Abstract
Objective:
For 3,670 stroke patients from the United Kingdom, United States, Australia, Belgium, and Italy, we performed a genome-wide meta-analysis of white matter hyperintensity volumes (WMHV) on data imputed to the 1000 Genomes reference dataset to provide insights into disease mechanisms.
Methods:
We first sought to identify genetic associations with white matter hyperintensities in a stroke population, and then examined whether genetic loci previously linked to WMHV in community populations are also associated in stroke patients. Having established that genetic associations are shared between the 2 populations, we performed a meta-analysis testing which associations with WMHV in stroke-free populations are associated overall when combined with stroke populations.
Results:
There were no associations at genome-wide significance with WMHV in stroke patients. All previously reported genome-wide significant associations with WMHV in community populations shared direction of effect in stroke patients. In a meta-analysis of the genome-wide significant and suggestive loci (p < 5 × 10−6) from community populations (15 single nucleotide polymorphisms in total) and from stroke patients, 6 independent loci were associated with WMHV in both populations. Four of these are novel associations at the genome-wide level (rs72934505 [NBEAL1], p = 2.2 × 10−8; rs941898 [EVL], p = 4.0 × 10−8; rs962888 [C1QL1], p = 1.1 × 10−8; rs9515201 [COL4A2], p = 6.9 × 10−9).
Conclusions:
Genetic associations with WMHV are shared in otherwise healthy individuals and patients with stroke, indicating common genetic susceptibility in cerebral small vessel disease.
White matter hyperintensities (WMH) on T2-weighted MRI are associated with increasing age and cardiovascular risk factors, particularly hypertension, and are predictive of both stroke and dementia in prospective community populations.1 Severe confluent WMH are often found in patients presenting with stroke, and are more common in patients with the small vessel stroke subtype.2 Furthermore, in these patients, WMH burden is linked to poor clinical outcomes after stroke.3,4 Understanding disease mechanisms that contribute to WMH could lead to advances in prevention, treatment, and rehabilitation of disability related to vascular cognitive impairment, age-related functional decline, and stroke.
Twin and family history studies suggest a significant genetic component to WMH. Heritability estimates range from 55% to 80%,5–8 suggesting that a moderate to large proportion of the disease risk can be attributed to genetic effects. The heritability attributed to common single nucleotide polymorphisms (SNPs) has been estimated to be between 13% and 45%.9 Previous genome-wide analyses have focused on the genetic influence on WMH in community populations,10,11 and a recent meta-analysis identified 8 regions associated with the disease.12 One might expect genetic risk factors for WMH in community populations to be similar to those that confer increased risk of WMH in stroke patients. However, the underlying pathology of WMH is heterogeneous, with small punctate lesions being associated with mixed causes, whereas more confluent areas often seen in stroke patients correspond primarily to small vessel disease.13 Therefore, it is unclear whether the lesions underlying WMH in the general population are pathologically distinct from the confluent lesions frequently observed in patients with stroke.
In this analysis, we investigated the role of the genetic contribution to WMH volumes (WMHV) in patients with ischemic stroke. We initially performed a genome-wide meta-analysis of WMHV in stroke patients with the aim of identifying novel associations. Second, we determined whether similar genetic factors contributed to WMHV in community populations and stroke patients. Finally, having established shared genetic factors in the 2 datasets, we performed a meta-analysis of the published associations from community populations with our dataset to identify genetic associations that are in common in the 2 populations.
METHODS
Study populations.
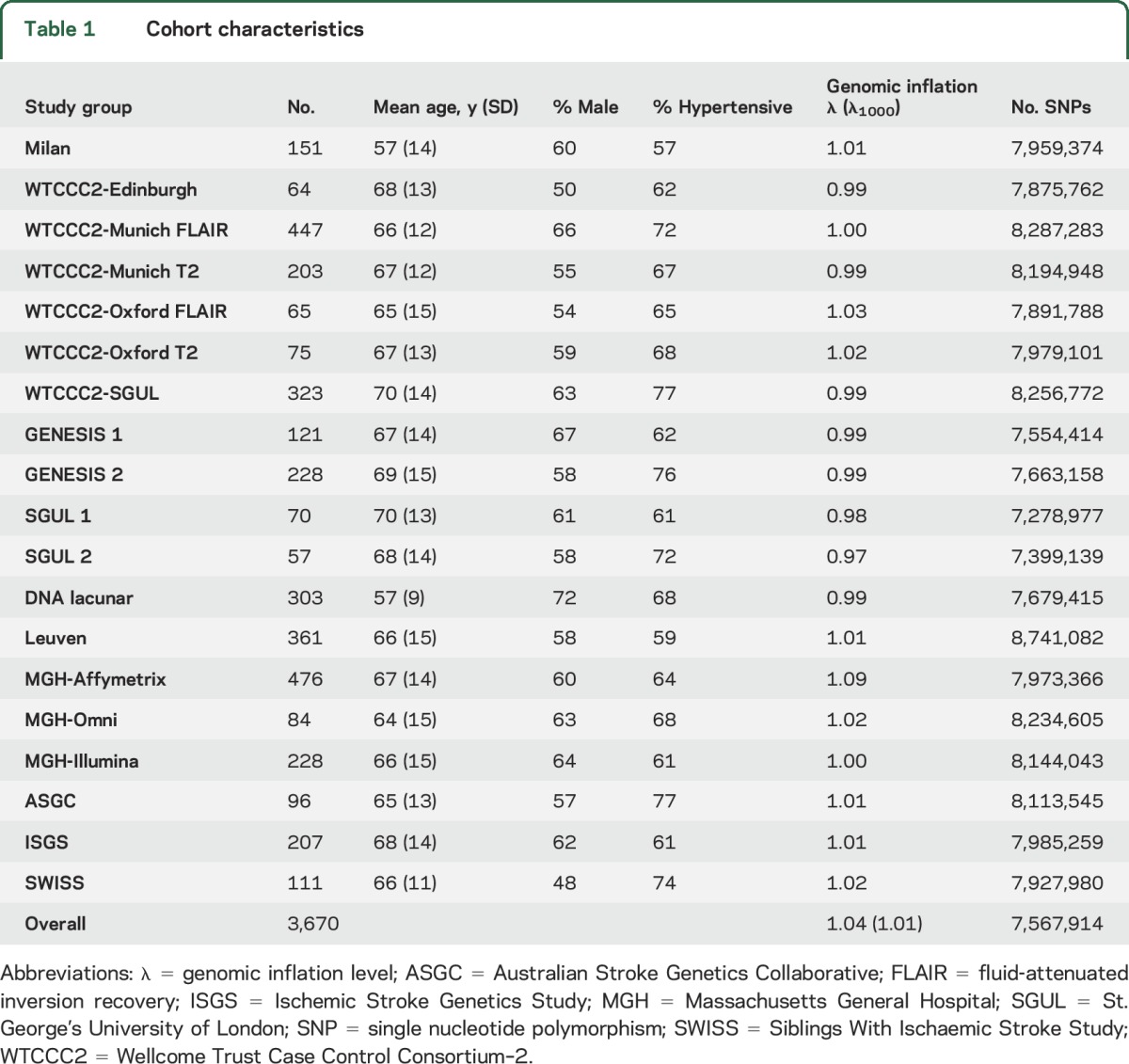
Ischemic stroke populations were enrolled through hospital-based studies between 1995 and 2013. Characteristics of the study populations are given in table 1; full details are given in the supplementary material on the Neurology® Web site at Neurology.org. Patients with cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy or any other suspected monogenic cause of stroke, vasculitis, or any other nonischemic cause of WMH including demyelinating and mitochondrial disorders were excluded from analyses.
