Abstract
Protein arginine methyltransferase 1 (PRMT1) is involved in cell proliferation, DNA damage response, and transcriptional regulation. Although PRMT1 is extensively expressed in the CNS at embryonic and perinatal stages, the physiological role of PRMT1 has been poorly understood. Here, to investigate the primary function of PRMT1 in the CNS, we generated CNS-specific PRMT1 knock-out mice by the Cre-loxP system. These mice exhibited postnatal growth retardation with tremors, and most of them died within 2 weeks after birth. Brain histological analyses revealed prominent cell reduction in the white matter tracts of the mutant mice. Furthermore, ultrastructural analysis demonstrated that myelin sheath was almost completely ablated in the CNS of these animals. In agreement with hypomyelination, we also observed that most major myelin proteins including myelin basic protein (MBP), 2′,3′-cyclic-nucleotide 3′-phosphodiesterase (CNPase), and myelin-associated glycoprotein (MAG) were dramatically decreased, although neuronal and astrocytic markers were preserved in the brain of CNS-specific PRMT1 knock-out mice. These animals had a reduced number of OLIG2+ oligodendrocyte lineage cells in the white matter. We found that expressions of transcription factors essential for oligodendrocyte specification and further maturation were significantly suppressed in the brain of the mutant mice. Our findings provide evidence that PRMT1 is required for CNS development, especially for oligodendrocyte maturation processes.
Keywords: cell differentiation, central nervous system (CNS), myelin, oligodendrocyte, post-translational modification (PTM)
Introduction
CNS development is achieved by proliferation of progenitor cells followed by the transition from a proliferative state to differentiation. This process is considered to be tightly regulated at multiple levels such as gene transcription, translation, and protein modification. In the oligodendrocyte lineage, transcription factors are known to determine cell fate and timing of differentiation by inducing the target genes that are critical for myelination (1–3). Moreover, emerging evidence suggests that post-translational modification of proteins is one of the important determinants for oligodendrocyte lineage progression (4). For example, phosphorylation of retinoblastoma protein by AMP-activated protein kinase is essential for cell cycle progression in neural stem cells (NSCs)2 and their differentiation into cell types including neurons and oligodendrocytes (5). On the other hand, a histone lysine deacetylase, SIRT1, has a role in limiting the expansion of oligodendrocyte precursor cells (OPCs) (6). However, there is no in vivo evidence for the involvement of methylation enzymes in oligodendrocyte development and myelination.
Protein arginine methylation is now widely accepted as one of the major post-translational modifications observed in both histone and non-histone proteins. Protein arginine methyltransferase (PRMT) 1, one of the type I PRMTs that catalyze monomethylation and asymmetric dimethylation of proteins, regulates transcription, cell death, DNA damage responses, and signal transduction as reviewed elsewhere (7). In mammals, PRMT1 is ubiquitously expressed in various tissues (8–11).
Previous studies demonstrated that mouse embryos that lack PRMT1 were arrested in development before embryonic day 6.5, suggesting that arginine methylation by PRMT1 is crucial for early embryonic development (9). Interestingly, the distribution pattern of PRMT1 in mouse embryo has shown high expression in neural tube during embryonic stage (9). After birth, PRMT1 is still strongly expressed in the brain during the perinatal stage (12). It should be noted that PRMT5, the most characterized member of type II PRMTs that catalyze the monomethylation and symmetric dimethylation of proteins, also shows substantial expression in the brain (12). Specific deletion of PRMT5 in CNS results in postnatal lethality with extensive apoptosis of NSCs (13). In addition, Chittka et al. (14) have demonstrated that PRMT5 controls NSC proliferation and differentiation. These studies indicate the important role of PRMTs in the CNS; however, little is known about PRMT1 in the development of nervous tissues.
Here, our results demonstrate the essential function of PRMT1 for proper organization of the CNS by using PRMT1 conditional knock-out mice and highlights the crucial role in oligodendrocyte lineage.
Experimental Procedures
Animals
PRMT1 knock-in mice (PRMT1-KI) that carry Prmt1tm1a(EUCOMM)Wtsi allele were obtained from the European Conditional Mouse Mutagenesis (EUCOMM). For generation of Prmt1flox/flox mice, the gene trap cassette was removed by intercrossing β-actin-Flp transgenic mouse (strain name: B6.SJL-Tg(ACTFLPe)9205Dym/J, stock number: 003800, The Jackson Laboratory). Prmt1flox/flox mice were further crossed with Nestin-Cre transgenic mice (B6.Cg-Tg(Nes-cre)1Kln/J, stock number: 003771, The Jackson Laboratory) to create Prmt1flox/flox; Nes-Cre mice.
Genotyping of mice was performed by PCR using genomic DNA from tail as described previously (15). The primers were Prmt1 forward primer (5′-GTGCTTGCCATACAAGAGATCC-3′) and reverse primer (5′-ACAGCCGAGTAGCAAGGAGG-3′). The primer set amplifies 410- and 277-bp fragments for the floxed and wild-type alleles, respectively. Nestin-Cre transgene was amplified using forward primer (5′-ACTGAACGCTAAAGGGTTAAG-3′) and reverse primer (5′-GTGAAACAGCATTGCTGTCACTT-3′).
All animal experiments were done in compliance with and approved by the Institutional Animal Experiment Committee of the University of Tsukuba. All these experiments were conducted in accordance with the Regulations for Animal Experiments at our institution and with the Fundamental Guidelines for the Proper Conduct of Animal Experiments and Related Activities in Academic Research Institutions under the jurisdiction of the Ministry of Education, Culture, Sports, Science and Technology of Japan.
RNA Analysis
Whole brains were harvested at postnatal day 0 (P0) or P10 and frozen in liquid nitrogen and then stored in a deep freezer until use. Frozen tissues were crushed into powder with a Multi-beads Shocker (Yasui Kikai Co., Osaka, Japan). From a subset of the tissue powder, brain total RNA was isolated with ISOGEN II (Nippon Gene, Ltd., Tokyo, Japan) following the manufacturer's instructions. 5 μg of the total RNA were treated with RQ1 RNase-Free DNase (Promega) and then reverse-transcribed into cDNA with ReverTra Ace (Toyobo Co., Ltd., Osaka, Japan). Relative gene expression level was determined by SYBR Green-based quantitative RT-PCR (Takara Bio Inc., Shiga, Japan). Expression levels of target gene were corrected for Gapdh expression levels by using ΔΔCt method. The amplification efficiency of primers for each target gene was confirmed to be equal by using serial dilutions of cDNA. The primer sequences were as follows: Olig1 (5′-CTCGCCCAGGTGTTTTGTTG-3′ and 5′-TAAGTCCGAACACCGATGGC-3′), Olig2 (5′-GAACCCCGAAAGGTGTGGAT-3′ and 5′-TTCCGAATGTGAATTAGATTTGAGG-3′),Nkx2.2 (5′-CCTTTCTACGACAGCAGCGA-3′ and 5′-CCGTGCAGGGAGTATTGGAG-3′), Sox10 (5′-GCAGAAAGCTAGCCGACCA-3′ and 5′-CTTTCGTTCAGCAACCTCCAGA-3′), and Gapdh (5′-TCACTGGCATGGCCTTCC-3′ and 5′-CAGGCGGCACGTCAGATC-3′).
