Abstract
Oak tree death caused by symbiosis of an ambrosia beetle, Platypus koryoensis, and an ophiostomatoid filamentous fungus, Raffaelea quercus-mongolicae, has been a nationwide problem in Korea since 2004. In this study, we surveyed the yeast species associated with P. koryoensis to better understand the diversity of fungal associates of the beetle pest. In 2009, a total of 195 yeast isolates were sampled from larvae and adult beetles (female and male) of P. koryoensis in Cheonan, Goyang, and Paju; 8 species were identified by based on their morphological, biochemical and molecular analyses. Meyerozyma guilliermondii and Candida kashinagacola were found to be the two dominant species. Among the 8 species, Candida homilentoma was a newly recorded yeast species in Korea, and thus, its mycological characteristics were described. The P. koryoensis symbiont R. quercusmongolicae did not show extracelluar CM-cellulase, xylanase and avicelase activity that are responsible for degradation of wood structure; however, C. kashinagacola and M. guilliermondii did show the three extracellular enzymatic activities. Extracelluar CM-cellulase activity was also found in Ambrosiozyma sp., C. homilentoma, C. kashinagacola, and Candida sp. Extracelluar pectinase activity was detected in Ambrosiozyma sp., C. homilentoma, Candida sp., and M. guilliermondii. All the 8 yeast species displayed compatible relationships with R. quercus-mongolicae when they were co-cultivated on yeast extract-malt extract plates. Overall, our results demonstrated that P. koryoensis carries the yeast species as a symbiotic fungal associate. This is first report of yeast diversity associated with P. koryoensis.
Keywords: Oak tree death, Platypus koryoensis, Raffaelea quercus-mongolicae, Yeast
Ambrosia beetles are members of the insect genus Platypus, which attack both broadleaf and coniferous trees and breed in the wood of hosts [1]. These beetles vector their associated ambrosia fungi, which develop on the walls of the beetle galleries. The larvae of the beetles feed on the developed ambrosia fungi. Several yeasts and filamentous fungi have been reported as ambrosia fungi. For instance, it is well known that species of the genus Ambrosiozyma can be isolated from sources associated with the bark or ambrosia beetles [2]. The ascomycete genus Raffaelea is known to be intimately associated with ambrosia beetles [3,4]. Raffaelea canadensis is associated with Platypus wilsoni [5], Raffaelea santoroi is associated with Platypus mutatus [6], and Raffaelea ambrosiae is associated with the oak ambrosia beetle, Platypus cylindrus [7]. In addition, tree diseases caused by these fungi could lead to reduction in the number and quality of timber products.
Oak trees have been dying in Japan since the 1980s, and incidences of oak wilt have become gradually more noticeable [8]. The extensive mortality of oak forests was referred to as the Japanese oak disease [9]. Dead oak trees are always found to be infested by the ambrosia beetle Platypus quercivorus. The two ophiostomatoid fungi, Ophiostoma longicollum and Raffaelea quercivora have been recovered from trees attacked by P. quercivorus and are described as new species [10,11]. Especially, R. quercivora has been isolated from the mycangium, body surface, and galleries of the beetle [8,12]. These indicated that R. quercivora is a fungal associate of P. quercivorus and attributed to the Japanese oak disease [11].
Recently, oak tree death caused by infestation of a wood boring beetle Platypus koryoensis has become a serious problem in forests and city landscapes in Korea [13]. Tree wilting of the foliage throughout the entire crown after mass attack of P. koryoensis resulted in tree death quickly. This beetle is one of the ambrosia beetles that are well known to have a symbiotic relation with microorganisms in other countries [14,15]. The beetle and microbial associates that accompany the beetle's infestation process are considered to contribute to oak tree mortality. Thus, to understand the mechanism of tree mortality, the diversity of microbial associates of the beetle needs to be clearly elucidated. The beetle is found to carry the ambrosia fungus Raffaelea quercus-mongolicae that has recently been identified as a new species in Korea [16]. This ophiostomatoid filamentous fungus has been considered as a pathogenic organism because it is the major fungus that has been found frequently in beetle-infested and dead oak trees (e.g., Quercus mongolica, Q. aliena, and Q. serrata) [16]. Although pathogenicity of R. quercus-mongolicae has not been proven yet, this fungus showed the ability to colonize sapwood, contribute to sapwood discoloration and disrupt sap flows around the inoculation sites of Q. mongolica [17]. In addition to R. quercus-mongolicae, other filamentous fungi, yeast and bacteria were isolated from the body of P. koryoensis [18]. Regarding filamentous fungi, 14 genera belonging to Ascomycota and Basidiomycota were identified from the body of P. koryoensis and some of them were found to be able to produce wood degrading enzymes [19]. However, no yeast has so far been identified from the beetle. Therefore, this study was carried out to identify and characterize the 8 yeast species isolated from the larvae and adults of P. koryoensis. This study also reports Candida homilentoma as a newly recorded yeast species in Korea, the different ability of producing extracellular enzymes by P. koryoensis-associated yeasts and their compatible relationships with R. quercus-mongolicae.
