Abstract
A striking feature of white-nose syndrome, a fungal infection of hibernating bats, is the difference in infection outcome between North America and Europe. Here we show high WNS prevalence both in Europe and on the West Siberian Plain in Asia. Palearctic bat communities tolerate similar fungal loads of Pseudogymnoascus destructans infection as their Nearctic counterparts and histopathology indicates equal focal skin tissue invasiveness pathognomonic for WNS lesions. Fungal load positively correlates with disease intensity and it reaches highest values at intermediate latitudes. Prevalence and fungal load dynamics in Palearctic bats remained persistent and high between 2012 and 2014. Dominant haplotypes of five genes are widespread in North America, Europe and Asia, expanding the source region of white-nose syndrome to non-European hibernacula. Our data provides evidence for both endemicity and tolerance to this persistent virulent fungus in the Palearctic, suggesting that host-pathogen interaction equilibrium has been established.
Emerging wildlife infections are a threat to global biodiversity1, yet the emergence and transmission of such infectious diseases may also be a function of ecosystem quality and biodiversity2. Moreover, anthropogenic disturbance and introductions have been highlighted as contributing considerably to such disease outbreaks3,4. The majority of emerging wildlife pathogens are of viral origin but fungal infections have also been recognised across a diverse range of taxa, including plants and poikilothermic animals5. Relatively few fungal species cause severe diseases in mammals due to their high body temperature and effective immune system6,7.
Prevalence of pathogens may be high in bat populations8,9. Despite this, bats have been reported to survive infections that are lethal to other taxa10,11,12. Bat’s capacity to carry pathogens may result from its ability to tolerate damage caused by the agent or the associated immune response13,14,15. Understanding the mechanisms that underlie any trade-off between tolerance and resistance to infectious agents in reservoir hosts is of utmost importance for effective disease control16.
Most bats in temperate regions enter seasonal hibernation, at which time they reduce their metabolic rate and decrease body temperature until it approaches the ambient temperature of the hibernaculum, which may alter immune response to pathogens17,18. During hibernation, bats are vulnerable to infection by the psychrophilic fungus Pseudogymnoascus destructans [formerly Geomyces destructans]19, which emerged as a novel pathogen in eastern North America in 200620. The P. destructans fungus is the causative agent of white-nose syndrome (WNS)20,21,22,23. Considered an epidemic of major conservation concern in North America, WNS is responsible for an unprecedented decline in bat populations24,25,26. WNS combines some of the worst possible epidemiological characteristics, including a highly virulent pathogen23,27 with density- and frequency-dependent transmission28, an environmental reservoir19, long-term persistence in hibernacula29,30 and susceptibility of multiple hosts19,20,31.
The origin of WNS in North America remains unknown and current research focuses on identifying the infectious agent’s source and identifying whether the agent is an introduced pathogen. Six main lines of evidence support introduction of WNS into North America: 1) Fungal communities associated with bats and their hibernacula in eastern North America comprise a diverse range of P. destructans allies22,32; phylogenetic evaluation, however, indicates that none of these is closely related to P. destructans22. 2) Previous studies have demonstrated clonal dispersal of a single P. destructans genotype among WNS-infected bats in the United States33,34. 3) Spread of WNS in North America follows a clear invasion front with varying survival rate since first appearance of the disease24,35,36,37. 4) Presence of P. destructans has been confirmed in many European countries38,39,40,41 but with no reports of mass mortality38. 5) European P. destructans isolates are pathogenic for North American bats23. Further, virulent skin infections producing focally severe lesions pathognomonic for WNS have been documented in European bats under natural infection conditions42. 6) While only one heterothallic fungal mating type has been recorded in North American P. destructans populations, two types have been found coexisting in European hibernacula. Effective recombination during sexual reproduction results in genetic variability and may be linked with virulence43. These indirect sources of evidence tend to support the introduced pathogen hypothesis, suggesting WNS may have originated outside of North America22,23,43,44. Comparative studies between North America and Europe have been proposed to explain the origin and differential manifestation of the fungal infection4,40,42,45.
Although some authors speculate on a non-European origin4,25, Europe is believed to be the likely source of WNS44. Supporting evidence for long exposure to the WNS fungus based on old European photographs, which appear to show a white cutaneous fungal infection38,40, lacks specificity for WNS. Another dermatophyte, Trichophyton redellii, produces a similar gross appearance in hibernating bats46 and the photographs might represent either. On the contrary, presence of WNS in multiple and diverse hosts31 indicates that the WNS fungus could persist across their ranges in the Palearctic.
To date, WNS diagnostic skin lesions have only been reported from the Czech Republic, Europe, with almost half of all species being WNS positive and prevalence based on histopathology reaching 55% in bats emerging from hibernacula in spring31,42. Occurrence of WNS in distantly related bat species with diverse ecologies, however, suggests the pathogen is a generalist and that all bats hibernating within the distribution range of P. destructans may be at risk of infection31. If we assume that P. destructans range expansion and its concurrent WNS epidemic occurred unnoticed in Europe in the past, we expect that WNS should be found at any site in Europe with conditions favourable for the pathogen and could extend to non-European hibernacula of the Palearctic region. We tested the following four hypotheses. First, P. destructans is present in European and non-European Palearctic hibernacula and, if the fungus is present in a hibernaculum, bats would test positive for WNS under histopathology. Second, an endemic steady-state of P. destructans infection in the Palearctic would be reflected in a persistent high prevalence among bats with no associated population declines. Third, given that the differences in population size changes in WNS-positive regions24,38 are explained as WNS resistance in Europe, Palearctic bats will display a lower pathogen load and smaller size of cupping erosions diagnostic for WNS than Nearctic species. Fourth, in regions with endemic pathogen occurrence, P. destructans load will be influenced by geographic location, sampling date and bat community structure.
Results
WNS in the Palearctic
We sampled and examined bats from sites in geographically distant regions (the Czech Republic, Slovenia, Latvia and Russia; n = 481) to assess occurrence of WNS in the Palearctic (Table 1, Supplementary Table S1). We found WNS-positive bats from multiple species at all sites, including the West Siberian Plain in Russia. At each site, all of the following criteria categorised the site as WNS positive: suspected fungal growth (Fig. 1), characteristic curved conidia of P. destructans observed on an adhesive tape imprint (Fig. 2A), P. destructans confirmed by DNA sequencing and qPCR using a species-specific probe, and definitive diagnosis confirmed through histopathology of biopsy punches from wing membrane lesions targeted by UV trans-illumination (Fig. 2B).
Table 1. Prevalence of white-nose syndrome and Pseudogymnoascus destructans infection in bat species from the Holarctic region.