Table 1.
Cohort characteristics
Standard protocol approvals, registrations, and patient consents.
An institutional review board or regional review board approved the use of human subjects in each of the study populations. All patients gave informed consent.
Neuroimaging analysis.
MRI scans were acquired as part of routine clinical practice for evaluation of ischemic stroke (table e-1). Fluid-attenuated inversion recovery (FLAIR) sequences were primarily used for WMH volumetric analysis; however, in their absence, T2-weighted sequences were used (Wellcome Trust Case Control Consortium–2 [WTCCC2], Oxford, and WTCCC2, Munich, only). In all scans, to avoid confounding by hyperintense signal due to acute stroke, WMHV was assessed quantitatively in the hemisphere contralateral to the acute infarction. Chronic lacunar infarcts were identified using standard criteria as low signal on T1 or FLAIR images and were excluded from WMHV estimates.14 Trained raters blinded to all patient information analyzed anonymized MRI scans. All supratentorial white matter and deep gray matter lesions were included in WMHV with the exception of WMH corresponding to infarcts, both lacunar and territorial.2 MRIs with excessive movement artefact, incomplete brain coverage, or bihemispheric infarcts (other than lacunar) were excluded.
To account for interindividual variability in head size, an estimate of total intracranial volume (TICV) was derived using site-specific volumetric methodology, as follows. MRIs from the Massachusetts General Hospital, Ischemic Stroke Genetics Study, and Australian Stroke Genetics Collaborative studies were analyzed in Boston. Scans from the Siblings with Ischaemic Stroke Study were analyzed in the same way at the University of Virginia by the Boston-trained rater. FLAIR sequences were analyzed using an MRIcro semiautomated method as previously described.2 Using operator-mediated quality assurances, overlapping regions of interest (ROIs) corresponding to WMH produced the final maps for WMHV calculation. Intracranial area was derived as a validated marker of TICV as the average of 2 midsagittal slices traced using anatomical landmarks on T1 sequences.15
The WTCCC2, GENESIS, SGUL, Leuven, and Milan cohorts were analyzed in London using DISPunc semiautomated lesion drawing software.16 A seed at the lesion border was first marked manually, and then outlined automatically based on the signal intensity gradient. Each WMH ROI was visually inspected and manually corrected as required. To estimate TICV, T2-weighted and, in their absence, FLAIR sequences were analyzed using an automated segmentation program, SIENAX,17 which calculates the total volume of CSF and gray and white matter volumes.
WMHV quantification agreement across the 2 main rating centers was performed for 50 randomly selected scans; agreement was very good (intraclass correlation coefficient 0.95, confidence interval 0.91–0.97, n = 50).
Phenotype definition.
To calculate the phenotype used in the genetic analysis, WMHV were doubled to obtain a whole brain estimate. This volume was then multiplied by the ratio of TICV (or intracranial area) to the mean TICV (or intracranial area) for the study, thereby correcting for natural differences in head size. The values were natural log transformed and the resulting ln (WMHV) values were entered into a linear regression model including age, sex, and the first 2 ancestry-informative principal components. To ensure the phenotype was normally distributed, the residuals from the model were then z-transformed and used as the WMHV phenotype in the genetic analysis.
Genome-wide genotyping and imputation.
Genotyping of all cohorts was performed on commercially available arrays from Affymetrix (Santa Clara, CA) or Illumina (San Diego, CA) (table e-2). All cohorts performed extensive quality control steps prior to imputation, removing SNPs showing significant departure from Hardy-Weinberg equilibrium, high levels of missingness, or low minor allele frequency. Individuals were removed who did not segregate with Hapmap II European populations based on ancestry informative principal component analysis using EIGENSTRAT or multidimensional scaling in PLINK.18,19 Additionally, individuals showing cryptic relatedness or having high levels of missingness or heterozygosity were excluded. All datasets were imputed to 1000 Genomes integrated variant set (March 2012) using IMPUTE v2.20
Genome-wide association analysis of WMHV in stroke patients.
