Abstract
Nitrogen is one of the most essential plant nutrients and one of the major factors limiting crop productivity. Having the goal to perform a more sustainable agriculture, there is a need to maximize biological nitrogen fixation, a feature of legumes. To enhance our understanding of the molecular mechanisms controlling the interaction between legumes and rhizobia, the symbiotic partner fixing and assimilating the atmospheric nitrogen for the plant, researchers took advantage of genetic and genomic resources developed across different legume models (e.g., Medicago truncatula, Lotus japonicus, Glycine max, and Phaseolus vulgaris) to identify key regulatory protein coding genes of the nodulation process. In this study, we are presenting the results of a comprehensive comparative genomic analysis to highlight orthologous and paralogous relationships between the legume genes controlling nodulation. Mining large transcriptomic datasets, we also identified several orthologous and paralogous genes characterized by the induction of their expression during nodulation across legume plant species. This comprehensive study prompts new insights into the evolution of the nodulation process in legume plant and will benefit the scientific community interested in the transfer of functional genomic information between species.
Keywords: legume, nodulation, comparative genomic, comparative transcriptomic, paralogs, orthologs, neo-/sub-functionalization, root hair cell
Introduction
Legumes (family Fabaceae) are the 2nd most important crop family after grass species. Legumes are an important source of oil and proteins for human and animal consumption, and they also fix atmospheric nitrogen leading to a sturdy supply in nitrogen fertilizers, which can benefit other plant species. This unique feature of legumes is the result of their symbiotic relationship with soil bacteria involved in nodulation (e.g., Bradyrhizobium japonicum and Sinorhizobium meliloti are the Glycine max and Medicago truncatula symbiotic partners, respectively). Ultimately, upon infection of the legume by the symbiotic bacteria, a novel plant root lateral organ called a nodule is formed where rhizobia fix and convert atmospheric nitrogen into nitrite/ammonia that can be used by plants. In exchange, acting as a carbon sink, the symbionts receive plant photosynthates.
Legume nodulation is initiated by the recognition of plant flavonoids by rhizobia. In response, rhizobia secretes the Nod Factor (NF), a lipochito-oligosaccharide signal molecule which is recognized by plant lysine motif (LysM) receptor kinase, such as Lotus japonicus NFR1/NFR5 (Madsen et al., 2003; Radutoiu et al., 2003), Pisum sativum SYM10 (Madsen et al., 2003), and M. truncatula NFP (Amor et al., 2003) and LYK3 (Limpens et al., 2003). Downstream of the recognition of the NF, a signaling cascade is activated leading to oscillations in calcium concentration within the nucleus of the root hair cell (Miwa et al., 2006; Capoen et al., 2009; Sieberer et al., 2009). Ultimately, this molecular recognition of the two partners will lead to root hair cell deformation, curling and formation of a shepherd hook (Kijne, 1992). Upon root hair curling. rhizobia infect the root hair cell through the formation of a tube-like apoplastic compartment called infection thread (VandenBosch et al., 1989). Concomitantly, the root cortex cells are actively dividing leading to the formation of the nodule primordium (Yang et al., 1994).
Through an examination of legume mutants defective in nodulation, root hair deformation and calcium spiking, the root hair regulatory pathway activated in response to rhizobia inoculation or NF treatment was characterized. For instance, while the M. truncatula mutants dmi1 and dmi2 (does not make infections) are not affected in their calcium flux and root hair deformation, the nfp (nod factor perception) mutant is impaired for both phenotypes suggesting that the DMI1 and DMI2 genes are acting downstream to NFP (Amor et al., 2003). As a part of this regulatory pathway, CCaMK protein, a nuclear protein sensitive to the calcium oscillations, interacts with and phosphorylates CYCLOPS, a nuclear coiled-coil transcription factor, directly inducing the expression of NODULE INCEPTION (NIN), encoding a RWP-RK transcription factor (Marsh et al., 2007; Singh et al., 2014). In another model legume, L. japonicus, LjNIN targets two Nuclear Factor-Y (NF-Y) genes, LjNF-YA1 and LjNF-YB1, to control nodule development (Soyano et al., 2013). NF-Y TFs play a central role during the nodulation process. For instance, in M. truncatula, MtNF-YA1, and MtNF-YA2 redundantly act to control the early stage of rhizobial infection via the transcriptional activation of MtERN1 (Ethylene Response Factor Required for Nodulation 1; Laloum et al., 2014). In Phaseolus vulgaris, PvNF-YC1 is also controlling nodule organogenesis as well as the selection process of the symbiosis partner (Zanetti et al., 2010). More recently, Baudin et al. (2015) demonstrated that both MtNF-YA1, MtNF-YB16 and MtNF-YC2, and PvNF-YA1, PvNF-YB7, and PvNF-YC1 proteins form heterotrimers recognizing the CCAAT box of the MtERN1 and PvERN1 promoter sequences, respectively, (Baudin et al., 2015). Interestingly, while MtERN2, the MtERN1 paralogous gene, also regulates the nodulation process, its activity does not overlap with MtERN1 supporting the existence of additional regulatory pathways (Cerri et al., 2012). Together, these legume TFs control the expression of various genes regulating the infection of the plant root hair cell by rhizobia then its progression in the root cells through the development of the infection thread. MtFLOT4 and LjSYMREM1, plasma membrane microdomain proteins, seem also to play integral roles during the early stages of infection of the root hair cells by rhizobia such as the formation of the infection thread (Haney and Long, 2010; Toth et al., 2012).
While the genetic resources developed on model legumes, such as M. truncatula have a considerable impact on the characterization of nodulation genes, additional resources have been more recently developed taking advantage of –omic technologies. For instance, the access to large quantities of M. truncatula (Breakspear et al., 2014) and soybean root hair cells inoculated by rhizobia (Brechenmacher et al., 2010, 2012; Libault et al., 2010a,b; Nguyen et al., 2012; Yan et al., 2015) are now allowing a more global understanding of the molecular processes controlling the early stages of legume nodulation. In this manuscript, we are taking advantage of the knowledge gained during the past two decades and the more recent release of genomic and transcriptomic datasets to perform a comprehensive analysis of the evolution of legume protein coding genes controlling the nodulation process. Our results confirm the strong conservation of a core set of legume protein coding genes controlling nodulation between species and also highlight the divergence of a subset of the paralogs. Interestingly, the nodulation genes are also characterized by their high density along the legume chromosomes suggesting that these genes share the same biological function and are physically co-localized on the chromosomes. This study represents a new resource to better understand the evolution of legume nodulation, maximize the transfer of the scientific information between legumes and to open perspectives regarding the role and the conservation of gene modules in controlling the nodulation process.
Materials and methods
Identification of the nodulation genes across legume species
To properly update the annotation of the functionally characterized nodulation genes, published nucleotidic sequences were used as a query for a BLAST search against the four different genome sequences [i.e., M. truncatula v4.0 (Young et al., 2011), G. max Wm82.a2.v1 (Schmutz et al., 2010), P. vulgaris v1.0 (Schmutz et al., 2014), and L. japonicus v2.5 (Sato et al., 2008) available on the Phytozome v10.3 (http://phytozome.jgi.doe.gov/pz/portal.html) and Miyakogusa v3.0 (http://www.kazusa.or.jp/lotus/) websites]. Hits with an e-value <10−12 and a score > 100 were considered for further analysis.
Syntenic analysis between nodulation genes
The Accelerating Comparative Genomic database [CoGe (https://genomevolution.org/coge/; Lyons and Freeling, 2008; Lyons et al., 2008)] was mined to characterize microsynteny relationships between legume genes. The most recent versions of the genome sequences available on the CoGe database were selected when highlighting microsynteny relationships (i.e.,M. truncatula v4.0, G. max v9.0, P. vulgaris v1.0 and L. japonicus v2.5). Only syntelog genes were used for further analysis. To better connect the v9.0 annotations of the soybean nodulation genes characterized by CoGe with the most recent release of the soybean genome (Wm82.a2.v1), we included both annotation systems in our analysis (Supplemental Table 1). The GEvo (genome evolution analysis) tool was applied to visualize the collinearity and/or rearrangement between syntenic regions.
Transcriptomic analysis of soybean and M. truncatula nodulation genes
Gene expression atlases were mined to characterize the transcriptional patterns of the G. max, M. truncatula, L. japonicas, and P. vulgaris nodulation genes including in root hairs in response to rhizobia inoculation (Benedito et al., 2008; Libault et al., 2010b,c; Verdier et al., 2013; Breakspear et al., 2014; O'Rourke et al., 2014). The expression pattern of orthologous genes were compared based on the induction of gene expression in root hair cells upon rhizobia inoculation and based on their specific expression in nodules compared to other plant tissues.
Gene density analysis
For each species, the annotation file (gff3) were collected from the Phytozome v10.3 (http://phytozome.jgi.doe.gov/pz/portal.html#) and the Miyakogusa v3.0 websites (http://www.kazusa.or.jp/lotus/).