Protein Analysis
Brain and kidney tissue powder was prepared as described above and was homogenized in ice-cold radioimmunoprecipitation assay buffer containing 20 mm Tris-HCl, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, and a proteinase inhibitor mixture (Nacalai Tesque, Inc., Kyoto, Japan). Protein extracts were centrifuged at 14,000 rpm for 15 min at 4 °C, and then concentration was determined by Bio-Rad protein assay dye reagent concentrate (Bio-Rad). Quantified total protein extracts were denatured with Laemmli sample buffer supplemented with 100 mm DTT, and then subjected to 10 or 12% SDS-PAGE and analyzed by Western blot with various antibodies, according to standard procedures. The antibodies are as follows: anti-PRMT1 (1:1000, rabbit polyclonal; catalog number 07-404, Millipore), anti-MBP (1:100, goat polyclonal; catalog number sc-13914, Santa Cruz Biotechnology), anti-CNPase (1:1000, rabbit monoclonal; catalog number 5664, Cell Signaling Technology), anti-MAG (1:1000, rabbit monoclonal; catalog number 9043, Cell Signaling Technology), anti-CRMP2 (1:1000, rabbit polyclonal; catalog number 9393, Cell Signaling Technology), anti-βIII-tubulin (1:1000, rabbit monoclonal; catalog number 5568, Cell Signaling Technology), anti-glial fibrillary acidic protein (GFAP) (1:1000, mouse monoclonal; catalog number G3893, Sigma), anti-asymmetric dimethyl arginine, Asym26 (1:1000, rabbit polyclonal; catalog number 13-0011, EpiCypher), anti-mono-methyl arginine (R*GG) (D5A12) (1:1000, rabbit monoclonal; catalog number 8711, Cell Signaling Technology), anti-symmetric dimethyl arginine (SDMA) motif (1:1000, rabbit monoclonal mix; catalog number 13222, Cell Signaling Technology), anti-GAPDH (1:10000, mouse monoclonal; catalog number 05-50118, American Research Products), and anti-β-actin (1:10000, mouse monoclonal; catalog number A5316, Sigma). Secondary antibodies are corresponding HRP-conjugated secondary antibodies. Visualization via chemiluminescent detection was performed using SuperSignal West Femto maximum sensitivity substrate (Thermo Fisher Scientific). Data images were obtained from an LAS-3000 (Fujifilm, Tokyo, Japan).
Histology
Mice were transcardially perfused with 4% paraformaldehyde, immersed in the same fixative for 24 h at 4 °C, and embedded in paraffin. Tissues were sectioned into 7-μm thickness using a rotary microtome (HM340E; Microm International GmbH, Walldorf, Germany). Brains were sectioned either in the midline sagittal plane or in the coronal section at the level of the mid-cerebellum and the forebrain. Spinal cord tissues were sectioned at the thoracic level with the surrounding bones. Deparaffinized sections were stained with H&E. Data images were obtained using a SZ61 microscope, a BX53 microscope, and a DP21 digital camera (Olympus, Tokyo, Japan). Three to five mice per each genotype were used for histological studies.
Immunohistochemistry
For immunohistochemistry, tissue sections were deparaffinized, followed by antigen retrieval with citrate buffer (pH 6.0), and then incubated in 3% H2O2 for 30 min to quench an endogenous peroxidase. Sections were incubated with TSA blocking reagent (catalog number FP1020, PerkinElmer) for 30 min and with each primary antibody for 1 h at room temperature. The antibodies used were as follows: anti-MBP (1:1000, goat polyclonal; catalog number sc-13914, Santa Cruz Biotechnology) and anti-OLIG2 (1:1000–1:2000, rabbit polyclonal; catalog number 18953, Immuno-Biological Laboratories, Gunma, Japan). Biotinylated horse anti-goat IgG (catalog number BA-9500, Vector Laboratories) or biotinylated goat anti-rabbit IgG (catalog number BA-1000, Vector Laboratories) was incubated for 30 min, and then reacted with streptavidin-HRP (catalog number NEL750001EA, PerkinElmer) for 30 min. Signal was amplified with the TSA Plus fluorescein system or TSA Plus tetramethylrhodamine (TMR) system (catalog number NEL756001KT PerkinElmer) for 10 min, followed by Hoechst counterstaining. Fluorescence images were obtained with a fluorescence microscope (BIOREVO BZ-9000, Keyence, Osaka, Japan) and FluoView FV10i confocal laser-scanning microscope (Olympus, Tokyo, Japan). For the quantification of immunohistochemical data, three mice per each genotype were evaluated with the ImageJ software.
Transmission Electron Microscopy
For ultrastructure analysis, mice were perfused with 2% glutaraldehyde/2% paraformaldehyde, and then brain and spinal cord were dissected, followed by immersion fixation in 2.5% glutaraldehyde in 0.1 m phosphate buffer. Tissues were then processed as standard procedure, and ultrathin sections were subjected to transmission electron microscopy analysis with a JEM-1400 electron microscope (JEOL USA) at the electron microscopy facility at the University of Tsukuba. For transmission electron microscope analysis, multiple numbers of sections from 2–3 mice per each genotype were analyzed.
LC-MS/MS Analysis
Brain tissue powder was prepared as described above and was homogenized in ice-cold deionized water. Protein extracts were centrifuged at 14,000 rpm for 10 min at 4 °C, and then concentration was determined by the Bio-Rad DC protein assay (Bio-Rad). Quantified total protein extracts were further precipitated with 10% TCA in cold acetone. The total proteins were hydrolyzed with 6 n HCl at 110 °C for 24 h. After hydrolysis, the residues were evaporated in vacuo, and then reconstituted in 100 μl of deionized water for the following LC-MS/MS analysis.
The LC-MS/MS analysis was performed on a Shimadzu LCMS-8050 triple quadrupole mass spectrometer coupled with a Shimadzu Nexera X2 ultra-high pressure liquid chromatography (UHPLC) system (Shimadzu, Kyoto, Japan). The instrument was operated under positive electrospray ionization and multiple reaction monitoring modes. The chromatography separation was performed with a SeQuant ZIC-HILIC column, 2.1 × 150 mm, 3.5 μm (Millipore) with gradient elution comprising distilled water containing 1% formic acid and 1% acetonitrile (mobile phase A) and acetonitrile containing 1% formic acid and 1% water (mobile phase B). A ZIC-HILIC guard-fitting column, 1.0 × 14 mm (Millipore), was installed before the SeQuant ZIC-HILIC column for the sake of sample cleanup. The MS operating conditions were optimized as follows: interface voltage, 4.0 kV; interface temperature, 300 °C; desolvation line temperature, 250 °C; heating block temperature, 400 °C; drying gas (N2); 10 liters min−1; nebulizing gas (N2); 3 liters min−1; heating gas (air), 10 liters min−1; and collision-induced dissociation gas (argon), 230 kilopascals. For the detection of asymmetric dimethyl arginine (ADMA), monomethyl arginine (MMA), and SDMA, the gradient program for positive ionization method started from 95% mobile phase B at 0–1 min, was decreased at 5–10 min, and then was stopped at 14 min. The mobile phase B content was further increased to 95% at 14.10 min and stopped at 21 min with a flow rate of 0.2 ml/min. The m/z values for each analyte are as follow: ADMA, 203.15 > 158.05; MMA, 189.15 > 70.15; SDMA, 203.25 > 171.95. All analyses and data processing were completed with the LabSolutions V5.60 software (Shimadzu Scientific Instruments, Inc., Columbia, MD). Four mice per each genotype were used for LC-MS/MS analysis.
