Abstract
The proprotein convertase subtilisin kexin isozyme-1 (SKI-1)/site-1 protease (S1P) is implicated in lipid homeostasis, the unfolded protein response, and lysosome biogenesis. The protease is further hijacked by highly pathogenic emerging viruses for the processing of their envelope glycoproteins. Zymogen activation of SKI-1/S1P requires removal of an N-terminal prodomain, by a multistep process, generating the mature enzyme. Here, we uncover a modular structure of the human SKI-1/S1P prodomain and define its function in folding and activation. We provide evidence that the N-terminal AB fragment of the prodomain represents an autonomous structural and functional unit that is necessary and sufficient for folding and partial activation. In contrast, the C-terminal BC fragment lacks a defined structure but is crucial for autoprocessing and full catalytic activity. Phylogenetic analysis revealed that the sequence of the AB domain is highly conserved, whereas the BC fragment shows considerable variation and seems even absent in some species. Notably, SKI-1/S1P of arthropods, like the fruit fly Drosophila melanogaster, contains a shorter prodomain comprised of full-length AB and truncated BC regions. Swapping the prodomain fragments between fly and human resulted in a fully mature and active SKI-1/S1P chimera. Our study suggests that primordial SKI-1/S1P likely contained a simpler prodomain consisting of the highly conserved AB fragment that represents an independent folding unit. The BC region appears as a later evolutionary acquisition, possibly allowing more subtle fine-tuning of the maturation process.
Keywords: catalysis, enzyme, protein folding, proteolysis, transport, prodomain, proprotein convertase
Introduction
Proteolytic processing by endoproteases represents a major post-translational modification and is essential to regulate and diversify protein functions. Proteolysis is tightly regulated and often spatially confined to specific intracellular compartments. Subtilisin/kexin type proprotein convertases (PCs)3 constitute a family of nine conserved calcium-dependent serine endoproteases and include the basic PCs: PC1/3, PC2, furin, PC4, PACE4, PC5/6, and PC7, as well as the nonbasic PCs: SKI-1/S1P, and PCSK9 (1). PCs share homology to the kexin subfamily of subtilases with a distinctive Ser/His/Asp catalytic triad that mediates peptide bond scission. Basic PCs have similar but not identical consensus sequences (K/R)Xn(K/R)↓ (where Xn = 0, 2, 4, or 6 spacer residues) that may result in overlapping patterns of substrate cleavage. In contrast, SKI-1/S1P and PCSK9 cleave after hydrophobic or small residues, basic-X-(hydrophobic)-X↓ (2) and VFAQ↓, respectively (3). Processing by PCs is essential for the proper function of a plethora of cellular proteins, including pro-hormones, growth factor precursors, transcription factors, proteases, and cell adhesion molecules. Because of their crucial function, PCs are linked to a wide range of human pathologies, including vascular disease, metabolic disorders, cancer, and infectious diseases.
In normal physiology, SKI-1/S1P is known to activate sterol element-binding proteins (SREBPs) that control gene expression associated with cholesterol and fatty acid biosynthesis (4). Other SKI-1/S1P substrates include activating transcription factor 6 and members of the CREB3 family that are implicated in the regulation of the cellular ER/Golgi stress response (5). Recently, SKI-1/S1P has been linked to lysosome biogenesis via maturation of the N-acetylglucosamine-1-phosphotransferase precursor, which is crucial for mannose-6-phosphate labeling of lysosomal proteins required for correct trafficking (6). SKI-1/S1P has also been hijacked by several pathogenic emerging viruses of the Arenavirus and Bunyavirus family for the processing of their envelope glycoproteins (7, 8). The Arenavirus Lassa virus (LASV) represents the most important of these pathogens and causes several hundred thousand infections each year in Africa. Processing of the viral envelope glycoprotein precursor (GPC) by SKI-1/S1P is crucial for productive viral infection (7, 9–11), and inhibitors of SKI-1/S1P show potent anti-viral activity (12, 13). SKI-1/S1P is initially synthesized as an inactive precursor of 1,052 amino acids, comprised of a signal peptide, an N-terminal prodomain, and the catalytic domain. The transmembrane domain of SKI-1/S1P is followed by a basic cytosolic tail. Upon translocation into the ER, SKI-1/S1P undergoes autocatalytic maturation by sequential cleavages of the N-terminal prodomain, first at sites B′/B (RKVF↓RSLK137↓), followed by site C (RRLL186↓) and the newly described site C′ (RRAS166↓) (14). The end product SKI-1/S1P-C represents the fully mature enzyme (15, 16). A significant proportion of membrane-bound SKI-1/S1P undergoes shedding, leaving a membrane-associated stub (amino acids 954–1052) behind (16). Membrane-associated SKI-1/S1P localizes predominantly in the Golgi and partially in endosomal compartments, but not at the cell surface (17). Our recent studies on the mechanisms underlying SKI-1/S1P maturation revealed that immature forms retaining fragments of the prodomain are already enzymatically active (14), which appears very different from the maturation of the prototypic basic PC furin. As a consequence of the peculiar mechanism of maturation, SKI-1/S1P can act in at least three different compartments of the secretory pathway, depending on the substrate. Processing of LASV GPC occurs already in the ER/cis-Golgi, whereas cleavage of the cellular substrates SREBP and N-acetylglucosamine-1-phosphotransferase precursor occurs in the median or late Golgi, respectively (18). Intrigued by the unusual mechanism of SKI-1/S1P maturation, we herein sought to gain insights into the structural organization of the prodomain and to define the relative contributions of the prodomain fragments in the maturation process.
Experimental Procedures
Antibodies
mAb 83.6 mouse anti-LCMV glycoprotein was produced and purified as described previously (19). Mouse anti-V5 mAb was purchased from Invitrogen, rabbit anti-Gaussia luciferase (GLuc) was from New England Biolabs, and mouse anti-α-tubulin mAb was obtained from Sigma. Mouse mAb anti-eukaryotic translation initiation factor 2α (elF2α), rabbit mAb anti-phospho-elF2α (Ser51), and mouse mAb to binding immunoglobulin protein (BiP)/78-kDa glucose-regulated protein (GRP78) were from Cell Signaling (Danvers, MA). Secondary antibodies conjugated to HRP, polyclonal rabbit anti-mouse, and polyclonal goat anti-rabbit were purchased from Dako.
Plasmids and Constructs
Plasmids coding for WT full-length SKI-1/S1P and the mutant H249A were previously described (16). The truncated SKI-1/S1P Δpro mutants were generated by deletion mutagenesis using PCR amplification with specific primers to delete the nucleotides encoding for residues 17–186 (ΔPro AC), 17–137 (ΔPro AB), and 140–186 (ΔPro BC) of the SKI-1/S1P full-length construct. The construct encoding for SKI-1/S1P proAC (1–189) has been described before (20). Site-directed mutagenesis was carried out in Pro AC to introduce a stop codon to generate the proAB (1–140) using a pair of specific oligonucleotides. The generation of soluble V5-tagged SKI-1/S1P truncated before-the-transmembrane domain (BTMD) has been reported (14). Expression plasmids for LASV and LCMV GPC are described elsewhere (11).
The SKI-1/S1P sensor (SS-LASV) has been described in a previous report (14). The chimeric construct SKI-1/S1P containing the prodomain fragment of Drosophila melanogaster (proDmel) was generated by introducing unique restriction sites at the 3′ and 5′ end of the prodomain in the human SKI-1/S1P WT construct by synonymous mutations using the QuikChange site-directed mutagenesis kit from Stratagene following the manufacturer's protocol. The D. melanogaster prodomain sequence was derived from the SKI-1/S1P mRNA sequence with the accession ID NM_141080.3. The proDmel sequence containing the same unique restriction site in overlapping regions was custom synthesized by GenScript (Piscataway, NJ). The final construct contains the following amino acid residues: 1–22 (human), 19–141 (D. melanogaster), and 187–1052 (human). All constructs were verified by double-stranded DNA sequencing.
