Abstract
Objective
Farnesoid X receptor (FXR) plays a prominent role in hepatic lipid metabolism. The FXR gene encodes four proteins with structural differences suggestive of discrete biological functions about which little is known.
Methods
We expressed each FXR variant in primary hepatocytes and evaluated global gene expression, lipid profile, and metabolic fluxes. Gene delivery of FXR variants to Fxr−/− mouse liver was performed to evaluate their role in vivo. The effects of fasting and physical exercise on hepatic Fxr splicing were determined.
Results
We show that FXR splice isoforms regulate largely different gene sets and have specific effects on hepatic metabolism. FXRα2 (but not α1) activates a broad transcriptional program in hepatocytes conducive to lipolysis, fatty acid oxidation, and ketogenesis. Consequently, FXRα2 decreases cellular lipid accumulation and improves cellular insulin signaling to AKT. FXRα2 expression in Fxr−/− mouse liver activates a similar gene program and robustly decreases hepatic triglyceride levels. On the other hand, FXRα1 reduces hepatic triglyceride content to a lesser extent and does so through regulation of lipogenic gene expression. Bioenergetic cues, such as fasting and exercise, dynamically regulate Fxr splicing in mouse liver to increase Fxrα2 expression.
Conclusions
Our results show that the main FXR variants in human liver (α1 and α2) reduce hepatic lipid accumulation through distinct mechanisms and to different degrees. Taking this novel mechanism into account could greatly improve the pharmacological targeting and therapeutic efficacy of FXR agonists.
Keywords: FXR isoforms, Splicing, NAFLD, Insulin resistance, Energy metabolism
Highlights
-
•
FXR variants regulate discrete gene programs with distinct biological outcomes.
-
•
FXRα2 (but not α1) enhances fatty acid handling and insulin responsiveness.
-
•
FXRα1 and α2 reduce liver lipid content through different mechanisms.
-
•
Fasting and physical exercise dynamically regulate Fxr splicing in liver.
1. Introduction
The regulation of hepatic energy storage and metabolism is essential for an appropriate adaptation to fluctuating metabolic demands. Impaired control over hepatic lipid homeostasis can lead to excessive lipid accumulation, a prominent feature of non-alcoholic fatty liver disease (NAFLD), the most prevalent liver disease in Western societies [1]. NAFLD is considered the hepatic hallmark of the metabolic syndrome and is strongly associated with hepatic insulin resistance and type 2 diabetes. Farnesoid X receptor (FXR) is a bile acid-activated nuclear receptor abundantly expressed in liver [2], [3]. FXR expression is markedly decreased in obese rodents and NAFLD patients [4], [5], [6]. Genetic FXR ablation promotes hepatic steatosis and hyperlipidemia [7], [8], whereas hepatic FXR expression and activation decrease local and circulating triacylglycerol (TAG) levels and improve glucose homeostasis [9], [10].
Four biologically active variants are expressed from the FXR gene (FXRα1 through 4), resulting from alternative promoter usage and mRNA splicing (Figure 1A) [11], [12]. FXR isoforms display distinct developmental and tissue-specific expression patterns and are differentially regulated by metabolic cues [11], [13], [14]. FXRα3 and α4 possess extended N-termini with an A/B domain different from the one in FXRα1 and α2. In addition, FXRα1 and α3 have a four amino acid insert (MYTG) in the hinge region. The structural disparity between FXR variants has functional implications that may influence the therapeutic outcome of pan-FXR agonists. Indeed, FXR agonists promote distinct gene expression profiles in hepatocytes, although the contribution of FXR isoform-specificity remains unknown [15]. Structural differences in the A/B domain of FXR isoforms modulate coactivator interaction [16], whereas the MYTG insert affects DNA-binding and target gene specificity [11], [14], [17], [18]. For example, the human bile salt export pump (BSEP) is regulated by FXRs in an isoform-dependent manner, predominantly by FXRα2 [14], [18]. Notably, hepatic FXRα2/α1 ratios and BSEP levels are decreased in hepatocellular carcinoma patients and by pro-inflammatory stimulation [14]. To date, the regulation of FXR splicing and its implications on hepatic energy metabolism remain unknown.
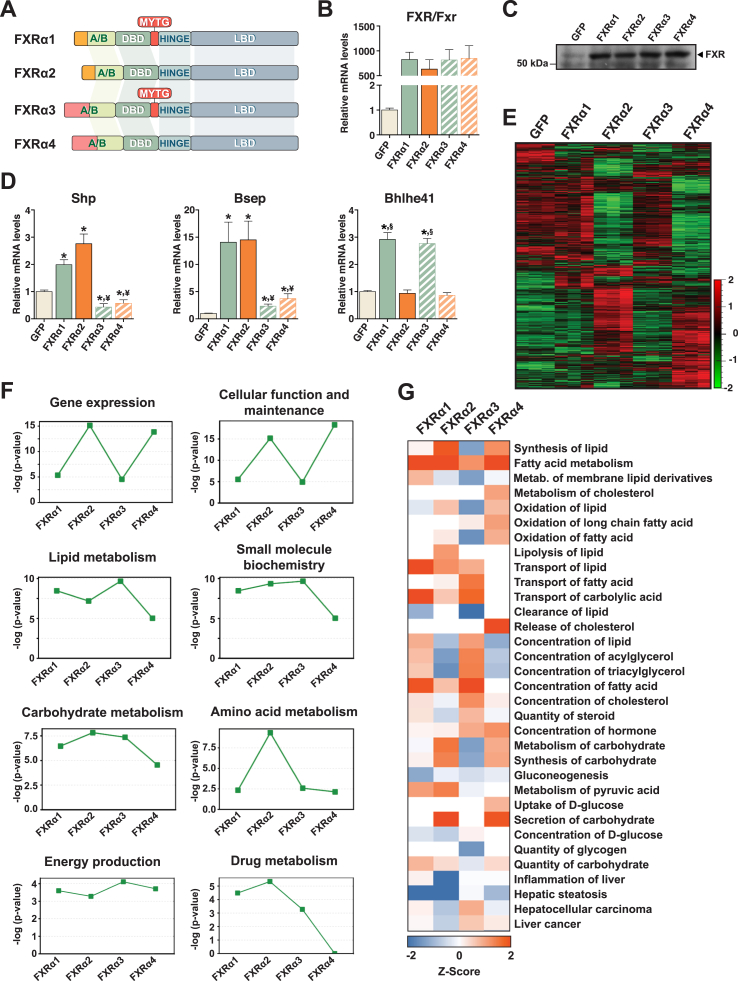
Figure 1.
FXR isoform-specific gene regulation. (A) Human FXR isoforms. The different N-termini (orange in FXRα1/α2; pink in FXRα3/α4) and the MYTG insert in FXRα1/α3 are indicated. DBD, DNA-binding domain; LBD, ligand-binding domain. (B–C) FXR mRNA (B) and protein (C) levels in hepatocytes expressing each FXR variant or GFP alone (control). (D) Gene expression analysis by qRT-PCR. (E) Heatmap of gene expression profiles in hepatocytes expressing each FXR isoform or GFP (F) Bioinformatic pathway analysis of gene expression profiling. Represented are biological functions significantly associated with one or more FXR isoforms. (G) Predicted effects on hepatic metabolism for each FXR isoform. Z-score values indicate induction (orange) or repression (blue). Bars depict mean values and error bars represent SEM (n ≥ 3). *p < 0.05 vs. control. ¥p < 0.05 vs. FXRα1/α2. §p < 0.05 vs. FXRα1/α3.
Here, we show that FXR variants regulate hepatic lipid metabolism in an isoform-dependent manner. This constitutes a novel mechanism by which alternative FXR splicing in the liver integrates systemic energetic demands with a gene program of enhanced lipid handling/utilization and ketogenesis that positively impacts hepatic steatosis and insulin sensitivity.
2. Materials and methods
2.1. Generation of recombinant adenoviruses
Human FXRα1, α2, α3, and α4 were amplified by PCR and cloned into the NotI/XhoI sites of a pcDNA3.1 plasmid (Life Technologies) containing a FLAG tag between the BamHI and NotI sites. The cDNAs encoding each FLAG-tagged FXR were subcloned into the pAdTrack-CMV vector (Stratagene). Adenoviruses expressing each FXR variant were generated using the AdEasy adenoviral vector system (Stratagene), amplified in Ad-293 cells and purified by CsCl gradient centrifugation as previously described [19]. All adenoviruses generated also express GFP from an independent CMV promoter. An adenovirus expressing only GFP was used as a control in all experiments.
2.2. Cell culture, transduction, and treatments
Primary hepatocytes from male C57BL/6J mice were isolated, cultured and transduced as previously described [20]. Unless otherwise stated, cells were processed for downstream analysis 36 h post-transduction. For insulin treatment, cells were incubated in 5 mM glucose DMEM for 2 h prior to stimulation with 80 nM insulin for 20 min. For agonist treatments, hepatocytes were treated with 250 μM CA for 24 h following an 8–10 h transduction.
2.3. Gene expression analysis
Total RNA was isolated from frozen tissues or cultured cells, DNase-treated, and reverse transcribed. Gene expression was analyzed using Applied Biosystems' Power SYBR Green PCR Master Mix and ViiA 7 Real-Time PCR System. Gene expression was normalized to hypoxanthine phosphoribosyltransferase 1 (Hprt) expression and expressed relative to experimental controls. Primer sequences are listed in Table S1. Microarray analysis was performed using Affymetrix Mouse Gene 1.1 ST Array and subsequent data analysis was conducted using Qlucore Omics Explorer, Partek Genomics and Ingenuity Pathway Analysis software suites. Affymetrix gene expression profiling experiments were performed at the Bioninformatics and Gene expression Analysis (BEA) core facility of the Karolinska Institutet (www.bea.ki.se). Microarray data are deposited at GEO with the accession number GSE73035.
2.4. Western blot analysis
Protein extracts were prepared in RIPA buffer (50 mM Tris–HCl pH 7.4, 150 mM NaCl, 0.1% SDS, 1% NP-40, 1% Na deoxycholate, 1 mM EDTA) supplemented with 1 mM DTT, 0.5 mM PMSF, and 1 mM sodium orthovanadate. Protein extracts (50 μg) were resolved by SDS polyacrylamide gel electrophoresis and transferred into polyvinylidene difluoride membranes. Immunoblotting was performed with antibodies against FXR (Santa Cruz Biotechnologies), AKT (Cell Signaling), and phospho-AKT (Ser473) (Cell Signaling), diluted in 0.1% BSA. Quantification was performed using ImageJ [21].
