Abstract
Urate transporter 1 (URAT1/SLC22A12), a urate transporter gene, is a causative gene for renal hypouricemia type 1. Among several reported nonsynonymous URAT1 variants, R90H (rs121907896) and W258X (rs121907892) are frequent causative mutations for renal hypouricemia. However, no case-control study has evaluated the relationship between gout and these two variants. Additionally, the effect size of these two variants on serum uric acid (SUA) levels remains to be clarified. Here, 1,993 primary gout patients and 4,902 health examination participants (3,305 males and 1,597 females) were genotyped with R90H and W258X. These URAT1 variants were not observed in any gout cases, while 174 subjects had the URAT1 variant in 2,499 health examination participants, respectively (P = 8.3 × 10−46). Moreover, in 4,902 health examination participants, the URAT1 nonfunctional variants significantly reduce the risk of hyperuricemia (P = 6.7 × 10−19; risk ratio = 0.036 in males). Males, having 1 or 2 nonfunctional variants of URAT1, show a marked decrease of 2.19 or 5.42 mg/dl SUA, respectively. Similarly, females, having 1 or 2 nonfunctional variants, also evidence a decrease of 1.08 or 3.89 mg/dl SUA, respectively. We show that URAT1 nonfunctional variants are protective genetic factors for gout/hyperuricemia, and also demonstrated the sex-dependent effect size of these URAT1 variants on SUA (P for interaction = 1.5 × 10−12).
Gout (MIM 138900) is one of the most common types of inflammatory arthritis as a consequence of hyperuricemia. Gout and hyperuricemia increase the risk of other common diseases, such as kidney diseases, cerebrovascular diseases, hypertension and cardiovascular diseases1. Several transporter genes associated with gout and serum uric acid (SUA) levels were previously reported, such as ATP-binding cassette transporter, subfamily G, member 2 (ABCG2/BCRP [MIM 603756])2,3,4,5,6, glucose transporter 9 (GLUT9/SLC2A9 [MIM 606142])2,7,8, sodium-dependent phosphate cotransporter type 1 (NPT1/SLC17A1 [MIM 182308])9, organic anion transporter 4 (OAT4/SLC22A11 [MIM 607097])10,11, and urate transporter 1 (URAT1/SLC22A12 [MIM 607096])12,13.
Among them, URAT1, which is a well-known urate transporter gene, has been identified as a causative gene for renal hypouricemia type 1 (MIM 220150)14. Among several reported nonsynonymous variants in URAT1, R90H (rs121907896) and W258X (rs121907892) are frequent causative mutations for renal hypouricemia15. Previous in vitro functional studies showed that R90H variant diminishes the urate transport activity of URAT115 as the other common variant, W258X14. It has been also reported that nonfunctional variants in URAT1 were not detected in 77 Spanish gout patients16, and W258X in URAT1 suppressed the development of gout17. However, to our knowledge, no large-scale case-control study has evaluated the relationship between gout/hyperuricemia and both variants (R90H and W258X). In this study, therefore, we investigated the association between gout and two URAT1 variants with large-scale Japanese primary gout cases and controls. Moreover, the risk ratio (RR) of these two nonfunctional variants for hyperuricemia was evaluated in approximately 5,000 Japanese health examination participants. Although there is a gender difference in SUA due to sex hormones18,19, the effect size of these two variants on SUA in each sex remains to be clarified. Furthermore, these URAT1 variants (R90H and W258X) are frequently observed especially in a Japanese population; thus, it is particularly important to analyze the sex-dependent effect size of these URAT1 variants on SUA in a general Japanese population. Therefore, we also evaluated the effect size of these URAT1 variants on SUA in each sex with a large number of Japanese health examination participants.
Results
Case-control study of gout
The genotyping results of URAT1 nonfunctional variants (R90H and W258X) for 1,993 gout cases and 2,499 controls were shown in Table 1. The two variants were in Hardy-Weinberg equilibrium (P > 0.05). The URAT1 nonfunctional variants (R90H and W258X) were not observed in any gout cases (n = 1,993), while R90H heterozygotes (G/A), W258X heterozygotes (G/A) and W258X homozygotes (A/A) were observed in 22, 150 and 2 subjects, respectively, among 2,499 control subjects (P = 8.3 × 10−46; Table 1). This result is compatible with previous studies16,17, and indicates that these URAT1 variants are protective factors of gout.
