Abstract
Two new fungal species of the genus Talaromyces, Talaromyces purpurogenus and Talaromyces trachyspermus from the Trichocomaceae family, were recovered during an investigation of fungal communities in soil collected from the Gangwon-do and Jeollanam-do provinces of Korea. These two species have not been previously officially reported from Korea. In this study, detailed descriptions of internal transcribed spacer rDNA and beta-tubulin gene regions of these two fungi are presented. Morphological features of the two fungi in five agar media, potato dextrose, oatmeal, malt extract, czapek yeast extract, and yeast extract sucrose, are also reported. The species were identified on the basis of molecular and morphological analysis, and herein we present data with detailed descriptions and figures.
Keywords: Molecular identification, Morphology, Talaromyces purpurogenus, Talaromyces trachyspermus
Talaromyces is a member of the order Eurotiales within the family Trichocomaceae, recorded from soil, indoor, food waste and has a worldwide distribution. The genus Talaromyces was introduced by Benjamin and Talaromyces vermiculatus is the first species of the type genus that produces soft, cottony fruit bodies (ascocarps) with cell walls made of tightly interwoven hyphae [1].
Soil fungi are one of the important components of microbial communities in the terrestrial ecosystem and they play a vital role in the decomposition of organic matter and nutrient cycling [2]. However, despite their immense importance in agro-ecosystems, they are probably the least-studied organisms. The microbial composition of soil is strongly affected by edaphic factors and cropping systems [3]. In the current study, an investigation was undertaken to assess the diversity of fungi in crop field soil from Gangwon-do and Jeollanam-do, and 120 morphologically distinct isolates were isolated. Of these, two species of Talaromyces were discovered which are new to Korea.
The present study compares these newly recorded species with previously described Talaromyces spp. with respect to their morphological and phylogenetic characteristics.
MATERIALS AND METHODS
Soil sampling and isolation of fungi
Soil samples were collected in 2014 from agricultural fields at various locations in Gangwon-do (37°09'50.46'' N, 129°11'52.32'' E) and Jeollanam-do (34°57'04.71'' N, 127°10'09.11'' E), Korea. Soil samples were taken from a depth of 0~15 cm, air dried and stored in plastic bags at 4℃ until use. Fungi were isolated using a conventional dilution plating technique [4] and cultured on potato dextrose agar (PDA; Difco Laboratories, Detroit, MI, USA) supplemented with 100 µg/L chloramphenicol (a bacteriostatic agent) for 5~7 days at 25℃ until fungal colony growth was observed. The pure cultures were maintained on PDA slants at 4℃ for future use.
Morphological characterization
Morphological characteristics of isolates KNU14-10 and KNU14-24-1 were observed on PDA (MB Cell, Los Angeles, CA, USA), oatmeal agar (OA; MB Cell), malt extract agar (MEA), czapek yeast extract agar (CYA), and yeast extract sucrose agar (YES). The strains were inoculated at three points on 9-cm petri dishes and incubated at 25℃ in the dark for 7 days. All media were prepared as described by Samson et al. [5]. After incubation, the diameter of the colonies on the various agar media was measured and the degree of sporulation was determined. Colony color (obverse and reverse sides) was described using Kornerup and Wanscher [6]. Photomicrographs were taken with an HK 3.1 CMOS digital camera (KOPTIC Korea Optics, Seoul, Korea) attached to an Olympus BX50F-3 microscope (Olympus Optical Co., Ltd., Tokyo, Japan) and a scanning electron microscope (LEO Model 1450VP Variable Pressure Scanning Electron Microscope; Carl Zeiss, Cambridge, MA, USA).