Results
PRMT1 Deletion in the CNS Causes Postnatal Lethality of Mice
To address the primary function of PRMT1 in the CNS, we produced CNS-specific PRMT1 knock-out mice. Prmt1KI/KI mice were mated with β-actin-Flp transgenic mice to produce Prmt1flox/flox mice. In Prmt1flox allele, exons 4 and 5, which encode a part of the methyltransferase domains, are flanked by two loxP sites, enabling us to obtain functional null allele by Cre-mediated recombination. Prmt1flox/flox mice were crossed to animals expressing Nestin-Cre (Nes-Cre) to generate Prmt1flox/flox; Nes-Cre animals (referred to as CKO in the figures) (Fig. 1, A and B) (16). In the whole brain lysates from embryos and postnatal mice, we confirmed the marked reduction of PRMT1 protein level by Western blot, whereas other tissues such as kidney expressed PRMT1 normally (Fig. 1, C and D). In the brain, there were faint remnants of expression that were possibly from Nestin-Cre inactive components such as vessels or from incomplete recombination at the Prmt1 gene locus.
FIGURE 1.
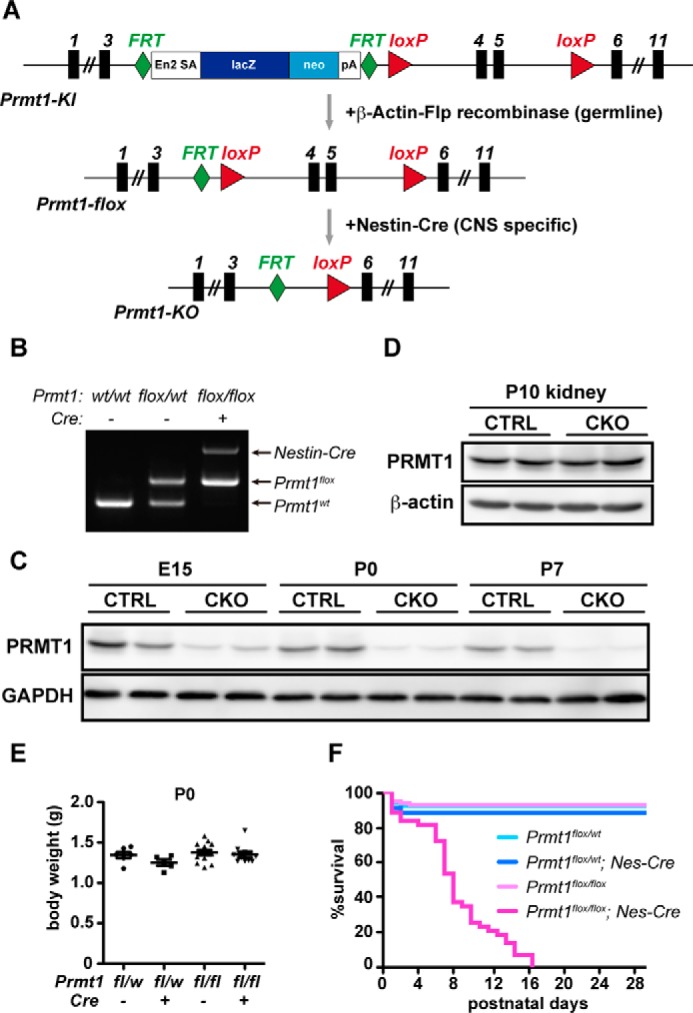
PRMT1 deletion in the CNS causes postnatal lethality of mice. A, schematic view of Prmt1 gene targeting design used in this study. Black rectangles represent exons, and gray arrows indicate mating with other transgenic mice for gene recombination. B, genotyping PCR for Prmt1 allele and Nestin-Cre (Nes-Cre) allele. C, Western blot showing developmental time course of PRMT1 protein expression in the brain of Prmt1flox/flox (hereafter, CTRL) and Prmt1flox/flox; Nes-Cre (hereafter, CKO) mice. D, Western blot showing PRMT1 protein expression in the kidney of CTRL and CKO mice at P10. E, body weights of Prmt1flox/wt (n = 6), Prmt1flox/wt; Nes-Cre (n = 5), Prmt1flox/flox (n = 13), and Prmt1flox/flox; Nes-Cre mice (n = 10) at P0. The points in the graph show individual data sets from each animal with lines and error bars indicating mean ± S.E. F, Kaplan-Meier survival analysis of Prmt1flox/flox (n = 106), Prmt1flox/wt; Nes-Cre (n = 88), Prmt1flox/wt (n = 57), and Prmt1flox/flox; Nes-Cre mice (n = 47).
Prmt1flox/flox; Nes-Cre animals were born close to the normal Mendelian ratio (Table 1) and appeared to be normal at birth. Their body weights were similar when compared with other littermates including Prmt1flox/flox, Prmt1flox/wt; Nes-Cre, and Prmt1flox/wt on the day of birth (Fig. 1E). However, Prmt1flox/flox; Nes-Cre animals displayed postnatal lethality, and all mice in this group died within 17 days after birth (Fig. 1F).
TABLE 1.
Genotypes of offspring obtained from the cross of Prmt1flox/wt; Nes-Cre mice and Prmt1flox/flox mice
The numbers and percentages (in parentheses) of mice of various genotypes obtained by crosses of Prmt1flox/wt; Nes-Cre and Prmt1flox/flox at P0 are shown. The expected numbers and percentages of mice were calculated according to the total number of mice born and based on the expected Mendelian 1:1:1:1 ratio.
| Prmt1 | Cre | Expected no. (%) | Observed no. (%) |
|---|---|---|---|
| fl/w | − | 86 (25) | 71 (21) |
| fl/w | + | 86 (25) | 90 (26) |
| fl/fl | − | 86 (25) | 109 (32) |
| fl/fl | + | 86 (25) | 73 (21) |
| Total 343 (100) |
Loss of PRMT1 in the CNS Results in Dynamic Changes in Methyl Arginine Levels in the Brain
Because PRMT1 catalyzes the formation of ADMA in proteins, we performed acid hydrolysis of proteins from the whole brain tissue and measured the level of ADMA by LC-MS/MS. As shown in Fig. 2A, ADMA levels in Prmt1flox/flox; Nes-Cre mice were almost 2-fold lower than those of Prmt1flox/flox mice. The decrease of tissue ADMA levels was also confirmed by Western blot using an antibody against ADMA (Fig. 2B). Altogether, these results indicate that PRMT1 serves as a major methyltransferase that catalyzes the formation of ADMA in the mouse brain.
FIGURE 2.