To discover novel associations between WMHV and each autosomal SNP, we performed linear regression of WMHV on genotype dosages using PLINK v1.07.19 SNPs with PLINK INFO <0.7 or MAF <0.01 were removed from further analyses. We used genomic inflation to evaluate inflation of test statistics in each study group.21 Results across all study groups were combined using a fixed-effects inverse variance weighted method using METAL.22 To control for any excess signal that might result from study-wise inflation of p values, we performed genomic control correction, multiplying the standard errors from each study by the square root of the genomic inflation factor.21 Heterogeneity was assessed using Cochran q statistic. Following the meta-analysis, we considered only SNPs present in more than 12 study groups, and with heterogeneity p > 0.001, for analysis. We set the significance threshold to p < 5 × 10−8. We used λ1000 to evaluate inflation at the meta-analysis level.23 We had 80% power to detect a variant explaining 1.1% of the trait variance (figure e-1).
Analysis of SNPs associated with WMH in community-based populations.
To determine whether SNPs contributing to WMHV in community populations were associated with WMHV in stroke patients, we evaluated each SNP reported as being associated with WMH in healthy adults in a recent publication,12 testing if the SNP was associated with WMHV in ischemic stroke patients. All 17,936 individuals in the previous study were stroke-free and nonoverlapping with the samples studied here. We performed this analysis first for all genome-wide associated loci from the publication, and second for loci reported at p < 1 ×10−5 in European populations or overall. We set a significance threshold at p = 0.0033, Bonferroni correcting for the 15 SNPs analyzed. We had 80% power to detect any associations that explain 0.4% of the trait variance. In addition, we tested whether there was evidence overall that genetic susceptibility factors were shared between the 2 populations. We used a binomial test to evaluate whether an excess of the 8 genome-wide significant SNPs shared direction of effect in community populations and stroke patients, and then extended this to the 15 genome-wide significant loci and loci reported at p < 1 × 10−5 in European populations or overall.
Meta-analysis of stroke samples and published population-based samples.
Having established that genetic factors were shared between community populations and stroke patients, we evaluated the overall evidence that each of the 15 previously reported SNPs (8 genome-wide significant, 7 suggestive) were associated with WMHV in both populations. We combined p values from the 2 sources using Stouffer z-score weighted method with equal weights, classifying SNPs with p < 0.05 in both populations and reaching p < 5 × 10−8 overall as significantly associated with WMH in both populations. We were not able to perform the reciprocal analysis, testing if suggestive associations with WMH in stroke patients were associated with WMH in stroke-free individuals, due to restrictions on access to the required summary level data. We then evaluated novel genome-wide associations in available databases to test for evidence that affects regulation of genes (RegulomeDB)24 or directly affects gene expression (GTEx).25
RESULTS
Study populations.
Clinical characteristics of all participating cohorts are given in table 1. In total, 3,670 individuals of European ancestry were included in the 19 study groups.
Genome-wide association analysis of WMHV in stroke patients.
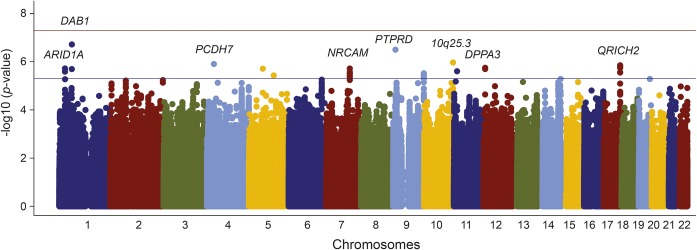
With the exception of one study group, genomic inflation was well-controlled (λ ≤ 1.03, table 1). Following quality control procedures, 7,567,914 autosomal SNPs remained for analysis. Genomic inflation was well-controlled at the meta-analysis level (λ = 1.04, λ1000 = 1.01; figure e-2). No SNP reached the significance level (figure 1), although a number of loci reached p < 5 × 10−6. These are detailed in table e-3, and regional plots of these loci are provided in figure e-3. All odds ratios reported are per 1 SD change in normally distributed WMHV after accounting for age, sex, and ancestry-informative principal components.
Figure 1. Association of genome-wide single nucleotide polymorphisms with white matter hyperintensity volume in ischemic stroke patients by genomic position.
Analysis of SNPs associated with WMH in community-based populations.
Eight independent SNPs have been associated with WMH in community populations.12 We evaluated each of these in our dataset of stroke patients. The direction of effect of all 8 associations was consistent with the direction in our study. This alone is unlikely to be due to chance (p = 7.8 × 10−3 from binomial test). For specific SNPs, no genome-wide associations from community populations reached our significance threshold, although all had p ≤ 0.24 for association with WMH in stroke patients, and 3 loci reached a nominal significance level (p < 0.05) in stroke patients (rs7214628 [TRIM65], p = 0.015; rs78857879 [EFEMP1], p = 0.0056; rs2984613 [PMF1-BGLAP], p = 0.017).
