Abstract
Rationale: Idiopathic pulmonary fibrosis (IPF) is a devastating lung disease of unknown etiology. The genes TOLLIP and MUC5B play important roles in lung host defense, which is an immune process influenced by oxidative signaling. Whether polymorphisms in TOLLIP and MUC5B modify the effect of immunosuppressive and antioxidant therapy in individuals with IPF is unknown.
Objectives: To determine whether single-nucleotide polymorphisms (SNPs) within TOLLIP and MUC5B modify the effect of interventions in subjects participating in the Evaluating the Effectiveness of Prednisone, Azathioprine, and N-Acetylcysteine in Patients with Idiopathic Pulmonary Fibrosis (PANTHER-IPF) clinical trial.
Methods: SNPs within TOLLIP (rs5743890/rs5743894/rs5743854/rs3750920) and MUC5B (rs35705950) were genotyped. Interaction modeling was conducted with multivariable Cox regression followed by genotype-stratified survival analysis using a composite endpoint of death, transplantation, hospitalization, or a decline of ≥10% in FVC.
Measurements and Main Results: Significant interaction was observed between N-acetylcysteine (NAC) therapy and rs3750920 within TOLLIP (Pinteraction = 0.001). After stratifying by rs3750920 genotype, NAC therapy was associated with a significant reduction in composite endpoint risk (hazard ratio, 0.14; 95% confidence interval, 0.02–0.83; P = 0.03) in those with a TT genotype, but a nonsignificant increase in composite endpoint risk (hazard ratio, 3.23; 95% confidence interval, 0.79–13.16; P = 0.10) was seen in those with a CC genotype. These findings were then replicated in an independent IPF cohort.
Conclusions: NAC may be an efficacious therapy for individuals with IPF with an rs3750920 (TOLLIP) TT genotype, but it was associated with a trend toward harm in those with a CC genotype. A genotype-stratified prospective clinical trial should be conducted before any recommendation regarding the use of off-label NAC to treat IPF.
Keywords: IPF, pharmacogenetics, N-acetylcysteine, drug–gene interaction, host defense
At a Glance
Scientific Knowledge on the Subject
Polymorphisms within two genes, TOLLIP and MUC5B, have been associated with idiopathic pulmonary fibrosis (IPF) susceptibility and survival. These genes are critical to lung host defense, an immunologic response sensitive to oxidative signaling and the presence of antioxidants. Whether polymorphisms in TOLLIP and MUC5B modify the effect of immunosuppressive and antioxidant therapy in patients with IPF is unknown.
What This Study Adds to the Field
This investigation demonstrates that the genetic makeup of individuals with IPF may influence their response to N-acetylcysteine therapy. These findings also highlight the importance of pharmacogenetics in IPF and support systematic biospecimen collection in IPF clinical trials so that genetic subgroups predisposed to treatment-related benefit or harm can be identified.
Idiopathic pulmonary fibrosis (IPF) is a devastating interstitial lung disease with high mortality and no known cure. Although its cause remains unknown, infectious agents, including bacterial and viral pathogens, together with particulate inhalation, have been implicated in IPF pathogenesis and progression (1–5). Much work has focused on characterizing lung host defense and the immune response to lung injury in IPF. Two genes involved in host defense are toll-interacting protein (TOLLIP) and mucin 5B (MUC5B). These genes reside in close proximity on chromosome 11p15.5 and have distinct biological functions. TOLLIP encodes toll-interacting protein (TOLLIP), which is an inhibitory adaptor protein acting downstream from the toll-like receptors (TLRs). TLRs are key mediators of the innate and adaptive immune response (6–10). MUC5B encodes a highly glycosylated mucin-5B precursor protein (Mucin-5B) that contributes to airway mucus production and is important in maintaining immune homeostasis (4, 7, 11).
Single-nucleotide polymorphisms (SNPs) within TOLLIP and MUC5B have recently been shown to be associated with IPF susceptibility and survival (12–14). SNPs within these genes have also been linked to alterations in the lung immune response (8, 11). Oxidative signaling plays a key role in the lung immune response, and can be modulated by the presence of antioxidants, such as N-acetylcysteine (NAC) (15–17). Although NAC therapy was recently shown to be no better than placebo in treating IPF (18), it remains a common off-label therapy for the disease (19, 20). Whether polymorphisms in TOLLIP and MUC5B influence antioxidant effects is unknown.
In this investigation, we conducted a post hoc exploratory analysis of subjects enrolled in the Idiopathic Pulmonary Fibrosis Clinical Research Network (IPFnet) clinical trial Evaluating the Effectiveness of Prednisone, Azathioprine, and N-Acetylcysteine in Patients with Idiopathic Pulmonary Fibrosis (PANTHER-IPF; NCT 00650091) (18, 21) to determine whether SNPs within TOLLIP and MUC5B modified the effect of trial interventions with regard to a composite clinical endpoint. We then analyzed an independent cohort of subjects drawn from the University of Chicago (UChicago) and The INSPIRE Trial: A Study of Interferon Gamma-1b for Idiopathic Pulmonary Fibrosis (INSPIRE; NCT 00075998) (22) to determine whether our findings could be replicated. Some of the results of these studies were previously reported in the form of an abstract (23).
Using a candidate gene approach, five SNPs were chosen to conduct this analysis. rs5743890 and rs5743894 reside within noncoding TOLLIP regulatory regions and are associated with IPF susceptibility and survival (rs5743890 only) (12). rs3750920 is a functional synonymous coding SNP residing within TOLLIP exon 3; it was marginally associated with IPF susceptibility in two genome-wide association study discovery cohorts (12, 13) and has been shown to mediate the pulmonary immune response vis-à-vis TLR2 and TLR4 signaling (8). rs5743854 is located within the TOLLIP promoter and has potential transcriptional consequences. rs35705950 resides within the promoter of MUC5B, is associated with IPF susceptibility and survival, and has been shown to reduce the risk of bacterial colonization in the lower airways of individuals with IPF (4, 12–14, 24).
