Abstract
Diabetes is accompanied by dysregulation of glucose, lipid, and protein metabolism. In recent years, much effort has been spent on understanding how insulin regulates glucose and lipid metabolism, whereas the effect of insulin on protein metabolism has received less attention. In diabetes, hepatic production of serum albumin decreases, and it has been long established that insulin positively controls albumin gene expression. In this study, we used a genetic approach in mice to identify the mechanism by which insulin regulates albumin gene transcription. Albumin expression was decreased significantly in livers with insulin signaling disrupted by ablation of the insulin receptor or Akt. Concomitant deletion of Forkhead Box O1 (Foxo1) in these livers rescued the decreased albumin secretion. Furthermore, activation of Foxo1 in the liver is sufficient to suppress albumin expression. These results suggest that Foxo1 acts as a repressor of albumin expression.
Keywords: albumin, FOXO, insulin, transcription, transcription repressor
Introduction
Diabetes is a growing pandemic, affecting about 29 million people in the United States and creating a huge economic toll on the health care system (1). The disease is caused by failed production of or dampened response to insulin, an important metabolic hormone that signals an absorptive state after feeding. In recent years, significant advances have been made in understanding how insulin signals the regulation of glucose and lipid metabolism. However, the control of protein metabolism by insulin has received little attention.
Albumin is the most abundant circulating protein, being synthesized solely in the liver and accounting for ∼60% of total serum proteins. In addition to representing the major determinant of oncotic pressure, albumin also functions as the carrier for many endogenous and exogenous compounds, including free fatty acids, ions, and drugs. Clinically, albumin is a crucial biomarker used to assess liver function (2). Multiple factors, including nutritional states, oncotic pressure, and hormonal factors, regulate albumin production (3–5). In diabetes, the concentration of albumin in blood is decreased, and administration of insulin is required to prevent hypoalbuminemia (6, 7). Early biochemical studies have shown that insulin stimulates albumin production in the liver by activating gene transcription (5–12). However, the detailed pathway by which insulin exerts this effect has not been described.
In the liver, insulin promotes protein production and lipid synthesis while turning off gluconeogenesis (13–15). The insulin signaling pathway has been well characterized. Insulin binds to the insulin receptor (IR),2 which leads to phosphorylation of the insulin receptor substrate. This then initiates a cascade of signaling events that results in the phosphorylation and activation of Akt protein kinases (14). Several pathways downstream of Akt mediate the effects of insulin on metabolism. Akt phosphorylates and inactivates the tuberous sclerosis 1/2 (TSC1-TSC2) complex, releasing the inhibition of mammalian target of rapamycin complex 1 (mTORC1) (16, 17). Activation of mTORC1 stimulates protein synthesis as well as lipogenesis (18–22). Akt also phosphorylates the transcription factor Forkhead Box O1 (Foxo1), causing its translocation out of the nucleus (23–26). Foxo1 binds directly to the insulin response elements in the promoters of key gluconeogenic enzymes to stimulate the expression of these genes under fasting conditions. Under postprandial conditions, when insulin is present, Foxo1 is located largely in the cytoplasm and, therefore, becomes inactive as a transcription factor (27, 28). In this study, we used a genetic approach to address the longstanding question of the mechanism by which insulin stimulates albumin transcription.
Experimental Procedures
Animals
All experiments were performed in male mice that were 10–12 weeks of age. The IrloxP/loxP, IrloxP/loxP; Foxo1loxP/loxP, Akt1loxP/loxP; Akt2loxP/loxP, Akt1loxP/loxP; Akt2loxP/loxP;Foxo1loxP/loxP, and Foxo1loxP/loxP mice have been described previously (13, 29, 30). To generate liver-specific knockouts, an adeno-associated virus expressing either GFP or Cre recombinase driven by the promoter of the liver-specific gene thyroxine binding globulin was injected into the above mice at 8–10 weeks of age (1 × 1011 genomic copies/mouse). Experiments were performed 2 weeks after virus injection. For fasting-refeeding experiments, mice were deprived of food for 16 h (4 pm to 9 am). The fasted group was sacrificed at 9 am, and the refed group was fed ad libitum for 4 h with normal chow (Laboratory Rodent Diet, catalog no. 5001) before sacrifice. All animal experiments were reviewed and approved by the University of Pennsylvania Institutional Animal Care and Use Committee in accordance with National Institutes of Health guidelines.