| Region | Bat species | Screened | WNS qPCR+ | WNS UV+ | WNS histo+ | |||
|---|---|---|---|---|---|---|---|---|
| n | n | qPCR+ (%) ± s.e. | n | UV+ (%) ± s.e. | n | histo+ (%) ± s.e. | ||
| Slovenia | Miniopterus schreibersii | 21 | 20 | 100 ± 7.1 | 21 | 47.62 ± 10.9 | 6 | 16.67 ± 20.05 |
| Myotis emarginatus | 3 | 0 | NA ± NA | 3 | 100 ± 32.22 | 3 | 100 ± 32.22 | |
| Myotis myotis | 11 | 10 | 100 ± 13.21 | 11 | 63.64 ± 12.16 | 4 | 0 ± 26.89 | |
| Rhinolophus euryale | 14 | 14 | 64.29 ± 9.83 | 14 | 28.57 ± 9.83 | 1 | 100 ± 48.47 | |
| Total | 49 | 44 | 88.1 ± 4.88 | 49 | 59.96 ± 7 | 14 | 54.17 ± 13.32 | |
| Czech Republic | Barbastella barbastellus | 18 | 17 | 64.71 ± 11.59 | 18 | 50 ± 11.79 | 4 | 75 ± 26.89 |
| Eptesicus nilssonii | 3 | 2 | 50 ± 39.61 | 2 | 0 ± 39.61 | 1 | 100 ± 48.47 | |
| Myotis alcathoe | 7 | 7 | 71.43 ± 17.76 | 7 | 14.29 ± 17.76 | 6 | 0 ± 20.05 | |
| Myotis bechsteinii | 23 | 23 | 86.96 ±6.23 | 23 | 39.13 ±10.18 | 9 | 44.44 ±14.45 | |
| Myotis brandtii | 17 | 16 | 68.75 ±8.71 | 17 | 23.53 ±8.24 | 1 | 100 ±48.47 | |
| Myotis dasycneme | 1 | 1 | 100 ±48.47 | 1 | 100 ±48.47 | 1 | 100 ±48.47 | |
| Myotis daubentonii | 33 | 30 | 86.67 ±4.85 | 31 | 51.61 ±8.98 | 14 | 35.71±12.81 | |
| Myotis emarginatus | 33 | 32 | 96.88 ±4.56 | 32 | 43.75 ±8.77 | 5 | 60 ±23 | |
| Myotis myotis | 156 | 150 | 99.33 ±1.01 | 108 | 96.3 ±1.4 | 61 | 73.77 ± 5.63 | |
| Myotis nattereri | 22 | 21 | 95.24 ±6.78 | 22 | 54.55 ±10.62 | 9 | 33.33 ±14.45 | |
| Plecotus auritus | 23 | 23 | 91.3 ± 6.23 | 22 | 68.18 ± 9.93 | 11 | 54.55 ± 12.16 | |
| Plecotus austriacus | 3 | 3 | 66.67 ± 32.22 | 3 | 33.33 ± 32.22 | 1 | 0 ± 48.47 | |
| Rhinolophus hipposideros | 35 | 29 | 79.31 ± 7.52 | 35 | 34.29 ± 8.02 | 8 | 50 ± 15.94 | |
| Total | 374 | 354 | 81.33 ± 2.07 | 321 | 46.84 ± 2.79 | 131 | 55.91 ± 4.34 | |
| Latvia | Eptesicus nilssonii | 4 | 4 | 25 ± 26.89 | 4 | 25 ± 26.89 | 1 | 0 ± 48.47 |
| Myotis brandtii | 4 | 4 | 50 ± 26.89 | 4 | 50 ± 26.89 | 0 | NA ± NA | |
| Myotis dasycneme | 8 | 8 | 100 ± 15.94 | 8 | 100 ± 15.94 | 6 | 50 ± 20.05 | |
| Myotis daubentonii | 9 | 9 | 100 ± 14.45 | 9 | 88.89 ± 14.45 | 6 | 66.67 ± 20.05 | |
| Total | 25 | 25 | 68.75 ± 9.27 | 25 | 65.97 ± 9.48 | 13 | 38.89 ± 13.52 | |
| Russia | Eptesicus nilssonii | 5 | 5 | 100 ± 23 | 5 | 100 ± 23 | 2 | 100 ± 39.61 |
| Myotis brandtii | 9 | 9 | 100 ±14.45 | 9 | 100 ± 14.45 | 2 | 0 ± 39.61 | |
| Myotis dasycneme | 17 | 17 | 100 ±8.24 | 17 | 100 ±8.24 | 12 | 75 ±11.27 | |
| Myotis daubentonii | 1 | 1 | 100 ± 48.47 | 1 | 100 ±48.47 | 0 | NA ± NA | |
| Plecotus auritus | 1 | 1 | 100 ± 48.47 | 1 | 100 ±48.47 | 1 | 100 ± 48.47 | |
| Total | 33 | 33 | 100 ± 4.43 | 33 | 100 ± 4.43 | 17 | 68.75 ± 11.24 | |
| USA | Myotis lucifugus | 10 | 0 | NA±NA | 10 | 100 ±13.21 | 10 | 100 ±13.21 |
| Total | 10 | 0 | NA ± NA | 10 | 100 ± 13.21 | 10 | 100 ± 13.21 | |
Screened = number of bats captured and examined by PCR (to detect the pathogen), UV light trans-illumination or histopathology (to detect WNS lesions). + = percentage of positive bats from the number screened by the method. NA = not available. Samples subjected for histopathology examination were suspect lesions selected under field conditions based on UV trans-illumination and thus prevalence on histopathology is not based on a randomized sample. It rather reflects qualitative information that WNS was confirmed in the species with histopathology.
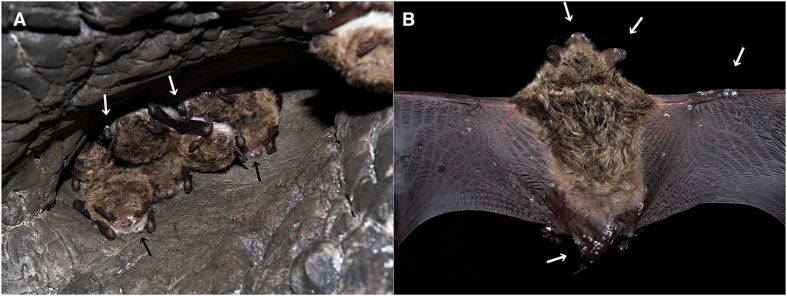
Figure 1. Fungal growth on hibernating bats from Russia.
(a) A hibernating cluster of pond bats Myotis dasycneme in a cave near Yekaterinburg, Russia, in May 2014. Black and white arrows indicate fungal growth on the muzzle and forearm, respectively. (b) A pond bat from the same hibernaculum showing visible fungal growths on the uropatagium, pelvic limb toes, plagio- and pro-patagium and the ears and muzzle (white arrows). Photo: Jiri Pikula.
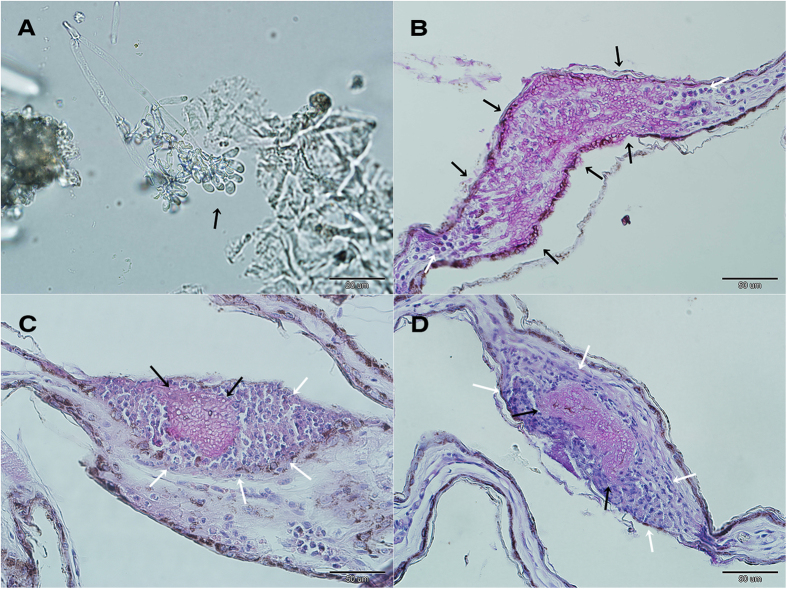
Figure 2. White-nose syndrome on a pond bat Myotis dasycneme near Yekaterinburg, Russia, in May 2014.
(A) Microscopic identification of the characteristic curved conidia of Pseudogymnoascus destructans (black arrow). (B) Invasive fungal growth penetrating the full-thickness of the wing membrane (black arrows), with several inflammatory cells (neutrophils) situated at both margins of the lesion (white arrows). (C) Packed fungal hyphae of cupping erosions (black arrows) sequestered by neutrophils (white arrows). (D) Histopathological finding from a WNS-positive Nearctic Myotis lucifugus, identical to that found in Palearctic Asia. Skin sections stained with periodic acid-Schiff stain.