Genomic DNA extraction, sequencing, and data analysis
Total genomic DNA was extracted from isolates KNU14-10 and KNU14-24-1 using a DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following manufacturer instructions. The internal transcribed spacer region (ITS) and β-tubulin gene were amplified using primers ITS1 (5'-TCCGTAGGTGAACCTGCG-3') and ITS4 (5'-TCCTCCGCTTATTGATATGC-3') [7], and Bt2a (5'-GGTAACCAAATCGGTGCTTTC-3') and Bt2b (5'-ACCCTCAGTGTAGTGACCCTT-3'), respectively [8]. The amplified PCR products were sequenced using an ABI Prism 3730 DNA analyzer (Applied Biosystems, Foster City, CA, USA). The sequences were compared with reference ITS and β-tubulin sequences from GenBank at National Center for Biotechnology Information (NCBI) using the basic local alignment search tool [9]. The nucleotide sequences were deposited in GenBank and assigned accession numbers KP055602 and KP055603 for isolates KNU14-10 and KNU14-24-1, respectively. Phylogenetic relationships were analyzed using molecular evolutionary genetic analysis (MEGA 6) software [10]. A neighbor-joining tree was constructed using the Kimura 2-parameter substitution model [11]. Bootstrap analysis was performed with 1,000 replications to determine the support for each clade.
RESULTS
Morphology of studied fungal isolates
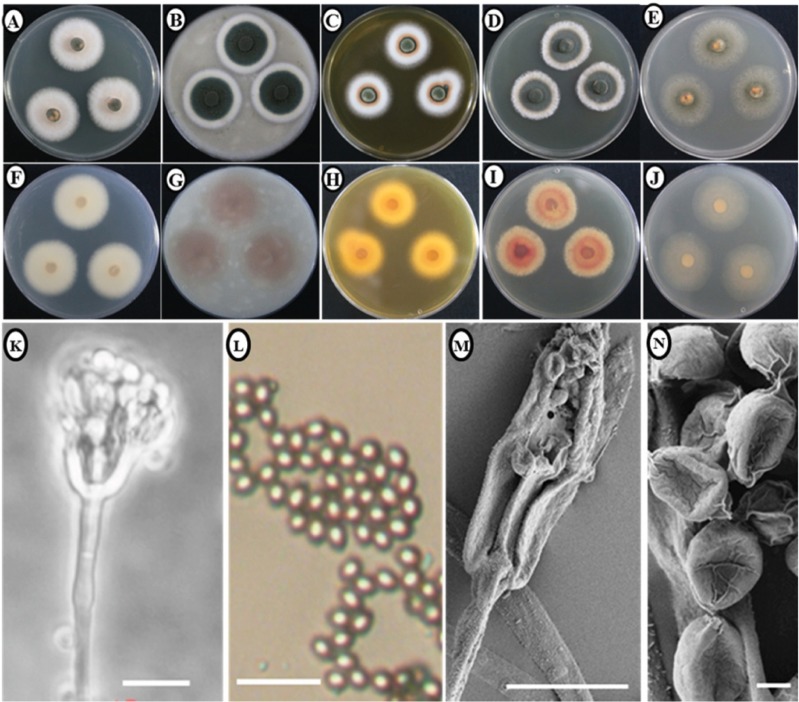
Morphology of the KNU14-10 isolate: Colony morphology: Photomicrographs of morphological structures of the isolates are shown in Fig. 1. Colonies grew moderately on PDA, attaining a diameter of 30~33 mm after 7 days at 25℃. The front- and back-side of the mycelium was white in color (Fig. 1A and 1F). The texture of the colony was floccose. Sporulation was dense and conidia were numerous. On OA, colonies grew moderately, reaching a diameter of 30~36 mm after 7 days at 25℃. The front side of the colony was green with a wide white margin (6~9 mm), while the reverse side of the colony was light brown (Fig. 1B and 1G). The texture of colony was floccose, especially near the center. Sporulation was moderately dense to dense. On YES, colonies grew slowly, attaining a diameter of 24~29 mm after 7 days at 25℃. The front side of the colony was white and reverse side was yellowish-brown in color (Fig. 1C and 1H). The texture of colony was floccose at the margin, sporulation was dense, and conidia were numerous. Colonies grew moderately on CYA, reaching a diameter of 27~32 mm after 7 days at 25℃. The front side of the colony was green with white margins. The reverse side of the colony was light brown (Fig. 1D and 1I). The texture of colony was floccose. Sporulation was sparse to moderately dense and conidia were numerous. On MEA, colonies grew moderately, attaining a diameter of 32~35 mm after 7 days at 25℃. The front- and back-sides of the colony were slightly green in color (Fig. 1E and 1J). The colony texture was floccose, sporulation was dense, and conidia were numerous.