Liver Lysates/Nuclear Extract Extraction and Western Blotting
After sacrifice, livers were dissected, freeze-clamped, and stored at −80 °C. Whole-cell lysates were prepared by homogenizing frozen liver samples in radioimmuno precipitation assay buffer (150 mm NaCl, 50 mm Tris (pH 7.6), 1% Triton X-100, 0.5% sodium deoxycholate, and 0.1% SDS, supplemented with protease and phosphatase inhibitors). To detect Foxo1, liver nuclear extracts were prepared using the NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific, catalog no. 78833). Cleared lysates and nuclear extracts were resolved by SDS-PAGE (10–12% acrylamide gel, constant voltage of 100 V), transferred onto nitrocellulose membranes, probed with various antibodies (IR, Cell Signaling Technology, catalog no. 3025S; Foxo1, Cell Signaling Technology, catalog no. 9454S; Akt1, Cell Signaling Technology, catalog no. 2967; Akt2, Cell Signaling Technology, catalog no. 2964S; and Actin, Abcam, catalog no. ab6276) and visualized with either IRDye secondary antibodies (LI-COR Biosciences, catalog no. 926-32213 and 926-68022) or ECL Western blotting detection reagents (Thermo Scientific, catalog no. 32106).
Primary Hepatocyte Isolation and in Vitro Albumin Secretion Assay
Primary hepatocytes were isolated as described previously (31). Cells were plated on collagen-treated plates in DMEM supplemented with 10% fetal bovine serum. After a 2- to 3-h attachment period, the cells were washed twice with PBS and incubated in serum-free Krebs-Ringer bicarbonate buffer (Sigma-Aldrich, catalog no. K4002) supplemented with 20 mm HEPES (pH 7.4) and 0.5% BSA for 2 h. The medium was collected, and hemoglobin (Sigma-Aldrich, catalog no. H2625) was added as a carrier protein (final concentration of 0.1%, w/v). For trichloroacetic acid (TCA) precipitation, 1 volume of 100% TCA (w/v) was added to 4 volumes of sample to precipitate total protein. The protein pellet was washed twice in ice-cold acetone, dried, and resuspended in Laemmli sample buffer (volume adjusted on the basis of cellular protein content). Albumin in the samples was then measured by Western blotting (anti-Alb, Nordic Immunology, catalog no. RAM/Alb/7s).
mRNA Isolation and Real-time PCR
Total RNA was isolated from frozen livers or primary hepatocytes using the Nucleospin RNA mini kit (Clontech, catalog no. 740955.250). cDNA was synthesized using Moloney Murine Leukemia Virus reverse transcriptase (New England Biolabs, catalog no. M0253S). Liver cDNA from transgenic mice expressing a constitutively active Foxo1 was a gift from Dr. Terry G. Unterman (University of Illinois at Chicago College of Medicine) (32). The relative expression of genes of interest was quantified by real-time PCR using the SYBR Green dye-based assay.
Serum Albumin Measurement
Blood samples were collected after sacrifice by cardiac puncture. After allowing the blood to clot, the samples were centrifuged to separate the serum. Albumin levels were measured using the Bromcresol Green Albumin Assay Kit (Sigma-Aldrich, catalog no. MAK124).
