Abstract
The post-translational protein modification O-linked β-N-acetylglucosamine (O-GlcNAc) is a proposed nutrient sensor that has been shown to regulate multiple biological pathways. This dynamic and inducible enzymatic modification to intracellular proteins utilizes the end product of the nutrient sensing hexosamine biosynthetic pathway, UDP-GlcNAc, as its substrate donor. Type II diabetic patients have elevated O-GlcNAc-modified proteins within pancreatic beta cells due to chronic hyperglycemia-induced glucose overload, but a molecular role for O-GlcNAc within beta cells remains unclear. Using directed pharmacological approaches in the mouse insulinoma-6 (Min6) cell line, we demonstrate that elevating nuclear O-GlcNAc increases intracellular insulin levels and preserves glucose-stimulated insulin secretion during chronic hyperglycemia. The molecular mechanism for these observed changes appears to be, at least in part, due to elevated O-GlcNAc-dependent increases in Ins1 and Ins2 mRNA levels via elevations in histone H3 transcriptional activation marks. Furthermore, RNA deep sequencing reveals that this mechanism of altered gene transcription is restricted and that the majority of genes regulated by elevated O-GlcNAc levels are similarly regulated by a shift from euglycemic to hyperglycemic conditions. These findings implicate the O-GlcNAc modification as a potential mechanism for hyperglycemic-regulated gene expression in the beta cell.
Keywords: beta cell (B-cell), diabetes, epigenetics, insulin, O-linked N-acetylglucosamine (O-GlcNAc), post-translational modification (PTM), RNASeq
Introduction
Type II diabetes-related complications continue to be a leading cause of death in the United States (1). In the diabetic condition, hyperglycemia results in insulin resistance in muscle, liver, and adipose tissues that are responsible for glucose clearance. The inability of peripheral tissues to absorb glucose leads to hyperinsulinemia and glucose toxicity, serving as the proposed main contributors toward disease progression (2). In attempts to cope with the elevated glucose levels, pancreatic β-cells respond by increasing insulin hormone production and secretion. However, continual stimulation via elevated glucose levels eventually leads to β-cell fatigue and decreased insulin release (3).
Numerous environmental factors act in combination to influence insulin regulation both at the transcriptional and translational levels (4–7). Post-translational protein modifications have emerged as key regulatory avenues dictating diverse cellular activities. Terminal hydroxyls of serine and threonine residues can be modified by O-linked β-N-acetylglucosamine (O-GlcNAc)3 to influence intracellular processes (8–12). This dynamic and inducible post-translational modification more closely mimics phosphorylation in that it is a single molecule addition cycling on and off intracellular proteins in response to the cellular environment (13–15). The donor substrate for O-GlcNAc is the sugar nucleotide UDP-GlcNAc, which is the end product of the HBP (16, 17). In mammals only a single gene encodes for each enzyme responsible for the addition and removal of O-GlcNAc, O-GlcNAc transferase, and O-GlcNAcase (OGA) (18–20). Due to the involvement of UDP-GlcNAc in multiple metabolic pathways and the diversity of proteins modified by O-GlcNAc within these systems, O-GlcNAc has been implicated in the etiology of several metabolic diseases, including diabetes, and been hypothesized to be a nutrient sensor (21–24).
High glucose stimulation in cultured mouse insulinoma 6 (Min6) cells is known to elevate O-GlcNAc levels (25), whereas direct modulation of either the HBP or O-GlcNAc regulatory enzymes appears to influence various pancreatic β-cell functions (25–28). Despite this link, the molecular mechanism through which this modification works remains elusive. Here we use pharmacological approaches to show that O-GlcNAc enables Min6 cells to secrete higher levels of insulin in response to chronic glucose stimulation. Increased O-GlcNAc levels also elevate intracellular insulin levels even when glucose flux is low. We also show that O-GlcNAc increases insulin 1 (Ins1) and insulin 2 (Ins2) mRNA levels in Min6 cells above physiological conditions in a glucose-independent manner. The observed elevation in insulin mRNA is caused at least in part by histone H3 regulation as demonstrated by elevations in epigenetic marks promoting transcriptional activation at the Ins2 promoter. O-GlcNAc also appears to modulate the expression of an abundance of genes in a parallel manner to high glucose. Together these results suggest that the O-GlcNAc modification serves as a glucose sensor in Min6 cells, regulating glucose-mediated gene expression in the pancreatic β cell.
Experimental Procedures
Tissue Culture
Min6 cells were a generous gift from Gerald Hart (Johns Hopkins University). Cells were cultured in Dulbecco's modified Eagle's medium containing 25 mm glucose, 15% (v/v) fetal bovine serum, 2 mm glutamine, and 100 μm β-mercaptoethanol. Upon reaching 70% confluency, cells were harvested using 0.25% trypsin, PBS solution (1:1) and split at a 1:3 ratio. All experiments were performed using cells between passages 20 and 30.
Pharmacological Incubation Conditions
Min6 cells were introduced to either low (2 mm) or high (25 mm) glucose media and pretreated with 25 μm O-(2-acetamido-2-deoxy-d-glucopyranosylidene)-amido N-phenylcarbamate (PUGNAc (PUG); Toronto Research Chemicals, Inc.) and 500 pm GlcNAcstatin (Dr. Daan Van Aalten, University of Dundee). Cells were incubated in these conditions for 6 h before a 1× PBS wash and 1-h starvation (0.5 mm glucose) period with the above-mentioned inhibitors. Afterward, cells were again washed with 1× PBS and returned to pharmacological conditions as outlined above for a 1-h acute treatment before harvesting media and/or cells for experimental procedures. Experiments requiring chronic 3-h pharmacological exposure were washed with 1× PBS and fed fresh conditioned media at 1-h increments until conclusion.
Western Blotting
Min6 cells from pharmacological conditions were separated into cytoplasmic and nuclear fractions as outlined previously (29). Proteins were resolved on SDS-PAGE gel by electrophoresis and transferred to PVDF membrane (Bio-Rad). Membranes were blocked for 1 h at room temperature in 1× TBST, 0.1% Tween 20 supplemented with BSA. After blocking, membranes were incubated overnight at 4 °C using the O-GlcNAc-specific monoclonal 10 (Mab10), anti-histone H3 tri-methyl K4 (Abcam, ab8580), acetyl-histone H3 Lys-9/Lys-14 (Cell Signaling, 9677L), or anti-histone H3 (Abcam, ab1791) antibody. Membranes were washed 3 times with 1× TBST, 0.1% Tween 20 and incubated at room temperature for 1 h with a secondary horseradish peroxidase-conjugated antibody. Afterward, membranes were washed 4 times with 1× TBST, 0.1% Tween 20 and subjected to ECL chemiluminescence (Thermo Scientific) via autoradiography for detection.
Quantification of Insulin Secretion
Media samples harvested from pharmacologically manipulated Min6 cells were diluted 1:100 in 1× PBS the same day. Insulin levels in diluted samples were determined using the rat insulin RIA kit (Millipore) following directions outlined in the manufacturer's protocol. Radioisotope quantification was performed using a Packard Cobra Quantum E5002 Gamma Counter (PerkinElmer Life Sciences).
Quantification of Intracellular Insulin Levels
Harvested Min6 cell samples were resuspended in lysis buffer (50 mm Tris-HCl, pH 7.5, 26.5 mm NaCl, 1% Nonidet P-40, 1% SDS, 50 mm EDTA, 0.1% protease inhibitor mixture (Calbiochem) and 0.1% phosphatase inhibitor mixture (Calbiochem)) and syringe-ruptured with a 23-gauge needle. Samples were centrifuged at 14,000 rpm for 30 min at 4 °C. Whole cell extracts were then diluted 1:1200 in 1× PBS and used for insulin determination with the rat insulin RIA kit as described above. Radioisotope quantification was performed using a Packard Cobra Quantum E5002 Gamma Counter. Results were normalized by protein concentrations between conditions.
Immunofluorescence and Confocal Microscopy
Pharmacologically treated Min6 were fixed using 3.7% paraformaldehyde at room temperature for 20 min. After fixation, cells were washed 3 times with 1× PBS and incubated in anti-insulin (Dako, A0564) primary antibody supplemented with 1 mg/ml BSA and 0.2% Triton X-100 for 1 h. Primary antibody was removed, and cells were washed 3 times in 1× PBS and incubated in Alexa-fluor 488 fluorophore-conjugated secondary antibody and TO-PRO 3 nuclear stain (Invitrogen) supplemented with 1 mg/ml BSA and 0.2% Triton X-100 for 45 min. Cells were washed 3 times with 1× PBS, mounted onto glass coverslips using Prolong Gold Antifade Reagent (Invitrogen), and allowed to dry overnight at 4 °C. Samples were imaged with a 60× oil immersion objective (N.A. 1.4) on an Olympus FV1000 laser scanning confocal microscope. Multiple images were acquired through a z-series, with a z-step of 0.45 μm. The resulting images were analyzed and compressed with Image J software. In most cases the presented images are a maximum intensity projection that includes the entire z-stack.
RNA Isolation and Quantitative RT-PCR
Total RNA was isolated from Min6 cells using the RNeasy Plus Minikit (Qiagen, 170-8840) as described in manufacturer's protocol. Template mRNA was diluted 1:1000 in 1× PBS for Ins1 and Ins2 analysis and undiluted for GalnT7 and Fbxo43. First-strand cDNA synthesis was performed using the iScript Reverse Transcription Supermix kit (Bio-Rad) according to the manufacturer's instructions. The resulting cDNA was used as a template for PCR amplification of the mouse Ins1 (Qiagen, QT01660855), mouse Ins2 (Qiagen, QT00114289), mouse Galnt7 (Qiagen, QT00125034), mouse Fbxo43 (Qiagen, QT01037862), and mouse Tbp1 (Qiagen, QT00198443) genes. Insulin gene mRNA levels were quantified using MyIQ Single Color Real-time PCR detection instrument (Bio-Rad) and normalized to Tbp1 expression levels.
