Abstract
Protein 4.1G is a membrane skeletal protein that can serve as an adapter between transmembrane proteins and the underlying membrane skeleton. The function of 4.1G remains largely unexplored. Here, using 4.1G knockout mouse embryonic fibroblasts (MEFs) as a model system, we explored the function of 4.1G in motile cells. We show that the adhesion, spreading, and migration of 4.1G−/− MEF cells are impaired significantly. We further show that, although the total cellular expression of β1 integrin is unchanged, the surface expression of β1 integrin and its active form are decreased significantly in 4.1G−/− MEF cells. Moreover, the phosphorylation of focal adhesion kinase, a downstream component of the integrin-mediated signal transduction pathway, is suppressed in 4.1G−/− MEF cells. Co-immunoprecipitation experiments and in vitro binding assays showed that 4.1G binds directly to β1 integrin via its membrane-binding domain. These findings identified a novel role of 4.1G in cell adhesion, spreading, and migration in MEF cells by modulating the surface expression of β1 integrin and subsequent downstream signal transduction.
Keywords: adhesion; cytoskeleton; membrane; migration; phosphorylation; β1 integrin; focal adhesion kinase; mouse embryonic, fibroblast; protein 4.1G; spreading
Introduction
Cell adhesion, spreading, and migration are inseparable features of many biological and pathological processes, including normal development, angiogenesis, wound repair, tumor invasion, and metastasis. The process of cell adhesion and the subsequent spreading and migration on the extracellular matrix involves dynamic changes in the cytoskeleton through the action of integrins, which transduce signals from the outside to the inside of the cell and vice versa (1, 2).
Integrins are heterodimeric transmembrane cell adhesion molecules comprising α and β subunits (3). As receptors for the extracellular matrix, integrins play important roles in mediating the signals from the extracellular matrix (4). The signals propagated by extracellular matrix-integrin interactions result in the activation of a number of signaling pathways (5). These pathways include protein tyrosine kinases, such as focal adhesion kinase (FAK)3 (6), and members of the Rho family of small GTP-binding proteins, such as Cdc42, Rac1, and RhoA (7), which play important roles in regulating the organization of the cytoskeleton. Activated FAK and Rho-GTPase regulate cell adhesion, spreading, and migration (8, 9).
One important feature of integrins is that they can shift between low- and high-affinity conformations for ligand binding. The shift from a low- to a high-affinity state is termed “integrin activation” (10). Because altered integrin activation is associated with many diseases, such as bleeding disorders, leukocyte adhesion deficiencies, and skin blistering, integrin activation has to be controlled stringently (11). It was originally thought that talin is the only master regulator of integrin activation (12). Later works have shown that the kindlin family of proteins is as important as talin in mediating integrin function (13, 14). Both talin and kindlins belong to a family of evolutionarily conserved FERM (four-point-one, ezrin, radixin, moesin) domain-containing proteins (15). They regulate integrin function by binding directly to the cytoplasmic tail of integrin via their FERM domain, which triggers a conformational change in the extracellular ligand-binding domain, increasing its affinity for its ligand (10, 16). These findings suggest that other FERM domain-containing proteins may also associate with integrin and regulate integrin function.
Protein 4.1 family members (which includes 4.1R, 4.1B, 4.1G, and 4.1N) are the prototypical members of the FERM domain-containing superfamily of proteins. We have shown recently that 4.1R binds to β1 integrin and modulates the surface expression of β1 integrin in keratinocytes (17). A study by McCarty et al. (18) has also documented the association of 4.1B with β8 integrin in cultured astrocytes and in the brain. In this study, we identified a novel role of 4.1G in cell adhesion, spreading, and migration of mouse embryonic fibroblasts by modulating the surface expression of β1 integrin through a direct association between 4.1G and β1 integrin.
Experimental Procedures
Antibodies
All anti-4.1 antibodies were generated in our laboratory and used in our published studies (17, 19, 20). Other antibodies used in this study were as follows: rat 9EG7 monoclonal antibody, which preferentially recognizes the active conformation of mouse β1 integrins (21) (BD Biosciences); conformation-independent MB1.2 rat monoclonal antibody against mouse β1 integrin (22, 23) (Millipore, Billerica, MA); anti-FAK and anti-phosphotyrosine (4G10) (Millipore); anti-α2-integrin, anti-α5-integrin, and anti-α6-integrin (Abcam, Cambridge, MA); and anti-α3-integrin and β4-integrin (BD Biosciences). Affinity-purified rabbit polyclonal antibodies against GST and His were prepared by our laboratory. Alexa Fluor 488-conjugated and Alexa Fluor 594-conjugated secondary antibody to mouse and rabbit IgG, TO-PRO3 for nuclear staining, and Alexa Fluor 488-labeled wheat germ agglutinin for membrane staining were from Invitrogen. Goat anti-mouse HRP and goat anti-rabbit HRP were obtained from Jackson ImmunoResearch Laboratories (West Grove, PA).
Cell Culture
Isolation of primary mouse embryonic fibroblast (MEF) cells from 4.1G+/+ and 4.1G−/− C57Bl/6 mice (20) was performed as described before (24). MEF cells were prepared from embryonic day 13.5 embryos. The head and internal organs were removed. The remaining embryonic tissue was minced using a pair of scissors and immersed in 0.25% trypsin overnight at 4 °C. After 24 h, MEF cells were collected after centrifugation at 1500 rpm and maintained in DMEM containing 10% FBS (Gibco) and 100 μg/ml penicillin/streptomycin. After two passages, the MEF cells were immortalized by retroviral transduction of the SV40 large T antigen. For serum starvation experiments, MEF cells were plated in DMEM containing 0.1% FBS and then incubated at 37 °C for 18 h.
Cloning of 4.1G cDNA from MEF Cells
Total RNA was isolated from 4.1G+/+ and 4.1G−/− MEF cells with the RNeasy mini kit (Qiagen). RNA (1 μg) was reverse-transcribed into cDNA using random nonamers and M-MuLV reverse transcriptase (New England Biolabs) for 60 min at 42 °C. An equivalent of 5 ng of cDNA was used for PCR. PCR was performed using Accuprime Platinum Pfx DNA polymerase (Invitrogen). The PCR primers used were as follows: forward, ATGACTACTGAAGTTGGCT-CTGCATCTGAA; reverse, TTATTCTTCTC-CTTCCTCCGCCAACTCTG. Primers were designed to incorporate recognition sequences for the restriction enzymes SacII and XmaI at the 5′ and 3′ ends of the PCR product, respectively. N-terminal GFP fusion constructs were created by ligating SacII/XmaI-digested 4.1G cDNAs downstream of the GFP coding sequence in the pEGFP-C3 vector. The fidelity of the constructs was confirmed by sequencing.
Immunofluorescence Staining
For confocal immunofluorescence microscopy, cells were grown on MatTek glass-bottom microwell cell culture dishes (MatTek) coated with 10 μg/ml fibronectin (FN), and we let the cells grow into sparse density or to ∼90% confluence. Then the cells were fixed with 1% paraformaldehyde for 15 min and permeabilized with 0.1% Triton X-100 in 0.25% paraformaldehyde-PBS. Cells were then incubated in 10% horse serum and 0.1% Triton X-100 in PBS for 30 min to minimize nonspecific antibody binding. The cells were incubated with primary antibodies at 4 °C overnight, washed three times with PBS, and incubated with the appropriate second antibody at room temperature for 30 min. The following primary antibodies were used: rabbit polyclonal antibodies to 4.1G-U3, rat monoclonal antibody against β1 integrin (clone 9EG7), and mouse monoclonal antibody against FAK and paxillin. Alexa Fluor-conjugated secondary antibodies were purchased from Molecular Probes and diluted 1/700. The secondary antibodies were donkey anti-rabbit, donkey anti-rat, and donkey anti-mouse IgG labeled with Alexa Fluor 488 or Alexa Fluor 594. Actin was counterstained with Rhodamine-phalloidin (red). Images were collected on a Zeiss LSM510 META confocal microscope using a ×63 oil immersion objective.