Fig. 1. Morphological characteristics of Talaromyces purpurogenus KNU14-10 grown for 7 days on potato dextrose agar (PDA), oatmeal agar (OA), czapek yeast extract agar (CYA), yeast extract sucrose agar (YES), and malt extract agar (MEA) at 25℃. Obverse colony from left to right (A~E) and reverse colony from left to right (F~J) grown on PDA, OA, CYA, YES, and MEA. Conidiophores and conidia produced on PDA and MEA (K, L). Scanning electron microscope (SEM) of conidiophore (M) and SEM of conidia (N) (scale bars: K~N = 10 µm).
Micromorphology: The conidiophores were monoverticillate, around 6~28 µm in length, and subterminal branches were absent. The stipes was smooth-walled. Phialides were acerose and 5.2~7.9 × 1.9~2.6 µm in size. Metulae were in verticils of 2~3 range, 6~7 × 1~3 µm in size and inflated at the apex to about 3.2 µm. Conidia were slightly ellipsoidal and 1.46~1.95 µm in diameter.
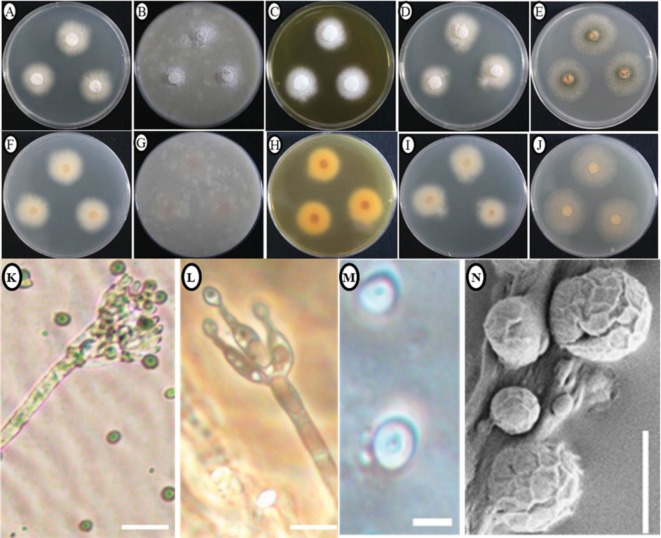
Morphology of the KNU14-24-1 isolate: Colony morphology: Photomicrographs of morphological structures of the isolate are shown in Fig. 2. On PDA, colonies grew moderately, attaining a diameter of 24~26 mm after 7 days at 25℃. The front and the reverse sides of the colony were white in color (Fig. 2A and 2F). Texture was floccose, sporulation was moderately dense to dense, and conidia were numerous. On OA, colonies grew slowly, reaching a diameter of 25~27 mm after 7 days at 25℃. The front side of the colony was white and reverse side was light brown in color (Fig. 2B and 2G). Texture was floccose, sporulation was moderate, and conidia were numerous. On YES, colonies grew slowly, attaining a diameter of 23~25 mm after 7 days at 25℃. The frontside of the colony was snowwhite in color while the backside was yellow in color (Fig. 2C and 2H). Texture was floccose, with moderate to dense sporulation. Conidia were numerous. On CYA, colonies grew slowly, reaching a diameter of 25~30 mm after 7 days at 25℃. Texture was floccose. The front and reverse sides of the colony were white in color (Fig. 2D and 2I). Sporulation was moderate and conidia were numerous. On MEA, colonies grew fast compared to the other media reaching a diameter of 44~47 mm after 7 days at 25℃. The front- and back-sides of the colony were light green in color (Fig. 2E and 2J). Texture was floccose, sporulation was moderate to dense, and conidia were numerous.
Fig. 2. Morphological characteristics of Talaromyces trachyspermus KNU14-24-1 grown for 7 days on potato dextrose agar (PDA), oatmeal agar (OA), czapek yeast extract agar (CYA), yeast extract sucrose agar (YES), and malt extract agar (MEA) at 25℃. A~J, Obverse colony from left to right (A~E) and reverse colony from left to right (F~J) grown on PDA, OA, CYA, YES, and MEA. K, Conidia produced on PDA; L, Conidiophores; M, Scanning electron microscope (SEM) of conidiophore; N, SEM of conidia (scale bars: K~N = 10 µm).