The distribution of nodulation genes on the chromosomes was performed with a sliding window sizes of 10, 6, 5 Mb and a step of 1, 0.6, and 0.5 Mb for G. max, P. vulgaris and L. japonicus/M. truncatula, respectively. The gene density was normalized with a Z-score calculation [Z-score = (gene density-u)/st, u and st are the mean and the standard deviation of the gene density for each chromosome, respectively], and R package ggplot2 (http://ggplot2.org/) was used to draw plot. The sample function in R was used to produce random nodulation gene distribution. The Kolmogorov-Smirnov test was used to validate the specific distributions of the genes on the chromosomes.
Results
Syntenic relationships reveal the orthologous relationships between legume nodulation-related protein-coding genes
To perform the most comprehensive evolutionary analysis of the protein coding genes involved in nodulation, we mined the scientific literature allowing us to list 110 functionally characterized genes from M. truncatula, L. japonicus, G. max, and P. vulgaris (Supplemental Table 1). To date, most protein coding genes involved in nodulation genes have been characterized in the model legumes G. max and M. truncatula notably upon the development of the Tnt1 retrotransposon insertion mutant population (Tadege et al., 2008; Pislariu et al., 2012; Cui et al., 2013). This observation supports the need to identify orthologous genes between legume species to facilitate the transfer of the scientific knowledge.
To update the annotation of these 110 genes, we first blasted their published nucleotidic sequences against the most recent release of the legume genome sequences [i.e., Phytozome v10.3 (http://phytozome.jgi.doe.gov/pz/portal.html) for M. truncatula (Mt4.0v1), G. max (Wm82.a2.v1), and P. vulgaris (v1.0); 2- Miyakogusa v3.0 (http://www.kazusa.or.jp/lotus/) for L. japonicus (v2.5); e-value <10−12 and score > 100; Supplemental Table 1].
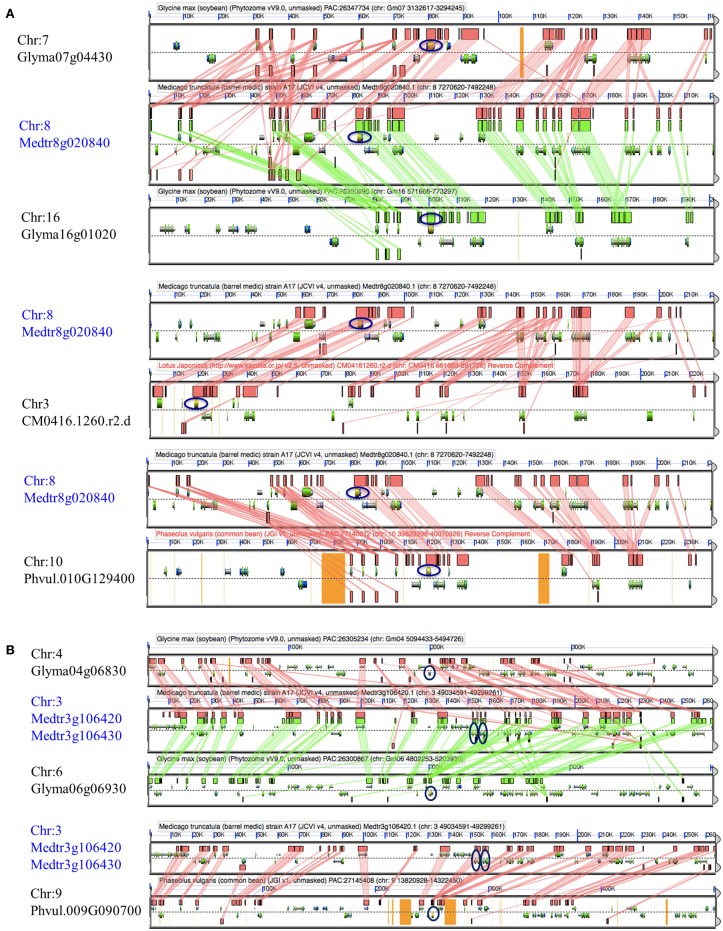
To identify orthologous and paralogous genes in and between L. japonicus, M. truncatula, G. max, and P. vulgaris, we examined syntenic relationships between corresponding genomic regions based on gene content, order, and orientation. To perform this analysis, we took advantage of the release of the sequence of various legume genomes and the development of comparative genomic resources such as CoGe (Lyons and Freeling, 2008; Lyons et al., 2008). Upon our evolutionary analysis, we repetitively observed strong syntenic relationships between genes from the four legume species including a large number of soybean paralogs, a consequence of the most recent whole genome duplication (WGD) of the soybean genome (Schmutz et al., 2010; Figure 1; Supplemental Table 1 and Supplemental Figure 1 for access to the entire datasets). Together, this comparative genomic analysis led to the characterization of 191, 92, 65 and 91 soybean, M. truncatula, L. japonicas, and common bean genes orthologous and paralogous to functionally described nodulation genes, respectively. These genes belong to 81 orthologous/paralogous groups (Supplemental Table 1).
Figure 1.
Syntenic relationships between MtNSP1 (A) and MtFLOT2/4 (B) loci and chromosome regions from Glycine max, Medicago truncatula, Lotus japonicas, and Phaseolus vulgaris. Each panel is a visualization of chromosome region showing the gene models on positive and negative strands. Green and red blocks in each panel highlight microsyntenic regions between legumes based on gene function and orientation. Genes highlighted in blue (e.g., Medtr8g020840 and Medtr3g106420/Medtr3g106430 for NSP1 and FLOT2/4 genes, respectively) were used as query when performing microsynteny analysis against the four legume species. The NSP1 and FLOT2/4 orthologs based on their microsyntenic relationships are highlighted with blue circles.
Comparative transcriptomic analysis reveals conservation and neo-/sub-functionalization between M. truncatula, G. max, L. japonicus and P. vulgaris nodulation genes
While microsynteny relationships clearly revealed the orthology existing between nodulation genes from different species, they are not sufficient to conclude about the conservation of their function. To provide a first insight into the conservation and divergence of the function between orthologous genes, we mined transcriptomic databases and integrated them into our comparative genomic analysis. Specifically, we took advantage of the release of the M. truncatula and soybean root hair transcriptomes and their perturbation in response to rhizobia inoculation (Libault et al., 2010b; Breakspear et al., 2014) as well as the access to the M. truncatula, G. max, L. japonicas, and P. vulgaris transcriptome atlases (Benedito et al., 2008; Libault et al., 2010c; Verdier et al., 2013; O'Rourke et al., 2014). In addition, we included in our analysis more focused transcriptomic studies on the nodulation process in the model plant M. truncatula (Roux et al., 2014; Larrainzar et al., 2015).
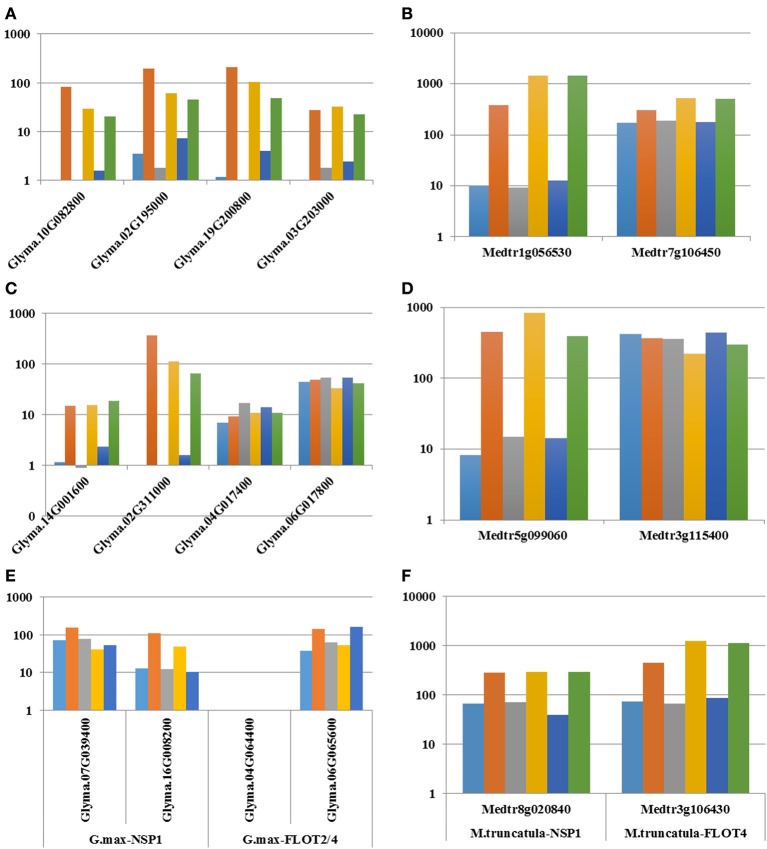
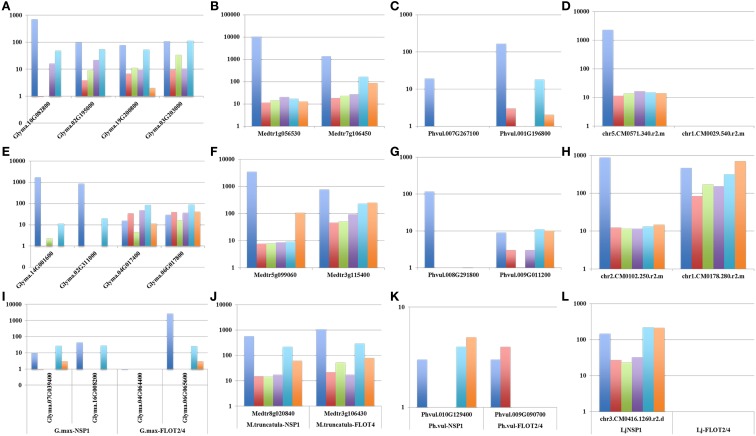
To identify the entire set of legume genes transcriptionally induced during nodulation, we independently analyzed their expression pattern during both the early (i.e., root hair response to rhizobia inoculation) and late events of the nodulation process (i.e., nodule specific expression compared to other plant organs). A total of 18 and 19 M. truncatula genes were induced in root hair cells in response to wild-type Sinorhizobium meliloti inoculation or were preferentially expressed in nodules compared to other plant tissues (fold-change > 2; Supplemental Table 2), respectively. Among those genes, eleven were both induced in root hair cells in response to S. meliloti and preferentially expressed in nodules including MtNF-YA1/2 (Figures 2B, 3B), MtNIN (Figures 2D, 3F), MtFLOT2/4, and MtNSP1 (Figures 2F, 3J). To provide a more complete understanding of the M. truncatula genes transcriptionally induced during nodulation, we mined recently published M. truncatula RNA-seq data sets (Roux et al., 2014; Larrainzar et al., 2015). We identified a total of 30 and 15 M. truncatula genes differentially expressed in specific zones of the M. truncatula nodule (i.e., Roux et al., 2014) and during the early stages of nodulation in root tissue (i.e., from 30 min to 2 days after S. meliloti inoculation; Larrainzar et al., 2015), respectively, (Supplemental Table 2). Analyzing the soybean transcriptome, the expression of 47 genes was induced in root hair cells in response to rhizobia inoculation while 38 soybean genes were preferentially expressed in nodules vs. other soybean tissues. Among those, 20 were both up-regulated in root hairs in response to B. japonicum and preferentially expressed in nodules such as GmNF-YA1/2 (Figures 2A, 3A), GmNIN (Figures 2C, 3E), and GmFLOT2/4 (Figures 2E, 3I). Mining the common bean and L. japonicus transcriptome atlases, we also characterized 19 and 16 genes preferentially expressed in common bean and L. japonicus nodules, respectively, such as PvNF-YA1/2 (Figure 3C), PvNIN (Figure 3G), LjNF-YA1/2 (Figure 3D), LjNIN (Figure 3H). Unexpectedly, PvNSP1, PvFLOT2/4, and LjNSP1 are not preferentially expressed in common bean and L. japonicus nodules (Figures 3D,H,K,L).