Chromatin Immunoprecipitation (ChIP)
ChIP was performed as previously described (30). Briefly, DNA and protein were cross-linked using 2% formaldehyde. Sonicated DNA extract was precleared using protein A/G-agarose beads and the corresponding agarose conjugate linked IgG. Chromatin from 3 × 106 cells were used for each immunoprecipitation. Lysates were incubated with previously mentioned anti-histone H3 tri-methyl K4, acetyl-histone H3 Lys-9/Lys-14, anti-histone H3, or anti-O-GlcNAc Mab10 antibody at 2 μg per reaction overnight at 4 °C with rotation. Protein-DNA complexes were incubated with protein-agarose A/G beads for 2 h and washed 3 times using buffers containing 0.1% SDS, 1& Triton X-100, 2 mm EDTA, 20 mm Tris, 150–500 mm NaCl, and protease inhibitors. DNA was eluted from beads using elution buffer containing 0.1% SDS and 100 mm NaHCO3. Cross-linking was reversed by the addition of NaCl to a final concentration of 325 mm, and DNA was incubated overnight at 65 °C. DNA was extracted using phenol-chloroform after RNase and proteinase K treatment and analyzed by quantitative real-time PCR (RT-PCR) against the mouse insulin 2 promoter region (Ins2 forward, 5′-TGACCTACCCCACCTGGAGC-3′; Ins2 reverse, 5′-CTGGTGGTTACTGGGTCCCC-3′).
RNA Sequencing Analysis and Bioinformatics
RNA extraction was performed using the RNeasy Plus Minikit (Qiagen, 170-8840) from LG, HG and LG + GlcNAcstatin (GNS) samples after the previously described 1-h incubation. Samples were sent to HudsonAlpha Genomic Services Laboratory (Huntsville, AL) for RNA-Seq library prep and sequencing. Briefly, the concentration and integrity of the extracted total RNA was estimated by Qubit® 2.0 Fluorometer (Invitrogen) and Agilent 2100 Bioanalyzer (Applied Biosystems, Carlsbad, CA), respectively. RNA-Seq library prep was performed with 500 ng of total RNA from each sample followed by enrichment for polyadenylated RNA sequences using the poly(A) selection technique. Each sample was individually barcoded with unique in-house Genomic Services Laboratory primers and amplified through eight cycles of PCR using KAPA HiFi HotStart Ready Mix (Kapa Biosystems, Inc., Woburn, MA). The quality of the libraries was assessed by a Qubit® 2.0 fluorometer, and the concentration of the libraries was estimated by utilizing a DNA 1000 chip on an Agilent 2100 Bioanalyzer.
Accurate quantification for sequencing applications was determined using the qPCR-based KAPA Biosystems Library Quantification kit (Kapa Biosystems, Inc., Woburn, MA). Each library was then diluted to a final concentration of 12.5 nm and pooled equimolar before clustering. Paired End sequencing was performed to generate approximately twenty-five million reads per sample using a 200-cycle TruSeq SBS HS v3 kit on an Illumina HiSeq2000 running HiSeq Control Software (HCS) v1.5.15.1 (Illumina, Inc., San Diego, CA). Image Raw reads were demultiplexed using bcl2fastq conversion software v1.8.3 (Illumina) with default settings. After RNA-Seq, raw reads were mapped to reference mouse genome mm9 using TopHat v2.0 (Trapnell 2009). Aligned reads were imported onto the Avadis NGS data analysis platform (Strand Life Sciences, San Francisco, CA). Reads were first filtered on their quality metrics, then duplicate reads were removed. Normalized gene expression was quantified using the TMM (trimmed mean of M values) algorithm (31). The transcriptional profile from each sample group (LG, HG, and LG+GNS) was compared by principle component analysis and hierarchal clustering analysis to determine the layout and spread of the expression data. Differential expression of genes was calculated on the basis of -fold change (using default cut-off ≥ ±2.0) observed between defined conditions, and the p value of the differentially expressed gene list was estimated by z-score calculations using a default cutoff of 0.05 as determined by Benjamini Hochberg FDR (32) correction.
Gene Ontology (GO) analysis was performed on the list of differentially expressed mRNAs between sample groups. Database for Annotation, Visualization, and Integrated Discovery (DAVID) v6.7 was used for this analysis. Prediction of affected protein classes from up and down-regulated genes sets were made on Panther gene list analysis (33).
Statistical Analysis
Data are expressed as the mean ± S.E. Statistical significance was determined using an ordinary one-way ANOVA comparing the means between each group (p < 0.05). A two-way ANOVA was used to determine significance in the extended insulin secretion experiment (Fig. 1D). All statistically significant values are included in the figures and figure legends. Analysis was performed using GraphPad software.
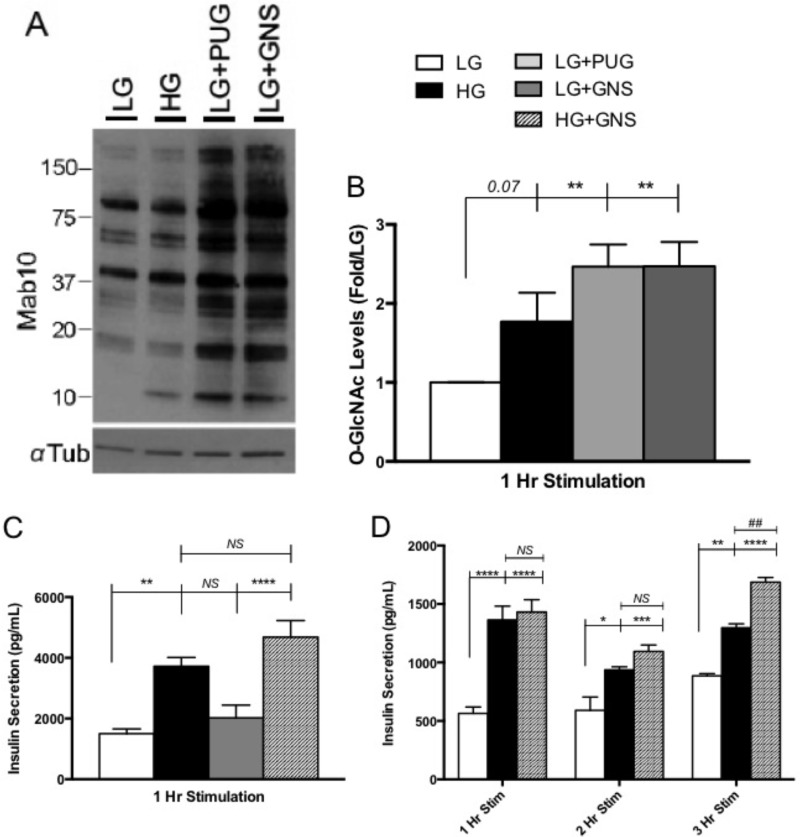
FIGURE 1.
O-GlcNAc maintained glucose-responsive insulin secretion in Min6 cells. A and B, nuclear enrichment from 1-h pharmacologically treated Min6 cells was immunoblotted with the O-GlcNAc monoclonal antibody Mab10. α-Tubulin served as the loading control (n = 6.). C, insulin secretion levels after 1 h of pharmacological stimulation from Min6 cells, quantified using RIA. Values are presented as insulin secretion (pg/ml) over time (n = 6). D, 1-h and 3-h insulin secretion in response to LG, HG, and HG+GNS treatments in the media from Min6 cells quantified using RIA. Values are presented as insulin secretion (pg/ml) over time (n = 3). All data are shown as the mean ± S.E. Significance was determined using an ordinary one-way ANOVA (B and C) or an ordinary two-way ANOVA (D). *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001 versus LG; ##, p < 0.01 versus HG. NS, not significant.
Results
Elevated O-GlcNAc Levels Maintain Insulin Secretion in Min6 Cells under Chronic Glucose Exposure
Increased glucose flux through the HBP increases O-GlcNAc protein modification in the nucleus of Min6 cells (25). To examine the effects of elevating O-GlcNAc levels in Min6 cells, we elevated HBP flux using high glucose stimulation and through two different pharmacological inhibitors. PUG and GNS are OGA inhibitors used to elevate intracellular O-GlcNAc. These inhibitors work by preventing the enzymatic removal of O-GlcNAc by OGA. Previous studies have shown that although PUG is a global hexosaminidase inhibitor, GNS is much more specific for OGA (34, 35). O-GlcNAc levels in Min6 cells after treatments were not statistically different from one another in whole cell extracts (data not shown). However, LG media supplemented with PUG or GNS increased nuclear O-GlcNAc compared with LG alone (2.7-fold ± 0.3 and 2.7-fold ± 0.2, respectively), as did cells exposed to HG (1.7-fold ± 0.4, not significant) to a lesser extent (Fig. 1, A and B). Due to the lack of specificity of PUG and the fact that GNS elevated nuclear O-GlcNAc levels in a similar manner, LG+PUG treatment was not investigated in downstream experiments.