Histopathological findings matching WNS diagnostic criteria were present in 100 bats of 13 different species (Table 1). Two species were newly identified as positive for WNS, Miniopterus schreibersii and Rhinolophus euryale. Species-specific prevalence, using qPCR to detect P. destructans and UV trans-illumination to detect lesions indicative of WNS, ranged from 64 to 100% (overall prevalence = 83.1%) and 14 to 100% (overall prevalence = 51.9%), respectively (Table 1).
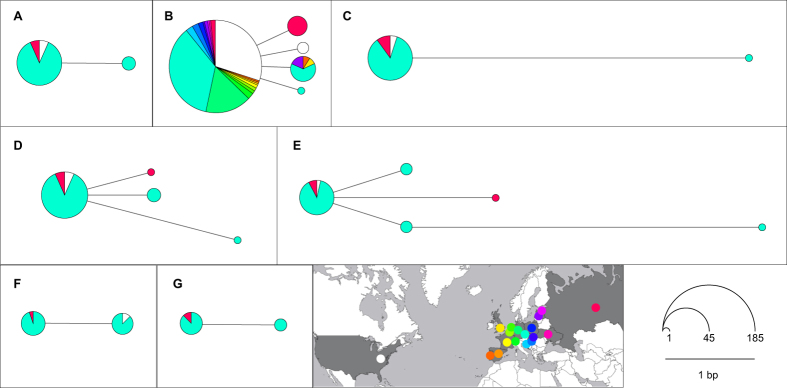
In order to evaluate P. destructans genetic variability, we sequenced six loci resulting in 254 new sequences. Two alleles were found in TUB2, MAT1-1-1, MAT1-2-1 and CAM loci, four alleles in TEF1α and three in ITS (Fig. 3). The most frequent alleles from all loci were found in both European and Asian P. destructans isolates. In ITS and TEF1α from Asia, we sampled isolates with one substitution difference from the most frequent allele. The dominant haplotype from the concatenated sequence of four genes (TUB2, CAM, TEF1α, ITS) is widespread in North America, Europe and Asia (Fig. 3E). Three divergent concatenated haplotypes were found in the Czech Republic and one local to Asian Russia. The P. destructans mating type proportion was 27 (MAT1-1-1) to 19 (MAT1-2-1) (χ2 = 1.391, P = 0.238).
Figure 3. Median-joining networks for Pseudogymnoascus destructans DNA sequences.
Node centroid distances correspond to the number of substitutions between haplotypes and the node size to the number of isolates sharing the haplotype. Isolate origin is signified through different colours based on the inset map. The map was modified from http://www.amcharts.com/visited_countries/ (last accessed on 9 October, 2015). (A) TUB2 (n = 46), (B) ITS (n = 203), (C) CAM (n = 47), (D) TEF1α (n = 51), (E) sequences concatenated with TUB2, ITS, CAM and TEF1α (n = 34); haplotypes pooled according to the available sequence and gaps treated as missing data, (F) MAT1-1-1 (n = 27), (G) MAT1-2-1 (n = 19).
Quantitative comparison of WNS on bats
The fungal load on qPCR-positive bats ranged from 0.21 pg to 3.41 μg across the surface of the left wing (Supplementary Fig. S1, see Table 1 for sample sizes). This range included fungal loads reported from Nearctic bat species (Supplementary Fig. S1). Log-transformed fungal load was not lower on Palearctic bats than on Nearctic bats (Wilcoxon W = 68030, P = 1, n = 413 and 247, respectively), even assuming that fungal load on Nearctic bats could vary up to ten-times due to the difference in sample collection method between continents (Wilcoxon W = 50910, P = 0.484).
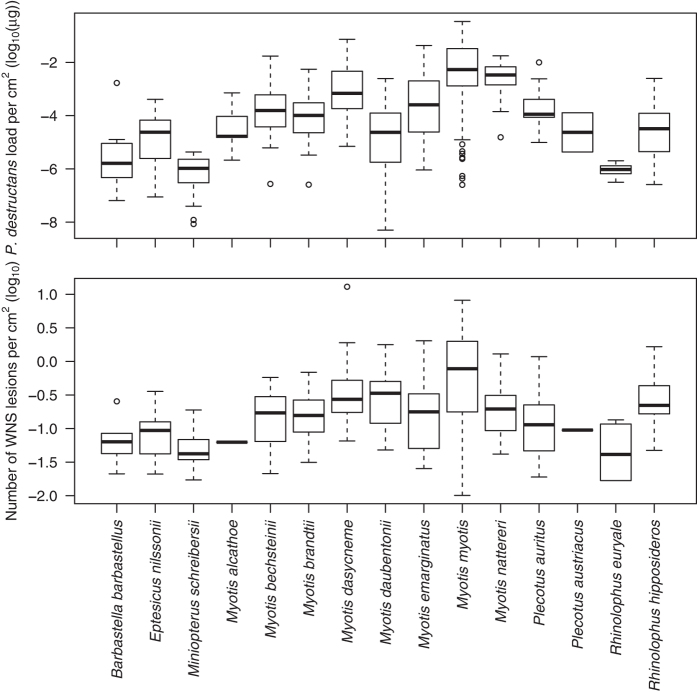
In order to account for body size differences between Palearctic bat species, we used log-transformed fungal load per cm2 of wing area (henceforth referred to as fungal load, unless specified otherwise). Fungal load differed significantly between regions (Kruskal-Wallis χ2 = 94.2, P < 0.001; Fig. 4) and species (Kruskal-Wallis χ2 = 221.2, P < 0.001; Fig. 5), with highest values recorded in the Czech Republic and two euryvalent bat species, Myotis myotis and Myotis nattereri, respectively. In frequently captured M. myotis, fungal load did not change between 2012 and 2014 (ANOVA: F2,146 = 1.209, P = 0.301). Prevalence dynamics of nine species captured in multiple years was statistically equal between years (χ2 test: P > 0.05; Czech Republic). The sampled regions contained multiple species with varying fungal loads (Fig. 5), and some species were present in multiple regions (Table 1). The observed difference of fungal load can be caused by a different set of species or different environmental conditions in sampled regions. Therefore, we separated the effect of the two variables on fungal load by correcting for the random effect of species and region. This had no impact on significance of the comparison between regions or species. The phylogenetic signal of mean species fungal load was significant (Blomberg’s K = 0.892, P = 0.002), meaning that closely related taxa had more similar mean fungal loads in the Palearctic than expected by chance.
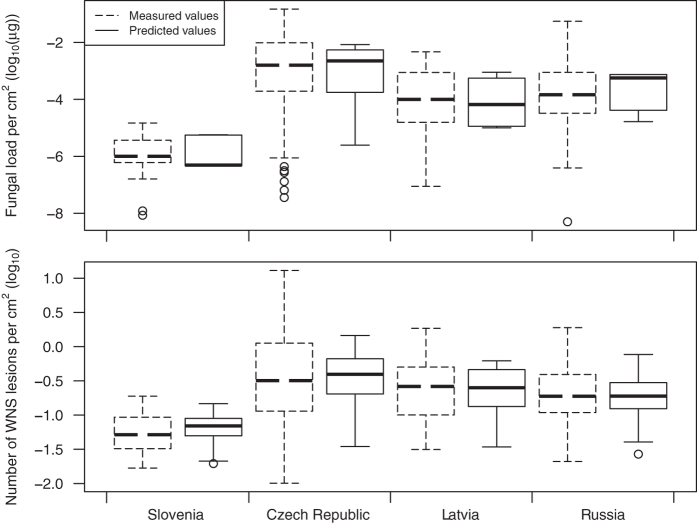
Figure 4. Fungal load and number of WNS lesions in Europe (Czech Republic, Latvia, Slovenia) and in Asia (Russia).
Fungal load is quantified as P. destructans-specific DNA per cm2 of wing area (established through quantitative PCR) and number of WNS lesions is counted on the same wing (using UV light trans-illumination).
Figure 5. Fungal load and number of WNS lesions on bats from Europe (Czech Republic, Latvia, Slovenia) and Asia (Russia).