Figure 2.
Relative expression levels of NF-YA1/2 (A, B), NIN (C, D), NSP1, and FLOT2/4 (E, F) M. truncatula genes (B, D, F), and their G. max orthologs (A, C, E) in inoculated and mock-inoculated root hair cells. Gene IDs are highlighted on the x-axis. The relative expression levels of the genes (log10 scale) are indicated on the y-axis. Gene expression datasets were mined from the G. max and M. truncatula root hair transcriptomic datasets (Libault et al., 2010b; Breakspear et al., 2014). For G. max: blue, 12H UN; orange, 12H IN; gray, 24H UN; yellow, 24H IN; dark blue, 48H UN; and green, 48H IN; For M. truncatula: blue, 1D UN; orange, 1D IN; gray, 2D UN; yellow, 2D IN; dark blue, 3D UN; and green, 3D IN.
Figure 3.
Relative expression levels of NF-YA1/2 (A–D), NIN (E–H), NSP1 and FLOT2/4 (I–L) of M. truncatula genes (B, F, J), and their G. max orthologs (A, E, I), P. vulgaris orthologs (C, G, K) and L. japonicus (D, H, L) in various plant organs (i.e., blue, nodule; orange, flower; gray, pod; yellow, leaf; dark blue, root; and green, root tip). Gene IDs are highlighted on the x-axis. The relative expression levels of the genes (log10 scale) are indicated on the y-axis. Gene expression datasets were mined from the G. max, M. truncatula, P. vulgaris, and L. japonicus atlases datasets (Benedito et al., 2008; Libault et al., 2010c (http://mtgea.noble.org/v3/); Verdier et al., 2013; O'Rourke et al., 2014).
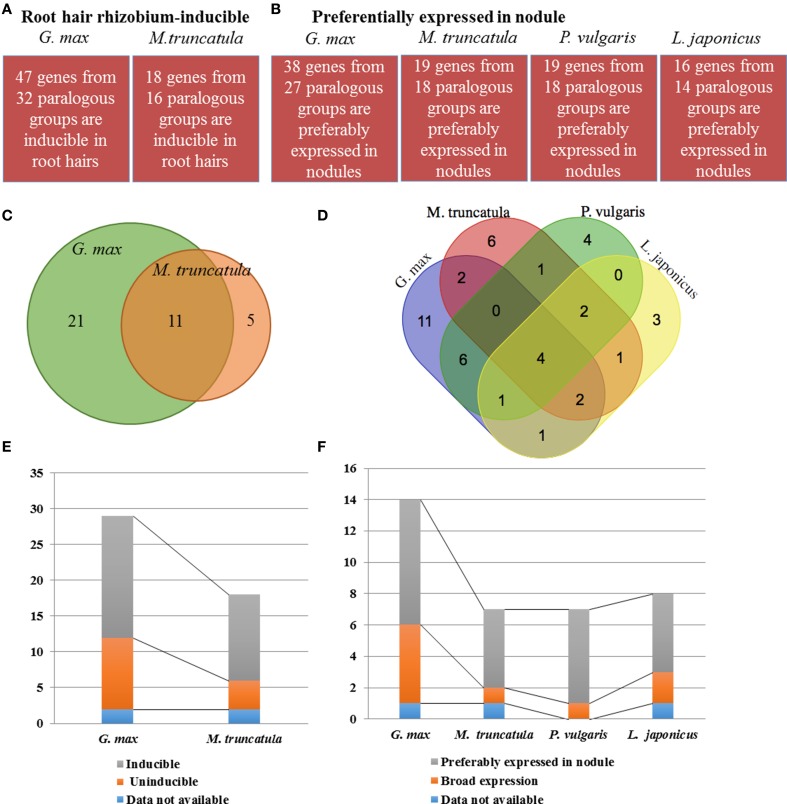
To better evaluate the impact of soybean WGDs on the population of genes controlling nodulation, we classified and compared the 47 and 18 soybean and M. truncatula genes induced in root hair cells in response to rhizobia inoculation based on their paralogous relationships. A total of 32 and 16 paralogous groups were identified for each plant species, respectively, (Figure 4A; Supplemental Table 1, Supplemental Table 3). Performing a similar analysis on the 38, 19, 19 and 16 soybean, M. truncatula, common bean and L. japonicus genes preferentially expressed in nodules, we characterized 27, 18 18 and 14 groups of paralogous genes, respectively, (Figure 4B, Supplemental Table 3). These results suggest that the large number of differentially expressed soybean genes during nodulation is not just the consequence of the WGDs.
Figure 4.
Comparison of the expression profiles between G. max, M. truncatula, P. vulgaris, and L. japonicus orthologous genes and groups during the early (i.e., root hair response to rhizobia inoculation; A, C, E) and late stages of the nodulation process (i.e., preferential expression in mature nodules; B, D, F). (A) and (B), numbers of G. max and M. truncatula genes and orthologous groups induced in root hairs upon rhizobia inoculation (A) and G. max, M. truncatula, P. vulgaris, and L. japonicus genes preferentially expressed in nodules (B; fold-change > 2; Supplemental Table 3 for details). (C) and (D): Venn diagrams showing the overlaps between G. max and M. truncatula root hair inducible (C) and overlaps between G. max, M. truncatula, P. vulgaris, and L. japonicus nodule-specific (D) orthologous groups; (E) and (F): Distribution of the number of G. max and M. truncatula genes which belong to the 11 root hair-inducible (E) and the number of G. max, M. truncatula, P. vulgaris, and L. japonicus genes which belong to four nodule preferential (F) orthologous groups, respectively, according to their expression patterns.
Upon WGDs, paralogs can share the same expression profiles leading to functional redundancy [e.g., induction of the expression of the four soybean NF-YA1/2 paralogs in root hair cells in response to B. japonicum inoculation (Figure 2A)], or could be a source for sub- and neo-functionalization [(e.g., GmNINs orthologs can be divided into two groups: Glyma.14G001600 and Glyma.02G311000, which are induced during nodulation and Glyma.04G017400 and Glyma.06G017800, characterized by their more constitutive expression Figures 2C, 3E)]. To further explore the consequences of WGDs on the transcriptional regulation of paralogous genes, we compared the expression of soybean and M. truncatula genes that belong to paralogous groups that are induced during nodulation.