Insulin secretion within the mammalian system follows a pulsatory pattern. The first phase consists of the immediate release of preloaded insulin from membrane-docked granules followed by an environmentally stimulated second phase dependent on gene transcription and new protein synthesis (36, 37). To address whether increasing O-GlcNAc influenced β cell secretion, media from pharmacologically treated Min6 cells were harvested and tested for secreted mature insulin levels. Using the standard insulin radioimmunoassay in the field, acute insulin secretion from Min6 cells was not altered by elevations in O-GlcNAc alone but was altered by HG treatment (Fig. 1C), suggesting that the O-GlcNAc modification is likely not involved in the depolarization process of Min6 cells. The combination of HG+GNS trended toward increased release compared with HG at 1 h (GNS = 1.3-fold ± 0.2) but failed to reach statistical significance (Fig. 1C).
It has been reported that hyperglycemia-induced insulin secretion from pancreatic β cells diminishes over time (38, 39), so we examined whether O-GlcNAc affects more prolonged secretory events. Min6 cells exposed to HG+GNS for extended periods of time appear to maintain glucose-responsive insulin secretion compared with HG alone, as observed 3 h post- stimulation (1.3-fold ± 0.1, Fig. 1D). Together these data suggest that elevating O-GlcNAc levels enables Min6 cells to maintain a higher glucose-stimulated insulin secretion response over time.
Intracellular Insulin Content Is Higher in Min6 Cells upon Elevating O-GlcNAc
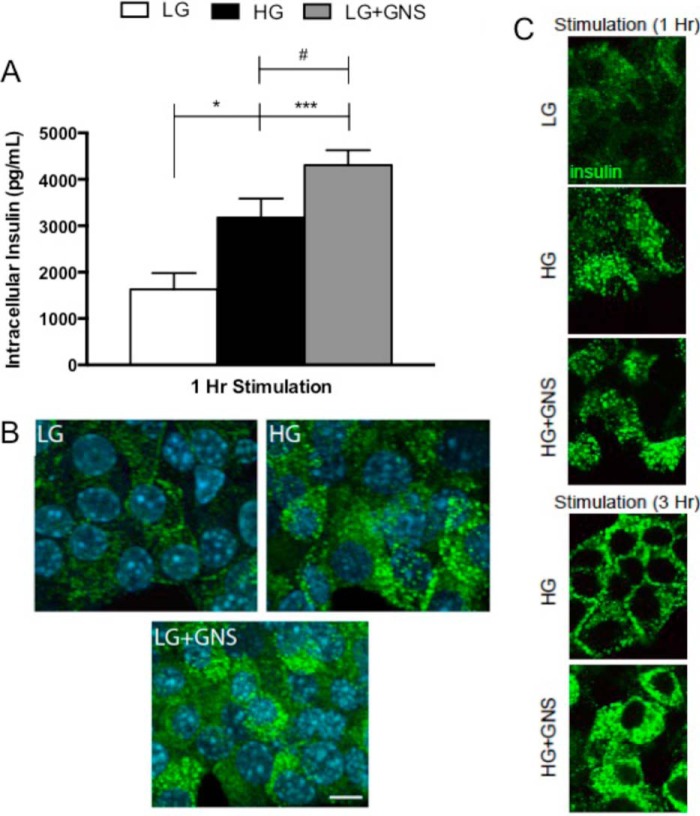
To explain this prolonged secretion of insulin over time and investigate whether increased O-GlcNAc affects insulin protein levels, Min6 cells were pharmacologically stimulated for 1 h, harvested, and lysed for intracellular mature insulin quantification via radioimmunoassay. As expected, the amount of intracellular insulin was higher in cells treated with HG compared with LG (1.9-fold ± 0.6, Fig. 2A). OGA inhibition using GNS raised intracellular insulin levels significantly above LG alone (2.6-fold ± 0.5) and was 1.4-fold ± 0.4 higher in comparison to HG treatment alone (Fig. 2A). These results were confirmed by immunohistochemical staining for intracellular insulin. After treatments, cells were fixed, stained, and subsequently analyzed by confocal microscopy (Fig. 2B). In contrast to cells treated with LG, cells incubated in either HG or in LG+GNS showed increases in the number of insulin-positive puncta. These results suggest that increasing O-GlcNAc in Min6 cells elevates intracellular insulin content.
FIGURE 2.
O-GlcNAc increased Min6 intracellular insulin levels. A, cells treated in pharmacological conditions for 1 h were harvested and lysed for intracellular insulin measurements using RIA. Values are presented as intracellular insulin levels (pg/ml) over time, and data show the mean ± S.E. Significance was determined using an ordinary one-way ANOVA (n = 6; *, p < 0.05; ***, p < 0.001 versus LG; #, p < 0.05 versus HG). B, Min6 were incubated in LG, HG, and LG+GNS for 1 h and fixed for immunohistochemical and confocal microscopy analysis. Preproinsulin-, proinsulin, and mature insulin are stained green, and the nucleus is stained blue (n = 3). C, Min6 cells treated in LG, HG, and HG+GNS were fixed for immunohistochemical and confocal microscopy analysis after 1 h incubation (top panel). Min6 cells treated for 3 h in HG or HG+GNS were prepared and analyzed as described (bottom panel). Preproinsulin, proinsulin, and mature insulin are again stained green (n = 3).
To further determine whether elevating O-GlcNAc levels impacts glucose responsiveness, Min6 cells were treated for 1 or 3 h in LG, HG, or HG+GNS. After treatment cells were fixed, stained, and subsequently analyzed by confocal microscopy. After 1 h, no differences in the content of intracellular insulin were detected between HG and HG+GNS (Fig. 2C). However, after 3 h of chronic treatment with HG+GNS, Min6 cells maintained increased intracellular insulin levels compared with HG alone (Fig. 2C). This pattern of staining likely represents both the ready releasable and reserve insulin granule pools, although specific localization was not confirmed. These findings suggest that elevations in O-GlcNAc levels help maintain glucose-stimulated insulin protein levels in Min6 cells under chronic hyperglycemia.
Elevating O-GlcNAc Increases Mouse Ins1 and Ins2 Steady State mRNA
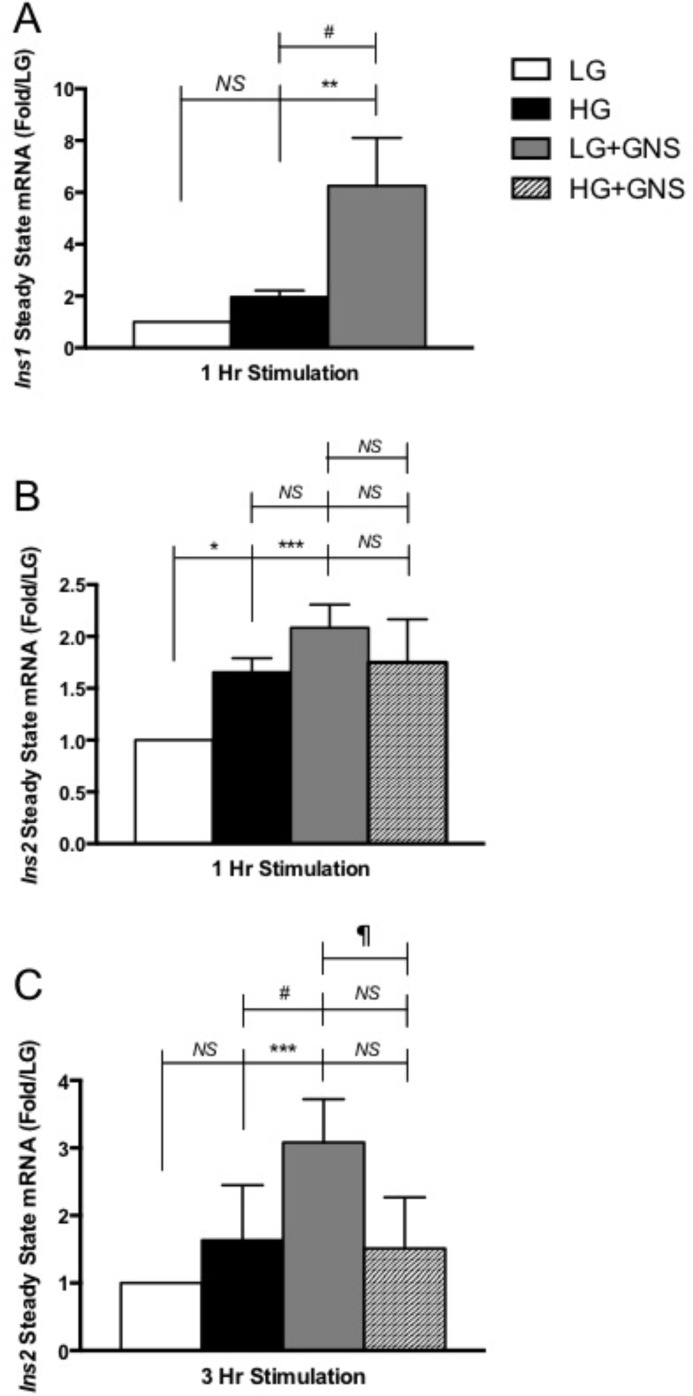
Because O-GlcNAc elevations correlated with increased insulin hormone levels in Min6 cells, we next asked whether this was due to insulin gene regulation. RNA was extracted from Min6 cells after 1 h of treatment and used for steady state mouse Ins1 and Ins2 quantitative mRNA measurements. Incubating Min6 cells in HG elevates both Ins1 (2.0-fold ± 0.2, not significant) and Ins2 (1.5-fold ± 0.1) mRNA levels compared with LG-treated cells (Fig. 3, A and B, respectively). Interestingly, elevating O-GlcNAc levels using GNS under LG conditions significantly raised mouse Ins1 (6.3-fold ± 1.8, Fig. 3A) and Ins2 (2.2-fold ± 0.2, Fig. 3B) mRNA levels compared with LG alone. Although LG+GNS significantly elevated Ins1 mRNA compared with HG (3.2-fold ± 0.5, Fig. 3A), a similar increase was not observed for Ins2 expression (Fig. 3B). This varying gene responsiveness may be due to the fact that Ins1 originated from a reverse-transcribed partially processed mRNA of Ins2 (40, 41). Although both genes contain the regulatory region required for preproinsulin mRNA synthesis (40), the majority of insulin in the rodent system is produced through Ins2 gene activation, making it more homologous to the human insulin gene (42). This agrees with our insulin synthesis and secretion data where we saw a modest O-GlcNAc-correlated increase in both experiments that is more representative of the Ins2 mRNA results versus Ins1.
