Abstract
Background and objective: Our previous studies reported that miR-451 could protect against erythroid oxidant stress target gene-Ywhaz (14-3-3zeta) via inhibiting FoxO3 in the erythropoiesis. This study aimed to investigate the potential mechanism underlying the regulatory effect of miR-451 on human colorectal cancer (CRC) cells. Methods: In this study, expressions of miR-451 and Ywhaz in CRC tissues and adjacent normal tissues were detected by quantitative real-time PCR (qRT-PCR) and immunohistochemistry respectively. Human colon cancer cell lines were transfected with miR-451-MSCV-PIG retroviral vector to restore miR-451 expression. Ywhaz-3’UTR luciferase reporter assay confirmed Ywhaz as a direct target gene of miR-451. HCT116 cells and H29 cells were transfected with -shRNA-Ywhaz (pSGU6-Ywahz-shRNA-GFP) and the protein level of FoxO3 in the nucleus and cytoplasm was detected via Western blot assay. The anti-tumor effects of miR-451 were further verified in nude mice. Results: miR-451 was significantly down-regulated in human colon cancer tissues and cell lines (HCT116 and HT29), and inversely correlated with Dukes stage of colon cancer. Ywhaz was a candidate target gene of miR-451 and able to stimulate tumor growth via binding to FoxO3, inhibiting the FoxO3 nuclear accumulation. Conclusion: miR-451 may inhibit the colon cancer growth in vitro and in vivo, likely through directly targeting Ywhaz and indirectly regulating the nuclear accumulation of FoxO3.
Keywords: Colorectal cancer, microRNA-451, Ywhaz (14-3-3zeta), FoxO3
Introduction
Colorectal cancer (CRC) is the third most common malignancy and the fourth leading cause of cancer related death [1]. Worldwide, approximately 50% of CRC patients die of distant metastases [2,3]. In 2008, there were more than 1.2 million CRC patients worldwide. Despite the improvements in the diagnosis and treatment of CRC, more than 600,000 patients die from CRC yearly. The 5-year survival rate has reached almost 65% in high-income countries, and is about 90% in patients with stage I CRC and slightly greater than 10% in patients with stage IV disease [4,5]. However, the mechanism underlying the metastasis of CRC remains unclear [6].
MicroRNAs (miRNAs) are small, highly conserved non-coding RNAs that are able to regulate the protein synthesis post-transcriptionally via imperfect Watson-Crick base pairing, typically within the 3’untranslated region (UTR) [3,7]. These mRNAs are involved in the progress of differentiation, development, apoptosis and proliferation of cells in physiological and pathological conditions including tumorigenesis. In recent years, studies have shown that miRNAs dysregulation takes part in the progression of human CRC, and novel therapeutic approaches may be developed by targeting miRNAs that are altered in CRC [8-12]. Several studies have evidenced that there is dysregulation of several miRNAs in human CRC. These miRNAs, including miR-451, may either regulate the expression of tumour suppressor genes and oncogenes or function as oncogenes or tumour suppressor in the progression of CRC [13-15]. miR-451 is mapped to chromosome 17q11.2 in human, and was first identified in the human pituitary glands in 2005 [16]. Our previous results have showed that miR-451 is highly expressed and plays a vital role in the development of erythrocytes [17]. In addition, there is evidence showing that miR-451 is able to inhibit cell growth, proliferation, invasion and migration in breast cancer, glioma and non-small cell lung cancer [18-21].
In this study, we confirmed that the level of miR-451 was down regulated, but the level of Ywhaz (14-3-3zeta) increased in human CRC. Luciferase activity assay showed Ywhaz 3’UTR was a direct target of miR-451. Furthermore, Ywhaz was found to inhibit the accumulation of FoxO3 and repress the transcription of FoxO3 in the nucleus of CRC cell lines. Above findings suggest that miR-451 may act as a novel tumor suppressor by directly targeting Ywhaz in human CRC, and indirectly regulating the relocation of FoxO3 between cytoplasm and nucleus to inhibit the proliferation and viability of CRC.
Materials and methods
Patient recruitment and sample collection
The clinical information was obtained from patients hospitalized in the Departments of General Surgery of Subei People’s Hospital of Jiangsu province and Taixing People’s Hospital affiliated to Yangzhou University, China. The CRC tissues and adjacent normal tissues were collected from 46 CRC patients and the pathological examination was conducted by two independent pathologists blind to this study. The Duke’s stage of CRC was also determined. Dukes’ A, B, C and D CRC were found in 10, 16, 12 and 8 patients, respectively. All the samples were obtained during surgery and stored in liquid nitrogen immediately. Informed consent was obtained from patients before operation and the study was approved by the Institute Research Medical Ethics Committee of the Medical College of Yangzhou University.
Cell culture and transfection
Human CRC cell lines (HCT116 and HT29) were kindly provided by the Department of Chinese and Western Integrative Medicine (Medical College of Yangzhou University). 293T cells were purchased from the Peking Union Medical College Cell Bank (Beijing, China). All the cell lines were maintained in Dulbecco modified Eagle medium (DMEM, Gibco, USA) containing 10% fetal bovine serum (FBS, Gibco, USA), penicillin, streptomycin and 4 mM L-glutamine (Sigma, St. Louis, MO) at 37°C with 5% CO2.
miR-451 over-expression and transfection with Ywhaz shRNA
The miR-451 was amplified by PCR from human DNA templates and cloned into murine stem cell virus-PIG retroviral vector (MSCV-PIG) via T4-clone [17], and sequenced in the Sangon Biotech Co., Ltd. (Shanghai, China). The Ywhaz shRNA plasmid vector was purchased from Sangon Biotech Co., Ltd with Bpi I (Bbs I) and Hind III restriction enzyme sites in pSGU6/GFP/Neo vector. Both HCT116 cells and HT29 cells were independently transfected with pSGU6-Ywahz-shRNA-GFP in the presence of Lipofectamine 2000 (Invitrogen, San Diego, USA). shRNA negative control (shRNA-NC) vector was used as a control. In order to increase miR-451 expression, HT29 cells and HCT116 cells were transfected according to the manufacturer’s instructions with 8 μg of miR-451-MSCV plasmid or MSCV-only. The miR-451 expression in above cell lines was detected by qRT-PCR. Western-blot and qRT-PCR were used to detect the expression of Ywhaz and Foxo3.