Flow Cytometry
4.1G+/+ and 4.1G−/− MEF cells were serum-starved for 18 h. The cells were trypsinized and washed twice with 0.5% BSA in PBS. Primary antibodies against total β1 integrin (catalog no. MAB1997, Millipore) and against active-form β1 integrin (clone 9EG7, BD Biosciences) were used to stain the cells in 0.5% BSA in PBS for 30 min on ice. The cells were washed twice and incubated with allophycocyanin-conjugated anti-rat or anti-mouse secondary antibody for an additional 30 min on ice. After further washing, flow cytometric analysis was performed on a FACSCanto flow cytometer (BD Biosciences), and flow data overlay plots were produced using CellQuest Pro software (BD Biosciences).
Immunoblot Analysis
Cells were trypsinized, washed with PBS, and lysed with ice-cold lysis buffer (50 mm HEPES (pH 8.3), 420 mm KCl, 0.1% Nonidet P-40, and 1 mm EDTA) for 30 min on ice in the presence of proteinase inhibitor mixture (Sigma) and phosphatase inhibitor (Roche). After centrifugation at 16,000 × g at 4 °C for 10 min, the supernatant was collected. Protein concentration was measured by the Bradford method using BSA as standard. 30 μg of protein was separated by 10% SDS-PAGE and transferred to a nitrocellulose membrane. After blocking for 1 h in blocking buffer (10 mm Tris-HCl (pH 7.4), 150 mm NaCl, 0.5% Tween 20, and 5% nonfat dried milk powder), the blot was probed for 1 h with the desired primary antibodies. After several washes, the blot was incubated with anti-rabbit or anti-mouse IgG coupled to HRP and developed with the SuperSignal West Pico chemiluminescence detection kit (Molecular Probes). All steps were performed at room temperature.
Co-immunoprecipitation
MEF cells were lysed with ice-cold lysis buffer (50 mm HEPES (pH 8.3), 420 mm KCl, 0.1% Nonidet P-40, and 1 mm EDTA) for 30 min on ice. The supernatant was collected after centrifugation at 16,000 × g at 4 °C for 10 min, and the concentration of protein in the supernatant was determined by the Bradford method using BSA as standard (Bio-Rad). 500 μg of extract was incubated with either 5 μg of anti-4.1G-HP or anti-β1 integrin antibody or preimmune IgG in 500 μl of co-immunoprecipitation buffer (Active Motif) at 4 °C overnight with rotation. The immunoprecipitates were isolated on protein G beads and separated by 10% SDS-PAGE, followed by transfer to a nitrocellulose membrane. The membrane was probed with antibodies against β1 integrin, α6 integrin, or 4.1G-HP.
Wound Healing Assay
MEF cells were grown at equivalent confluence for 18 h on a MatTek glass-bottomed chamber precoated with 10 μg/ml FN. Confluent 4.1G+/+ and 4.1G−/− MEF cells were arrested mitotically by incubation with 8 μg/ml mitomycin C (Roche) in DMEM for 2 h under normal culture conditions. Mitomycin C was removed by three washes in PBS. A pseudowound was introduced in an equivalent confluent monolayer of cells by lightly scratching with a 10-μl pipette tip across the cell layer. Cell debris was removed by two washes with culture medium. A minimum of six “wounded areas” was filmed for each sample by obtaining images every 15 min for 16 h. Images were collected on a Zeiss LSM510 META confocal microscope using a ×25 phase-contrast objective. The wounded area was measured using LSM510 software for each representative time point.
Cell Spreading Assay
MEF cells were trypsinized and replated on coverslips precoated with 10 μg/ml FN and allowed to spread for 1 or 3 h at 37 °C in the presence of complete media. Cells were fixed and labeled with Alexa Fluor 488-conjugated wheat germ agglutinin (Invitrogen) for 30 min to better visualize the cell outlines. Cell surface boundaries were outlined for 35 individual cells chosen randomly, and LSM 510 software was used to calculate the mean surface area and standard deviation of each population. One-tailed Student's t tests were applied to test the statistical significance of the data. We would like to note that, in our preliminary experiments, we checked the spreading area of MEF cells at 1, 3, 6, and 12 h. Although the spreading areas increased from 1 to 3 h, there was no significant difference between 3, 6, and 12 h, demonstrating that MEF cells are fully spread after 3 h. Therefore, in this study, we chose the 1- and 3-h time points to perform the cell spreading assay.
Transwell Migration Assay
For migration assays, 8-μm-diameter pore transwell cell culture inserts (BD Biosciences) were placed in 6-well plates. The underside of the insert and the bottom of the well were coated with 10 μg/ml of FN at 37 °C for 1 h. Cells suspended in serum-free media were seeded into the upper chamber of the insert (4 × 105/well), and complete medium was added to the lower chamber. Cells were then incubated for another 6 h, during which cells migrated through the pores in the insert to the lower side of the membrane insert. At the end of cell migration, we cleansed the upper side of the chamber with a cotton swab, stained the filter for 1 h with crystal violet (Sigma) in 2% ethanol, and then rinsed it in water. The filters were then imaged with a Leica inverted microscope. Five representative images (×10 magnification) were captured randomly for each insert and used to manually count the number of cells present. The results were presented as mean number of cells per field ± S.D.
Preparation of Recombinant Proteins
The plasmid DNA encoding various recombinant proteins was transformed into Escherichia coli BL21(DE3) for protein expression. The recombinant proteins were expressed at 16 °C in the presence of 0.1 mm isopropyl-β-d-thiogalactopyranoside. GST-tagged 4.1G domains were purified by a glutathione-Sepharose 4B affinity column, and the maltose binding protein (MBP)-tagged cytoplasmic domain of β1 integrin was purified by amylose resin.
GST Pulldown Assay
For the pulldown assay, various domains of 4.1G were cloned into pGEX 4T-2, and the cytoplasmic domain of β1 integrin was cloned into the pMal-c2X vector. GST-tagged proteins were coupled to glutathione-Sepharose 4B beads, and the MBP-tagged cytoplasmic domain of β1 integrin was coupled to amylose resin at room temperature for 1 h. The beads were pelleted and washed. GST-tagged 4.1G domains or the MBP-tagged cytoplasmic domain of β1 integrin were added to the coupled beads in a final volume of 100 μl. The final concentration of the coupled protein was 2 μm. The mixture was incubated for 1 h at room temperature, pelleted, washed, and eluted with 10% SDS. The pellet was analyzed by SDS-PAGE. The binding of GST-tagged 4.1G domains to the MBP-tagged cytoplasmic domain of the β1 integrin fragment was detected by Western blot using anti-GST or anti-MBP antibody.