We identified 11 overlapping root hair inducible groups between the two plant species (34 and 69% of the G. max and M. truncatula root hair-inducible groups, respectively; Figure 4C, Supplemental Table 3). Twenty-nine and 18 soybean and M. truncatula genes are represented in these 11 groups, respectively, including the NOD100, NMNa, RIP1, MtPUB1, NODULIN-26a, NIN, FLOT2/4, NSP1, MtNramp1, NF-YA1/2, and ERN1/2 genes (Supplemental Table 3). Performing a similar analysis on the 27, 18, 18 and 14 G. max, M. truncatula, P. vulgaris, and L. japonicus groups preferentially expressed in nodules, we characterized four overlapping orthologous groups (11, 22, 22, and 29% of the G. max, M. truncatula, P. vulgaris, and L. japonicus groups preferentially expressed in nodules, respectively; Figure 4D). These four groups are composed of 14, 7, 7, and 8 G. max, M. truncatula, P. vulgaris, and L. japonicus genes, respectively, such as ENOD20, NIN, NIN2, and NF-YA1/2 genes (Supplemental Table 3).
Analyzing the expression patterns of these genes, we only observed a slight increase in the number of soybean genes differentially expressed during the early (Figure 4E; gray bar) and late events of the nodulation process compared to M. truncatula, P. vulgaris, and L. japonicus genes (Figure 4F, gray bar). Oppositely, many soybean paralogs are not transcriptionally responsive to rhizobium, but are characterized by their broad expression patterns (Figures 4E,F, orange bars). These results suggest that upon soybean WGDs, a subset of the paralogous genes were transcriptionally restricted to their role during nodulation based on the conservation of their induction in response to rhizobia while other paralogs gained new function after alteration of their expression patterns.
Legume genes specialized in the nodulation processes are forming gene modules on legume chromosomes
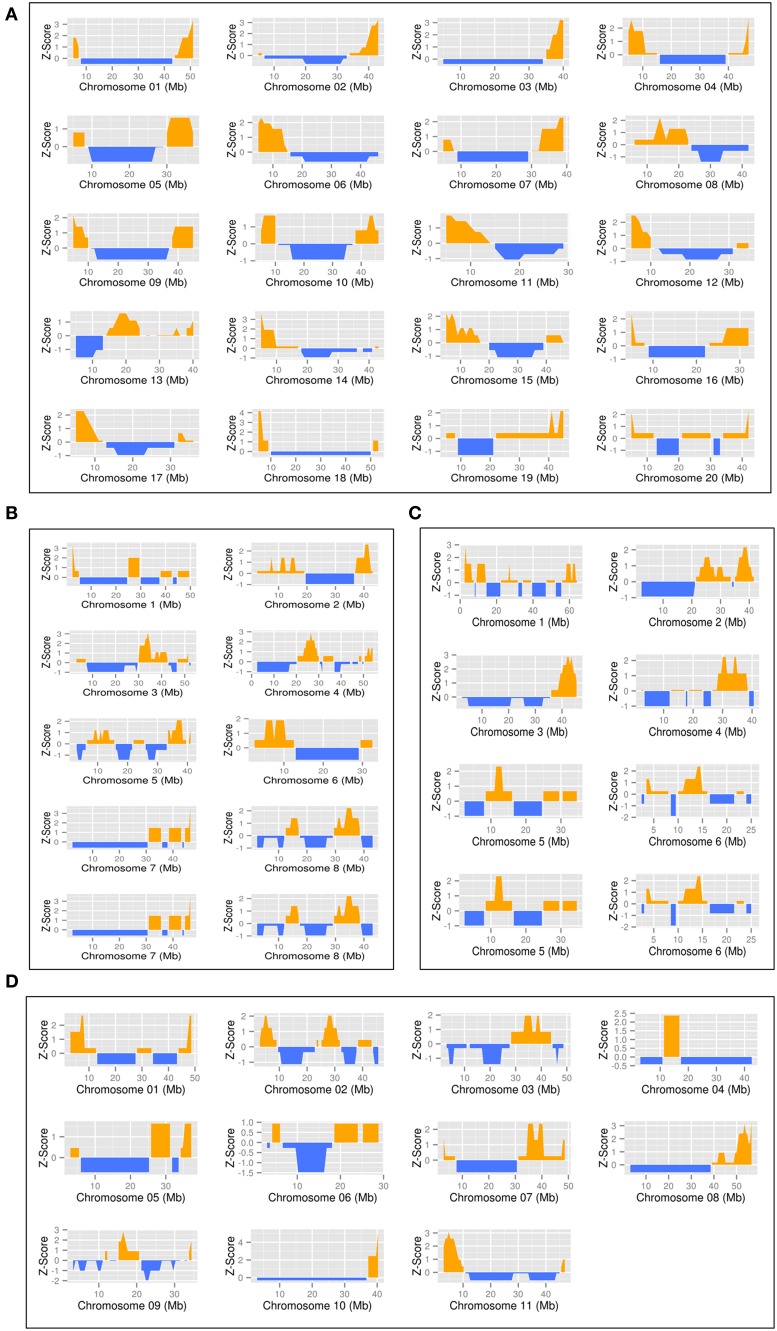
Previous studies revealed the correlation existing between gene function and the organization of the euchromatin. Specifically, gene modules on chromosomes are characterized by the presence of genes involved in the same biological function and sharing similar expression patterns (Williams and Bowles, 2004; Zhan et al., 2006; Pingault et al., 2015). Hypothesizing that nodulation genes are also located in modules along the legume chromosomes, we analyzed their chromosomal distribution and density. Accordingly, we mapped the annotated legume genes controlling nodulation (Supplemental Table 1) on the 20, 8, 6, and 11 chromosomes of G. max, M. truncatula, L. japonicas, and P. vulgaris, respectively. To take in consideration the differences in the size of the genomes of the four legume plants, we analyzed the distribution of nodulation genes on the chromosomes generating window sizes of 10, 6, 5 Mb, and steps of 1, 0.6, and 0.5 Mb for G. max, P.vulgaris and L. japonicus/M. truncatula, respectively. To highlight the significant increase of the density of nodulation genes on chromosomes as reflected by their Z-score, we also normalized these results based on a random distribution of the nodulation genes (Figure 5). A significant enrichment in nodulation genes was repetitively observed on the M. truncatula, L. japonicas, and P. vulgaris chromosomes and on 12 out of the 20 soybean chromosomes (Kolmogorov-Smirnov test; p-value <0.05; Supplemental Table 4). Together, these results confirmed the clustering of nodulation genes in functional modules on the chromosomes. The fact that these modules are conserved between model legumes suggests their essential roles in controlling the nodulation process.
Figure 5.
Representation of the density of the legume nodulation genes along each legume chromosome. Gene density is represented by a z-score in a 10 Mb sliding window (step 1 Mb) along each chromosomes for each legume genome. Positive values are in orange, negative value in blue. The position on the chromosomes are indicated in Mb. The genomes represented are Glycine max (A), Medicago truncatula (B), Lotus japonicus (C), and Phaseolus vulgaris (D).
Discussion
Applying comparative genomic and transcriptomic analyses to reveal the conservation and divergence between nodulation orthologous/paralogous genes
Functional genomic studies led to the characterization of 110 genes controlling the early and late events of the nodulation process. As described by Chen et al. (2015), a core set of M. truncatula genes such as ERN1/2, NSP2, and several NF-Y genes are playing a central role in plant cell infection by rhizobia, both early (i.e., root hair cell infection) and late (i.e., nodule cell infection) during the nodulation process (Chen et al., 2015). In this study, our analyses have been conducted in four different legume models having the objective to better transfer scientific knowledge between species. Accordingly, we took advantage of the genomic information now available to perform a global and comprehensive analysis of the evolution of nodulation genes based on: 1- their orthology and paralogy upon demonstration of their microsyntenic relationships and; 2- the conservation and divergence of their transcriptomic patterns.
Applying various bioinformatics tools available on the CoGe platform (https://genomevolution.org/coge/) such as Synfind and GEvo, we repetitively analyzed the evolutionary relationship between nodulation genes across four model legume plants. Upon the availability of functional genomic datasets, we were able to confirm the orthologous relationships existing between genes known to control nodulation across different species. For instance, using the soybean NF receptor gene GmNFR1a (Indrasumunar et al., 2011) as query, we clearly highlighted its orthology with the M. truncatula MtLYK3/HCL (Limpens et al., 2003) and the L. japonicus LjNFR1 genes (Radutoiu et al., 2003). To further support our analysis, we revealed the orthology existing between 65 functionally characterized genes across different species [e.g., SYMRK / NORK, SUNN/NARK/HAR1, RDN2, POLLUX/DMI1, NSP2, NSP1, NIN, NFR/NFP, NFR1/LYK3/HCL, MtENOD20/ENOD55-2. ENOD16/ENOD55-1 and CCaMK/DMI3 genes (Supplemental Figure 1)].
While the conservation of the function between orthologous genes is often assumed, biological evidences to validate these assumptions are limited. Transcriptomes provide a first insight into the conservation of the function of orthologous and paralogous genes based on the conservation of a similar expression pattern. For instance, previous studies focusing on the fate of soybean paralogs, products of the successive duplications of the soybean genome, concluded about the differential expression of 50% of the paralogs leading to their sub-functionalization (Roulin et al., 2013). Taking advantage of the release of legume transcriptome atlases as well as the analysis of the transcriptomic response of the root hair cells to rhizobia inoculation, we compared the nodulation related protein coding genes' expression patterns between soybean, M. truncatula, common bean and L. japonicus orthologous genes. These analyses were also conducted under the context of paralogy. Our study clearly revealed the conservation of the transcriptomic patterns between M. truncatula, G. max, L. japonicas, and P. vulgaris orthologous genes during the nodulation process supporting the conservation of the biological function of at least one paralog. Interestingly, a significant number of soybean paralogs can display very distinctive expression patterns suggesting a gain of function or a sub-functionalization (e.g., soybean NIN genes; Figures 2, 3, 4). Differential epigenome and the evolution of the promoter regions of paralogous genes (i.e., presence and absence of cis- and trans-regulatory elements) should be carefully investigated to reveal the evolutionary mechanisms controlling these transcriptomic changes.