Quantitative real-time PCR
Total RNA was extracted from HT29 cells and HCT116 cells, as well as of human CRC tissues using Trizol reagent (Invitrogen Life Technologies, San Diego, USA) according to the manufacturer’s instructions. Then, total RNA (2 μg) was purified with the purification kit (Thermo Fisher scientific), and reversely transcribed into cDNA using the PrimeScript™ II 1st Strand cDNA synthesis kit (Takara Bio, Inc., Dalian, Japan) and All-in-One™ miRNA First Strand cDNA synthesis kit (GeneCopoeia, Rockville, MD, USA) according to the manufacturer’s instructions. qRT-PCR was performed using the PrimeScript™ RT Master Mix (Takara Bio, Inc., Dalian, Japan) and All-in-One™ miRNA Qrt-PCR Detection kit (GeneCopoeia, Rockville, MD, USA) on an Applied BioSystems 7500 Real-Time PCR system (Applied Biosystems, White Plains, NY, USA). The 2-step PCR was performed: 95°C for 10 min and 40 cycles of 95°C for 10 sec and 60.5°C for 30 sec. The RNA U6 and GAPDH mRNA were used as internal controls. All the reactions were conducted in triplicate. Forward primers for miR-451 and U6 were 5’-AAACCGTTACCATTACTGAGTT-3’ and 5’-CGCTTCGGCAGCACATATAC-3’, respectively, and the reverse primer was Universal Adaptor PCR Primer (GeneCopoeia, Rockville, MD, USA).Primers for Ywhaz were: 5’-AGGAGCCCGTAGGTCATCTT-3’ (forward) and 5’-TGCTTGTGAAGCATTGGGGA-3’ (reverse). Primers for Foxo3 were 5’-TCACGCACCAATTCTAACGC-3’ (forward) and 5’-CACGGCTTGCTTACTGAAGG-3’ (reverse). Primers for GAPDH were 5’-AAGGTGAAGGTCGGAGTCAAC-3’ (forward) and 5’-GGGGTCATTGATGGCAACAATA-3’ (reverse). Data are shown as the fold change.
Luciferase activity assay
The sequence with 429 nucleotides (including of the binding sites with miR-451) of 3’-UTR wild type (wt) and 3’-UTR mutation (mut) of Ywhaz were amplified by PCR from human genomic DNA and cloned into the EcoRI and XbaI sites of pGL3-BS luciferase reporter vector (Promega, WI, USA) respectively. The primers for 3’-UTR wt of Ywhaz were 5’-ACACGAATTCAAAGGGATCATTGGTTCC-3’ (forward) and 5’-ACACTCTAGAAGTTAAATAGATGGGTTT-3’ (reverse). The primers of mutant Ywhaz 3’UTR were 5’-CTGGATAAGGGCAGAGGTTACCTCACAT-3’ (forward) and 5’-ACAAATAATGGAATGTGGGTAACCTCTG-3’ (reverse). 293T cells were cultured in 24-well plates and transfected with 0.5 μg of pGL3-BS luciferase reporters, 0.01 μg of Renilla and 0.5 μg of miR-451 MSCV-PIG in the presence of Lip2000. All the reactions were conducted in triplicate. After 48-h transfection, dual luciferase activity was measured using Luciferase Assay System according to the manufacturer’s instructions (Promega, WI, USA).
Proliferation and viability assays
Proliferation and viability assays of CRC cells were performed with Cell Counting Kit-8 (CCK-8, Biosharp, China) and CASY measurement (Reutlingen, Germany) in vitro. For the proliferation assay, HCT116 cells and HT29 cells were transfected with miR-451-MSCV or MSCV-only and seeded in triplicate into 96-well plates at a density of 7×103 cells/well. Then, cells were cultured for 24, 48 and 72 h and 10 μl of CCK-8 reagent was added into each well, followed by incubation for 4 h. Absorbance was measured at 450 nm with a Microplate Reader. For the viability assay, 2×105 cells were added to the 24-well plates and cell counting was conducted at 24 h, 48 h and 72 h. All the experiments were repeated three times.
Apoptosis assay
In order to investigate the role of miR-451 in the apoptosis of CRC cells, miR-451 was overexpressed in HT29 cells and HCT116 cells via transfection with miR-451-MSCV-PIG. After 48-h transfection, Annexin V-APC (BD Biosciences) and Propidium Iodide (PI, BD Biosciences) were added and apoptotic cells were determined by flow cytometry (FCM, BD Biosciences).