Fungal load is quantified as P. destructans-specific DNA per cm2 of wing area (established through quantitative PCR) and number of WNS lesions is counted on the same wing (using UV light trans-illumination).
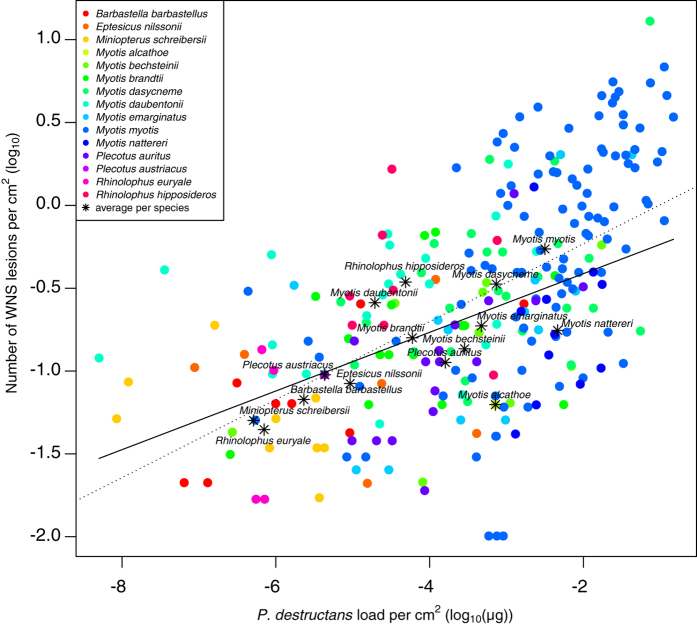
Fungal load was significantly correlated with the number of WNS lesions (Spearman’s rank correlation: r = 0.61, P < 0.001). Phylogenetic generalised least-squares, which accounts for intra-specific variability and species phylogenetic relationships, indicated that the number of WNS lesions increased with increasing fungal load across bat diversity in this study (intercept = –0.058, slope = 0.177; Fig. 6). No significant phylogenetic signal was detected in the mean number of WNS lesions by species (Blomberg’s K = 0.511, P = 0.073).
Figure 6. Phylogenetic generalised least-squares for number of WNS lesions, dependent on fungal load and accounting for within-species variation and relatedness.
The dotted line represents the linear regression without phylogenetic correction.
Microscopic WNS cupping erosion width ranged between 28.6 and 397.2 μm (median = 86.34 μm, n = 105) and depth between 11.3 and 91.8 μm (median = 31.29 μm), this also being the WNS-lesion size range detectable photographically as individual spots using UV trans-illumination. Size of WNS lesion (log-transformed) did not differ significantly between species (Kruskal-Wallis χ2 = 20.7, P = 0.079, n = 14 species across the Holarctic; Fig. 7, see Table 1 for sample sizes). Phylogenetic signal for mean species-specific WNS lesion size was also not significant (Blomberg’s K = 0.448, P = 0.177) and there was no significant difference observed between Palearctic and Nearctic bats (phylogenetic ANOVA: F = 0.43, P = 0.292).
Figure 7. Variation in focal tissue invasiveness among hibernating bat species infected by Pseudogymnoascus destructans.
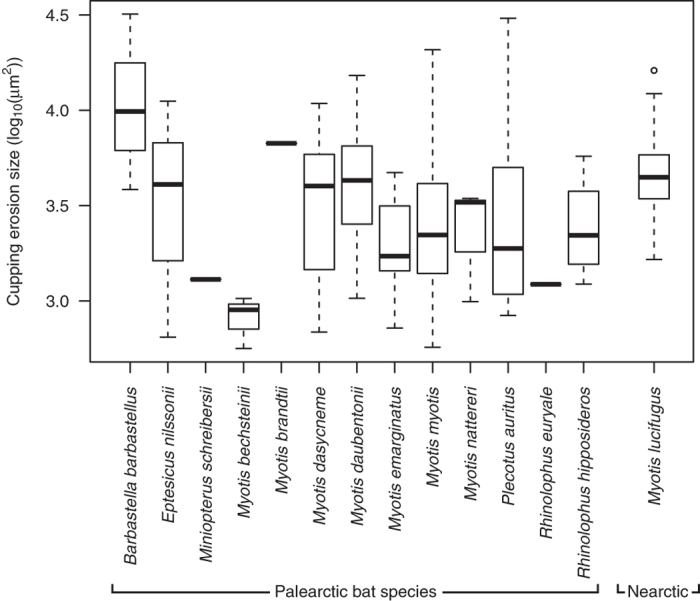
A series of 80 periodic acid–Schiff stained sections from each bat’s wing membrane biopsy were used to determine mean maximum size of WNS-diagnostic cupping erosions (log10(μm2)). See Table 1 for sample sizes.
The number of WNS lesions in animals positive over UV ranged from 1 to 805 (median = 13) and differed significantly between Palearctic regions (log-transformed per cm2; Kruskal-Wallis χ2 = 33.6, P < 0.001; Fig. 4) and species (Kruskal-Wallis χ2 = 76.7, P < 0.001; Fig. 5; correction did not influence significance). The fungal load from UV-negative individuals (median = 3.78 × 10−5 μg) overlapped that from UV-positive individuals (median = 7.46 × 10−4 μg; Fig. 8).
Figure 8. Frequency of Pseudogymnoascus destructans fungal load per cm2 of wing area.
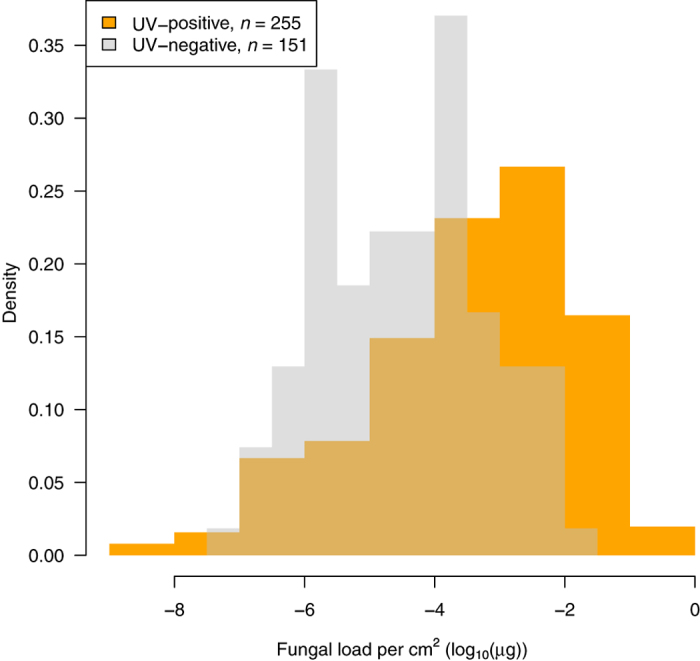
Bats were identified as positive (grey; n = 255) and negative (orange; n = 151) using UV trans-illumination.
Comparison of AIC values indicated that the best fitting model for fungal load across regions included geographic coordinates (95% confidence interval of parameter estimates: longitude: –0.07–(–0.03), latitude: 0.16–0.36), sampling date (0.002–0.02) and structure of common bat species community found in the respective region (–3.86−(−2.24)) (Supplementary Table S2). On the other hand, disease intensity, quantified as number of WNS lesions, was best modelled using fungal load (0.18–0.28) and sampling date (–0.007–0.0003), though addition or removal of other variables did not change the relationship between WNS-lesion number and fungal load significantly (0.18–0.27). In both cases, species was treated as a random effect. Values predicted from the optimal model for both fungal load and number of WNS lesions were highest in the Czech Republic (Fig. 4). Lowest values were predicted in Slovenia and intermediate values in Latvia and Russia.
Discussion
Here we show WNS infection in Palearctic bat communities within and beyond the borders of Europe, greatly extending the distribution range of P. destructans and confirming its generalist nature. Furthermore, we confirm WNS skin lesions in two more bat species, belonging to families Miniopteridae and Rhinolophidae.