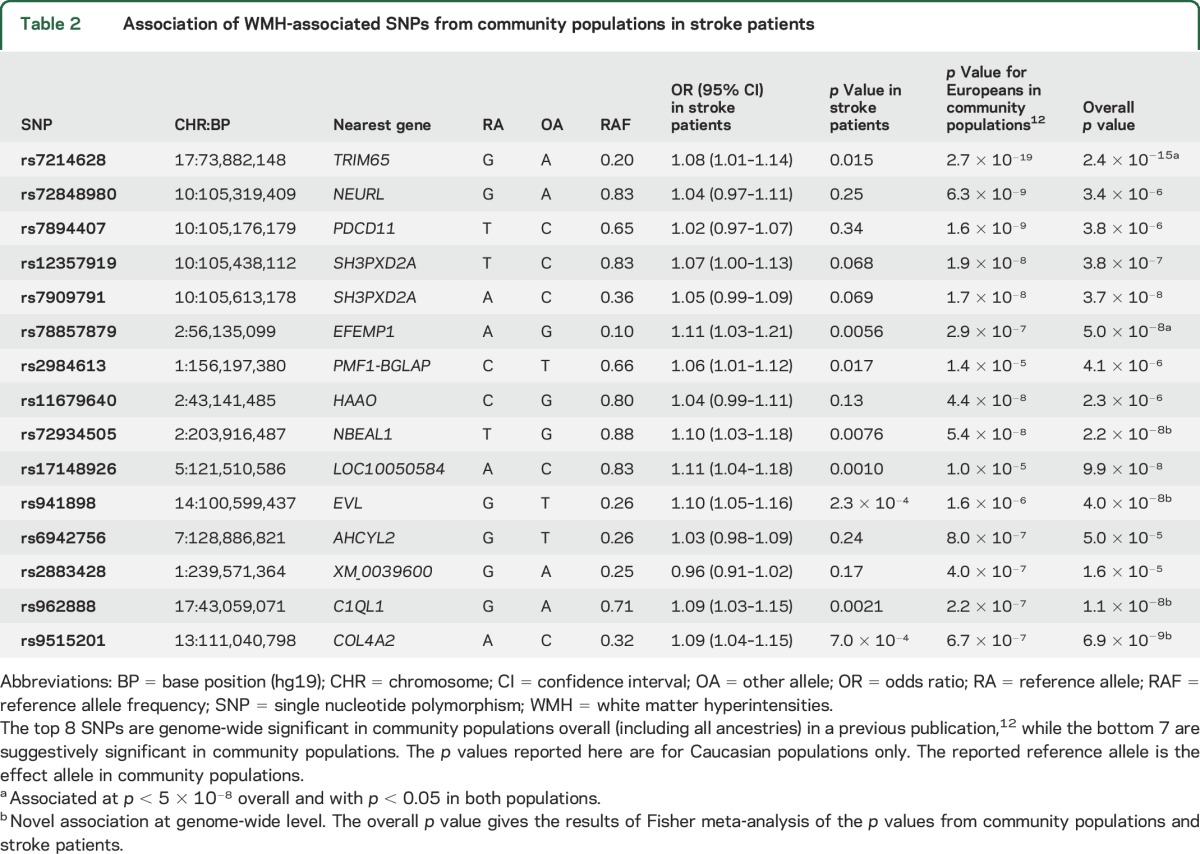
Additionally, 10 loci were reported as suggestively significant in the same recent publication,12 with p < 1 × 10−5 in Europeans or overall. Three of these were rare (MAF ≤ 0.02), and were not imputed with enough accuracy to be analyzed in our dataset (rs186314186, rs150695384, rs117126031). We evaluated each of the 7 remaining associations in our population. Of these, 4 passed our significance threshold (table 2). One locus was nonsignificant and in the opposite direction in our study (rs2883428, p = 0.17). In total, 14 of the 15 genome-wide and suggestively significant loci shared direction between community individuals and stroke patients (p = 9.8 × 10−4 from binomial test).
Table 2.
Association of WMH-associated SNPs from community populations in stroke patients
In addition, we searched for other publications describing associations with any of the SNPs or genes studied using the following search terms in PubMed: (SNP or gene) and (white matter or leukoaraiosis or small vessel disease). No relevant publications were identified.
Meta-analysis of stroke samples and published population-based samples.
When combining our results in stroke patients with the 15 previously reported associations using Stouffer z-score meta-analysis, 6 associations reached genome-wide significance overall and had p < 0.05 in both studies (table 2). Four of these are novel associations at genome-wide significance (rs72934505 [NBEAL1], p = 2.2 × 10−8; rs941898 [EVL], p = 4.0 × 10−8; rs962888 [C1QL1], p = 1.1 × 10−8; rs9515201 [COL4A2], p = 6.9 × 10−9), all of which showed good consistency across the 19 cohorts (figure e-4). The same 6 associations reached genome-wide significance using an alternative meta-analysis approach (Fisher method). The association with COL4A2 (rs9515201) is in strong LD (r2 > 0.8) with SNPs previously reported to be associated with cerebral small vessel disease, and is therefore likely to represent the same locus.26
For each of these 4 novel associations, we queried the RegulomeDB database and GTEx portal for evidence that the SNPs affect DNA binding or expression of any mRNA molecule (figure e-5).24,25 rs962888 lies 25 Kb downstream from C1QL1; however, interrogation of GTEx portal showed that the common allele (G, risk allele) of the SNP decreases expression of elongation factor tu GTP binding domain containing 2 (EFTUD2) in tibial arteries, 100 kb away (p = 5.3 × 10−6). Data from RegulomeDB support this observation, as the SNP overlies DNase-seq, FAIRE-seq, and CHIP-seq peaks in numerous tissues from ENCODE.27 Similarly, the common allele (T, risk allele) of rs72934505 increases expression of the nearby gene NBEAL1 in tibial arteries in GTEx (p = 2.5 × 10−11), and also decreases expression of islet cell autoantigen 1.69 kDa-Like (ICA1L) in the thyroid (p = 6.6 × 10−6), 200 kb away. No significant eQTLs were identified for rs941898 or rs9515201, but both overlap numerous CHIP-seq and DNAse-seq peaks from ENCODE, indicating they may have a regulatory function.