Western blot assay
Human CRC tissues were grounded in liquid nitrogen, and CRC cell lines were lysed in ice-cold lysis buffer (Beyotime Institute of Biotechnology) after transfection according to the manufacturer’s instructions. The Enhanced BCA Protein Assay kit (Beyotime Institute of Biotechnology) was used to measure the protein concentrations. For western blot assay, equal amount of total protein (20 μg) was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 12% SDS polyacrylamide gel and then transferred onto a poly-vinylidene fluoride (PVDF) membrane (Millipore, Billerica, MA) which was subsequently blocked for 1 h with 5% bovine serum albumin (BSA, Sigma, St. Louis, MO, USA) at room temperature. The membrane was incubated with human monoclonal primary Ywhaz, p-Ywhaz (1:1000, Santa Cruz Biotechnology) or FoxO3 antibody (1:1000, Cell Signaling Technologies, Boston, MA, USA) overnight. The specific protein was detected using a FluorChem FC2 Imaging System (Alpha Innotech, San Leandro, CA, USA) after incubation with horseradish peroxidase (HRP)-conjugated secondary antibody (1:1000, Cell Signaling Technologies, Boston, MA, USA) for 1 h at 37°C. GAPDH (1:4000, Santa Cruz, USA) was used as a control.
Tumorigenesis in nude mice
Eight-week-old BALB/c-A nude mice were purchased from the Experimental Animal Center of Yangzhou University and given ad libitum access to water and food. The mice were randomly divided into 3 groups (n=5 per group): HCT116 cells, HCT116 cells transfected with MSCV-only, and HCT116 cells transfected with miR-451-MSCV-PIG. Cells (n=6×106 cells) in each group were inoculated subcutaneously into the left back of nude mice. Two weeks later, the tumor volume and body weight were measured once every 2 days. The tumor width and length were measured with a digital caliper and tumor volume was calculated as follow: tumor volume =1/2×(length×width2). After 24-day observation, the tumors were harvested from mouse, fixed in formalin, embedded in paraffin and sectioned for immunohistochemistry.
Immunohistochemistry
The paraffin-embedded tissues (CRC tissues and tumor tissues from nude mice) were examined for Ywhaz (14-3-3zeta), p-Ywhaz and FoxO3 expressions. Sections were deparaffinized, treated with 3% H2O2 for 10 min and 5% BSA for 30 min and incubated with primary antibodies (1:100; Santa Cruz Biotechnology; 1:200, Cell Signaling Technology) overnight at 4°C. After incubation with HRP-conjugated secondary antibody (1:100) for 1 h at 37°C, sections were washed in Tris buffer, counterstained with hematoxylin, and visualized under a microscope (Olympus, Japan).
Statistical analysis
SPSS version 19.0 (SPSS Inc., Chicago, IL, USA) was used for statistical analysis. Data from three independent experiments are presented as mean ± standard deviation (SD). One-way analysis of variance (ANOVA) among groups and two-tailed Student’s T-test were used for comparisons between groups. Kaplan-Meier survival curve was plotted and survival rate was compared. A value of P<0.05 was considered statistically significant.
Results
miR-451 expression was downregulated in CRC tissues and negatively correlated with Duke’s stage
In order to identify the potential role of miR-451 in the progression of human CRC and investigate the potential mechanism, the miR-451 expression was measured in human CRC tissues (n=46) and adjacent normal tissues (n=46) by qRT-PCR. In addition, the mRNA expressions of Ywhaz and FoxO3 were also detected in CRC tissues, adjacent normal tissues and CRC cell lines. As shown in Figure 1A, miR-451 expression was significantly down-regulated in colon cancer tissues and this decrease was accompanied by an increase in the Duke’s stage. Meanwhile, the miR-451 expression was also down-regulated in both CRC cell lines (Figure 1A). Furthermore, Ywhaz expression was up-regulated in CRC significantly (Figure 1B), but Foxo3 expression remarkably decreased (Figure 1B), which were also confirmed by immunohistochemistry (Figure 1C).
Figure 1.
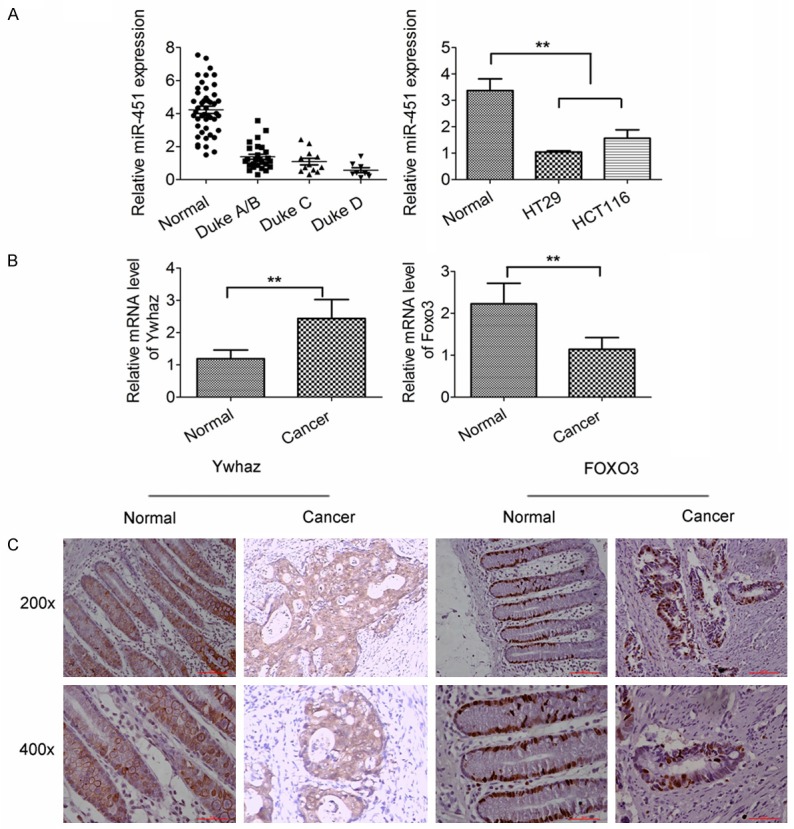
Dysregulation expression of miR-451, Ywhaz and FoxO3 in human CRC tissues. A. miR-451 expression was down-regulated in CRC tissues (Duke A: n=10, Duke B: n=16, Duke C: n=12, Duke D: n=8) and CRC cell lines (HT-29 cells and HCT-116 cells) when compare with adjacent normal tissues (n=46) (qRT-PCR; normalized by U6). B. When compared with normal colon tissues (n=46), Ywhaz mRNA expression was up-regulated in CRC tissues significantly, but FoxO3 mRNA expression decreased (qRT-PCR; normalized to GAPDH). C. Immunohistochemistry showed Ywhaz expression in the cytoplasm and membrane and FoxO3 expression in the nucleus were up-regulated and down-regulated, respectively in CRC tissues. *P<0.05, **P<0.01 vs. control. All experiments were repeated three times.