Methods
Study Cohorts
The study was approved by the University of Chicago Institutional Review Board (IRB#15177 and 14163A). A detailed description of the PANTHER, INSPIRE, and UChicago cohorts can be found in the online supplement. All subjects in this investigation met criteria for IPF based on American Thoracic Society/European Respiratory Society guidelines (25). To minimize the effects of population stratification, only self-reported non-Hispanic white participants were included in the analysis.
SNP Selection and Genotyping
Five SNPs at the chr11p15.5 locus were chosen for this analysis: rs5743890, rs5743894, rs5743854, rs3750920, and rs35705950. Eight SNPs were initially genotyped, but three were excluded due to the presence of moderate-to-strong linkage disequilibrium with another SNP based on a SNP annotation and proxy search program R2 approximation using 1,000 genomes project data (see Figure E1 in the online supplement) (26, 27). A detailed description of genotyping methods for each cohort can be found in the online supplement.
Statistical Analysis
The primary analysis tested whether statistical interaction was present between SNPs of interest and two therapeutic interventions—NAC therapy and a combination therapy composed of prednisone, azathioprine, and NAC—with regard to composite endpoint-free survival, which was defined as time from trial enrollment to death, transplantation, hospitalization, or a decline of ≥10% in FVC. This composite endpoint was chosen as part of an enrichment strategy to improve statistical power (28) and because these endpoints have previously been shown to be clinically meaningful (29–31). SNP–treatment interaction was formally tested with a multivariable Cox regression model that included variables for SNP genotype, treatment, and a SNP–treatment interaction term. A multiplicative model was used because relative risks were used to measure the treatment effect (32). Interaction was considered present when the Wald z-statistic for SNP–treatment interaction term corresponded to a P value <0.01, which adjusted for multiple SNP testing by Bonferroni correction. To demonstrate the interaction identified by this approach (32), composite endpoint-free survival was then compared between treatment groups after stratification by genotype with unadjusted log-rank testing and multivariable Cox regression adjusted for baseline age, sex, FVC (percentage predicted), and diffusion capacity of the lung for carbon monoxide (DlCO) (percentage predicted). Survival was plotted using the Kaplan-Meier estimator. With the exception of the interaction modeling outlined previously, statistical significance was considered present when P < 0.05.
Continuous variables are reported as means (SD) and are compared using a Student’s t test or analysis of variance, as appropriate. Categorical variables are reported as counts and percentages and compared using a χ2 or Fisher’s exact test, as appropriate. SNP correlation was determined using Pearson’s correlation coefficient. Cox regression models were checked to ensure that the proportional hazards assumption was met. Based on SNP distributions, an additive allelic model was assumed for rs3750920 (TOLLIP) and a dominant model for rs5743890 (TOLLIP), rs5743894 (TOLLIP), rs5743854 (TOLLIP), and rs35705950 (MUC5B).
Analysis of the individual and combined replication cohorts was performed using similar methods. It was not possible to obtain hospitalization data for these cohorts; therefore, composite endpoint-free survival was defined as time from trial enrollment (INSPIRE) or blood draw (UChicago) to death, transplantation, or a decline of ≥10% in FVC in those who did not receive NAC therapy and time from NAC initiation to death, transplantation, or a decline of ≥10% in FVC in those who received NAC therapy. All statistical analysis was conducted using Stata (StataCorp, 2011, release 12. College Station, TX).
Results
PANTHER Cohort
Of the 341 subjects enrolled in the PANTHER trial, 315 were eligible for analysis based on self-reported race and ethnicity (see Figure E2). Of these subjects, 154 consented to genetic testing and were included in the primary analysis. Fifty-four subjects received placebo therapy, 60 subjects received NAC therapy, and 40 received prednisone, azathioprine, and NAC therapy. A comparison of baseline characteristics, outcomes, and SNP genotypes between treatment arms and genotyped/nongenotyped trial subjects is shown in Table 1. No differences were observed between groups with regard to age, sex, smoking history, and pulmonary function. Groups were similar with regard to deaths, transplantations, hospitalizations, and FVC decline. SNP genotype frequencies were also similar between treatment arms.
Table 1.
PANTHER Cohort Baseline Characteristics and Genotypes*
| Characteristic | Genotyped (n = 154) |
Nongenotyped (n = 161) |
P Value | ||||
|---|---|---|---|---|---|---|---|
| Placebo Arm (n = 54) | NAC Arm (n = 60) | PAN Arm (n = 40) | Placebo Arm (n = 66) | NAC Arm (n = 61) | PAN Arm (n = 34) | ||
| Age, mean (SD) | 66.1 (7.9) | 67.9 (8.7) | 69.7 (6.8) | 66.8 (8.2) | 67.8 (8.2) | 67 (7.7) | 0.39 |
| Male, n (%) | 39 (72.2) | 47 (78.3) | 31 (77.5) | 53 (80.3) | 51 (83.6) | 27 (79.4) | 0.79 |
| Ever-smoker, n (%) | 41 (75.9) | 44 (73.3) | 28 (70) | 50 (75.8) | 46 (76.7) | 24 (70.6) | 0.96 |
| FVC, % predicted, mean (SD) | 73.2 (14.7) | 73 (15.7) | 71.2 (15.2) | 74 (13.8) | 73 (15.8) | 67.6 (15.5) | 0.43 |
| DlCO, % predicted, mean (SD) | 46.4 (11.7) | 43.9 (10.9) | 43 (10.6) | 44.7 (12.1) | 46.2 (10.8) | 39.9 (9.4) | 0.11 |
| Death, n (%) | 2 (3.7) | 1 (1.7) | 4 (10) | 1 (1.5) | 3 (4.9) | 4 (11.8) | 0.1 |
| FVC decline ≥ 10%, n (%) | 12 (22.2) | 11 (18.3) | 6 (15) | 17 (25.8) | 17 (27.9) | 3 (8.8) | 0.23 |
| Hospitalization, n (%) | 8 (14.8) | 8 (13.3) | 13 (32.5) | 10 (15.2) | 9 (14.8) | 8 (23.5) | 0.17 |
| Transplant, n (%) | 1 (1.9) | 3 (5) | 1 (2.5) | 1 (1.5) | 1 (1.6) | 1 (2.9) | 0.45 |
| Composite endpoint,† n (%) | 17 (31.5) | 19 (31.7) | 19 (47.5) | 24 (36.4) | 26 (42.6) | 12 (35.3) | 0.52 |
| SNP (gene) genotype, n (%) | |||||||
| rs5743890 (TOLLIP) AA/AG/GG | 43/10/0 (81/19/0) | 52/8/0 (87/13/0) | 30/6/0 (83/17/0) | — | — | — | 0.72 |
| rs5743894 (TOLLIP) AA/AG/GG | 29/21/4 (54/39/7) | 31/25/4 (52/42/6) | 18/17/5 (45/43/12) | — | — | — | 0.83 |
| rs3750920 (TOLLIP) CC/CT/TT | 14/23/17 (26/43/31) | 13/30/16 (22/51/27) | 6/24/10 (15/60/25) | — | — | — | 0.55 |
| rs5743854 (TOLLIP) CC/CG/GG | 44/10/0 (81/19/0) | 55/5/0 (92/8/0) | 37/3/0 (93/7/0) | 0.19 | |||
| rs35705950 (MUC5B) GG/GT/TT | 20/29/5 (37/54/9) | 14/42/4 (23/70/7) | 11/23/6 (28/57/15) | — | — | — | 0.3 |
Definition of abbreviations: DlCO = diffusion capacity of the lung for carbon monoxide; NAC = N-acetylcysteine; PAN = prednisone/azathioprine/N-acetylcysteine; PANTHER = Evaluating the Effectiveness of Prednisone, Azathioprine, and N-Acetylcysteine in Patients with Idiopathic Pulmonary Fibrosis; SNP = single-nucleotide polymorphism.