DISCUSSION
We report the first phase of a collaborative genome-wide meta-analysis of WMHV in stroke patients. We did not identify any associations with WMHV in ischemic stroke patients at the genome-wide significance level. The most likely explanation for this is lack of power. We had 80% power to identify a variant explaining 1.1% of the trait variance (supplementary material), suggesting that it is unlikely that any common variants explain more than this proportion of the variance of WMH in stroke patients. However, we cannot rule out the existence of rare variants conferring a considerable proportion of disease risk.
We found strong evidence that many of the same genome-wide associations with WMHV in healthy individuals influence WMHV in stroke patients. All genome-wide significant associations with WMHV shared direction of effect in our study and 3 reached a nominal significance threshold. More convincing is that of the 7 suggestive associations reported with WMH in healthy individuals, 4 were significantly associated with WMH in stroke patients. A meta-analysis of these SNPs in 21,606 subjects suggests that 4 of these loci are linked to WMH in community populations and stroke patients at genome-wide significance. Two of these associations influence expression of nearby gene products (NBEAL1/ICA1L [rs72934505] and EFTUD2 [rs962888]). A genome-wide significant association with rs9515201, located in an intron of COL4A2, which encodes collagen 4 subunit 2, was also identified. This association is particularly interesting as rare mutations in COL4A2 and the closely related COL4A1 protein lead to small vessel disease and hemorrhagic stroke,28–30 and common variants in close LD with this SNP (r2 > 0.8) have been linked to sporadic small vessel disease.26
The observation that genetic risk factors for WMH in community populations also influence WMH in stroke patients has implications. It suggests that the white matter changes seen on the brain MRI scans of otherwise healthy elderly reflect a similar disease process as the more severe forms that underlie cerebral small vessel disease in patients with stroke. Previous studies have indicated heterogeneity in WMH pathology: our results do not preclude this possibility, but suggest that many of the same genetic factors contribute to both pathologies.
Our study has several strengths. Protocols were uniformly employed across analyses, including imputation to the same reference build across all study groups, using the same software. Similarly, analyses were performed using the same software on the same phenotype, derived in the same way. We performed volumetric analysis of all MRI scans to quantify WMHV, which has strengths over rating scales, which are known to have ceiling effects.14 Inter-rater agreement between the 2 coordinating centers was shown to be good. WMHV was quantified using semiautomated volumetric protocols validated for use in patients with stroke and clinical grade MRI scans.
Our study also has limitations. Large-scale collaborative GWAS such as that undertaken here necessarily combine studies with some degree of phenotypic variability. Differences in environmental exposures, possibly resulting in epigenetic modifications, may contribute to such variability, which could alter the results. We identified 4 novel associations at genome-wide significance when combined with previous publications. However, we have not provided replication of these findings and therefore further evidence will be necessary to verify these associations with WMHV. MRI used in the analyses were drawn from a number of centers, with varying image quality. Therefore, to minimize bias arising from differing image quality, we quantified WMHV per study group and meta-analyzed the results. This approach may limit our ability to detect associations with low frequency variants due to small sample sizes in some study groups. The majority of MRI scans used were from FLAIR sequences. However, where these were unavailable, we used T2-weighted images, which are less sensitive to white matter changes. Such differences in sensitivity may affect quantification of WMHV across study groups, although future studies that involve centralized volumetric MRI analysis pipelines, such as those currently in development, may account for this variability. In this analysis, we considered all subtypes of stroke together as we were underpowered to investigate subtype-specific influences on WMH. It is possible that causes of WMH may differ by stroke subtype, but larger studies with sufficient power will be required before this issue can be addressed adequately. Similarly, it has been hypothesized that periventricular and deep WMH might have distinct underlying pathophysiology. In this analysis, we considered total WMHV, rather than treating these regions separately; our lesion volume analysis did not differentiate into these 2 regions. Further analyses should address this area.
We have shown that the age-related white matter changes seen in otherwise healthy populations share genetic susceptibility with the extensive lesions that underlie cerebral small vessel disease. We report 6 independent loci that are associated with WMHV in healthy individuals as well as stroke patients, 4 of which are novel associations at the genome-wide level. Our results suggest that a full genome-wide meta-analysis of available cohorts of WMH in ischemic stroke patients and community populations is likely to uncover further associations.
Supplementary Material
ACKNOWLEDGMENT
The authors thank Kristiina Rannikmae for comments on the manuscript; and Wellcome Trust Case Control Consortium 2.