miR-451 overexpression inhibited the proliferation and viability of CRC cells
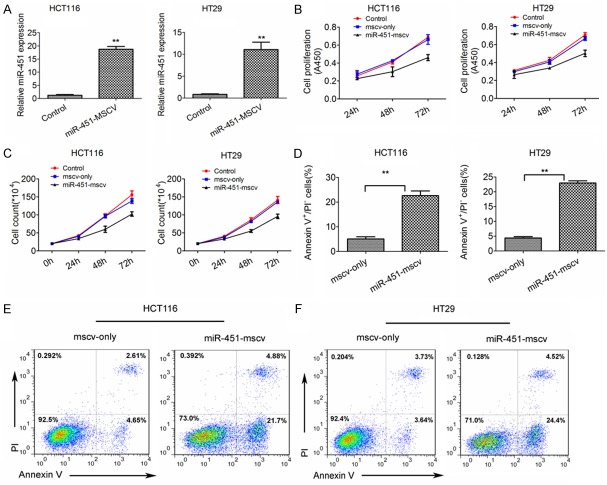
qPCR showed that miR-451 expression increased significantly in HCT-116 cells and HT-29 cells (18.72-fold and 11.09-fold, respectively) after transfection with miR-451-MSCV-PIG as compared to cells transfected with MSCV empty vector (Figure 2A). Moreover, miR-451 overexpression was found to inhibit the proliferation and viability of two cancer cell lines when compared with control cells (Figure 2B, 2C). Detection of cell apoptosis revealed that miR-451 could promote the apoptosis of CRC cells (Figure 2D-F). According to these findings we speculate that miR-451 may act as a tumor suppressor in the progression of CRC.
Figure 2.
Over-expression of miR-451 in CRC cells transfected with miR-451-MSCV inhibits the proliferation and viability and promotes the apoptosis of CRC cells. A. miR-451 expression in HT-29 cells and HCT-116 cells transfected with miR-451-MSCV or MSCV-only. miR-451 was over-expressed in HT-29 cells and HCT-116 cells transfected with miR-451-MSCV when compared with negative controls (qRT-PCR). B, C. miR-451 over-expression in HCT116 cells and HT29 cells inhibited the proliferation and viability in vitro. D. miR-451 over-expression promotes the apoptosis of CRC cells (HCT-116 cells and HT-29 cells) as compared to negative controls (FCM). E, F. FCM of apoptotic CRC cells. **P<0.01 vs. control. All experiments were repeated three times.
miR-451 inhibited Ywhaz expression via targeting 3’UTR of Ywhaz
It is well known that miRNAs are able to negatively regulate hundreds of mRNAs through the 3’UTR [18,19]. Previous studies showed that dysregulated miR-451 expression in several types of cancers was involved in the suppressed or accelerated progression of cancers [22,23], but the specific mechanism remains unclear due to little understanding of miR-451 target genes. Our previous study showed miR-451 could protect against erythroid oxidant stress via targeting Ywhaz (14-3-3zeta) in the development of erythrocytes [17]. In recent years, several studies also report the role of Ywhaz in the tumor progression [24-26]. Therefore, we hypothesized that Ywhaz acted as a potential target for miR-451 in human CRC. Our results showed miR-451 over-expression significantly down-regulated the mRNA and protein expressions of Ywhaz in CRC cells (Figure 3A-C). Furthermore, dual luciferase reporter activity assay indicated Ywhaz was a target gene of miR-451 in 293T cell lines (Figure 3D-F).
Figure 3.