Streptozotocin-induced Type I Diabetes
At 8–10 weeks of age, Foxo1loxP/loxP mice received a retro-orbital injection of adeno-associated virus encoding either GFP or Cre recombinase at 1 × 1011 genomic copies/mouse. 5 days after virus injection, the mice received an intraperitoneal injection of either control buffer (0.1 m citrate (pH 4.5)) or streptozotocin (EMD Chemicals, catalog no. 572201) at 200 mg/kg of body weight. The mice were sacrificed 2 weeks after virus injection (9 days after STZ injection).
ChIP Assay
Liver chromatin was prepared as described previously (33). Immunoprecipitations were performed using anti-C/EBPα (Santa Cruz Biotechnology, catalog no. sc-61, 10 μg/immunoprecipitation). Real-time PCR oligos used to measure C/EBPα occupancy at the albumin promoter were as follows: site 1, 5′-CGCAAGGGATTTAGTCAAACAAC-3′ and 5′-AACCATACTTACCTCGCATTTCA-3′; site 2, 5′-TCCCAGACCCATCAATTGTG-3′ and 5′-TCCTGGCTCTTAGATTGCTCA-3′; site 3, 5′-AGCTAACCTTCTGTCCTAGTGG-3′ and 5′-TGAACTCTGACTCACGATGGA-3′; and site 4, 5′-ACAGAGGGTTGGATGGACAC-3′ and 5′-CCTCATTACCTTTGTGCACCA-3′.
Statistical Analysis
All values are expressed as mean ± S.E. Two-way ANOVA with Bonferroni post test was used when multiple conditions were involved when comparing different genotypes. Two-tailed, unpaired Student's t test was used when only two groups of data were concerned.
Results
Reduced Albumin Expression in Diabetic Livers
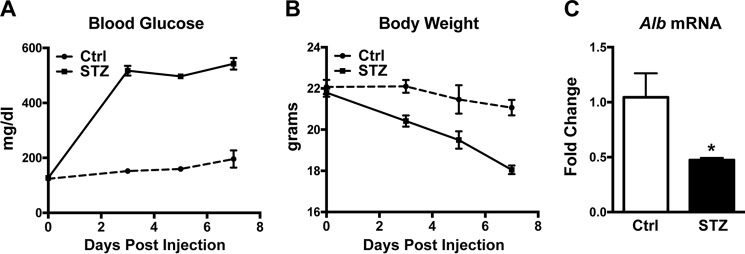
To assess the effect of diabetes on albumin expression in mice, we used animals treated with streptozotocin (STZ), a compound that induces β cell death and is employed frequently to induce diabetes in animal models. Mice injected with STZ developed severe hyperglycemia (Fig. 1A) and lost a significant amount of body weight (Fig. 1B). Importantly, albumin expression in the liver was decreased significantly compared with control animals (Fig. 1C). This result is consistent with early studies in rats showing that insulin positively regulates hepatic albumin production at the transcriptional level. We did not observe a difference in serum albumin level between STZ-treated and control animals (data not shown), possibly because the half-life of albumin protein is longer than the duration of the experiment (21 versus 11 days, respectively) (2).
FIGURE 1.
Albumin expression is decreased in diabetic livers. Mice received intraperitoneal injections of either control buffer (Ctrl, n = 3) or STZ (n = 4) at 200 mg/kg of body weight. A, blood glucose levels after injection. B, body weight after injection. C, hepatic albumin mRNA level measured 7 days after injection. *, p < 0.05 by two-tailed Student's t test. All values are expressed as mean ± S.E.
Direct Effect of Insulin in the Liver on Albumin Production
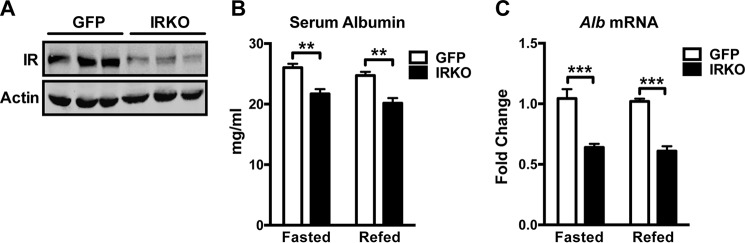
To address whether insulin signals directly on the liver to regulate albumin expression, we deleted IR specifically in the liver (Fig. 2A, IRKO). The level of serum albumin was reduced significantly in IRKO animals compared with controls (Fig. 2B, GFP). Consistent with the reduced circulating albumin, albumin expression in IRKO livers was decreased significantly compared with controls (Fig. 2C). These results suggest that insulin controls albumin production by signaling directly on the liver. Notably, a short-term fast did not affect serum albumin level or albumin expression because there was no significant difference in circulating albumin protein or albumin mRNA between the overnight-fasted state and the fed state (Fig. 2, B and C).
