To the Editor:
Scicluna and colleagues recently reported the gene expression ratio of Fas apoptotic inhibitory molecule 3 (FAIM3) to placenta-specific 8 (PLAC8) as a new sepsis diagnostic biomarker (1). Accurate sepsis diagnosis is critical, as the mortality rate of sepsis increases for each hour that antibiotics are not administered (2). The FAIM3:PLAC8 ratio was discovered in a cohort comparing critically ill patients within 24 hours of admission for community-acquired pneumonia with noninfected patients. Scicluna and colleagues found that the FAIM3:PLAC8 ratio had areas under the receiver operating characteristic curve (AUCs) of 0.845 and 0.784 for diagnosing community-acquired pneumonia in their discovery and validation cohorts, respectively (1).
Of paramount importance in the further study of any proposed biomarker is its validation in independent heterogeneous cohorts, as estimates in discovery cohorts are often overfit. As we recently reported, there are multiple public gene expression datasets that compare noninfected systemic inflammatory response syndrome (SIRS)/trauma patients with patients with sepsis from various sources; we comprehensively organized all publicly available cohorts into 13 time-matched subcohorts and performed gene expression meta-analysis to derive an 11-gene set diagnostic for sepsis compared with sterile SIRS/trauma (3). These public datasets can be reanalyzed to test the diagnostic power of new gene expression biomarkers as well.
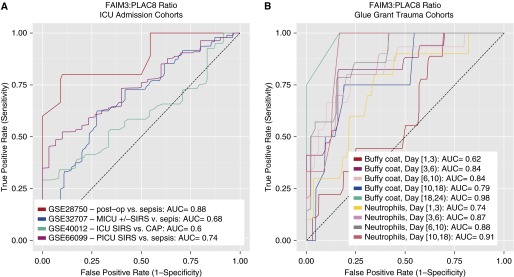
We thus tested the FAIM3:PLAC8 ratio in the previously described time-matched gene expression cohorts (four intensive care unit admission cohorts, nine Glue Grant trauma subcohorts; total n = 881). In intensive care unit admission cohorts, the FAIM3:PLAC8 mean AUC was 0.72 (range, 0.60–0.88; Figure 1A). In the Glue Grant trauma cohorts (4), which were split into time-matched subcohorts comparing patients within ±24 hours of diagnosis of infection with time-matched trauma patients who were never infected, the mean AUC was 0.83 (range, 0.62–0.98; Figure 1B) and generally increased over time since injury. In comparison, our 11-gene set had mean AUCs of 0.82 and 0.87 in the intensive care unit admission and Glue Grant cohorts, respectively. Finally, we tested our 11-gene set for diagnostic power in the 134 discovery samples from the study of Scicluna and colleagues (GEO dataset GSE65682; AUC = 0.79).
Figure 1.
Receiver operating characteristic curves showing the diagnostic power of sterile SIRS/trauma patients versus patients with sepsis in multiple public gene expression datasets. (A) Datasets comparing patients at intensive care unit admission. (B) Subcohorts of time-matched trauma patients, split out by days since initial injury. Buffy coat and neutrophil data were from separate patient cohorts. AUC = area under the receiver operating characteristic curve; CAP = community-acquired pneumonia; FAIM3 = Fas apoptotic inhibitory molecule 3; ICU = intensive care unit; MICU = medical ICU; PICU = pediatric ICU; PLAC8 = placenta-specific 8; SIRS = systemic inflammatory response syndrome.
Perhaps the greatest surprise is poor performance of the FAIM3:PLAC8 ratio in GSE40012 (5), another dataset that specifically studied community-acquired pneumonia. However, both GSE40012 and GSE32707 (6) were run on Illumina platforms and performed worse than almost all other tested cohorts; technical differences may thus be contributing to the observed decrease in AUC. In addition, the performance of FAIM3:PLAC8 in the Glue Grant data (mean AUC, 0.83) was nearly as good as in the original discovery cohort.