FAK Phosphorylation Assay
MEF cells were serum-starved by culturing in DMEM without FBS for 24 h. Following trypsinization and replating on coverslips precoated with 10 μg/ml FN, cells were allowed to recover for various time periods. The cells were then collected and lysed with ice-cold lysis buffer (50 mm HEPES (pH 8.3), 420 mm KCl, 0.1% Nonidet P-40, and 1 mm EDTA) for 30 min on ice. The supernatant was collected, and protein concentration was determined by the Bradford method using BSA as standard (Bio-Rad). 500 μg of extract was incubated with 5 μg of anti-FAK in 500 μl of co-immunoprecipitation buffer (Active Motif) at 4 °C overnight with rotation. The immunoprecipitated proteins were isolated on protein G beads and separated by 10% SDS-PAGE, followed by transfer to a nitrocellulose membrane. The membrane was probed with anti-FAK antibody or 4G10 antibody, which specifically recognizes phosphorylated tyrosine.
Results
Expression of 4.1G in MEF Cells
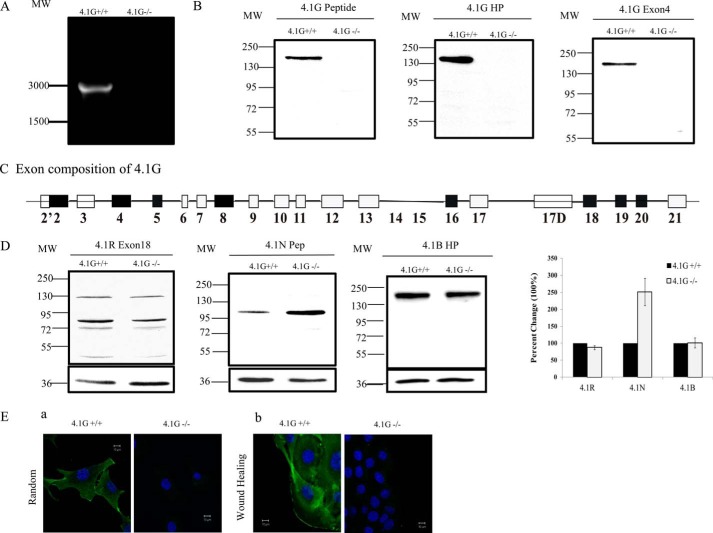
It has been well established that all four genes encoding the family of 4.1 proteins undergo extensive alternative splicing that leads to the generation of multiple isoforms. As the first step toward investigating the function of 4.1G in MEF cells, we examined the expression of 4.1G by RT-PCR and Western blotting. RT-PCR using a 4.1G-specific primer set amplified a band around 3000 bp from 4.1G+/+ MEF cells but not 4.1G−/− MEF cells (Fig. 1A). Sequencing of the PCR product of six distinct clones revealed that it contains 2967 bp. The sequence was identical to the coding sequence of RefSeq entry NM_001199265.1. The corresponding protein database entry is NP_001186194.1. The exon composition of this isoform is shown in Fig. 1B. The 4.1G transcript contains exons encoding the head piece (U1, exon 2), the FERM domain (exons 4–12), the U2 region (exon 13), spectrin-actin binding domain (exons 16, 17), the U3 region (exon 17D), and the C-terminal domain (exons 18–21).
FIGURE 1.
Expression and localization of 4.1G in MEF cells. A, RT-PCR analysis of the expression of 4.1G in 4.1G+/+ and 4.1G −/− MEF cells. MW, molecular weight. B, immunoblot analysis of the expression of 4.1G in 4.1G+/+ and 4.1G−/− MEF cells. Total lysates (35 μg of protein) were probed with polyclonal rabbit antibodies against the 4.1G peptide, the 4.1G head piece, and 4.1G exon4. C, schematic of the 4.1G protein structure and exon organization in MEF cells. D, immunoblot analysis of protein 4.1 members in 4.1G+/+ and 4.1G−/− MEF cells. Total lysates (35 μg of protein) were probed with polyclonal goat antibody against 4.1R exon13 and rabbit antibodies against the 4.1N peptide and 4.1B head piece. Quantitative analysis of immunoblot results from three independent experiments is shown in the right panel. GAPDH was used as a loading control. E, immunofluorescence staining of endogenous 4.1G in randomly migrated and directionally migrated 4.1G+/+ and 4.1G−/− MEF cells. Subconfluent cells were used as randomly migrated cells, and confluent cells were checked 4 h after wounding as directionally migrated cells. Cells were fixed and stained using anti-4.1G-U3 antibody (green) and DAPI (blue).
The expression of 4.1G protein was examined by Western blot analysis using three 4.1G-specific antibodies. Fig. 1C shows that all antibodies detected one band with a molecular mass of ∼160 KD. The specificity of the band was validated by the finding that it is not detected in 4.1G−/− MEF cells. We also examined the expression of the other three protein 4.1 family members by Western blot analysis. Fig. 1D shows that 4.1R, 4.1B, and 4.1N are all expressed in MEF cells. Interestingly, although the expression levels of 4.1R and 4.1B are similar in 4.1G+/+ and 4.1G−/− MEF cells, the expression of 4.1N is up-regulated markedly in 4.1G−/− MEF cells. Quantitative analysis from three independent experiments revealed a ∼2.5-fold increase of 4.1N in 4.1G−/− cells. GAPDH was used as control in all Western blot analyses. These findings suggest that 4.1N may partially compensate for 4.1G function.
Localization of 4.1G in MEF Cells
We then examined the localization of endogenous 4.1G by immunofluorescence staining using anti-4.1G U3 antibody. Fig. 1E, a, shows both membrane and cytoplasmic localization of 4.1G. Furthermore, as shown in Fig. 1E, b, 4.1G appears to localize at the leading edge of a motile cell. No staining is seen in 4.1G−/− MEF cells. The localization profile of 4.1G strongly suggests the potential involvement of 4.1G in cell spreading and migration.
Impaired Adhesion and Spreading of 4.1G−/− MEF Cells
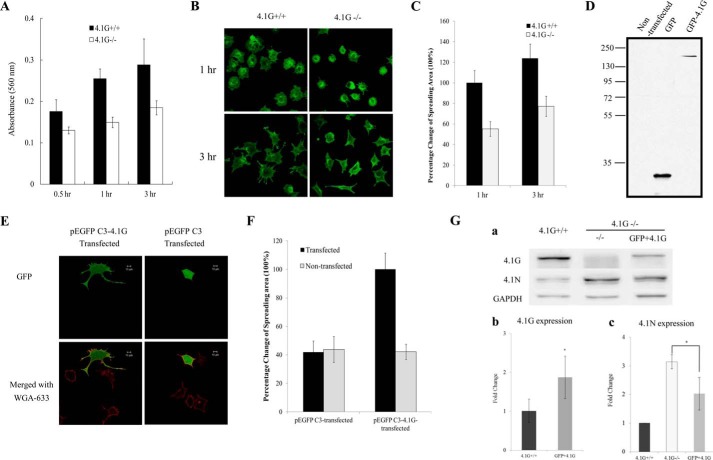
Having characterized the expression and localization of 4.1G in MEF cells, we further explored the function of 4.1G in these cells. First we compared the adhesion of 4.1G+/+ and 4.1G−/− MEF cells to the FN-coated surface. The adhesion was examined 0.5, 1, and 3 h after plating. Fig. 2A shows that, at all time points, the adhesion of 4.1G−/− cells to the FN-coated surface was less than that of 4.1G+/+ MEF cells (∼20% less after 0.5 h and ∼35% less after 1 and 3 h). We also examined the extent of cell spreading on an FN-coated surface. Fig. 2B shows that, after 1 or 3 h of incubation, the extent of spreading of 4.1G−/− cells was much less than that of 4.1G+/+ cells. Quantitative analysis revealed an ∼50% reduction in the spreading area of 4.1G−/− cells compared with that of 4.1G+/+ cells at both time points (Fig. 2C).