Table 1. Genotype distributions of nonfunctional variants in URAT1/SLC22A12 in gout patients and controls.
| Number | R90H |
W258X |
Number of URAT1 nonfunctional alleles* |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| G/G | G/A | A/A | G/G | G/A | A/A | 0 | 1 | 2 | P value† | ||
| Gout | 1,993 | 1,993 | 0 | 0 | 1,993 | 0 | 0 | 1,993 | 0 | 0 | |
| Control | 2,499 | 2,477 | 22 | 0 | 2,347 | 150 | 2 | 2,325 | 172 | 2 | 8.3 × 10−46 |
*The nonfunctional alleles mean A allele of R90H or W258X.
†2 × 3 Fisher’s exact test.
Risk ratio for hyperuricemia
Next, Table 2 and Supplementary Table S1 show the genotype distributions of URAT1 nonfunctional variants in 4,902 health examination participants of the Japan Multi-Institutional Collaborative Cohort (J-MICC) study (3,305 males and 1,597 females). Among the 4,902 participants, the nonfunctional allele frequencies of R90H and W258X were 0.28% and 2.24%, respectively. All of the participants were divided into hyperuricemia (SUA > 7.0 mg/dl) or control (SUA ≤ 7.0 mg/dl).
Table 2. Genotype distributions of URAT1 nonfunctional variants in 3,305 males and risk ratio for hyperuricemia.
| Hyperuricemia | Control* | P value† | RR (95% CI) | Reciprocal RR (95% CI) | ||
|---|---|---|---|---|---|---|
| R90H | G/G | 806 | 2,477 | |||
| G/A | 0 | 22 | ||||
| A/A | 0 | 0 | 4.3 × 10−3 | — | — | |
| W258X | G/G | 804 | 2,347 | |||
| G/A | 2 | 150 | ||||
| A/A | 0 | 2 | 3.3 × 10−16 | 0.041 (0.010–0.164)‡ | 24.5 (6.1–98.7)‡ | |
| Number of nonfunctional alleles(R90H or W258X) | 0 | 804 | 2,325 | |||
| 1 | 2 | 172 | ||||
| 2 | 0 | 2 | 6.7 × 10−19 | 0.036 (0.009–0.143)§ | 28.1 (7.0–112.8)§ | |
3,305 males (806 hyperuricemia and 2,499 controls) are health examination participants of the J-MICC study.
Abbreviation: RR = risk ratio; CI = confidence interval.
*Control group is comprised of individuals with serum uric acid levels ≤ 7.0 mg/dl, no gout history and no treatments for gout/hyperuricemia.
†3 × 2 Fisher’s exact test.
‡Dominant model (G/G versus G/A or A/A).
§Dominant model (0 versus 1 or 2).
In 3,305 males (Table 2), there were significant differences between hyperuricemia and control in both R90H (P = 4.3 × 10−3) and W258X genotype distributions (P = 3.3 × 10−16). Additionally, the number of R90H or W258X nonfunctional alleles in each group was calculated. Then, the proportion of nonfunctional alleles was more frequent in control than that in hyperuricemia (P = 6.7 × 10−19; RR = 0.036; 95% confidence interval [CI]: 0.009–0.143, Table 2).
In 1,597 females, both R90H and W258X were observed only in controls (Supplementary Table S1). However, because the female hyperuricemia group was comprised of very small sample size (24 individuals), no association analysis was performed.
Effect size of URAT1 variants on SUA levels
We also investigated SUA for each number of URAT1 nonfunctional alleles using 4,753 individuals (3,158 males and 1,595 females), who received no medication for gout and/or hyperuricemia among 4,902 health examination participants of the J-MICC Study. The mean SUA levels with standard error of the mean (SEM) of having 0, 1 and 2 nonfunctional alleles were 6.22 ± 0.02, 4.03 ± 0.07 and 0.80 ± 0.10 mg/dl in males, respectively, and were 4.49 ± 0.02, 3.48 ± 0.15 and 0.60 ± 0.06 mg/dl in females, respectively (Fig. 1). Then, the nonfunctional alleles of two URAT1 variants significantly decreased SUA in both males and females (P = 2.2 × 10−138 and 2.6 × 10−24, respectively).
Figure 1. Changes of serum uric acid levels of URAT1 nonfunctional alleles in health examination participants.