Non-Hispanic white individuals by self-report.
Death, 10% FVC decline, hospitalization, or transplantation.
When considering subjects enrolled before and after the PANTHER clinical trial, a nonstatistically significant imbalance in genotype frequencies was observed between the placebo and NAC arms with regard to rs5743894 (TOLLIP) (P = 0.17), rs3750920 (TOLLIP) (P = 0.12), and rs35705950 (MUC5B) (P = 0.08) (see Table E1). When comparing the minor allele frequency for each SNP between the PANTHER cohort and a recent genome-wide association study (12) stage II replication cohort (see Table E2), the minor allele frequency of rs35705950_T (MUC5B) was significantly higher in the PANTHER cohort compared with the genome-wide association study cohort (0.4 vs 0.33, respectively; P = 0.02). None of the chosen SNPs had a strong linkage disequilibrium based on R2 criteria (26, 27), nor were any significantly correlated in the PANTHER dataset (see Table E3).
No SNP independently predicted composite endpoint-free survival in multivariable Cox regression after adjusting for treatment arm assignment, age, sex, FVC (percentage predicted), and DlCO (percentage predicted) (see Table E4). The proportional hazards assumption for rs5743854 was not met in this model, so this SNP was omitted from subsequent analyses. Interaction modeling (see Table E5) showed significant interaction between NAC therapy and the T allele of rs3750920 within TOLLIP (Pinteraction = 0.001). A suggestion of possible interaction was observed between NAC therapy and the T allele of rs35705950 (MUC5B) (Pinteraction = 0.02) and the G allele of rs5743894 (TOLLIP) (Pinteraction = 0.03), but these did not cross the significance threshold adjusted for multiple testing. No significant interaction was detected between prednisone, azathioprine, and NAC therapy and any of the SNPs of interest.
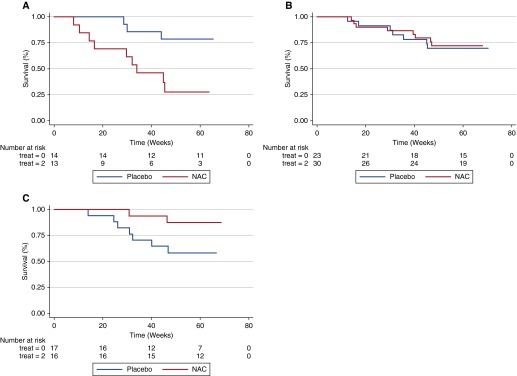
Survival analysis stratified by rs3750920 (TOLLIP) genotype is shown in Figures 1A–C. Among subjects with a CC genotype, NAC therapy was associated with significantly worse survival than placebo in unadjusted analysis (P = 0.01), but not after multivariable adjustment (hazard ratio [HR], 3.23; 95% confidence interval [CI], 0.79–13.16; P = 0.10). There was no difference in survival between NAC and placebo groups in subjects with a CT genotype in unadjusted analysis (P = 0.82) or after multivariable adjustment (HR, 0.76; 95% CI, 0.27–2.19; P = 0.62). Among subjects with a TT genotype, NAC therapy was associated with borderline improved survival compared with placebo in unadjusted analysis (P = 0.06), and with significantly reduced endpoint risk after multivariable adjustment (HR, 0.14; 95% CI, 0.02–0.83; P = 0.03). Because of the potential for misspecification of Cox models when using a composite endpoint (33), a Wei-Lin-Weissfeld (34) analysis of multiple failure data was conducted and showed similar results (see Table E6).
Figure 1.
Composite endpoint-free survival between N-acetylcysteine (NAC) and placebo groups after stratification by rs3750920 (TOLLIP) genotype. In those with a CC genotype (A), NAC therapy is associated with worse survival than placebo (Plogrank = 0.01; hazard ratio [HR], 3.23; 95% confidence interval [CI], 0.79–13.16; P = 0.10). In those with a CT genotype (B), survival is similar between groups (Plogrank = 0.82; HR 0.76; 95% CI 0.27–2.19; P = 0.62). In those with a TT genotype (C), NAC therapy is associated with improved survival compared with placebo (Plogrank = 0.06; HR 0.14 ; 95% CI 0.02–0.83; P = 0.03). Multivariable Cox regression models adjusted for age, sex, FVC (percentage predicted), and diffusion capacity of the lung for carbon monoxide (percentage predicted) at time of study enrollment.