FIGURE 3.
Elevations in O-GlcNAc increased Ins1 and Ins2 steady state mRNA levels in Min6 cells. A, extracted RNA from Min6 cells following 1 h treatment was used for mouse Ins1 mRNA quantification. Values are presented as -fold change to LG. B, RNA extraction was performed from Min6 cells as described above but used for mouse Ins2 mRNA quantification. Values are presented as -fold change to LG. C, extracted RNA from Min6 cells after 3 h of treatment was used for mouse Ins2 mRNA quantification. Values are presented as -fold change to LG (n ≥ 3). All data are shown as the mean ± S.E. Significance was determined using an ordinary one-way ANOVA. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus LG; #, p < 0.05 versus HG; ‖, p < 0.05 LG+GNS versus HG+GNS. NS, not significant.
Chronic elevations in O-GlcNAc levels show similar trends observed during 1 h of incubation. Min6 cells incubated in LG+GNS for 3 h contain significantly higher Ins2 mRNA levels than LG and HG treatments alone (3.1-fold ± 0.4 and 1.9-fold ± 0.1 respectively, Fig. 3C). However, supplementing GNS with HG stimulation does not appear to increase Ins2 mRNA compared with HG treatment alone (Fig. 3C). These results suggest that the elevation of intracellular insulin levels during LG+GNS treatment is at least partially caused by an O-GlcNAc-mediated insulin gene regulatory mechanism. They also imply that the increased insulin secretion and intracellular accumulation observed after 3 h of HG+GNS stimulation cannot be explained by increased steady state Ins2 transcription.
Increasing O-GlcNAc Directly Correlates to Elevated Histone H3 Epigenetic Marks Promoting Transcriptional Activation
Recent studies demonstrate that O-GlcNAc can directly modify histone proteins and several other components of the nucleosome complex (43). To investigate whether the increases in steady state Ins2 mRNA were due to epigenetic events, we performed ChIP experiments in combination with quantitative PCR toward the mouse Ins2 promoter sequence. Histone H3 acetylation and trimethylation are epigenetic modifications serving as global marks to indicate active areas of gene transcription (44, 45). We used antibodies designed against histone H3 trimethylated lysine 4 (H3K4me3) and histone H3 bi-acetylated lysine 9 and 14 (H3K9,14Ac) to probe for O-GlcNAc-associated chromatin changes at the mouse Ins2 promoter.
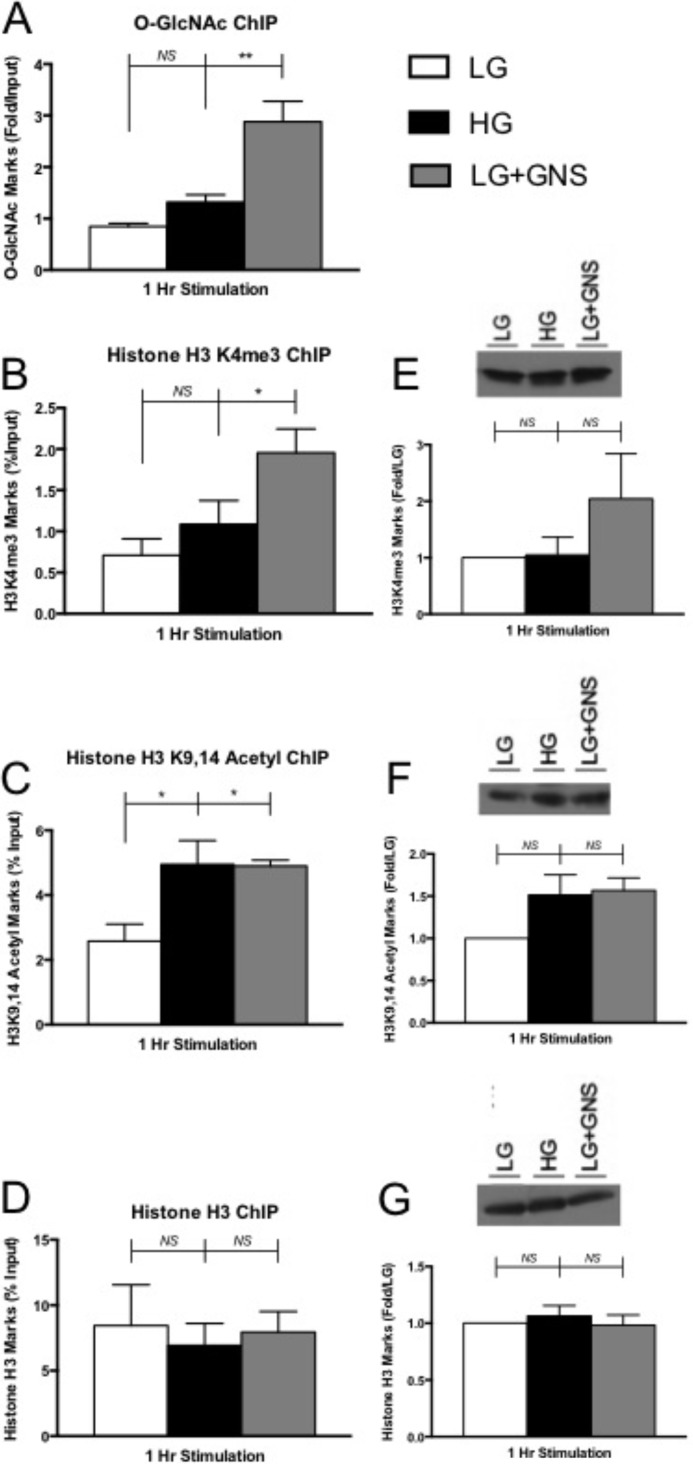
GNS treatment significantly increased the amount of O-GlcNAc levels at the Ins2 promoter region (3.4-fold ± 0.4), whereas HG only showed a modest increase (1.6-fold ± 0.2, Fig. 4A). When investigating H3K4me3 at the Ins2 promoter, HG samples always seemed to contain more marks than LG (1.5-fold ± 0.3) but never reached statistical significance (Fig. 4B). This was previously observed when investigating genome-wide histone modifications in human pancreatic islets and may be true for the more homologous mouse Ins2 gene (46). Conversely, GNS-treated cells displayed abundantly more H3K4me3 marks (2.8-fold ± 0.3) at the Ins2 promoter versus LG (Fig. 4B).
FIGURE 4.
Histone H3 activation marked and O-GlcNAc levels increases at the Ins2 promoter. After 1 h of treatment, Min6 cells were fixed and prepared to analyze binding occupancy of several marks within a 300-base pair region of the Ins2 promoter. ChIP was performed using Mab10 (A), and the values were determined using sample qPCR-fold change over IgG control (n ≥ 4). ChIP was also performed for H3K4me3 (B), H3K9,14Ac (C), and histone H3 (D) levels. These values were determined using qPCR relative to the % input (n ≥ 3). Corresponding H3K4me3 (E), H3K9,14Ac (F), and histone H3 (G) immunoblot analysis on global histone extracts is displayed next to ChIP data. H3K4me3 and H3K9,14Ac were normalized to total histone H3 levels, and histone H3 was normalized to α tubulin (n ≥ 3). All data are shown as the mean ± S.E. Significance was determined using an ordinary one-way ANOVA. *, p < 0.05; **, p < 0.01 versus LG. NS, not significant.
Min6 cells incubated in HG and GNS conditions also displayed more H3K9,14Ac marks (1.9-fold ± 0.7 and 1.9-fold ± 0.2, respectively) at the mouse Ins2 promoter compared with LG-treated cells (Fig. 4C). Histone H3 data revealed there was no significant change in the amount of H3 at the Ins2 promoter between our conditions, ensuring the O-GlcNAc-directed increases in H3 activation marks was not due to nucleosome degradation (Fig. 4D).
We also wanted to investigate the degree to which increasing O-GlcNAc levels contributed to global histone modification. Western blot analysis investigating global H3K4me3, H3K9,14Ac, and histone H3 levels showed no significant differences between pharmacological conditions in acid-extracted histone samples (Fig. 4, E–G, respectively). Together, these results showed that O-GlcNAc accumulation at the Ins2 gene promoter directly correlates with elevations in H3K9,14Ac and H3K4me3 chromatin-associated transcriptional activating marks but did not appear to affect these H3 marks on a whole genome scale.