MATERIALS AND METHODS
Yeast isolation and culture conditions
For yeast sampling, wood disks with a diameter of 20 cm and thickness of 13 cm were cut from P. koryensis infested Mongolian oak trees located in Cheonan, Goyang, and Paju, Korea from 2007 to 2008 (Fig. 1A and 1B). The wood disks were brought to the laboratory under humid conditions in a plastic container, and broken into small wood pieces using a sterile hammer and chisel. A total of 30 larvae and adult beetles of P. koryoensis were sampled from the wood disks of each geographic region, by chipping out the wood pieces along the insect galleries and used for yeast isolation. The genders of the sampled adult beetles were differentiated by observing the presence of mycangia which is well developed in the female beetle of P. koryoensis [13].
Fig. 1. Photos of Platypus koryoensis captured from oak wilt diseased trees. A, Oak wilt diseased trees; B, The beetle infested oak with entrance holes; C, A larvae of P. koryoensis; D, Adult beetles (left, male; right, female). Black arrows indicate entrance holes (scale bars: B = 5 cm, C, D = 1 mm).

For yeast isolation from P. koryoensis, the standard plating methods were used. The captured beetles were ground with a mortar and pestle using sterile distilled water. The ground beetle samples were spread on yeast extract-malt extract (YEME) agar (BD Science, San Jose, CA, USA) plates using a sterile glass rod. The inoculated YEME plates were incubated at 28℃ for 3~7 days. Yeast colony formed on the YEME plates were confirmed by observing yeast cell type structure using a SZ61 stereo microscope (Olympus Opitical Co., Tokyo, Japan). Single colony isolation was performed at least three times from the grown colonies. Yeast colonies were initially grouped based on the colony morphology and randomly selected colonies from each group were subjected to species identification. Yeast cell were morphology observed using an optical microscope (ZEISS AXIOSKOP40; Carl Zeiss, Jena, Germany) and a scanning electron microscope (SEM; Hitachi, Tokyo, Japan). Cell sizes were measured by SEM observation. Yeast identification kit of API 20C AUX (BioMerieux, Marcyl'Etoile, France) was used for manual yeast identification, according to the manufacturer's instructions. Test results were interpreted through internetbased APIwe service (https://apiweb.biomerieux.com). The obtained yeast isolates were maintained on YEME for the entire duration of the experiment.
Genomic DNA extraction
For the preparation of yeast genomic DNA, each isolate was cultured in YEME liquid media for 3 days at 28℃, from where 1 mL of the cultured yeast broth was transferred to a 2-mL microcentrifuge tube. After spinning at 10,000 rpm for 1 min, the supernatant was decanted. The cell pellet was re-suspended in 700 µL lysis buffer (0.5M NaCl, 0.2M Tris-Cl, 0.01M ethylenediaminetetraacetic acid, 1% sodium dodecyl sulfate), and 200 µL phenol : chlorforom : isoamylalcohol (25 : 24 : 1), followed by addition of 200 mg of acid washed glass beads. The tube was then vigorously vortexed and centrifuged at 14,000 rpm for 10 min at 4℃ [20]. The top layer was transferred to a new microcentrifuge tube and mixed with 0.55 volume of cold isopropanol. After being spun at 14,000 rpm for 7 min at 4℃, the supernatant was decanted and the pellet was washed twice with ice-cold 70% ethanol, and then dried. The dried pellet was resuspended in 50 µL TE-buffer and kept at -20℃ until used.
PCR amplification of large subunit ribosomal DNA (LSU rDNA) D1/D2 region and nucleotide sequencing
The LSU rDNA D1/D2 region was amplified by PCR using universal primer set LROR (5'-ACCCGCTGAACTTAAGC-3') and LR 4 (5'-ACCAGAGTTTCCTCTGG-3') [21,22]. PCR was performed in 50 µL volumes of a reaction mixture containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2·6H2O, 200mM of each dNTPs, 20 pmol of each primer, 20~50 ng of genomic DNA, and 0.5 U Taq DNA polymerase (Roche Ltd., Basel, Swiss). Amplification of the LSU rDNA was performed as follows: 95℃ for 5 min, followed by 29 cycles consisting of 94℃ for 50 sec, 52℃ for 50 sec and 72℃ for 1 min. Amplification was done in a GeneAmp-950 thermal cycler (Thermo Fisher Scientific Inc., Waltham, MA, USA). The PCR products were purified with a High Pure PCR Purification Kit (Roche Ltd.) and then ligated into pGEM T-easy vectors (Promega Corp., Madison, WI, USA). The recombinant plasmids were transformed into competent Escherichia coli DH5α cells according to the manufacturer's instructions. Nucleotide sequencing was performed by Macrogen Inc. (Seoul, Korea). The determined nucleotide sequences were searched for homologous sequences through BLASTN at the GenBank database of National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/genbank). The determined LSU rDNA D1/D2 region sequences were deposited to GenBank using the Sequin program.
Phylogenetic analysis
The determined nucleotide sequences of LSU rDNA D1/D2 were manually edited and aligned using the biological sequence alignment editor v7.0.5. Reference LSU rDNA D1/D2 sequences of related taxa were obtained from the GenBank database. Phylogenetic analysis was performed with related species using PAUP v.4.0b10 [23]. Parsimony tree was constructed by the neighbor-joining method [24]. Bootstrap values were generated with 1,000 replicates.