SKI-1/S1P Prodomain Synthesis and Purification
The SKI-1/S1P prodomain construct was synthesized as optimized gene for Escherichia coli and subcloned in the expression vector pETite containing a C-terminal His tag fusion protein. The prodomain was expressed in BL21 (DE3) E. coli (Lucigen) cultured in ECPM-I medium for 14 h at 18 °C. The sample was lysed by sonication in phosphate buffer (20 mm Na2HPO4, pH 7.5, 300 mm NaCl, 0.2 mm PMSF) supplemented with 0.01% Nonidet P-40 (w/v) and centrifuged for 30 min at 14,000 rpm. The supernatant was loaded to an immobilized metal ion affinity chromatography column and eluted with an imidazole gradient (0–500 mm imidazole) for purification. The fractions of highest purity were pooled and further subjected to gel filtration chromatography using a Superdex 200 column (10/300 GE Healthcare).
Cell Culture and Transfection
HEK293T cells were maintained in DMEM (Invitrogen) completed with 10% (v/v) FBS and 100 units/ml penicillin and 0.1 mg/ml streptomycin. Chinese hamster ovary (CHO)-K1 were maintained in DMEM/Ham's F12 1:1 (Biochrom AG) supplemented with 10% (v/v) FBS and penicillin/streptomycin. CHO-K1-derived SRD12B cells (SKI-1/S1P-deficient) (21) were provided by Michael S. Brown and Joseph L. Goldstein (University of Texas Southwestern Medical Center, Dallas, TX) and grown in CHO-K1 medium supplemented with 5 μg/ml cholesterol (Sigma), 20 μm sodium oleate (Sigma), and 1 mm sodium mevalonate (Sigma). All cell lines were incubated at 37 °C and 5% CO2. Transfections were performed as previously described (14), and efficiencies were evaluated after the indicated time points by detection of the EGFP reporter fluorescence when present.
Drug Treatments
Induction of genes regulated by SREBP2 was triggered by treatment with Ham's F-12 DMEM 1:1 containing 5% (v/v) delipidated FBS, 50 μm sodium mevalonate, and 50 μm mevastatin (Enzo Lifescience) for 18 h, as reported previously (20). DMSO (Sigma-Aldrich) was used as a negative control. At the indicated time points, conditioned medium was collected for analysis, and cell lysates were analyzed by Western blotting. To artificially trigger the unfolded protein response (UPR) in positive controls, cells were treated 4 h with 600 nm thapsigargin (Sigma-Aldrich).
Western Blot Analysis
Media were collected, and cells were washed twice with cold PBS and lysed with cell lytic buffer (Sigma) supplemented with Mini cOmplete® protease inhibitor mixture (Roche) following the manufacturer's instructions. Cell lysates were cleared by centrifugation (15,000 rpm, 10 min), and supernatants were transferred to new tubes. Conditioned media were centrifuged twice (1,500 rpm, 5 min) to remove cellular debris, and the supernatants were transferred to new tubes. Samples were mixed 1:1 with 2× SDS-PAGE sample buffer containing 100 mm DTT and boiled (5 min at 95 °C). Samples were separated by SDS-PAGE and blotted to nitrocellulose membranes. Membranes were blocked in 3% (w/v) skim milk in TBS, and proteins were detected with primary antibody after overnight incubation at 4 °C using HRP-conjugated secondary antibodies as described (11). Antibodies were used in the following dilutions: mAb 83.6 mouse anti-LCMV GP (1:1,000), anti-V5 mAb (1:5,000), rabbit anti-GLuc (1:6,000), anti-α-tubulin mAb (1:10,000), polyclonal rabbit anti-mouse-HRP (1:3,000), and polyclonal rat anti-rabbit-HRP (1:3,000). The membranes were developed by chemiluminescence using LiteABlot kit (EuroClone) or Amersham Biosciences ECL Prime Western blotting detection reagent (GE Healthcare Lifesciences). Signal acquisition was performed with ImageQuant LAS 4000Mini (GE Healthcare Lifesciences) or by exposure to x-ray films.
SKI-1/S1P Sensor Assays
To quantify secreted GLuc activity in conditioned media following sensor cleavage, we followed the protocol of transfection, collection, and analysis as reported (14). Briefly, a TriStar LB 941 (Berthold®) microplate reader was used to measure the luminescence of cell media samples. Conditioned media (3.5 μl) were manually distributed in a white half-volume 96-well plate and mixed with 60 μl of substrate solution composed of coelenterazine stock (Molecular Probes®) diluted in PBS (1:1,000). Stock solution was prepared dissolving coelenterazine in acidified methanol at 160 μg/ml and stored at 80 °C. Substrate injection was performed automatically and immediately followed by luminescence reading. Substrate solution was freshly prepared each time.
Circular Dichroism Analysis
Circular dichroism data were acquired using a JASCO J-715 spectropolarimeter in phosphate buffer, pH 7.5, 298 K, using a 0.05-cm-path length cell in the wavelength interval 190–260 nm. The buffer contribution was subtracted using the buffer spectrum collected in the same conditions. Protein concentration was estimated by UV-visible spectroscopy and theoretical absorption coefficient calculated by ProtParam software (ExPASy server).
Determination of in Vitro Activities of Soluble SKI-1/S1P
To assess the in vitro activities of the soluble SKI-1/S1P BTMD variants used in this study, constructs were expressed in HEK293T cells, and enzymatic activity was detected in conditioned medium via a reliable enzymatic assay as described previously (22). Briefly, conditioned medium containing enzyme was added to an assay buffer followed by the addition of the SKI-1/S1P-cleavable substrate (25 μm Ac-YISRRLL-MCA) just prior to the reaction start. The reactions were carried out at room temperature in buffer solution (25 mm Tris, pH 7.5, 25 mm MES, 1 mm CaCl2). The peptides were purchased from GenScript Inc. at >92% HPLC purity. Fluorescence of the cleaved aminocoumarine moiety was detected in a TriStar LB 941 multimode microplate spectrofluorometer (Berthold Technologies, Bad Wildbad, Germany). Assays where performed in biological triplicate, and statistical analysis was performed using the GraphPad Prism software package. One-way analysis of variance was used for multiple comparisons, and a p value of 0.05 was set as threshold for significance.
Homology-based Modeling of SKI-1/S1P Prodomain and Secondary Structure Prediction
For the search for templates and homology-based modeling of the SKI-1/S1P prodomain, the human SKI-1/S1P sequence amino acids 18–186 (Uniprot Q14703) was defined as a query sequence. The HHpred tool was used to identify homologous templates by searching the structural database of protein sequences in the Protein Data Bank. Default settings were used, except for the scoring of secondary structure where “predicted versus predicted” was used. Secondary structure predictions were performed with the tool PSIPRED to aid manual alignment corrections and loop modeling. Resulting query-template alignments from HHpred were submitted to Modeler V9.10 for homology-based modeling (23). The discrete optimized protein energy scores were calculated for each model, and averages were calculated for each query-template group. Except where otherwise stated, 100 models were created in each Modeler run. As a starting model we choose the lowest scored model generated using 2P4E, chain P as template. Region optimization was performed by analysis of discrete optimized protein energy profiles of the produced three-dimensional models structural alignments with the templates and also based in protein secondary structure prediction. Refinements were also performed using Modeler software V9.10. Molecular graphics images were produced with the UCSF Chimera V1.8 package (24). Secondary structure predictions where made using the psipred (25) and PredictProtein servers (26).
Phylogenetic Analysis of SKI-1/S1P
SKI-1/S1P orthologue protein sequence data used for phylogenetic analysis were collected from the UniProt database (27) by performing a blast search using the human SKI-1/S1P sequence as a query. Nonredundant protein sequences that contained both a signal peptide and a fully conserved catalytic triad where kept for further analysis, resulting in a total of 143 sequences from unique species. This protein sequence data set was used to construct a multiple sequence alignment using the MAFFT V.017 (28) algorithm from inside the Geneious 7.19 software (29). The generated multiple sequence alignment was then inspected manually, and major gaps were refined and used to generate conservation plots. The multiple sequence alignment was also used to map residue conservation in the SKI-1/S1P homology-based models.
