al. Resources for methylome analysis suitable for gene knockout studies of potential epigenome modifiers. GigaScience. 2012;1(1):3 10.1186/2047-217X-1-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dietrich LCaJ. MEDIPS: MeDIP-Seq data analysis. R package version 1.6.0. 2010.
- 41.Nix DA, Courdy SJ, Boucher KM. Empirical methods for controlling false positives and estimating confidence in ChIP-Seq peaks. BMC bioinformatics. 2008;9:523 Epub 2008/12/09. 10.1186/1471-2105-9-523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nature Biotechnology. 2008;26(12):1351–9. 10.1038/nbt.1508 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Q, Brown JB, Huang H, Bickel PJ. MEASURING REPRODUCIBILITY OF HIGH-THROUGHPUT EXPERIMENTS. Annals of Applied Statistics. 2011;5(3):1752–79. 10.1214/11-aoas466 . [DOI] [Google Scholar]
- 44.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–2. 10.1093/bioinformatics/btq033 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ji X, Li W, Song J, Wei L, Liu XS. CEAS: cis-regulatory element annotation system. Nucleic Acids Research. 2006;34:W551–W4. 10.1093/nar/gkl322 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salmon-Divon M, Dvinge H, Tammoja K, Bertone P. PeakAnalyzer: Genome-wide annotation of chromatin binding and modification loci. Bmc Bioinformatics. 2010;11 10.1186/1471-2105-11-415 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–11. Epub 2009/03/18. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Breese MR, Liu Y. NGSUtils: a software suite for analyzing and manipulating next-generation sequencing datasets. Bioinformatics. 2013;29(4):494–6. Epub 2013/01/15. 10.1093/bioinformatics/bts731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. Epub 2009/06/10. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. Epub 2009/11/17. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tabas-Madrid D. GeneCodis3: a non- redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Research. 2012;40(Web Server issue):W478 10.1093/nar/gks402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Landt SG. ChIP- seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Research. 2012;22(9):1813 10.1101/gr.136184.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marinov GK, Kundaje A, Park PJ, Wold BJ. Large-Scale Quality Analysis of Published ChIP-seq Data. G3-Genes Genomes Genetics. 2014;4(2):209–23. 10.1534/g3.113.008680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, et al. GREAT improves functional interpretation of cis-regulatory regions. Nature Biotechnology. 2010;28(5):495–U155. 10.1038/nbt.1630 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He X-J, Chen T, Zhu J-K. Regulation and function of DNA methylation in plants and animals. Cell Research. 2011;21(3):442–65. 10.1038/cr.2011.23 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Degan P, Cesarone CF, Ottaggio L, Galleri G, Meloni MA, Zunino A, et al. Effects of simulated microgravity on metabolic activities related to DNA damage and repair in lymphoblastoid cells. Journal of gravitational physiology: a journal of the International Society for Gravitational Physiology. 2001;8(1):P21–2. . [PubMed] [Google Scholar]
- 57.Yu YJ, Little JB. p53 is involved in but not required for ionizing radiation-induced caspase-3 activation and apoptosis in human lymphoblast cell lines. Cancer Research. 1998;58(19):4277–81. . [PubMed] [Google Scholar]
- 58.Rodriguez J, Frigola J, Vendrell E, Risques R-A, Fraga M, Morales C, et al. Chromosomal Instability Correlates with Genome- wide DNA Demethylation in Human Primary Colorectal Cancers. Cancer Research. 2006;66(17):8462–9468. [DOI] [PubMed] [Google Scholar]
- 59.Schwartz JL, Jordan R, Evans HH, Lenarczyk M, Liber HL. Baseline levels of chromosome instability in the human lymphoblastoid cell TK6. Mutagenesis. 2004;19(6):477–82. 10.1093/mutage/geh060 . [DOI] [PubMed] [Google Scholar]
- 60.Levy JA, Virolain M, Defendi V. HUMAN LYMPHOBLASTOID LINES FROM LYMPH NODE AND SPLEEN. Cancer. 1968;22(3):517–&. . [DOI] [PubMed] [Google Scholar]
- 61.Londin ER, Keller MA, D'Andrea MR, Delgrosso K, Ertel A, Surrey S, et al. Whole-exome sequencing of DNA from peripheral blood mononuclear cells (PBMC) and EBV-transformed lymphocytes from the same donor. Bmc Genomics. 2011;12 10.1186/1471-2164-12-464 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.style = "font-size:10.5pt s, line-height:115%, font-family:"Georgia" s, mso-fareast-font-family:Calibri, mso-bidi-font-family:, Roman" TN, et al. Identification of Putative Major Space Genes Using Genome-Wide Literature Data. In: Hammond Timothy G WBL, Birdsall Holly H and Clement Jade Q, editor. Biotechnology: InTech; 2015.