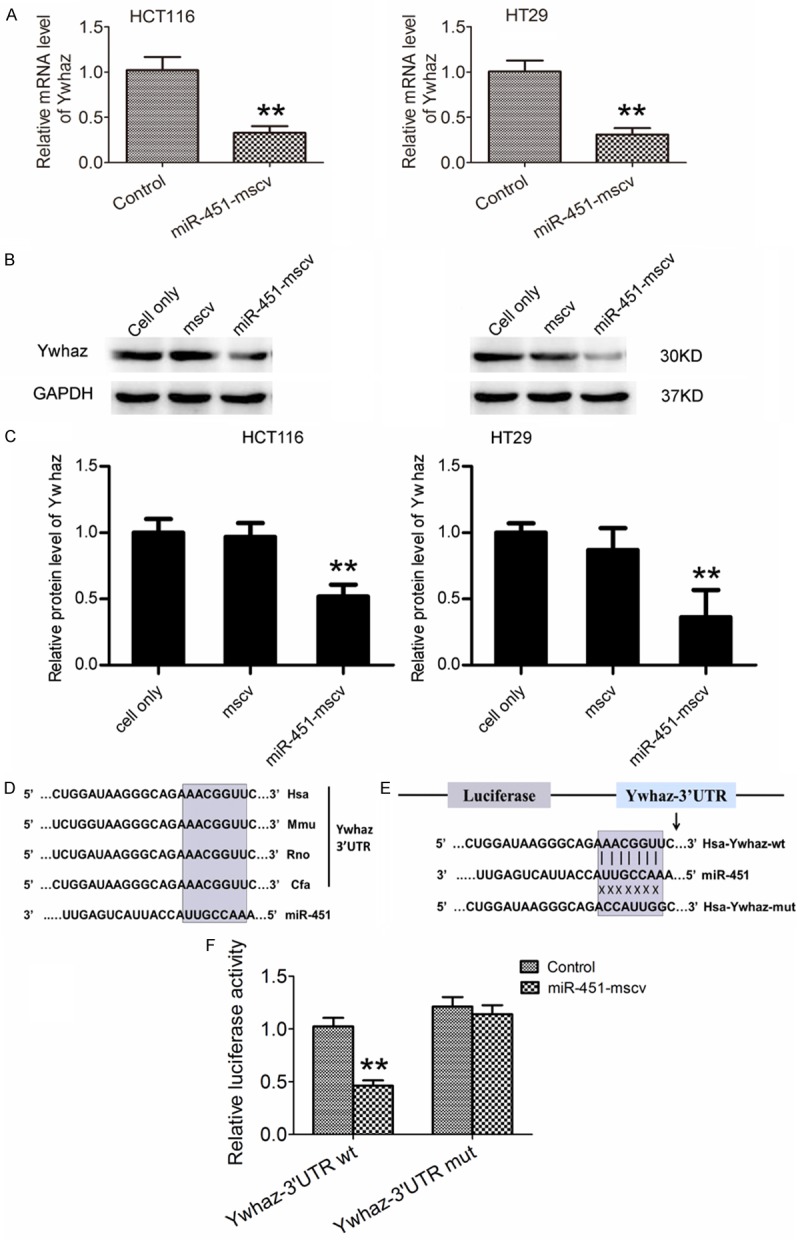
miR-451 negatively regulates Ywhaz expression via targeting Ywhaz 3’UTR. A. Ywhaz mRNA expression was down-regulated in HCT116 cells and HT29 cells after transfection of miR-451-MSCV (qRT-PCR, *P<0.05, **P<0.01). B, C. Ywhaz protein expression was down-regulated in HCT-116 cells and HT-29 cells transfected with miR-451-MSCV as compare to control cells (Western blot assay). D. The potential binding sites for miR-451 within the Ywhaz 3’UTR of human (Hsa), mouse (Mmu), Rat (Rno), and dog (cfa). Seed sequences of miR-451 are highlighted with gray. E. The wild type and mutant sequences of pGL3-Ywhaz 3’UTR in luciferase reporter constructs. F. Relative luciferase activity in 293T cells after transfection with pGL3-Ywhaz-3’UTR and miR-451-mscv plasmid (**P=0.0065). All experiments were repeated three times.
miR-451 regulated FoxO3 nuclear accumulation via inhibiting Ywhaz
Several studies have reported that FoxO3 is involved in the suppression of cancer progression [27-29]. To identify the role of FoxO3 in human CRC and the relationship between Ywhaz and FoxO3, FoxO3 expression was detected by Western blot assay in CRC cell lines with increased miR-451 expression. Results showed that the total FoxO3 protein expression was similar to control cells (Figure 4A). According to our previous findings in erythroid development [17], Ywhaz could inhibit the FoxO3 nuclear accumulation. In the present study, Ywhaz expression was knocked down by transfection with pSGU6-Ywhaz-shRNA-GFP-plasmid (Figure 4B), and the total, cytoplasmic and nuclear protein expressions of Ywhaz and FoxO3 were detected (Figure 4C). Results showed that Ywhaz could inhibit FoxO3 nuclear accumulation in colon cancer cells.
Figure 4.
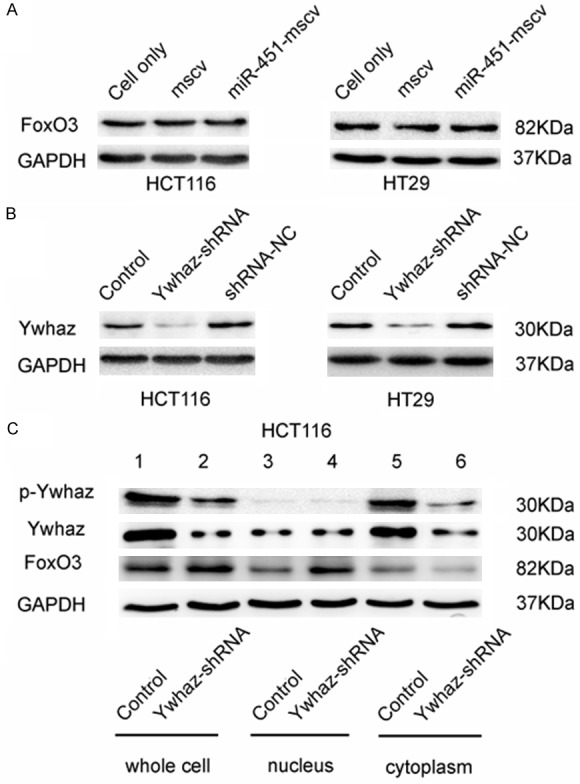
FoxO3 expression in HCT116 cells and HT29 cells after transfection with Ywhaz-shRNA. A. FoxO3 protein expression in HCT116 cells and HT29 cells transfected with MSCV-only was similar to that in cells after transfection with miR-451-MSCV-pig (Western-blot assay). B. Reduced Ywhaz protein expression in HCT116 cells and HT29 cells after Ywhaz-shRNA transfection as compared to untreated HCT116 cells and HCT116 cells trasnfected with shRNA-NC. C. Increased FoxO3 accumulation in the nucleus of HCT116 cells transfected with pSGU6-Ywhaz-shRNA in vitro. HCT116 cells and HT29 cells were transfected with pSGU6-Ywhaz-shRNA or shRNA-NC, and the total (lanes 1, 2), nuclear (lanes 3, 4) and cytoplasmic (lanes 5, 6) FoxO3 protein expressions were detected by Western blot assay. GAPDH served as a loading control. Data were from three independent experiments.