Micromorphology: The conidiophores were monoverticillate, around 6~28 µm in length, and subterminal branches were absent. The stipes was smooth-walled. Phialides were lanceolate, 2~6 in a verticil and 11~18 × 2~2.3 µm in size. Metulae in small verticils of 2~3 range 12~14 × 1.4~2.1 µm in size. Conidia were ellipsoidal to ovoidal and 5.90~ 7.87 µm in diameter and smooth-walled.
Molecular phylogeny of studied fungal isolates
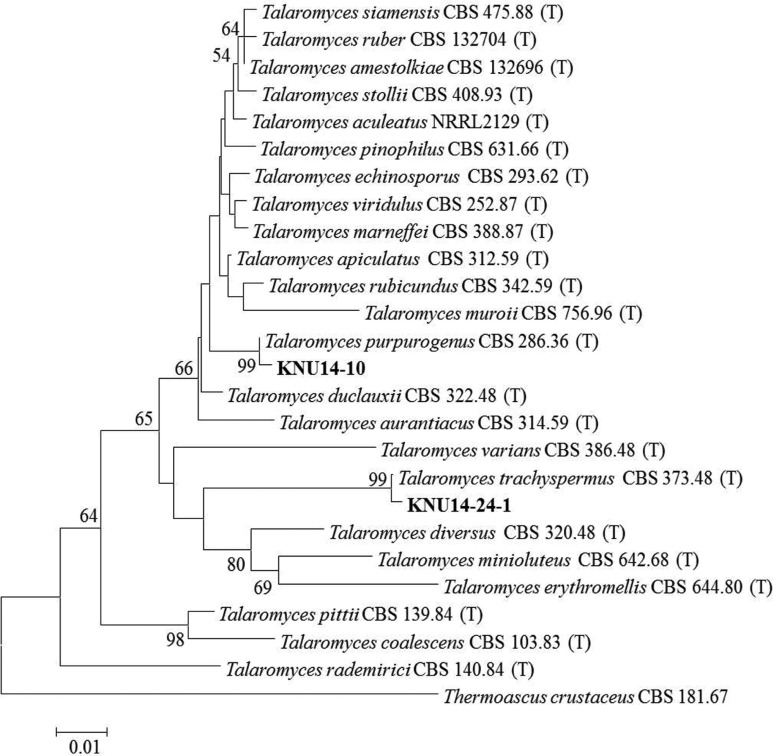
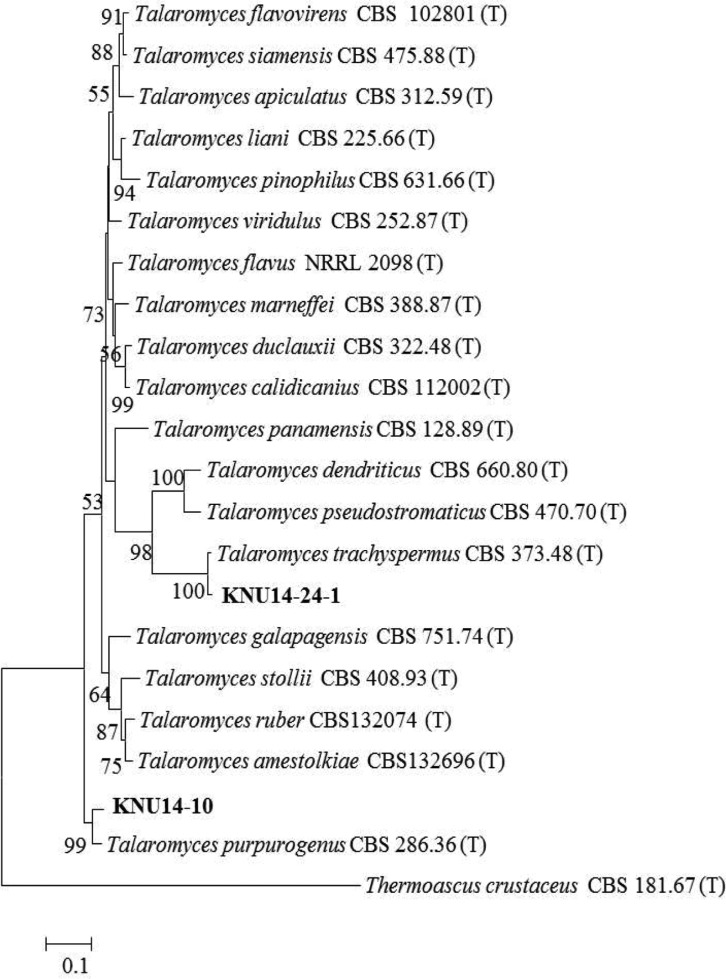
Molecular phylogeny of the KNU14-10 isolate: The ITS region and β-tubulin sequences were compared to determine the phylogenetic relationship between the KNU14-10 isolate and previously described Talaromyces species. The isolate was most closely related to T. purpurogenus CBS286.36, and formed a monophyletic group with bootstrap values of 99% and 99% (Figs. 3 and 4). The phylogenetic analysis showed that the isolate is T. purpurogenus. This is a common species, found especially in soil, but this is the first report of the isolation of T. purpurogenus in Korea.