Plant genes acting together in the same biological process are physically closely located on the chromosomes (Williams and Bowles, 2004; Zhan et al., 2006; Pingault et al., 2015). Taking advantage of the identification of a large number of nodulation genes from four different species (i.e., 191, 92, 65 and 91 G. max, M. truncatula, L. japonicus, P. vulgaris genes, respectively), we analyzed their distribution along the 20, 8, 6, and 11 chromosomes of each species, respectively. We observed that the legumes genes controlling the nodulation process are characterized by their high density on the chromosomes rather than being randomly located on the chromosomes. This close relationship might be beneficial to their co-expression during nodulation. The relationships existing between chromosome territories, position of gene modules on the chromosome, their epigenomic context and their transcriptional activities should be investigated.
Author contributions
ML designed research. MN characterized and updated the annotation of the legume nodulation genes. ZQ performed the syntenic and transcriptomic analysis between legume genes. LP analyzed the distribution of the nodulation genes on the legume chromosomes. ML wrote the article.
Funding
This work was funded by a grant from the National Science Foundation-Plant Genome Research Program (#IOS-1339194) and by the National Science Foundation-CAREER program (#IOS-1453613).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00034
References
- Amor B. B., Shaw S. L., Oldroyd G. E., Maillet F., Penmetsa R. V., Cook D., et al. (2003). The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J. 34, 495–506. 10.1046/j.1365-313X.2003.01743.x [DOI] [PubMed] [Google Scholar]
- Ané J.-M., Kiss G. B., Riely B. K., Penmetsa R. V., Oldroyd G. E., Ayax C., et al. (2004). Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303, 1364–1367. 10.1126/science.1092986 [DOI] [PubMed] [Google Scholar]
- Andriankaja A., Boisson-Dernier A., Frances L., Sauviac L., Jauneau A., Barker D. G., et al. (2007). AP2-ERF transcription factors mediate Nod factor–dependent Mt ENOD11 activation in root hairs via a novel cis-regulatory motif. Plant Cell 19, 2866–2885. 10.1105/tpc.107.052944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrighi J.-F., Barre A., Amor B. B., Bersoult A., Soriano L. C., Mirabella R., et al. (2006). The Medicago truncatula lysine motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 142, 265–279. 10.1104/pp.106.084657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia M., Rípodas C., Clúa J., Baudin M., Aguilar O. M., Niebel A., et al. (2014). A nuclear factor Y interacting protein of the GRAS family is required for nodule organogenesis, infection thread progression, and lateral root growth. Plant Physiol. 164, 1430–1442. 10.1104/pp.113.230896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin M., Laloum T., Lepage A., Ripodas C., Ariel F., Frances L., et al. (2015). A phylogenetically conserved group of NF-Y transcription factors interact to control nodulation in legumes. Plant Physiol. 169, 2761–2773. 10.1104/pp.15.01144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedito V. A., Torres-Jerez I., Murray J. D., Andriankaja A., Allen S., Kakar K., et al. (2008). A gene expression atlas of the model legume Medicago truncatula. Plant J. 55, 504–513. 10.1111/j.1365-313X.2008.03519.x [DOI] [PubMed] [Google Scholar]
- Blank C., Mylona P., Yang W. C., Katinakis P., Bisseling T., Franssen H. (1993). Characterization of the soybean early nodulin cDNA clone GmENOD55. Plant Mol. Biol. 22, 1167–1171. 10.1007/BF00028987 [DOI] [PubMed] [Google Scholar]
- Borisov A. Y., Madsen L. H., Tsyganov V. E., Umehara Y., Voroshilova V. A., Batagov A. O., et al. (2003). The Sym35 gene required for root nodule development in pea is an ortholog of Nin from Lotus japonicus. Plant Physiol. 131, 1009–1017. 10.1104/pp.102.016071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breakspear A., Liu C., Roy S., Stacey N., Rogers C., Trick M., et al. (2014). The root hair “infectome” of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for Auxin signaling in rhizobial infection. Plant Cell 26, 4680–4701. 10.1105/tpc.114.133496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brechenmacher L., Lei Z., Libault M., Findley S., Sugawara M., Sadowsky M. J., et al. (2010). Soybean metabolites regulated in root hairs in response to the symbiotic bacterium Bradyrhizobium japonicum. Plant Physiol. 153, 1808–1822. 10.1104/pp.110.157800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brechenmacher L., Nguyen T. H., Hixson K., Libault M., Aldrich J., Pasa-Tolic L., et al. (2012). Identification of soybean proteins from a single cell type: the root hair. Proteomics 12, 3365–3373. 10.1002/pmic.201200160 [DOI] [PubMed] [Google Scholar]
- Capoen W., Den Herder J., Sun J., Verplancke C., De Keyser A., De Rycke R., et al. (2009). Calcium spiking patterns and the role of the calcium/calmodulin-dependent kinase CCaMK in lateral root base nodulation of Sesbania rostrata. Plant Cell 21, 1526–1540. 10.1105/tpc.109.066233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebolla A., Vinardell J. M., Kiss E., Oláh B., Roudier F., Kondorosi A., et al. (1999). The mitotic inhibitor ccs52 is required for endoreduplication and ploidy−dependent cell enlargement in plants. EMBO J. 18, 4476–4484. 10.1093/emboj/18.16.4476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerri M. R., Frances L., Laloum T., Auriac M.-C., Niebel A., Oldroyd G. E., et al. (2012). Medicago truncatula ERN transcription factors: regulatory interplay with NSP1/NSP2 GRAS factors and expression dynamics throughout rhizobial infection. Plant Physiol. 160, 2155–2172. 10.1104/pp.112.203190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D.-S., Liu C.-W., Roy S., Cousins D., Stacey N., Murray J. D. (2015). Identification of a core set of rhizobial infection genes using data from single cell-types. Front. Plant Sci. 6:575. 10.3389/fpls.2015.00575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D., Dreyer D., Bonnet D., Howell M., Nony E., VandenBosch K. (1995). Transient induction of a peroxidase gene in Medicago truncatula precedes infection by Rhizobium meliloti. Plant Cell 7, 43–55. 10.1105/tpc.7.1.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Barampuram S., Stacey M. G., Hancock C. N., Findley S., Mathieu M., et al. (2013). Tnt1 retrotransposon mutagenesis: a tool for soybean functional genomics. Plant Physiol. 161, 36–47. 10.1104/pp.112.205369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Billy F., Grosjean C., May S., Bennett M., Cullimore J. V. (2001). Expression studies on AUX1-like genes in Medicago truncatula suggest that auxin is required at two steps in early nodule development. Mol. Plant Microbe Interact. 14, 267–277. 10.1094/MPMI.2001.14.3.267 [DOI] [PubMed] [Google Scholar]
- de Carvalho Niebel F., Lescure N., Cullimore J. V., Gamas P. (1998). The Medicago truncatula MtAnn1 gene encoding an annexin is induced by Nod factors and during the symbiotic interaction with Rhizobium meliloti. Mol. Plant Microbe Interact. 11, 504–513. 10.1094/MPMI.1998.11.6.504 [DOI] [PubMed] [Google Scholar]
- Delauney A., Cheon C.-I., Snyder P., Verma D. (1990). A nodule-specific sequence encoding a methionine-rich polypeptide, nodulin-21. Plant Mol. Biol. 14, 449–451. 10.1007/BF00028782 [DOI] [PubMed] [Google Scholar]
- Edwards A., Heckmann A. B., Yousafzai F., Duc G., Downie J. A. (2007). Structural implications of mutations in the pea SYM8 symbiosis gene, the DMI1 ortholog, encoding a predicted ion channel. Mol. Plant Microbe Interact. 20, 1183–1191. 10.1094/MPMI-20-10-1183 [DOI] [PubMed] [Google Scholar]
- Elise S., Etienne-Pascal J., de Fernanda C.-N., Gérard D., Julia F. (2005). The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Mol. Biol. 58, 809–822. 10.1007/s11103-005-8102-y [DOI] [PubMed] [Google Scholar]
- Endre G., Kereszt A., Kevei Z., Mihacea S., Kaló P., Kiss G. B. (2002). A receptor kinase gene regulating symbiotic nodule development. Nature 417, 962–966. 10.1038/nature00842 [DOI] [PubMed] [Google Scholar]
- Fortin M. G., Morrison N. A., Verma D. P. S. (1987). Nodulin-26, a peribacteroid membrane nodulin is expressed independently of the development of the peribacteroid compartment. Nucleic Acids Res. 15, 813–824. 10.1093/nar/15.2.813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamas P., de Billy F., Truchet G. (1998). Symbiosis-specific expression of two Medicago truncatula nodulin genes, MtN1 and MtN13, encoding products homologous to plant defense proteins. Mol. Plant Microbe Interact. 11, 393–403. 10.1094/MPMI.1998.11.5.393 [DOI] [PubMed] [Google Scholar]