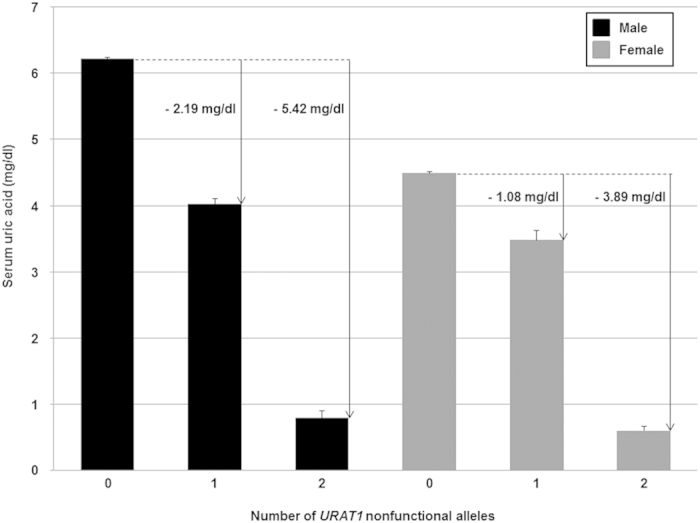
4,753 health examination participants, who received no medication for gout and/or hyperuricemia, were analyzed. Among 3,158 male participants (left, black bars), 0, 1 and 2 nonfunctional alleles (R90H or W258X) were detected in 2,982, 174 and 2 males, respectively. Among 1,595 female participants (right, grey bars), 0, 1 and 2 nonfunctional alleles (R90H or W258X) were detected in 1,529, 63 and 3 females, respectively. Serum uric acid (SUA) levels of participants having 0, 1 and 2 nonfunctional alleles were shown in each sex. The sex-dependent effect size of SUA decrease by nonfunctional alleles (arrow) was also shown. All bars are expressed as means ± SEM.
Furthermore, a multiple regression analysis, which focused on the statistical significance of the interaction term, revealed that there was an interaction between URAT1 nonfunctional variants and sex (P for interaction = 1.5 × 10−12, Table 3).
Table 3. Multiple regression analysis focused on the interaction between URAT1 variants and sex.
| Partial regression coefficient | P value | |
|---|---|---|
| b1 | −2.21 | 8.2 × 10−155 |
| b2 | −1.72 | <1.0 × 10−323* |
| b3 | 1.05 | 1.5 × 10−12 |
y = b0 + b1 x1 + b2 x2 + b3x1 x2, where y is SUA level, x1 is an ordinal variable representing the number of nonfunctional alleles of two URAT1 variants (R90H and W258X), and x2 is a dummy variable representing the sex (male = 0 and female = 1). x1 x2 is an interaction term.
*P value was extremely low and the calculation was impossible by the software R.
Discussion
URAT1 has been identified as a urate-anion exchanger which regulates SUA levels by playing an important role in the reabsorption of urate in human kidney14. In this study, we performed the genotyping of the two URAT1 nonfunctional variants (R90H and W258X), and demonstrated the association with gout (Table 1), and the significant effect on hyperuricemia progression (Table 2) and that on SUA (Fig. 1).
Consistent with previous reports (on 77 Spanish16 and 185 Japanese17 gout patients, respectively), no URAT1 nonfunctional variants (R90H or W258X) were found even in our large number of gout patients (n = 1,993). Our results indicate that these URAT1 variants prevent the development of gout by the large-scale case-control study (case = 1,993 and control = 2,499).
Moreover, we revealed that the URAT1 nonfunctional alleles of R90H and W258X markedly reduce the risk of hyperuricemia (RR = 0.036 in males; Table 2) and severely decrease SUA (Fig. 1) using 4,902 health examination participants. Males, having 1 or 2 nonfunctional alleles of URAT1 exhibit a marked decrease of 2.19 or 5.42 mg/dl SUA, respectively (Fig. 1). Similarly, females, having 1 or 2 nonfunctional alleles of URAT1 also show a decrease of 1.08 or 3.89 mg/dl SUA, respectively (Fig. 1). Moreover, the interaction between URAT1 nonfunctional variants and sex was present (P for interaction = 1.5 × 10−12, Table 3). Thus, for the first time, we demonstrated the sex-dependent effect size of SUA by URAT1 nonfunctional variants, which is also important for understanding the pathogenesis of renal hypouricemia because mild renal hypouricemia (SUA ≤ 3.0 mg/dl) could be caused by a heterozygous nonfunctional variant of URAT120 or GLUT921. Our data clearly demonstrated that some individuals with a heterozygous URAT1 nonfunctional variant exhibit renal hypouricemia.