Results
Partial or Complete Deletion of the N-terminal Prodomain Impairs SKI-1/S1P Autocatalytic Cleavage Both in cis and in trans
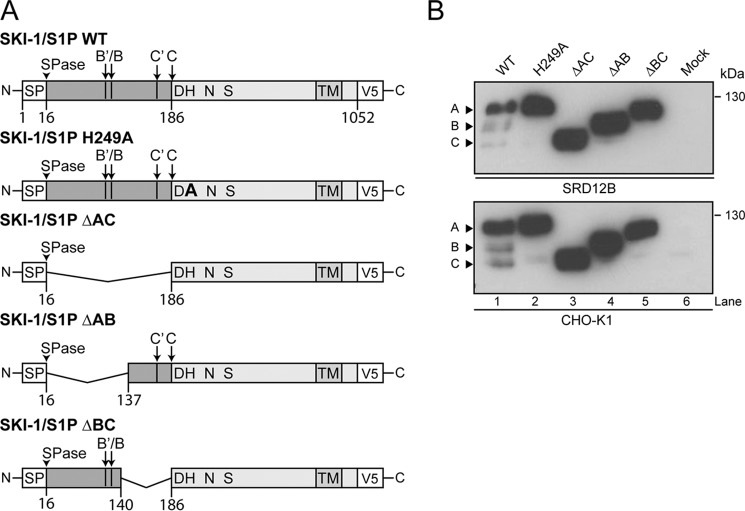
Previous studies indicated that in bacterial subtilases (30) and in SKI-1/S1P (14), the prodomain is crucial for folding, maturation, and acquisition of catalytic activity of the enzyme (14, 30), similar to other PCs. To define the specific roles of prodomain regions, we generated SKI-1/S1P constructs lacking either the entire prodomain (aa 17–186, ΔAC), the AB fragment (aa 17–137, ΔAB), or the BC portion (aa 140–186, ΔBC) (16). The mutant SKI-1/S1P ΔAC lacks all autoprocessing sites (B′/B and C′/C), whereas ΔBC and ΔAB contain either sites B′/B or C′/C, respectively. Of note, the deletion mutant ΔAC mimics the fully mature C and ΔAB the B′/B forms. SKI-1/S1P WT, which undergoes normal maturation, was included as positive control, whereas the “catalytically dead” mutant SKI-1/S1P H249A and empty vector were added as negative controls, respectively. All constructs were C-terminally labeled with a V5 tag and inserted in the expression vector pIRES-EGFP (Fig. 1A). The SKI-1/S1P prodomain deletion mutants were analyzed for their expression and maturation by transient overexpression in the SKI-1/S1P-deficient CHO cell line SRD12B (21). Western blot analysis revealed that all SKI-1/S1P prodomain deletion mutants were well expressed and showed the expected apparent molecular masses, without undergoing detectable cleavage (Fig. 1B). We and others have shown that maturation at the C′/C but not B′/B sites can occur in trans (14, 31). We therefore tested whether the prodomain deletion mutants could undergo cleavage in trans by endogenous SKI-1/S1P upon expression in parental CHOK1 cells. As shown in Fig. 1B, the ΔAB and ΔAC mutants did not undergo detectable maturation, suggesting that the endogenous enzyme was unable to efficiently process the C′/C sites present in ΔAB mutant in trans. In contrast, the catalytically dead mutant H249A was cleaved to a low extent (Fig. 1B), in agreement with published reports (31). Upon expression of the ΔBC mutant in CHOK1 (Fig. 1B) and HEK293T cells (not shown), a faint band at lower molecular mass was observed in some of our experiments, suggesting that, perhaps depending on the expression level, the B/B′ sites may undergo processing to a very low degree in trans in the context of this mutant. Taken together, the data show that partial or complete deletion of the N-terminal prodomain impairs SKI-1/S1P autocatalytic cleavage both in cis (B′/B) and in trans (C′/C).
FIGURE 1.
Characterization of the prodomain deletion mutants. A, schematic of the SKI-1/S1P prodomain deletion mutants. Signal peptide cleavage (SPase) and SKI-1/S1P autoprocessing sites (B′/B and C′/C) are indicated. The mutants ΔAC (aa 17–189), ΔAB (aa 17–137), and ΔBC (aa 140–189) are displayed. All mutants contain the catalytic and transmembrane domains and are C-terminally V5-tagged. B, expression and processing of mutant and WT SKI-1/S1P in SRD12B and CHOK1 cells. At 48 h post-transfection, total cell lysates were analyzed by Western blot using an anti-V5 antibody. The relative positions of the SKI-1/S1P precursor (form A) and the processed forms B and C are indicated by arrows.
Upon overexpression in CHOK1, SRD12B, and HEK293T cells, we detected significant amounts of proSKI-1/S1P (Fig. 1B). Previous studies likewise revealed accumulation of proSKI-1/S1P in nontransfected cells (17), suggesting that the presence of the A form of SKI-1/S1P seems not an artifact of our overexpression system. To address this question in a complementary manner, we examined the induction of the UPR of the cell upon overexpression of SKI-1/S1P. To this end, SKI-1/S1P WT was transiently expressed in CHOK1 and HEK293T cells, and the induction of the UPR was monitored by detection of the ER stress markers GRP78/BiP and phosphorylation of eIF2α that represent well characterized downstream targets of the UPR sensors activation transcription factor 6 and protein kinase RNA-like endoplasmic reticulum kinase, respectively. Treatment with 600 nm thapsigargin for 4 h was used as a positive control. Overexpression of SKI-1/S1P in CHOK1 cells over 48 h, corresponding to the duration of our experiments, did not result in marked induction of GRP78/BiP and phospho-eIF2α (Fig. 2). However, in HEK293T cells, we observed enhanced expression of GRP78/BiP and elevated phospho-eIF2α levels at 48 h post-transfection, corresponding to the peak of SKI-1/S1P expression. The data indicate induction of UPR and suggest that the rates and extents of processing and exit from the ER for our overexpressed SKI-1/S1P constructs may be altered by the level of expression.
FIGURE 2.
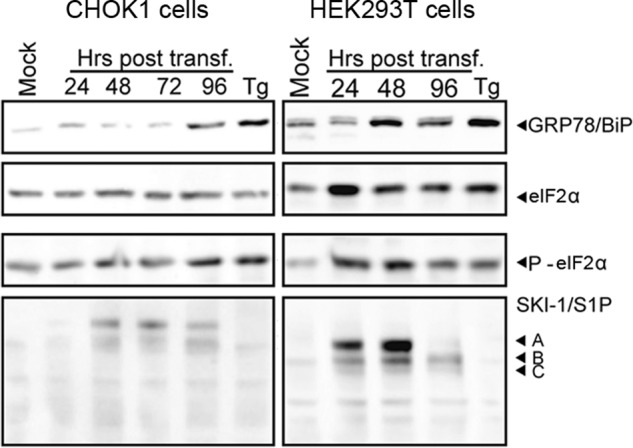
Induction of GRP78/BiP and phosphorylation of eIF2α after overexpression of SKI-1/S1P. CHOK1 and HEK293T cells were transfected with recombinant WT SKI-1/S1P. At the indicated time points, cells were lysed, and proteins were separated by SDS-PAGE and blotted to nitrocellulose. The expression of GRP78/BiP was detected with a specific antibody. To assess the phosphorylation of eIF2α, blots were probed with an antibody to phospho-eIF2α and an antibody that detects eIF2α protein independently of phosphorylation. The expression of SKI-1/S1P was detected as described in the legend to Fig. 1B. The positions of GRP78/BiP, phospho-eIF2α (P-eIF2α), total eIF2α, and the different forms of SKI-1/S1P (forms A, B, and C) are indicated. Please note that all experiments in this study were performed at 48 h post-transfection.