FIGURE 2.
Insulin acts on the liver directly to positively regulate albumin expression. A, Western blotting analyses for IR and actin in liver homogenates of control (GFP) and IRKO animals. B and C, serum albumin concentration (B) and hepatic albumin mRNA level (C) in GFP (n = 4–6) and IRKO (n = 3–5) animals that were fasted overnight or fasted overnight and refed for 4 h. **, p < 0.01; ***, p < 0.001 by two-way ANOVA. All values are expressed as mean ± S.E.
Requirement of Akt for Albumin Production
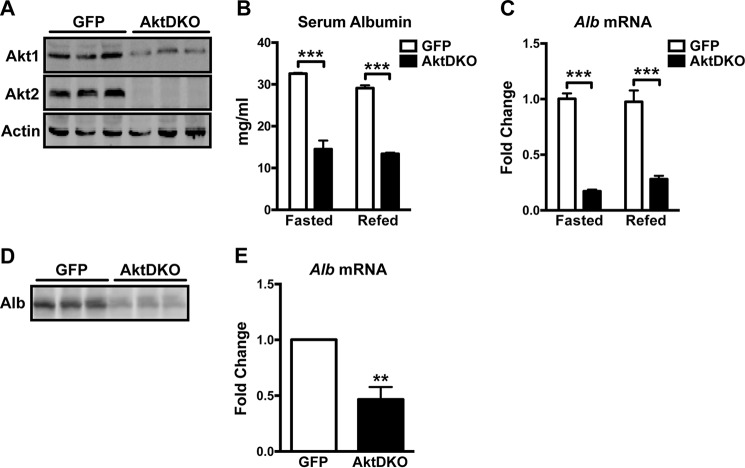
Akt, also known as protein kinase B, is a ubiquitous serine/threonine protein kinase that mediates many of the metabolic effects of insulin (34, 35). We investigated whether Akt is required for the regulation of albumin production downstream of IR. To this end, we deleted the only isoforms of Akt expressed in liver, Akt1 and Akt2 (Fig. 3A). Deletion of Akt1 and Akt2 specifically in the liver did not increase apoptosis (data not shown). Mice with Akt-null livers (AktDKO) displayed a 50% reduction in serum albumin (Fig. 3B). This severe hypoalbuminemia correlated well with a dramatic decrease in albumin expression (Fig. 3C). To address whether this albumin production defect is cell-autonomous, we isolated primary hepatocytes from control (GFP) and AktDKO mice and measured albumin secretion in vitro. AktDKO hepatocytes secreted significantly less albumin compared with controls (Fig. 3D). Consistent with the reduced albumin secretion, albumin expression in AktDKO hepatocytes was decreased significantly (Fig. 3E). These results suggest that insulin signals through the Akt pathway to control albumin transcription and secretion.
FIGURE 3.
Akt is required for the effect of insulin on albumin production. A, Western blotting analyses for Akt1, Akt2, and actin in liver homogenates of control (GFP) and AktDKO animals. B and C, serum albumin concentration (B) and hepatic albumin mRNA level (C) in GFP (n = 4) and AktDKO (n = 3–4) animals that were fasted overnight or fasted overnight and refed for 4 h. ***, p < 0.001 by two-way ANOVA. D, primary hepatocytes isolated from GFP and AktDKO livers were cultured in serum-free media for 2 h. Secreted proteins were TCA-precipitated and subjected to Western blotting for albumin. E, albumin mRNA levels in primary hepatocytes isolated from GFP (n = 3) and AktDKO (n = 4) livers were assayed by quantitative RT-PCR. **, p < 0.01 by two-tailed Student's t test. All values are expressed as mean ± S.E.