Interestingly, although the sex difference in SUA is well-known18,19, SUA of the individuals having 2 nonfunctional alleles is similar between males (0.80 mg/dl) and females (0.60 mg/dl). Moreover, the sex difference in SUA is smaller in the individuals having 1 nonfunctional allele (0.55 mg/dl) than in individuals without nonfunctional alleles (1.73 mg/dl). In other words, our data show that the sex difference of SUA becomes greater as the number of functional alleles (wild-type alleles) of URAT1 increases, which suggests that the presence of functional URAT1 transporter is strongly related to the sex difference in SUA.
Previously, the sex difference in the expression of URAT1 had been found in a mouse model22. In addition, testosterone reportedly enhances the mRNA of Urat1 in a mouse model23 and increases promoter activity of human URAT124. Combined with these previous reports, our data suggest that one of the main causes of the sex difference in SUA is the different expression levels of functional URAT1 transporters between males and females due to sex hormones.
In summary, we demonstrated that the URAT1 nonfunctional variants are protective genetic factors for gout and hyperuricemia, and showed the sex-dependent effect size of these URAT1 variants on SUA. These findings provide a better understanding of genetic factors for SUA and gout/hyperuricemia progression.
Methods
Patients and controls
This study was approved by the institutional ethical committee of the National Defense Medical College. All procedures were performed in accordance with the Declaration of Helsinki, and written informed consent was obtained from each subject participating in the present study.
In a case-control study of gout, 1,993 Japanese male patients with primary gout were recruited from the outpatients of Midorigaoka Hospital (Osaka, Japan), Kyoto Industrial Health Association (Kyoto, Japan) and Ryougoku East Gate Clinic (Tokyo, Japan). All of the gout patients were diagnosed according to the criteria established by the American College of Rheumatology25. Hyperuricemia was defined as the SUA level that exceeds 7.0 mg/dl (=416.36 mol/l) according to the guideline of the Japanese Society of Gout and Nucleic Acid Metabolism26. As the control group, 2,499 male Japanese individuals without hyperuricemia and gout history were selected from participants in the Shizuoka area in the J-MICC Study27,28.
For evaluation of the influence of two URAT1 variants on SUA, 4,902 Japanese individuals (3,305 males including above 2,499 controls, and 1,597 females) were also recruited from health examination participants in the J-MICC Study. The details of participants in this study are shown in Supplementary Tables S2 and S3.
Genotyping
Genomic DNA was extracted from whole peripheral blood cells21. Genotyping of R90H and W258X variants in URAT1 was performed by TaqMan method (Life Technologies Corporation, Carlsbad, CA, USA) with a LightCycler 480 (Roche Diagnostics, Mannheim, Germany)29. Custom TaqMan assay probes were designed as follows: for R90H in URAT1, VIC-CCGCCACTTCCGC and FAM-CGCCGCTTCCGC; for W258X in URAT1, VIC-CGGGACTGAACACTG and FAM-CGGGACTGGACACTG. All of R90H heterozygotes (G/A), W258X heterozygotes (G/A) and W258X homozygotes (A/A) were confirmed by direct sequencing with a 3130xl Genetic Analyzer (Life Technologies Corporation)29 and the following primers: for R90H in URAT1, forward 5′-GTTGGAGCCACCCCAAGTGAC-3′ and reverse 5′-GTCTGACCCACCGTGATCCATG-3′; for W258X in URAT1, forward 5′-TGATGAACACGGGCACTCTC-3′ and reverse 5′-CTTTCCACTCGCTCCCCTAG-3′.
Data analysis
For all calculations in the statistical analysis, the software R (version 3.1.1) (http://www.r-project.org/) was used30. The association analyses were examined with the Fisher’s exact tests. RRs were calculated under a dominant model: i.e. G/G versus G/A or A/A in W258X, 0 versus 1 or 2 in the number of nonfunctional alleles, respectively. Linear regression analyses were performed to evaluate the influence of two URAT1 variants on SUA. Furthermore, we carried out a multiple regression analysis with an interaction term (x1 x2): y = b0 + b1 x1 + b2 x2 + b3x1 x2, where y is SUA level, x1 is an ordinal variable representing the number f nonfunctional alleles of two URAT1 variants, and x2 is a dummy variable representing the sex (male = 0 and female = 1). For the robustness of the statistical test, random re-sampling methods with computer simulation are often applied31,32. In this study, the permutation test32 was used for random re-sampling in a case-control study with replacement for 1,000,000 times, and the robustness of statistics was confirmed. All P values were two-tailed and P value < 0.05 was considered statistically significant.