SKI-1/S1P Lacking the BC Fragment Retains Partial Enzymatic Activity and Exits the ER
In a next step, we assessed the catalytic activity of our SKI-1/S1P prodomain deletion mutants. First, we studied SKI-1/S1P-mediated processing of the transcription factor SREBP-2, which represents a well described cellular substrate. To this end, SRD12B cells were transiently transfected with mutant and WT SKI-1/S1P, followed by cholesterol depletion with mevastatin to induce endogenous SREBP-2 processing. Activation of SREBP-2 was detected by monitoring the transcription of the downstream gene 3-hydroxy-3-methylglutaryl-CoA synthase 1 (HMGCS1). Only WT SKI-1/S1P, but none of the mutants was able to significantly activate SREBP-2 and induce HMGCS1 transcription (Fig. 3A). Examination of protein expression levels in Western blot revealed comparable levels for all SKI-1/S1P variants in the presence of mevastatin (Fig. 3B). Previous studies revealed that some pathogen-derived SKI-1/S1P substrates, in particular the envelope GPC of arenaviruses, are excellent substrates for SKI-1/S1P (2, 22). Therefore, we next tested the ability of the SKI-1/S1P variants to process the GPCs of LASV and the lymphocytic choriomeningitis virus (LCMV) isolate clone 13. Upon co-expression in SRD12B cells, the mutant SKI-1/S1P ΔBCconsistently showed low but detectable processing of LASV and LCMV GPC, whereas the mutants ΔAC and ΔAB seemed inactive (Fig. 3, C and D). Because our Western blot analysis used for the detection of GPC cleavage was semiquantitative, we sought to confirm the partial activity of SKI-1/S1P ΔBC using a recently developed robust and quantitative cell-based molecular sensor for SKI-1/S1P activity (14). The assay is based on a chimeric protein composed of a GLuc reporter anchored to the membrane by the stump region of SKI-1/S1P through a cleavable 9-mer sequence IYISRRLL↓G, derived from the SKI-1/S1P recognition site of LASV GPC (Fig. 3E). Upon processing, this sensor releases GLuc into the medium, where it can be detected using a sensitive luciferase assay. Our previous studies revealed that this sensor recapitulates key features of authentic substrates and is suitable for the detection of incremental changes in SKI-1/S1P activity. Using SKI-1/S1P deficient cells, we co-expressed the sensor with the SKI-1/S1P mutants and detected cleavage by Western blot and detection of secreted GLuc via luciferase assay. In line with the previous results with the viral GPCs (Fig. 3, C and D), we found that the mutant ΔBC is capable of mediating significant cleavage of the sensor (Fig. 3F). Taken together, our data suggest that removing the AB region from the prodomain results in SKI-1/S1P forms lacking activity against all tested substrates. In contrast, deletion of the BC region results in a form that retains at least some enzymatic activity, as revealed by the processing of viral GPCs and the SKI-1/S1P sensor. Because LASV and LCMV GPC undergo SKI-1/S1P processing in the ER/cis-Golgi and in the late Golgi, respectively (7, 9, 18), efficient processing of both GPCs suggests that SKI-1/S1P ΔBC is partially active throughout the ER/Golgi compartments of the secretory pathway.
FIGURE 3.
Characterization of the activity of the prodomain deletion mutants. A, SREBP-2 processing by SKI-1/S1P mutants. SRD12B cells were transfected with the indicated SKI-1/S1P constructs. After 32 h, the SREBP-2 pathway was induced by mevastatin treatment for 18 h, whereas control cells were treated with DMSO. After treatment, total RNA was extracted to analyze HGMCS1 gene induction by quantitative RT-PCR. The results were analyzed by the ΔΔCT method, and the data were normalized against hydroxymethylbilane synthase gene expression. Gene expression is represented as fold induction above levels for control treatment (pIR-HA DMSO) (mean ± S.D.; n = 3). B, expression of SKI-1/S1P variants in A was assessed by Western blot as in Fig. 1B. C and D, LASV and LCMV GPC processing by the SKI-1/S1P mutants. SRD12B cell were co-transfected with either LASV GPC (C) or LCMV GPC (D) and the indicated SKI-1/S1P variants. At 48 h post-transfection, the cell lysates were analyzed by Western blot to assess GPC processing. SKI-1/S1P expression was detected with anti-V5 antibody and tubulin (Tub) detected as loading control. Tubulin, the precursor GPC, mature GP2, and maturation forms of SKI-1/S1P (forms A, B, and C) are indicated. E, schematic of the SKI-1/S1P sensor bearing the cleavage motif of LASV GPC. The signal peptide (SP), the GLuc reporter, the LASV GPC-derived cleavage motif, and the SKI-1/S1P-derived stump region are indicated. F, processing of the SKI-1/S1P sensor by SKI-1/S1P mutants. SRD12B cells were co-transfected with the SKI-1/S1P sensor (SS-LASV) and the indicated SKI-1/S1P variants. At 48 h post-transfection, cell lysates and supernatants were collected and analyzed by Western blot using an anti-GLuc antibody (bottom). Cleaved sensor (cGLuc) and the uncleaved precursor (pGLuc) are indicated. Conditioned media were analyzed for GLuc activity by addition of coelenterazine substrate. The data are shown as relative light units (RLU) (means ± S.D.; n = 3).
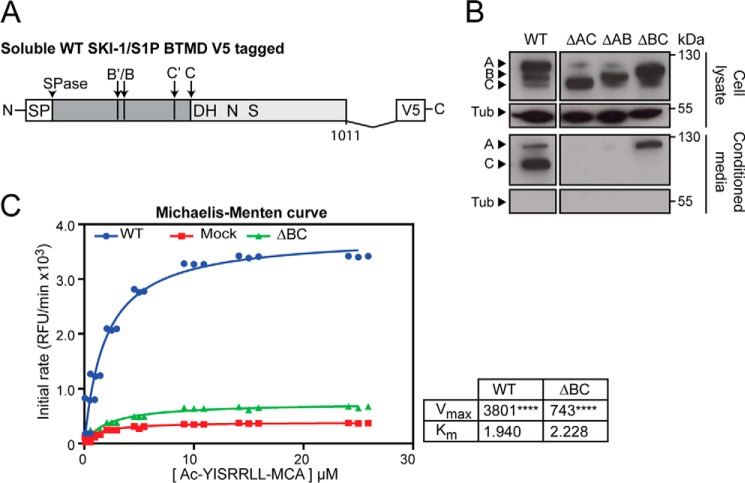
The partial activity of SKI-1/S1P ΔBC against LCMV GPC, which undergoes processing in the late Golgi, suggested that the mutant can exit the ER in absence of its processing at the B′/B site. This seems in stark contrast to maturation of other subtilases in which the intact prodomain functions as a chaperone and is required for proper folding, as well as ER exit (33, 34). To study possible chaperone-like activities of the SKI-1/S1P prodomain and fragments thereof, we constructed soluble versions of SKI-1/S1P WT and the prodomain deletion mutants by truncation BTMD (SKI-1/S1P WT-BTMD, BTMD-ΔAC, BTMD-ΔAB, and BTMD-ΔBC) (Fig. 4A). The soluble enzyme was previously shown to behave similar in maturation and activation as the full-length form (16, 18). To monitor transport of the mutants, the soluble SKI-1/S1P variants were expressed in SRD12B cells followed by detection in total cell lysates and culture supernatants. In line with previous studies, WT SKI-1/S1P-BTMD was abundant in supernatants, where it was detected mainly in the fully mature C-form together with lower amounts of unprocessed species (Fig. 4B). Although SKI-1/S1P BTMD-ΔBC was clearly detected in the supernatant, this was not the case for SKI-1/S1P BTMD-ΔAC and SKI-1/S1P BTMD-ΔAB. The absence of detectable amounts of tubulin in the supernatants excluded cellular contamination as a possible source of SKI-1/S1P BTMD-ΔBC (Fig. 4B), indicating that this mutant is capable of exiting the ER to undergo secretion. Using a well established in vitro assay for SKI-1/S1P activity based on fluorogenic peptides (for details, please see “Experimental Procedures”), we measured the catalytic activity of secreted SKI-1/S1P WT and ΔBC. As shown in Fig. 4C, secreted SKI-1/S1P BTMD-ΔBC showed a detectable catalytic activity, albeit much lower than the WT.