GLOSSARY
- FLAIR
fluid-attenuated inversion recovery
- ROI
region of interest
- SNP
single nucleotide polymorphism
- TICV
total intracranial volume
- WMH
white matter hyperintensities
- WMHV
white matter hyperintensity volume
- WTCCC2
Wellcome Trust Case Control Consortium–2
Footnotes
Supplemental data at Neurology.org
Contributor Information
Collaborators: International Stroke Genetics Consortium, Sylvia Smoller, John Sorkin, Xingwu Wang, Magdy Selim, Aleksandra Pikula, Philip Wolf, Stephanie Debette, Sudha Seshadri, Paul de Bakker, Daniel Chasman, Kathryn Rexrode, Ida Chen, Jerome Rotter, May Luke, Michelle Sale, Tsong-Hai Lee, Ku-Chou Chang, Mitchell Elkind, Larry Goldstein, Michael (Luke) James, Monique Breteler, Chris O'Donnell, Didier Leys, Cara Carty, Chelsea Kidwell, Jes Olesen, Pankaj Sharma, Stephen Rich, Turgot Tatlisumak, Olli Happola, Philippe Bijlenga, Carolina Soriano, Eva Giralt, Jaume Roquer, Jordi Jimenez-Conde, Ioana Cotlarcius, John Hardy, Michal Korostynski, Giorgio Boncoraglio, Elena Ballabio, Eugenio Parati, Adamski Mateusz, Andrzej Urbanik, Tomasz Dziedzic, Jeremiasz Jagiella, Jerzy Gasowski, Marcin Wnuk, Rafał Olszanecki, Joanna Pera, Agnieszka Slowik, Karol Józef Juchniewicz, Christopher Levi, Paul Nyquist, Iscia Cendes, Norberto Cabral, Paulo Franca, Anderson Goncalves, Lina Keller, Milita Crisby, Konstantinos Kostulas, Robin Lemmens, Kourosh Ahmadi, Christian Opherk, Marco Duering, Martin Dichgans, Rainer Malik, Mariya Gonik, Julie Staals, Olle Melander, Philippe Burri, Ariane Sadr-Nabavi, Javier Romero, Alessandro Biffi, Chris Anderson, Guido Falcone, Bart Brouwers, Jonathan Rosand, Natalia Rost, Rose Du, Christina Kourkoulis, Thomas Battey, Steven Lubitz, Bertram Mueller-Myhsok, James Meschia, Thomas Brott, Guillaume Pare, Alexander Pichler, Christian Enzinger, Helena Schmidt, Reinhold Schmidt, Stephan Seiler, Susan Blanton, Yoshiji Yamada, Anna Bersano, Tatjana Rundek, Ralph Sacco, Yu-Feng Yvonne Chan, Andreas Gschwendtner, Zhen Deng, Taura Barr, Katrina Gwinn, Roderick Corriveau, Andrew Singleton, Salina Waddy, Lenore Launer, Christopher Chen, Kim En Le, Wei Ling Lee, Eng King Tan, Akintomi Olugbodi, Peter Rothwell, Sabrina Schilling, Vincent Mok, Elena Lebedeva, Christina Jern, Katarina Jood, Sandra Olsson, Helen Kim, Chaeyoung Lee, Laura Kilarski, Hugh Markus, Jennifer Peycke, Steve Bevan, Wayne Sheu, Hung Yi Chiou, Joseph Chern, Elias Giraldo, Muhammad Taqi, Vivek Jain, Olivia Lam, George Howard, Daniel Woo, Steven Kittner, Braxton Mitchell, John Cole, Jeff O'Connell, Dianna Milewicz, Kachikwu Illoh, Bradford Worrall, Colin Stine, Bartosz Karaszewski, David Werring, Reecha Sofat, June Smalley, Arne Lindgren, Bjorn Hansen, Bo Norrving, Gustav Smith, Juan José Martín, Vincent Thijs, Karin Klijn, Femke van't Hof, Ale Algra, Mary Macleod, Rodney Perry, Donna Arnett, Alessandro Pezzini, Alessandro Padovani, Steve Cramer, Mark Fisher, Danish Saleheen, Joseph Broderick, Brett Kissela, Alex Doney, Cathie Sudlow, Kristiina Rannikmae, Scott Silliman, Caitrin McDonough, Matthew Walters, Annie Pedersen, Kazuma Nakagawa, Christy Chang, Mark Dobbins, Patrick McArdle, Yu-Ching Chang, Robert Brown, Devin Brown, Elizabeth Holliday, Raj Kalaria, Jane Maguire, John Attia, Martin Farrall, Anne-Katrin Giese, Myriam Fornage, Jennifer Majersik, Mary Cushman, Keith Keene, Siiri Bennett, David Tirschwell, Bruce Psaty, Alex Reiner, Will Longstreth, David Spence, Joan Montaner, Israel Fernandez-Cadenas, Carl Langefeld, Cheryl Bushnell, Laura Heitsch, Jin-Moo Lee, and Kevin Sheth
AUTHOR AFFILIATIONS
From the Department of Clinical Neurosciences (M.T., H.S.M.) and the Autism Research Centre (O.E.P.), University of Cambridge, UK; the J. Philip Kistler Stroke Research Center (C.R.Z., W.J.D., L.C., G.J.F., K.F., A.K., N.R.) and the Center for Human Genetic Research (W.J.D., G.J.F., F.R., J.R., N.R.), Department of Neurology, Massachusetts General Hospital, Boston; the Neurosciences Research Centre (P.A.-S., S.G., T.R.B., B.M.), St George's, University of London, UK; the Neurology Unit (S.L., J.R.), IRCCS Ca' Granda Ospedale Maggiore Policlinico, Milan, Italy; Program in Medical and Population Genetics (F.R.), Broad Institute, Cambridge, MA; the Department of Medical & Molecular Genetics (C.M.L.) and the Social, Genetic and Developmental Psychiatry Centre, Institute of Psychiatry (C.M.L.), King's College London, UK; the Department of Cerebrovascular Diseases (G.B.B.), Fondazione IRCCS Istituto Neurologico “Carlo Besta,” Milan, Italy; KU Leuven, University of Leuven, the Department of Neurosciences, Experimental Neurology and Leuven Research Institute for Neuroscience and Disease (R.L., V.T.); University Hospitals Leuven, the Department of Neurology (R.L., V.T.); VIB, Vesalius Research Center, Laboratory of Neurobiology (R.L., V.T.), Leuven, Belgium; Austin Health and Melbourne Brain Center (V.T.), Florey Institute of Neuroscience and Mental Health, University of Melbourne, Heidelberg, Victoria, Australia; the Division of Clinical Neurosciences, Neuroimaging Sciences and Institute of Genetics and Molecular Medicine (C.S.), and Neuroimaging Sciences, Centre for Clinical Brain Sciences (J.W.), University of Edinburgh, UK; the Stroke Prevention Research Unit (P.M.R.), Nuffield Department of Neuroscience, University of Oxford, UK; the Department of Neurology (J.F.M.), Mayo Clinic, Jacksonville, FL; the Departments of Neurology and Public Health Sciences (B.B.W.), University of Virginia, Charlottesville; the Centre for Clinical Epidemiology and Biostatistics (C.L.), Hunter Medical Research Institute and School of Medicine and Public Health, University of Newcastle, NSW, Australia; School of Life Science (S.B.), University of Lincoln, Lincoln, UK; the Department of Neurology (K.L.F.), Rhode Island Hospital, Alpert Medical School of Brown University, Providence; the Institute for Stroke and Dementia Research (M.D.), Klinikum der Universität München, Ludwig-Maximilians-University Munich; and the Munich Cluster for Systems Neurology (SyNergy) (M.D.), Munich, Germany.