Fig. 3. Neighbor-joining phylogenetic analysis of the partial 18S-ITS1-5.8S-ITS2-28S rDNA sequences of Talaromyces purpurogenus KNU14-10 and Talaromyces trachyspermus KNU14-24-1 obtained from crop field soil in Korea. A phylogenetic tree was constructed using the MEGA ver. 6 program. Sequences obtained in the study are shown in boldface. The mark (T) indicates type strain. Numerical values (> 50) on branches are the bootstrap values as percentage of bootstrap replication from a 1,000 replicate analysis. Thermoascus crustaceus was used as an outgroup. Scale bar represents the number of substitutions per site.
Fig. 4. Neighbor-joining phylogenetic analysis of β-tubulin gene sequences of Talaromyces purpurogenus KNU14-10 and Talaromyces trachyspermus KNU14-24-1 obtained from crop field soil in Korea. A phylogenetic tree was constructed using the MEGA ver. 6 program. Sequences obtained in the study are shown in boldface. The mark (T) indicates type strain. Numerical values (> 50) on branches are the bootstrap values as percentage of bootstrap replication from a 1,000 replicate analysis. Thermoascus crustaceus was used as an outgroup. Scale bar represents the number of substitutions per site.
Molecular phylogeny of the KNU14-24-1 isolate: To determine the evolutionary status between isolate KNU14-24-1 and previously identified Talaromyces species, ITS rDNA and β-tubulin sequences were compared. The isolate was most closely related to T. trachyspermus and formed a monophyletic group with bootstrap values of 99% and 100% (Figs. 3 and 4). The phylogenetic analysis showed that the study isolate is T. trachyspermus.
DISCUSSION
The KNU14-10 isolate from crop field soil was presumed to be a Talaromyces, based on its conidiophores, conidia and its colony appearance, and it reasonably fits the description of T. purpurogenus [12]. The study isolate KNU14-10 only differs slightly from the original description by the size of the conidiophores, and the conidia. It has been reported that the acerose phialides characteristics are more definitive morphological characteristics of Talaromyces anamorphs [13], and we found these to be similar in isolate KNU14-10. It has been reported that the growth pattern and colony morphology of T. purpurogenus differs according to the growth media used [14], and we observed this in our isolate. Thus, it can be concluded that isolate KNU14-10 is T. purpurogenus. Several studies have shown that T. purpurogenus has been used in the biodeterioration of cellulose materials such as textiles, paper and adhesives, as well as the ability to grow on plant material such as corn [15]. In addition, T. purpurogenus is important for biotechnology, because of its ability to produce enzymes and extrolites [16,17]. However, further studies on production of enzymes and extrolites by the isolate KNU14-10 are needed. Additional sequence similarity analysis of ITS regions and β-tubulin revealed that the studied isolates, KNU14-10 and KNU14-24-1, belong to T. purpurogenus and T. trachyspermus, respectively (Table 1). Based on the aforementioned taxonomical properties, isolate KNU14-24-1 from crop field soil was considered T. trachyspermus. This study isolate differs only slightly from the original description by the size of the conidiophores and the conidia. Our findings are in accordance with those of Stock and Samson [13]. Thus, it can be concluded that the isolate KNU14-24-1 is T. trachyspermus. In addition, T. trachyspermus has been reported as a food-borne, heat resistant, potential food spoilage mold [18]. Therefore, it would be valuable to perform further studies on the isolate in this context.