Histopathology of skin sections is presently considered the ‘gold standard’ for diagnosing WNS47. The microscopically identified WNS in bat species sampled in Russia was consistent with WNS histopathological criteria42,47. These included characteristic cup-shaped epidermal erosions, P. destructans-infected hair follicles, associated sebaceous glands and regional connective tissues, and invasive fungal growth throughout the wing-membrane thickness (Fig. 2). Our results demonstrate that P. destructans is virulent for Palearctic bats under natural infectious conditions, as previously shown in the Czech Republic31,42. Local invasiveness and WNS-lesion severity in wing membranes of bats collected in the Palearctic was comparable with that of Nearctic bats (Figs 2 and 8), with diagnostic features ranging from cupping erosions to full-thickness fungal invasion found on all continents. In our experience, detection of WNS-positive bats was greatly enhanced by the use of a UV lamp combined with non-lethal punch biopsies of suspected skin lesions48. The effectiveness of this method enabled us to identify seven species as WNS-positive in multiple regions, i.e. Myotis brandtii, Myotis dasycneme, Myotis daubentonii, Myotis emarginatus, M. myotis, Eptesicus nilssonii and Plecotus auritus (Table 1). As predicted31, M. schreibersii and R. euryale were confirmed positive for skin lesions. These are thermophilic species that only hibernate in the northern part of their ranges. Miniopterus schreibersii, a long-distance migrant, forms large mixed colonies with other cave-dwelling bat species49 and such characteristics make the species another effective candidate for pathogen dispersal.
Theoretically, WNS spread is limited by conditions prevalent in underground hibernacula being favourable to P. destructans and presence of susceptible hibernating bats50. Pseudogymnoascus destructans, being a generalist pathogen, could infect any bat species hibernating under the right microclimatic conditions and ecological and evolutionary differences in the hibernating bats would not pose a barrier31. Distribution ranges of multiple WNS-positive bat species (E. nilssonii, M. dasycneme, M. daubentonii, M. brandtii, P. auritus) extend across Palearctic Asia and overlap with their sister species or ecological counterparts (e.g. Myotis petax, Myotis sibiricus). As bats can form multi-species clusters at hibernation sites, there is a good chance that P. destructans could switch host species to presently WNS-negative taxa (e.g. Myotis ikonnikovi, M. petax, M. sibiricus). This suggests that WNS may be present throughout the Palearctic where suitable environmental conditions occur and in all bat species that utilise such sites. In fact, a paper published ahead of print during review of this article corroborates our conclusions with one M. petax found positive for WNS in North China51.
Given the tragic impact of WNS on bat biodiversity in North America24, it is important to identify the source of possible introduction4. However, P. destructans exhibits notoriously low genetic diversity in both the clonal North American populations33,34,43 and the sexually reproducing populations of Europe44, effectively thwarting efforts to pinpoint the source region. Our study further exacerbates the enigma of where the North American P. destructans strain originates as the North American haplotype was found in all non-mating gene types in Palearctic Asia, thereby expanding the putative source region (Fig. 3). Additionally, the search for a source region should shift to markers with a faster mutation rate and better resolution for P. destructans phylogeography, as the low genetic variability in markers routinely used in fungal population studies indicates a relatively recent origin or expansion of the Palearctic population.
Theory suggests that if a disease is detectable at high prevalence it is probably mild and unlikely to be a major problem52. Two methods were used to establish prevalence in this study: qPCR, used to quantify the pathogen on wing skin53, and UV trans-illumination of the wing membrane, which enables detection of fluorescing lesions associated with WNS skin infection48. Both methods provided comparable results of high prevalence, and, together with stable or increasing host population sizes54, our data strongly suggest endemicity of P. destructans within Palearctic bat populations. Persistent high prevalence and pathogen load with absence of population declines in the Palearctic are in sharp contrast to the situation in the Nearctic. In Midwestern United States, high prevalence of P. destructans infection was followed by a decrease in number of hibernating bats55. Some form of host-pathogen equilibrium, analogous to the amphibian chytrid fungus observed in post-decline frog communities56, may already have occurred in the Palearctic but not in the Nearctic.
Pathogen load indicates both exposure to the infectious agent and suitability of the host for pathogen replication57. Pseudogymnoascus destructans fungal load can be evaluated using swab samples from wing membranes53. Although swabs do not sample fungus invasion deep in the wing tissue, our results show that fungal load sampled in this way is positively correlated with the number of WNS lesions (Fig. 6). Hence, fungal load from swabs may be assumed to approximate the total load associated with the skin infection. Fungal loads determined across the Palearctic were similar to those observed in Nearctic species (Supplementary Fig. S1). The swabbing technique used in the North American studies19,58,59, however, does not allow standardisation of P. destructans load per cm2 and thus detailed statistical comparisons cannot be made. Nevertheless, fungal loads in Nearctic bats are similar to the observed fungal load of Palearctic bats (Supplementary Fig. S1), even if the former were underestimated ten-fold with the different swabbing technique.
In the Palearctic study area, fungal load differed regionally (Fig. 4) and between species (Fig. 5) with no reported mass decline in bat populations attributable to WNS38,54. Fungal load increased with increasing northing and westing, sampling day (load increasing later in the year) and in regions where phylogenetically closely-related hibernating bats predominated (Fig. 4). The driving factor that best modelled the number of WNS lesions observed using UV trans-illumination in the Palearctic was the fungal load. Increasing fungal load positively correlated with disease intensity across species diversity indicates that hyphae are more likely to invade deep tissues and cause WNS lesions with heavy fungal growth on bat wings (Fig. 5). However, with overlap of fungal loads in UV-negative and UV-positive bats, no clear fungal load threshold defines the pathogen pressure where development of WNS lesions starts.
With persistent and similar fungal load (Supplementary Fig. S1) and cupping erosion size (Fig. 7) on bats from the Palearctic and the Nearctic, the difference in population size response is striking. While population sizes dropped dramatically under the WNS epidemic in the Nearctic24, population size changes remained within normal inter-annual fluctuation levels in the Palearctic38,54,60. Our data suggest that, once the hibernacula are contaminated by the fungus, Palearctic bats are exposed to high pathogen pressure and the infection is continuously present to a high level, i.e. the so-called hyperendemic condition. The difference in infection outcome is therefore a function of adaptation to pathogen pressure.
While reduction of pathogen load is a function of host resistance, the ability to limit the harm caused by a given load is a result of tolerance61,62. These two alternative and complementary forms of defence may have profound effects on the epidemiology of infectious diseases and on host-pathogen coevolution62. Resistance protects the host at the expense of the pathogen and tolerance saves the host from harm without direct negative effects on the infectious agent. Evolution of resistance, therefore, should also reduce the prevalence of the pathogen in host populations. On the other hand, tolerance is expected to have a neutral, or even positive, effect on prevalence of the pathogen. Lack of resistance to WNS infection in hibernating European bats has been demonstrated in 13 species31,42 that exhibit stable or increasing population sizes54. Yet, prevalence of P. destructans-positive bats reached 100% in the Palearctic. In light of the above arguments, it would appear that mechanisms promoting tolerance to P. destructans infection are in operation in the area studied. Palearctic bat species tolerate comparable fungal loads and WNS-lesion size to their Nearctic counterparts suffering population declines. We hypothesise that the balance between tolerance and resistance mechanisms changes with transition of bats from hibernation to euthermia.
Our results provide evidence of P. destructans and pathognomonic WNS skin lesions in hibernating bats sampled from the West Siberian Plain of Russian Asia and indicate endemicity of this virulent fungus in the Palearctic region. Data suggesting bat tolerance imply lowered risk following establishment of equilibrium in the host-pathogen interaction. The extensive spatial distribution of the agent may pose a threat, however, representing a continued source of introduction to other regions with naïve bats not yet exposed to the pathogen. Classical models employing the disease triangle concept suggest that the epidemiological outcome of an infection depends on determinants of the pathogen, host(s) and the environment. Alterations in any of these determinants may trigger shifts in the complex host-pathogen system, as seen here by infection tolerance in hibernating Palearctic bats.