Extracellular enzyme activity test
The ability of producing extracellular enzymes was evaluated on chromogenic reaction medium containing different substrates: 0.5% CM-cellulose (Sigma-Aldrich, St. Louis, MO, USA), D-cellobiose (Sigma-Aldrich), polygalacturonic acid (MP Biomedicals, Sanata Ana, CA, USA), starch (Sigma-Aldrich), xylan (Sigma-Aldrich), and avicel cellulose (Fluka, Cork, Ireland) as a carbon source, 0.1% yeast nitrogen base without amino acid (BD Biosciences, Franklin Lakes, NJ, USA) as a nitrogen source, 1.5% agar, and 0.5% Congo Red (Sigma-Aldrich) as a dye [25]. The yeast species isolated from this study were pre-cultured in YEME broth at 28℃ for 18 hr. Ten microliters of each species' cultured broth was spotted on chromogenic media. After 5 days of incubation at 28℃, the presence of a clear zone, formed on the surface of the chromogenic media due to the reaction between extracellular enzyme produced by yeast and substrate in the media was observed.
Dual-culture experiments
The relationships between R. quercus-mongolicae and each of 8 yeast species isolated from this study and Exophiala alcalophila isolated from another study, was examined by dual-culture experiments on YEME plates. Agar block of the R. quercus-mongolicae was placed on the center of a YEME plate. Each yeast species was inoculated by streak on the YEME plate at the position which was 20 mm apart from the agar block of R. quercus-mongolicae. The existence of antagonistic growth inhibition or no hindering of growth between the dual-cultures was examined after incubating the cultures at 28℃ for 5 days. The experiment was repeated with three replicates.
RESULTS AND DISCUSSION
Yeast isolation and identification
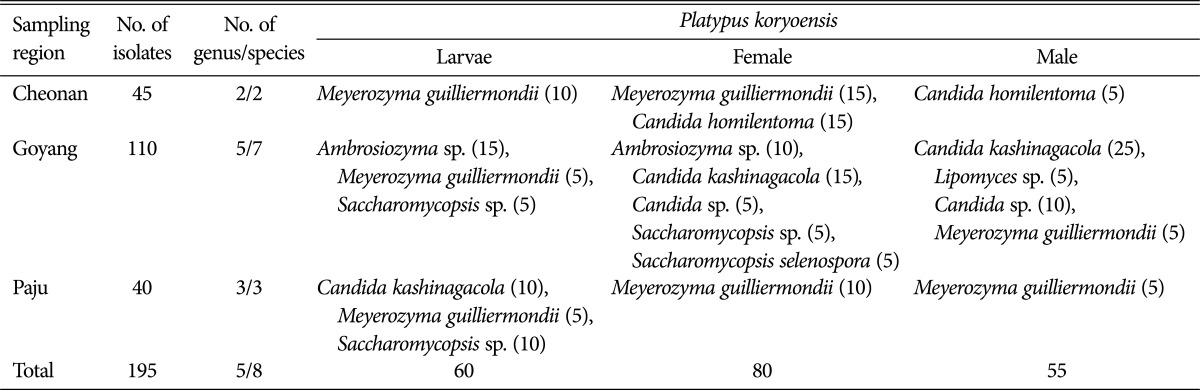
A total of 195 yeast isolates were obtained from the samples of P. koryoensis in this study (Table 1). The highest number of yeast isolates were isolated from P. koryoensis sampled in oak wood disks originated from Goyang region. When we looked at the isolation source of yeast from P. koryoensis, 60 isolates were from larvae, 80 from female adult beetles, and 55 from male adult beetles (Table 1). It is interesting that more number of yeast isolates were obtained from female rather than male and larvae. This result could be attributed to the fact that the female of P. koryoensis is known to have mycangia on the surface of the body, which carry the symbiotic fungi [13].
Table 1. Yeast species isolated from larvae, female and male of Platypus koryoensis sampled from the beetle-infested oak trees that were located in three different regions in Korea.
Number of obtained isolates is indicated in parentheses.
To identify the yeast isolates from oak infesting insect pest P. koryoensis, the LSU rDNA D1/D2 regions that are commonly used in yeast taxonomy were amplified, sequenced, and analyzed. Nucleotide sequence similarity analysis of the 195 yeast isolates through GenBank search showed that they had 94 to 100% homology in LSU rDNA sequences of known yeast species in the GenBank database. Among the 195 yeast isolates, 94% to 96% homology was found in Saccharomycopsis sp. and Lipomyces sp., respectively. In the near future, isolates of these species need to be further analyzed for classification. Based on the LSU rDNA D1/D2 analyses, a total of 8 species belonging to 5 genera were identified from the 195 yeast isolates (Table 1). All of these yeast species belonged to Ascomycota; no basidiomycete yeast were found (Table 1). Two yeast species, Meyerozyma guilliermondii (from larvae and female) and Candida homilentoma (female and male), were identified in Cheonan. Seven yeast species, Ambrosiozyma sp. (larva and female), M. guilliermondii (larvae and male), Saccharomycopsis sp. (larvae and female), S. selenospora (female), Candida kashinagacola (female and male), Candida sp. (female and male), and Lipomyces sp. (male), were identified in Goyang. Three yeast species, C. kashinagacola (larva), M. guilliermondii (larvae, female, and male), and Saccharomycopsis sp. (larvae) were identified in Paju. M. guilliermondii and C. kashinagacola were the most dominant species in all three locations. The highest species diversity was found from female-derived isolates.