miR-451 inhibited the progression of human CRC in vivo
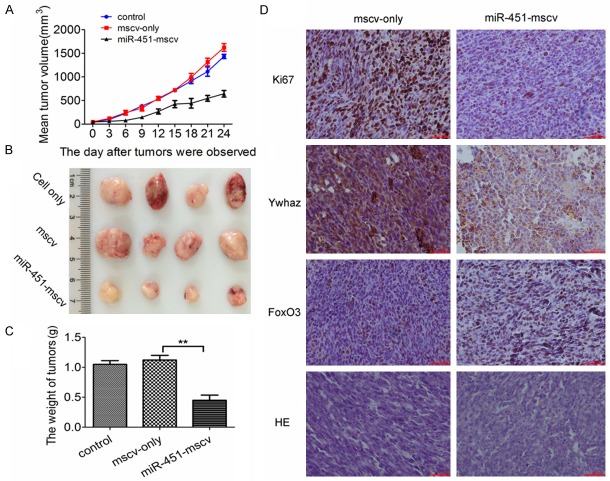
To further study the role of miR-451 in the inhibition of CRC progression in vivo, HCT116 xenograft model was established in nude mice. The tumors were palpable after 2 weeks. The tumors in miR-451-mscv group grew more slowly than in control groups (HCT116 cells group and mscv-only group) (Figure 5A). There was no difference in the tumor volume between HCT116 cells group and mscv-only group (Figure 5A). At the end of study, significant differences were observed in the tumor volume and weight between miR-451-MSCV group and two control groups (P<0.05) (Figure 5B, 5C). To determine whether miR-451 affected the expression of Ywhaz and Foxo3, the expressions of Ywhaz and Foxo3 were tested by immunohistochemistry. Results showed that Ki67 expression increased in cell nucleus after miR-451 over-expression. Ywhaz expression was distributed in the cytoplasm of cancer cells and significantly down-regulated in miR-451-MSCV group when compared with MSCV-only group. Meanwhile, the FoxO3 expression increased in the nucleus of cancer cells (Figure 5D).
Figure 5.
Effect of miR-451 over-expression on the growth of CRC in vivo. A. The mean tumor volume in miR-451-MSCV group, untreated cells group and MSCV-only group. B, C. Tumorxenografts from 3 groups at the end of study. There were significant differences in tumor volume and weight between miR-451 treated group and control group (P<0.05). D. Immunohistochemistry for Ki67, Ywhaz and FoxO3 in tumor xenografts. Ki67 reflects the proliferation activity of cells. Ywhaz expression decreased in miR-451-mscv group but FoxO3 expression increased in the nucleus of cancer cells in tumor xenografts.
Discussion
miRNAs are key regulators in the tumorigenesis and tumor metastasis. Several miRNAs involved in the cancer metastasis have been identified in breast cancer cell lines [2,30]. Abnormal expression has been found in a number of miRNAs in human CRC including miR-451 [11]. Meanwhile, miR-451 has been proven in several studies to act as a suppressor in the progression of gastrointestinal cancer [12]. The clinicopathological analysis showed that miR-451 was associated with a poor prognosis of gastric cancer. Nerea and his colleagues found that miR-451 expression was down-regulated in paraffin-embedded human CRC sections via in-situ hybridization and miR-451 was able to suppress the tumor formation by regulating the expression of macrophage migration inhibitory factor (MIF). They also found this in colon cancer stem cells tumor sphere in vitro [12,31]. However, whether miR-451 participates in the tumorigenesis of human CRC trough other signal pathways are still unclear. As a target gene of miR-451, the role of Ywhaz in the tumorigenesis and metastasis of human CRC is poorly investigated.
In the present study, qPCR showed the miR-451 expression was down-regulated in human colonic cancer cell lines and human CRC tissues and this decrease in CRC tissues was accompanied by an increase in the Duke’s stage. In addition, increased expression of miR-451 inhibited the proliferation and promoted the apoptosis of two human colonic cancer cell lines. Both mRNA and protein expressions of Ywhaz increased remarkably in CRC tissues as compared to normal tissues, but FoxO3 mRNA expression reduced. According to these findings, we postulated that miR-451 may act as a tumor suppressor in the progression of CRC by inhibiting its target gene Ywhaz and then promoting the nuclear translocation of FoxO3.