FIGURE 2.
Impaired adhesion and spreading of 4.1G−/− MEF cells on fibronectin. A, cells were plated on fibronectin-coated 96-well plates and incubated for 0.5, 1, and 3 h. The adherent cells were stained with crystal violet, and the staining intensity was quantified by spectrophotometry at 560 nm. The results are mean ± S.E. of three independent experiments. B, cells were plated on fibronectin-coated 4-well chambers and allowed to spread for 3 h. The cells were labeled with Alexa Fluor 488-conjugated wheat germ agglutinin (Invitrogen), and the images were collected using a Zeiss Axiophot wide-field epifluorescence microscope. C, the mean surface area from 35 individual cells was calculated using LSM 5 Pascal software. The data are shown as mean ± S.E. of three experiments. One-tailed Student's t tests were applied to test the statistical significance of the data, with the p value for 4.1G+/+ cells versus 4.1G−/− cells at 1 h being p < 0.0001 and of 4.1G+/+versus 4.1G−/− cells at 3 h being p < 0.0001. D, immunoblot analysis of 4.1G−/− MEF cells transfected with GFP or GFP-4.1G. Total lysates (35 μg of protein) were probed with polyclonal rabbit antibodies against GFP. E, 4.1G−/− MEF cells transfected with GFP or GFP-4.1G were plated on FN-coated coverslips and allowed to spread for 3 h. The cells were labeled with Alexa Fluor 488-conjugated wheat germ agglutinin (Invitrogen), and the images were collected using a Zeiss Axiophot wide-field epifluorescence microscope. Scale bars = 20 μm. F, the mean surface area of 35 individual cells was calculated using LSM 5 Pascal software. The data shown are mean ± S.E. of three experiments. One-tailed Student's t tests were applied to test the statistical significance of the data, with the p value for 4.1G −/− cells transfected with GFP-4.1G versus 4.1G−/− cells being p < 0.0001 and of 4.1G−/− cells transfected with GFP-4.1G versus 4.1G −/− cells transfected with GFP being p < 0.0001. G, analysis of 4.1G and 4.1N upon GFP-4.1G rescue. a, immunoblot. Total lysates were probed with antibodies as indicated. b, quantitative analysis of endogenous 4.1G and GFP-4.1G. GFP-4.1G was calculated on the basis of band intensity and transfection efficiency. c, quantitative analysis of 4.1N. *, p < 0.05.
To confirm a direct role of 4.1G in MEF cell spreading, we transfected 4.1G−/− MEF cells with a plasmid encoding GFP-4.1G or GFP only. Western blot analysis showed that both GFP and GFP-4.1G proteins are expressed in the transfected cells (Fig. 2D). Fig. 2E shows that the spreading area of 4.1G−/− MEF cells transfected with a GFP-4.1G plasmid is increased significantly compared with the neighboring untransfected 4.1G−/− MEF cells, whereas the spreading area of cells transfected with GFP showed no significant change. Quantitative analysis revealed an ∼220% increase in the spreading area of 4.1G−/− cells transfected with GFP-4.1G compared with that of non-transfected cells or cells transfected with GFP (Fig. 2F).
Western blot analysis was also performed to compare the expression of GFP-4.1G in 4.1G−/− cells with endogenous 4.1G in wild-type cells and to examine whether the levels of 4.1N decrease upon 4.1G rescue. These results are shown in Fig. 2G, a. In the situation that the transfection efficiency of GFP-4.1G−/− is about 20–30%, the expression level of GRP-4.1G is about 40% of that of the 4.1F+/+ cells. We estimated that GFP-4.1G, when the transfection efficiency is 100%, is about 1.5 to 2 times higher than that of endogenous 4.1G (Fig. 2G, b). Fig. 2G, c, shows that, upon rescue of 4.1G levels in 4.1G−/− MEFs, the levels of 4.1N decreased.
Impaired Directional Migration of 4.1G−/− MEF Cells
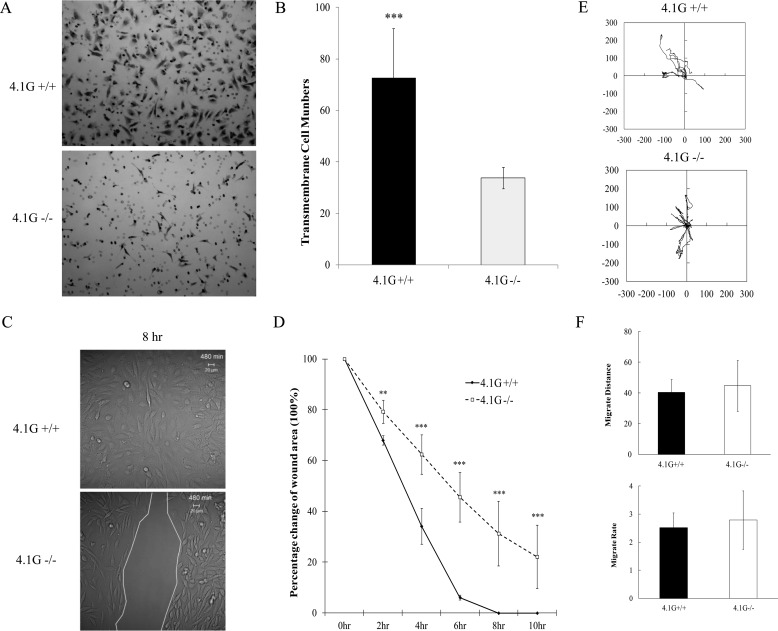
The directional migration of 4.1G+/+ and 4.1G−/− MEF cells was evaluated by transwell migration assay and by wound healing assay. The transwell migration assay measures the migration of cells toward FN through the pores of the transwell inserts. Representative images of the cells that migrated through the pores of the insert are shown in Fig. 3A, and quantitation of the number of cells that migrated through the pores from three independent experiments are shown in Fig. 3B. The number of 4.1G−/− cells migrating toward FN through the pores of the transwell cell inserts was reduced by ∼60% relative to 4.1G+/+ cells. The results of cell migration during wound healing are shown in Fig. 3, C and D. Representative images of cell migration 8 h after wounding are shown in Fig. 3C, and the rate of wound closure is shown in Fig. 3D. Although the closure of the wound area of 4.1G+/+ cells was nearly complete 8 h after wounding, more than 30% of the wounded area of 4.1G−/− cells was still not closed. We also examined the effect of 4.1G on random motility of MEF cells using live-cell video microscopy. Track plots of the randomly migrating MEF cells revealed no detectable difference in either distance moved or velocity of migration between 4.1G+/+ and 4.1G−/− MEF cells (Fig. 3, E and F). Therefore, 4.1G plays a role in directional migration but not in random motility of MEF cells.
FIGURE 3.