A total of 8 species representing 40 yeast isolates were subject to phylogenetic analysis, to verify their position. The phylogenetic relationships of the 40 yeast isolates to the reference strains were inferred based the LSU rDNA D1/D2 region sequences by the neighbor-joining analysis, and the resulting phylogenetic tree is presented in Fig. 2. The yeasts from this study were placed into 6 different phylogenetic clades (Fig. 2). The phylogenetic tree supported the molecular identification of the 8 species. The sequences of the 40 isolates used for phylogenetic analysis were deposited on the GenBank with accession Nos. HQ454938~HQ454942 (Ambrosiozyma sp.), HQ454943~HQ454946 (C. homilentoma), HQ454947~HQ454956 (Candida kashinagacola), HQ454958 (Lipomyces sp.), HQ454959~HQ454961 (Candida sp.), HQ454962~HQ454972 (Meyerozyma guilliermondii), HQ454973~HQ454976 (Saccharomycopsis sp.), and HQ454958 (Saccharomycopsis selenospora).
Fig. 2. Phylogenetic relationships of the yeast isolates sampled from Platypus koryoensis in this study. The tree was constructed based on partial large subunit ribosomal DNA D1/D2 sequence using the neighbor-joining method. Sequences of reference species were included for comparison. Numbers at nodes represent the percentage of bootstrap resampling based on 1,000 replicates. Schizosaccharomyces pombi was used as the outgroup. Isolates in this study are given in bold. Type strains are indicated with superscript T.
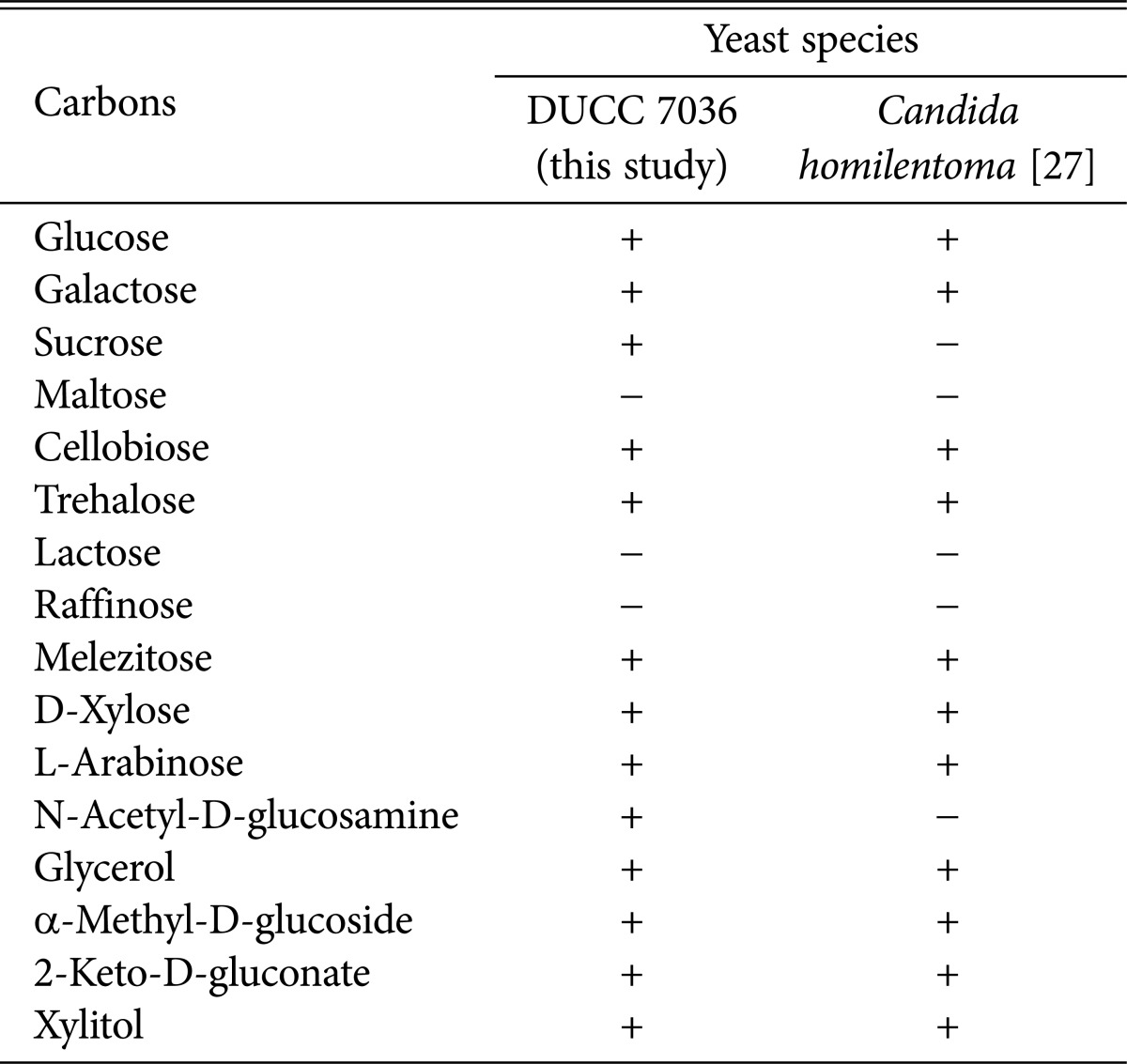
Among the identified 8 species, C. homilentoma was found to be an unrecorded yeast in Korea. The GenBank registered C. homilentoma isolates DUCC 7036~7039 shared 100% (504/504 bp) sequence identities in LSU rDNA D1/D2 region with its type strain, C. homilentoma NRRL Y10941. The DUCC 7036 was deposited on the National Institute of Biological Resources with accession No. NIBRFGC000140844. Moreover, morphological characteristics of C. homilentoma DUCC 7036 (Fig. 3) corresponded to the type strain [26]. The results of API 20 AUX kit for yeast growth in Table 2 revealed that C. homilentoma isolate DUCC 7036 could assimilate diverse carbohydrate sources, similar to the known C. homilentoma [27], the only difference being N-Acetyl-D-glucosamine assimilation. C. homilentoma isolate DUCC 7036 could assimilate Nacetyl-L-glucosamine, whereas the kown strain was incapable of assimilation.