As noted by Scicluna and colleagues, biomarkers often perform better in combination than alone. In the cohorts tested here, the FAIM3:PLAC8 ratio was generally highly correlated to our sepsis diagnostic 11-gene set (3) (mean Spearman correlation, 0.68; SD, 0.10), and combining the two separate scores into a single diagnostic did not yield a synergistic increase in AUC (not shown).
We wish to congratulate Scicluna and colleagues for a well-performed study and for their derivation of the FAIM3:PLAC8 sepsis diagnostic biomarker. The AUCs of the FAIM3:PLAC8 ratio may not be high enough to bring it into practice in sole use; however, the team demonstrated superiority to procalcitonin in their cohort. Thus, it may be worthy of further study in combination with other infection markers. In addition, the use of a simple gene expression ratio (similar to other popular methods such as differences of arithmetic or geometric means) allows for easy testing and comparison without constructing a complicated model. Overall, given the relative ease with which researchers can now test a gene expression biomarker in multiple public cohorts, we recommend that further gene expression biomarker studies should include these comparisons as a basic benchmark.
Acknowledgments
Acknowledgment
The authors thank the authors of the deposited public datasets, without whom none of this would be possible. Use of the Glue Grant data was approved by both the Glue Grant Consortium and the Stanford University IRB (protocol 29798). The Glue Grant investigators receive ongoing support for their database from NIGMS Glue Grant Legacy Award R24GM102656.
Footnotes
T.E.S. is funded by a Stanford Child Health Research Institute Young Investigator Award (through the Institute for Immunity, Transplantation, and Infection), a Society for University Surgeons Resident Research Award, and the Stanford Department of Surgery. P.K. is funded by the Bill and Melinda Gates Foundation and National Institute of Allergy and Infectious Diseases grants 1 U19AI109662, U19AI057229, U54I117925, and U01AI089859.
Author disclosures are available with the text of this letter at www.atsjournals.org.
References
- 1.Scicluna BP, Klein Klouwenberg PM, van Vught LA, Wiewel MA, Ong DS, Zwinderman AH, Franitza M, Toliat MR, Nürnberg P, Hoogendijk AJ, et al. A molecular biomarker to diagnose community-acquired pneumonia on intensive care unit admission. Am J Respir Crit Care Med. 2015;192:826–835. doi: 10.1164/rccm.201502-0355OC. [DOI] [PubMed] [Google Scholar]
- 2.Ferrer R, Martin-Loeches I, Phillips G, Osborn TM, Townsend S, Dellinger RP, Artigas A, Schorr C, Levy MM. Empiric antibiotic treatment reduces mortality in severe sepsis and septic shock from the first hour: results from a guideline-based performance improvement program. Crit Care Med. 2014;42:1749–1755. doi: 10.1097/CCM.0000000000000330. [DOI] [PubMed] [Google Scholar]
- 3.Sweeney TE, Shidham A, Wong HR, Khatri P. A comprehensive time-course-based multicohort analysis of sepsis and sterile inflammation reveals a robust diagnostic gene set. Sci Transl Med. 2015;7:287ra71. doi: 10.1126/scitranslmed.aaa5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao W, Mindrinos MN, Seok J, Cuschieri J, Cuenca AG, Gao H, Hayden DL, Hennessy L, Moore EE, Minei JP, et al. Inflammation and Host Response to Injury Large-Scale Collaborative Research Program. A genomic storm in critically injured humans. J Exp Med. 2011;208:2581–2590. doi: 10.1084/jem.20111354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parnell GP, McLean AS, Booth DR, Armstrong NJ, Nalos M, Huang SJ, Manak J, Tang W, Tam OY, Chan S, et al. A distinct influenza infection signature in the blood transcriptome of patients with severe community-acquired pneumonia. Crit Care. 2012;16:R157. doi: 10.1186/cc11477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dolinay T, Kim YS, Howrylak J, Hunninghake GM, An CH, Fredenburgh L, Massaro AF, Rogers A, Gazourian L, Nakahira K, et al. Inflammasome-regulated cytokines are critical mediators of acute lung injury. Am J Respir Crit Care Med. 2012;185:1225–1234. doi: 10.1164/rccm.201201-0003OC. [DOI] [PMC free article] [PubMed] [Google Scholar]