Impaired directional migration and motility of 4.1G−/− MEF cells. A, 8-μm-diameter pore Transwell cell culture inserts were placed in 6-well plates, the bottoms of which were coated with fibronectin. Equal numbers of 4.1G+/+ and 4.1G−/− MEF cells were seeded on top of the inserts and incubated for 4 h. The cells migrated to the bottom of the well were fixed and stained with crystal violet. B, absorbance was read using a multiwell plate reader at 560-nm wavelength. The mean from three experiments are shown ± S.E. Standard deviations are depicted by the error bars, and Student's t test values for significance were calculated (***, p < 0.0001). C, 4.1G+/+ and 4.1G−/− MEF cells were plated on fibronectin-coated coverslips and left to grow to ∼90% confluence. Scratches of ∼700 μm were introduced on the coverslips. Living cells were observed using confocal microscopy. Representative differential interference contrast images are shown at 8-h time points after making scratches. D, LSM 5 Pascal software was used to calculate the mean scratch area of 4.1G+/+ and 4.1G−/− MEF cells for each representative time point. The data shown are from three experiments. Standard deviations are depicted by the error bars, and Student's t test values for significance were as follows: **, p < 0.01; *** p < 0.001. E, live-cell images were obtained every 5 min over a period of 3 h (60 images in total). Each track represents an individual cell. F, migration distance and migration rate were measured using the cell migration and chemotaxis plug-in (Ibidi) for ImageJ. Standard deviations are depicted by the error bars and Student's t test values for significance calculation, with p > 0.1.
Decreased Surface Expression of β1 Integrin in 4.1G−/− MEF Cells
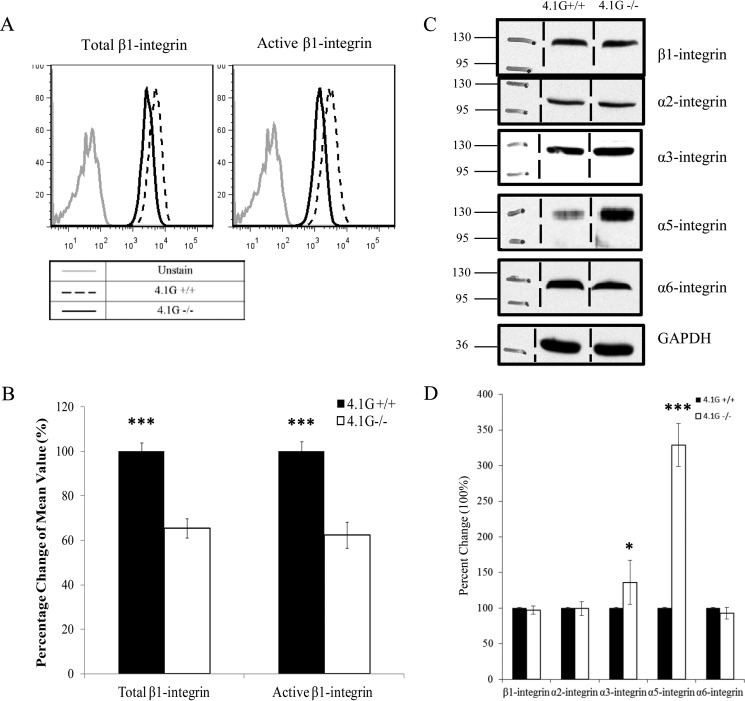
Integrins play important roles in cell adhesion, spreading, and migration. Members of FERM domain-containing proteins such as talin (25), kindlin (13), 4.1B (18), and 4.1R (17) have been shown to interact with integrins and modulate their functions. These findings promoted us to examine how deletion of 4.1G in MEF cells affects integrins. We first examined the surface expression of β1 integrin by flow cytometry, using antibodies specific for the total or activated form of β1 integrin. Representative flow cytometric profiles and the quantitative data from three independent experiments are shown in Fig. 4, A and B. These results reveal that the surface expression of total β1 integrin was decreased by ∼40% and that the active form of β1 integrin was decreased by ∼50% in 4.1G−/− MEF cells. Interestingly, Western blot analyses showed no significant change in the expression levels of β1 integrin in 4.1G+/+ and 4.1G−/− MEF cells. These findings suggest that 4.1G, although not affecting the expression levels of β1 integrin, plays an important role in the surface expression β1 integrin. Because β integrin pairs with α integrin to form α/β heterodimers, we also examined the expression of several α integrins (potential β1 partners) by Western blot analysis. Although no significant difference in the expression levels of α2 and α6 integrin, but a significant difference in the expression level of α3 integrin, was observed between 4.1G+/+ and 4.1G−/− MEF cells, surprisingly, a more than 3-fold increase in α5 integrin expression was observed in 4.1G−/− cells (Fig. 4, C and D).
FIGURE 4.
Decreased surface expression and activity of β1 integrin in 4.1G−/− MEF cells. A and B, surface expression of total and active-form β1 integrins in 4.1G+/+ and 4.1G−/− MEF cells was measured by flow cytometry. The representative profiles and quantitative analysis (the mean fluorescence intensity ± S.E. from three independent experiments) are shown in A and B, respectively (***, p < 0.001). For simplicity, an autofluorescence control from only wild-type cells is shown. C, Western blot analysis of integrins in 4.1G+/+ and 4.1G−/− MEF cells. 35 μg of protein of total lysates was probed with the indicated antibodies. A GAPDH immunoblot served as a loading control. D, quantitative analysis from three independent experiments. Standard deviations are depicted by the error bars, and Student's t test values for significance were as follows: *, p < 0.1; ***, p < 0.001.
Association of Protein 4.1G with β1 Integrin
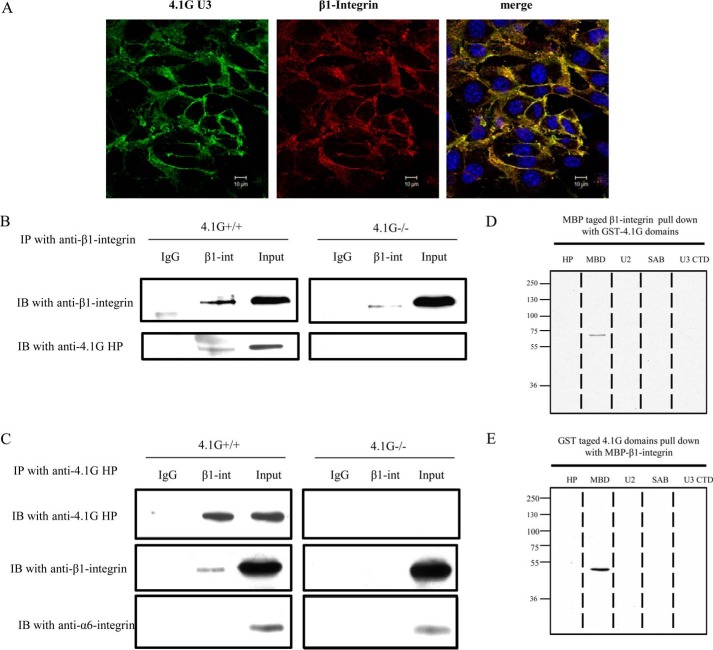
Members of the protein 4.1 superfamily have been shown to bind to integrins and to engage in regulation of the integrin-mediated signaling pathway (13, 17, 18, 25). We searched for a similar activity on the part of 4.1G. Double staining of 4.1G and β1 integrin was performed to examine whether the two molecules co-localize. As shown in Fig. 5A, under confluent conditions, 4.1G and β1 integrin co-localized in the cytoplasm and on the plasma membrane. Co-immunoprecipitation and GST pulldown assays were performed to test for direct binding of 4.1G to β1 integrin. As shown in Fig. 5B, protein 4.1G could be co-immunoprecipitated with β1 integrin by anti-β1 integrin antibody. Conversely, β1 integrin was pulled down with protein 4.1G by anti-4.1G HP antibody. In a negative control, α6 integrin did not co-immunoprecipitate with 4.1G (Fig. 5C). GST pulldown assays were performed with a set of distinct 4.1G domains and the MBP-tagged β1 integrin cytoplasmic domain (Fig. 5D). Binding of the 4.1G MBD (membrane binding domain) to the β1 integrin cytoplasmic domain was demonstrated by the capture of the GST-4.1G domains by the immobilized integrin domain and the integrin domain by the immobilized 4.1G domain (Fig. 5E). These results demonstrate that protein 4.1G interacts directly with the cytoplasmic domain of β1 integrin in MEF cells through its membrane binding domain.