Fig. 3. Morphological images of the unrecorded strain Candida homilentoma isolated from Platypus koryoensis. Colony on yeast extract-malt extract grown at 28℃ for 3 days (A); scanning electron microscope images of pseudohyphae and budding yeast cells (B) and blastoconidia (C) (scale bars: B = 10 µm, C = 2 µm).
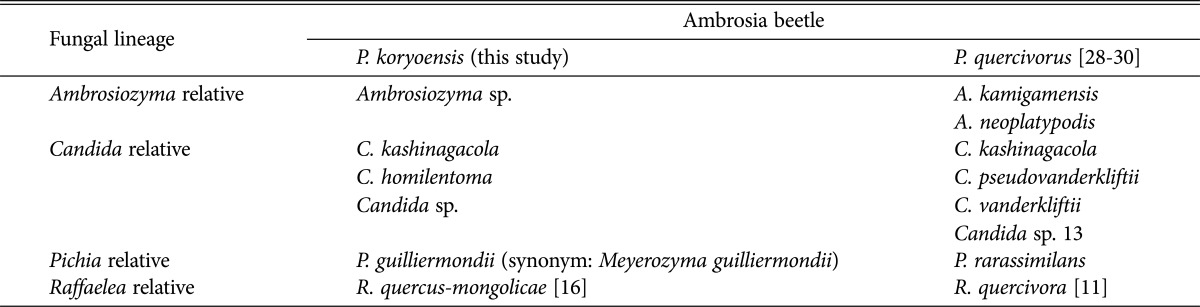
Table 2. Comparison of yeast species isolated from Platypus koryoensis in Korea and Platypus quercivorus in Japan.
The morphological characteristics of C. homilentoma DUCC 7036 are described as below. Taxonomy: Candida homilentoma Van der Walt & Nakase, Antonie van Leeuwenhoek 39: 450 (1973). Etymology: This species was isolated from adult P. koryeonsis in Cheonan, Korea. Description: The colony was white to cream and butyrous, when cultured on YEME at 28℃ for 3 days (Fig. 3A). After 3 days at 28℃, pseudohyphae of long cylindrical cells and septate hyphae with yeast cells are present (Fig. 3B). Yeast cells (blastoconidia) are ellipsoidal, ovoid to cylindrical and occur singly, in pairs, short chains or small clusters, and measure 1.5~3.0 × 1.6~6.0 µm (Fig. 3C). Bud scar is shown in a cell (Fig. 3C).
Yeasts isolated from oak infesting Platypus quercivora beetle were reported in Japan [28] and C. kashinagacola, C. pseudovanderkliftii, C. vanderkliftii, Ambrosiozyma kamigamensis, and Ambrosiozyma neoplatypodis were reported as new species [29,30,31]. When we compared yeast species from P. koryoensis in Korea to those from P. quercivora in Japan, C. kashinagacola and Candida sp. (that is corresponded Candida sp. 13 in Japan as seen in phyogenetic tree in Fig. 2) were commonly isolated (Table 3). Recently, C. kashinagacola has been reported as a newly recorded species which was associated with oak wilt disease in Korea [32]. This species was also isolated from the Korean oak wilt disease vector, P. koryoensis, in this study. Isolates of C. kashinagacola from this study was placed in the same clade with the species isolated from Japan (Fig. 2). The comparison of species in Table 3 shows that the yeast diversity in the two Platypus beetle are similar at the genus level, but different at species level. It seems that Ambrosiozyma, Candida, and Pichia are major yeast groups associated with Platypus beetles that attack oak trees in Korea and Japan.
Table 3. The assimilation ability of different carbon sources by the isolate DUCC 7036.
DUCC, Dankook University Culture Collection; +, assimilation positive; -, assimilation negative.