FIGURE 4.
Role of the SKI-1/S1P prodomain in maturation and folding of the enzyme. A, schematic of the soluble WT SKI-1/S1P BTMD. The signal peptide cleavage (SPase) and SKI-1/S1P autoprocessing sites (B′/B and C′/C) are indicated. Soluble SKI-1/S1P forms are truncated BTMD at residue Glu997. Constructs are C-terminally V5-tagged. B, expression and analysis of soluble SKI-1/S1P forms. SRD12B cells were transfected with the indicated soluble SKI-1/S1P variants. After 48 h, conditioned media and cell lysates were collected and analyzed as in Fig. 1B. C, SKI-1/S1P enzymatic activity in conditioned supernatants of SRD12B cells transfected with soluble SKI-1/S1P variants. SRD12B cells were transfected with recombinant SKI-1/S1P-BTMD and conditioned cell culture supernatants subjected to enzymatic assay as detailed under “Experimental Procedures.” Shown are the Michaelis-Menten curves obtained for WT and ΔBC SKI-1/S1P-BTMD compared with mock samples. Vmax and Km values are also shown. Statistical significance between Mock and SKI-1/S1P forms Vmax was assessed by one-way analysis of variance with p < 0.001 (****) indicated.
The Prodomain Provided in trans Can Rescue Folding, Autoprocessing, and Activity of SKI-1/S1P
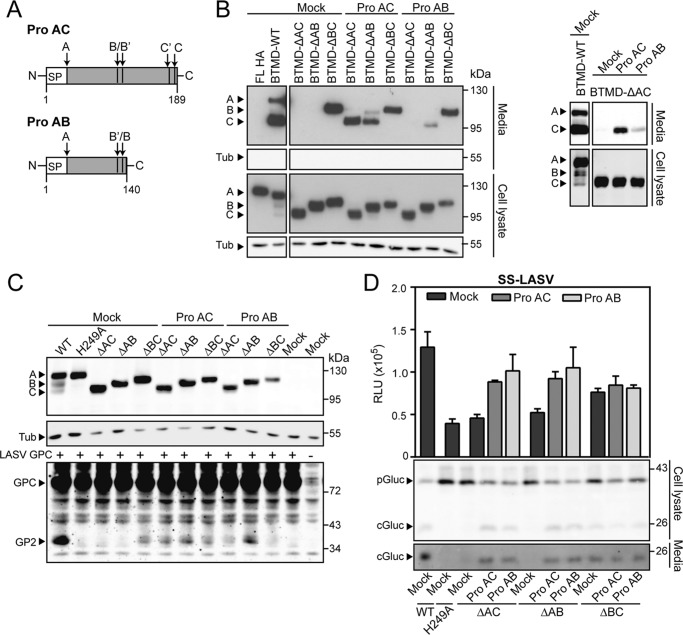
SKI-1/S1P lacking the entire prodomain is inactive and retained in the ER, highlighting its crucial role in maturation. In the case of bacterial subtilisins (35), furin (36), and PCSK9 (37), folding of the prodomain and the catalytic domain are independent events. As a consequence, expression of the prodomain in trans restores both folding and activity of the cognate truncated protease. To address this issue in the context of SKI-1/S1P, we generated a recombinant form of the full-length AC fragment of the SKI-1/S1P prodomain (aa 1–189, proAC) and a variant corresponding to the AB segment (aa 1–140, proAB) (Fig. 5A). To test our prodomain variants for their ability to facilitate maturation in trans, proAC and proAB were co-expressed with N-terminally truncated SKI-1/S1P-BTMD variants in SRD12B cells. When provided in trans, proAC was able to restore autoprocessing of SKI-1/S1P BTMD-ΔAC and BTMD-ΔAB at the B′/B and C′/C sites, as well as their secretion. In contrast, the efficiency of the shorter proAB to support ΔAC SKI-1/S1P folding was much less pronounced, though some effects were visible (Fig. 5B). Notably, both proAC and proAB failed to rescue maturation of SKI-1/S1P ΔBC-BTMD (Fig. 5B).
FIGURE 5.
Complementation studies with recombinant prodomain constructs. A, schematic of the prodomain constructs used for the complementation studies. Signal peptide cleavage and SKI-1/S1P autoprocessing (B′/B and C′/C) sites are indicated as in Fig. 1A. The full-length pro AC (aa 1–186) and the fragment proAB (aa 1–140) are displayed. B, complementation of N-terminally truncated SKI-1/S1P with proAC and proAB. SRD12B cells were co-transfected with the indicated truncated SKI-1/S1P-BTMD variants in combination with proAC, proAB, or empty vector (pIR). After 48 h, maturation of SKI-/S1P was detected in cell lysates and culture supernatants as in Fig. 1B. The A, B, and C forms of the protease are indicated. Detection of tubulin (Tub) served as loading control (lysates) and to exclude cellular contamination (supernatants). The right panel corresponds to a more exposed film, allowing the detection of BTMD-ΔAC in the supernatant. C, to assess whether proAC and proAB were able to restore enzymatic activity toward LASV GPC, SRD12B cells were co-transfected with the indicated SKI-1/S1P mutants, proAC or proAB and LASV GPC encoding plasmids. At 48 h post transfection, cells were lysed, and GPC processing, as well as SKI-1/S1P expression, was analyzed by Western blot. D, detection of SKI-1/S1P activity rescue upon prodomain complementation using the SKI-1/S1P sensor. The indicated sensor constructs were transfected in SRD12B cells, and samples were collected after 48 h. Analysis was performed as in Fig. 3F.
To investigate whether maturation by proAC and proAB in trans results in active enzymes, we tested the ability of the truncated SKI-1/S1P variants to cleave the viral substrate LASV GPC. To this end, SRD12B cells were co-transfected with proAC and proAB together with the N-terminally truncated forms of the enzyme and LASV GPC. Wild-type SKI-1/S1P and the mutant SKI-1/S1P H249A were included as positive and negative controls, respectively, and processing of LASV GPC was monitored by Western blot. In line with their capacity to mediate enzyme maturation, proAC and proAB were able to partially rescue GPC processing by SKI-1/S1P ΔAC and ΔAB (Fig. 5C, bottom panel). In contrast, co-expression of proAC and proAB did not further enhance the previously observed partial GPC processing by SKI-1/S1P ΔBC (Fig. 5C, bottom panel). In a complementary approach using our SKI-1/S1P sensor, we were able to confirm some rescue of catalytic activity of SKI-1/S1P ΔAC and ΔAB by expression of proAC and proAB in trans, whereas SKI-1/S1P ΔBC seemed unaffected (Fig. 5D). In sum, our data show that the enzymatic activity can be at least partially restored when the inactive ΔAC SKI-1/S1P is complemented in trans by the full-length AC prodomain. Our data provide evidence that the proAB fragment alone can restore activity as efficiently as proAC (Fig. 5, C and D), suggesting that proAB represents an independent functional unit with chaperone-like activity.
Phylogenic Analysis of SKI-1/S1P Prodomains Reveals Conservation of the AB Segment
Our recently developed homology model of the human SKI-1/S1P enzyme (14, 32) and the prodomain homology model presented here predict that the AB region folds with a defined structure, having high contents of β-sheet and α-helix. In contrast, no specific folding is predicted for the BC region (Fig. 6A) (14). To complement our modeling data, we sought to obtain information about the secondary structures present in the SKI-1/S1P prodomain. For this purpose, the recombinant prodomain construct proAC was expressed as a C-terminal His-tagged protein in E. coli and purified as described under “Experimental Procedures” (Fig. 6B). For initial characterization, the recombinant prodomain was subjected to CD analysis, which revealed folding with 37% β-sheets and 10% α-helices (Table 1), which is in line with our model (Fig. 6A). The relative contribution of the predicted secondary structures within AB to the overall folding of the prodomain in our molecular model (Fig. 6A) was consistent with the CD spectroscopy data. Specifically, random conformation accounts for ∼35%, whereas the BC region covers ∼20% of the sequence (Table 1). Next, we examined the sequences of the SKI-1/S1P prodomains of different species and performed a phylogenetic analysis of reported catalytic domains and proregions, as detailed under “Experimental Procedures” and summarized in Fig. 6C. In sum, we found 70% homology for the catalytic pocket with the catalytic triad His/Asp/Ser strictly conserved across all species. In contrast, the prodomain turned out to be more variable, with only 30% homology. We have also observed that at least one autoprocessing site seems to be present, but the site positions are not fixed, resulting in different lengths of the BC segment. Furthermore, analyses of the prodomain secondary structure found in our homology-based model (Fig. 6A) and as predicted by different prediction servers (Fig. 6C, top panel) are consistent with each other, and structured regions overlap with highly conserved motifs of the prodomain (Fig. 6C, bottom panel).