Sensitivity analysis was conducted to determine the endpoints driving the observed interaction between NAC therapy and rs3750920 (TOLLIP) genotype (Table 2). rs5743894 within TOLLIP and rs35705950 within MUC5B were also included in this analysis because of the borderline interaction detected in the previously discussed model. Based on this analysis, the potential harm associated with NAC therapy in those with an rs3750920 (TOLLIP) CC genotype was driven primarily by hospitalizations, because this association was substantially weaker when considering only death, transplantation, or a decline of ≥10% in FVC. The benefit associated with NAC therapy in those with an rs3750920 (TOLLIP) TT genotype was consistent, irrespective of the composite endpoint chosen. This analysis also showed that NAC therapy in the setting of one or more rs5743894 (TOLLIP) and rs35705950 (MUC5B) minor alleles was associated with a significant reduction in risk when considering a composite endpoint of death hospitalization, or decline of ≥10% in FVC.
Table 2.
Sensitivity Analysis of NAC-associated Endpoint Risk after rs3750920, rs5743894, and rs35705950 Genotype Stratification*
| SNP (Gene) Genotype | Death/Transplant/Hosp/FVC Decline |
Death/Transplant/Hosp |
Death/Transplant/FVC Decline |
|||
|---|---|---|---|---|---|---|
| HR (95% CI) | P Value | HR (95% CI) | P Value | HR (95% CI) | P Value | |
| rs3750920 (TOLLIP) CC | 3.22 (0.79–13.16) | 0.1 | 3.74 (0.35–39.80) | 0.27 | 1.27 (0.21–7.55) | 0.79 |
| rs3750920 (TOLLIP) CT | 0.76 (0.27–2.19) | 0.62 | 0.45 (0.10–2.15) | 0.32 | 0.63 (0.17–2.30) | 0.48 |
| rs3750920 (TOLLIP) TT | 0.14 (0.02–0.83) | 0.03 | † | † | 0.09 (0.01–0.76) | 0.03 |
| rs5743894 (TOLLIP) AA | 1.67 (0.71–3.95) | 0.24 | 1.43 (0.44–4.70) | 0.56 | 1.68 (0.58–4.90) | 0.34 |
| rs5743894 (TOLLIP) AG/GG | 0.33 (0.10–1.14) | 0.08 | 0.21 (0.02–1.99) | 0.18 | 0.18 (0.04–0.90) | 0.04 |
| rs35705950 (MUC5B) GG | 2.02 (0.57–7.20) | 0.28 | 1.25 (0.15–10.30) | 0.84 | 3.72 (0.69–20.16) | 0.13 |
| rs35705950 (MUC5B) GT/TT | 0.60 (0.27–1.35) | 0.22 | 0.49 (0.15–1.58) | 0.23 | 0.32 (0.11–0.94) | 0.04 |
Definition of abbreviations: CI = confidence interval; Hosp = hospitalization; HR = hazard ratio; NAC = N-acetylcysteine; SNP = single-nucleotide polymorphism.
Models adjusted for age, sex, FVC (percentage predicted) and diffusion capacity of the lung for carbon monoxide (percentage predicted).
HR cannot be estimated owing to absence of events.
Replication Cohort
Based on the preceding findings, interaction modeling was performed in an independent cohort of subjects with IPF. In addition to rs3750920 (TOLLIP), genotypes were also determined for rs5743894 (TOLLIP) and rs35705950 (MUC5B) because of the borderline interaction observed in the PANTHER cohort. Of the 771 non-Hispanic white subjects enrolled in the INSPIRE trial, DNA was available for 314. In the UChicago cohort, of 151 non-Hispanic white subjects with sequencing data, 91 had longitudinal pulmonary function testing data and were included in the analysis. A summary of baseline demographic and clinical characteristics, together with genotype counts for the INSPIRE, UChicago, and combined cohorts is shown in Table 3. NAC recipients, compared with those who did not receive NAC, had increased use of concurrent azathioprine (10.8% vs 2.8%, respectively, P = 0.02), lower mean FVC (percentage predicted) (62.6% vs 71.1%, respectively; P < 0.001), lower mean DlCO (percentage predicted) (39.1% vs 48.0%, respectively; P = 0.003), and more deaths (40.4% vs 21.8%, respectively; P = 0.01). There was no difference in genotype counts between groups.
Table 3.