FIGURE 5.
Direct association of β1 integrin with 4.1G. A, immunofluorescence staining showing the co-localization of 4.1G with β1 integrin in confluent 4.1G+/+MEF cells. Cells were fixed and stained using anti-4.1G-U3 antibody (green), anti-β1 integrin antibody (MB1.2), and DAPI (blue). B and C, 4.1G and β1 integrin associate in situ. B, immunoprecipitation (IP) of β1 integrin. β1 integrin (β1 int) was immunoprecipitated from 4.1G+/+ and 4.1G−/− MEF cells using anti-β1 integrin. β1 integrin or 4.1G in the immunoprecipitate was detected using anti-β1 integrin antibody or anti-4.1G HP antibody. IB, immunoblot. C, immunoprecipitation of 4.1G. 4.1G was immunoprecipitated from MEF cells using anti-4.1G HP antibody. 4.1G, β1 integrin, and α6 integrin in the immunoprecipitate were detected using the corresponding antibodies. D, binding of 4.1G to the cytoplasmic domain of β1 integrin. GST-tagged 4.1G-HP, MBD, U2, SAB, and the C-terminal domain (CTD) were incubated for 30 min at room temperature with the MBP-tagged cytoplasmic domain of β1 integrin, and binding was assessed by pulldown assay. β1 integrin binding was detected by blotting with anti-MBP antibody. E, the MBP-tagged cytoplasmic domain of β1 integrin was incubated for 30 min at room temperature with GST-tagged 4.1G HP, MBD, U2, SAB, and C-terminal domains, and binding was assessed by pulldown assay. 4.1G binding was detected by blotting with anti-GST antibody.
Diminished Phosphorylation of FAK in 4.1G−/− MEF Cells
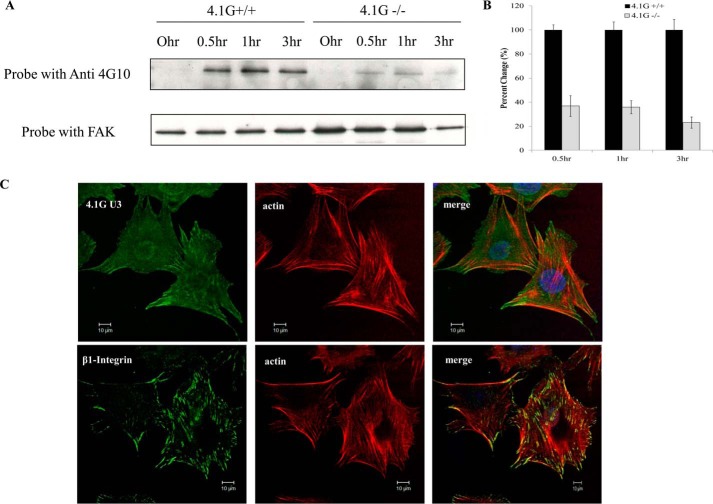
FAK is known to be a crucial component in the transduction of signaling pathways initiated by integrin ligation (26). This, in turn, regulates cell spreading and migration (27). The impaired cell spreading and motility of cells lacking 4.1G raises the question of whether the protein plays a role in the phosphorylation of FAK. We therefore compared the phosphorylation levels of FAK in 4.1G+/+ and 4.1G−/− MEF cells at different times after FN stimulation. Fig. 6 shows that, in 4.1G+/+ cells, phosphorylation of FAK is increased in response to FN stimulation. In 4.1G−/− cells, by contrast, the phosphorylation level upon FN stimulation is significantly less than that in 4.1G+/+ cells (Fig. 6, A and B). Therefore, protein 4.1G is required for the phosphorylation of FAK. In support of a role of 4.1G in FAK phosphorylation, Fig. 6C shows that, similar to β1 integrin, 4.1G co-localizes at the focal adhesion site.
FIGURE 6.
Impaired FAK phosphorylation in 4.1G−/− MEF cells. A, immunoprecipitation to check the expression of total FAK and phosphorylated FAK of 4.1G+/+ and 4.1G−/− MEF cells. Cells that were serum-starved for 24 h were plated on an FN-coated cell culture dish and harvested at different time points. Cell lysates were immunoprecipitated with FAK antibody and then probed with 4G10 antibody to detect phosphorylated FAK or FAK antibody to check total FAK expression. B, quantitative analysis from three independent experiments. C, immunofluorescence staining showing the localization of 4.1G and β1 integrin at the focal adhesion site of 4.1G+/+ MEF cells. Cells were fixed and stained using anti-4.1G-U3 antibody or anti-β1 integrin antibody (green), Rhodamine-phalloidin (red), and DAPI (blue).
Discussion
4.1G is a member of the protein 4.1 family, which includes 4.1R, 4.1G, 4.1B, and 4.1N (28, 29). In contrast to an extensive understanding of the function of the prototypical member 4.1R, the knowledge regarding 4.1G is very limited. Earlier in vitro biochemical studies have shown the association of 4.1G with several transmembrane receptors (30–32). Using 4.1G−/− mice, we and others have recently documented the role of 4.1G in male fertility (20) and in the organization of the internodes in peripheral myelinated nerves (33). In this study, using MEF cells derived from 4.1G−/− mice, we identified a previously unrecognized role for 4.1G in the motile behavior of MEF cells. We further documented that 4.1G affects cell adhesion, spreading, and migration through the β1 integrin pathway.
Members of the FERM protein superfamily have been reported to participate in integrin-linked downstream functions. For example, by binding to the cytoplasmic tails of β1 or β3 integrin, talin induces conformational changes in the extracellular domain of integrins, increasing their affinity for ligands (10). Members of another group of FERM proteins, the kindlin family, are also known to be involved in integrin transmembrane signaling, and expression of kindlin is necessary for integrin activation (13, 34). Recent studies have revealed that members of the protein 4.1 family also play important modulatory roles in integrin-related processes. Protein 4.1B has been found to interact selectively with αvβ8 integrin and plays an important functional role in the development and maintenance of the CNS (18). We recently documented that 4.1R plays an important role in cell adhesion, spreading, and migration of keratinocytes by modulating the surface expression of β1 integrin (17).
Although several members of the protein 4.1 superfamily have been implicated in mediating integrin functions, it is interesting to note that they probably do so through different mechanisms. Although the binding of talin or kindlin to cytoplasmic tails of integrin induces conformational changes in the extracellular domain of integrin, leading to integrin activation, the association of integrin to members of protein 4.1 is required for the surface expression of integrin.