FIGURE 6.
A homology model of the SKI-1/S1P prodomain structure. A, the proposed SKI-1/S1P prodomain model is depicted and colored with a gradient from blue (N terminus) to red (C terminus). For simplicity, nonstructured regions on the N and C termini of the prodomain model were omitted, and only the region from Gly37 to Lys137 is shown. A schematic of a typical αβ-plaid is also displayed. Circles represent α-helixes, and the rounded boxes represent the β-strands. Regions pointing in the opposite direction from the viewer are marked with a plus sign (N terminus), and those pointing toward the viewer are marked with an asterisk (C terminus). B, Coomassie-stained gel of purified recombinant prodomain AC. C, schematic representation of the human SKI-1/S1P prodomain in scale. Maturation sites B′/B and C′/C are indicated by yellow and magenta circles, respectively. On the upper panel, the secondary structures are indicated, as predicted by the psipred and RePROF prediction servers. The secondary structures of the model in Fig. 6A are added for comparison. The bottom part of the panel represents a conservation plot of the prodomain over 143 SKI-1/S1P orthologues. The y axis indicates the degree of conservation with a sliding window of five residues, green colored residues indicate 100% identity, brown colored residues indicate ≥30% <100%, and red colored residues indicate <30% identity.
TABLE 1.
Secondary structure assignment of the recombinant proAC construct
The values have been obtained through the deconvolution (CDNN software) of circular dichroism data acquired in the 190–260-nm range. Prediction values are from the indicated secondary structure prediction servers and our SKI-1/S1P prodomain homology-based model.
| Deconvolution values (CDNN) | psipred server | PredictProtein server | Model | |
|---|---|---|---|---|
| Interval | 190–260 nm | |||
| Helix | 10% | 12% | 12% | 11% |
| Beta sheets | 37% | 20% | 23% | 15% |
| Beta turn | 18% | |||
| Other | 35% | 68% | 65% | 74% |
The SKI-1/S1P Prodomain of D. melanogaster Can Replace the Human One: Proof of Concept
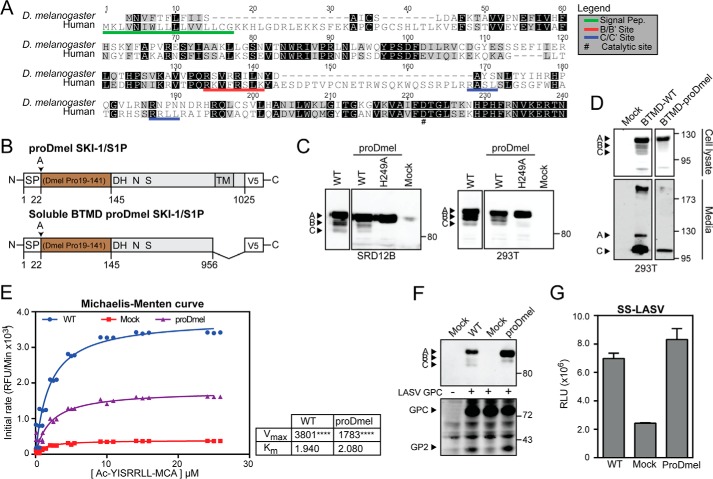
Our phylogenetic analysis revealed that SKI-1/S1P of some species, including the fruit fly D. melanogaster, lacks most of the BC region (Fig. 7A), suggesting that AB is necessary and sufficient for maturation. To test this hypothesis, we inserted the prodomain of Drosophila into human SKI-1/S1P. The chimeric SKI-1/S1P protein (proDmel SKI-1/S1P) contained the signal peptide of the human SKI-1/S1P and the proregion of Drosophila SKI-1/S1P fused to the catalytic and transmembrane domains of human SKI-1/S1P (Fig. 7B). In addition, we generated an inactive chimera bearing the catalytic H249A mutation (SKI-1/S1P proDmel H249A). The proDmel SKI-1/S1P, its catalytic dead mutant, and the WT form of human SKI-1/S1P were transiently expressed in SRD12B cells. Examination by Western blot revealed that SKI-1/S1P proDmel undergoes automaturation with the generation of a single processed band (Fig. 7C), whereas the H249A catalytically inactive version seemed unable to autoprocess (Fig. 7C). Generating a soluble version of the chimeric form (SKI-1/S1P proDmel-BTMD), we assessed its ability to traffic through the secretory pathway. The enzyme was secreted and detectable in the supernatant with the expected apparent molecular mass (Fig. 7D). Despite lower expression levels when compared with the WT, the enzymatic activity of SKI-1/S1P proDmel-BTMB was enhanced, as assessed by in vitro assay (Fig. 7E). Finally, we addressed the enzymatic activity of SKI-1/S1P proDmel using LASV GPC and our sensor system. As shown in Fig. 7F, LASV GPC was cleaved by SKI-1/S1P proDmel at similar extents as by WT SKI-1/S1P. These data were confirmed by the activity of SKI-1/S1P proDmel-BTMD against our SKI-1/S1P sensor (Fig. 7G). In sum, our data show that replacement of the human prodomain by the shorter Drosophila version results in a fully active enzyme.
FIGURE 7.
The prodomain of D. melanogaster SKI-1/S1P is able to complement the human SKI-1/S1P form. A, alignment of the prodomains of human and D. melanogaster SKI-1/S1P. Alignment was performed with ClustalW running from the Geneious software package. Identical and similar residues are indicated by black and gray shading. Maturation sites are underlined in red and blue. B, schematic of the chimeric construct. The prodomain of D. melanogaster (aa 19–141) was fused to the human signal peptide (aa 1–22) and human SKI-1/S1P C terminus (aa 186–1052). The construct was tagged with a C-terminal V5 tag. C, maturation of chimeric and WT SKI-1/S1P. SRD12B and HEK293T cells were transfected with the indicated constructs and SKI-1/S1P detected in cell lysates by Western blot as in Fig. 1B. D, secretion of the SKI-1/S1P chimera. To verify that our chimeric construct underwent proper maturation and folding, a soluble BTMD form was generated. Constructs were expressed in HEK293T cells, and the presence of the SKI-1/S1P variants was detected in cell lysates and media. E, enzymatic activity of the chimera SKI-1/S1P proDmel-BTMD. Conditioned media from HEK293T cells expressing SKI-1/S1P proDmel-BTMD and WT were assayed for enzymatic activity as in Fig. 4C. Shown are Michaelis-Menten curves, Vmax, and Km values. Statistical significance between Mock and SKI-1/S1P forms Vmax was assessed by one-way analysis of variance with p < 0.001 (****) indicated. F, processing of LASV GPC by SKI-1/S1P proDmel-BTMD detected by Western blot as in Fig. 5C. G, activity of SKI-1/S1P proDmel against the SKI-1/S1P sensor was assessed by co-transfection into SRD12B cells. Conditioned media were analyzed for GLuc activity as in Fig. 5D. The data are shown as relative light units (RLU) (means ± S.D.; n = 3).