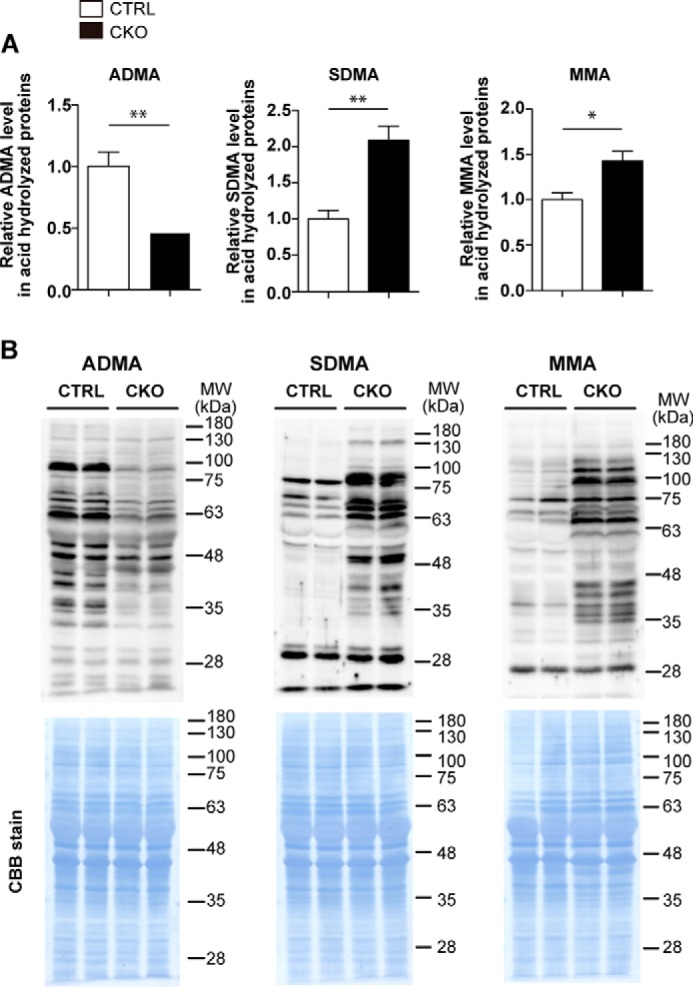
Loss of PRMT1 in the CNS results in dynamic change in methyl arginine levels in the brain. A, ADMA, SDMA, and MMA levels in the brain of CTRL and CKO mice were quantitated by LC-MS/MS at P10. Data were obtained from acid hydrolysate of proteins from each brain sample. The data shown are mean ± S.E. and were analyzed by a two-tailed Student's t test. n = 4 per each genotype.*, p < 0.05, **, p < 0.01. B, ADMA, SDMA, and MMA levels in the whole brains were detected by Western blot (upper panels) at P10, and loaded protein samples on membrane were confirmed to be equal by Coomassie Brilliant Blue (CBB) stain (lower panels). Molecular weight (MW) markers are indicated in kDa on the right of each panel.
Deletion of PRMT1 in mouse embryonic fibroblasts induces up-regulation of MMA and SDMA levels (17). We therefore examined the level of MMA and SDMA in brain tissues by LC-MS/MS and Western blot. Both MMA and SDMA levels showed dramatic elevation in Prmt1flox/flox; Nes-Cre pups when compared with Prmt1flox/flox mice (Fig. 2, A and B), suggesting that the loss of PRMT1 actually affected the status of protein arginine methylation on multiple proteins in agreement with the previous in vitro work (17).
Growth Retardation and Reduced Nuclei in the White Matter in Prmt1flox/flox; Nes-Cre Mice
We then characterized the brain morphology and the behavioral outcomes to clarify the effect of PRMT1 deletion in the CNS. Prmt1flox/flox; Nes-Cre animals started to show growth retardation at about P4 (Fig. 3A); their body size was remarkably small at P10 with reduced brain size and weight, and they were noticeably emaciated (Fig. 3, B and C). Histological analysis of nervous tissues revealed that all white matter tracts in the CNS including corpus callosum, fimbria, anterior commissure, and spinal white matter had a reduced number of nuclei as indicated by hematoxylin staining. Although nuclei in both white and gray matter areas were decreased in the spinal cords, a more prominent difference was observed in the white matter (Fig. 4). Considering that white matter tracts consist of predominantly oligodendrocytes and a much lower number of nuclei of neuron and astrocytes, our data imply that cellular organizations are changed in the CNS tissues of Prmt1flox/flox; Nes-Cre mice.
FIGURE 3.
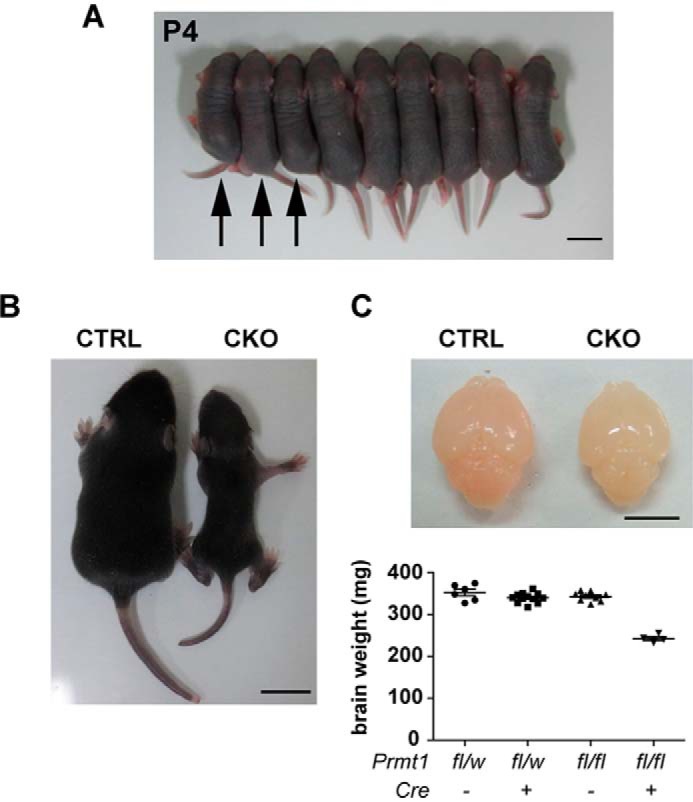
Growth retardation in Prmt1flox/flox; Nes-Cre mice. A, photograph of representative CKO mice and other littermate controls at P4. The left three pups (arrows) are confirmed to be Prmt1flox/flox; Nes-Cre by PCR analysis and smaller than other littermates. Other control littermates include Prmt1flox/flox and Prmt1flox/wt; Nes-Cre mice. Scale bar represents 1 cm. B, representative example of body sizes of CTRL and CKO mice at P10. Scale bar represents 1 cm. C, the upper panel shows the representative brain size, and the lower graph shows the fresh brain weights from littermate controls at P10. Scale bar represents 5 mm. The points in the graph show individual data sets from each animal with lines and error bars indicating mean ± S.E. n = 4–12 per each genotype.
FIGURE 4.