Foxo1-dependent Repression of Albumin Expression
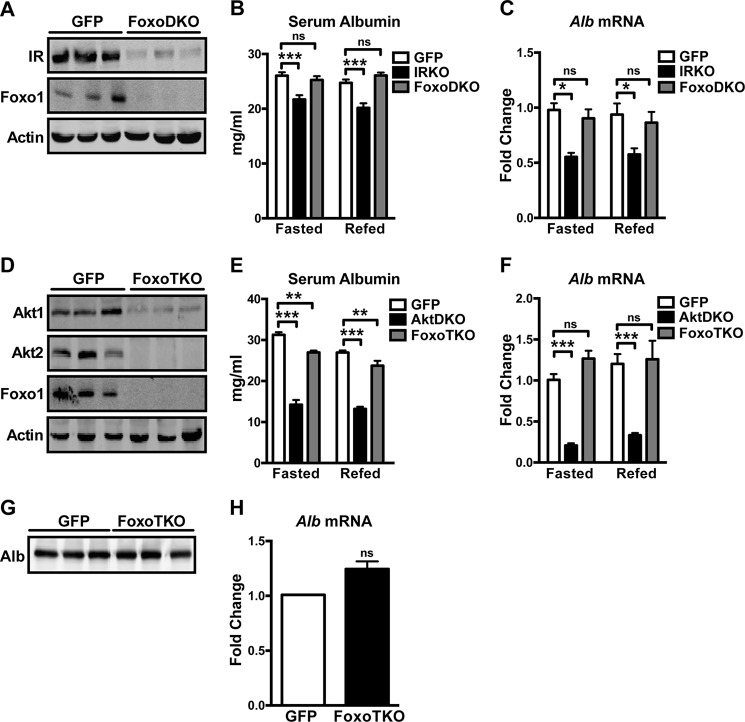
Foxo1, the transcription factor downstream of Akt, becomes constitutively active when insulin signaling is disrupted. We asked whether inactivation of Foxo1 downstream of insulin signaling is required for maintaining albumin transcription. Deletion of Foxo1 from IRKO livers (Fig. 4A, FoxoDKO) fully rescued the reduced circulating albumin level observed in IRKO mice (Fig. 4B). In addition, albumin expression in FoxoDKO livers completely returned to control levels (Fig. 4C). Similarly, concomitant deletion of Foxo1 in AktDKO livers (Fig. 4D, FoxoTKO) almost completely normalized albumin levels in serum (Fig. 4E) and completely rescued the reduced albumin expression observed in AktDKO livers (Fig. 4F). This rescue was cell-autonomous because primary hepatocytes isolated fromFoxoTKO livers displayed comparable albumin secretion (Fig. 4G) and albumin expression (Fig. 4H) as control cells. These results suggest that insulin controls albumin transcription and secretion at least in part via inhibition of Foxo1.
FIGURE 4.
Inhibition of Foxo1 rescues reduced albumin production in models with defective hepatic insulin signaling. A, Western blotting analyses for IR, Foxo1, and actin in liver homogenates of control (GFP) and FoxoDKO animals. B and C, serum albumin concentration (B) and hepatic albumin mRNA level (C) in GFP (n = 4–5), IRKO (n = 4–5), and FoxoDKO (n = 5) animals that were fasted overnight or fasted overnight and refed for 4 h. *, p < 0.05; **, p < 0.01; ns, not significant by two-way ANOVA. D, Western blotting analyses for Akt1, Akt2, Foxo1, and actin in liver homogenates of GFP and FoxoTKO animals. E and F, serum albumin concentration (E) and hepatic albumin mRNA level (F) in GFP (n = 3–5), AktDKO (n = 2–4), and FoxoTKO (n = 3–4) animals that were fasted overnight or fasted overnight and refed for 4 h. **, p < 0.01; ***, p < 0.001; two-way ANOVA. G, primary hepatocytes isolated from GFP and FoxoTKO livers were cultured in serum-free media for 2 h. Secreted proteins were TCA-precipitated and subjected to Western blotting for albumin. H, albumin mRNA levels in primary hepatocytes isolated from GFP (n = 3) and FoxoTKO (n = 4) livers were assayed by quantitative RT-PCR (two-tailed Student's t test). All values are expressed as mean ± S.E.