- Greene E. A., Erard M., Dedieu A., Barker D. G. (1998). MtENOD16 and 20 are members of a family of phytocyanin-related early nodulins. Plant Mol. Biol. 36, 775–783. 10.1023/A:1005916821224 [DOI] [PubMed] [Google Scholar]
- Groth M., Takeda N., Perry J., Uchida H., Dräxl S., Brachmann A., et al. (2010). NENA, a Lotus japonicus homolog of Sec13, is required for rhizodermal infection by arbuscular mycorrhiza fungi and rhizobia but dispensable for cortical endosymbiotic development. Plant Cell 22, 2509–2526. 10.1105/tpc.109.069807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haney C. H., Long S. R. (2010). Plant flotillins are required for infection by nitrogen-fixing bacteria. Proc. Natl. Acad. Sci. U.S.A. 107, 478–483. 10.1073/pnas.0910081107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann A. B., Lombardo F., Miwa H., Perry J. A., Bunnewell S., Parniske M., et al. (2006). Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol. 142, 1739–1750. 10.1104/pp.106.089508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi-Anraku H., Takeda N., Charpentier M., Perry J., Miwa H., Umehara Y., et al. (2005). Plastid proteins crucial for symbiotic fungal and bacterial entry into plant roots. Nature 433, 527–531. 10.1038/nature03237 [DOI] [PubMed] [Google Scholar]
- Indrasumunar A., Kereszt A., Searle I., Miyagi M., Li D., Nguyen C. D., et al. (2010). Inactivation of duplicated nod factor receptor 5 (NFR5) genes in recessive loss-of-function non-nodulation mutants of allotetraploid soybean (Glycine max L. Merr.). Plant Cell Physiol. 51, 201–214. 10.1093/pcp/pcp178 [DOI] [PubMed] [Google Scholar]
- Indrasumunar A., Searle I., Lin M. H., Kereszt A., Men A., Carroll B. J., et al. (2011). Nodulation factor receptor kinase 1α controls nodule organ number in soybean (Glycine max L. Merr). Plant J. 65, 39–50. 10.1111/j.1365-313X.2010.04398.x [DOI] [PubMed] [Google Scholar]
- Jing R., Johnson R., Seres A., Kiss G., Ambrose M. J., Knox M. R., et al. (2007). Gene-based sequence diversity analysis of field pea (Pisum). Genetics 177, 2263–2275. 10.1534/genetics.107.081323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet E.-P., El-Gachtouli N., Vernoud V., de Billy F., Pichon M., Dedieu A., et al. (2001). Medicago truncatula ENOD11: a novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol. Plant Microbe Interact. 14, 737–748. 10.1094/MPMI.2001.14.6.737 [DOI] [PubMed] [Google Scholar]
- Kanamori N., Madsen L. H., Radutoiu S., Frantescu M., Quistgaard E. M., Miwa H., et al. (2006). A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc. Natl. Acad. Sci. U.S.A. 103, 359–364. 10.1073/pnas.0508883103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katinakis P., Verma D. P. S. (1985). Nodulin-24 gene of soybean codes for a peptide of the peribacteroid membrane and was generated by tandem duplication of a sequence resembling an insertion element. Proc. Natl. Acad. Sci. U.S.A. 82, 4157–4161. 10.1073/pnas.82.12.4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevei Z., Lougnon G., Mergaert P., Horváth G. V., Kereszt A., Jayaraman D., et al. (2007). 3-Hydroxy-3-methylglutaryl coenzyme A reductase1 interacts with NORK and is crucial for nodulation in Medicago truncatula. Plant Cell 19, 3974–3989. 10.1105/tpc.107.053975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijne J. W. (1992). The rhizobium infection process. Biol. Nitrogen Fixation 1349, 349–398. [Google Scholar]
- Kistner C., Winzer T., Pitzschke A., Mulder L., Sato S., Kaneko T., et al. (2005). Seven Lotus japonicus genes required for transcriptional reprogramming of the root during fungal and bacterial symbiosis. Plant Cell 17, 2217–2229. 10.1105/tpc.105.032714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouchi H., Hata S. (1993). Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Mol. Gen. Genet. MGG 238, 106–119. [DOI] [PubMed] [Google Scholar]
- Kouchi H., Hata S. (1995). GmN56, a novel nodule-specific cDNA from soybean root nodules encodes a protein homologous to isopropylmalate synthase and homocitrate synthase. MPMI Mol. Plant Microbe Interact. 8, 172–176. 10.1094/MPMI-8-0172 [DOI] [PubMed] [Google Scholar]
- Krusell L., Madsen L. H., Sato S., Aubert G., Genua A., Szczyglowski K., et al. (2002). Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature 420, 422–426. 10.1038/nature01207 [DOI] [PubMed] [Google Scholar]
- Kuppusamy K. T., Endre G., Prabhu R., Penmetsa R. V., Veereshlingam H., Cook D. R., et al. (2004). LIN, a Medicago truncatula gene required for nodule differentiation and persistence of rhizobial infections. Plant Physiol. 136, 3682–3691. 10.1104/pp.104.045575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloum T., Baudin M., Frances L., Lepage A., Billault-Penneteau B., Cerri M. R., et al. (2014). Two CCAAT-box-binding transcription factors redundantly regulate early steps of the legume-rhizobia endosymbiosis. Plant J. 79, 757–768. 10.1111/tpj.12587 [DOI] [PubMed] [Google Scholar]
- Larrainzar E., Riely B. K., Kim S. C., Carrasquilla-Garcia N., Yu H. J., Hwang H. J., et al. (2015). Deep sequencing of the Medicago truncatula root transcriptome reveals a massive and early interaction between nodulation factor and ethylene signals. Plant Physiol. 169, 233–265. 10.1104/pp.15.00350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M.-J., Wang Q., Li X., Chen A., Luo L., Xie Y., et al. (2015). The small GTPase ROP10 of Medicago truncatula is required for both tip growth of root hairs and nod factor-induced root hair deformation. Plant Cell 27, 806–822. 10.1105/tpc.114.135210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévy J., Bres C., Geurts R., Chalhoub B., Kulikova O., Duc G., et al. (2004). A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303, 1361–1364. 10.1126/science.1093038 [DOI] [PubMed] [Google Scholar]
- Libault M., Brechenmacher L., Cheng J., Xu D., Stacey G. (2010a). Root hair systems biology. Trends Plant Sci. 15, 641–650. 10.1016/j.tplants.2010.08.010 [DOI] [PubMed] [Google Scholar]
- Libault M., Farmer A., Brechenmacher L., Drnevich J., Langley R. J., Bilgin D. D., et al. (2010b). Complete transcriptome of the soybean root hair cell, a single-cell model, and its alteration in response to Bradyrhizobium japonicum infection. Plant Physiol. 152, 541–552. 10.1104/pp.109.148379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M., Farmer A., Joshi T., Takahashi K., Langley R. J., Franklin L. D., et al. (2010c). An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J. 63, 86–99. 10.1111/j.1365-313x.2010.04222.x [DOI] [PubMed] [Google Scholar]
- Libault M., Govindarajulu M., Berg R. H., Ong Y. T., Puricelli K., Taylor C. G., et al. (2011). A dual-targeted soybean protein is involved in Bradyrhizobium japonicum infection of soybean root hair and cortical cells. Mol. Plant Microbe Interact. 24, 1051–1060. 10.1094/MPMI-12-10-0281 [DOI] [PubMed] [Google Scholar]
- Libault M., Zhang X. C., Govindarajulu M., Qiu J., Ong Y. T., Brechenmacher L., et al. (2010d). A member of the highly conserved FWL (tomato FW2. 2-like) gene family is essential for soybean nodule organogenesis. Plant J. 62, 852–864. 10.1111/j.1365-313X.2010.04201.x [DOI] [PubMed] [Google Scholar]
- Limpens E., Franken C., Smit P., Willemse J., Bisseling T., Geurts R. (2003). LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302, 630–633. 10.1126/science.1090074 [DOI] [PubMed] [Google Scholar]
- Liu C., Yeung A., Dickstein R. (1998). The cDNA sequence of Medicago truncatula cv. Jemalong Enod8, a gene associated with nitrogen fixing root nodule organogenesis. Plant Physiol. 117, 116.113. [Google Scholar]
- Lyons E., Freeling M. (2008). How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J. 53, 661–673. 10.1111/j.1365-313X.2007.03326.x [DOI] [PubMed] [Google Scholar]
- Lyons E., Pedersen B., Kane J., Alam M., Ming R., Tang H., et al. (2008). Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar, and grape: CoGe with rosids. Plant Physiol. 148, 1772–1781. 10.1104/pp.108.124867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen E. B., Madsen L. H., Radutoiu S., Olbryt M., Rakwalska M., Szczyglowski K., et al. (2003). A receptor kinase gene of the LysM type is involved in legumeperception of rhizobial signals. Nature 425, 637–640. 10.1038/nature02045 [DOI] [PubMed] [Google Scholar]