AUTHOR CONTRIBUTIONS
Matthew Traylor: drafting/revising the manuscript for content, study concept or design, analysis or interpretation of data, statistical analysis, accepts responsibility for conduct of research and final approval. Cathy Zhang: drafting/revising the manuscript for content, study concept or design, analysis or interpretation of data, statistical analysis, accepts responsibility for conduct of research and final approval. Poneh Adib-Samii: drafting/revising the manuscript for content, study concept or design, analysis or interpretation of data, statistical analysis, accepts responsibility for conduct of research and final approval. William Devan: drafting/revising the manuscript for content, study concept or design, analysis or interpretation of data, statistical analysis, accepts responsibility for conduct of research and final approval. Owen Parsons: drafting/revising the manuscript for content, analysis or interpretation of data, accepts responsibility for conduct of research and final approval. Silvia Lanfranconi: drafting/revising the manuscript for content, analysis or interpretation of data, accepts responsibility for conduct of research and final approval. Sarah Gregory: drafting/revising the manuscript for content, analysis or interpretation of data, accepts responsibility for conduct of research and final approval. Lisa Cloonan: drafting/revising the manuscript for content, analysis or interpretation of data, accepts responsibility for conduct of research and final approval. Guido Falcone: drafting/revising the manuscript for content, accepts responsibility for conduct of research and final approval. Farid Radmanesh: drafting/revising the manuscript for content, accepts responsibility for conduct of research and final approval. Kaitlin Fitzpatrick: drafting/revising the manuscript for content, accepts responsibility for conduct of research and final approval. Alison Kanakis: drafting/revising the manuscript for content, accepts responsibility for conduct of research and final approval. Tom Barrick: drafting/revising the manuscript for content, accepts responsibility for conduct of research and final approval. Barry Moynihan: drafting/revising the manuscript for content, acquisition of data, accepts responsibility for conduct of research and final approval. Cathryn Lewis: drafting/revising the manuscript for content, analysis or interpretation of data, accepts responsibility for conduct of research and final approval. Giorgio Boncoraglio: drafting/revising the manuscript for content, acquisition of data, accepts responsibility for conduct of research and final approval. Robin Lemmens: drafting/revising the manuscript for content, acquisition of data, accepts responsibility for conduct of research and final approval. Vincent Thijs: drafting/revising the manuscript for content, acquisition of data, accepts responsibility for conduct of research and final approval. Cathie Sudlow: drafting/revising the manuscript for content, acquisition of data, accepts responsibility for conduct of research and final approval. Joanna Wardlaw: drafting/revising the manuscript for content, acquisition of data, accepts responsibility for conduct of research and final approval. Peter Rothwell: drafting/revising the manuscript for content, acquisition of data, accepts responsibility for conduct of research and final approval. James Meschia: drafting/revising the manuscript for content, acquisition of data, accepts responsibility for conduct of research and final approval. Bradford Worall: drafting/revising the manuscript for content, acquisition of data, accepts responsibility for conduct of research and final approval. Chris Levi: drafting/revising the manuscript for content, acquisition of data, accepts responsibility for conduct of research and final approval. Karen Furie: drafting/revising the manuscript for content, acquisition of data, accepts responsibility for conduct of research and final approval. Steve Bevan: drafting/revising the manuscript for content, analysis or interpretation of data, accepts responsibility for conduct of research and final approval. Martin Dichgans: drafting/revising the manuscript for content, acquisition of data, accepts responsibility for conduct of research and final approval. Jonathan Rosand: drafting/revising the manuscript for content, study concept or design, acquisition of data, analysis or interpretation of data, study supervision or coordination, accepts responsibility for conduct of research and final approval. Hugh Markus: drafting/revising the manuscript for content, study concept or design, acquisition of data, analysis or interpretation of data, study supervision or coordination, accepts responsibility for conduct of research and final approval. Natalia Rost: drafting/revising the manuscript for content, study concept or design, acquisition of data, analysis or interpretation of data, study supervision or coordination, accepts responsibility for conduct of research and final approval.
STUDY FUNDING
Funding for collection, genotyping, and analysis of stroke samples was provided by Wellcome Trust Case Control Consortium–2, a functional genomics grant from the Wellcome Trust (DNA-Lacunar), the Stroke Association (DNA-Lacunar), the Intramural Research Program of National Institute of Ageing (Massachusetts General Hospital [MGH] and Ischemic Stroke Genetics Study [ISGS]), National Institute of Neurological Disorders and Stroke (Siblings With Ischemic Stroke Study, ISGS, and MGH), the American Heart Association/Bugher Foundation Centers for Stroke Prevention Research (MGH), Deane Institute for Integrative Study of Atrial Fibrillation and Stroke (MGH), National Health and Medical Research Council (Australian Stroke Genetics Collaborative), and Italian Ministry of Health (Milan). Additional support for sample collection came from the Medical Research Council, National Institute of Health Research Biomedical Research Centre and Acute Vascular Imaging Centre (Oxford), Wellcome Trust and Binks Trust (Edinburgh), and Vascular Dementia Research Foundation (Munich). M.T. is supported by a project grant from the Stroke Association (TSA 2013/01). H.S.M. is supported by an NIHR Senior Investigator award. H.S.M. and S.B. are supported by the NIHR Cambridge University Hospitals Comprehensive Biomedical Research Centre. V.T. and R.L. are supported by grants from FWO Flanders. P.R. holds NIHR and Wellcome Trust Senior Investigator Awards. P.A.S. is supported by an MRC Fellowship. C.M.L.'s research is supported by the National Institute for Health Research Biomedical Research Centre (BRC) based at Guy's and St Thomas' NHS Foundation Trust and King's College London, and the BRC for Mental Health at South London and Maudsley NHS Foundation Trust and King's College London.