Discussion
The maturation of basic PCs is characterized by an on/off mechanism of activation and temporally defined autoprocessing steps. In contrast, recent studies revealed unique features of SKI-1/S1P maturation during which autoprocessing is not required for activity and the proregion remains attached to the mature enzyme (14). As a consequence, the protease seems to exist as different forms that vary in the length of the prodomain, cleaved at multiple sites including B′/B and/or C′/C (14). Autoprocessing seems limited to the BC region of the prodomain, whereas the AB part is always found intact. The reasons for this complex and unusual maturation pattern are currently unclear. One might speculate that the different mature forms of SKI-1/S1P present throughout the secretory pathway may show some selectivity for specific substrates, possibly in a compartment-dependent manner. Intrigued by the unusual mechanism of SKI-1/S1P activation, we investigated the role of different regions of the enzyme prodomain in the maturation process. Performing structure-function analysis, we were able to identify the N-terminal AB fragment of the prodomain as a functional unit necessary and sufficient for maturation and partial activation. In contrast, the BC segment of the prodomain seemed dispensable for folding but was required for optimal activation of the enzyme. How the BC segment contributes to full activation is currently unclear. The reduced activity of SKI-1/S1P lacking BC may be the result of suboptimal substrate recognition caused by obstruction of the catalytic pocket by the uncleaved prodomain. Alternatively, the absence of the BC sequence may result in an overall conformational change of the enzyme that prevents full activity.
In trans complementation of prodomains has previously been shown to rescue maturation and activation of bacterial subtilisin and PCSK9. Our studies revealed that the full-length SKI-1/S1P prodomain and the AB portion were able to significantly restore maturation and enzymatic activity when provided in trans. This further supports our hypothesis of the AB fragment representing a functional unit with chaperone-like properties. Molecular modeling combined with CD spectroscopy performed on bacterially expressed recombinant prodomain supported the notion that the AB fragment represents an independent folding unit, whereas the BC moiety seems to lack a defined structure. The data at hand suggest that the AB region contains chaperonin-like functions, likely facilitating folding of the adjacent catalytic domain. The BC domain may be required to assume an unstructured conformation to be optimally autocleaved at multiple motifs. This autoprocessing may promote physical detachment of the N terminus and provide substrates access to the catalytic pocket. In this model, the minimal AB folding unit would seem strictly required, whereas the length of the “joining” BC peptide may vary. To validate our working hypothesis, we performed phylogenetic analysis of over 140 SKI-1/S1P metazoan orthologues. Across species, we noted a high degree of conservation in the AB region but considerable variation in length and sequence of the BC moiety. Interestingly, the SKI-1/S1P prodomain of some arthropod species, e.g. D. melanogaster, partially lacked the BC sequence. In a proof of concept study, we replaced the human SKI-1/S1P prodomain with that of Drosophila and obtained a fully functional chimera that underwent normal autoprocessing. We hypothesize that the prodomain of Drosophila SKI-1/S1P, which shares 37% identity with the human sequence, contains the minimal necessary chaperonin-like function together with the autocleavage site(s) to activate the human enzyme. The ability of intramolecular chaperones to efficiently function across species despite low sequence conservation has to the best of our knowledge not yet been reported. The capacity of the prodomain of Drosophila SKI-1/S1P to act on the human catalytic domain may be due to conservation of amino acids located at their interface. Moreover, the overall folding of Drosophila and human prodomains may be similar, facilitating formation of matching molecular surfaces. Our sequence alignment further revealed that the conserved amino acids of the AB segment lie within the highly structured regions predicted for the interface with the catalytic domain (Fig. 8). The BC region in contrast shows a markedly lower level of conservation. For the catalytic domain, residues surrounding the catalytic groove, as well as sites predicted to interact with the prodomain AB fragment, also show significantly higher conservation (Fig. 8). The observed variations in length and sequence of the BC region support our model of BC being a “joining peptide” between the cleavage sites B/B′ and C/C′ that can vary between species. It is conceivable that the BC joining peptide may affect the exact positioning of the autoprocessing sites and therefore modulate the kinetics of enzyme maturation. In line with this, deletion of the entire BC segment results in an uncleavable precursor that retains some activity. The exact sequence and length of the BC “joining peptide” may thus affect SKI-1/S1P activity. The apparent redundancy of the multiple autoprocessing (B/B′ and C/C′) may be required to generate forms of the protease with different activities. In sum, our study provides a novel model for the structure and function of the SKI-1/S1P prodomain that helps to understand the peculiar mechanism of maturation of this enzyme that is involved in many basic physiological processes and is at the cross-roads of several human disorders.
FIGURE 8.
Homology-based model of human SKI-1/S1P with mapped sequence conservation. The top panel shows sequence conservation in the prodomain, and the bottom panel shows sequence conservation in the catalytic domain (catalytic grove face on the left and opposite side on the right). Highly conserved residues are depicted in blue, whereas low conservation residues are colored in red.
Author Contributions
A. P., N. G. S., L. C., and S. K. conceived and coordinated the study and wrote the paper. J. R. d. P., L. C., and A. P. designed, performed, and analyzed the experiments. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Michael S. Brown and Joseph L. Goldstein (University of Texas Southwestern Medical Center, Dallas, TX) for the S1P-deficient SRD12B cells.
This work was supported by Swiss National Science Foundation Grants FN 31003A-135536 and FN 310030_149746 (to S. K.), Grant 14B086 from the Novartis Stiftung für Medizinisch-Biologische Forschung, and funds from the University of Lausanne (to S. K.), as well as by Canadian Institutes of Health Research Grant 93792 and Canada Chair Grant 216684 (to N. G. S.). The authors declare that they have no conflicts of interest with the contents of this article.
- PC
- proprotein convertase
- SREBP
- sterol element-binding protein
- ER
- endoplasmic reticulum
- GPC
- glycoprotein precursor
- LASV
- Arenavirus Lassa virus
- GLuc
- Gaussia luciferase
- UPR
- unfolded protein response
- aa
- amino acid(s)
- LCMV
- lymphocytic choriomeningitis virus
- BTMD
- before-the-transmembrane domain.