Methods
Material collection
Between 2012 and 2014, we sampled 481 bats (15 species) at 20 sites in Slovenia, the Czech Republic, Latvia and Russia (West Siberian Plain, Asia; Table 1, Fig. 3). Samples were taken as late in the hibernation or as early in the post-hibernation season as possible (February-May) to minimise the impact of disturbance to specific bat species. Following capture, the wings, ears and muzzle were swabbed with a nylon (FLOQ Swabs, Copan Flock Technologies srl, Brescia, Italy) or cotton swab (Plain swab sterile plastic applicator, Copan) in a standardised manner in order to collect fungal biomass from the whole skin area for fungal detection (dorsal side of left wing only for qPCR). The bats were photographed over a 368 nm wavelength UV lamp and wing punch biopsies of suspect tissue collected in 10% formalin for histopathological examination. All bats were then released at the site.
WNS was diagnosed according to current standards47, i.e. P. destructans presence was confirmed with qPCR53 and selected orange-yellow spots observed over UV were sampled. A series of 80 periodic acid-Schiff stained sections embedded in paraffin were obtained from each wing membrane biopsy. These were then observed under an Olympus BX51 light microscope (Olympus Corporation, Tokyo, Japan). Fungal cell walls were stained magenta under the periodic acid-Schiff stain, which allowed measurement of cupping erosions packed with hyphae. Using cellSense Software tools (Olympus Soft Imaging, GmbH, Münster, Germany), we measured total area (size) of cupping erosions. Trans-illuminated photographs of the left wing membrane stretched over a UV lamp were used to manually enumerate yellow-orange fluorescing pinpoints indicative of WNS lesions48, using the individual object counting tool of ImageJ63.
Bats were considered WNS-positive if qPCR confirmed P. destructans infection, wings exhibited characteristic UV fluorescence48 and cupping erosions packed with fungal hyphae or a full thickness fungal invasion of the wing membrane were observed under histopathology47. Additional swabs taken from the wings and muzzle after sampling for qPCR were used for cultivation on Sabouraud dextrose agar plates and isolation of the fungus in pure cultures38. Representative isolates were deposited at the Culture Collection of Fungi, Charles University in Prague, Czech Republic.
Field work and sampling in the Czech Republic was performed in accordance with Czech Law No. 114/1992 on Nature and Landscape Protection, based on permits 01662/MK/2012S/00775/MK/2012, 866/JS/2012 and 00356/KK/2008/AOPK issued by the Agency for Nature Conservation and Landscape Protection of the Czech Republic. Experimental procedures were approved by the Ethical Committee of the Academy of Sciences of the Czech Republic (No. 169/2011). Sampling in Latvia was approved by the Nature Conservation Agency (No. 3.15/146/2014-N), in Slovenia by the Ministry of Environment and Spatial Planning of the Slovenian Republic, Slovenian Environment Agency (No. 35601-35/2010-6) and in Russia by the Institute of Plant and Animal Ecology, Ural Division of the Russian Academy of Sciences (No. 16353–2115/325). The authors were authorised to handle free-living bats according to the Czech Certificate of Competency (No. CZ01341; §17, Act No. 246/1992 Coll.) and a permit approved by the Latvian Nature Conservation Agency (No. 05/2014).
DNA isolation
Total DNA was isolated from swabs using the QIAamp DNA Mini Kit (Qiagen, Halden, Germany). Swabs were placed in tissue lysis buffer with proteinase K and incubated at 56 °C for two hours. Lysis buffer was added and the samples incubated for a further 10 min. After this step, we followed the Qiagen Buccal Swab Spin Protocol according to the manufacturer’s recommendations.
Quantitative PCR
We performed quantitative PCR53 (qPCR) using TaqMan® Universal Master Mix II with UNG (Life Technologies, Foster City, CA, USA). To optimise the PCR reaction, we added bovine serum albumin and Platinum® Taq DNA Polymerase (Life Technologies) in final concentrations of 0.05 mg/μl and 0.025 U/μl, respectively. We used both forward and reverse primers (0.3 μM each) and species-specific and genus-specific fluorescently-labelled custom probes were used for quantification of the PCR product. The reaction mix was prepared on ice and three replicates were prepared for each DNA sample. We performed real-time PCR reaction on a LightCycler 480 PCR platform (Life Technologies) with initial inactivation at 50 °C for 2 min and a hot start at 96 °C for 10 min. Nine cycles with a denaturation step at 95 °C for 15 sec and annealing at 62 °C for 1 min were followed by 43 identical cycles with quantification detection. qPCR was finalised by dissociation at 95-60-95 °C for 15 sec each and cooling to 40 °C for 10 min. DNA isolated from CCF3937 culture38 and water were used as positive and negative controls and as concentration references for each run.
Fungal load on Palearctic bats
We calculated a DNA concentration calibration curve using a CCF3937 dilution series. Exact DNA concentrations (ng μl−1) in the dilution series were determined using Qubit HS fluorometry via the manufacturer’s protocol. Effectivity of qPCR was 1.96. We used this dilution series to estimate the relationship between fungal load and qPCR cycle, calculating P. destructans DNA concentration in the sample using custom scripts in R64 with the equation log(qPdDNA) = 3.194 – 0.287 Cp (R2 = 0.9719), where q is the DNA concentration and Cp the cycle. Each result was converted to fungal load based on the positive control and overall elution of DNA.
Fungal load on Nearctic bats
We downloaded previously published P. destructans loads from natural infections19,58 or recalculated loads from mean Cp values according to the authors’ equation59. In one case, we plotted means as individual data points as the authors19 included species means per site (plus standard error) but fungal loads per individual were not published. The sample sizes for Nearctic bat species sampled within the dates used in this study were: Corynorhinus rafinesquii (n = 2), Eptesicus fuscus (n = 6), Lasiurus borealis (n = 2), Myotis grisescens (n = 11), M. leibii (n = 1), M. lucifugus (n = 36), M. septentrionalis (n = 4), M. sodalis (n = 13) and Perimyotis fuscus (n = 172). None of the North American studies specified sampled area on the bat with sufficient precision to enable standardisation to cm2 or universal statistical comparison.
Molecular genetic variability of P. destructans
To assess genetic variability, we used a set of isolates collected between 2009 and 2014 in the Czech Republic38,65 and isolates from this study. The isolates were characterised using ITS and five other nuclear gene sequences using previously published primers (Supplementary Table S3). Chromatograms were assembled to DNA sequences in Geneious 6 (Biomatters, Auckland, New Zealand) and submitted to the European Nucleotide Archive (LN871244-LN871428). These were checked with blast and P. destructans was confirmed at all sites with a sequence identity to previously sequenced strains ≥99% in all markers. We then downloaded previously published P. destructans sequences of the respective genes from GenBank. Sequence haplotypes were identified based on available nucleotide residues, with gaps and unresolved bases treated as missing data. We estimated relationships between haplotypes using median-joining networks66.
Individual-based linear mixed models
We modelled intensity of WNS infection using fungal load on the dorsal side of the left wing and the number of WNS lesions on the same wing visualised over UV in R64. UV-detectable lesions correspond with cupping erosions diagnostic for WNS in Europe and North America, thereby reflecting disease intensity47,48. Our previous experience in the Czech Republic has shown that wing damage tends to be localised and that lesions do not usually merge31, allowing them to be counted. The yellow-orange fluorescent spots were counted by a researcher with no knowledge of the qPCR and histopathology results for the samples. We scaled variables to 1 cm2 of wing area67 and log-transformed them in order to account for body-size differences between species. We included latitude and longitude, sampling day (from the beginning of the calendar year) and scores for the first two principal components (characterising dominant hibernating bat community in each region; Supplementary Fig. S2) as fixed-effect variables and species as a random effect. Microclimate at a specific bat roost might influence fungal load and disease intensity on an individual through a compound effect of optimizing growth conditions for the pathogen and hibernation conditions for the host. We used geographic coordinates as a proxy for possible general site microclimate (mean annual temperature, elevation, humidity) under the assumption that fungal load and number of WNS lesions would increase at higher latitudes and with longitudinal shift from oceanic to more continental climate. Similarly, we expected day of sampling to be reflected in the dependent variables as increased time for fungus propagation might increase its loading. As such, geographic location, combined with sampling date, can be understood as a proxy for cave microclimate, which would influence growth of the WNS fungus50,68.