2.5. Neutral lipid staining
Hepatocytes were fixed with 4% paraformaldehyde for 10 min at room temperature. After 3 washes with PBS, cells were incubated for 2 min in 60% isopropanol and stained with freshly prepared Oil Red-O solution (0.4% in 60% isopropanol) for 30 min at room temperature. Cells were washed with PBS, incubated for 2 min with 300 nM 4′,6-diamidino-2-phenylindole, dihydrochloride (DAPI) (Life Technologies), and washed with PBS again. Images were taken at a 200× magnification. Total red and blue pixels were quantified using the Color Range Select tool in Adobe Photoshop CS5.1.
2.6. Lipid profiling
Hepatocytes were incubated with 0.2 μCi/mL [U-14C] palmitate (Perkin Elmer, USA) and 25 nM non-radioactive palmitate for 4 h. Lipids were extracted, separated by thin layer chromatography, detected by autoradiography, and quantified as previously described [22].
2.7. FA oxidation assay
Hepatocytes were incubated in DMEM supplemented with 1 g/L glucose, 12.5 μM BSA-conjugated palmitic acid (Sigma Aldrich), and 2.5 μCi palmitic acid [9,10-3H(N)] (Perkin Elmer, USA). Four hours later, 0.2 mL of media were collected to 2 mL tubes containing 0.8 mL of activated charcoal slurry. Samples incubated at room temperature for 30 min with frequent mixing. After centrifugation at 13,000 rpm for 15 min, 0.2 mL of supernatant were transferred to 4 mL scintillation vials containing 2.8 mL of Ultima FLO M scintillation liquid (Perkin Elmer, USA) and measured in a Wallac Winspectral 1414 liquid scintillation counter. Cells were lysed in 0.03% SDS and protein concentration was determined for each lysate. The FA oxidation rate was normalized by cellular protein content.
2.8. Metabolite analysis by mass spectrometry
Metabolite analysis was done according to standard mass spectrometry procedures [23]. Briefly, metabolites were isolated from N2 frozen hepatocytes by a FastPrep (3 × 6.5 M/s for 60 s) in a MeOH-Chlorophorm-H2O suspension according to Wu et al. [24]. Extracted metabolites were mixed with an internal standard consisting of chloramphenicol; l-proline 1,2,3,4,5–13C5-, 2-15N-; l-glutamine 13C5; l-arginine C13N15; uracil 1,3–15N2-, 2-13C- and l-valine, 2,3,4,4,4,5,5,5-d8-, 100 μM each. Three MRM's of each metabolite were run as triplicates on an online coupled LC-MS/MS system (1290 Infinity UHPLC, Agilent, USA; QTrap 6500, ABSciex, Canada) featuring a Reprosil-PUR C18-AQ (1.9 μm, 120 Å, 150 × 2 mm ID; Dr. Maisch, Germany) column and a zicHILIC (3.5 μm, 100 Å, 150 × 2.1 mm ID; di2chrom, Germany) column at 30 °C. Transitions were monitored and acquired at low resolution in quadrupole Q1 and unit resolution in Q3. Data acquisition was performed with an ion spray voltage of 5.5 kV in positive and 4.5 kV in negative mode of the ESI source, N2 as collision gas was set to high, curtain gas to 30 psi, ion source gas 1 and 2 to 50 and 70 psi, and an interface heater temperature of 350 °C, operated with AB Sciex Analyst 1.6.1 software with components for 6500 series instruments. Quantification was performed using MultiQuantTM software v.2.1.1 (AB Sciex, USA) using appropriate internal controls for technical validation. Integration settings were a Gaussian smooth width of 2 points and a peak splitting factor of 2. Peak integrations were reviewed manually and subsequent data analysis was done using Ingenuity Pathway Analysis software suite.
2.9. Glucose and β-OHB output assays
Glucose output was measured as previously described [25]. For β-OHB output, media was changed to phenol red-free DMEM (5 g/L glucose) 24 h post-transduction and cells incubated for 16 h. Media was collected and β-OHB levels were measured with the β-OHB Assay Kit (Abcam) according to the manufacturer's instructions. Both glucose and β-OHB levels were normalized by cellular protein content (extracted in 0.03% SDS).
2.10. Animal experimentation
C57BL/6J male mice were purchased from Charles River Laboratories (Sulzfeld, Germany) and housed in a 12 h light/dark cycle with free access to food and water. Unless otherwise stated, we used 9–10 week-old male C57BL/6J mice for all experiments. In all experiments, mice were euthanized by cervical dislocation and tissues were rapidly harvested and snap-frozen in liquid nitrogen. All experimental protocols were approved by the Regional Animal Ethics Committee of Northern Stockholm or the Ethical Committee for Animal Experiments of the University of Groningen.
2.10.1. Fasting/Refeeding
Mice (n = 5) were fasted for 14 h, beginning at 17:00. Mice in the refeeding group (n = 7) were fasted at the same time but given free access to food for 3 h following fasting. The respective controls (n = 5 each) were euthanized at the same time but had continuous access to food.
2.10.2. High fat diet
Mice (n = 5 each) were fed normal chow or a high fat diet (TD.93075, Harlan, USA) for one week.
2.10.3. Treadmill running
Mice (n = 6) did not follow any exercise protocol. Treadmill runners (n = 8) followed the acclimation protocol detailed in Table S2. Acclimation and running sessions began at 18:30. After acclimation, mice were placed on a motorized treadmill (Columbus Instruments, USA), with a slope of 5% and warmed-up for 5 min at 5 m/min. The speed was increased (5 m/min every 5 min) up to 20 m/min. Speeds varied between 15 and 20 m/min during the experiment to allow mice to keep up with the protocol. Mice ran for 1 h or until exhaustion. We considered exhaustion as the total incapacity to keep up with the running pace even when the speed was lowered to 15 m/min. Mice were euthanized immediately after running.
2.10.4. Chronic exercise
Mice (n = 6 each) were given free access to a running wheel for 8 weeks as previously described [26].
2.10.5. Adenovirus-mediated gene delivery to mouse liver
Age-matched, male whole-body Fxr−/− mice [27] on a C57BL/6J background received 5 × 109 adenoviral particles in PBS into the retro-orbital sinus (n = 5–6 per group). Five days post-transduction, mice were fasted for 8 h prior to termination.
2.11. Analytical procedures
Plasma TAG and β-OHB levels were determined using commercially available kits (Abcam). Liver TAGs were extracted in 5% NP-40 and quantified with the same kit (Abcam).
2.12. Quantification of Fxrα2/Fxrα1 relative expression
The method used to quantify the Fxrα2/Fxrα1 ratios was based on the one described by Chen, Y et al. [14]. A fragment from Fxrα1+Fxrα2 was amplified by PCR using a forward primer specific for Fxrα1/α2 (5′-TCTCTGGCCCAAAGCAATCCAA-3′) and a reverse primer specific for Fxr (all isoforms; 5′-TGAAAATCTCCGCCGAACGA-3′). The amplified fragment encompasses the 12 bp insert present in Fxrα1 (but not α2), which creates a BstZ17I restriction site that discriminates Fxrα1 from α2. The amplification products were digested with BstZ17I and the resulting fragment pattern was resolved by agarose electrophoresis and quantified using ImageJ [21].
2.13. Statistical analysis
Statistical analysis was performed using Prism software. Two-tailed, unpaired Student's t-test was used to analyze statistical differences between groups. Statistical significance was defined as p < 0.05
3. Results
3.1. FXR isoform-specific target gene regulation
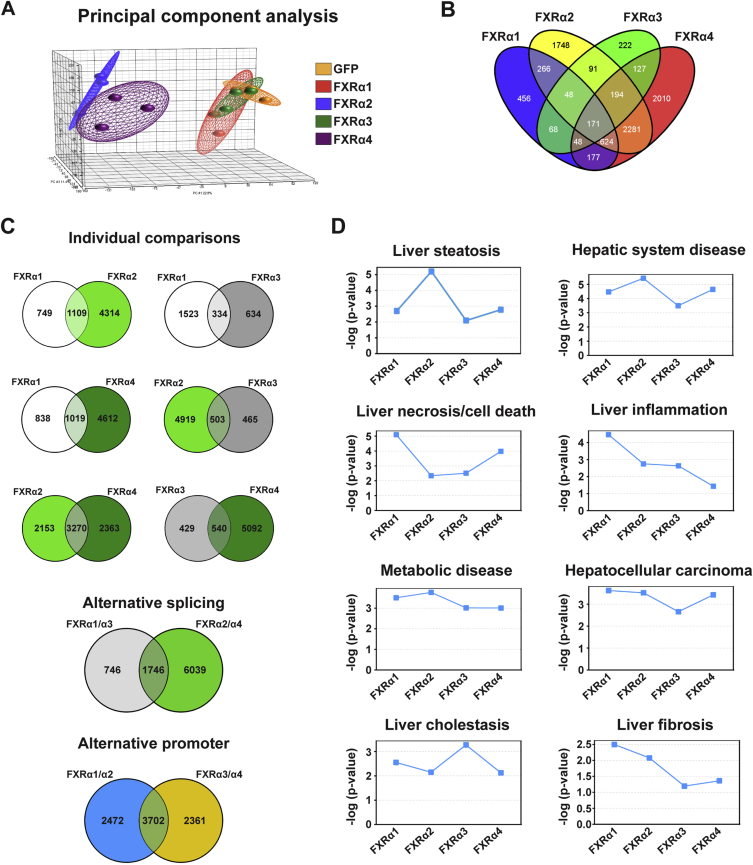
FXR variants possess structural differences suggestive of non-overlapping biological functions (Figure 1A). To investigate the effect(s) of such variation on FXR-mediated transcriptional regulation and hepatic metabolism, we expressed each human FXR isoform in mouse primary hepatocytes. To this end, we generated recombinant adenovirus expressing from independent promoters GFP and each human FXR isoform, or GFP alone (control). Transduction of hepatocytes with the different adenovirus elevated total FXR mRNA and protein to similar levels (Figure 1B,C). Remarkably, even well characterized FXR target genes, including small heterodimer partner (Shp) or Bsep [28], [29], were regulated in an isoform-dependent manner (Figure 1D). The expression of basic helix-loop-helix family member e41 (Bhlhe41/Dec2/Sharp1), shown to repress FXR transactivation [30], was increased by isoforms containing the MYTG motif in the hinge region (FXRα1 and α3) but not by variants lacking this feature (FXRα2 and α4). To evaluate the transcriptional networks regulated by each FXR isoform, we profiled global gene expression in hepatocytes expressing each FXR variant. By comparing genes regulated by each FXR isoform with the GFP control, we generated the heat map presented in Figure 1E and the principal component analysis plot in Figure S1A. Both analyses indicated that FXR isoforms lacking the MYTG motif (α2 and α4) regulated much larger gene sets than their MYTG-containing counterparts. Strikingly, from over 8,500 genes regulated by FXRs, only 171 genes were coregulated by all isoforms (Figure S1B). Gene set comparison revealed that together FXRα2 and α4 modulate the expression of ∼7,800 genes, 42% of which are coregulated by both variants (Figure S1C). Bioinformatic pathway analysis identified significant associations between all FXR variants and key hepatic metabolic functions and pathologies (Figure 1F and S1D). Further analysis of the gene expression data, taking into consideration whether a particular pathway or function was predicted to be activated or repressed, suggested contrasting effects of FXR splice variants on hepatic lipid metabolism (Figure 1G). The transcriptional programs driven by FXRα2 and α4 were predicted to increase lipid/fatty acid (FA) oxidation and decrease lipid/acylglycerol concentration, whereas the opposite effect was predicted for the gene programs regulated by FXRα1 and α3. These results show that FXR splice variants regulate distinct transcriptional programs that can impact hepatic energy metabolism.