We then investigated whether ablating Foxo1 is sufficient to rescue the reduced albumin expression in diabetic livers. To this end, we used STZ to induce diabetes in either control (GFP) or liver-specific Foxo1 knockout mice (Fig. 5A, FoxoKO). Both genotypes developed severe hyperglycemia following injection of STZ (Fig. 5B). Importantly, STZ treatment in control mice caused a 50% decrease in albumin expression in the liver, and this reduction was absent in FoxoKO mice (Fig. 5C). In contrast, transgenic mice expressing a constitutively active Foxo1 (CA-Foxo1) significantly reduced hepatic albumin expression compared with WT controls (Fig. 5D). These results suggest that aberrant activation of hepatic Foxo1 is sufficient to repress albumin expression and that it contributes to decreased albumin production in diabetes.
FIGURE 5.
Inhibition of Foxo1 rescues reduced albumin expression in streptozotocin-induced type 1 diabetic livers. A, Western blotting analyses for Foxo1 and lamin in liver nuclear extracts of control (GFP) and FoxoKO animals. B and C, GFP (n = 5–7) and FoxoKO (n = 5–7) mice received intraperitoneal injections of either control buffer (Ctrl) or STZ (200 mg/kg of body weight). Blood glucose (B) and hepatic albumin mRNA levels (C) were measured 9 days after injection. **, p < 0.01; ***, p < 0.001; two-way ANOVA. D, hepatic albumin mRNA level of the WT (n = 3) and liver expressing CA-Foxo1 (n = 3). **, p < 0.01 by two-tailed Student's t test. All values are expressed as mean ± S.E.
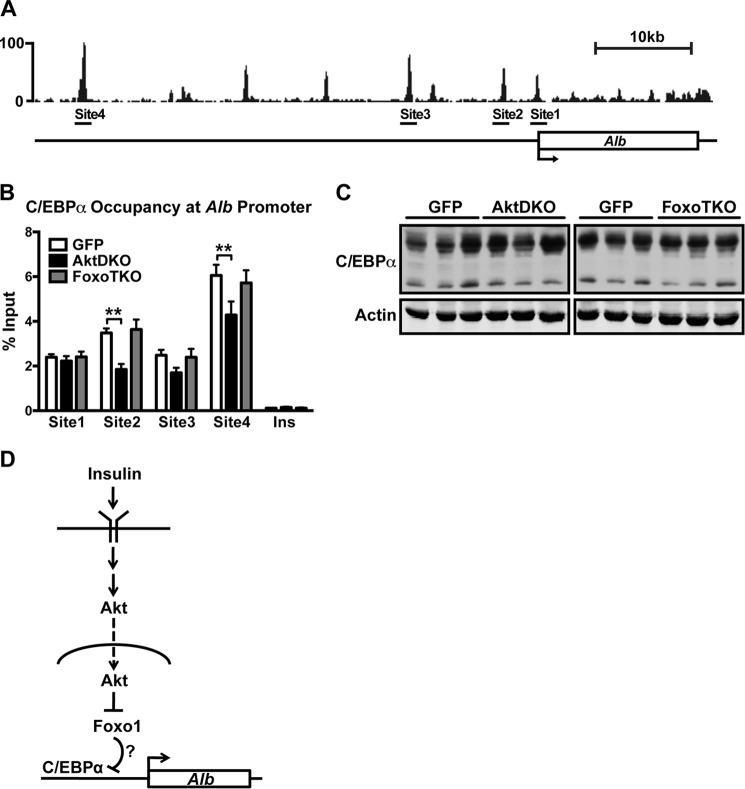
C/EBPα Occupancy at the Albumin Promoter
Transcription factor CCAAT/enhancer binding protein α (C/EBPα) is known to regulate albumin expression in the liver (36). Recent studies have suggested that Foxo1 directly interacts with C/EBPα and interferes with its binding to DNA (37, 38). To test whether Foxo1 activation interferes with C/EBPα DNA binding at the albumin promoter, we performed ChIP of C/EBPα in GFP, AktDKO, and FoxoTKO livers. Fig. 6A shows the C/EBPβ binding peaks identified by ChIP sequencing in wild-type mouse livers at the albumin promoter (39). Because C/EBPα and C/EBPβ often bind to DNA as a heterodimer, the peaks were used to predict C/EBPα binding sites, which were confirmed by ChIP using an antibody directed against C/EBPα (Fig. 6B). Interestingly, C/EBPα occupancy at the albumin promoter was reduced in AktDKO livers compared with controls, and additional deletion of Foxo1 in the liver completely reversed this decrease (Fig. 6B). Notably, the decline in DNA binding was not due to a decrease in protein levels of C/EBPα, which was unchanged, as assessed by Western blotting (Fig. 6C). These results suggest that activation of Foxo1 is negatively correlated with C/EBPα binding to the albumin promoter.
FIGURE 6.