DISCLOSURE
The authors report no disclosures relevant to the manuscript. Go to Neurology.org for full disclosures.
REFERENCES
- 1.Debette S, Markus HS. The clinical importance of white matter hyperintensities on brain magnetic resonance imaging: systematic review and meta-analysis. BMJ 2010;341:c3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rost NS, Rahman RM, Biffi A, et al. White matter hyperintensity volume is increased in small vessel stroke subtypes. Neurology 2010;75:1670–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ay H, Arsava EM, Rosand J, et al. Severity of leukoaraiosis and susceptibility to infarct growth in acute stroke. Stroke 2008;39:1409–1413. [DOI] [PubMed] [Google Scholar]
- 4.Arsava EM, Rahman R, Rosand J, et al. Severity of leukoaraiosis correlates with clinical outcome after ischemic stroke. Neurology 2009;72:1403–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atwood LD, Wolf PA, Heard-Costa NL, et al. Genetic variation in white matter hyperintensity volume in the Framingham Study. Stroke 2004;35:1609–1613. [DOI] [PubMed] [Google Scholar]
- 6.Turner ST, Jack CR, Fornage M, Mosley TH, Boerwinkle E, de Andrade M. Heritability of leukoaraiosis in hypertensive sibships. Hypertension 2004;43:483–487. [DOI] [PubMed] [Google Scholar]
- 7.Carmelli D, DeCarli C, Swan GE, et al. Evidence for genetic variance in white matter hyperintensity volume in normal elderly male twins. Stroke 1998;29:1177–1181. [DOI] [PubMed] [Google Scholar]
- 8.Opherk C, Peters N, Holtmannspotter M, Gschwendtner A, Muller-Myhsok B, Dichgans M. Heritability of MRI lesion volume in CADASIL: evidence for genetic modifiers. Stroke 2006;37:2684–2689. [DOI] [PubMed] [Google Scholar]
- 9.Adib-Samii P, Devan W, Traylor M, et al. Genetic architecture of white matter hyperintensities differs in hypertensive and nonhypertensive ischemic stroke. Stroke 2015;46:348–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fornage M, Debette S, Bis JC, et al. Genome-wide association studies of cerebral white matter lesion burden: the CHARGE consortium. Ann Neurol 2011;69:928–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez LM, Hill WD, Harris SE, et al. Genes from a translational analysis support a multifactorial nature of white matter hyperintensities. Stroke 2015;46:341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verhaaren BF, Debette S, Bis JC, et al. Multi-ethnic genome-wide association study of cerebral white matter hyperintensities on MRI. Circ Cardiovasc Genet 2015;8:398–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidt R, Schmidt H, Haybaeck J, et al. Heterogeneity in age-related white matter changes. Acta Neuropathol 2011;122:171–185. [DOI] [PubMed] [Google Scholar]
- 14.Wardlaw JM, Smith EE, Biessels GJ, et al. Neuroimaging standards for research into small vessel disease and its contribution to ageing and neurodegeneration. Lancet Neurol 2013;12:822–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen YW, Gurol ME, Rosand J, et al. Progression of white matter lesions and hemorrhages in cerebral amyloid angiopathy. Neurology 2006;67:83–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grimaud J, Lai M, Thorpe J, et al. Quantification of MRI lesion load in multiple sclerosis: a comparison of three computer-assisted techniques. Magn Reson Imaging 1996;14:495–505. [DOI] [PubMed] [Google Scholar]
- 17.Smith SM, Zhang Y, Jenkinson M, et al. Accurate, robust, and automated longitudinal and cross-sectional brain change analysis. Neuroimage 2002;17:479–489. [DOI] [PubMed] [Google Scholar]
- 18.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006;38:904–909. [DOI] [PubMed] [Google Scholar]
- 19.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abecasis GR, Auton A, Brooks LD, et al. An integrated map of genetic variation from 1,092 human genomes. Nature 2012;491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Devlin B, Roeder K. Genomic control for association studies. Biometrics 1999;55:997–1004. [DOI] [PubMed] [Google Scholar]
- 22.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010;26:2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Bakker PI, Ferreira MA, Jia X, Neale BM, Raychaudhuri S, Voight BF. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet 2008;17:R122–R128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boyle AP, Hong EL, Hariharan M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 2012;22:1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.The Genotype-Tissue Expression (GTEx) Project. Nat Genet 2013;45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rannikmae K, Davies G, Thomson PA, et al. Common variation in COL4A1/COL4A2 is associated with sporadic cerebral small vessel disease. Neurology 2015;84:918–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dunham I, Kundaje A, Aldred SF, et al. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gould DB, Phalan FC, van Mil SE, et al. Role of COL4A1 in small-vessel disease and hemorrhagic stroke. N Engl J Med 2006;354:1489–1496. [DOI] [PubMed] [Google Scholar]
- 29.Lanfranconi S, Markus HS. COL4A1 mutations as a monogenic cause of cerebral small vessel disease: a systematic review. Stroke 2010;41:e513–e518. [DOI] [PubMed] [Google Scholar]
- 30.Jeanne M, Labelle-Dumais C, Jorgensen J, et al. COL4A2 mutations impair COL4A1 and COL4A2 secretion and cause hemorrhagic stroke. Am J Hum Genet 2012;90:91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.