3.2. FXRα2 and α4 reduce hepatocyte lipid accumulation and increase insulin responsiveness
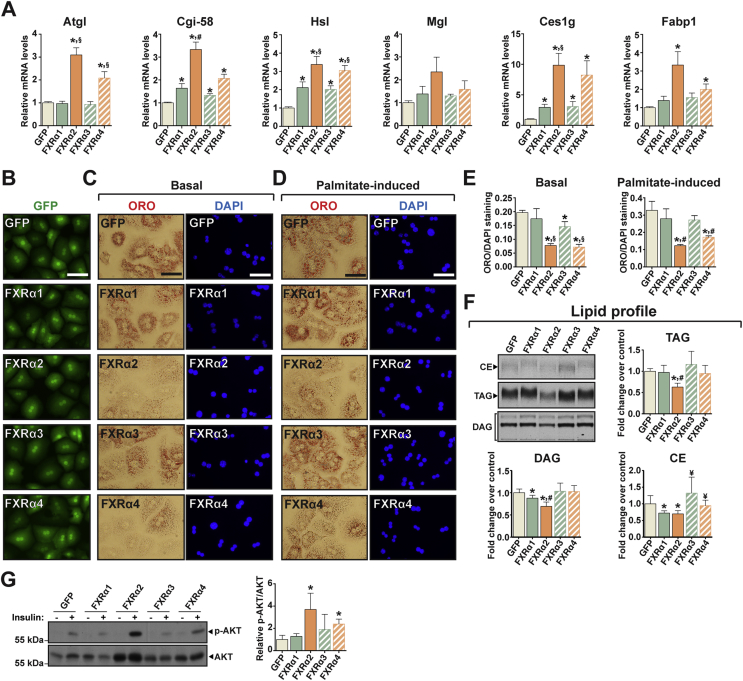
Gene expression profiling suggested that FXR splicing variants may have contrasting effects on hepatic lipid metabolism, with possible repercussions on intracellular lipid accumulation. To pinpoint the gene program(s) mediating the putative effects of FXRs on hepatic fat storage, we analyzed the expression of key mediators of liver lipid metabolism. We observed minor isoform specificity over lipogenic gene expression (Figure S2A). However, FXRα2 and α4 increased the expression of several hepatic lipases (Figure 2A). These include adipose triglyceride lipase (Atgl), hormone-sensitive lipase (Hsl), and carboxylesterase 1 g (Ces1g), together with Atgl's coactivator comparative gene identification 58 (Cgi-58), all shown to decrease hepatic lipid accumulation [31], [32], [33]. In addition, these FXR variants also increased the expression FA binding protein 1 (Fabp1), which regulates intracellular FA trafficking and reduces free fatty acid (FFA) lipotoxicity [34]. To determine whether the changes in gene expression elicited by FXRα2 and α4 effectively translate into a biological outcome of reduced lipid accumulation, we evaluated intracellular lipid storage in hepatocytes expressing each FXR isoform or GFP alone. This revealed a robust reduction in lipid accumulation only in hepatocytes expressing FXRα2 and α4 (Figure 2B,C). A sustained increase in circulating FFA levels can lead to hepatic steatosis and insulin resistance [35]. To investigate whether FXRs exert a protective effect against hepatic lipid accumulation upon elevated FFA supply, we treated hepatocytes expressing each FXR variant with palmitate. Indeed, FXRα2 and α4 largely protected hepatocytes against palmitate-induced lipid accumulation, reducing neutral lipid staining by 62% and 47%, respectively (Figure 2B,C). To determine changes in the lipid profile of hepatocytes expressing each FXR variant, we used 14C-palmitate thin layer chromatography (Figure 2D). In line with the reduced lipid content, FXRα2 decreased 14C-palmitate incorporation into TAG and diacylglycerol (DAG). This is unlikely to stem from reduced palmitate uptake or DAG/TAG biosynthesis, since the main FA transport proteins and TAG biosynthesis enzymes were expressed to similar or higher levels in hepatocytes expressing FXRα2 (Figure S2B) On the other hand, FXRα1 induced a small reduction in radiolabeled DAG content without affecting radiolabeled TAG levels. Both FXRα1 and α2 decreased radiolabeled cholesteryl ester levels, which tended to increase in cells expressing FXRα3 (32%, p = 0.17). Under these conditions, FXRα4 did not affect 14C-palmitate incorporation into any of the lipid species analyzed. Hepatic lipid accumulation has been extensively implicated in the development of hepatic insulin resistance [35], [36], which prompted us to investigate possible FXR effects on insulin sensitivity. In line with decreased intracellular lipid content and radiolabeled DAG and TAG levels, FXRα2 expression increased insulin-stimulated AKT phosphorylation, indicative of enhanced insulin sensitivity (Figure 2E). FXRα4 expression also resulted in enhanced insulin-stimulated AKT phosphorylation, although to a lesser extent than FXRα2. These results show that FXRα2 (and to a lesser extent FXRα4) exert a positive effect on cellular insulin signaling to AKT, likely due to reduced lipid accumulation.
Figure 2.
FXRα2 and FXRα4 reduce hepatocyte lipid content and increase insulin sensitivity. (A–G) Hepatocytes were transduced with adenoviruses expressing GFP alone (control) or together with each FXR variant. (A) Gene expression analysis by qRT-PCR. (B) Transduction monitoring by fluorescence microscopy. Scale bar, 50 μm. (C–E) Analysis of lipid accumulation by Oil Red O (ORO) staining. Hepatocytes were cultured without (C) or with (D) palmitate treatment and stained for neutral lipids (ORO, red) and genomic DNA (DAPI, blue), followed by quantification (E). Scale bar, 50 μm. (F) Lipid profiling by TLC. (G) Immunoblot for total AKT and p-AKT (ser473) in response to insulin. Bars depict mean values and error bars represent SEM (n ≥ 3). *p < 0.05 vs. control. #p < 0.05 vs. all other isoforms. ¥p < 0.05 vs. FXRα1/α2. §p < 0.05 vs. FXRα1/α3.
3.3. FXRα2 promotes FA oxidation and ketogenesis
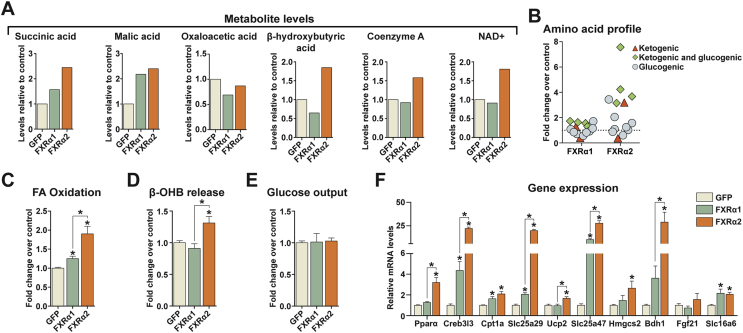
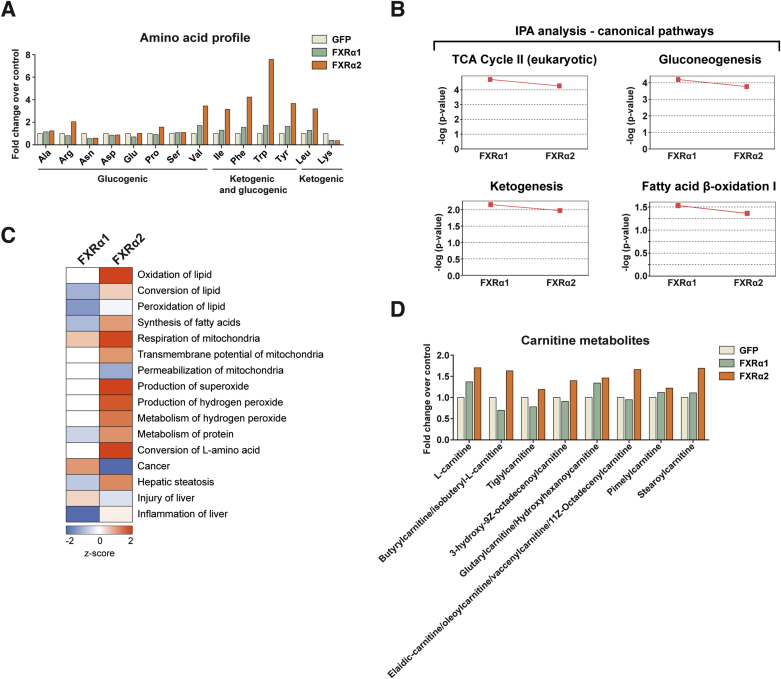
The FXR isoform-specific effects on hepatic lipid metabolism correlate with the presence or absence of the splicing-regulated hinge region MYTG motif. Since FXRα1 and α2 are the main isoforms in human liver [11], [14], we focused on these variants and further investigated their role in hepatic metabolism. To better understand their effects on cellular metabolic fluxes, we performed unbiased metabolomics analysis of hepatocytes expressing FXRα1, FXRα2, or GFP alone. We observed increased levels of tricarboxylic acid (TCA) cycle intermediaries (succinic acid and malic acid) in hepatocytes expressing FXRα2 and to a lesser extent FXRα1 (Figure 3A). Furthermore, FXRα2 expression increased intracellular β-hydroxybutyric acid content, whereas FXRα1 had the opposite effect. Free coenzyme A and NAD+, two byproducts of the ketogenesis pathway, were also increased by FXRα2, but not α1. Adding to this, FXRα2 promoted marked changes in the hepatocyte amino acid profile, particularly in amino acids involved in ketone body production (Appendix A, Figure 3A). From bioinformatic pathway analysis we identified significant associations between both FXR variants and metabolic pathways with predominant roles in hepatic bioenergetics, including TCA cycle, gluconeogenesis, ketogenesis, and FA β-oxidation (Figure S3B). Furthermore, this analysis predicted increased lipid oxidation, mitochondrial respiration, and reactive oxygen species production/metabolism in hepatocytes expressing FXRα2 (Figure S3C). Thus, our metabolomics analysis suggested that the gene program activated by FXRα2 leads to increased FA oxidation and that the acetyl-CoA generated is not only channeled through the TCA cycle but also directed towards ketone body production. In agreement, we could verify that FXRα2 expression increased cellular FA oxidation (Figure 3C) and β-hydroxybutyrate (β-OHB) release (Figure 3D), whereas FXRα1 enhanced FA oxidation only marginally and did not affect β-OHB release. Glucose output was not affected by either FXR variant (Figure 3E). Consistent with these results, FXRα2 activated the expression of key transcriptional regulators of FA metabolism, namely peroxisome proliferator-activated receptor-α (Pparα) and cAMP-responsive element binding protein 3-like 3 (Creb3l3) (Figure 3F). Moreover, hepatocytes expressing FXRα2 had increased levels of several acylcarnitines (Figure S3D), accompanied by elevated carnitine palmitoyltransferase 1a (Cpt1a) and carnitine/acylcarnitine translocase-like (Slc25a29) expression (Figure 3F). FXRα2 also induced the expression of the mitochondrial uncoupling proteins Ucp2 and Slc25a47, as well as the major ketogenic enzymes 3-hydroxy-3-methylglutaryl-CoA synthase 2 (Hmgcs2) and 3-hydroxybutyrate dehydrogenase type 1 (Bdh1). On the other hand, FXRα1 activated the transcription of some of the above-mentioned genes, but to a much lesser extent (Figure 3F). Taken together, our results indicate that FXRα2 (but not α1) activates a comprehensive gene program that promotes mobilization cellular lipid stores for β-oxidation and, at least in part, ketone body production.