O-GlcNAc Regulates a Subset of Genes in Min6 Cells
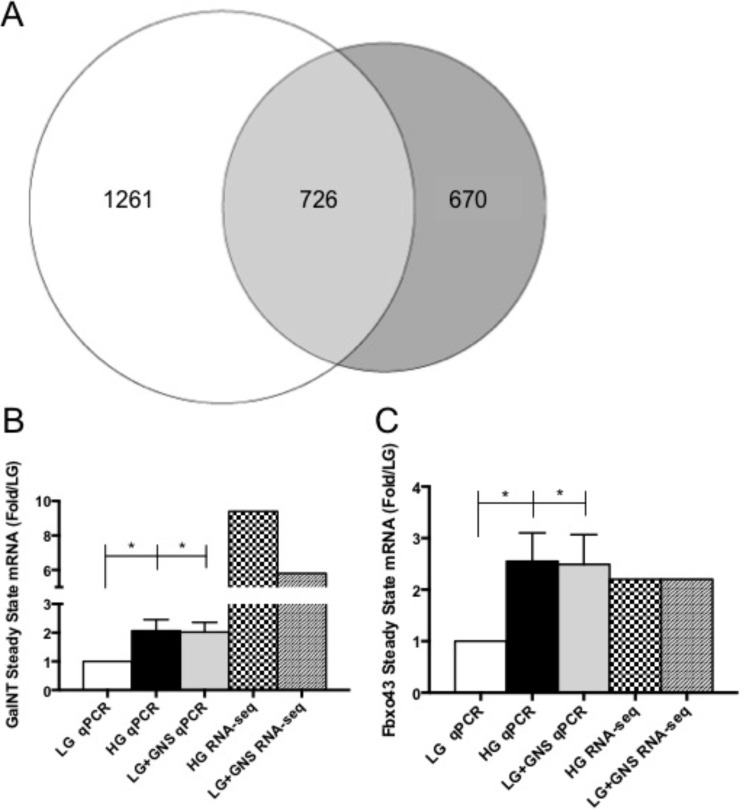
Considering global histone activating marks were unaffected between our conditions, we next investigated the degree to which O-GlcNAc levels regulated gene expression in Min6 cells. Illumina Hiseq 2000 RNA sequencing confirmed numerous genes exhibited altered expression patterns in response to hyperglycemia and/or OGA inhibition (Fig. 5, supplemental Datasets 1–3). Comprehensive bioinformatics showed that 2657 genes (16%) were affected (altered by 2-fold or more) in at least one of our conditions of a total of 16,638 expressed. Based on these data, 84% of the genes in Min6 were unchanged by our pharmacological conditions, which is likely the reason we were unable to see changes in total H3 activating marks (Fig. 4, E–G). HG treatment altered the expression of 1987 (11.9%) of the total genes expressed and LG+GNS influenced 1369 (8.4%). If the two conditions (hyperglycemia and O-GlcNAcase inhibition) were completely independent, one would expect an ∼1% overlap in gene expression changes based on these two datasets. However, bioinformatics analysis revealed that 726 genes were similarly affected (changed in the same direction by at least 2-fold) in both HG- and LG+GNS-treated samples, representing 27.3% of the total altered subset of genes (Fig. 5A, supplemental Datasets 2 and 3). Over half of all gene expression profiles altered by OGA inhibition overlapped with hyperglycemia-induced changes, and more than one-third of all hyperglycemia-induced changes overlapped with OGA inhibition (Fig. 5A), providing evidence in support of the O-GlcNAc modification being a glucose sensor in Min6 cells.
FIGURE 5.
HG stimulation and O-GlcNAc elevation regulated overlapping gene expression profiles in Min6 cells. A, in all, the expressions of 2657 genes were altered in either HG or LG+GNS treatment. HG alone affected 1261 genes in Min6 cells, whereas LG+GNS influenced 670. Combined, HG and LG+GNS changed the expression of 726 common genes that represent ∼27.3% of total genes affected (n = 1). RNA-sequencing expression changes were validated and compared using qPCR for Galnt7 (B; n ≥ 5) and Fbxo43 (C; n ≥ 5). All data are shown as the mean ± S.E. Significance was determined using an ordinary one-way ANOVA. *, p < 0.05 versus LG.
We next wanted to validate several genes of interest identified in the RNA-seq data by qPCR (Fig. 5, B and C). UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNT7) is a glycosyl transferase that was shown to be drastically up-regulated by HG (9.4-fold) and GNS (5.8-fold) in the RNA-seq data (Table 1). qPCR analysis confirms these results under HG (2.2-fold ± 0.4)- and GNS (2.1-fold ± 0.4)-treated cells (Fig. 5B) although to a lesser extent of up-regulation. F-box protein 43 (Fbxo43) is part of the forkhead box transcription factor family of which several members have been shown to be functionally O-GlcNAc-modified in other cell types and influence the diabetic phenotype (47, 48). Both the RNA-seq (2.2-fold each) and qPCR (HG = 2.6-fold ± 0.5; GNS = 2.5-fold ± 0.6) data displayed very similar gene expression changes (Fig. 5C), validating the 2-fold expression parameters used to determine gene changes. Together, our sequencing results demonstrate that the impact of OGA inhibition on gene expression correlates significantly with hyperglycemia-stimulated gene expression.
TABLE 1.
Most up regulated and down regulated genes by HG and O-GlcNAcase inhibition (GNS) in Min6 cells presented as -fold change to LG
| Gene | Up-regulated genes |
Down-regulated genes |
Average -fold change | ||||
|---|---|---|---|---|---|---|---|
| HG/LG | GNS/LG | Average -fold change | Gene | LG/HG | LG/GNS | ||
| Bfsp1 | 25.9 | 26.0 | 25.9 | Myo1h | 236.1 | 94.1 | 165.1 |
| Slfn3 | 14.1 | 15.0 | 14.5 | Tcte3 | 14.2 | 14.2 | 14.2 |
| Gpr21 | 17.6 | 7.5 | 12.5 | Cyp4f40 | 23.7 | 3.1 | 13.4 |
| Ngef | 14.6 | 8.6 | 11.6 | Ip6k3 | 11.1 | 11.1 | 11.1 |
| Abcc9 | 14.1 | 7.5 | 10.8 | Krt25 | 9.6 | 12.2 | 10.9 |
| Adamts5 | 10.1 | 11.3 | 10.7 | Csf2rb2 | 10.1 | 10.1 | 10.1 |
| Grin3a | 4.0 | 16.9 | 10.5 | Creg2 | 2.7 | 17.2 | 9.9 |
| Trp73 | 13.1 | 7.5 | 10.3 | Saa3 | 6.1 | 11.9 | 9.0 |
| Srpk3 | 10.1 | 10.3 | 10.2 | Cntnap3 | 13.1 | 3.5 | 8.3 |
| Micall2 | 10.9 | 7.4 | 9.2 | Pla2g4a | 8.1 | 8.1 | 8.1 |
| Wnt16 | 12.1 | 5.6 | 8.9 | Col10a1 | 8.1 | 8.1 | 8.1 |
| Zfp459 | 8.6 | 8.4 | 8.5 | Fscn3 | 8.0 | 8.0 | 8.0 |
| Ccbe1 | 8.5 | 8.4 | 8.5 | Zmynd10 | 7.7 | 8.4 | 8.0 |
| Foxg1 | 5.0 | 11.1 | 8.0 | Odf312 | 13.2 | 2.0 | 7.6 |
| Gm 10335 | 2.8 | 12.8 | 7.8 | Lif | 5.0 | 10.1 | 7.6 |
| Gm 16485 | 9.9 | 5.6 | 7.8 | Slamf9 | 9.7 | 5.2 | 7.5 |
| Ptgds | 6.0 | 9.4 | 7.7 | Trh | 3.7 | 10.8 | 7.3 |
| Cdh15 | 10.1 | 5.3 | 7.7 | Gbgt1 | 4.5 | 9.1 | 6.8 |
| C1rl | 5.9 | 9.4 | 7.7 | Ifit3 | 8.8 | 4.7 | 6.8 |
| Smtnl1 | 8.0 | 7.4 | 7.7 | Gm14085 | 10.8 | 2.7 | 6.8 |
| Galnt7 | 9.4 | 5.8 | 7.6 | Nphs1 | 8.8 | 4.7 | 6.7 |
| Zfp114 | 8.6 | 6.5 | 7.5 | Tekt5 | 4.5 | 9.0 | 6.7 |
| Gp9 | 4.0 | 10.7 | 7.3 | Colec11 | 10.9 | 2.3 | 6.6 |
| Tal2 | 6.0 | 7.9 | 7.0 | Mmrn2 | 8.7 | 3.8 | 6.3 |
| Higd1a | 10.0 | 3.8 | 6.9 | Gdf10 | 8.1 | 4.3 | 6.2 |
Discussion
Using pharmacological strategies manipulating intracellular O-GlcNAc levels, we propose that this post-translational modification serves as a glucose sensor in the pancreatic β cell and influences gene expression in Min6 cells. Nuclear enrichment clearly displays elevated O-GlcNAc levels during OGA enzyme inhibition, whereas hyperglycemia slightly elevated modification levels (Fig. 1, A and B). This difference could be attributed to the relatively small amount of intracellular glucose entering the HBP (3–5%) compared with other metabolic pathways (16). GNS treatment on the other hand is highly specific for OGA, directly influencing the O-GlcNAc status within the cell. Elevating O-GlcNAc in the absence of glucose stimulation did not appear to mediate insulin secretory events after 1 h of stimulation (Fig. 1C). This suggests that although proteins within the exocytosis pathway are known to be O-GlcNAc-modified (49, 50), it does not alone affect depolarization and acute insulin hormone release in Min6 cells. However, we were able to show that when inhibiting OGA and chronically stimulating insulin release, Min6 cells trended toward secreting more insulin compared with HG alone (Fig. 1D).