Additional Information
How to cite this article: Sakiyama, M. et al. The effects of URAT1/SLC22A12 nonfunctional variants, R90H and W258X, on serum uric acid levels and gout/hyperuricemia progression. Sci. Rep. 6, 20148; doi: 10.1038/srep20148 (2016).
Supplementary Material
Acknowledgments
We thank all the participants involved in this study. We are especially indebted to K. Gotanda, Y. Morimoto, J. Abe, M. Miyazawa, H. Inoue, Y. Kawamura, T. Chiba and Y. Takada for genetic analysis. We are indebted to A. Tokumasu, K. Wakai and N. Hamajima, for sample collection. We also thank M. Hosoyamada and T. Hosoya for their helpful discussion. This study was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan including the MEXT KAKENHI (Grant numbers 221S0001, 221S0002, 25293145, 22689021, 25670307), the Ministry of Health, Labour and Welfare of Japan, the Ministry of Defense of Japan, the Kawano Masanori Memorial Foundation for Promotion of Pediatrics, and the Gout Research Foundation of Japan.
Footnotes
Yes, there is potential competing interest: H.M., T.T. and N.S. have a patent pending based on the work reported in this paper. The other authors declare that they have no conflict of interest.
Author Contributions M.S., H.M. and N.S. conceived and designed this study. M.S., H.M., S. Shimizu, A.N. and T.H. performed genetic analysis. M.S., H.M., H.N. and T.N. performed statistical analyses. M.N., S. Suma, A.H., H.O. and T. Shimizu. collected samples and analyzed clinical data. M.S. and H.M. wrote the manuscript. T. Satoh, Y.S., T.T., K.I. and N.S. provided intellectual input and assisted with the preparation of the manuscript. M.S. and H.M. contributed equally to this work.
References
- Feig D. I., Kang D. H. & Johnson R. J. Uric acid and cardiovascular risk. N. Engl. J. Med. 359, 1811–1821 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehghan A. et al. Association of three genetic loci with uric acid concentration and risk of gout: a genome-wide association study. Lancet. 372, 1953–1961 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward O. M. et al. Identification of a urate transporter, ABCG2, with a common functional polymorphism causing gout. Proc. Natl. Acad. Sci. USA. 106, 10338–10342 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo H. et al. Common defects of ABCG2, a high-capacity urate exporter, cause gout: a function-based genetic analysis in a Japanese population. Sci. Transl. Med. 1, 5ra11 (2009). [DOI] [PubMed] [Google Scholar]
- Ichida K. et al. Decreased extra-renal urate excretion is a common cause of hyperuricemia. Nat. Commun. 3, 764 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama A. et al. Common dysfunctional variants of ABCG2 have stronger impact on hyperuricemia progression than typical environmental risk factors. Sci. Rep. 4, 5227 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döring A. et al. SLC2A9 influences uric acid concentrations with pronounced sex-specific effects. Nat. Genet. 40, 430–436 (2008). [DOI] [PubMed] [Google Scholar]
- Matsuo H. et al. Genome-wide association study of clinically defined gout identifies multiple risk loci and its association with clinical subtypes. Ann. Rheum. Dis. 10.1136/annrheumdis-2014-206191 (2015). (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba T. et al. NPT1/SLC17A1 is a renal urate exporter in humans and its common gain-of-function variant decreases the risk of renal underexcretion gout. Arthritis Rheum. 67, 281–287 (2015). [DOI] [PubMed] [Google Scholar]
- Kolz M. et al. Meta-analysis of 28,141 individuals identifies common variants within five new loci that influence uric acid concentrations. PLoS Genet. 5, e1000504 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakiyama M. et al. A common variant of organic anion transporter 4 (OAT4/SLC22A11) gene is associated with renal underexcretion type gout. Drug Metab. Pharmacokinet. 29, 208–210 (2014). [DOI] [PubMed] [Google Scholar]