Figure 3.
FXRα2 promotes FA oxidation and β-OHB production. (A–F) Hepatocytes were transduced with GFP alone (control) or together with FXRα1 or FXRα2. (A–B) Metabolomics analysis. Metabolite levels (A) and amino acid profile (B) determined by mass spectrometry. Graphs show metabolite levels in cells expressing FXRα1 or FXRα2 relative to control. Amino acids are grouped according to their potential for gluconeogenesis (blue), ketogenesis (red), or both (green) (n = 1). (C–E) Metabolic fluxes. FA oxidation (C), β-OHB release (D), and glucose output (E). (F) Gene expression analysis by qRT-PCR of key mediators of hepatic FA oxidation, mitochondrial uncoupling and ketogenesis. Bars depict mean values and error bars represent SEM (n ≥ 3). *p < 0.05 compared to control (GFP) or between the indicated groups.
3.4. Changing FXRα2/α1 ratios promotes transcriptional reprogramming
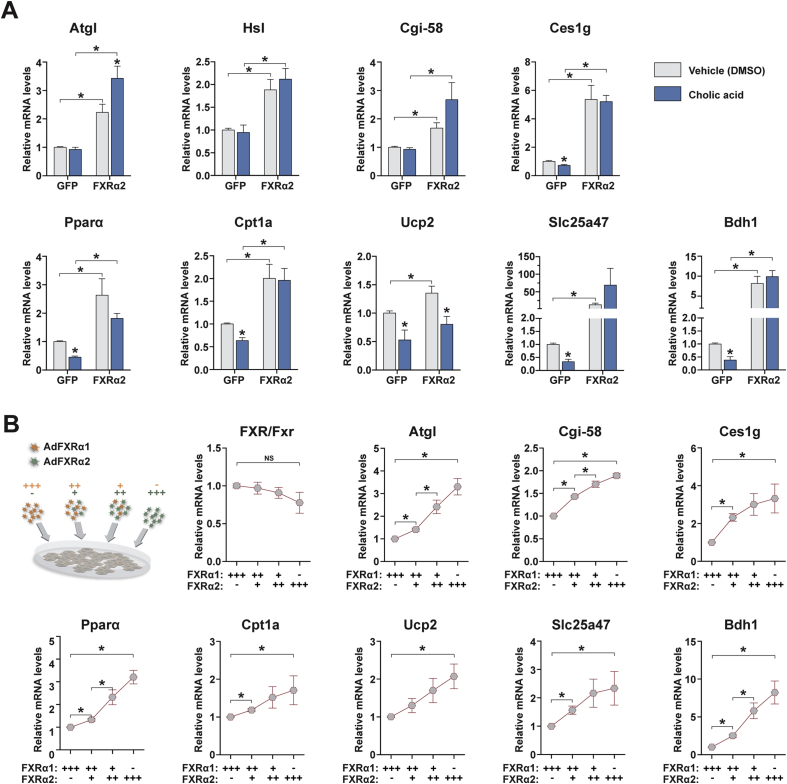
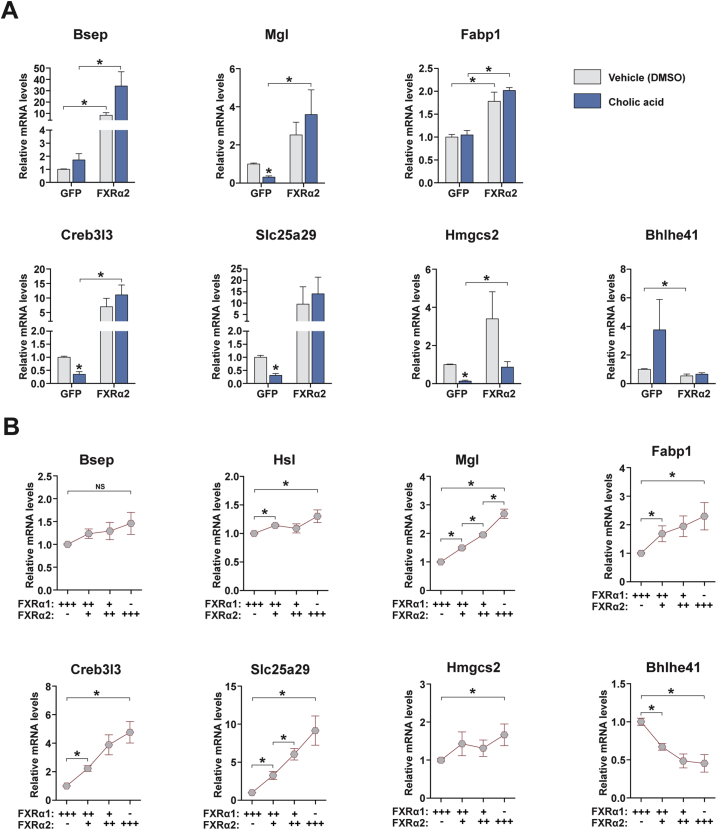
Bile acids function as endogenous FXRs ligands and greatly increase their capacity to transcriptionally activate a vast array of target genes [2], [3]. However, alternative endogenous ligands and ligand-independent FXR activity have also been reported [13], [37]. To investigate whether bile acids affect the gene program under FXRα2 control, we treated hepatocytes expressing FXRα2 or GFP alone with 250 μM cholic acid (CA), a concentration shown to yield moderate FXR activation [38]. CA treatment in GFP-transduced cells decreased the expression of most FXRα2 target genes involved in lipolysis, FA oxidation, mitochondrial uncoupling, and ketogenesis (Figure 4A and Figure S4A). The exceptions were Atgl, Hsl, Cgi-58, and Fabp1, whose expression was not affected by CA treatment. In contrast, when FXRα2 expression was increased, only Ucp2, Hmgcs2 and to a lesser extent Pparα displayed reduced mRNA levels upon CA treatment, when compared to vehicle-treated cells. For all other genes analyzed, expression was unchanged or in some cases (e.g. Atgl and Slc25a47) enhanced by CA treatment. Interestingly, increased FXRα2 levels blunted the CA-mediated effects on Bhlhe41 expression.
Figure 4.
Changing FXRα2/α1 ratios promotes transcriptional reprogramming. (A) Target gene expression analysis by qRT-PCR in hepatocytes expressing GFP or FXRα2 and treated with vehicle (DMSO) or 250 μM cholic acid (CA) for 24 h. Bars depict mean values and error bars represent SEM (n = 5). *p < 0.05 compared to vehicle-treated or between the indicated groups. (B) Gene expression analysis by qRT-PCR in hepatocytes co-transduced with FXRα1 and FXRα2 in different proportions (+++, 100%; ++, 70%; +, 30%; −, 0%). Graphs represent expression levels relative to hepatocytes transduced with FXRα1 alone. Graphs depict mean values and error bars represent SEM (n = 5). *p < 0.05 compared to FXRα1 alone or between the indicated groups.
In addition to ligand-mediated activation, the relative expression of FXR splice isoforms has important implications on hepatic metabolism [11], [14], [18]. To verify whether changing FXRα2/α1 ratios is sufficient to promote transcriptional reprogramming, we co-expressed different proportions of FXRα1:FXRα2 in hepatocytes. For clarity, the relative amounts of FXRα1 and α2 adenoviral particles used are illustrated in Figure 4B. While maintaining similar total FXR expression levels, increasing FXRα2/α1 ratios gradually induced the FXRα2-mediated gene program conducive to increased lipolysis, FA oxidation and ketogenesis (Appendix A, Figure 4B). Bhlhe41 expression showed the opposite pattern. These data indicate that changes in FXR isoform composition through alternative splicing can promote transcriptional reprogramming and alter hepatic lipid handling and ketogenic capacity.
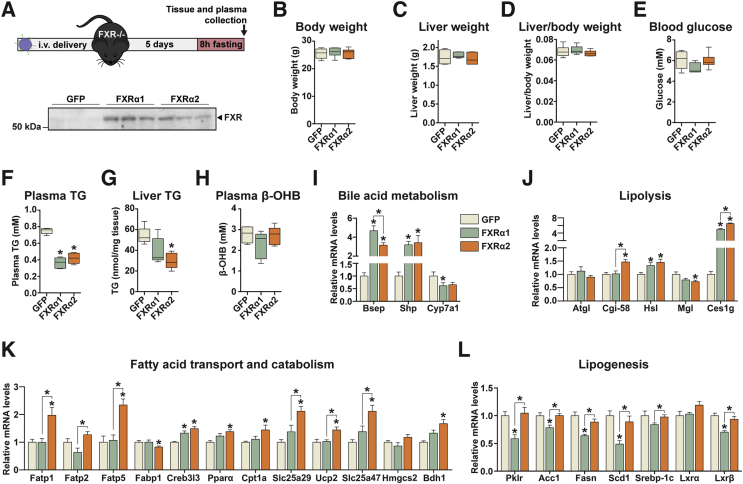
3.5. FXRα1 and α2 regulate discrete gene programs in vivo
We then sought to determine the gene programs regulated by each FXR variant in mouse liver. As expected, both FXR variants robustly activated Bsep and Shp transcription and reduced Cyp7a1 expression (Figure 5I). In terms of lipolytic factors, Hsl and Ces1g expression was increased by both FXRs, whereas Cgi-58 transcription was only activated by FXRα2 (Figure 5J). We observed no differences in Atgl mRNA levels and Mgl expression was decreased by both FXR variants. This likely reflects a decreased reliance on intracellular fat stores in face of a constant FFA supply from circulation, unlike cultured hepatocytes, which rely heavily on intracellular lipids to sustain metabolic fluxes. In line with the results obtained in hepatocytes, FXRα2 activated the expression of a vast array of genes with key roles in FA uptake (Fabp1, 2 and 5) and oxidation (Creb3l3, Pparα, Cpt1a, and Slc25a29), mitochondrial uncoupling (Ucp2 and Slc25a47), and ketogenesis (Bdh1) (Figure 5K). On the other hand, FXRα1 had marginal regulatory effects on this gene program, since only Creb3l3 was significantly induced by this variant. Instead, FXRα1 markedly reduced lipogenic gene expression, as evidenced by decreased pyruvate kinase (Pklr), acetyl-CoA carboxylase alpha (Acc-1), FA synthase (Fasn), stearoyl-CoA desaturase 1 (Scd1), and liver X receptor β (Lxrβ) mRNA levels (Figure 5L). In contrast, FXRα2 did not regulate lipogenic gene expression. Our results show that FXRα1 and α2 regulate distinct transcriptional programs in mouse liver, which influences their capacity to reduce hepatic steatosis.