Replication Cohort Baseline Characteristics and Genotypes*
| Characteristic | INSPIRE (n = 314) |
UChicago (n = 91) |
Combined Cohort |
P Value | |||
|---|---|---|---|---|---|---|---|
| NAC (n = 29) | Non-NAC (n = 285) | NAC (n = 18) | Non-NAC (n = 73) | NAC (n = 47) | Non-NAC (n = 358) | ||
| Age, mean (SD) | 65.8 (7.5) | 66.2 (7.7) | 72.4 (8.0) | 67.3 (8.1) | 68.3 (8.3) | 66.4 (7.8) | 0.12 |
| Male, n (%) | 18 (62.1) | 210 (73.7) | 15 (83.3) | 57 (78.1) | 33 (70.2) | 267 (74.6) | 0.52 |
| Ever-smoker, n (%) | 18 (62.1) | 199 (69.8) | 15 (83.3) | 36 (73.5) | 33 (70.2) | 235 (70.4) | 0.98 |
| Azathioprine therapy, n (%) | 4 (13.8) | 9 (3.2) | 1 (5.9) | 1 (1.4) | 5 (10.9) | 10 (2.8) | 0.02 |
| Prednisone therapy, n (%) | 19 (65.5) | 121 (42.5) | 4 (23.5) | 11 (15.1) | 23 (50.0) | 132 (36.9) | 0.09 |
| FVC, % predicted, mean (SD) | 64.7 (20.7) | 71.3 (12.6) | 58.9 (15.0) | 70.3 (19.4) | 62.6 (18.6) | 71.1 (14.2) | <0.001 |
| DlCO, % predicted, mean (SD) | 40.2 (10.4) | 47.3 (9.2) | 37 (10.8) | 50.7 (19.1) | 39.1 (10.5) | 48.0 (11.9) | 0.003 |
| Death, n (%) | 6 (20.7) | 49 (17.2) | 13 (72.2) | 29 (39.7) | 19 (40.4) | 78 (21.8) | 0.01 |
| FVC decline ≥ 10%, n (%) | 13 (44.8) | 115 (40.4) | 4 (22.2) | 17 (23.3) | 17 (36.2) | 132 (36.9) | 0.93 |
| Transplant, n (%) | 2 (6.9) | 4 (1.4) | 0 (0) | 7 (9.6) | 2 (4.3) | 11 (3.1) | 0.46 |
| Composite endpoint,† n (%) | 14 (48.3) | 146 (51.2) | 13 (72.2) | 45 (61.6) | 27 (57.5) | 167 (46.7) | 0.6 |
| SNP (gene) genotype, n (%) |
|||||||
| rs5743894 (TOLLIP) AA/AG/GG | 18/11/0 (62/38/0) | 143/129/14 (50/45/5) | 11/8/2 (52/38/10) | 60/31/10 (59/31/10) | 29/16/2 (62/34/4) | 188/151/19 (53/42/5) | 0.53 |
| rs3750920 (TOLLIP) CC/CT/TT | 6/15/8 (21/52/27) | 61/157/66 (22/55/23) | 3/12/6 (14/57/29) | 21/49/31 (21/48/31) | 9/25/13 (19/53/28) | 77/194/86 (22/54/24) | 0.84 |
| rs35705950 (MUC5B) GG/GT/TT | 10/16/3 (35/55/10) | 82/185/15 (29/66/5) | 8/13/0 (38/62/0) | 31/62/8 (31/61/8) | 17/27/3 (36/57/7) | 105/230/20 (30/65/5) | 0.57 |
Definition of abbreviations: DlCO = diffusion capacity of the lung for carbon monoxide; INSPIRE = The INSPIRE Trial: A Study of Interferon Gamma-1b for Idiopathic Pulmonary Fibrosis; NAC = N-acetylcysteine; SNP = single-nucleotide polymorphism; UChicago = University of Chicago.
Non-Hispanic white individuals by self-report.
Death, transplantation, or 10% FVC decline.
Multivariable Cox interaction models (see Table E6), adjusted for prednisone use, azathioprine use, FVC (percentage predicted), and DlCO (percentage predicted) due to baseline differences between cohorts, showed significant interaction between NAC therapy and rs3750920 (TOLLIP) (Pinteraction = 0.01). A suggestion of interaction was observed between NAC therapy and rs35705950 (MUC5B) (Pinteraction = 0.02), but this did not cross the predetermined significance threshold after adjustment for multiple testing. No interaction was observed between NAC therapy and rs5743894 (TOLLIP) (Pinteraction = 0.41).
After stratification by rs3750920 (TOLLIP) genotype (Table 4), multivariable Cox modeling adjusted for age, sex, azathioprine use, prednisone use, FVC (percentage predicted), DlCO (percentage predicted), and study location was performed. Compared with those who did not receive NAC therapy, the composite endpoint risk associated with NAC therapy was significantly reduced among those with a rs3750920 (TOLLIP) TT genotype (HR, 0.23; 95% CI, 0.06–0.93; P = 0.04), but was significantly increased among those with a CT genotype (HR, 2.18; 95% CI, 1.14–4.13; P = 0.02) and CC genotype (HR, 3.11; 95% CI, 1.25–7.72; P = 0.01). The harm associated with NAC therapy in those with a CC or CT was driven primarily by those in the INSPIRE cohort, but also qualitatively increased in the UChicago cohort. The benefit associated with NAC therapy in those with a TT genotype was qualitatively similar across both cohorts.
Table 4.
Replication Cohort Genotype-stratified Composite Endpoint Risk* Associated with NAC Therapy
| Genotype | INSPIRE |
UChicago |
Combined |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Log-Rank P Value | HR | 95% CI | P Value | Log-Rank P Value | HR | 95% CI | P Value | Log-Rank P Value | HR | 95% CI | P Value | |
| rs3750920 CC | <0.001 | 5.1 | 1.70–15.3 | 0.004 | 0.61 | 2.1 | 0.22–20.3 | 0.52 | 0.004 | 3.11 | 1.25–7.72 | 0.01 |
| rs3750920 CT | 0.0002 | 5.63 | 2.45–12.9 | <0.001 | 0.16 | 1.97 | 0.69–5.60 | 0.2 | 0.002 | 2.18 | 1.14–4.13 | 0.02 |
| rs3750920 TT | 0.39 | 0.22 | 0.04–1.34 | 0.1 | 0.68 | 0.36 | 0.04–3.46 | 0.38 | 0.51 | 0.23 | 0.06–0.94 | 0.04 |
Definition of abbreviations: CI = confidence interval; HR = hazard ratio; INSPIRE = The INSPIRE Trial: A Study of Interferon Gamma-1b for Idiopathic Pulmonary Fibrosis; NAC = N-acetylcysteine; UChicago = University of Chicago.
Adjusted for age, sex, prednisone use, azathioprine use, FVC (percentage predicted), diffusion capacity of the lung for carbon monoxide (percentage predicted), and trial/center.
Discussion
In this study, we showed that the rs3750920 polymorphisms within TOLLIP might influence the response to NAC therapy in individuals with IPF. To our knowledge, this represents the first evidence of a significant drug–gene interaction in individuals with IPF. Although PANTHER investigators reported previously that NAC therapy did not provide benefit for individuals with IPF (18), we demonstrated that NAC might reduce clinically meaningful (29, 30) endpoint risk in genetically predisposed individuals, specifically those carrying an rs3750920 (TOLLIP) TT genotype. Because this genotype is found in approximately 25% of individuals with IPF, those who may benefit from NAC therapy represent a significant minority. However, because of the potential harm observed with NAC therapy among those with an rs3750920 (TOLLIP) CC genotype, the off-label use of NAC should be reconsidered until a genotype-stratified clinical trial can be conducted.