References
- 1. Seidah N. G., and Prat A. (2002) Precursor convertases in the secretory pathway, cytosol and extracellular milieu. Essays Biochem. 38, 79–94 [DOI] [PubMed] [Google Scholar]
- 2. Pasquato A., Pullikotil P., Asselin M. C., Vacatello M., Paolillo L., Ghezzo F., Basso F., Di Bello C., Dettin M., and Seidah N. G. (2006) The proprotein convertase SKI-1/S1P: in vitro analysis of Lassa virus glycoprotein-derived substrates and ex vivo validation of irreversible peptide inhibitors. J. Biol. Chem. 281, 23471–23481 [DOI] [PubMed] [Google Scholar]
- 3. Benjannet S., Rhainds D., Essalmani R., Mayne J., Wickham L., Jin W., Asselin M. C., Hamelin J., Varret M., Allard D., Trillard M., Abifadel M., Tebon A., Attie A. D., Rader D. J., Boileau C., Brissette L., Chrétien M., Prat A., and Seidah N. G. (2004) NARC-1/PCSK9 and its natural mutants: zymogen cleavage and effects on the low density lipoprotein (LDL) receptor and LDL cholesterol. J. Biol. Chem. 279, 48865–48875 [DOI] [PubMed] [Google Scholar]
- 4. Sakai J., Rawson R. B., Espenshade P. J., Cheng D., Seegmiller A. C., Goldstein J. L., and Brown M. S. (1998) Molecular identification of the sterol-regulated luminal protease that cleaves SREBPs and controls lipid composition of animal cells. Mol. Cell 2, 505–514 [DOI] [PubMed] [Google Scholar]
- 5. Seidah N. G., and Prat A. (2012) The biology and therapeutic targeting of the proprotein convertases. Nat. Rev. Drug Discov. 11, 367–383 [DOI] [PubMed] [Google Scholar]
- 6. Marschner K., Kollmann K., Schweizer M., Braulke T., and Pohl S. (2011) A key enzyme in the biogenesis of lysosomes is a protease that regulates cholesterol metabolism. Science 333, 87–90 [DOI] [PubMed] [Google Scholar]
- 7. Lenz O., ter Meulen J., Klenk H. D., Seidah N. G., and Garten W. (2001) The Lassa virus glycoprotein precursor GP-C is proteolytically processed by subtilase SKI-1/S1P. Proc. Natl. Acad. Sci. U.S.A. 98, 12701–12705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vincent M. J., Sanchez A. J., Erickson B. R., Basak A., Chretien M., Seidah N. G., and Nichol S. T. (2003) Crimean-Congo hemorrhagic fever virus glycoprotein proteolytic processing by subtilase SKI-1. J. Virol. 77, 8640–8649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Beyer W. R., Pöpplau D., Garten W., von Laer D., and Lenz O. (2003) Endoproteolytic processing of the lymphocytic choriomeningitis virus glycoprotein by the subtilase SKI-1/S1P. J. Virol. 77, 2866–2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rojek J. M., Lee A. M., Nguyen N., Spiropoulou C. F., and Kunz S. (2008) Site 1 protease is required for proteolytic processing of the glycoproteins of the South American hemorrhagic fever viruses Junin, Machupo, and Guanarito. J. Virol. 82, 6045–6051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kunz S., Edelmann K. H., de la Torre J. C., Gorney R., and Oldstone M. B. (2003) Mechanisms for lymphocytic choriomeningitis virus glycoprotein cleavage, transport, and incorporation into virions. Virology 314, 168–178 [DOI] [PubMed] [Google Scholar]
- 12. Rojek J. M., Pasqual G., Sanchez A. B., Nguyen N. T., de la Torre J. C., and Kunz S. (2010) Targeting the proteolytic processing of the viral glycoprotein precursor is a promising novel antiviral strategy against arenaviruses. J. Virol. 84, 573–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Blanchet M., Sureau C., Guévin C., Seidah N. G., and Labonté P. (2015) SKI-1/S1P inhibitor PF-429242 impairs the onset of HCV infection. Antiviral Res. 115, 94–104 [DOI] [PubMed] [Google Scholar]
- 14. da Palma J. R., Burri D. J., Oppliger J., Salamina M., Cendron L., de Laureto P. P., Seidah N. G., Kunz S., and Pasquato A. (2014) Zymogen activation and subcellular activity of subtilisin kexin isozyme 1/site 1 protease. J. Biol. Chem. 289, 35743–35756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Touré B. B., Munzer J. S., Basak A., Benjannet S., Rochemont J., Lazure C., Chrétien M., and Seidah N. G. (2000) Biosynthesis and enzymatic characterization of human SKI-1/S1P and the processing of its inhibitory prosegment. J. Biol. Chem. 275, 2349–2358 [DOI] [PubMed] [Google Scholar]
- 16. Elagoz A., Benjannet S., Mammarbassi A., Wickham L., and Seidah N. G. (2002) Biosynthesis and cellular trafficking of the convertase SKI-1/S1P: ectodomain shedding requires SKI-1 activity. J. Biol. Chem. 277, 11265–11275 [DOI] [PubMed] [Google Scholar]
- 17. Pullikotil P., Benjannet S., Mayne J., and Seidah N. G. (2007) The proprotein convertase SKI-1/S1P: alternate translation and subcellular localization. J. Biol. Chem. 282, 27402–27413 [DOI] [PubMed] [Google Scholar]
- 18. Burri D. J., Pasqual G., Rochat C., Seidah N. G., Pasquato A., and Kunz S. (2012) Molecular characterization of the processing of arenavirus envelope glycoprotein precursors by subtilisin kexin isozyme-1/site-1 protease. J. Virol. 86, 4935–4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Weber E. L., and Buchmeier M. J. (1988) Fine mapping of a peptide sequence containing an antigenic site conserved among arenaviruses. Virology 164, 30–38 [DOI] [PubMed] [Google Scholar]
- 20. Pullikotil P., Vincent M., Nichol S. T., and Seidah N. G. (2004) Development of protein-based inhibitors of the proprotein of convertase SKI-1/S1P: processing of SREBP-2, ATF6, and a viral glycoprotein. J. Biol. Chem. 279, 17338–17347 [DOI] [PubMed] [Google Scholar]
- 21. Rawson R. B., Cheng D., Brown M. S., and Goldstein J. L. (1998) Isolation of cholesterol-requiring mutant Chinese hamster ovary cells with defects in cleavage of sterol regulatory element-binding proteins at site 1. J. Biol. Chem. 273, 28261–28269 [DOI] [PubMed] [Google Scholar]
- 22. Pasquato A., Burri D. J., Traba E. G., Hanna-El-Daher L., Seidah N. G., and Kunz S. (2011) Arenavirus envelope glycoproteins mimic autoprocessing sites of the cellular proprotein convertase subtilisin kexin isozyme-1/site-1 protease. Virology 417, 18–26 [DOI] [PubMed] [Google Scholar]
- 23. Eswar N., Eramian D., Webb B., Shen M. Y., and Sali A. (2008) Protein structure modeling with MODELLER. Methods Mol. Biol. 426, 145–159 [DOI] [PubMed] [Google Scholar]
- 24. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 25. Buchan D. W., Minneci F., Nugent T. C., Bryson K., and Jones D. T. (2013) Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 41, W349–W357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yachdav G., Kloppmann E., Kajan L., Hecht M., Goldberg T., Hamp T., Hönigschmid P., Schafferhans A., Roos M., Bernhofer M., Richter L., Ashkenazy H., Punta M., Schlessinger A., Bromberg Y., Schneider R., Vriend G., Sander C., Ben-Tal N., and Rost B. (2014) PredictProtein: an open resource for online prediction of protein structural and functional features. Nucleic Acids Res. 42, W337–W343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. UniProt Consortium (2015) UniProt: a hub for protein information. Nucleic Acids Res. 43, D204–D212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Katoh K., Misawa K., Kuma K., and Miyata T. (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., Thierer T., Ashton B., Meintjes P., and Drummond A. (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhu X. L., Ohta Y., Jordan F., and Inouye M. (1989) Pro-sequence of subtilisin can guide the refolding of denatured subtilisin in an intermolecular process. Nature 339, 483–484 [DOI] [PubMed] [Google Scholar]
- 31. Espenshade P. J., Cheng D., Goldstein J. L., and Brown M. S. (1999) Autocatalytic processing of site-1 protease removes propeptide and permits cleavage of sterol regulatory element-binding proteins. J. Biol. Chem. 274, 22795–22804 [DOI] [PubMed] [Google Scholar]
- 32. Burri D. J., da Palma J. R., Seidah N. G., Zanotti G., Cendron L., Pasquato A., and Kunz S. (2013) Differential recognition of Old World and New World arenavirus envelope glycoproteins by subtilisin kexin isozyme 1 (SKI-1)/site 1 protease (S1P). J. Virol. 87, 6406–6414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lesage G., Prat A., Lacombe J., Thomas D. Y., Seidah N. G., and Boileau G. (2000) The Kex2p proregion is essential for the biosynthesis of an active enzyme and requires a C-terminal basic residue for its function. Mol. Biol. Cell 11, 1947–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Inouye M. (1991) Intramolecular chaperone: the role of the pro-peptide in protein folding. Enzyme 45, 314–321 [DOI] [PubMed] [Google Scholar]
- 35. Shinde U., and Thomas G. (2011) Insights from bacterial subtilases into the mechanisms of intramolecular chaperone-mediated activation of furin. Methods Mol. Biol. 768, 59–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Anderson E. D., Molloy S. S., Jean F., Fei H., Shimamura S., and Thomas G. (2002) The ordered and compartment-specfific autoproteolytic removal of the furin intramolecular chaperone is required for enzyme activation. J. Biol. Chem. 277, 12879–12890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li J., Tumanut C., Gavigan J. A., Huang W. J., Hampton E. N., Tumanut R., Suen K. F., Trauger J. W., Spraggon G., Lesley S. A., Liau G., Yowe D., and Harris J. L. (2007) Secreted PCSK9 promotes LDL receptor degradation independently of proteolytic activity. Biochem. J. 406, 203–207 [DOI] [PMC free article] [PubMed] [Google Scholar]