It was clear by our prolonged secretion experiments that O-GlcNAc levels influenced insulin within Min6 cells but not through direct secretory pathway modulation. We were able to demonstrate that increasing levels of O-GlcNAc in Min6 cells drastically elevates the intracellular insulin content in a glucose-independent manner (Fig. 2, A and B). We hypothesize that the increased insulin hormone localizes predominantly in the perinuclear region of the cells, representative of the reserve vesicle pool. Although we did not confirm this experimentally, recent work shows that O-GlcNAc may play an important role in regulating synapsin I interaction with synaptic vesicles to establish the reserve pool of synaptic vesicles (51) and may similarly affect the reserve insulin pool in β cells. Furthermore, inhibiting OGA under prolonged glucose stimulation enabled Min6 cells to maintain high levels of evenly dispersed intracellular insulin levels compared with HG treatment alone (Fig. 2C). These results indicate that elevating O-GlcNAc levels in Min6 cells increases insulin hormone levels and preserves glucose-induced insulin release under chronic hyperglycemic conditions.
Insulin production is governed by gene transcription in response to environmental stimuli (52), and because O-GlcNAcylated proteins are heavily involved in nuclear events (8), it was logical to think that the intracellular insulin elevation was due to transcriptional control. Our data clearly display increasing O-GlcNAc in Min6 cells directly correlates with elevations in mouse Ins1 and Ins2 steady state mRNA levels (Fig. 3, A and B). We also show that elevating O-GlcNAc levels during LG treatment enable Min6 cells to maintain higher levels of Ins2 transcripts compared with HG treatment alone (Fig. 3C). Several glucose-responsive transcription factors that bind the insulin promoter are O-GlcNAc-modified (25–27). But we found that O-GlcNAc did not impact Ins1 promoter activity in luciferase-reporter assays (data not shown), prompting investigation of alternative molecular mechanisms.
Surprisingly, Ins2 mRNA levels were not increased in response to 3 h of stimulation with HG+GNS compared with HG alone (Fig. 3C), suggesting the observed increase in secretion and intracellular protein accumulation is not caused by an O-GlcNAc-mediated transcriptional mechanism. Insulin mRNA synthesis and translation can be regulated through a variety of mechanisms (3, 53, 54). Previous work has demonstrated that O-GlcNAc is capable of regulating protein and mRNA stability (55, 56). However, this mechanism is unlikely to be at work here, as the half-life of Ins2 mRNA in Min6 cells is greater than 24 h (57). Considering the time frame of our experiment (8 h) and the -fold increases observed, transcript stabilization is unlikely to account for the observed changes. Other O-GlcNAc-mediated processes that may account for our observation include ribosomal protein activity (58), attenuation of the unfolded protein response, and endoplasmic reticulum stress (59), decreased proteasomal degradation (60), or blunted cellular autophagy (61, 62). Although mechanistically reasonable, further studies are needed to determine the process by which HG+GNS increases chronic insulin secretion and protein levels.
Recently, groups have shown that O-GlcNAc plays a role in epigenetics either by directly modifying histone tails (63) or through O-GlcNAc transferase and OGA incorporation into chromatin complexes (64–66). We used several histone H3 transcriptional activation marks to demonstrate that the O-GlcNAc-mediated increase in insulin mRNA is likely through a chromatin mechanism. OGA inhibition dramatically increases the amount of H3K4me3 and H3K9,14 bi-acetyl marks at the Ins2 promoter compared with LG (Fig. 4, B and C), whereas hyperglycemia only trended toward elevating these activation marks (Fig. 4, B and C). An O-GlcNAc-specific antibody indicated that there is more O-GlcNAc modification at the Ins2 promoter when OGA is inhibited but only slightly higher upon elevated glucose exposure (Fig. 4A). These results suggest that a modest increase in O-GlcNAc levels (hyperglycemia) slightly elevates H3 activation marks, whereas a more pronounced increase in the post-translational modification results in a significant increase on H3 activation marks. Importantly, we also demonstrate that the observed elevation in H3 transcriptional activating marks is not simply due to O-GlcNAc-directed nucleosome degradation, as all three conditions contain similar histone H3 levels at the Ins2 promoter (Fig. 4D). Of interest, immunoblot experiments demonstrate that O-GlcNAc levels do not influence H3K4me3 and H3K9,14 bi-acetyl marks at a global level (Fig. 4, E–G), leading us to investigate the gene specificity of hyperglycemia and O-GlcNAcase inhibition.
Deep sequencing revealed numerous genes that were modulated by O-GlcNAc, many of which greatly overlap with HG stimulation (Fig. 5A, supplemental Datasets S1-S3). The 25 most influenced genes (Table 1) belong to several protein-coding gene families, including: immune cell function (Slfn3 (67) and Ifit3 (68)), cardiac function (Itgb1bp2 (69) and Grem2 (70)), development (Tal2; Ref. 71) and markers of cellular stress (Saa3; Ref. 72). Interestingly, all of these families are regulated by O-GlcNAc to varying degrees (73–76).
At first glance the fact that there are no changes in either Ins1 or Ins2 between our conditions using RNA sequencing was concerning. However, our qPCR results depict mRNA changes after diluting the total RNA input 1000-fold as these are two of the more highly expressed transcripts at the basal LG condition (Dataset 1). The samples used to generate the RNA-sequencing results were undiluted to prevent masking potential changes in lower abundance genes. Unfortunately, this also means we may have missed changes in more highly expressed genes between our conditions, i.e. insulin, which will need to be addressed in further work.
Considering changes in cellular glycosylation (77) and O-GlcNAc modification of F-box transcription factor members (47, 78) proposed to contribute to the diabetes phenotype, the observed Galnt7 and Fbxo43 changes are extremely interesting (Fig. 5, B and C). HBP flux and increased O-GlcNAcylation may impact glycosyltransferase gene activity in β cells, such as Galnt7, to influence glycosylation, stability, and activity of cell surface receptors and transporters. Additionally, our data provide further evidence to suggest that elevating glucose and the O-GlcNAc modification levels similarly affect expression of F-box transcription factor-mediated genes. In all, the overlapping genes between HG stimulation and OGA inhibition suggest O-GlcNAc greatly contributes to the hyperglycemic-induced gene expression changes in pancreatic β cells and provides a substantial dataset for further targeted investigation.
Our results support the hypothesis that glucose-stimulated insulin gene activation in the Min6 cell line utilizes the O-GlcNAc protein modification in an epigenetic mechanism to mediate transcription. Future experiments will be aimed at determining the precise mechanism through which O-GlcNAc epigenetically regulates insulin gene activity and to investigate if the results in our cell culture model are recapitulated in vivo. We also plan to address how chronic exposure to HG+GNS increases insulin secretion and intracellular accumulation. Of significant future interest and study is the large overlap in regulated genes by both hyperglycemia and O-GlcNAcase inhibition. Our current working model to be further tested is that the O-GlcNAc modification is a major effector of hyperglycemia-induced transcriptome regulation in pancreatic β cells.
Author Contributions
S. P. D. participated in the conception and design of experiments and carried out all data collection with the exception of the confocal microscopy and wrote the initial draft. H. F.-S. aided in conception and performed the confocal microscopy-based experiments and assisted in editing of the manuscript. N. P. performed much of the bioinformatics analysis of RNA-Seq data. L. W. participated in the conception and overall design of the experiments and generated the final document for submission. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Gerald Hart for supplying the Min6 cells and Daan Van Aalten for the GlcNAcStatin used in our studies. We also thank Shawn Levy of HudsonAlpha for assistance with RNA sequencing and bioinformatics analysis.
This work was supported, in whole or in part, by National Institutes of Health Grants P41GM103490 and U01CA128454 (both to L. W.). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Datasets 1–3.
RNA-Seq datasets have been deposited at the NCBI GEO website.
- O-GlcNAc
- O-linked β-N-acetylglucosamine
- Min6
- mouse insulinoma 6
- HBP
- hexosamine biosynthetic pathway
- OGA
- O-GlcNAcase
- PUG
- O-(2-acetamido-2-deoxy-d-glucopyranosylidine)amino-N-phenylcarbamate
- GNS
- GlcNAcstatin
- LG
- low glucose
- HG
- high glucose
- qPCR
- quantitative PCR
- ANOVA
- analysis of variance
- H3K4me3
- histone H3 trimethylated lysine 4
- H3K9,14Ac
- histone H3 bi-acetylated lysine 9 and 14
- GalNT7
- UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7
- Fbxo43
- F-box protein 43.