Figure 5.
FXRα2 improves hepatic lipid handling in vivo. (A–L) Adenovirus-mediated FXRα1 and FXRα2 gene delivery to Fxr−/− mice liver. (A) Experimental design and FXR protein levels in liver determined by western blot. (B–H) Box plots of body weight (B), liver weight (C), liver/body weight ratio (D), blood glucose (E), plasma TAG (F), liver TAG (G), and plasma β-OHB (H) levels in Fxr−/− following FXRs gene delivery. (I–L) Gene expression analysis by qRT-PCR of genes involved in bile acid metabolism (I), lipolysis (J), FA transport and catabolism (K) and lipogenesis (L). Bars depict mean values and error bars represent SEM (n = 5–6). *p < 0.05 compared to control (GFP) or between the indicated groups.
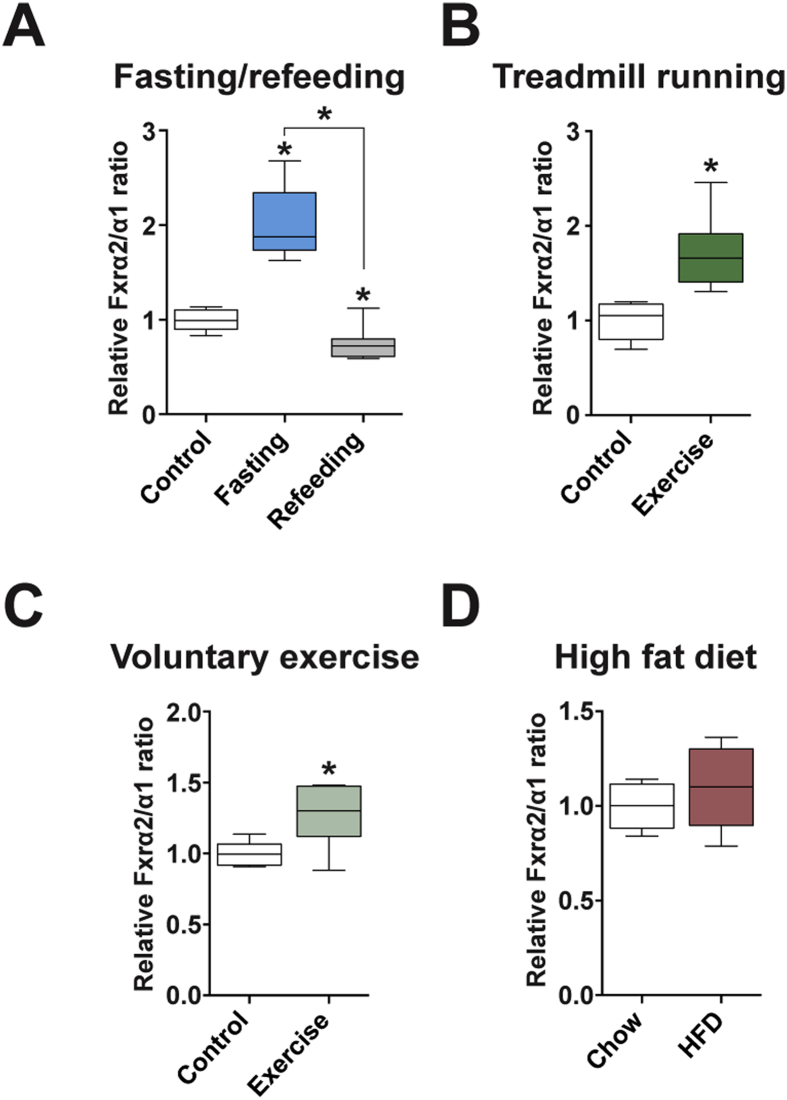
3.6. Liver Fxr splicing is dynamically regulated by systemic metabolic demands
Our data indicate that FXRα1 and α2 regulate discrete gene programs in liver and that changing the FXRα2/α1 ratio is sufficient to promote transcriptional reprogramming and alter hepatic lipid metabolism. This could constitute a molecular mechanism of hepatic adaptation to increased energetic demands. To test this hypothesis, we investigated if Fxr splicing is altered by fasting when hepatic metabolism is modulated to handle higher lipid influx and meet systemic energetic demands. Indeed, fasting increased liver Fxrα2/α1 ratios, which dropped below fed control levels upon refeeding (Figure 6A). Physical exercise also promotes rapid and profound changes in hepatic metabolism, by increasing FA oxidation and ketogenesis [39], [40]. To test whether Fxr splicing plays a role in hepatic adaptions to exercise, we evaluated whether Fxrα2/α1 ratios changed in response to acute and chronic exercise. A single bout of strenuous exercise was sufficient to change the relative levels of Fxr variants in the liver (Figure 6B). Mice challenged with a treadmill run to exhaustion had higher Fxrα2/α1 ratios than non-exercised controls. To investigate if regular, moderate exercise exerts similar effects on liver Fxr splicing, mice were given access to a running wheel for 8 weeks. After this milder but longer exercise protocol, the exercised group had higher Fxrα2/α1 ratios than sedentary controls (Figure 6C). Elevated FFA efflux from adipose tissue is characteristic of both fasting and physical exercise. Hence, Fxr splicing could be responsive to elevated FA supply and elicit an adaptive response to improve liver lipid handling capacity. Indeed, increased hepatic FA oxidation and TCA cycle turnover have been reported in obese mice and humans, likely reflecting an adaptive response to elevated hepatic lipid influx [41], [42]. Thus, we investigated whether high fat feeding modulates liver Fxr splicing. After one week of high fat feeding, sufficient to induce hepatic steatosis in rodents [36], we observed no significant changes in liver Fxrα2/α1 ratios (Figure 6D). These results indicate that Fxr splicing changes in response to elevated energetic demands and coordinates an adaptive response conducive to improved lipid handling/disposal through enhanced lipolysis, FA oxidation and ketogenesis.
Figure 6.
Systemic bioenergetic demands change Fxrα2/α1 ratios. (A–D) Hepatic Fxrα2/α1 splicing in response to metabolic stressors. Graphs represent box plots of Fxrα2/α1 ratios relative to controls upon overnight fasting and 3 h after refeeding (n = 7) (A), a single bout of intensive exercise on a treadmill for 1 h or until exhaustion (n = 6–8) (B), eight weeks of voluntary exercise in running wheels (n = 6) (C), and one week on a high fat diet (n = 5) (D). *p < 0.05 compared to control or between the indicated groups.
4. Discussion
Hepatic lipid accumulation is a hallmark of NAFLD and associated pathologies. The molecular mechanisms mediating this metabolic disarray remain largely unknown. Consequently, current NAFLD treatments target the metabolic symptoms rather than the underlying mechanisms. Due to its beneficial effects on hepatic lipid homeostasis, FXR is a promising target for the treatment of NAFLD. However, FXR consists of four distinct proteins and little is known about the biological functions of each variant.
We show here that FXR isoforms drive distinct gene programs in hepatocytes and liver, with specific biological outcomes. Our data indicate that a “MYTG” amino acid sequence in the hinge region of some FXR variants is a major determinant of transcriptional and biological activity. We observed that variants lacking this motif (FXRα2 and α4) regulate much larger gene sets than the MYTG-containing counterparts. In line with this observation, the MYTG motif has previously been shown to impair FXRs' ability to transactivate specific reporter constructs [11], [17], [18], [43]. The different N-termini of FXRα1/α2 and FXRα3/α4 do not seem to significantly influence the metabolic pathways addressed in this study but also underlie important changes in FXR target gene activation. For example, FXRα1 and α2 activated Bsep expression to a much greater extent than FXRα3 and α4. Accordingly, Fxrα2 and α4, which differ only in the N-termini, have been shown to differentially modulate Bsep expression in mouse liver but decrease hepatic TAG levels to similar extents [43]. Notably, only FXRα2 and α4 induced Atgl and Hsl expression, two lipases with key roles in hepatic lipid homeostasis. Hepatic overexpression of Atgl or Hsl reduces liver TAG levels in obese mice, without increasing plasma TAGs or apolipoprotein B secretion [31]. Conversely, Atgl loss-of-function aggravates hepatic steatosis and inflammation [44], [45], whereas Hsl deficiency promotes liver cholesteryl ester accumulation [46]. FXRα2 and α4 also elevated Cgi-58 expression, an Atgl activator [32]. Activating this transcriptional program reduced hepatocyte basal and palmitate-induced lipid accumulation and improved insulin responsiveness.
Our metabolomics analysis suggested that the FA mobilized from intracellular lipid stores were being channeled to mitochondrial FA oxidation. Indeed, hepatocytes expressing FXRα2 oxidized 90% more palmitate than controls. In support of this observation, FXRα2 activated the expression of Pparα and Creb3l3, two important regulators of hepatic FA metabolism with protective roles against obesity-associated metabolic disarrays [47], [48], [49], [50]. Moreover, FXRα2 increased acylcarnitine levels and Cpt1a and Slc25a29 expression, indicating increased shuttling of FA to mitochondrial β-oxidation. Excessive TCA cycle fluxes have been reported in NAFLD patients and correlated with increased gluconeogenesis [41]. However, neither FXR variant affected hepatocyte glucose output. Instead, FXRα2 enhanced β-OHB production and release by increasing both ketogenic gene expression and substrate supply. This mechanism is in accordance with the reported plasma β-OHB increase in mice expressing ectopic Atgl and Hsl [31]. In addition, hepatocytes expressing FXRα2 had higher mRNA levels of the mitochondrial uncoupling proteins Ucp2 and Slc25a47, suggesting that mitochondrial uncoupling may take place to sustain FA oxidation fluxes and alleviate oxidative stress [51], [52].