- Köttgen A. et al. Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat. Genet. 45, 145–154 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y. et al. Meta-analysis identifies multiple loci associated with kidney function-related traits in east Asian populations. Nat. Genet. 44, 904–909 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto A. et al. Molecular identification of a renal urate anion exchanger that regulates blood urate levels. Nature. 417, 447–452 (2002). [DOI] [PubMed] [Google Scholar]
- Ichida K. et al. Clinical and molecular analysis of patients with renal hypouricemia in Japan-influence of URAT1 gene on urinary urate excretion. J. Am. Soc. Nephrol. 15, 164–173 (2004). [DOI] [PubMed] [Google Scholar]
- Torres R. J., De Miguel E., Bailen R. & Puig J. G. Absence of SLC22A12/URAT1 gene mutations in patients with primary gout. J. Rheumatol. 39, 1901 (2012). [DOI] [PubMed] [Google Scholar]
- Taniguchi A. et al. A common mutation in an organic anion transporter gene, SLC22A12, is a suppressing factor for the development of gout. Arthritis Rheum. 52, 2576–2577 (2005). [DOI] [PubMed] [Google Scholar]
- Adamopoulos D., Vlassopoulos C., Seitanides B., Contoyiannis P. & Vassilopoulos P. The relationship of sex steroids to uric acid levels in plasma and urine. Acta Endocrinol. (Copenh). 85, 198–208 (1977). [DOI] [PubMed] [Google Scholar]
- Yahyaoui R. et al. Effect of long-term administration of cross-sex hormone therapy on serum and urinary uric acid in transsexual persons. J. Clin. Endocrinol. Metab. 93, 2230–2233 (2008). [DOI] [PubMed] [Google Scholar]
- Wakida N. et al. Mutations in human urate transporter 1 gene in presecretory reabsorption defect type of familial renal hypouricemia. J. Clin. Endocrinol. Metab. 90, 2169–2174 (2005). [DOI] [PubMed] [Google Scholar]
- Matsuo H. et al. Mutations in glucose transporter 9 gene SLC2A9 cause renal hypouricemia. Am. J. Hum. Genet. 83, 744–751 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoyamada M., Ichida K., Enomoto A., Hosoya T. & Endou H. Function and localization of urate transporter 1 in mouse kidney. J. Am. Soc. Nephrol. 15, 261–268 (2004). [DOI] [PubMed] [Google Scholar]
- Hosoyamada M., Takiue Y., Shibasaki T. & Saito H. The effect of testosterone upon the urate reabsorptive transport system in mouse kidney. Nucleosides Nucleotides Nucleic Acids. 29, 574–579 (2010). [DOI] [PubMed] [Google Scholar]
- Li T., Walsh J. R., Ghishan F. K. & Bai L. Molecular cloning and characterization of a human urate transporter (hURAT1) gene promoter. Biochim. Biophys. Acta. 1681, 53–58 (2004). [DOI] [PubMed] [Google Scholar]
- Wallace S. L. et al. Preliminary criteria for the classification of the acute arthritis of primary gout. Arthritis Rheum. 20, 895–900 (1977). [DOI] [PubMed] [Google Scholar]
- The guideline revising committee of the Japanese Society of Gout and Nucleic Acid Metabolism. In Guideline for the Management of Hyperuricemia and Gout 2nd edn, (ed The guideline revising committee of the Japanese Society of Gout and Nucleic Acid Metabolism) Ch. 2, 60–72 (Medical Review, 2010). [Google Scholar]
- Asai Y. et al. Baseline data of Shizuoka area in the Japan Multi-Institutional Collaborative Cohort Study (J-MICC Study). Nagoya J. Med. Sci. 71, 137–144 (2009). [PMC free article] [PubMed] [Google Scholar]
- Hamajima N. & J-MICC Study Group. The Japan Multi-Institutional Collaborative Cohort Study (J-MICC Study) to detect gene-environment interactions for cancer. Asian Pac. J. Cancer Prev. 8, 317–323 (2007). [PubMed] [Google Scholar]
- Sakiyama M. et al. Common variant of leucine-rich repeat-containing 16A (LRRC16A) gene is associated with gout susceptibility. Hum. Cell. 27, 1–4 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core Team R. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria, (2013). URL http://www.R-project.org/
- Li J. et al. Identification of high-quality cancer prognostic markers and metastasis network modules. Nat. Commun. 1, 34 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron B. & Tibshirani R. J. In An Introduction to the Bootstrap (eds Cox. D. R. et al. ) 202–219 (Chapman & Hall, 1993). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