References
- 1. Kochanek K. D., Murphy S. L., and Xu J. (2015) Deaths: final data for 2011. Centers for Disease Control and Prevention, National Center for Health Statistics, National Vital Statistics System. Natl. Vital Stat. Rep. 63, 1–120 [PubMed] [Google Scholar]
- 2. Marshall S., and Olefsky J. M. (1980) Effects of insulin incubation on insulin binding, glucose transport, and insulin degradation by isolated rat adipocytes. Evidence for hormone-induced desensitization at the receptor and postreceptor level. J. Clin. Invest. 66, 763–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Poitout V., and Robertson R. P. (1996) An integrated view of beta cell dysfunction in type II diabetes. Annu. Rev. Med. 47, 69–83 [DOI] [PubMed] [Google Scholar]
- 4. Nielsen D. A., Welsh M., Casadaban M. J., and Steiner D. F. (1985) Control of insulin gene expression in pancreatic beta cells and in an insulin-producing cell line, RIN-5F cells. I. Effects of glucose and cyclic AMP on the transcription of insulin mRNA. J. Biol. Chem. 260, 13585–13589 [PubMed] [Google Scholar]
- 5. Hammonds P., Schofield P. N., Ashcroft S. J., Sutton R., and Gray D. W. (1987) Regulation and specificity of glucose-stimulated insulin gene expression in human islets of Langerhans. FEBS lett. 223, 131–137 [DOI] [PubMed] [Google Scholar]
- 6. Jahr H., Schröder D., Ziegler B., Ziegler M., and Zühlke H. (1980) Transcriptional and translational control of glucose-stimulated (pro)insulin biosynthesis. Eur. J. Biochem. 110, 499–505 [DOI] [PubMed] [Google Scholar]
- 7. Nagamatsu S., and Steiner D. F. (1992) Altered glucose regulation of insulin biosynthesis in insulinoma cells: mouse beta TC3 cells secrete insulin-related peptides predominantly via a constitutive pathway. Endocrinology 130, 748–754 [DOI] [PubMed] [Google Scholar]
- 8. Comer F. I., and Hart G. W. (1999) O-GlcNAc and the control of gene expression. Biochim. Biophys. Acta 1473, 161–171 [DOI] [PubMed] [Google Scholar]
- 9. Wells L., Vosseller K., and Hart G. W. (2001) Glycosylation of nucleocytoplasmic proteins: signal transduction and O-GlcNAc. Science 291, 2376–2378 [DOI] [PubMed] [Google Scholar]
- 10. Zachara N. E., and Hart G. W. (2002) The emerging significance of O-GlcNAc in cellular regulation. Chem. Rev. 102, 431–438 [DOI] [PubMed] [Google Scholar]
- 11. Zachara N. E., and Hart G. W. (2006) Cell signaling, the essential role of O-GlcNAc! Biochim. Biophys. Acta 1761, 599–617 [DOI] [PubMed] [Google Scholar]
- 12. Ozcan S., Andrali S. S., and Cantrell J. E. (2010) Modulation of transcription factor function by O-GlcNAc modification. Biochim. Biophys. Acta 1799, 353–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zeidan Q., and Hart G. W. (2010) The intersections between O-GlcNAcylation and phosphorylation: implications for multiple signaling pathways. J. Cell Sci. 123, 13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Haltiwanger R. S., Kelly W. G., Roquemore E. P., Blomberg M. A., Dong L. Y., Kreppel L., Chou T. Y., and Hart G. W. (1992) Glycosylation of nuclear and cytoplasmic proteins is ubiquitous and dynamic. Biochem. Soc. Trans. 20, 264–269 [DOI] [PubMed] [Google Scholar]
- 15. Zachara N. E., and Hart G. W. (2004) O-GlcNAc a sensor of cellular state: the role of nucleocytoplasmic glycosylation in modulating cellular function in response to nutrition and stress. Biochim. Biophys. Acta 1673, 13–28 [DOI] [PubMed] [Google Scholar]
- 16. Hanover J. A., Lai Z., Lee G., Lubas W. A., and Sato S. M. (1999) Elevated O-linked N-acetylglucosamine metabolism in pancreatic beta cells. Arch. Biochem. Biophys. 362, 38–45 [DOI] [PubMed] [Google Scholar]
- 17. Hart G. W., Haltiwanger R. S., Holt G. D., and Kelly W. G. (1989) Nucleoplasmic and cytoplasmic glycoproteins. Ciba Found. Symp. 145, 102–112, discussion 112–108 [DOI] [PubMed] [Google Scholar]
- 18. Kreppel L. K., Blomberg M. A., and Hart G. W. (1997) Dynamic glycosylation of nuclear and cytosolic proteins: cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J. Biol. Chem. 272, 9308–9315 [DOI] [PubMed] [Google Scholar]
- 19. Dong D. L., and Hart G. W. (1994) Purification and characterization of an O-GlcNAc selective N-acetyl-β-d-glucosaminidase from rat spleen cytosol. J. Biol. Chem. 269, 19321–19330 [PubMed] [Google Scholar]
- 20. Hart G. W., Housley M. P., and Slawson C. (2007) Cycling of O-linked β-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 446, 1017–1022 [DOI] [PubMed] [Google Scholar]
- 21. Konrad R. J., and Kudlow J. E. (2002) The role of O-linked protein glycosylation in beta cell dysfunction. Int J Mol Med 10, 535–539 [PubMed] [Google Scholar]
- 22. Akimoto Y., Hart G. W., Hirano H., and Kawakami H. (2005) O-GlcNAc modification of nucleocytoplasmic proteins and diabetes. Med. Mol. Morphol. 38, 84–91 [DOI] [PubMed] [Google Scholar]
- 23. Dias W. B., and Hart G. W. (2007) O-GlcNAc modification in diabetes and Alzheimer's disease. Mol. Biosyst. 3, 766–772 [DOI] [PubMed] [Google Scholar]
- 24. Wells L., Vosseller K., and Hart G. W. (2003) A role for N-acetylglucosamine as a nutrient sensor and mediator of insulin resistance. Cell. Mol. Life Sci. 60, 222–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gao Y., Miyazaki J., and Hart G. W. (2003) The transcription factor PDX-1 is post-translationally modified by O-linked N-acetylglucosamine, and this modification is correlated with its DNA binding activity and insulin secretion in min6 beta cells. Arch. Biochem. Biophys. 415, 155–163 [DOI] [PubMed] [Google Scholar]
- 26. Andrali S. S., Qian Q., and Ozcan S. (2007) Glucose mediates the translocation of NeuroD1 by O-linked glycosylation. J. Biol. Chem. 282, 15589–15596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vanderford N. L., Andrali S. S., and Ozcan S. (2007) Glucose induces MafA expression in pancreatic beta cell lines via the hexosamine biosynthetic pathway. J. Biol. Chem. 282, 1577–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Soesanto Y., Luo B., Parker G., Jones D., Cooksey R. C., and McClain D. A. (2011) Pleiotropic and age-dependent effects of decreased protein modification by O-linked N-acetylglucosamine on pancreatic beta-cell function and vascularization. J. Biol. Chem. 286, 26118–26126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schreiber E., Matthias P., Müller M. M., and Schaffner W. (1989) Rapid detection of octamer binding proteins with “mini-extracts,” prepared from a small number of cells. Nucleic Acids Res. 17, 6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ekanayake D., and Sabatini R. (2011) Epigenetic regulation of polymerase II transcription initiation in Trypanosoma cruzi: modulation of nucleosome abundance, histone modification, and polymerase occupancy by O-linked thymine DNA glucosylation. Eukaryot. Cell 10, 1465–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Oshlack A., Robinson M. D., and Young M. D. (2010) From RNA-seq reads to differential expression results. Genome Biology 11, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Benjamini Y., Drai D., Elmer G., Kafkafi N., and Golani I. (2001) Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 125, 279–284 [DOI] [PubMed] [Google Scholar]
- 33. Mi H., Muruganujan A., Casagrande J. T., and Thomas P. D. (2013) Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 8, 1551–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dorfmueller H. C., Borodkin V. S., Schimpl M., Shepherd S. M., Shpiro N. A., and van Aalten D. M. (2006) GlcNAcstatin: a picomolar, selective O-GlcNAcase inhibitor that modulates intracellular O-glcNAcylation levels. J. Am. Chem. Soc. 128, 16484–16485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Haltiwanger R. S., Grove K., and Philipsberg G. A. (1998) Modulation of O-linked N-acetylglucosamine levels on nuclear and cytoplasmic proteins in vivo using the peptide O-GlcNAc-β-N-acetylglucosaminidase inhibitor O-(2-acetamido-2-deoxy-d-glucopyranosylidene)amino-N-phenylcarbamate. J. Biol. Chem. 273, 3611–3617 [DOI] [PubMed] [Google Scholar]
- 36. Curry D. L., Bennett L. L., and Grodsky G. M. (1968) Dynamics of insulin secretion by the perfused rat pancreas. Endocrinology 83, 572–584 [DOI] [PubMed] [Google Scholar]
- 37. Porte D. Jr., and Pupo A. A. (1969) Insulin responses to glucose: evidence for a two pool system in man. J. Clin. Invest. 48, 2309–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Colella R. M., May J. M., Bonner-Weir S., Leahy J. L., and Weir G. C. (1987) Glucose utilization in islets of hyperglycemic rat models with impaired glucose-induced insulin secretion. Metab. Clin. Exp. 36, 335–337 [DOI] [PubMed] [Google Scholar]
- 39. Beck-Nielsen H., Nielsen O. H., Pedersen O., Bak J., Faber O., and Schmitz O. (1988) Insulin action and insulin secretion in identical twins with MODY: evidence for defects in both insulin action and secretion. Diabetes 37, 730–735 [DOI] [PubMed] [Google Scholar]
- 40. Soares M. B., Schon E., Henderson A., Karathanasis S. K., Cate R., Zeitlin S., Chirgwin J., and Efstratiadis A. (1985) RNA-mediated gene duplication: the rat preproinsulin I gene is a functional retroposon. Mol. Cell. Biol. 5, 2090–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wentworth B. M., Schaefer I. M., Villa-Komaroff L., and Chirgwin J. M. (1986) Characterization of the two nonallelic genes encoding mouse preproinsulin. J. Mol. Evol. 23, 305–312 [DOI] [PubMed] [Google Scholar]