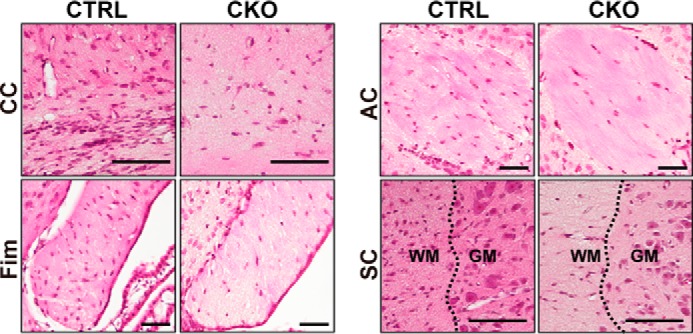
Histological abnormalities of the white matter parts in Prmt1flox/flox; Nes-Cre mice. Hematoxylin and eosin staining images of brains and spinal cords from CTRL and CKO mice at P10 obtained from coronal sections of tissues are shown. CC, corpus callosum; Fim, fimbria; AC, anterior commissure; SC, spinal cord. Dotted lines represent the boundary of white matter (WM) and gray matter (GM). Scale bars represent 100 μm.
Behavioral Deficits in Prmt1flox/flox; Nes-Cre Mice
Remarkably, Prmt1flox/flox; Nes-Cre mice manifested ataxic behavior and tremors, which became evident after 1 week of birth (supplemental Video S1). This behavioral defect is reminiscent of mouse models with hypomyelination such as myelin-deficient shiverer mice (shi) (18) or quaking viable mice (qkv) (19, 20). The similar behavioral phenotype in Prmt1flox/flox; Nes-Cre animals prompted us to analyze the level of myelination in the CNS.
Severe Hypomyelination and Dramatic Decrease of Mature Oligodendrocytes in Prmt1flox/flox; Nes-Cre Mice
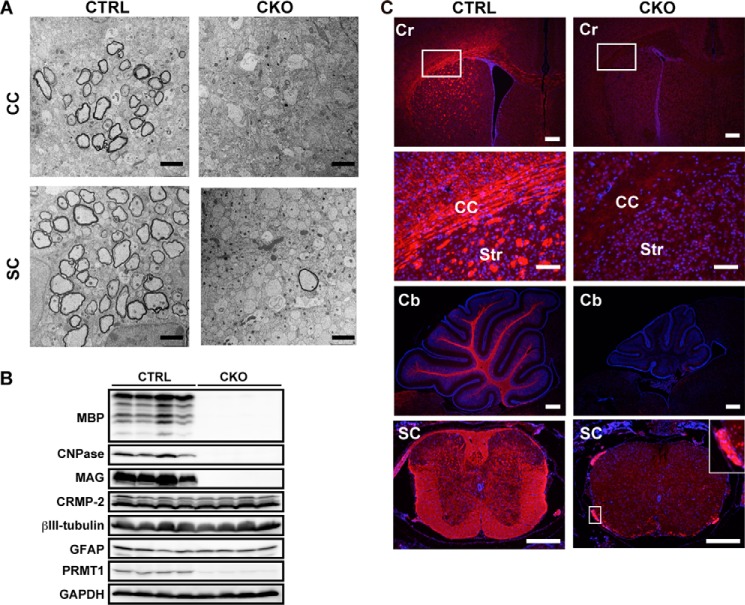
Myelin is a multilamellar structure that is formed by mature oligodendrocytes and is wrapping around a number of axons, allowing rapid saltatory conduction in the CNS. We evaluated myelin formation at P10 because myelination actively occurs from the first week to the third week after birth in rodents (21), and we had difficulties in obtaining tissues after the second week due to the short life span of Prmt1flox/flox; Nes-Cre mice. To examine the ultrastructure of myelin sheaths, we performed transmission electron microscope analysis and confirmed that a substantial number of axons were myelinated in Prmt1flox/flox mice spinal cord and brain. By contrast, in the spinal cord, only a few axons were myelinated in Prmt1flox/flox; Nes-Cre mice. Similarly, myelin structure was totally ablated in the corpus callosum of Prmt1flox/flox; Nes-Cre mice (Fig. 5A). Thus, transmission electron microscope analysis clearly showed hypomyelination in the CNS tissues of Prmt1flox/flox; Nes-Cre mice.
FIGURE 5.
Severe hypomyelination and dramatic decrease of mature oligodendrocytes in Prmt1flox/flox; Nes-Cre mice. A, representative electron micrograph images of corpus callosum (CC) and spinal cord (SC) of CTRL and CKO mice at P10. B, Western blot of MBP, CNPase, MAG, CRMP-2, βIII-tubulin, glial fibrillary acidic protein (GFAP), and PRMT1 expression in the brain of CTRL and CKO mice at P10. C, immunostaining images of MBP (red) with nuclear counterstain Hoechst (blue) in the cerebrum (Cr), cerebellum (Cb), and spinal cord (SC) of CTRL and CKO mice at P10. The white boxed region from the Cr panels is magnified in the CC panels. The white boxed region from the SC panels is magnified and shown in the inset (right side). Scale bars represent 2 μm in A; 300 μm in cerebrum, cerebellum, and spinal cord in C; and 100 μm in magnified images of corpus callosum in C. Str, striatum.
Mature oligodendrocytes produce myelin-specific proteins such as myelin basic protein (MBP), 2′,3′-cyclic-nucleotide 3′-phosphodiesterase (CNPase), and myelin-associated glycoprotein (MAG) (22, 23), allowing compact myelin formation and axon stability. The myelin protein levels were dramatically decreased in the Prmt1flox/flox; Nes-Cre mice when compared with Prmt1flox/flox mice as confirmed by Western blot. On the other hand, neuronal and astrocytic markers were observed in the same level between Prmt1flox/flox and Prmt1flox/flox; Nes-Cre brain (Fig. 5B). Consistent with those findings, immunohistochemical analysis revealed the near complete loss of MBP-positive signals in whole brain regions including the white matter tracts in the corpus callosum, striatum, cerebellum, and spinal cord in Prmt1flox/flox; Nes-Cre mice (Fig. 5C), although there were still intense signals in the spinal roots that are composed of peripheral nerves (Fig. 5C, inset). Collectively, these data demonstrated extensive dysmyelination caused by the dramatic reduction of mature oligodendrocytes in the CNS tissues of Prmt1flox/flox; Nes-Cre mice.
Oligodendrocyte Lineage Cells Were Reduced in Prmt1flox/flox; Nes-Cre Mice
Myelinating mature oligodendrocytes are developmentally derived from highly proliferative OPCs that were originally from NSCs. OPCs develop into premyelinating oligodendrocytes and then terminally differentiate into mature myelinating oligodendrocytes (1). Because we observed the near complete loss of mature oligodendrocytes, we investigated whether OPCs and premyelinating oligodendrocytes are also absent by immunohistochemistry for OLIG2, which indicates all oligodendrocyte lineage cells, and MBP, which indicates only mature oligodendrocytes. To exclude mature oligodendrocytes from all OLIG2+ cells, we counted the number of OLIG2+MBP− cells in the striatum and assumed them to be OPCs or immature oligodendrocytes. We detected a reduced number of OLIG2+ cells not only in the striatum but also in the cerebellum and spinal cord in Prmt1flox/flox; Nes-Cre mice, suggesting that oligodendrocyte lineage progression is not completely but only partially suppressed (Fig. 6, A and B, upper panels). Furthermore, Prmt1flox/flox; Nes-Cre mice had less than half the number of OLIG2+MBP− cells relative to Prmt1flox/flox mice in the striatum (Fig. 6A, lower panels and graph). Therefore, these results show that OPCs and/or premyelinating oligodendrocytes are dramatically reduced in PRMT1 mutant mice.