Both relatedness and species assemblage differences at hibernacula in a given region could skew regional fungal load and disease intensity in favour of those areas where severely infested species co-occur. We estimated bat communities present at hibernacula from local surveys. Relatedness of species recorded was assessed using mean phylogenetic distance and mean nearest taxon distance (both with weighted abundance) and UniFrac metric measuring the percentage of phylogeny shared by a given community. We used these metrics for principal components analysis. We used backward stepwise variable selection to develop the model, which was assessed using the Akaike Information Criterion (AIC).
Species-level phylogenetically-informed model
Previously published phylogenetic trees for European bats31 based on multilocus sequence data were rescaled as an ultrametric tree using penalised likelihood69,70 with λ and root height = 1. We used R70,71 to calculate phylogenetic signal in species means and phylogenetic ANOVA71,72. We used the phylogenetic generalised least-squares method72,73 to explain the number of visible WNS lesions on a UV trans-illuminated wing based on P. destructans fungal load.
Additional Information
How to cite this article: Zukal, J. et al. White-nose syndrome without borders: Pseudogymnoascus destructans infection tolerated in Europe and Palearctic Asia but not in North America. Sci. Rep. 6, 19829; doi: 10.1038/srep19829 (2016).
Supplementary Material
Acknowledgments
This study was supported through a grant from the Czech Science Foundation (Grant No. P506-12-1064). We are grateful to Tomáš Bartonička, Hana Berková and Masha Orlova for invaluable field assistance, to Matej Dolinay, Jiří C. Moravec, Patrícia Pečnerová and Aneta Reichová for laboratory assistance and to Gregory G. Turner for biopsy samples from Nearctic bats.
Footnotes
Author Contributions J.Z., J.P. and N.M. designed the study and participated in field research, with help from H.B., J.B., V.K., O.O., P.P. and J.S. H.B., A.C., A.K., A.N., J.P. and A.Z. performed the laboratory analyses. K.S.J., M.K., J.P. and N.M. analysed the data. J.Z., J.P. and N.M. wrote the manuscript, with contributions from all authors.
References
- Daszak P., Cunningham A. A. & Hyatt A. D. Emerging infectious diseases of wildlife - Threats to biodiversity and human health. Science 287, 443–449 (2000). [DOI] [PubMed] [Google Scholar]
- Keesing F. et al. Impacts of biodiversity on the emergence and transmission of infectious diseases. Nature 468, 647–652 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson A. & Foufopoulos J. Emerging infectious pathogens of wildlife. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356, 1001–1012 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puechmaille S. J. et al. White-nose syndrome: is this emerging disease a threat to European bats? Trends Ecol. Evol. 26, 570–576 (2011). [DOI] [PubMed] [Google Scholar]
- Fisher M. C. et al. Emerging fungal threats to animal, plant and ecosystem health. Nature 484, 186–194 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman A. & Casadevall A. Mammalian endothermy optimally restricts fungi and metabolic costs. mBio 1, e00212–00210 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Solache M. A. & Casadevall A. Global warming will bring new fungal diseases for mammals. mBio 1, e00061–00010 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-González A. E. et al. Histoplasma capsulatum and Pneumocystis spp. co-infection in wild bats from Argentina, French Guyana, and Mexico. BMC Microbiology 14, 23 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johara M. Y. et al. Nipah virus infection in bats (order Chiroptera) in peninsular Malaysia. Emerg. Infect. Dis. 7, 439–441 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman D. T. S. et al. Long-term survival of an urban fruit bat seropositive for Ebola and Lagos bat viruses. PLoS ONE 5, e11978 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton D. J. et al. Experimental Nipah virus infection in Pteropid bats (Pteropus poliocephalus). J. Comp. Pathol. 136, 266–272 (2007). [DOI] [PubMed] [Google Scholar]
- Olival K. J. & Hayman D. T. S. Filoviruses in bats: Current knowledge and future directions. Viruses 6, 1759–1788 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calisher C. H., Childs J. E., Field H. E., Holmes K. V. & Schountz T. Bats: Important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 19, 531–545 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moratelli R. & Calisher C. H. Bats and zoonotic viruses: can we confidently link bats with emerging deadly viruses? Mem. Inst. Oswaldo Cruz. 110, 1–22 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea T. J. et al. Bat flight and zoonotic viruses. Emerg. Infect. Dis. 20, 741–745 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandl J. N. et al. Reservoir host immune responses to emerging zoonotic viruses. Cell 160, 20–35 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meteyer C. U., Barber D. & Mandl J. N. Pathology in euthermic bats with white nose syndrome suggests a natural manifestation of immune reconstitution inflammatory syndrome. Virulence 3, 583–588 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouma H. R., Carey H. V. & Kroese F. G. M. Hibernation: the immune system at rest? J. Leukoc. Biol. 88, 619–624 (2010). [DOI] [PubMed] [Google Scholar]
- Langwig K. E. et al. Host and pathogen ecology drive the seasonal dynamics of a fungal disease, white-nose syndrome. Proc. R. Soc. B Biol. Sci. 282, 20142335 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blehert D. S. et al. Bat white-nose syndrome: An emerging fungal pathogen? Science 323, 227 (2009). [DOI] [PubMed] [Google Scholar]
- Gargas A., Trest M. T., Christensen M., Volk T. J. & Blehert D. S. Geomyces destructans sp. nov. associated with bat white-nose syndrome. Mycotaxon 108, 147–154 (2009). [Google Scholar]
- Minnis A. M. & Lindner D. L. Phylogenetic evaluation of Geomyces and allies reveals no close relatives of Pseudogymnoascus destructans, comb. nov., in bat hibernacula of eastern North America. Fungal Biol. 117, 638–649 (2013). [DOI] [PubMed] [Google Scholar]
- Warnecke L. et al. Inoculation of bats with European Geomyces destructans supports the novel pathogen hypothesis for the origin of white-nose syndrome. Proc. Nat. Acad. Sci. USA 109, 6999–7003 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick W. F. et al. An emerging disease causes regional population collapse of a common North American bat species. Science 329, 679–682 (2010). [DOI] [PubMed] [Google Scholar]
- Coleman J. T. H. & Reichard J. D. Bat white-nose syndrome in 2014: A brief assessment seven years after discovery of a virulent fungal pathogen in North America. Outlooks in Pest Management 25, 374–377 (2014). [Google Scholar]
- Turner G. G., Reeder D. M. & Coleman J. T. H. A five-year assessment of mortality and geographic spread of white-nose syndrome in North American bats, with a look at the future. Update of white-nose syndrome in bats. Bat Research News 52, 13–27 (2011). [Google Scholar]
- Lorch J. M. et al. Experimental infection of bats with Geomyces destructans causes white-nose syndrome. Nature 480, 376–378 (2011). [DOI] [PubMed] [Google Scholar]
- Langwig K. E. et al. Sociality, density-dependence and microclimates determine the persistence of populations suffering from a novel fungal disease, white-nose syndrome. Ecol. Lett. 15, 1050–1057 (2012). [DOI] [PubMed] [Google Scholar]
- Lorch J. M. et al. Distribution and environmental persistence of the causative agent of white-nose syndrome, Geomyces destructans, in bat hibernacula of the Eastern United States. Appl. Environ. Microbiol. 79, 1293–1301 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt J. R. et al. Long-term persistence of Pseudogymnoascus destructans, the causative agent of white-nose syndrome, in the absence of bats. EcoHealth, 10.1007/s10393-014-0981-4 (2014). [DOI] [PubMed] [Google Scholar]