Extracellular enzyme activity
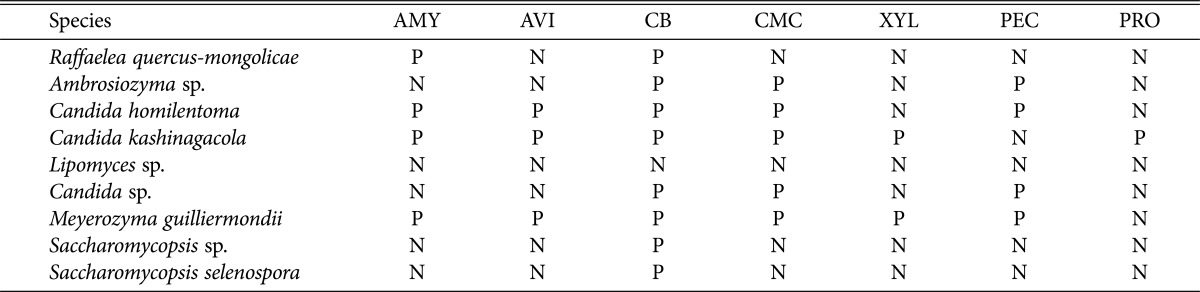
To grow on wood environment (such as beetle gallery) by using the wood components as nutrient sources, yeast species are expected to have the ability of producing extracellular enzymes that degrade polymeric components of wood cells into monomers. Cellobiose, cellulose, protein, pectin, starch, and xylan are known as the major polymeric components of wood cells in trees. Thus, we investigated the 8 yeast species from P. koryoensis including R. quercus-mongolicae known as oak wilt pathogen in Korea whether they could use the major polymeric components of wood cells as substrates. Chromogenic media-based tests with 7 different substrates allowed us to detect the extracellular activity of amylase, avicelase (avicel-cellulase), β-glucosidase, CM-cellulase, pectinase, protease, and xylanase from the tested yeast species (Table 4). The oak wilt pathogen R. quercusmongolicae showed just the activities of amylase and β-glucosidase. Regarding yeast species, amylase and pectinase activities were found in 4 species, β-glucosidase activity in 8 species, CM-cellulase activity in 5 species, avicelase activity in 3 species, xylanase activity in 2 species, and protease activity in 1 species. Among the tested eight species, M. guilliermondii and C. kashinagacola, the 2 dominant species derived from P. koryoensis showed 6 kinds of extracellular enzyme activities. C. homilentoma, C. kashinagacola, and M. guilliermondii showed avicelase, CM-cellulase and xylanase activity that are responsible for decomposition of structural components of wood. Ambrosiozyma sp. showed CM-cellulase and pectinase activities that could also degrade structural parts of wood. P. koryoensis beetle is known to make gallery in oak trees not only near sapwood which is the living, softer part of the wood which is more abundant, but also deep in the heart wood which is older, nonfunctioning, and usually darker and harder than the younger sapwood. Thus, the possession of the ability to degrade cellulose, pectin, and/or xylan is likely to be greatly beneficial to these 4 yeast species when they grow on heartwood as well as near the sapwood. Lipomyces sp. showed none of the extracellular enzymatic activities tested. This species is assumed to be less important in the wood environment of the beetle. It is noticeable that none of the yeast species could produce all 7 kinds of extracellular enzymes. Overall, the results in Table 4 showed that each yeast species has differing ability to produce extracellular enzymes. It means that the polymeric wood components would be better utlized not by a single yeast species, but by diverse yeast species together. Namely, yeast diversity is needed to make better use of the wood substrates. Since Platypodid ambrosia beetles prepare galleries into the wood of host trees, and use their fungal symbionts to exploit the nutrient-poor xylem [30,31], it is obvious that association of diverse yeast species that are able to colonize wood is beneficial to P. koryoensis.
Table 4. The results of chromogenic media-based assessment of the ability to produce extracellular enzymatic activity by the oak wilt pathogen Raffaelea quercus-mongolicae and 8 yeast species isolated from Platypus koryoensis in this study.
AMY, amylase; AVI, avicelase; CB, β-glucosidase; CMC, CM-cellulase; XYL, xylanase; PEC, pectinase; PRO, protease; P, activity; N, no activity.
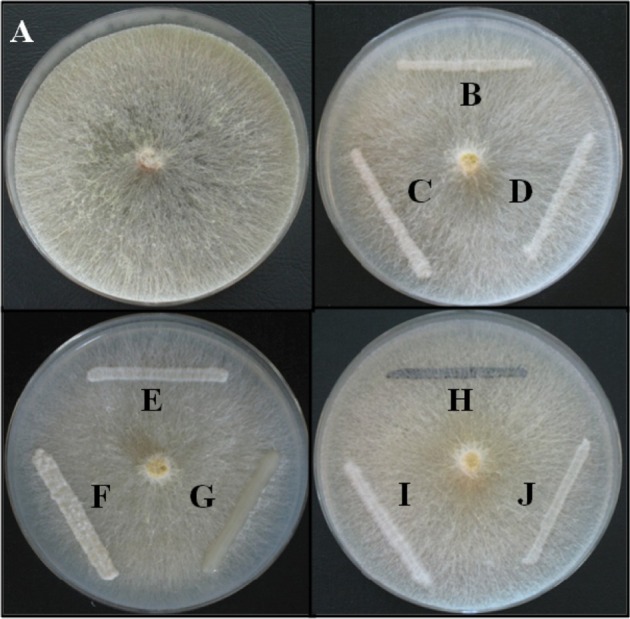
Dual-culture experiments
Although the names of yeast found in Platypus beetles were reported, their relationships to ambrosia fungi such as Raffaelea, one of the well-studied fungi as a symbiont of Platypus beetles, was unknown. Thus, in this study the relationships between P. koryoensis-associated yeasts and P. koryoensis-vectored the oak wilt pathogen R. quercus-mongolicae was examined by co-culturing them on YEME plate (Fig. 4). No antagonistic ability to R. quercus-mongolicae was found in all the yeast species tested. All yeast species were overgrown by R. quercus-mongolicae and all the yeast species could grow on the overgrown position of R. quercus-mongolicae. These results demonstrated that a compatible relationship exists between yeast species and R. quercus-mongolicae. This compatible relationship would allow diverse yeast and R. quercus-mongolicae to grow together as co-inhabitants in the galleries of P. koryoensis in oak trees. We assume that to be co-inhabitants on wood environment, sharing of nutrients in wood is necessary for both diverse yeast and R. quercus-mongolicae. This necessity might explain why each yeast species has different abilities in producing extracellular enzymes, as seen in Table 4.