Table 1. Identification of fungal isolates at the species level, with reference species based on the analyses of internal transcribed spacer (ITS) and β-tubulin gene sequences.
ACKNOWLEDGEMENTS
This work was supported by a grant (NIBR2014-01205) from the National Institute of Biological Resources (NIBR), funded by the Ministry of environment (MOE) of the Republic of Korea for the project on Survey and Discovery of Indigenous Fungal Species of Korea.
References
- 1.Benjamin CR. Ascocarps of Aspergillus and Penicillium. Mycologia. 1955;47:669–687. [Google Scholar]
- 2.Warcup JH. Soil-steaming: a selective method for the isolation of ascomycetes from soil. Trans Br Mycol Soc. 1951;34:515–518. [Google Scholar]
- 3.Hargreaves SK, Williams RJ, Hofmockel KS. Environmental filtering of microbial communities in agricultural soil shifts with crop growth. PLoS One. 2015;10:e0134345. doi: 10.1371/journal.pone.0134345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davet P, Rouxel F. Detection and isolation of soil fungi. Enfield (NH): Science Publishers; 2000. [Google Scholar]
- 5.Samson RA, Houbraken J, Thrane U, Frisvad JC, Andersen B. Food and indoor fungi. CBS laboratory manual series 2. Utrecht: CBS-KNAW Fungal Biodiversity Centre; 2010. [Google Scholar]
- 6.Kornerup A, Wanscher JH. Methuen handbook of color. 2nd ed. Copenhagen: Sankt Jorgen Tryk; 1967. [Google Scholar]
- 7.White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego (CA): Academic Press; 1990. pp. 315–322. [Google Scholar]
- 8.Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol. 1995;61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.National Center for Biotechnology Information. GenBank overview [Internet] Bethesda (MD): National Center for Biotechnology Information; 2015. [cited 2015 May 5]. Available from: http://www.ncbi.nlm.nih.gov/Blast. [Google Scholar]
- 10.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimura M. Simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 12.Yilmaz N, Houbraken J, Hoekstra ES, Frisvad JC, Visagie CM, Samson RA. Delimitation and characterization of Talaromyces purpurogenus and related species. Persoonia. 2012;29:39–54. doi: 10.3767/003158512X659500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stolk AC, Samson RA. Studies on Talaromyces and related genera II. The genus Talaromyces. Stud Mycol. 1972;2:1–65. [Google Scholar]
- 14.Visagie CM, Jacobs K. Three new additions to the genus Talaromyces isolated from Atlantis sandveld fynbos soils. Persoonia. 2012;28:14–24. doi: 10.3767/003158512X632455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moss MO, Hill IW. Strain variation in the production of rubratoxins by Penicillium rubrum Stoll. Mycopathol Mycol Appl. 1970;40:81–88. doi: 10.1007/BF02051985. [DOI] [PubMed] [Google Scholar]
- 16.Carvallo M, de Ioannes P, Navarro C, Chavez R, Peirano A, Bull P, Eyzaguirre J. Characterization of an alpha-L-arabinofuranosidase gene (abf1) from Penicillium purpurogenum and its expression. Mycol Res. 2003;107(Pt 4):388–394. doi: 10.1017/s0953756203007603. [DOI] [PubMed] [Google Scholar]
- 17.Jeya M, Joo AR, Lee KM, Tiwari MK, Lee KM, Kim SH, Lee JK. Characeterization of β-glucosidase from a strain of Penicillium purpurogenum KJS506. Appl Microbiol Biotechnol. 2010;86:1473–1484. doi: 10.1007/s00253-009-2395-8. [DOI] [PubMed] [Google Scholar]
- 18.Enigl DC, King AD, Török T. Talaromyces trachyspermus, a heat-resistant mold isolated from fruit juice. J Food Prot. 1993;56:1039–1042. doi: 10.4315/0362-028X-56.12.1039. [DOI] [PubMed] [Google Scholar]