- Marsh J. F., Rakocevic A., Mitra R. M., Brocard L., Sun J., Eschstruth A., et al. (2007). Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol. 144, 324–335. 10.1104/pp.106.093021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis R., Grosjean C., De Billy F., Huguet T., Gamas P. (1999). The early nodulin gene MtN6 is a novel marker for events preceding infection of Medicago truncatula roots by Sinorhizobium meliloti. Mol. Plant Microbe Interact. 12, 544–555. 10.1094/MPMI.1999.12.6.544 [DOI] [PubMed] [Google Scholar]
- Mazziotta L., Reynoso M., Aguilar O., Blanco F., Zanetti M. (2013). Transcriptional and functional variation of NF-YC1 in genetically diverse accessions of Phaseolus vulgaris during the symbiotic association with Rhizobium etli. Plant Biol. 15, 808–818. 10.1111/j.1438-8677.2012.00683.x [DOI] [PubMed] [Google Scholar]
- Mbengue M., Camut S., de Carvalho-Niebel F., Deslandes L., Froidure S., Klaus-Heisen D., et al. (2010). The Medicago truncatula E3 ubiquitin ligase PUB1 interacts with the LYK3 symbiotic receptor and negatively regulates infection and nodulation. Plant Cell 22, 3474–3488. 10.1105/tpc.110.075861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra R. M., Gleason C. A., Edwards A., Hadfield J., Downie J. A., Oldroyd G. E., et al. (2004). A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: gene identification by transcript-based cloning. Proc. Natl. Acad. Sci. U.S.A. 101, 4701–4705. 10.1073/pnas.0400595101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa H., Sun J., Oldroyd G. E., Downie J. A. (2006). Analysis of calcium spiking using a cameleon calcium sensor reveals that nodulation gene expression is regulated by calcium spike number and the developmental status of the cell. Plant J. 48, 883–894. 10.1111/j.1365-313X.2006.02926.x [DOI] [PubMed] [Google Scholar]
- Miyahara A., Hirani T. A., Oakes M., Kereszt A., Kobe B., Djordjevic M. A., et al. (2008). Soybean nodule autoregulation receptor kinase phosphorylates two kinase-associated protein phosphatases in vitro. J. Biol. Chem. 283, 25381–25391. 10.1074/jbc.M800400200 [DOI] [PubMed] [Google Scholar]
- Miyazawa H., Oka-Kira E., Sato N., Takahashi H., Wu G.-J., Sato S., et al. (2010). The receptor-like kinase KLAVIER mediates systemic regulation of nodulation and non-symbiotic shoot development in Lotus japonicus. Development 137, 4317–4325. 10.1242/dev.058891 [DOI] [PubMed] [Google Scholar]
- Moreau S., Fromentin J., Vailleau F., Vernié T., Huguet S., Balzergue S., et al. (2014). The symbiotic transcription factor MtEFD and cytokinins are positively acting in the Medicago truncatula and Ralstonia solanacearum pathogenic interaction. New Phytol. 201, 1343–1357. 10.1111/nph.12636 [DOI] [PubMed] [Google Scholar]
- Murakami Y., Miwa H., Imaizumi-Anraku H., Kouchi H., Downie J. A., Kawaguchi M., et al. (2007). Positional cloning identifies Lotus japonicus NSP2, a putative transcription factor of the GRAS family, required for NIN and ENOD40 gene expression in nodule initiation. DNA Res. 13, 255–265. 10.1093/dnares/dsl017 [DOI] [PubMed] [Google Scholar]
- Nguyen T. H., Brechenmacher L., Aldrich J. T., Clauss T. R., Gritsenko M. A., Hixson K. K., et al. (2012). Quantitative phosphoproteomic analysis of soybean root hairs inoculated with Bradyrhizobium japonicum. Mol. Cell. Proteomics 11, 1140–1155. 10.1074/mcp.M112.018028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T., Zelechowska M., Foster V., Bergmann H., Verma D. (1985). Primary structure of the soybean nodulin-35 gene encoding uricase II localized in the peroxisomes of uninfected cells of nodules. Proc. Natl. Acad. Sci. U.S.A. 82, 5040–5044. 10.1073/pnas.82.15.5040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirunsuksiri W., Sengupta-Gopalan C. (1990). Characterization of a novel nodulin gene in soybean that shares sequence similarity to the gene for nodulin-24. Plant Mol. Biol. 15, 835–849. 10.1007/BF00039424 [DOI] [PubMed] [Google Scholar]
- Nishimura R., Ohmori M., Kawaguchi M. (2002). The novel symbiotic phenotype of enhanced-nodulating mutant of Lotus japonicus: astray mutant is an early nodulating mutant with wider nodulation zone. Plant Cell Physiol. 43, 853–859. 10.1093/pcp/pcf098 [DOI] [PubMed] [Google Scholar]
- Oka-Kira E., Tateno K., Miura K. I., Haga T., Hayashi M., Harada K., et al. (2005). klavier (klv), a novel hypernodulation mutant of Lotus japonicus affected in vascular tissue organization and floral induction. Plant J. 44, 505–515. 10.1111/j.1365-313X.2005.02543.x [DOI] [PubMed] [Google Scholar]
- O'Rourke J. A., Iniguez L. P., Fu F., Bucciarelli B., Miller S. S., Jackson S. A., et al. (2014). An RNA-Seq based gene expression atlas of the common bean. BMC Genomics 15:866. 10.1186/1471-2164-15-866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penmetsa R. V., Uribe P., Anderson J., Lichtenzveig J., Gish J. C., Nam Y. W., et al. (2008). The Medicago truncatula ortholog of Arabidopsis EIN2, sickle, is a negative regulator of symbiotic and pathogenic microbial associations. Plant J. 55, 580–595. 10.1111/j.1365-313X.2008.03531.x [DOI] [PubMed] [Google Scholar]
- Pingault L., Choulet F., Alberti A., Glover N., Wincker P., Feuillet C., et al. (2015). Deep transcriptome sequencing provides new insights into the structural and functional organization of the wheat genome. Genome Biol. 16, 29. 10.1186/s13059-015-0601-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pislariu C. I., Murray J. D., Wen J., Cosson V., Muni R. R. D., Wang M., et al. (2012). A Medicago truncatula tobacco retrotransposon insertion mutant collection with defects in nodule development and symbiotic nitrogen fixation. Plant Physiol. 159, 1686–1699. 10.1104/pp.112.197061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plet J., Wasson A., Ariel F., Le Signor C., Baker D., Mathesius U., et al. (2011). MtCRE1−dependent cytokinin signaling integrates bacterial and plant cues to coordinate symbiotic nodule organogenesis in Medicago truncatula. Plant J. 65, 622–633. 10.1111/j.1365-313X.2010.04447.x [DOI] [PubMed] [Google Scholar]
- Radutoiu S., Madsen L. H., Madsen E. B., Felle H. H., Umehara Y., Grønlund M., et al. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425, 585–592. 10.1038/nature02039 [DOI] [PubMed] [Google Scholar]
- Reid D. E., Ferguson B. J., Gresshoff P. M. (2011). Inoculation- and nitrate-induced CLE peptides of soybean control NARK-dependent nodule formation. Mol. Plant Microbe Interact. 24, 606–618. 10.1094/MPMI-09-10-0207 [DOI] [PubMed] [Google Scholar]
- Roulin A., Auer P. L., Libault M., Schlueter J., Farmer A., May G., et al. (2013). The fate of duplicated genes in a polyploid plant genome. Plant J. 73, 143–153. 10.1111/tpj.12026 [DOI] [PubMed] [Google Scholar]
- Roussis A., Flemetakis E., Dimou M., Kavroulakis N., Venieraki A., Aivalakis G., et al. (2003). Nodulin PvNOD33, a putative phosphatase whose expression is induced during Phaseolus vulgaris nodule development. Plant Physiol. Biochem. 41, 719–725. 10.1016/S0981-9428(03)00096-2 [DOI] [Google Scholar]
- Roux B., Rodde N., Jardinaud M. F., Timmers T., Sauviac L., Cottret L., et al. (2014). An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J. 77, 817–837. 10.1111/tpj.12442 [DOI] [PubMed] [Google Scholar]
- Saito K., Yoshikawa M., Yano K., Miwa H., Uchida H., Asamizu E., et al. (2007). NUCLEOPORIN85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in Lotus japonicus. Plant Cell 19, 610–624. 10.1105/tpc.106.046938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-López R., Jáuregui D., Nava N., Alvarado-Affantranger X., Montiel J., Santana O., et al. (2011). Down-regulation of SymRK correlates with a deficiency in vascular bundle development in Phaseolus vulgaris nodules. Plant Cell Environ. 34, 2109–2121. 10.1111/j.1365-3040.2011.02408.x [DOI] [PubMed] [Google Scholar]
- Sandal N. N., Bojsen K., Marcker K. A. (1987). A small family of nodule specific genes from soybean. Nucleic Acids Res. 15, 1507–1519. 10.1093/nar/15.4.1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S., Nakamura Y., Kaneko T., Asamizu E., Kato T., Nakao M., et al. (2008). Genome structure of the legume, Lotus japonicus. DNA Res. 15, 227–239. 10.1093/dnares/dsn008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauser L., Roussis A., Stiller J., Stougaard J. (1999). A plant regulator controlling development of symbiotic root nodules. Nature 402, 191–195. 10.1038/46058 [DOI] [PubMed] [Google Scholar]
- Schauser L., Wieloch W., Stougaard J. (2005). Evolution of NIN-like proteins in Arabidopsis, rice, and Lotus japonicus. J. Mol. Evol. 60, 229–237. 10.1007/s00239-004-0144-2 [DOI] [PubMed] [Google Scholar]