The biology underpinning an interaction between NAC therapy and TOLLIP may lie in lung host defense. TOLLIP encodes the TOLLIP protein, which is a negative inhibitor of TLRs, including TLR2 and TLR4, which are active in the lung (8, 35). TOLLIP increases antiinflammatory IL-10 production and suppresses tumor necrosis factor-α and IL-6 production after interaction with TLR2 and TLR4 ligands (8). TLR4 signaling is oxidant-dependent and can lead to lung injury (15, 17). NAC has been shown to block the TLR4-mediated inflammatory response in vitro, together with increasing antiinflammatory IL-10 production in liver transplantation recipients (36). The rs3750920 polymorphism is a functional synonymous coding SNP within TOLLIP exon 3 and has been linked to decreased TOLLIP mRNA production (8), which potentially leads to unmitigated TLR4-mediated inflammatory cytokine production. NAC, in the setting of the rs3750920 polymorphism, may improve immune homeostasis vis-à-vis decreased TLR-4 signaling and increased IL-10 production. However, why NAC therapy would be potentially harmful in those with an rs3750920 CC genotype remains unclear.
Evidence for potential interaction between NAC therapy and rs35705950 within MUC5B was also observed, but failed to reach the predetermined significance threshold adjusted for multiple testing in both discovery and replication cohorts. This was due in part to the composite endpoint chosen, because the rs35705950 (MUC5B) minor allele was associated with a reduced composite endpoint risk when considering death, transplantation, or a decline in FVC in sensitivity analysis. Like TOLLIP, MUC5B also plays a critical role in airway defense. Reduced MUC5B expression has been linked to impaired airway mucociliary clearance and chronic bacterial infection (11), whereas increased expression, which occurs with the rs35705950 polymorphism (24), has been linked to improved host defense (4, 11). Staphylococcal species, together with a high bacterial burden, has been associated with reduced survival in IPF (2, 4). NAC has been shown to reduce biofilm formation and enhance intracellular killing of Staphylococcus (37, 38), which may further enhance airway host defense in the presence of the rs35705950 polymorphism.
Despite the novel findings of this investigation, it has several limitations. First, this was an exploratory, post hoc secondary analysis of a clinical trial dataset with negative findings. As such, subjects were not randomized by genotype, which resulted in unequal genotype frequencies between treatment groups, and possibly biased the results. Second, adjustment for genetic ancestry was not possible because race and ethnicity were self-reported. Finally, <50% of individuals consented to genetic analysis in the PANTHER cohort, which significantly limited the power of this analysis, both in terms of interaction analysis and the ability to detect differences between individual clinical endpoints. The small number of events within our genotype-stratified models also introduced significant variability, leaving these models susceptible to large changes in point estimates, with a small change in the number of events. These observations made it unclear whether our findings would be consistent throughout the entire PANTHER cohort. No systematic baseline or endpoint differences were observed between genotyped and nongenotyped subjects (Table 1), and the nongenotype-adjusted composite endpoint risk associated with NAC therapy was not statistically significant in either group (see Table E8). Furthermore, replication of these findings helps assure this interaction is not unique to the PANTHER dataset. The weaker interaction observed in the replication cohort might be due in part to the advanced disease of those treated with NAC. The small sample size also limited our ability to use the additive allelic model for each SNP. An additive model may better characterize the dose-dependent nature of gene polymorphisms, and has been used to model genotypes in previous IPF genetic and genomic investigations (12, 14, 24, 39).
In conclusion, the genetic makeup of individuals with IPF may determine their response to NAC therapy. Despite the novelty of these findings, they should be viewed as hypothesis generating and as a rationale for a genotype-stratified randomized clinical trial to definitively answer this question. Replication of our findings in this manner would identify an inexpensive, readily available therapy for approximately 25% of individuals with IPF and determine whether NAC is detrimental to those carrying both rs3750920 ancestral alleles. Such a trial would also be an excellent opportunity to explore synergy between NAC and newly approved therapies for IPF, as has been recently suggested (19). Finally, our findings highlight the importance of pharmacogenetics in IPF and strongly support an effort to systematically acquire biospecimens from all individuals participating in clinical trials. Such practice may not only identify genetic subgroups likely to benefit from a particular therapy, but spare others significant side effects and adverse events.
Acknowledgments
Acknowledgment
The authors thank the Balbach family, whose donations made this investigation possible.
Footnotes
Supported by donations by the Balbach family, together with National Institutes of Health grants U10 HL080513 and T32 HL007605.