Foxo1 activity negatively correlates with C/EBPα occupancy at the albumin promoter. A, ChIP sequencing profile for C/EBPβ at the albumin promoter. C/EBPβ binding sites selected for analyzing C/EBPα enrichment are indicated. B, enrichment of C/EBPα at the indicated sites of the albumin promoter in control (GFP, n = 4), AktDKO (n = 4), and FoxoTKO (n = 4) livers. Insulin served as a negative control site not bound by C/EBPα. **, p < 0.01 by two-way ANOVA. C, Western blotting analyses for C/EBPα and actin in liver homogenates of GFP, AktDKO, and FoxoTKO livers. D, model showing how insulin regulates albumin expression. All values are expressed as mean ± S.E.
Discussion
In this study, we used genetic models to elucidate the pathway by which insulin controls albumin expression in the liver. Specifically, insulin acts directly on the liver through the IR/PI3K/Akt pathway to inhibit Foxo1, which functions as a repressor of albumin expression.
Deletion of the insulin receptor in the liver (IRKO) leads to a decrease in serum albumin, consistent with previous observations described by Michael et al. (29). The phenotype we observed was much milder, possibly because our knockout model was acute and the animals were much younger. Disruption of both isoforms of Akt specifically in the liver (AktDKO) caused a larger reduction in serum albumin, suggesting that basal hepatic Akt activity in IRKO mice maintained some albumin production. In both IRKO and AktDKO mice, albumin gene expression was decreased significantly. Taken together, these results suggest that insulin signals directly on the liver through IR and Akt to stimulate albumin gene expression.
Foxo1 is an important target whose inhibition mediates many of the actions of insulin. Liver-specific Foxo1 knockout mice phenocopy the effect of insulin in having impaired glucose production (30). In addition, inhibition of hepatic Foxo1 activity protects against high-fat diet-induced hepatic insulin resistance (40, 41). On the other hand, transgenic mice with liver-specific expression of constitutively active Foxo1 exhibit fasting hyperglycemia, reduced de novo lipogenesis, and hepatic insulin resistance (42). These studies highlight the fact that Foxo1 plays a significant role in regulating glucose and lipid metabolism in the liver. In this study, we found that Foxo1 also reduced albumin expression. In livers in which Foxo1 is constitutively active (IRKO, AktDKO, and STZ-induced diabetes), albumin expression was decreased, and genetic ablation of Foxo1 in these models completely rescued the decreased albumin expression. These observations suggest that active Foxo1 represses albumin expression. Furthermore, we demonstrated that constitutive activation of Foxo1 in the liver is sufficient to decrease albumin expression. Taken together, we conclude that insulin stimulates albumin production by inhibiting Foxo1, which represses albumin expression (Fig. 6D).