- 42. Babaya N., Nakayama M., Moriyama H., Gianani R., Still T., Miao D., Yu L., Hutton J. C., and Eisenbarth G. S. (2006) A new model of insulin-deficient diabetes: male NOD mice with a single copy of Ins1 and no Ins2. Diabetologia 49, 1222–1228 [DOI] [PubMed] [Google Scholar]
- 43. Hanover J. A., Krause M. W., and Love D. C. (2012) Bittersweet memories: linking metabolism to epigenetics through O-GlcNAcylation. Nat. Rev. Mol. Cell Biol. 13, 312–321 [DOI] [PubMed] [Google Scholar]
- 44. Martens J. H., Verlaan M., Kalkhoven E., and Zantema A. (2003) Cascade of distinct histone modifications during collagenase gene activation. Mol. Cell. Biol. 23, 1808–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Marushige K. (1976) Activation of chromatin by acetylation of histone side chains. Proc. Natl. Acad. Sci. U.S.A. 73, 3937–3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bhandare R., Schug J., Le Lay J., Fox A., Smirnova O., Liu C., Naji A., and Kaestner K. H. (2010) Genome-wide analysis of histone modifications in human pancreatic islets. Genome Res. 20, 428–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Housley M. P., Rodgers J. T., Udeshi N. D., Kelly T. J., Shabanowitz J., Hunt D. F., Puigserver P., and Hart G. W. (2008) O-GlcNAc regulates FoxO activation in response to glucose. J. Biol. Chem. 283, 16283–16292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Housley M. P., Udeshi N. D., Rodgers J. T., Shabanowitz J., Puigserver P., Hunt D. F., and Hart G. W. (2009) A PGC-1α-O-GlcNAc transferase complex regulates FoxO transcription factor activity in response to glucose. J. Biol. Chem. 284, 5148–5157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chalkley R. J., Thalhammer A., Schoepfer R., and Burlingame A. L. (2009) Identification of protein O-GlcNAcylation sites using electron transfer dissociation mass spectrometry on native peptides. Proc. Natl. Acad. Sci. U.S.A. 106, 8894–8899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vosseller K., Trinidad J. C., Chalkley R. J., Specht C. G., Thalhammer A., Lynn A. J., Snedecor J. O., Guan S., Medzihradszky K. F., Maltby D. A., Schoepfer R., and Burlingame A. L. (2006) O-Linked N-acetylglucosamine proteomics of postsynaptic density preparations using lectin weak affinity chromatography and mass spectrometry. Mol. Cell. Proteomics 5, 923–934 [DOI] [PubMed] [Google Scholar]
- 51. Skorobogatko Y., Landicho A., Chalkley R. J., Kossenkov A. V., Gallo G., and Vosseller K. (2014) O-GlcNAc site Thr-87 regulates synapsin I localization to synapses and size of the reserve pool of synaptic vesicles. J. Biol. Chem. 289, 3602–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Permutt M. A., and Kipnis D. M. (1975) Insulin biosynthesis and secretion. Fed. Proc. 34, 1549–1555 [PubMed] [Google Scholar]
- 53. Docherty K., and Clark A. R. (1994) Nutrient regulation of insulin gene expression. FASEB J. 8, 20–27 [DOI] [PubMed] [Google Scholar]
- 54. Fred R. G., and Welsh N. (2009) The importance of RNA binding proteins in preproinsulin mRNA stability. Mol. Cell. Endocrinol. 297, 28–33 [DOI] [PubMed] [Google Scholar]
- 55. Ohn T., Kedersha N., Hickman T., Tisdale S., and Anderson P. (2008) A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat. Cell Biol. 10, 1224–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Olivier-Van Stichelen S., Dehennaut V., Buzy A., Zachayus J. L., Guinez C., Mir A. M., El Yazidi-Belkoura I., Copin M. C., Boureme D., Loyaux D., Ferrara P., and Lefebvre T. (2014) O-GlcNAcylation stabilizes β-catenin through direct competition with phosphorylation at threonine 41. FASEB J. 28, 3325–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ritz-Laser B., Meda P., Constant I., Klages N., Charollais A., Morales A., Magnan C., Ktorza A., and Philippe J. (1999) Glucose-induced preproinsulin gene expression is inhibited by the free fatty acid palmitate. Endocrinology 140, 4005–4014 [DOI] [PubMed] [Google Scholar]
- 58. Zeidan Q., Wang Z., De Maio A., and Hart G. W. (2010) O-GlcNAc cycling enzymes associate with the translational machinery and modify core ribosomal proteins. Mol. Biol. Cell 21, 1922–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ngoh G. A., Hamid T., Prabhu S. D., and Jones S. P. (2009) O-GlcNAc signaling attenuates ER stress-induced cardiomyocyte death. Am. J. Physiol. Heart Circ. Physiol. 297, H1711–H1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang P., Lazarus B. D., Forsythe M. E., Love D. C., Krause M. W., and Hanover J. A. (2012) O-GlcNAc cycling mutants modulate proteotoxicity in Caenorhabditis elegans models of human neurodegenerative diseases. Proc. Natl. Acad. Sci. U.S.A. 109, 17669–17674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kumar A., Singh P. K., Parihar R., Dwivedi V., Lakhotia S. C., and Ganesh S. (2014) Decreased O-linked GlcNAcylation protects from cytotoxicity mediated by huntingtin exon1 protein fragment. J. Biol. Chem. 289, 13543–13553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Marsh S. A., Powell P. C., Dell'italia L. J., and Chatham J. C. (2013) Cardiac O-GlcNAcylation blunts autophagic signaling in the diabetic heart. Life sciences 92, 648–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sakabe K., Wang Z., and Hart G. W. (2010) β-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc. Natl. Acad. Sci. U.S.A. 107, 19915–19920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Deplus R., Delatte B., Schwinn M. K., Defrance M., Méndez J., Murphy N., Dawson M. A., Volkmar M., Putmans P., Calonne E., Shih A. H., Levine R. L., Bernard O., Mercher T., Solary E., Urh M., Daniels D. L., and Fuks F. (2013) TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 32, 645–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hayakawa K., Hirosawa M., Tabei Y., Arai D., Tanaka S., Murakami N., Yagi S., and Shiota K. (2013) Epigenetic switching by the metabolism-sensing factors in the generation of orexin neurons from mouse embryonic stem cells. J. Biol. Chem. 288, 17099–17110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ito R., Katsura S., Shimada H., Tsuchiya H., Hada M., Okumura T., Sugawara A., and Yokoyama A. (2014) TET3-OGT interaction increases the stability and the presence of OGT in chromatin. Genes Cells 19, 52–65 [DOI] [PubMed] [Google Scholar]
- 67. Condamine T., Le Luduec J. B., Chiffoleau E., Bériou G., Louvet C., Heslan M., Tilly G., and Cuturi M. C. (2010) Characterization of Schlafen-3 expression in effector and regulatory T cells. J. Leukoc. Biol. 87, 451–456 [DOI] [PubMed] [Google Scholar]
- 68. Chen H. W., King K., Tu J., Sanchez M., Luster A. D., and Shresta S. (2013) The roles of IRF-3 and IRF-7 in innate antiviral immunity against dengue virus. J. Immunol. 191, 4194–4201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ruppert V., Meyer T., Richter A., Maisch B., Pankuweit S., and German Competence Network of Heart, F. (2013) Identification of a missense mutation in the melusin-encoding ITGB1BP2 gene in a patient with dilated cardiomyopathy. Gene 512, 206–210 [DOI] [PubMed] [Google Scholar]
- 70. Müller I. I., Melville D. B., Tanwar V., Rybski W. M., Mukherjee A., Shoemaker M. B., Wang W. D., Schoenhard J. A., Roden D. M., Darbar D., Knapik E. W., and Hatzopoulos A. K. (2013) Functional modeling in zebrafish demonstrates that the atrial-fibrillation-associated gene GREM2 regulates cardiac laterality, cardiomyocyte differentiation, and atrial rhythm. Dis. Model Mech. 6, 332–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bucher K., Sofroniew M. V., Pannell R., Impey H., Smith A. J., Torres E. M., Dunnett S. B., Jin Y., Baer R., and Rabbitts T. H. (2000) The T cell oncogene Tal2 is necessary for normal development of the mouse brain. Dev. Biol. 227, 533–544 [DOI] [PubMed] [Google Scholar]
- 72. Imadome K., Iwakawa M., Nojiri K., Tamaki T., Sakai M., Nakawatari M., Moritake T., Yanagisawa M., Nakamura E., Tsujii H., and Imai T. (2008) Upregulation of stress-response genes with cell cycle arrest induced by carbon ion irradiation in multiple murine tumors models. Cancer Biol. Ther. 7, 208–217 [DOI] [PubMed] [Google Scholar]
- 73. Dassanayaka S., and Jones S. P. (2014) O-GlcNAc and the cardiovascular system. Pharmacol. Ther. 142, 62–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Groves J. A., Lee A., Yildirir G., and Zachara N. E. (2013) Dynamic O-GlcNAcylation and its roles in the cellular stress response and homeostasis. Cell Stress Chaperones 18, 535–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Love D. C., Krause M. W., and Hanover J. A. (2010) O-GlcNAc cycling: emerging roles in development and epigenetics. Semin. Cell Dev. Biol. 21, 646–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ramakrishnan P., Clark P. M., Mason D. E., Peters E. C., Hsieh-Wilson L. C., and Baltimore D. (2013) Activation of the transcriptional function of the NF-kappaB protein c-Rel by O-GlcNAc glycosylation. Sci. Signal. 6, ra75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ohtsubo K., Chen M. Z., Olefsky J. M., and Marth J. D. (2011) Pathway to diabetes through attenuation of pancreatic beta cell glycosylation and glucose transport. Nat. Med. 17, 1067–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Rahman M. M., Stuchlick O., El-Karim E. G., Stuart R., Kipreos E. T., and Wells L. (2010) Intracellular protein glycosylation modulates insulin mediated lifespan in C. elegans. Aging 2, 678–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Trapnell C., Pachter L., and Salzberg S. L. (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.