It is well known that miRNAs specifically inhibit protein expression by binding to the 3’UTR of target mRNAs. Dual luciferase reporter assay has been widely used to determine the targets of miRNA. Our previous findings also indicated that miR-451 was able to inhibit the Ywhaz protein expression via binding to its 3’UTR during the development of erythrocytes [17]. In this study, dual luciferase reporter assay showed Ywhaz was a target gene of miR-451. Ywhaz/14-3-3 zeta protein belongs to the 14-3-3 protein family, and is highly conservative in different species. Ywhaz is one of important factors inhibiting cell apoptosis [32]. Several studies have reported Ywhaz may promote tumorigenesis and metastasis in several cancers such as lung cancer, breast cancer and glioma [24-26].In addition, Ywhaz may regulate the activity of FoxO3. -FoxO3 is an inhibitory transcription factor and belongs to the forkhead family which is characterized by a distinct forkhead domain. FoxO3 participates in the inhibition of cancer progression [27-29]. However, the role of Ywhaz in human CRC and the relationship between Ywhaz and FoxO3 are still poorly understood.
Our results showed the FoxO3 protein expression in the nucleus was up-regulated markedly in HCT116 cells and HT29 cells after Ywhaz knockdown. Immunohistochemistry also revealed the Ywhaz expression was down-regulated in tumor xenografts of nude mice inoculated with cells with miR-451 over-expression, and FoxO3 expression increased reversely.
In conclusion, our results demonstrate that miR-451 is able to inhibit cell proliferation in vitro and in vivo. We postulate that miR-451 may act as a cancer suppressor via regulating FoxO3 nuclear accumulation through targeting Yhwaz in human CRC. Thus, miR-451 may be a potential target for the treatment of human CRC.
Acknowledgements
This study was supported by the National Natural Science Foundation of China [No. 81470277 (D.Y), No. 81501135 (X.W) and No. 81302041 (Y.Z)], the Natural Science Foundation of the Jiangsu province of China (No. BK20130454), the Jiangsu Higher Education Science Foundation [No. CXLX-1439 (YY.L)], and the -scientific research item of Taixing people’s hospital [No. try1401 (YY.L)].
Disclosure of conflict of interest
None.
References
- 1.Toward better control of colorectal cancer. Lancet. 2014;383:1437. doi: 10.1016/S0140-6736(14)60699-1. [DOI] [PubMed] [Google Scholar]
- 2.de Krijger I, Mekenkamp LJ, Punt CJ, Nagtegaal ID. MicroRNAs in colorectal cancer metastasis. J Pathol. 2011;224:438–447. doi: 10.1002/path.2922. [DOI] [PubMed] [Google Scholar]
- 3.Li HY, Zhang Y, Cai JH, Bian HL. MicroRNA-451 inhibits growth of human colorectal carcinoma cells via downregulation of Pi3k/Akt pathway. Asian Pac J Cancer Prev. 2013;14:3631–3634. doi: 10.7314/apjcp.2013.14.6.3631. [DOI] [PubMed] [Google Scholar]
- 4.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 5.Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 6.May M. Statistics: Attacking an epidemic. Nature. 2014;509:S50–51. doi: 10.1038/509S50a. [DOI] [PubMed] [Google Scholar]
- 7.Zhao G, Yu D, Weiss MJ. MicroRNAs in erythropoiesis. Curr Opin Hematol. 2010;17:155–162. doi: 10.1097/MOH.0b013e328337ba6c. [DOI] [PubMed] [Google Scholar]
- 8.Papapetrou EP, Korkola JE, Sadelain M. A genetic strategy for single and combinatorial analysis of miRNA function in mammalian hematopoietic stem cells. Stem Cells. 2010;28:287–296. doi: 10.1002/stem.257. [DOI] [PubMed] [Google Scholar]
- 9.Godlewski J, Nowicki MO, Bronisz A, Nuovo G, Palatini J, De Lay M, Van Brocklyn J, Ostrowski MC, Chiocca EA, Lawler SE. MicroRNA-451 regulates LKB1/AMPK signaling and allows adaptation to metabolic stress in glioma cells. Mol Cell. 2010;37:620–632. doi: 10.1016/j.molcel.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, Dirbas FM, Somlo G, Pera RA, Lao K, Clarke MF. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shivapurkar N, Weiner LM, Marshall JL, Madhavan S, Deslattes Mays A, Juhl H, Wellstein A. Recurrence of early stage colon cancer predicted by expression pattern of circulating microRNAs. PLoS One. 2014;9:e84686. doi: 10.1371/journal.pone.0084686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bandres E, Bitarte N, Arias F, Agorreta J, Fortes P, Agirre X, Zarate R, Diaz-Gonzalez JA, Ramirez N, Sola JJ, Jimenez P, Rodriguez J, Garcia-Foncillas J. microRNA-451 regulates macrophage migration inhibitory factor production and proliferation of gastrointestinal cancer cells. Clin Cancer Res. 2009;15:2281–2290. doi: 10.1158/1078-0432.CCR-08-1818. [DOI] [PubMed] [Google Scholar]
- 13.Vickers MM, Bar J, Gorn-Hondermann I, Yarom N, Daneshmand M, Hanson JE, Addison CL, Asmis TR, Jonker DJ, Maroun J, Lorimer IA, Goss GD, Dimitroulakos J. Stage-dependent differential expression of microRNAs in colorectal cancer: potential role as markers of metastatic disease. Clin Exp Metastasis. 2012;29:123–132. doi: 10.1007/s10585-011-9435-3. [DOI] [PubMed] [Google Scholar]