FIGURE 6.
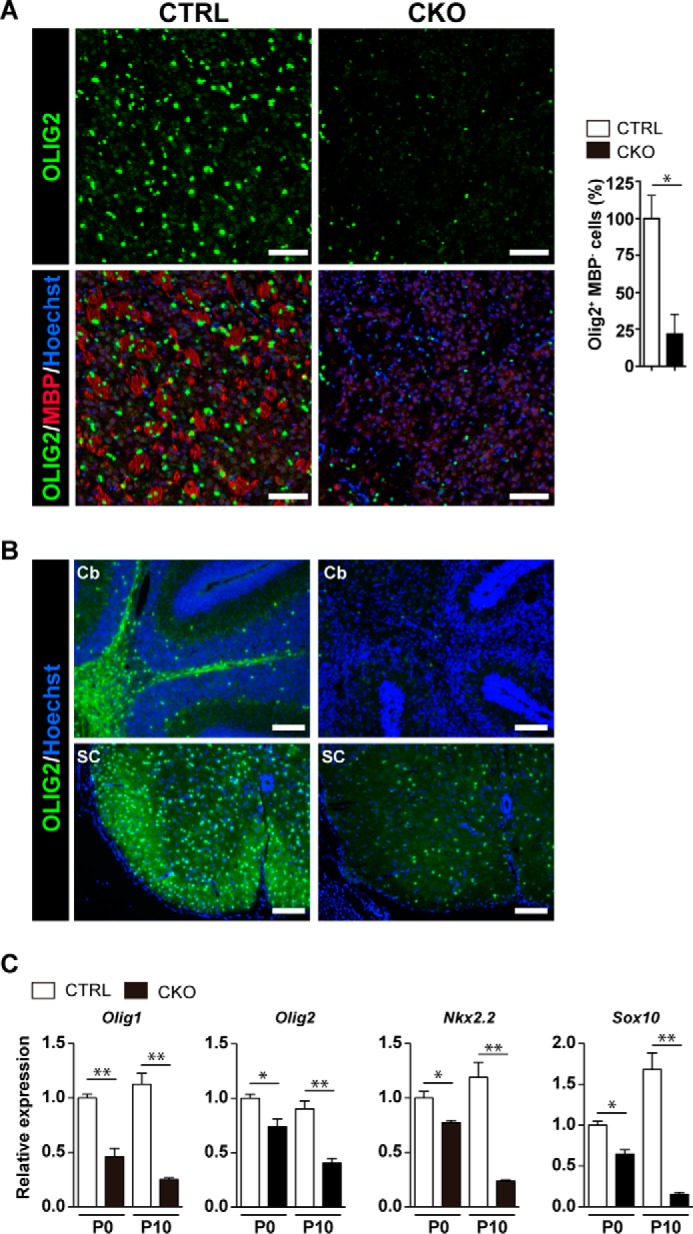
Oligodendrocyte lineage cells were reduced in Prmt1flox/flox; Nes-Cre mice. A, immunostaining of OLIG2 (green) and MBP (red) with nuclear counterstain Hoechst (blue) in the striatum of CTRL and CKO mice at P10. The graph on the right shows the quantification of the number of OLIG2+MBP− cells in the striatum. Numbers represent the average of three sections per each genotype from a single experiment. The data shown are mean ± S.E. and analyzed by a two-tailed Student's t test. n = 3 per each genotype. *, p < 0.05 B, immunostaining of OLIG2 (green) with nuclear counterstain Hoechst (blue) in the cerebellum (Cb) and the spinal cord (SC) of CTRL and CKO mice at P10. C, relative gene expression of Olig1, Olig2, Nkx2.2, and Sox10 in the brain of CTRL and CKO mice at P0 and P10 as determined by quantitative RT-PCR. All data were normalized to Gapdh. All data shown are mean ± S.E. and were analyzed by a two-tailed Student's t test. n = 4 per each genotype.*, p < 0.05, **, p < 0.01 Scale bars represent 100 μm.
Oligodendrocyte development is mainly promoted by several key transcription factors, and many of these proteins belong to the basic-helix-loop-helix (bHLH) protein family, homeodomain proteins, and high mobility group (HMG) domain proteins. In particular, Olig1, Olig2, Nkx2.2, and Sox10 are well characterized as positive regulators of oligodendrocyte differentiation (23–27). Although Olig2 is necessary for specification of oligodendrocyte lineages, Olig1, Nkx2.2, and Sox10 serve as modulators of later differentiation, for example by adjusting the timing of oligodendrocyte differentiation (3). Quantitative RT-PCR analysis revealed a significant reduction of these genes in Prmt1flox/flox; Nes-Cre brain at birth. Moreover, the differences became more evident at P10 when oligodendrocyte maturation is most active under normal conditions (Fig. 6C). Thus, the down-regulation of these transcription factors may explain the reason for suppressed differentiation of oligodendrocyte and/or the reduced number of OLIG2+ cells. Taken together, these data indicated that PRMT1 is a key regulator of oligodendrocyte differentiation during the CNS development.
Discussion
The aim of this study was to clarify the physiological role of PRMT1 in the CNS. CNS-specific PRMT1 knock-out mice exhibited morphological abnormalities of the brain, and many of them died within 2 weeks after birth. Because they displayed severe tremors, we analyzed CNS myelination and found that they exhibit nearly complete loss of myelinating oligodendrocytes. In addition, oligodendrocyte lineages were significantly decreased in CNS tissues. These results suggest that PRMT1 plays a crucial role in oligodendrocyte differentiation and CNS development.
We showed that MBP+ mature oligodendrocytes were almost lost in CNS-specific PRMT1 knock-out mice. Furthermore, we found only a small number of OLIG2+ oligodendrocyte lineages and OLIG2+MBP− OPCs/immature oligodendrocytes in the mutant animals. At least two possibilities may explain the decrease of these cells in CNS. First, it could be caused by the decreased level of OPCs proliferation. OPCs increase their number before differentiation into postmitotic oligodendrocytes (28). Thus, failure of this process will directly affect the number of OLIG2+ oligodendrocyte lineage cells. It is also likely that the increased cell death of OPCs affected the lower cell number, for example by cell cycle deregulation (5). By contrast, an alternative explanation is an altered differentiation potential of NSCs. In other words, PRMT1-deleted NSCs might have a low ability to differentiate into OPCs. From these possible mechanisms, PRMT1 may modulate oligodendrocyte development not only at terminal differentiation/myelination but also at the earlier point of differentiation.
In the present study, we found that PRMT1 plays a critical role in oligodendrocyte lineage progression. A previous study has shown that PRMT5 knockdown in C6 rat glioma cells and primary rat OPCs led to repressed maturation of oligodendrocyte, indicating that PRMT5 is important for oligodendrocyte differentiation (12). These studies suggest that both PRMT1 and PRMT5 positively regulate oligodendrocyte development. It is of note that PRMT1 and PRMT5 show distinct temporal expression patterns in the brain (12). Although PRMT1 shows a strong expression at birth and then decreases as mice grow, PRMT5 gradually increases after P11 and peaks at adult stage (12). Given these distinct expression patterns, it is likely that PRMT1 and PRMT5 control oligodendrocyte maturation in different time points during CNS development.