Fig. 4. Dual culture interactions between 9 yeast species isolated from Platypus koryoensis and Raffaelea quercusmongolicae. A, R. quercus-mongolicae; B, Candida kashinagacola; C, Meyerozyma guilliermondii; D, Ambrosiozyma sp.; E, Saccharomycopsis sp.; F, Saccharomycopsis selenospora; G, Candida sp.; H, Exophiala alcalophila; I, Lipomyces sp.; J, Candida homilentoma.
In conclusion, we identified and described P. koryoensisassociated yeasts in Korea. This study is the first report of yeast diversity associated with the ambrosia beetle. Also, we described C. homilentoma as a newly recorded species in Korea. Our study demonstrated that P. koryoensis-associated yeasts have different ability of producing extracellular enzymes involved in degradation of wood components. The presence of compatible relationships between P. koryoensis-associated yeasts and P. koryoensisvectored R. quercus-mongolicae was also explored. Although the names of yeast found in Platypus beetles were reported, the properties of these yeasts have not been well-studied. Therefore, information on the properties of P. koryoensisassociated yeasts generated in this study will be the groundwork for understanding of the ecological distribution and roles of yeasts in P. koryoensis.
ACKNOWLEDGEMENTS
This research was supported by the National Institute of Biological Resources (NIBR), Korea and the Korea Forest Research Institute, and National Research Foundation of Korea (616-2011-3-F00002).
References
- 1.Bright DE, Skidmore RE. A catalogue of Scolytidae and Platypodidae (Coleoptera), Supplement 2 (1995-1999) Ottawa: NRC Research Press; 2002. [Google Scholar]
- 2.Van der Walt JP. The yeast genus Ambrosiozyma gen. nov. (Ascomycetes) Mycopathol Mycol Appl. 1972;46:305–316. [Google Scholar]
- 3.Batra LR. Ambrosia fungi: a taxonomic revision, and nutritional studies of some species. Mycologia. 1967;59:976–1017. [Google Scholar]
- 4.Funk A. Fungal symbionts of the ambrosia beetle Gnathotrichus sulcatus. Can J Bot. 1970;48:1445–1448. [Google Scholar]
- 5.Furniss RL, Carolin VM. Western forest insects. Miscellaneous publication No. 1339. Washington, DC: US Department of Agriculture, Forest Service; 1977. [Google Scholar]
- 6.Giménez RA, Etiennot AE. Host range of Platypus mutatus (Chapois, 1865) (Coleoptera: Platypodidae) Entomotropica. 2003;18:89–94. [Google Scholar]
- 7.Babuder G, Pohleven F. Fungal succession in the tunnels of ambrosia beetles in oak wood (Quercus sp.) Res Rep For Wood Sci Technol. 1995;47:241–254. [Google Scholar]
- 8.Ito S. Mass mortality of Quercus species (Fagaceae)-mystery in symbiotic relationships between fungi and ambrosia beetle. Nishi Nippon Branch Mycol Soc Jpn. 2000;10:16–22. [Google Scholar]
- 9.Kaneko S. Mass death of oaks in Japan; IUFRO XX World Congress; 1995 Aug 6-12; Tampere, Finland. [Google Scholar]
- 10.Masuya H, Kaneko S, Yamaoka Y. A new Ophiostoma species isolated from Japanese oak infested by Platypus quercivorus. Mycoscience. 1998;39:347–350. [Google Scholar]
- 11.Murata M, Yamada T, Matsuda Y, Ito S. Discoloured and non-conductive sapwood among six Fagaceae species inoculated with Raffaelea quercivora. For Pathol. 2007;37:73–79. [Google Scholar]
- 12.Kinuura H, Kobayashi M. Death of Quercus crispula by inoculation with adult Platypus quercivorus (Coleoptera: Platypodidae) Appl Entomol Zool. 2006;41:123–128. [Google Scholar]
- 13.Hong KJ, Kwon YD, Park SW, Lyu DP. Platypus koryoensis (Murayama)(Platypodidae: Coleoptera), the vector of oak wilt disease. Korean J Appl Entomol. 2006;45:113–117. [Google Scholar]
- 14.Beaver RA. Insect-fungus relationships in the bark and ambrosia beetles. In: Wilding N, Collins NM, Hammond PM, Webber JF, editors. Insect-fungus interactions. London: Academic Press; 1989. pp. 121–143. [Google Scholar]
- 15.Paine TD, Raffa KF, Harrington TC. Interactions among scolytid bark beetles, their associated fungi, and live host conifers. Annu Rev Entomol. 1997;42:179–206. doi: 10.1146/annurev.ento.42.1.179. [DOI] [PubMed] [Google Scholar]