- Schmutz J., Cannon S. B., Schlueter J., Ma J., Mitros T., Nelson W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. 10.1038/nature08670 [DOI] [PubMed] [Google Scholar]
- Schmutz J., McClean P. E., Mamidi S., Wu G. A., Cannon S. B., Grimwood J., et al. (2014). A reference genome for common bean and genome-wide analysis of dual domestications. Nat. Genet. 46, 707–713. 10.1038/ng.3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel E. L., Frugoli J. (2004). The PIN and LAX families of auxin transport genes in Medicago truncatula. Mol. Genet. Genomics 272, 420–432. 10.1007/s00438-004-1057-x [DOI] [PubMed] [Google Scholar]
- Schnabel E. L., Kassaw T. K., Smith L. S., Marsh J. F., Oldroyd G. E., Long S. R., et al. (2011). The ROOT DETERMINED NODULATION1 gene regulates nodule number in roots of Medicago truncatula and defines a highly conserved, uncharacterized plant gene family. Plant Physiol. 157, 328–340. 10.1104/pp.111.178756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle I. R., Men A. E., Laniya T. S., Buzas D. M., Iturbe-Ormaetxe I., Carroll B. J., et al. (2003). Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science 299, 109–112. 10.1126/science.1077937 [DOI] [PubMed] [Google Scholar]
- Sengupta-Gopalan C., Pitas J. W., Thompson D. V., Hoffman L. M. (1986). Expression of host genes during root nodule development in soybeans. Mol. Gen. Genet. MGG 203, 410–420. 10.1007/BF00422065 [DOI] [Google Scholar]
- Sieberer B. J., Chabaud M., Timmers A. C., Monin A., Fournier J., Barker D. G. (2009). A nuclear-targeted cameleon demonstrates intranuclear Ca2+ spiking in Medicago truncatula root hairs in response to rhizobial nodulation factors. Plant Physiol. 151, 1197–1206. 10.1104/pp.109.142851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S., Katzer K., Lambert J., Cerri M., Parniske M. (2014). CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe 15, 139–152. 10.1016/j.chom.2014.01.011 [DOI] [PubMed] [Google Scholar]
- Smit P., Limpens E., Geurts R., Fedorova E., Dolgikh E., Gough C., et al. (2007). Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol. 145, 183–191. 10.1104/pp.107.100495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyano T., Kouchi H., Hirota A., Hayashi M. (2013). Nodule inception directly targets NF-Y subunit genes to regulate essential processes of root nodule development in Lotus japonicus. PLoS Genet. 9:e1003352. 10.1371/journal.pgen.1003352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke S., Kistner C., Yoshida S., Mulder L., Sato S., Kaneko T., et al. (2002). A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417, 959–962. 10.1038/nature00841 [DOI] [PubMed] [Google Scholar]
- Tadege M., Wen J., He J., Tu H., Kwak Y., Eschstruth A., et al. (2008). Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 54, 335–347. 10.1111/j.1365-313X.2008.03418.x [DOI] [PubMed] [Google Scholar]
- Tejada-Jiménez M., Castro-Rodríguez R., Kryvoruchko I., Lucas M. M., Udvardi M., Imperial J., et al. (2015). Medicago truncatula natural resistance-associated macrophage protein1 is required for iron uptake by rhizobia-infected nodule cells. Plant Physiol. 168, 258–272. 10.1104/pp.114.254672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirichine L., Imaizumi-Anraku H., Yoshida S., Murakami Y., Madsen L. H., Miwa H., et al. (2006). Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441, 1153–1156. 10.1038/nature04862 [DOI] [PubMed] [Google Scholar]
- Tóth K., Stratil T. F., Madsen E. B., Ye J., Popp C., Antolin-Llovera M., et al. (2012). Functional domain analysis of the Remorin protein LjSYMREM1 in Lotus japonicus. PLoS ONE 7:e30817. 10.1371/journal.pone.0030817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis B., Wandrey M., Colebatch G., Udvardi M. K. (2002). The soybean GmN6L gene encodes a late nodulin expressed in the infected zone of nitrogen-fixing nodules. Mol. Plant Microbe Interact. 15, 630–636. 10.1094/MPMI.2002.15.7.630 [DOI] [PubMed] [Google Scholar]
- Uehlein N., Fileschi K., Eckert M., Bienert G. P., Bertl A., Kaldenhoff R. (2007). Arbuscular mycorrhizal symbiosis and plant aquaporin expression. Phytochemistry 68, 122–129. 10.1016/j.phytochem.2006.09.033 [DOI] [PubMed] [Google Scholar]
- VandenBosch K. A., Bradley D. J., Knox J. P., Perotto S., Butcher G. W., Brewin N. J. (1989). Common components of the infection thread matrix and the intercellular space identified by immunocytochemical analysis of pea nodules and uninfected roots. EMBO J. 8, 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdier J., Torres-Jerez I., Wang M., Andriankaja A., Allen S. N., He J., et al. (2013). Establishment of the Lotus japonicus Gene Expression Atlas (LjGEA) and its use to explore legume seed maturation. Plant J. 74, 351–362. 10.1111/tpj.12119 [DOI] [PubMed] [Google Scholar]
- Wang C., Zhu M., Duan L., Yu H., Chang X., Li L., et al. (2015). Lotus japonicus clathrin heavy chain1 is associated with Rho-Like GTPase ROP6 and involved in nodule formation. Plant Physiol. 167, 1497–1510. 10.1104/pp.114.256107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb C. J., Chan-Weiher C., Johnson D. A. (2008). Isolation of a novel family of genes related to 2-oxoglutarate-dependent dioxygenases from soybean and analysis of their expression during root nodule senescence. J. Plant Physiol. 165, 1736–1744. 10.1016/j.jplph.2007.10.004 [DOI] [PubMed] [Google Scholar]
- Williams E. J., Bowles D. J. (2004). Coexpression of neighboring genes in the genome of Arabidopsis thaliana. Genome Res. 14, 1060–1067. 10.1101/gr.2131104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F., Murray J. D., Kim J., Heckmann A. B., Edwards A., Oldroyd G. E., et al. (2012). Legume pectate lyase required for root infection by rhizobia. Proc. Natl. Acad. Sci. U.S.A. 109, 633–638. 10.1073/pnas.1113992109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z., Hossain M. S., Valdés-López O., Hoang N. T., Zhai J., Wang J., et al. (2015). Identification and functional characterization of soybean root hair microRNAs expressed in response to Bradyrhizobium japonicum infection. Plant Biotechnol. J. 14, 332–341. 10.1111/pbi.12387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W.-C., de Blank C., Meskiene I., Hirt H., Bakker J., van Kammen A., et al. (1994). Rhizobium nod factors reactivate the cell cycle during infection and nodule primordium formation, but the cycle is only completed in primordium formation. Plant Cell 6, 1415–1426. 10.1105/tpc.6.10.1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. C., Katinakis P., Hendriks P., Smolders A., Vries F., Spee J., et al. (1993). Characterization of GmENOD40, a gene showing novel patterns of cell-specific expression during soybean nodule development. Plant J. 3, 573–585. 10.1046/j.1365-313X.1993.03040573.x [DOI] [PubMed] [Google Scholar]
- Yano K., Yoshida S., Müller J., Singh S., Banba M., Vickers K., et al. (2008). CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc. Natl. Acad. Sci. U.S.A. 105, 20540–20545. 10.1073/pnas.0806858105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young N. D., Debellé F., Oldroyd G. E., Geurts R., Cannon S. B., Udvardi M. K., et al. (2011). The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480, 520–524. 10.1038/nature10625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti M. E., Blanco F. A., Beker M. P., Battaglia M., Aguilar O. M. (2010). AC subunit of the plant nuclear factor NF-Y required for rhizobial infection and nodule development affects partner selection in the common bean–Rhizobium etli symbiosis. Plant Cell 22, 4142–4157. 10.1105/tpc.110.079137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan S., Horrocks J., Lukens L. N. (2006). Islands of co-expressed neighbouring genes in Arabidopsis thaliana suggest higher-order chromosome domains. Plant J. 45, 347–357. 10.1111/j.1365-313X.2005.02619.x [DOI] [PubMed] [Google Scholar]
- Zhang X.-Q., Lund A. A., Sarath G., Cerny R. L., Roberts D. M., Chollet R. (1999). Soybean nodule sucrose synthase (nodulin-100): further analysis of its phosphorylation using recombinant and authentic root-nodule enzymes. Arch. Biochem. Biophys. 371, 70–82. 10.1006/abbi.1999.1415 [DOI] [PubMed] [Google Scholar]
- Zhu H., Riely B. K., Burns N. J., Ané J.-M. (2006). Tracing nonlegume orthologs of legume genes required for nodulation and arbuscular mycorrhizal symbioses. Genetics 172, 2491–2499. 10.1534/genetics.105.051185 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.