- 14.Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, Liu CG, Calin GA, Croce CM, Harris CC. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan X, Wang R, Wang ZX. The potential role of miR-451 in cancer diagnosis, prognosis, and therapy. Mol Cancer Ther. 2013;12:1153–1162. doi: 10.1158/1535-7163.MCT-12-0802. [DOI] [PubMed] [Google Scholar]
- 16.Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, Brownstein MJ, Tuschl T, Margalit H. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005;33:2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu D, dos Santos CO, Zhao G, Jiang J, Amigo JD, Khandros E, Dore LC, Yao Y, D’Souza J, Zhang Z, Ghaffari S, Choi J, Friend S, Tong W, Orange JS, Paw BH, Weiss MJ. miR-451 protects against erythroid oxidant stress by repressing 14-3-3zeta. Genes Dev. 2010;24:1620–1633. doi: 10.1101/gad.1942110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang R, Wang ZX, Yang JS, Pan X, De W, Chen LB. MicroRNA-451 functions as a tumor suppressor in human non-small cell lung cancer by targeting ras-related protein 14 (RAB14) Oncogene. 2011;30:2644–2658. doi: 10.1038/onc.2010.642. [DOI] [PubMed] [Google Scholar]
- 19.Bergamaschi A, Katzenellenbogen BS. Tamoxifen downregulation of miR-451 increases 14-3-3zeta and promotes breast cancer cell survival and endocrine resistance. Oncogene. 2012;31:39–47. doi: 10.1038/onc.2011.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X, Sanda T, Look AT, Novina CD, von Boehmer H. Repression of tumor suppressor miR-451 is essential for NOTCH1-induced oncogenesis in T-ALL. J Exp Med. 2011;208:663–675. doi: 10.1084/jem.20102384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian Y, Nan Y, Han L, Zhang A, Wang G, Jia Z, Hao J, Pu P, Zhong Y, Kang C. MicroRNA miR-451 downregulates the PI3K/AKT pathway through CAB39 in human glioma. Int J Oncol. 2012;40:1105–1112. doi: 10.3892/ijo.2011.1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nan Y, Han L, Zhang A, Wang G, Jia Z, Yang Y, Yue X, Pu P, Zhong Y, Kang C. MiRNA-451 plays a role as tumor suppressor in human glioma cells. Brain Res. 2010;1359:14–21. doi: 10.1016/j.brainres.2010.08.074. [DOI] [PubMed] [Google Scholar]
- 23.Godlewski J, Bronisz A, Nowicki MO, Chiocca EA, Lawler S. microRNA-451: A conditional switch controlling glioma cell proliferation and migration. Cell Cycle. 2010;9:2742–2748. [PubMed] [Google Scholar]
- 24.Yang X, Cao W, Zhang L, Zhang W, Zhang X, Lin H. Targeting 14-3-3zeta in cancer therapy. Cancer Gene Ther. 2012;19:153–159. doi: 10.1038/cgt.2011.85. [DOI] [PubMed] [Google Scholar]
- 25.Matta A, Siu KW, Ralhan R. 14-3-3 zeta as novel molecular target for cancer therapy. Expert Opin Ther Targets. 2012;16:515–523. doi: 10.1517/14728222.2012.668185. [DOI] [PubMed] [Google Scholar]
- 26.Matta A, DeSouza LV, Ralhan R, Siu KW. Small interfering RNA targeting 14-3-3zeta increases efficacy of chemotherapeutic agents in head and neck cancer cells. Mol Cancer Ther. 2010;9:2676–2688. doi: 10.1158/1535-7163.MCT-10-0312. [DOI] [PubMed] [Google Scholar]
- 27.Sunayama J, Sato A, Matsuda K, Tachibana K, Watanabe E, Seino S, Suzuki K, Narita Y, Shibui S, Sakurada K, Kayama T, Tomiyama A, Kitanaka C. FoxO3a functions as a key integrator of cellular signals that control glioblastoma stem-like cell differentiation and tumorigenicity. Stem Cells. 2011;29:1327–1337. doi: 10.1002/stem.696. [DOI] [PubMed] [Google Scholar]
- 28.Lin C, Wu Z, Lin X, Yu C, Shi T, Zeng Y, Wang X, Li J, Song L. Knockdown of FLOT1 impairs cell proliferation and tumorigenicity in breast cancer through upregulation of FOXO3a. Clin Cancer Res. 2011;17:3089–3099. doi: 10.1158/1078-0432.CCR-10-3068. [DOI] [PubMed] [Google Scholar]
- 29.Wang S, Wang H, Davis BC, Liang J, Cui R, Chen SJ, Xu ZX. PARP1 inhibitors attenuate AKT phosphorylation via the upregulation of PHLPP1. Biochem Biophys Res Commun. 2011;412:379–384. doi: 10.1016/j.bbrc.2011.07.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang CW, Wu HC, Terry MB, Santella RM. microRNA Expression in Prospectively Collected Blood as a Potential Biomarker of Breast Cancer Risk in the BCFR. Anticancer Res. 2015;35:3969–3977. [PMC free article] [PubMed] [Google Scholar]
- 31.Bitarte N, Bandres E, Boni V, Zarate R, Rodriguez J, Gonzalez-Huarriz M, Lopez I, Javier Sola J, Alonso MM, Fortes P, Garcia-Foncillas J. MicroRNA-451 is involved in the self-renewal, tumorigenicity, and chemoresistance of colorectal cancer stem cells. Stem Cells. 2011;29:1661–1671. doi: 10.1002/stem.741. [DOI] [PubMed] [Google Scholar]
- 32.Fu H, Subramanian RR, Masters SC. 14-3-3 proteins: structure, function, and regulation. Annu Rev Pharmacol Toxicol. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]