- 16.Kim KH, Choi YJ, Seo ST, Shin HD. Raffaelea quercusmongolicae sp. nov. associated with Platypus koryoensis on oak in Korea. Mycotaxon. 2009;110:189–197. [Google Scholar]
- 17.Torii M, Matsuda Y, Seo ST, Kim KH, Ito S, Moon MJ, Kim SH, Yamada T. The effect of Raffaelea quercus-mongolicae inoculations on the formation of non-conductive sapwood of Quercus mongolica. Mycobiology. 2014;42:210–214. doi: 10.5941/MYCO.2014.42.2.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suh DY, Hyun MW, Choi IJ, Kim SH, Seo ST, Kim KH. Filamentous fungi and yeasts isolated from Platypus koryoensis and the beetle infested oak tree in Korea; Proceeding of Asian Mycological Congress 2011 & The 12th International Marine and Freshwater Mycology Symposium; 2011 Aug 7-11; Incheon, Korea. Seoul: The Korean Society of Mycology; 2011. p. 196. [Google Scholar]
- 19.Suh DY, Hyun MW, Kim SH, Seo ST, Kim KH. Filamentous fungi isolated from Platypus koryoensis, the insect vector of oak wilt disease in Korea. Mycobiology. 2014;39:313–316. doi: 10.5941/MYCO.2011.39.4.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Müller FM, Werner KE, Kasai M, Francesconi A, Chanock SJ, Walsh TJ. Rapid extraction of genomic DNA from medically important yeasts and filamentous fungi by high-speed cell disruption. J Clin Microbiol. 1998;36:1625–1629. doi: 10.1128/jcm.36.6.1625-1629.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York: Academic Press; 1990. pp. 315–322. [Google Scholar]
- 22.O'Donnell K, Nirenberg HI, Aoki T, Cigelnik E. A multigene phylogeny of the Gibberella fujikuroi species complex: detection of additional phylogenetically distinct species. Mycoscience. 2000;41:61–78. [Google Scholar]
- 23.Swofford DL. Paup*: phylogenetic analysis using parsimony (*and other methods). Ver. 4.0. B5. Sunderland (MA): Sinauer Associates; 2001. [Google Scholar]
- 24.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 25.Yoon JH, Park JE, Suh DY, Hong SB, Ko SJ, Kim SH. Comparison of dyes for easy detection of extracellular cellulases in fungi. Mycobiology. 2007;35:21–24. doi: 10.4489/MYCO.2007.35.1.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der, Nakase T. Candida homilentoma, a new yeast from South African insect sources. Antonie Van Leeuwenhoek. 1973;39:449–453. doi: 10.1007/BF02578887. [DOI] [PubMed] [Google Scholar]
- 27.Kurtzman CP, Fell JW, Boekhout T. The yeasts: a taxonomic study. 5th ed. London: Elsevier; 2011. pp. 1107–1108. [Google Scholar]
- 28.Endoh R, Suzuki M, Benno Y. Pichia rarassimilans sp. nov., a novel yeast species isolated from body surface of the ambrosia beetle Platypus quercivorus. J Gen Appl Microbiol. 2008;54:181–186. doi: 10.2323/jgam.54.181. [DOI] [PubMed] [Google Scholar]
- 29.Endoh R, Suzuki M, Benno Y. Ambrosiozyma kamigamensis sp. nov. and A. neoplatypodis sp. nov., two new ascomycetous yeasts from ambrosia beetle galleries. Antonie Van Leeuwenhoek. 2008;94:365–376. doi: 10.1007/s10482-008-9253-z. [DOI] [PubMed] [Google Scholar]
- 30.Endoh R, Suzuki M, Benno Y, Futai K. Candida kashinagacola sp. nov., C. pseudovanderkliftii sp. nov. and C. vanderkliftii sp. nov., three new yeasts from ambrosia beetle-associated sources. Antonie Van Leeuwenhoek. 2008;94:389–402. doi: 10.1007/s10482-008-9256-9. [DOI] [PubMed] [Google Scholar]
- 31.Endoh R, Suzuki M, Okada G, Takeuchi Y, Futai K. Fungus symbionts colonizing the galleries of the ambrosia beetle Platypus quercivorus. Microb Ecol. 2011;62:106–120. doi: 10.1007/s00248-011-9838-3. [DOI] [PubMed] [Google Scholar]
- 32.Suh DY, Kim SH, Son SY, Seo ST, Kim KH. A new record of Candida kashinagacola (synonym Ambrosiozyma kashinagacola) from galleries of Platypus koryoensis, the oak wilt disease vector, in Korea. Mycobiology. 2013;41:245–247. doi: 10.5941/MYCO.2013.41.4.245. [DOI] [PMC free article] [PubMed] [Google Scholar]