Oligodendrocyte differentiation is tightly regulated by various key transcription factors. Several studies have shown that their interplay is important for proper oligodendrocyte development (29–31). Id2 and Id4, which are known oligodendrocyte differentiation inhibitors, were up-regulated upon knockdown of PRMT5 in rat primary OPCs and the C6 glioma cell line (12). In C6 glioma cells, PRMT5 symmetrically dimethylated arginine 3 of histone H4 (H4R3) to lead to repression of these genes (12). In general, although symmetric dimethylation of H4R3 (H4R3me2s) causes transcriptional repression (32), asymmetric dimethylation of H4R3 (H4R3me2a) by PRMT1 induces gene transcriptional activation by permitting subsequent acetylation of chromatin, leading to the establishment of an active state of chromatin (33, 34). In addition, a growing amount of evidence suggests that PRMT1 positively regulates gene expression by methylating transcriptional factors and transcription coactivators (35–37). In light of these findings, PRMT1 may regulate the function of key transcription factors including Olig2, Sox10, or Nkx2.2. by methylation and modulate the timing of oligodendroglial maturation. Interestingly, although these positive regulators of differentiation were clearly decreased (Fig. 6C), differentiation inhibitors including Id2 and Id4 were not changed in the brain of CNS-specific PRMT1 knock-out mice at P0 and P10 (data not shown). This observation also supports our novel hypothetical view that PRMT1 and PRMT5 have different regulatory mechanisms of oligodendrocyte differentiation.
CNS-specific PRMT1 knock-out neonates exhibited small body size, and consequently, most of them died in 2 weeks. A famous hypomyelination model, shiverer mouse (shi), has autosomal recessive mutation in Mbp gene. Shi mutant mice are seemingly normal at birth but develop tremors later and die within 5 months (18). When compared with shi, our model has a shorter life span and is remarkably small and emaciated. Because both models show nearly complete absence of myelination, the difference in life span and growth may not be explained just by defective myelination. In many cases, gene deletion of positive regulator of oligodendrocyte differentiation factors results in perinatal death. For instance, Sox10-deficient mice show defective oligodendrocyte development and die either before birth or at birth (27, 38). Olig2 null mutant show the complete absence of oligodendrocyte lineages and die at the day of birth (24, 25). In the course of oligodendrocyte development, Olig2 and Sox10 regulate initial and later differentiation, and in comparison, Mbp is crucial for establishing myelin structure. Considering that these proteins temporally regulate oligodendrocyte development, PRMT1 may be important for cell commitment to oligodendrocyte lineage before myelination. If so, it is reasonable that our model dies earlier than the shi mutant. Another possible reason for postnatal death in CNS-specific PRMT1 knock-out animals may be attributed to further deficiency in other cell lineages besides OPC/oligodendrocytes, although we observed neuronal and astrocytic markers in a similar level between the mutant and control mice.
In conclusion, this study provides evidence that PRMT1 is critical for oligodendrocyte lineage progression. In addition, decreased numbers of oligodendrocyte lineage cells suggest that PRMT1 is a key regulator for the cell development of NSCs and OPCs/immature oligodendrocytes in the CNS. Finally, the present genetic model will enable future challenges on analyses of additional functions of PRMT1 in the CNS.
Author Contributions
M. H., K. M., J. I., and A. F. designed the research; M. H. and A. K. performed the experiments and analyzed the data; M. H., K. M., and A. F. wrote the manuscript; and M. H., K. M., J. I., Y. K., and A. F. discussed the results and commented on the manuscript.
Supplementary Material
Acknowledgment
We thank Yuko Jinzenji (University of Tsukuba) for technical assistance.
This work was supported by JSPS KAKENHI Grant Number 25252062. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Video S1.
- NSC
- neural stem cell
- OPC
- oligodendrocyte precursor cell
- PRMT
- protein arginine methyltransferase
- MBP
- myelin basic protein
- CNPase
- 2′,3′-cyclic-nucleotide 3′-phosphodiesterase
- MAG
- myelin-associated glycoprotein
- p
- postnatal day
- SDMA
- symmetric dimethyl arginine
- ADMA
- asymmetric dimethyl arginine
- MMA
- monomethyl arginine
- KI
- knock-in
- CKO
- conditional knock-out
- CTRL
- control.
References
- 1. Emery B. (2010) Regulation of oligodendrocyte differentiation and myelination. Science 330, 779–782 [DOI] [PubMed] [Google Scholar]
- 2. Ross S. E., Greenberg M. E., and Stiles C. D. (2003) Basic helix-loop-helix factors in cortical development. Neuron 39, 13–25 [DOI] [PubMed] [Google Scholar]
- 3. Rowitch D. H. (2004) Glial specification in the vertebrate neural tube. Nat. Rev. Neurosci. 5, 409–419 [DOI] [PubMed] [Google Scholar]
- 4. Copray S., Huynh J. L., Sher F., Casaccia-Bonnefil P., and Boddeke E. (2009) Epigenetic mechanisms facilitating oligodendrocyte development, maturation, and aging. Glia 57, 1579–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dasgupta B., and Milbrandt J. (2009) AMP-activated protein kinase phosphorylates retinoblastoma protein to control mammalian brain development. Dev. Cell 16, 256–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rafalski V. A., Ho P. P., Brett J. O., Ucar D., Dugas J. C., Pollina E. A., Chow L. M., Ibrahim A., Baker S. J., Barres B. A., Steinman L., and Brunet A. (2013) Expansion of oligodendrocyte progenitor cells following SIRT1 inactivation in the adult brain. Nat. Cell Biol. 15, 614–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bedford M. T., and Clarke S. G. (2009) Protein arginine methylation in mammals: who, what, and why. Mol. Cell 33, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bulau P., Zakrzewicz D., Kitowska K., Leiper J., Gunther A., Grimminger F., and Eickelberg O. (2007) Analysis of methylarginine metabolism in the cardiovascular system identifies the lung as a major source of ADMA. Am. J. Physiol. Lung Cell. Mol. Physiol. 292, L18–L24 [DOI] [PubMed] [Google Scholar]
- 9. Pawlak M. R., Scherer C. A., Chen J., Roshon M. J., and Ruley H. E. (2000) Arginine N-methyltransferase 1 is required for early postimplantation mouse development, but cells deficient in the enzyme are viable. Mol. Cell. Biol. 20, 4859–4869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lin W. J., Gary J. D., Yang M. C., Clarke S., and Herschman H. R. (1996) The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J. Biol. Chem. 271, 15034–15044 [DOI] [PubMed] [Google Scholar]
- 11. Hong E., Lim Y., Lee E., Oh M., and Kwon D. (2012) Tissue-specific and age-dependent expression of protein arginine methyltransferases (PRMTs) in male rat tissues. Biogerontology 13, 329–336 [DOI] [PubMed] [Google Scholar]