FXR is activated by bile acids at physiological concentrations [2], [3], and most of its reported effects are dependent on or potentiated by ligand-mediated activation. In line with the increasingly recognized role of bile acids as sensors of the feeding state [53], [54], CA reduced the expression of key mediators of FA oxidation, mitochondrial uncoupling, and ketogenesis in hepatocytes. However, FXRα2 expression blunted (or in some cases even reversed) most CA effects on these target genes, when compared to hepatocytes expressing GFP alone. Thus, our results indicate that in addition to bile acid-mediated activation, FXR expression greatly influences target gene activation and biological outcome. Indeed, reduced Fxr expression was shown to contribute to hepatic metabolic dysfunction even in the presence of elevated hepatic bile acid levels [4]. Conversely, Fxrα2 expression in mouse liver improves hepatic steatosis despite a slight decrease in the bile acid pool [43]. Although FXRs can interact with coactivators and transactivate target promoters in a ligand-independent manner [13], they have flexible ligand-binding pockets that accommodate ligands with diverse structures [15], [55] and alternative endogenous agonists have been described [37]. Thus, other endogenous ligands may contribute to FXRα2-mediated regulation of hepatic energy metabolism.
The relative expression of FXR splice variants affects its sum transcriptional activity and biological function [11], [14], [43]. Hepatic FXRα2/α1 ratios are decreased in hepatocellular carcinoma patients and by tumor necrosis factor α and interleukin 6 [14], two cytokines associated with hepatic steatosis and insulin resistance [56], [57], [58], [59]. It is interesting to note that bioinformatic pathway analysis of both gene microarray and metabolomics data predicted opposite roles for FXRα1 and α2 on liver cancer, with FXRα2 being protective. Our results indicate that elevating the FXRα2/FXRα1 ratio in hepatocytes without increasing total FXR levels is sufficient to activate a broad energy-mobilizing gene program, which can constitute an adaptive mechanism to physiological settings of high systemic energetic demands. In strong support of this notion, liver Fxrα2/α1 ratios were increased during fasting and rapidly decreased upon refeeding. Physical exercise modulates bile acid homeostasis, reduces hepatic lipid accumulation and is also associated with enhanced hepatic β-oxidation and ketogenesis [39], [40], [60]. We found that aerobic exercise increases the liver Fxrα2/α1 ratio, supporting the idea that the beneficial effects of exercise on hepatic lipid metabolism may involve changes in Fxr splicing.
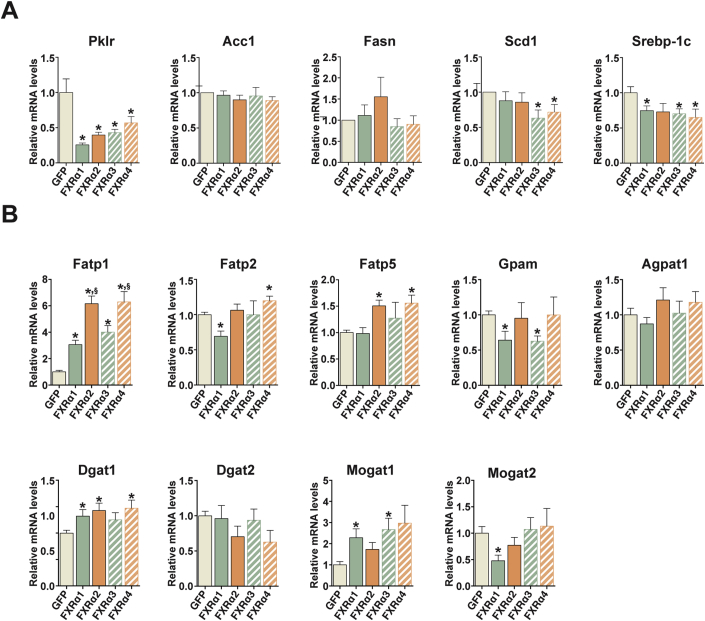
In agreement with the results obtained with cultured hepatocytes, FXRα2 expression in Fxr−/− mouse liver activated a broad set of genes involved in lipolysis, FA oxidation, mitochondrial uncoupling, and ketogenesis. Surprisingly, Atgl expression was not regulated by FXRα2 in vivo. Although the plentiful supply of FFA from circulation may alleviate the dependence on intracellular lipid stores, it is important to note that the expression of Cgi-58, which increases Atgl TAG hydrolase activity up to 20-fold [32], was induced by FXRα2 and may contribute for elevated lipolytic capacity. Indeed, Cgi-58 suppression alone causes hepatic steatosis, steatohepatitis, and fibrosis [61], [62], which demonstrates its pivotal role in hepatic lipid metabolism. As a result of these transcriptional adaptations, hepatic TAG levels were decreased in mice transduced with FXRα2. Noteworthy, FXRα2 induced Fatp1 and Fatp5 expression, whereas Fatp2 was expressed to similar levels compared to controls but higher than in livers transduced with FXRα1. This indicates that the reduced TAG content in FXRα2-expressing livers is not secondary to reduced hepatic FA uptake capacity. Although FXRa2 expression in Fxr−/− mouse liver was sufficient to regulate most of the pathways we observed in isolated hepatocytes, and reduce lipid accumulation, it did not increase plasma β-OHB levels, likely due to unchanged Hmgcs2 expression. FXRα1 decreased hepatic TAG levels to a lesser extent and did so through distinct mechanisms. This variant had marginal effects on FXRα2-regulated genes but decreased the expression of key mediators of de novo lipogenesis. Interestingly, in cultured hepatocytes we observed minor isoform dependence in the transcriptional control of de novo lipogenesis. FXR's transcriptional regulation of de novo lipogenesis has been previously shown to depend on bile acid-mediated activation [63]. Thus, the decrease in lipogenic gene expression observed in liver likely reflects FXRα1 activation by endogenous bile acids, which did not occur in isolated hepatocytes. These results clearly dissociate two different mechanisms by which FXRα1 and α2 reduce hepatic steatosis.
Due to its prominent role in hepatic lipid metabolism, FXR has long been suggested as a promising therapeutic target for NAFLD and other hepatic metabolic disorders. Indeed, obeticholic acid, a potent FXR agonist, improves insulin sensitivity and reduces liver inflammation and fibrosis in patients with NAFLD and type 2 diabetes [64]. More recently, the FLINT clinical trial reported improved liver histology in 45% of non-alcoholic steatohepatitis patients treated with obeticholic acid [65]. Our results show that the main FXR variants in human liver (α1 and α2) regulate distinct liver gene programs, which may have important clinical implications. In NAFLD, approximately 60% of hepatic TAG content is derived from plasma FFA and only 26% from de novo lipogenesis [66]. Therapeutic strategies specifically targeting FXRα2 should offer advantages over classical pan-FXR activation. Perhaps more importantly, our data show that FXR splicing in mouse liver is modulated by feeding status and physical exercise, which may modify the therapeutic response to pan-FXR agonists. This knowledge could be rapidly translated into clinic practice to improve the therapeutic efficacy of existing FXR agonists.
Acknowledgments
This project was supported by grants from the Diabetes Wellness Network Sweden (3848/2013SW) (JLR), Swedish Research Council (2014-3410, JLR; 2011-3550, JRZ), Strategic Research Programme in Diabetes at Karolinska Institutet (SRPD-KI) (JLR, JRZ), Novo Nordisk Foundation (Denmark, NNF12OC1016062) (JLR), Swedish Diabetes Foundation (DIA2013-076, JLR; DIA2012-082, JRZ), Swedish Foundation for Strategic Research (SRL10-0027) (JRZ), the European Research Council (233285) (JRZ) and the European Union (MC CIG 294232, JLR; FP7-HEALTH nr.305707, AKG). JCC was supported in part by a PhD fellowship from the Fundação para a Ciência e Tecnologia (FCT, Portugal, SFRH/BD/44825/2008), VMR by a postdoctoral fellowship from the Wenner-Gren Foundations (Sweden), JM by a Swedish Institute scholarship (00303/2012), and IS by the SRPD-KI. VR was supported by FCT (Portugal, project PEst-OE/EQB/LA0023/2013). DM and SS were supported by the German Ministry for Education and Research (BMBF, grant number 0315082 (01EA1303)) and the Max Planck Society. We thank Drs. Anna Krook, Alfredo Gimenez-Cassina and Peter Arner (KI) for valuable scientific discussions.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2015.09.005.
Conflicts of interest
None declared.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
Figure S1.
Isoform-specific FXR target gene regulation. (A) Principal component analysis of relative changes in gene expression induced by each FXR isoform. The ellipsoids show a distinct directionality in different groups based on similarities and differences. The axes correspond to principal component 1 (PC1, x-axis), PC2 (y-axis), and PC3 (z-axis). (B) Venn diagram represents the number of genes regulated by the four FXR isoforms and the number of common target genes between different isoforms. Venn diagrams were generated using Venny, freely available at http://bioinfogp.cnb.csic.es/tools/venny/[67]. (C) Venn diagrams represent the number of genes regulated by each FXR variant in common between two isoforms (individual comparisons). Also represented in Venn diagrams are the number of genes regulated by isoforms generated by alternative splicing (FXRα1 + FXRα3 vs. FXRα2 + FXRα4) and by different promoter usage (FXRα1 + FXRα2 vs. FXRα3 + FXRα4). (D) Analysis of gene expression profiling data by Ingenuity Pathway Analysis (IPA). Represented are hepatic pathologies significantly associated with the expression of one or more FXR isoforms.
Figure S2.
Transcriptional regulation of lipogenesis, fatty acid uptake and TAG synthesis by FXR isoforms. (A) Gene expression analysis by qRT-PCR of lipogenic factors in hepatocytes expressing each FXR variant or GFP alone. (B) Gene expression analysis by qRT-PCR of genes encoding fatty acid transport proteins and TAG synthesis enzymes in hepatocytes expressing each FXR variant or GFP alone. Graphs depict mean values and error bars represent SEM. *p < 0.05 vs. control. #p < 0.05 vs. all other isoforms. §p < 0.05 vs. FXRα1/α3. Acc1, Acetyl-CoA carboxylase alpha; Agpat1, 1-acylglycerol-3-phosphate O-acyltransferase 1; Dgat, Diacylglycerol O-acyltransferase; Fatp, Fatty acid transport protein; Fasn, Fatty acid synthase; Gpam, Glycerol-3-phosphate acyltransferase; Mogat, Monoacylglycerol O-acyltransferase; Pklr, Pyruvate kinase, liver and RBC; Scd1, Stearoyl-CoA desaturase 1; Srebp-1c, Sterol regulatory element-binding protein 1c.
Figure S3.
Metabolomics analysis of hepatocytes expressing FXRα1 or FXRα2. (A) Amino acid profile determined by mass spectrometry in hepatocytes expressing FXRα1, FXRα2, or GFP alone. The data presented is the same as in Figure 3B but plotted are the relative levels of each individual amino acid analyzed. (B) Analysis of metabolomics data by Ingenuity Pathway Analysis (IPA). Represented are canonical significantly associated with the expression of one or more FXR isoforms. (C) Predicted effects on hepatic metabolism for each FXR isoform. Z-score values indicate induction (orange) or repression (blue). (D) Levels of l-carnitine and acylcarnitines in hepatocytes expressing FXRα1, FXRα2, or GFP alone. Bars represent mean values (n = 1).