- 63.Petroski MD, Deshaies RJ. Function and regulation of Cullin-RING ubiquitin ligases. Nature Reviews Molecular Cell Biology. 2005;6(1):9–20. 10.1038/nrm1547 . [DOI] [PubMed] [Google Scholar]
- 64.Kaiser P, Su N-Y, Yen JL, Ouni I, Flick K. The yeast ubiquitin ligase SCFMet30: connecting environmental and intracellular conditions to cell division. Cell Division. 2006;1 10.1186/1747-1028-1-16 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yen JL, Flick K, Papagiannis CV, Mathur R, Tyrrell A, Ouni I, et al. Signal-Induced Disassembly of the SCF Ubiquitin Ligase Complex by Cdc48/p97. Molecular Cell. 2012;48(2):288–97. 10.1016/j.molcel.2012.08.015 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baudouin-Cornu P, Labarre J. Regulation of the cadmium stress response through SCF-like ubiquitin ligases: comparison between Saccharomyces cerevisiae, Schizosaccharomyces pombe and mammalian cells. Biochimie. 2006;88(11):1673–85. 10.1016/j.biochi.2006.03.001 . [DOI] [PubMed] [Google Scholar]
- 67.Michael W, Ines H, Michael BS, Liliana R, Svante P, Michael R, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nature Genetics. 2007;39(4):457 10.1038/ng1990 [DOI] [PubMed] [Google Scholar]
- 68.Bell JT, Pai AA, Pickrell JK, Gaffney DJ, Pique-Regi R, Degner JF, et al. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biology. 2011;12(1):R10–R. 10.1186/gb-2011-12-1-r10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pai AA, Bell JT, Marioni JC, Pritchard JK, Gilad Y. A Genome- Wide Study of DNA Methylation Patterns and Gene Expression Levels in Multiple Human and Chimpanzee Tissues (Gene Regulation by DNA Methylation in Primates). PLoS Genetics. 2011;7(2):e1001316 10.1371/journal.pgen.1001316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chowdhury B, McGovern A, Cui Y, Choudhury SR, Cho I-H, Cooper B, et al. The hypomethylating agent Decitabine causes a paradoxical increase in 5-hydroxymethylcytosine in human leukemia cells. Scientific Reports. 2015;5 10.1038/srep09281 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Madeleine PB, Jin Billy L, Yuan G, Je-Hyuk L, Emily ML, In-Hyun P, et al. Targeted and genome- scale strategies reveal gene- body methylation signatures in human cells. Nature Biotechnology. 2009;27(4):361 10.1038/nbt.1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rauch TA. A human B cell methylome at 100- base pair resolution. Proceedings Of The National Academy Of Sciences Of The United States Of America. 2009;106(3):671 10.1073/pnas.0812399106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–22. 10.1038/nature08514 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ryan L, Mattia P, Robert HD, Hawkins RD, Gary H, Julian T-F, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315 10.1038/nature08514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Matthew CL, David RD, Mike S, Mark G. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nature Structural & Molecular Biology. 2004;11(11):1068 10.1038/nsmb840 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
The total number of reads (white) and the total number of unique reads aligned to the human genome (blue) obtained by performing hmeDIP-seq and meDIP-seq on TK6 cells cultured under static (control) and simulated microgravity (12 rpm) conditions for 48 hours. S1 Table demonstrates the exact numbers and percentage of mapped reads.
(TIF)
(TIF)
Color of these lines represent the fold coverage of the CpGs as shown in the legend.
(TIF)
Color of these lines represent the fold coverage of the CpGs as shown in the legend.
(TIF)
(TIF)
(XLSX)
For every DMR identified, a description of the genomic features found in this region has been provided. The columns represent the following information: (A) Genomic coordinates of the region defined as a DMR and (B) ENCODE IDs of features (such as gene, transcript, pseudogene, non-coding RNA or other regulatory feature) present in the region.
(XLSX)
For every DMR identified, a description of the genomic features found in this region has been provided. The columns represent the following information: (A) Genomic coordinates of the region defined as a DMR and (B) ENCODE IDs of features (such as gene, transcript, pseudogene, non-coding RNA or other regulatory feature) present in the region.
(XLSX)
The columns represents the respective ontology name, term name / identifier, term description, binomial rank, binomial p-value (uncorrected), binomial Bonferroni corrected p-value, binomial FDR q-value and names of gene hits generated by GREAT version 3.0.0; Species assembly: hg19 and association rule: Basal+extension: 5000 bp upstream, 1000 bp downstream, 1000000 bp max extension, curated regulatory domains included.
(XLSX)
The columns represents the respective ontology name, term name / identifier, term description, binomial rank, binomial p-value (uncorrected), binomial Bonferroni corrected p-value, binomial FDR q-value and names of gene hits generated by GREAT version 3.0.0; Species assembly: hg19 and association rule: Basal+extension: 5000 bp upstream, 1000 bp downstream, 1000000 bp max extension, curated regulatory domains included.
(XLSX)
For every DHMR identified, a description of the genomic features found in this region has been provided. The columns represent the following information: (A) Genomic coordinates of the region defined as a DHMR and (B) ENCODE IDs of features (such as gene, transcript, pseudogene, non-coding RNA or other regulatory feature) present in the region.
(XLSX)
The columns represent the following information for each identified DHMR: (A) Genomic coordinates of the region defined as a DHMR and (B) ENCODE IDs of features (such as gene, transcript, pseudogene, non-coding RNA or other regulatory feature) present in the region.
(XLSX)
The columns represents the respective ontology name, term name / identifier, term description, binomial rank, binomial p-value (uncorrected), binomial Bonferroni corrected p-value, binomial FDR q-value and names of gene hits generated by GREAT version 3.0.0; Species assembly: hg19 and association rule: Basal+extension: 5000 bp upstream, 1000 bp downstream, 1000000 bp max extension, curated regulatory domains included.
(XLSX)
The columns represent geneID, name of gene from UCSC Genome Browser (duplicates exist because multiple geneID can map to same gene), chromosome location, strand: + or–, transcription start position, transcription end position, the log2-fold-change of gene expression, the average log2-counts-per-million of comparison, p-value of comparison, false discovery rate (corrected p-value) of comparison.
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. Raw sequencing data is deposited at GEO (Accession Number: GSE65944) and available in the database (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE65944).