- 12. Huang J., Vogel G., Yu Z., Almazan G., and Richard S. (2011) Type II arginine methyltransferase PRMT5 regulates gene expression of inhibitors of differentiation/DNA binding Id2 and Id4 during glial cell differentiation. J. Biol. Chem. 286, 44424–44432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bezzi M., Teo S. X., Muller J., Mok W. C., Sahu S. K., Vardy L. A., Bonday Z. Q., and Guccione E. (2013) Regulation of constitutive and alternative splicing by PRMT5 reveals a role for Mdm4 pre-mRNA in sensing defects in the spliceosomal machinery. Genes Dev. 27, 1903–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chittka A., Nitarska J., Grazini U., and Richardson W. D. (2012) Transcription factor positive regulatory domain 4 (PRDM4) recruits protein arginine methyltransferase 5 (PRMT5) to mediate histone arginine methylation and control neural stem cell proliferation and differentiation. J. Biol. Chem. 287, 42995–43006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hamada J., Baasanjav A., Ono N., Murata K., Kako K., Ishida J., and Fukamizu A. (2015) Possible involvement of downregulation of the apelin-APJ system in doxorubicin-induced cardiotoxicity. Am. J. Physiol. Heart Circ. Physiol. 308, H931–H941 [DOI] [PubMed] [Google Scholar]
- 16. Tronche F., Kellendonk C., Kretz O., Gass P., Anlag K., Orban P. C., Bock R., Klein R., and Schütz G. (1999) Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat. Genet. 23, 99–103 [DOI] [PubMed] [Google Scholar]
- 17. Dhar S., Vemulapalli V., Patananan A. N., Huang G. L., Di Lorenzo A., Richard S., Comb M. J., Guo A., Clarke S. G., and Bedford M. T. (2013) Loss of the major Type I arginine methyltransferase PRMT1 causes substrate scavenging by other PRMTs. Sci. Rep. 3, 1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Readhead C., Popko B., Takahashi N., Shine H. D., Saavedra R. A., Sidman R. L., and Hood L. (1987) Expression of a myelin basic protein gene in transgenic shiverer mice: correction of the dysmyelinating phenotype. Cell 48, 703–712 [DOI] [PubMed] [Google Scholar]
- 19. Sidman R. L., Dickie M. M., and Appel S. H. (1964) Mutant mice (Quaking and Jimpy) with deficient myelination in the central nervous system. Science 144, 309–311 [DOI] [PubMed] [Google Scholar]
- 20. Larocque D., Pilotte J., Chen T., Cloutier F., Massie B., Pedraza L., Couture R., Lasko P., Almazan G., and Richard S. (2002) Nuclear retention of MBP mRNAs in the quaking viable mice. Neuron 36, 815–829 [DOI] [PubMed] [Google Scholar]
- 21. He Y., Dupree J., Wang J., Sandoval J., Li J., Liu H., Shi Y., Nave K. A., and Casaccia-Bonnefil P. (2007) The transcription factor Yin Yang 1 is essential for oligodendrocyte progenitor differentiation. Neuron 55, 217–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nave K. (2010) Myelination and the trophic support of long axons. Nat. Rev. Neurosci. 11, 275–283 [DOI] [PubMed] [Google Scholar]
- 23. Jahn O., Tenzer S., and Werner H. B. (2009) Myelin proteomics: molecular anatomy of an insulating sheath. Mol. Neurobiol. 40, 55–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lu Q. R., Sun T., Zhu Z., Ma N., Garcia M., Stiles C. D., and Rowitch D. H. (2002) Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell 109, 75–86 [DOI] [PubMed] [Google Scholar]
- 25. Takebayashi H., Nabeshima Y., Yoshida S., Chisaka O., Ikenaka K., and Nabeshima Y. (2002) The basic helix-loop-helix factor Olig2 is essential for the development of motoneuron and oligodendrocyte lineages. Curr. Biol. 12, 1157–1163 [DOI] [PubMed] [Google Scholar]
- 26. Qi Y., Cai J., Wu Y., Wu R., Lee J., Fu H., Rao M., Sussel L., Rubenstein J., and Qiu M. (2001) Control of oligodendrocyte differentiation by the Nkx2.2 homeodomain transcription factor. Development 128, 2723–2733 [DOI] [PubMed] [Google Scholar]
- 27. Stolt C. C., Rehberg S., Ader M., Lommes P., Riethmacher D., Schachner M., Bartsch U., and Wegner M. (2002) Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev. 16, 165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nishiyama A., Komitova M., Suzuki R., and Zhu X. (2009) Polydendrocytes (NG2 cells): multifunctional cells with lineage plasticity. Nat. Rev. Neurosci. 10, 9–22 [DOI] [PubMed] [Google Scholar]
- 29. Liu Z., Hu X., Cai J., Liu B., Peng X., Wegner M., and Qiu M. (2007) Induction of oligodendrocyte differentiation by Olig2 and Sox10: evidence for reciprocal interactions and dosage-dependent mechanisms. Dev. Biol. 302, 683–693 [DOI] [PubMed] [Google Scholar]
- 30. Küspert M., Hammer A., Bösl M. R., and Wegner M. (2011) Olig2 regulates Sox10 expression in oligodendrocyte precursors through an evolutionary conserved distal enhancer. Nucleic Acids Res. 39, 1280–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhou Q., Choi G., and Anderson D. J. (2001) The bHLH transcription factor Olig2 promotes oligodendrocyte differentiation in collaboration with Nkx2.2. Neuron 31, 791–807 [DOI] [PubMed] [Google Scholar]
- 32. Zhao Q., Rank G., Tan Y. T., Li H., Moritz R. L., Simpson R. J., Cerruti L., Curtis D. J., Patel D. J., Allis C. D., Cunningham J. M., and Jane S. M. (2009) PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat. Struct. Mol. Biol. 16, 304–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huang S., Litt M., and Felsenfeld G. (2005) Methylation of histone H4 by arginine methyltransferase PRMT1 is essential in vivo for many subsequent histone modifications. Genes Dev. 19, 1885–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang H., Huang Z. Q., Xia L., Feng Q., Erdjument-Bromage H., Strahl B. D., Briggs S. D., Allis C. D., Wong J., Tempst P., and Zhang Y. (2001) Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science 293, 853–857 [DOI] [PubMed] [Google Scholar]
- 35. Yamagata K., Daitoku H., Takahashi Y., Namiki K., Hisatake K., Kako K., Mukai H., Kasuya Y., and Fukamizu A. (2008) Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol. Cell 32, 221–231 [DOI] [PubMed] [Google Scholar]
- 36. Zheng S., Moehlenbrink J., Lu Y. C., Zalmas L. P., Sagum C. A., Carr S., McGouran J. F., Alexander L., Fedorov O., Munro S., Kessler B., Bedford M. T., Yu Q., and La Thangue N. B. (2013) Arginine methylation-dependent reader-writer interplay governs growth control by E2F-1. Mol. Cell 52, 37–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Teyssier C., Ma H., Emter R., Kralli A., and Stallcup M. R. (2005) Activation of nuclear receptor coactivator PGC-1α by arginine methylation. Genes Dev. 19, 1466–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Britsch S., Goerich D. E., Riethmacher D., Peirano R. I., Rossner M., Nave K. A., Birchmeier C., and Wegner M. (2001) The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev. 15, 66–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.