Figure S4.
Changing FXRα2/α1 ratios promotes transcriptional reprogramming. (A) Target gene expression analysis by qRT-PCR in hepatocytes expressing GFP or FXRα2 and treated with vehicle (DMSO) or 250 μM cholic acid (CA) for 24h. Bars depict mean values and error bars represent SEM (n = 5). *p < 0.05 compared to vehicle-treated or between the indicated groups. (B) Gene expression analysis by qRT-PCR in hepatocytes co-transduced with FXRα1 and FXRα2 in different proportions (+++, 100%; ++, 70%; +, 30%; −, 0%). Graphs represent expression levels relative to hepatocytes transduced with FXRα1 alone. Bars depict mean values and error bars represent SEM (n = 5). *p < 0.05 compared to FXRα1 alone or between the indicated groups.
References
- 1.Loomba R., Sanyal A.J. The global NAFLD epidemic. Nature Reviews Endocrinology & Hepatology. 2013;10(11):686–690. doi: 10.1038/nrgastro.2013.171. [DOI] [PubMed] [Google Scholar]
- 2.Parks D.J., Blanchard S.G., Bledsoe R.K., Chandra G., Consler T.G., Kliewer S.A. Bile acids: natural ligands for an orphan nuclear receptor. Science. 1999;284(5418):1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- 3.Makishima M., Okamoto A.Y., Repa J.J., Tu H., Learned R.M., Luk A. Identification of a nuclear receptor for bile acids. Science. 1999;284(5418):1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- 4.Lu Y., Ma Z., Zhang Z., Xiong X., Wang X., Zhang H. Yin Yang 1 promotes hepatic steatosis through repression of farnesoid X receptor in obese mice. Gut. 2014;63(1):170–178. doi: 10.1136/gutjnl-2012-303150. [DOI] [PubMed] [Google Scholar]
- 5.Xiong X., Wang X., Lu Y., Wang E., Zhang Z., Yang J. Hepatic steatosis exacerbated by endoplasmic reticulum stress-mediated downregulation of FXR in aging mice. Journal of Hepatology. 2014;60(4):847–854. doi: 10.1016/j.jhep.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 6.Yang Z.X., Shen W., Sun H. Effects of nuclear receptor FXR on the regulation of liver lipid metabolism in patients with non-alcoholic fatty liver disease. Hepatology International. 2010;4(4):741–748. doi: 10.1007/s12072-010-9202-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sinal C.J., Tohkin M., Miyata M., Ward J.M., Lambert G., Gonzalez F.J. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. 2000;102(6):731–744. doi: 10.1016/s0092-8674(00)00062-3. [DOI] [PubMed] [Google Scholar]
- 8.Ma K., Saha P.K., Chan L., Moore D.D. Farnesoid X receptor is essential for normal glucose homeostasis. Journal of Clinical Investigation. 2006;116(4):1102–1109. doi: 10.1172/JCI25604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cipriani S., Mencarelli A., Palladino G., Fiorucci S. FXR activation reverses insulin resistance and lipid abnormalities and protects against liver steatosis in Zucker (fa/fa) obese rats. Journal of Lipid Research. 2010;51(4):771–784. doi: 10.1194/jlr.M001602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y., Lee F.Y., Barrera G., Lee H., Vales C., Gonzalez F.J. Activation of the nuclear receptor FXR improves hyperglycemia and hyperlipidemia in diabetic mice. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(4):1006–1011. doi: 10.1073/pnas.0506982103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y., Kast-Woelbern H.R., Edwards P.A. Natural structural variants of the nuclear receptor farnesoid X receptor affect transcriptional activation. The Journal of Biological Chemistry. 2003;278(1):104–110. doi: 10.1074/jbc.M209505200. [DOI] [PubMed] [Google Scholar]
- 12.Huber R.M., Murphy K., Miao B., Link J.R., Cunningham M.R., Rupar M.J. Generation of multiple farnesoid-X-receptor isoforms through the use of alternative promoters. Gene. 2002;290(1–2):35–43. doi: 10.1016/s0378-1119(02)00557-7. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y., Castellani L.W., Sinal C.J., Gonzalez F.J., Edwards P.A. Peroxisome proliferator-activated receptor-gamma coactivator 1alpha (PGC-1alpha) regulates triglyceride metabolism by activation of the nuclear receptor FXR. Genes & Development. 2004;18(2):157–169. doi: 10.1101/gad.1138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y., Song X., Valanejad L., Vasilenko A., More V., Qiu X. Bile salt export pump is dysregulated with altered farnesoid X receptor isoform expression in patients with hepatocellular carcinoma. Hepatology. 2013;57(4):1530–1541. doi: 10.1002/hep.26187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Downes M., Verdecia M.A., Roecker A.J., Hughes R., Hogenesch J.B., Kast-Woelbern H.R. A chemical, genetic, and structural analysis of the nuclear bile acid receptor FXR. Molecular Cell. 2003;11(4):1079–1092. doi: 10.1016/s1097-2765(03)00104-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howarth D.L., Hagey L.R., Law S.H., Ai N., Krasowski M.D., Ekins S. Two farnesoid X receptor alpha isoforms in Japanese medaka (Oryzias latipes) are differentially activated in vitro. Aquatic Toxicology. 2010;98(3):245–255. doi: 10.1016/j.aquatox.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anisfeld A.M., Kast-Woelbern H.R., Meyer M.E., Jones S.A., Zhang Y., Williams K.J. Syndecan-1 expression is regulated in an isoform-specific manner by the farnesoid-X receptor. The Journal of Biological Chemistry. 2003;278(22):20420–20428. doi: 10.1074/jbc.M302505200. [DOI] [PubMed] [Google Scholar]
- 18.Song X., Chen Y., Valanejad L., Kaimal R., Yan B., Stoner M. Mechanistic insights into isoform-dependent and species-specific regulation of bile salt export pump by farnesoid X receptor. Journal of Lipid Research. 2013;54(11):3030–3044. doi: 10.1194/jlr.M038323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo J., Deng Z.L., Luo X., Tang N., Song W.X., Chen J. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nature Protocols. 2007;2(5):1236–1247. doi: 10.1038/nprot.2007.135. [DOI] [PubMed] [Google Scholar]
- 20.Estall J.L., Ruas J.L., Choi C.S., Laznik D., Badman M., Maratos-Flier E. PGC-1alpha negatively regulates hepatic FGF21 expression by modulating the heme/Rev-Erb(alpha) axis. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(52):22510–22515. doi: 10.1073/pnas.0912533106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nature Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Massart J., Zierath J.R., Chibalin A.V. A simple and rapid method to characterize lipid fate in skeletal muscle. BMC Research Notes. 2014;7:391. doi: 10.1186/1756-0500-7-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo B., Chen B., Liu A., Zhu W., Yao S. Liquid chromatography-mass spectrometric multiple reaction monitoring-based strategies for expanding targeted profiling towards quantitative metabolomics. Current Drug Metabolism. 2012;13(9):1226–1243. doi: 10.2174/138920012803341401. [DOI] [PubMed] [Google Scholar]
- 24.Wu H., Southam A.D., Hines A., Viant M.R. High-throughput tissue extraction protocol for NMR- and MS-based metabolomics. Analytical Biochemistry. 2008;372(2):204–212. doi: 10.1016/j.ab.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Lustig Y., Ruas J.L., Estall J.L., Lo J.C., Devarakonda S., Laznik D. Separation of the gluconeogenic and mitochondrial functions of PGC-1{alpha} through S6 kinase. Genes & Development. 2011;25(12):1232–1244. doi: 10.1101/gad.2054711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agudelo L.Z., Femenia T., Orhan F., Porsmyr-Palmertz M., Goiny M., Martinez-Redondo V. Skeletal muscle PGC-1alpha1 modulates kynurenine metabolism and mediates resilience to stress-induced depression. Cell. 2014;159(1):33–45. doi: 10.1016/j.cell.2014.07.051. [DOI] [PubMed] [Google Scholar]
- 27.Kok T., Hulzebos C.V., Wolters H., Havinga R., Agellon L.B., Stellaard F. Enterohepatic circulation of bile salts in farnesoid X receptor-deficient mice: efficient intestinal bile salt absorption in the absence of ileal bile acid-binding protein. The Journal of Biological Chemistry. 2003;278(43):41930–41937. doi: 10.1074/jbc.M306309200. [DOI] [PubMed] [Google Scholar]
- 28.Lu T.T., Makishima M., Repa J.J., Schoonjans K., Kerr T.A., Auwerx J. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Molecular Cell. 2000;6(3):507–515. doi: 10.1016/s1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- 29.Ananthanarayanan M., Balasubramanian N., Makishima M., Mangelsdorf D.J., Suchy F.J. Human bile salt export pump promoter is transactivated by the farnesoid X receptor/bile acid receptor. The Journal of Biological Chemistry. 2001;276(31):28857–28865. doi: 10.1074/jbc.M011610200. [DOI] [PubMed] [Google Scholar]
- 30.Cho Y., Noshiro M., Choi M., Morita K., Kawamoto T., Fujimoto K. The basic helix-loop-helix proteins differentiated embryo chondrocyte (DEC) 1 and DEC2 function as corepressors of retinoid X receptors. Molecular Pharmacology. 2009;76(6):1360–1369. doi: 10.1124/mol.109.057000. [DOI] [PubMed] [Google Scholar]
- 31.Reid B.N., Ables G.P., Otlivanchik O.A., Schoiswohl G., Zechner R., Blaner W.S. Hepatic overexpression of hormone-sensitive lipase and adipose triglyceride lipase promotes fatty acid oxidation, stimulates direct release of free fatty acids, and ameliorates steatosis. The Journal of Biological Chemistry. 2008;283(19):13087–13099. doi: 10.1074/jbc.M800533200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lass A., Zimmermann R., Haemmerle G., Riederer M., Schoiswohl G., Schweiger M. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman syndrome. Cell Metabolism. 2006;3(5):309–319. doi: 10.1016/j.cmet.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 33.Quiroga A.D., Li L., Trotzmuller M., Nelson R., Proctor S.D., Kofeler H. Deficiency of carboxylesterase 1/esterase-x results in obesity, hepatic steatosis, and hyperlipidemia. Hepatology. 2012;56(6):2188–2198. doi: 10.1002/hep.25961. [DOI] [PubMed] [Google Scholar]