Author Contributions: J.M.O. contributed to the study design, analyzed the data, and wrote the manuscript. S.-F.M. contributed to the study design and performed the genotyping. F.J.M. contributed to the study design, interpretation of results, and manuscript preparation. K.J.A. assisted with data analysis and interpretation of results. G.R. contributed to the study design, interpretation of results, and manuscript preparation. D.A.S. contributed to interpretation of results and manuscript preparation. E.V., L.W., and C.L. collected data for the University of Chicago cohort analysis. R.V. contributed to the study design and manuscript preparation. Y.H. assisted with data analysis and interpretation of results. M.E.S. contributed to interpretation of results and manuscript preparation. I.N. designed the study and contributed to interpretation of results and manuscript preparation. All authors reviewed, revised, and approved the manuscript for submission.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201505-1010OC on September 2, 2015
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Tang YW, Johnson JE, Browning PJ, Cruz-Gervis RA, Davis A, Graham BS, Brigham KL, Oates JA, Jr, Loyd JE, Stecenko AA. Herpesvirus DNA is consistently detected in lungs of patients with idiopathic pulmonary fibrosis. J Clin Microbiol. 2003;41:2633–2640. doi: 10.1128/JCM.41.6.2633-2640.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Han MK, Zhou Y, Murray S, Tayob N, Noth I, Lama VN, Moore BB, White ES, Flaherty KR, Huffnagle GB, et al. COMET Investigators. Lung microbiome and disease progression in idiopathic pulmonary fibrosis: an analysis of the COMET study. Lancet Respir Med. 2014;2:548–556. doi: 10.1016/S2213-2600(14)70069-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hubbard R, Lewis S, Richards K, Johnston I, Britton J. Occupational exposure to metal or wood dust and aetiology of cryptogenic fibrosing alveolitis. Lancet. 1996;347:284–289. doi: 10.1016/s0140-6736(96)90465-1. [DOI] [PubMed] [Google Scholar]
- 4.Molyneaux PL, Cox MJ, Willis-Owen SA, Mallia P, Russell KE, Russell AM, Murphy E, Johnston SL, Schwartz DA, Wells AU, et al. The role of bacteria in the pathogenesis and progression of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2014;190:906–913. doi: 10.1164/rccm.201403-0541OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee SH, Kim DS, Kim YW, Chung MP, Uh ST, Park CS, Jeong SH, Park YB, Lee HL, Song JS, et al. Association between occupational dust exposure and prognosis of idiopathic pulmonary fibrosis: a Korean national survey. Chest. 2015;147:465–474. doi: 10.1378/chest.14-0994. [DOI] [PubMed] [Google Scholar]
- 6.Zhang G, Ghosh S. Negative regulation of toll-like receptor-mediated signaling by Tollip. J Biol Chem. 2002;277:7059–7065. doi: 10.1074/jbc.M109537200. [DOI] [PubMed] [Google Scholar]
- 7.Pruitt KD, Brown GR, Hiatt SM, Thibaud-Nissen F, Astashyn A, Ermolaeva O, Farrell CM, Hart J, Landrum MJ, McGarvey KM, et al. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 2014;42:D756–D763. doi: 10.1093/nar/gkt1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shah JA, Vary JC, Chau TT, Bang ND, Yen NT, Farrar JJ, Dunstan SJ, Hawn TR. Human TOLLIP regulates TLR2 and TLR4 signaling and its polymorphisms are associated with susceptibility to tuberculosis. J Immunol. 2012;189:1737–1746. doi: 10.4049/jimmunol.1103541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saito T, Yamamoto T, Kazawa T, Gejyo H, Naito M. Expression of toll-like receptor 2 and 4 in lipopolysaccharide-induced lung injury in mouse. Cell Tissue Res. 2005;321:75–88. doi: 10.1007/s00441-005-1113-9. [DOI] [PubMed] [Google Scholar]
- 10.Janardhan KS, McIsaac M, Fowlie J, Shrivastav A, Caldwell S, Sharma RK, Singh B. Toll like receptor-4 expression in lipopolysaccharide induced lung inflammation. Histol Histopathol. 2006;21:687–696. doi: 10.14670/HH-21.687. [DOI] [PubMed] [Google Scholar]
- 11.Roy MG, Livraghi-Butrico A, Fletcher AA, McElwee MM, Evans SE, Boerner RM, Alexander SN, Bellinghausen LK, Song AS, Petrova YM, et al. Muc5b is required for airway defence. Nature. 2014;505:412–416. doi: 10.1038/nature12807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noth I, Zhang Y, Ma S-F, Flores C, Barber M, Huang Y, Broderick SM, Wade MS, Hysi P, Scuirba J, et al. Genetic variants associated with idiopathic pulmonary fibrosis susceptibility and mortality: a genome-wide association study. Lancet Respir Med. 2013;1:309–317. doi: 10.1016/S2213-2600(13)70045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fingerlin TE, Murphy E, Zhang W, Peljto AL, Brown KK, Steele MP, Loyd JE, Cosgrove GP, Lynch D, Groshong S, et al. Genome-wide association study identifies multiple susceptibility loci for pulmonary fibrosis. Nat Genet. 2013;45:613–620. doi: 10.1038/ng.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peljto AL, Zhang Y, Fingerlin TE, Ma SF, Garcia JG, Richards TJ, Silveira LJ, Lindell KO, Steele MP, Loyd JE, et al. Association between the MUC5B promoter polymorphism and survival in patients with idiopathic pulmonary fibrosis. JAMA. 2013;309:2232–2239. doi: 10.1001/jama.2013.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Asehnoune K, Strassheim D, Mitra S, Kim JY, Abraham E. Involvement of reactive oxygen species in Toll-like receptor 4-dependent activation of NF-kappa B. J Immunol. 2004;172:2522–2529. doi: 10.4049/jimmunol.172.4.2522. [DOI] [PubMed] [Google Scholar]
- 16.De Flora S, Grassi C, Carati L. Attenuation of influenza-like symptomatology and improvement of cell-mediated immunity with long-term N-acetylcysteine treatment. Eur Respir J. 1997;10:1535–1541. doi: 10.1183/09031936.97.10071535. [DOI] [PubMed] [Google Scholar]