One consensus role of the protein 4.1 family members is their ability to associate with a variety of transmembrane proteins and regulate the expression of these proteins. However, it should be noted that there are some differences. In red cells, deficiency of 4.1R leads to decreased expression levels of its binding partners glycophorins C, band 3, XK, and Duffy (35). In stomach epithelial cells, lack of 4.1R resulted in decreased expression of the binding partner β-catenin (36). Similarly, the expression of nectin-like 4 is reduced in 4.1G-deficient testis Sertoli cells (20). On the other hand, lack of 4.1R in keratinocytes only impaired the surface expression of β1 integrin, which was accompanied by an increased in overall β1 integrin expression (17). Here we show that, in MEF cells, lack of 4.1G leads to decreased surface expression of β1 integrin without affecting overall β1 integrin expression. Given the findings that some other protein 4.1 members, such as 4.1N and 4.1R, play an important role in intracellular protein traffic (37, 38) and that integrins undergo extensive intracellular trafficking (39, 40), it is reasonable to speculate that protein 4.1 family members affect the expression of their binding partners through different mechanisms.
Another emerging role of protein 4.1 family members is in mediating intracellular signal transduction. For example, we have documented that, in CD4+ T cells, lack of 4.1R results in hyper-phosphorylation of the adapter protein linker of activation of T cells (LAT) and enhanced downstream signal transduction, implying that 4.1R negatively regulates signal transduction in CD4+ cells (38). In contrast, here we show that lack of 4.1G in MEF cells led to impaired phosphorylation of FAK.
Although in vitro studies have suggested that protein 4.1 family members may play many different functional roles (41, 42), the phenotypic changes in various individual 4.1 knockout mouse models are not as severe as would be expected from in vitro studies. It is possible that members of the protein 4.1 family may compensate function for each other. Indeed, we have shown previously that all 4.1 family members are expressed in CD4+ cells and that the expression of 4.1N is significantly up-regulated in 4.1R-deficient CD4+ T cells (41). We have also shown that all 4.1 family members are expressed in keratinocytes and that, in 4.1R-deficient keratinocytes, both 4.1N and 4.1G are up-regulated (17). In addition, all protein 4.1 members are expressed in the adrenal gland (43). We now show that all members of the protein 4.1 family are also expressed in MEF cells and that 4.1N is up-regulated significantly in 4.1G−/− MEF cells, suggesting that 4.1N may partially compensate for the loss of function of 4.1G in vivo. The compensatory effect of 4.1N was also supported by our findings showing that the up-regulation of 4.1N in 4.1G−/− MEF cells was decreased upon 4.1G rescue (Fig. 2G) and that the attempted knockdown of 4.1N in 4.1G-deficient MEF cells caused severe cell death (data not shown). To further clarify whether protein 4.1 family members have redundant or distinct function, studies on double or triple knockout mice or cells derived from these mice should help to address these issues.
Author Contributions
L. C., T. W., Y. Q., H. W., Q. K., J. Z., and X. G. performed the experiments and analyzed the data. A. J. B. analyzed and interpreted the data. A. J. B. and N. M. provided input into the design of the study and edited and critiqued the paper. X. A. designed the experiments, analyzed the data, and wrote the manuscript.
This work was supported in part by Grants 81171905 and 81272187 from the Natural Science Foundation of China and by National Institutes of Health Grants DK26263 and DK100180. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- FAK
- focal adhesion kinase
- MEF
- mouse embryonic fibroblast
- FN
- fibronectin
- HP
- head piece.
References
- 1. Fath K. R., Edgell C. J., and Burridge K. (1989) The distribution of distinct integrins in focal contacts is determined by the substratum composition. J. Cell Sci. 92, 67–75 [DOI] [PubMed] [Google Scholar]
- 2. Lauffenburger D. A., and Horwitz A. F. (1996) Cell migration: a physically integrated molecular process. Cell 84, 359–369 [DOI] [PubMed] [Google Scholar]
- 3. Hemler M. E. (1999) in Integrins: Guidebook to the Extracellular Matrix and Adhesion Proteins (Kreis T., and Vale R., ed.), pp. 196–216, Oxford University Press, Oxford [Google Scholar]
- 4. Plow E. F., Haas T. A., Zhang L., Loftus J., and Smith J. W. (2000) Ligand binding to integrins. J. Biol. Chem. 275, 21785–21788 [DOI] [PubMed] [Google Scholar]
- 5. Giancotti F. G., and Ruoslahti E. (1999) Integrin signaling. Science 285, 1028–1032 [DOI] [PubMed] [Google Scholar]
- 6. Michael K. E., Dumbauld D. W., Burns K. L., Hanks S. K., and García A. J. (2009) Focal adhesion kinase modulates cell adhesion strengthening via integrin activation. Mol. Biol. Cell 20, 2508–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clark E. A., King W. G., Brugge J. S., Symons M., and Hynes R. O. (1998) Integrin-mediated signals regulated by members of the rho family of GTPases. J. Cell Biol. 142, 573–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mitra S. K., Hanson D. A., and Schlaepfer D. D. (2005) Focal adhesion kinase: in command and control of cell motility. Nat. Rev. Mol. Cell Biol. 6, 56–68 [DOI] [PubMed] [Google Scholar]
- 9. Nobes C. D., and Hall A. (1999) Rho GTPases control polarity, protrusion, and adhesion during cell movement. J. Cell Biol. 144, 1235–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Calderwood D. A. (2004) Integrin activation. J. Cell Sci. 117, 657–666 [DOI] [PubMed] [Google Scholar]
- 11. Hynes R. O. (2002) Integrins: bidirectional, allosteric signaling machines. Cell 110, 673–687 [DOI] [PubMed] [Google Scholar]
- 12. Calderwood D. A., Zent R., Grant R., Rees D. J., Hynes R. O., and Ginsberg M. H. (1999) The Talin head domain binds to integrin β subunit cytoplasmic tails and regulates integrin activation. J. Biol. Chem. 274, 28071–28074 [DOI] [PubMed] [Google Scholar]
- 13. Harburger D. S., Bouaouina M., and Calderwood D. A. (2009) Kindlin-1 and -2 directly bind the C-terminal region of β integrin cytoplasmic tails and exert integrin-specific activation effects. J. Biol. Chem. 284, 11485–11497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Herz C., Aumailley M., Schulte C., Schlötzer-Schrehardt U., Bruckner-Tuderman L., and Has C. (2006) Kindlin-1 is a phosphoprotein involved in regulation of polarity, proliferation, and motility of epidermal keratinocytes. J. Biol. Chem. 281, 36082–36090 [DOI] [PubMed] [Google Scholar]
- 15. Chishti A. H., Kim A. C., Marfatia S. M., Lutchman M., Hanspal M., Jindal H., Liu S. C., Low P. S., Rouleau G. A., Mohandas N., Chasis J. A., Conboy J. G., Gascard P., Takakuwa Y., Huang S. C., Benz E. J. Jr., Bretscher A., Fehon R. G., Gusella J. F., Ramesh V., Solomon F., Marchesi V. T., Tsukita S., Tsukita S., and Hoover K. B. (1998) The FERM domain: a unique module involved in the linkage of cytoplasmic proteins to the membrane. Trends Biochem. Sci. 23, 281–282 [DOI] [PubMed] [Google Scholar]