- 34.Atshaves B.P., Martin G.G., Hostetler H.A., McIntosh A.L., Kier A.B., Schroeder F. Liver fatty acid-binding protein and obesity. The Journal of Nutritional Biochemistry. 2010;21(11):1015–1032. doi: 10.1016/j.jnutbio.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Samuel V.T., Liu Z.X., Qu X., Elder B.D., Bilz S., Befroy D. Mechanism of hepatic insulin resistance in non-alcoholic fatty liver disease. The Journal of Biological Chemistry. 2004;279(31):32345–32353. doi: 10.1074/jbc.M313478200. [DOI] [PubMed] [Google Scholar]
- 36.Samuel V.T., Liu Z.X., Wang A., Beddow S.A., Geisler J.G., Kahn M. Inhibition of protein kinase Cepsilon prevents hepatic insulin resistance in nonalcoholic fatty liver disease. Journal of Clinical Investigation. 2007;117(3):739–745. doi: 10.1172/JCI30400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao A., Yu J., Lew J.L., Huang L., Wright S.D., Cui J. Polyunsaturated fatty acids are FXR ligands and differentially regulate expression of FXR targets. DNA and Cell Biology. 2004;23(8):519–526. doi: 10.1089/1044549041562267. [DOI] [PubMed] [Google Scholar]
- 38.Lew J.L., Zhao A., Yu J., Huang L., De Pedro N., Pelaez F. The farnesoid X receptor controls gene expression in a ligand- and promoter-selective fashion. The Journal of Biological Chemistry. 2004;279(10):8856–8861. doi: 10.1074/jbc.M306422200. [DOI] [PubMed] [Google Scholar]
- 39.Rector R.S., Thyfault J.P., Morris R.T., Laye M.J., Borengasser S.J., Booth F.W. Daily exercise increases hepatic fatty acid oxidation and prevents steatosis in Otsuka Long-Evans Tokushima fatty rats. American Journal of Physiology - Gastrointestinal and Liver Physiology. 2008;294(3):G619–G626. doi: 10.1152/ajpgi.00428.2007. [DOI] [PubMed] [Google Scholar]
- 40.Kim K.H., Kim S.H., Min Y.K., Yang H.M., Lee J.B., Lee M.S. Acute exercise induces FGF21 expression in mice and in healthy humans. PLoS One. 2013;8(5):e63517. doi: 10.1371/journal.pone.0063517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sunny N.E., Parks E.J., Browning J.D., Burgess S.C. Excessive hepatic mitochondrial TCA cycle and gluconeogenesis in humans with nonalcoholic fatty liver disease. Cell Metabolism. 2011;14(6):804–810. doi: 10.1016/j.cmet.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Satapati S., Sunny N.E., Kucejova B., Fu X., He T.T., Mendez-Lucas A. Elevated TCA cycle function in the pathology of diet-induced hepatic insulin resistance and fatty liver. Journal of Lipid Research. 2012;53(6):1080–1092. doi: 10.1194/jlr.M023382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boesjes M., Bloks V.W., Hageman J., Bos T., van Dijk T.H., Havinga R. Hepatic farnesoid X-receptor isoforms alpha2 and alpha4 differentially modulate bile salt and lipoprotein metabolism in mice. PLoS One. 2014;9(12):e115028. doi: 10.1371/journal.pone.0115028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu J.W., Wang S.P., Alvarez F., Casavant S., Gauthier N., Abed L. Deficiency of liver adipose triglyceride lipase in mice causes progressive hepatic steatosis. Hepatology. 2011;54(1):122–132. doi: 10.1002/hep.24338. [DOI] [PubMed] [Google Scholar]
- 45.Jha P., Claudel T., Baghdasaryan A., Mueller M., Halilbasic E., Das S.K. Role of adipose triglyceride lipase (PNPLA2) in protection from hepatic inflammation in mouse models of steatohepatitis and endotoxemia. Hepatology. 2014;59(3):858–869. doi: 10.1002/hep.26732. [DOI] [PubMed] [Google Scholar]
- 46.Sekiya M., Osuga J., Yahagi N., Okazaki H., Tamura Y., Igarashi M. Hormone-sensitive lipase is involved in hepatic cholesteryl ester hydrolysis. Journal of Lipid Research. 2008;49(8):1829–1838. doi: 10.1194/jlr.M800198-JLR200. [DOI] [PubMed] [Google Scholar]
- 47.Kersten S., Seydoux J., Peters J.M., Gonzalez F.J., Desvergne B., Wahli W. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. Journal of Clinical Investigation. 1999;103(11):1489–1498. doi: 10.1172/JCI6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abdelmegeed M.A., Yoo S.H., Henderson L.E., Gonzalez F.J., Woodcroft K.J., Song B.J. PPARalpha expression protects male mice from high fat-induced nonalcoholic fatty liver. The Journal of Nutrition. 2011;141(4):603–610. doi: 10.3945/jn.110.135210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakagawa Y., Satoh A., Yabe S., Furusawa M., Tokushige N., Tezuka H. Hepatic CREB3L3 controls whole-body energy homeostasis and improves obesity and diabetes. Endocrinology. 2014;155(12):4706–4719. doi: 10.1210/en.2014-1113. [DOI] [PubMed] [Google Scholar]
- 50.Staels B., Rubenstrunk A., Noel B., Rigou G., Delataille P., Millatt L.J. Hepatoprotective effects of the dual peroxisome proliferator-activated receptor alpha/delta agonist, GFT505, in rodent models of nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. Hepatology. 2013;58(6):1941–1952. doi: 10.1002/hep.26461. [DOI] [PubMed] [Google Scholar]
- 51.Jin X., Yang Y.D., Chen K., Lv Z.Y., Zheng L., Liu Y.P. HDMCP uncouples yeast mitochondrial respiration and alleviates steatosis in L02 and hepG2 cells by decreasing ATP and H2O2 levels: a novel mechanism for NAFLD. Journal of Hepatology. 2009;50(5):1019–1028. doi: 10.1016/j.jhep.2008.10.034. [DOI] [PubMed] [Google Scholar]
- 52.Serviddio G., Bellanti F., Tamborra R., Rollo T., Capitanio N., Romano A.D. Uncoupling protein-2 (UCP2) induces mitochondrial proton leak and increases susceptibility of non-alcoholic steatohepatitis (NASH) liver to ischaemia-reperfusion injury. Gut. 2008;57(7):957–965. doi: 10.1136/gut.2007.147496. [DOI] [PubMed] [Google Scholar]
- 53.Seok S., Fu T., Choi S.E., Li Y., Zhu R., Kumar S. Transcriptional regulation of autophagy by an FXR-CREB axis. Nature. 2014;516(7529):108–111. doi: 10.1038/nature13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee J.M., Wagner M., Xiao R., Kim K.H., Feng D., Lazar M.A. Nutrient-sensing nuclear receptors coordinate autophagy. Nature. 2014;516(7529):112–115. doi: 10.1038/nature13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jin L., Feng X., Rong H., Pan Z., Inaba Y., Qiu L. The antiparasitic drug ivermectin is a novel FXR ligand that regulates metabolism. Nature Communications. 2013;4:1937. doi: 10.1038/ncomms2924. [DOI] [PubMed] [Google Scholar]
- 56.Li Z., Yang S., Lin H., Huang J., Watkins P.A., Moser A.B. Probiotics and antibodies to TNF inhibit inflammatory activity and improve nonalcoholic fatty liver disease. Hepatology. 2003;37(2):343–350. doi: 10.1053/jhep.2003.50048. [DOI] [PubMed] [Google Scholar]
- 57.Cheung A.T., Wang J., Ree D., Kolls J.K., Bryer-Ash M. Tumor necrosis factor-alpha induces hepatic insulin resistance in obese Zucker (fa/fa) rats via interaction of leukocyte antigen-related tyrosine phosphatase with focal adhesion kinase. Diabetes. 2000;49(5):810–819. doi: 10.2337/diabetes.49.5.810. [DOI] [PubMed] [Google Scholar]
- 58.Wieckowska A., Papouchado B.G., Li Z., Lopez R., Zein N.N., Feldstein A.E. Increased hepatic and circulating interleukin-6 levels in human nonalcoholic steatohepatitis. American Journal of Gastroenterology. 2008;103(6):1372–1379. doi: 10.1111/j.1572-0241.2007.01774.x. [DOI] [PubMed] [Google Scholar]
- 59.Klover P.J., Zimmers T.A., Koniaris L.G., Mooney R.A. Chronic exposure to interleukin-6 causes hepatic insulin resistance in mice. Diabetes. 2003;52(11):2784–2789. doi: 10.2337/diabetes.52.11.2784. [DOI] [PubMed] [Google Scholar]
- 60.Meissner M., Lombardo E., Havinga R., Tietge U.J., Kuipers F., Groen A.K. Voluntary wheel running increases bile acid as well as cholesterol excretion and decreases atherosclerosis in hypercholesterolemic mice. Atherosclerosis. 2011;218(2):323–329. doi: 10.1016/j.atherosclerosis.2011.06.040. [DOI] [PubMed] [Google Scholar]
- 61.Brown J.M., Betters J.L., Lord C., Ma Y., Han X., Yang K. CGI-58 knockdown in mice causes hepatic steatosis but prevents diet-induced obesity and glucose intolerance. Journal of Lipid Research. 2010;51(11):3306–3315. doi: 10.1194/jlr.M010256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guo F., Ma Y., Kadegowda A.K., Betters J.L., Xie P., Liu G. Deficiency of liver comparative gene Identification-58 causes steatohepatitis and fibrosis in mice. Journal of Lipid Research. 2013;54(8):2109–2120. doi: 10.1194/jlr.M035519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Watanabe M., Houten S.M., Wang L., Moschetta A., Mangelsdorf D.J., Heyman R.A. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. Journal of Clinical Investigation. 2004;113(10):1408–1418. doi: 10.1172/JCI21025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mudaliar S., Henry R.R., Sanyal A.J., Morrow L., Marschall H.U., Kipnes M. Efficacy and safety of the farnesoid X receptor agonist obeticholic acid in patients with type 2 diabetes and nonalcoholic fatty liver disease. Gastroenterology. 2013;145(3):574–582. doi: 10.1053/j.gastro.2013.05.042. e571. [DOI] [PubMed] [Google Scholar]
- 65.Neuschwander-Tetri B.A., Loomba R., Sanyal A.J., Lavine J.E., Van Natta M.L., Abdelmalek M.F. Farnesoid X nuclear receptor ligand obeticholic acid for non-cirrhotic, non-alcoholic steatohepatitis (FLINT): a multicentre, randomised, placebo-controlled trial. Lancet. 2015;385(9972):956–965. doi: 10.1016/S0140-6736(14)61933-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Donnelly K.L., Smith C.I., Schwarzenberg S.J., Jessurun J., Boldt M.D., Parks E.J. Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. Journal of Clinical Investigation. 2005;115(5):1343–1351. doi: 10.1172/JCI23621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oliveros J.C. 2007. VENNY. An interactive tool for comparing lists with Venn Diagrams. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.