- Zukal J. et al. White-nose syndrome fungus: A generalist pathogen of hibernating bats. PLoS ONE 9, e97224 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch J. M. et al. A culture-based survey of fungi in soil from bat hibernacula in the eastern United States and its implications for detection of Geomyces destructans, the causal agent of bat white-nose syndrome. Mycologia 105, 237–252 (2013). [DOI] [PubMed] [Google Scholar]
- Ren P. et al. Clonal spread of Geomyces destructans among bats, midwestern and southern United States. Emerg. Infect. Dis. 18, 883–885 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkumar S. S. et al. Clonal genotype of Geomyces destructans among bats with white nose syndrome, New York, USA. Emerg. Infect. Dis. 17, 1273–1276 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar L. E., Lira-Noriega A., Medina-Vogel G. & Townsend Peterson A. Potential for spread of the white-nose fungus (Pseudogymnoascus destructans) in the Americas: use of Maxent and NicheA to assure strict model transference. Geospat. Health 9, 221–229 (2014). [DOI] [PubMed] [Google Scholar]
- Maher S. P. et al. Spread of white-nose syndrome on a network regulated by geography and climate. Nat. Commun. 3, 1306 (2012). [DOI] [PubMed] [Google Scholar]
- Maslo B. & Fefferman N. H. A case study of bats and white-nose syndrome demonstrating how to model population viability with evolutionary effects. Conserv. Biol. 29, 1176–1185 (2015). [DOI] [PubMed] [Google Scholar]
- Martínková N. et al. Increasing incidence of Geomyces destructans fungus in bats from the Czech Republic and Slovakia. PLoS ONE 5, e13853 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puechmaille S. J. et al. Pan-European distribution of white-nose syndrome fungus (Geomyces destructans) not associated with mass mortality. PLoS ONE 6, e19167 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibbelt G. et al. White-nose syndrome fungus (Geomyces destructans) in bats, Europe. Emerg. Infect. Dis. 16, 1237–1243 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlinić I., Đaković M. & Lojkić I. Pseudogymnoascus destructans in Croatia confirmed. Eur. J. Wildl. Res. 61, 325–328 (2015). [Google Scholar]
- Bandouchova H. et al. Pseudogymnoascus destructans: evidence of virulent skin invasion for bats under natural conditions, Europe. Transbound. Emerg. Dis. 62, 1–5 (2015). [DOI] [PubMed] [Google Scholar]
- Palmer J. M. et al. Molecular characterization of a heterothallic mating system in Pseudogymnoascus destructans, the fungus causing white-nose syndrome of bats. G3 (Bethesda) 4, 1755–1763 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leopardi S., Blake D. & Puechmaille S. J. White-nose syndrome fungus introduced from Europe to North America. Curr. Biol. 25, R217–219 (2015). [DOI] [PubMed] [Google Scholar]
- Blehert D. S. Fungal disease and the developing story of bat white-nose syndrome. PLoS Pathog. 8, e1002779 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch J. M. et al. The fungus Trichophyton redellii sp. nov. causes skin infections that resemble white-nose syndrome of hibernating bats. J. Wildl. Dis. 51, 36–47 (2015). [DOI] [PubMed] [Google Scholar]
- Meteyer C. U. et al. Histopathologic criteria to confirm white-nose syndrome in bats. J. Vet. Diagn. Invest. 21, 411–414 (2009). [DOI] [PubMed] [Google Scholar]
- Turner G. G. et al. Nonlethal screening of bat-wing skin with the use of ultraviolet fluorescence to detect lesions indicative of white-nose syndrome. J. Wildl. Dis. 50, 566–573 (2014). [DOI] [PubMed] [Google Scholar]
- Hutterer R., Ivanova T., Meyer-Cords C. & Rodrigues L. Bat Migrations in Europe: A Review of Banding Data and Literature. (Federal Agency for Nature Conservation, 2005). [Google Scholar]
- Perry R. W. A review of factors affecting cave climates for hibernating bats in temperate North America. Environ. Rev. 21, 28–39 (2013). [Google Scholar]
- Hoyt J. R. et al. Widespread bat white-nose syndrome fungus, Northeastern China. Emerg. Infect. Dis. 22, 10.3201/eid2201.151314 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum H. & Dobson A. Detecting disease and parasite threats to endangered species and ecosystems. Trends Ecol. Evol. 10, 190–194 (1995). [DOI] [PubMed] [Google Scholar]
- Shuey M. M., Drees K. P., Lindner D. L., Keim P. & Foster J. T. Highly sensitive quantitative PCR for the detection and differentiation of Pseudogymnoascus destructans and other Pseudogymnoascus species. Appl. Environ. Microbiol. 80, 1726–1731 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Meij T. et al. Return of the bats? A prototype indicator of trends in European bat populations in underground hibernacula. Mammalian Biology - Zeitschrift für Säugetierkunde 80, 170–177 (2015). [Google Scholar]
- Langwig K. E. et al. Invasion dynamics of white-nose syndrome fungus, Midwestern United States, 2012–2014. Emerg. Infect. Dis. 21, 10.3201/eid2106.150123 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retallick R. W., McCallum H. & Speare R. Endemic infection of the amphibian chytrid fungus in a frog community post-decline. PLoS Biol. 2, e351 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horrocks N. P., Matson K. D. & Tieleman B. I. Pathogen pressure puts immune defense into perspective. Integr. Comp. Biol. 51, 563–576 (2011). [DOI] [PubMed] [Google Scholar]
- Bernard R. F., Foster J. T., Willcox E. V., Parise K. L. & McCracken G. F. Molecular detection of the causative agent of white-nose syndrome on Rafinesque’s big-eared bats (Corynorhinus rafinesquii) and two species of migratory bats in the southeastern USA. J. Wildl. Dis. 51, 519–522 (2015). [DOI] [PubMed] [Google Scholar]
- Janicki A. F. et al. Efficacy of visual surveys for White-nose syndrome at bat hibernacula. PLoS ONE 10, e0133390, 10.1371/journal.pone.0133390 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick W. F. et al. Disease alters macroecological patterns of North American bats. Glob. Ecol. Biogeogr. 24, 741–749, 10.1111/geb.12290 (2015). [DOI] [Google Scholar]
- Medzhitov R., Schneider D. S. & Soares M. P. Disease tolerance as a defense strategy. Science 335, 936–941 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Råberg L., Graham A. L. & Read A. F. Decomposing health: tolerance and resistance to parasites in animals. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 37–49 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Meth. 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core Team. R. R: A language and environment for statistical computing, http://www.R-project.org/ (2013).
- Kubátová A., Koukol O. & Nováková A. Geomyces destructans, phenotypic features of some Czech isolates. Czech Mycol. 63, 65–75 (2011). [Google Scholar]
- Bandelt H. J., Forster P. & Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48 (1999). [DOI] [PubMed] [Google Scholar]
- Norberg U. M. & Rayner J. M. V. Ecological morphology and flight in bats (Mammalia, Chiroptera): Wing adaptations, flight performance, foraging strategy and echolocation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 316, 335–427 (1987). [Google Scholar]
- Hallam T. G. & Federico P. The panzootic white-nose syndrome: an environmentally constrained disease? Transbound. Emerg. Dis. 59, 269–278 (2012). [DOI] [PubMed] [Google Scholar]
- Paradis E., Claude J. & Strimmer K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (2004). [DOI] [PubMed] [Google Scholar]
- Sanderson M. J. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Mol. Biol. Evol. 19, 101–109 (2002). [DOI] [PubMed] [Google Scholar]
- Harmon L. J., Weir J. T., Brock C. D., Glor R. E. & Challenger W. GEIGER: Investigating evolutionary radiations. Bioinformatics 24, 129–131 (2008). [DOI] [PubMed] [Google Scholar]
- Revell L. J. phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223 (2012). [Google Scholar]
- Ives A. R., Midford P. E. & Garland T. Jr. Within-species measurement error in phylogenetic comparative methods. Syst. Biol. 56, 252–270 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.