- 17.Imai Y, Kuba K, Neely GG, Yaghubian-Malhami R, Perkmann T, van Loo G, Ermolaeva M, Veldhuizen R, Leung YH, Wang H, et al. Identification of oxidative stress and Toll-like receptor 4 signaling as a key pathway of acute lung injury. Cell. 2008;133:235–249. doi: 10.1016/j.cell.2008.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Idiopathic Pulmonary Fibrosis Clinical Research Network. Martinez FJ, de Andrade JA, Anstrom KJ, King TE, Jr, Raghu G. Randomized trial of acetylcysteine in idiopathic pulmonary fibrosis. N Engl J Med. 2014;370:2093–2101. doi: 10.1056/NEJMoa1401739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakamoto S, Muramatsu Y, Satoh K, Ishida F, Kikuchi N, Sano G, Sugino K, Isobe K, Takai Y, Homma S. Effectiveness of combined therapy with pirfenidone and inhaled N-acetylcysteine for advanced idiopathic pulmonary fibrosis: a case-control study. Respirology. 2015;20:445–452. doi: 10.1111/resp.12477. [DOI] [PubMed] [Google Scholar]
- 20.Behr J, Kreuter M, Hoeper MM, Wirtz H, Klotsche J, Koschel D, Andreas S, Claussen M, Grohé C, Wilkens H, et al. Management of patients with idiopathic pulmonary fibrosis in clinical practice: the INSIGHTS-IPF registry. Eur Respir J. 2015;46:186–196. doi: 10.1183/09031936.00217614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Idiopathic Pulmonary Fibrosis Clinical Research Network. Raghu G, Anstrom KJ, King TE, Jr, Lasky JA, Martinez FJ. Prednisone, azathioprine, and N-acetylcysteine for pulmonary fibrosis. N Engl J Med. 2012;366:1968–1977. doi: 10.1056/NEJMoa1113354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.King TE, Jr, Albera C, Bradford WZ, Costabel U, Hormel P, Lancaster L, Noble PW, Sahn SA, Szwarcberg J, Thomeer M, et al. INSPIRE Study Group. Effect of interferon gamma-1b on survival in patients with idiopathic pulmonary fibrosis (INSPIRE): a multicentre, randomised, placebo-controlled trial. Lancet. 2009;374:222–228. doi: 10.1016/S0140-6736(09)60551-1. [DOI] [PubMed] [Google Scholar]
- 23.Oldham JM, Ma S-F, Vij R, Huang Y, Raghu G, Anstrom KJ, Martinez FJ, Noth I. Genetic heterogeneity among patients enrolled in the PANTHER-IPF clinical trial [abstract] Am J Respir Crit Care Med. 2015;191:A2162. [Google Scholar]
- 24.Seibold MA, Wise AL, Speer MC, Steele MP, Brown KK, Loyd JE, Fingerlin TE, Zhang W, Gudmundsson G, Groshong SD, et al. A common MUC5B promoter polymorphism and pulmonary fibrosis. N Engl J Med. 2011;364:1503–1512. doi: 10.1056/NEJMoa1013660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raghu G, Collard HR, Egan JJ, Martinez FJ, Behr J, Brown KK, Colby TV, Cordier JF, Flaherty KR, Lasky JA, et al. ATS/ERS/JRS/ALAT Committee on Idiopathic Pulmonary Fibrosis. An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med. 2011;183:788–824. doi: 10.1164/rccm.2009-040GL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collard HR, Brown KK, Martinez FJ, Raghu G, Roberts RS, Anstrom KJ IPFnet Investigators. Study design implications of death and hospitalization as end points in idiopathic pulmonary fibrosis. Chest. 2014;146:1256–1262. doi: 10.1378/chest.14-0492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raghu G, Collard HR, Anstrom KJ, Flaherty KR, Fleming TR, King TE, Jr, Martinez FJ, Brown KK. Idiopathic pulmonary fibrosis: clinically meaningful primary endpoints in phase 3 clinical trials. Am J Respir Crit Care Med. 2012;185:1044–1048. doi: 10.1164/rccm.201201-0006PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Durheim MT, Collard HR, Roberts RS, Brown KK, Flaherty KR, King TE, Jr, Palmer SM, Raghu G, Snyder LD, Anstrom KJ, et al. IPFnet investigators. Association of hospital admission and forced vital capacity endpoints with survival in patients with idiopathic pulmonary fibrosis: analysis of a pooled cohort from three clinical trials. Lancet Respir Med. 2015;3:388–396. doi: 10.1016/S2213-2600(15)00093-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt SL, Tayob N, Han MK, Zappala C, Kervitsky D, Murray S, Wells AU, Brown KK, Martinez FJ, Flaherty KR. Predicting pulmonary fibrosis disease course from past trends in pulmonary function. Chest. 2014;145:579–585. doi: 10.1378/chest.13-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smits KM, Schouten JS, Smits LJ, Stelma FF, Nelemans P, Prins MH. A review on the design and reporting of studies on drug-gene interaction. J Clin Epidemiol. 2005;58:651–654. doi: 10.1016/j.jclinepi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 33.Wu L, Cook RJ. Misspecification of Cox regression models with composite endpoints. Stat Med. 2012;31:3545–3562. doi: 10.1002/sim.5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei LJ, Lin DY, Weissfeld L. Regression analysis of multivariate incomplete failure time data by modeling marginal distributions. J Am Stat Assoc. 1989;84:1065–1073. [Google Scholar]
- 35.Jiang D, Liang J, Fan J, Yu S, Chen S, Luo Y, Prestwich GD, Mascarenhas MM, Garg HG, Quinn DA, et al. Regulation of lung injury and repair by Toll-like receptors and hyaluronan. Nat Med. 2005;11:1173–1179. doi: 10.1038/nm1315. [DOI] [PubMed] [Google Scholar]
- 36.Santiago FM, Bueno P, Olmedo C, Muffak-Granero K, Comino A, Serradilla M, Mansilla A, Villar JM, Garrote D, Ferrón JA. Effect of N-acetylcysteine administration on intraoperative plasma levels of interleukin-4 and interleukin-10 in liver transplant recipients. Transplant Proc. 2008;40:2978–2980. doi: 10.1016/j.transproceed.2008.08.103. [DOI] [PubMed] [Google Scholar]
- 37.Pérez-Giraldo C, Rodríguez-Benito A, Morán FJ, Hurtado C, Blanco MT, Gómez-García AC. Influence of N-acetylcysteine on the formation of biofilm by Staphylococcus epidermidis. J Antimicrob Chemother. 1997;39:643–646. doi: 10.1093/jac/39.5.643. [DOI] [PubMed] [Google Scholar]
- 38.Oddera S, Silvestri M, Sacco O, Eftimiadi C, Rossi GA. N-acetylcysteine enhances in vitro the intracellular killing of Staphylococcus aureus by human alveolar macrophages and blood polymorphonuclear leukocytes and partially protects phagocytes from self-killing. J Lab Clin Med. 1994;124:293–301. [PubMed] [Google Scholar]
- 39.Zhang Y, Noth I, Garcia JG, Kaminski N. A variant in the promoter of MUC5B and idiopathic pulmonary fibrosis. N Engl J Med. 2011;364:1576–1577. doi: 10.1056/NEJMc1013504. [DOI] [PMC free article] [PubMed] [Google Scholar]