- 16. Xu Z., Gao J., Hong J., and Ma Y. Q. (2013) Integrity of kindlin-2 FERM subdomains is required for supporting integrin activation. Biochem. Biophys. Res. Commun. 434, 382–387 [DOI] [PubMed] [Google Scholar]
- 17. Chen L., Hughes R. A., Baines A. J., Conboy J., Mohandas N., and An X. (2011) Protein 4.1R regulates cell adhesion, spreading, migration and motility of mouse keratinocytes by modulating surface expression of β1 integrin. J. Cell Sci. 124, 2478–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McCarty J. H., Cook A. A., and Hynes R. O. (2005) An interaction between αvβ8 integrin and Band 4.1B via a highly conserved region of the Band 4.1 C-terminal domain. Proc. Natl. Acad. Sci. U.S.A. 102, 13479–13483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kang Q., Wang T., Zhang H., Mohandas N., and An X. (2009) A Golgi-associated protein 4.1B variant is required for assimilation of proteins in the membrane. J. Cell Sci. 122, 1091–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yang S., Weng H., Chen L., Guo X., Parra M., Conboy J., Debnath G., Lambert A. J., Peters L. L., Baines A. J., Mohandas N., and An X. (2011) Lack of protein 4.1G causes altered expression and localization of the cell adhesion molecule nectin-like 4 in testis and can cause male infertility. Mol. Cell. Biol. 31, 2276–2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bazzoni G., Shih D. T., Buck C. A., and Hemler M. E. (1995) Monoclonal antibody 9EG7 defines a novel β 1 integrin epitope induced by soluble ligand and manganese, but inhibited by calcium. J. Biol. Chem. 270, 25570–25577 [DOI] [PubMed] [Google Scholar]
- 22. Sakai T., Zhang Q., Fässler R., and Mosher D. F. (1998) Modulation of β1A integrin functions by tyrosine residues in the beta1 cytoplasmic domain. J. Cell Biol. 141, 527–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Von Ballestrem C. G., Uniyal S., McCormick J. I., Chau T., Singh B., and Chan B. M. (1996) VLA-β 1 integrin subunit-specific monoclonal antibodies MB1.1 and MB1.2: binding to epitopes not dependent on thymocyte development or regulated by phorbol ester and divalent cations. Hybridoma 15, 125–132 [DOI] [PubMed] [Google Scholar]
- 24. Xu J. (2005) Preparation, culture, and immortalization of mouse embryonic fibroblasts. Curr. Protoc. Mol. Biol. Chapter 28, Unit 28.1 [DOI] [PubMed] [Google Scholar]
- 25. Tadokoro S., Shattil S. J., Eto K., Tai V., Liddington R. C., de Pereda J. M., Ginsberg M. H., and Calderwood D. A. (2003) Talin binding to integrin β tails: a final common step in integrin activation. Science 302, 103–106 [DOI] [PubMed] [Google Scholar]
- 26. Sieg D. J., Hauck C. R., and Schlaepfer D. D. (1999) Required role of focal adhesion kinase (FAK) for integrin-stimulated cell migration. J. Cell Sci. 112, 2677–2691 [DOI] [PubMed] [Google Scholar]
- 27. Ilić D., Furuta Y., Kanazawa S., Takeda N., Sobue K., Nakatsuji N., Nomura S., Fujimoto J., Okada M., and Yamamoto T. (1995) Reduced cell motility and enhanced focal adhesion contact formation in cells from FAK-deficient mice. Nature 377, 539–544 [DOI] [PubMed] [Google Scholar]
- 28. Parra M., Gascard P., Walensky L. D., Snyder S. H., Mohandas N., and Conboy J. G. (1998) Cloning and characterization of 4.1G (EPB41L2), a new member of the skeletal protein 4.1 (EPB41) gene family. Genomics 49, 298–306 [DOI] [PubMed] [Google Scholar]
- 29. Walensky L. D., Gascard P., Fields M. E., Blackshaw S., Conboy J. G., Mohandas N., and Snyder S. H. (1998) The 13-kD FK506 binding protein, FKBP13, interacts with a novel homologue of the erythrocyte membrane cytoskeletal protein 4.1. J. Cell Biol. 141, 143–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lu D., Yan H., Othman T., and Rivkees S. A. (2004) Cytoskeletal protein 4.1G is a binding partner of the metabotropic glutamate receptor subtype 1 α. J. Neurosci. Res. 78, 49–55 [DOI] [PubMed] [Google Scholar]
- 31. Lu D., Yan H., Othman T., Turner C. P., Woolf T., and Rivkees S. A. (2004) Cytoskeletal protein 4.1G binds to the third intracellular loop of the A1 adenosine receptor and inhibits receptor action. Biochem. J. 377, 51–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ralston K. J., Hird S. L., Zhang X., Scott J. L., Jin B., Thorne R. F., Berndt M. C., Boyd A. W., and Burns G. F. (2004) The LFA-1-associated molecule PTA-1 (CD226) on T cells forms a dynamic molecular complex with protein 4.1G and human discs large. J. Biol. Chem. 279, 33816–33828 [DOI] [PubMed] [Google Scholar]
- 33. Ivanovic A., Horresh I., Golan N., Spiegel I., Sabanay H., Frechter S., Ohno S., Terada N., Möbius W., Rosenbluth J., Brose N., and Peles E. (2012) The cytoskeletal adapter protein 4.1G organizes the internodes in peripheral myelinated nerves. J. Cell Biol. 196, 337–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Calderwood D. A., Campbell I. D., and Critchley D. R. (2013) Talins and kindlins: partners in integrin-mediated adhesion. Nat. Rev. Mol. Cell Biol. 14, 503–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Salomao M., Zhang X., Yang Y., Lee S., Hartwig J. H., Chasis J. A., Mohandas N., and An X. (2008) Protein 4.1R-dependent multiprotein complex: new insights into the structural organization of the red blood cell membrane. Proc. Natl. Acad. Sci. U.S.A. 105, 8026–8031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang S., Guo X., Debnath G., Mohandas N., and An X. (2009) Protein 4.1R links E-cadherin/β-catenin complex to the cytoskeleton through its direct interaction with β-catenin and modulates adherens junction integrity. Biochim. Biophys. Acta 1788, 1458–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Coleman S. K., Cai C., Mottershead D. G., Haapalahti J. P., and Keinänen K. (2003) Surface expression of GluR-D AMPA receptor is dependent on an interaction between its C-terminal domain and a 4.1 protein. J. Neurosci. 23, 798–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kang Q., Yu Y., Pei X., Hughes R., Heck S., Zhang X., Guo X., Halverson G., Mohandas N., and An X. (2009) Cytoskeletal protein 4.1R negatively regulates T-cell activation by inhibiting the phosphorylation of LAT. Blood 113, 6128–6137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Caswell P. T., and Norman J. C. (2006) Integrin trafficking and the control of cell migration. Traffic 7, 14–21 [DOI] [PubMed] [Google Scholar]
- 40. Pellinen T., and Ivaska J. (2006) Integrin traffic. J. Cell Sci. 119, 3723–3731 [DOI] [PubMed] [Google Scholar]
- 41. Krauss S. W., Chen C., Penman S., and Heald R. (2003) Nuclear actin and protein 4.1: essential interactions during nuclear assembly in vitro. Proc. Natl. Acad. Sci. U.S.A. 100, 10752–10757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Marfatia S. M., Lue R. A., Branton D., and Chishti A. H. (1994) In vitro binding studies suggest a membrane-associated complex between erythroid p55, protein 4.1, and glycophorin C. J. Biol. Chem. 269, 8631–8634 [PubMed] [Google Scholar]
- 43. Wang H., Liu C., Debnath G., Baines A. J., Conboy J. G., Mohandas N., and An X. (2010) Comprehensive characterization of expression patterns of protein 4.1 family members in mouse adrenal gland: implications for functions. Histochem. Cell Biol. 134, 411–420 [DOI] [PubMed] [Google Scholar]