There still remains considerable uncertainty about the mechanisms by which Foxo1 can function as a repressor. Genetic studies in Caenorhabditis elegans suggest that DAF-16, the Foxo ortholog, functions as a transcriptional activator downstream of the insulin/IGF-1 signaling pathway but does not bind to the promoter of repressed genes. Rather, genes that are down-regulated by DAF-16 (class II genes) are more likely indirect targets and regulated by the induction of a repressor protein by DAF-16 (43–45). Tepper et al. (45) recently described an elusive transcriptional activator, PQM-1, that is mutually antagonistic with DAF-16 with regard to subcellular localization in response to insulin/IGF-1 signaling, providing a mechanism for the regulation of Class II genes. To date, it is unclear whether a similar mechanism exists in mammals.
Alternatively, it has been demonstrated that Foxo1 interacts with a variety of transcription factors to repress gene expression by inhibiting the DNA binding of these transcription factors (32, 46–49). C/EBPα, a liver-enriched transcription factor known to activate albumin transcription (36), has been shown to interact directly with Foxo1 (37, 38). Interestingly, we found that the occupancy of C/EBPα is negatively correlated with Foxo1 activity. In AktDKO livers where Foxo1 is constitutively nuclear and active, C/EBPα binding to the albumin promoter is decreased, and genetic ablation of Foxo1 in this model completely restored the decreased occupancy to control levels. These data suggest that Foxo1 may repress albumin transcription by interacting directly with C/EBPα and inhibiting its DNA binding.
It is of note that, although deleting Foxo1 completely rescued the reduced albumin expression in AktDKO livers (Fig. 4F), there remains a small but statistically significant decrease in serum albumin (Fig. 4E), suggesting that insulin may also regulate albumin production by a post-transcriptional mechanism. Because insulin signaling acutely promotes protein translation by activating mTORC1 and its downstream targets and exerting long-term effects, such as promoting ribosome biogenesis to increase translation capacity, it is reasonable to speculate that insulin also promotes albumin translation.
Albumin mRNA and protein have a long half-life and, therefore, do not change with normal fasting and feeding but decrease during diabetes and prolonged starvation. Given that albumin is synthesized at a high rate (12–25 g/day in a young healthy adult) (2), this regulatory mechanism may have evolved as an adaptive mechanism to preserve limited amino acids during an extended fast. In this study, we showed that the reduction in albumin production in the absence of insulin is due to chronic activation of Foxo1.
Author Contributions
Q. C., M. L., and M. J. B. conceived the study. B. R. M. provided technical assistance. Q. C. and M. J. B. designed the study, analyzed the results, wrote the paper, and approved the final version of the manuscript.
Acknowledgments
We thank Dr. David J. Steger (University of Pennsylvania) for technical assistance with the chromatin immunoprecipitation assay and Dr. Mitch A. Lazar (University of Pennsylvania) for critical reading of the manuscript. We also thank Dr. Domenico Accili (Columbia University) and Dr. Terry G. Unterman (University of Illinois at Chicago) for sharing the Foxo1loxP/loxP mice and liver cDNA from CA-Foxo1 mice, respectively.
This work was supported by National Institutes of Health Grant RO1 DK56886. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- IR
- insulin receptor
- STZ
- streptozotocin
- DKO
- double knockout
- TKO
- triple knockout
- CA
- constitutionally active
- C/EBP
- CCAAT/enhancer binding protein.
References
- 1. Centers for Disease Control and Prevention. (2014) National Diabetes Statistics Report: Estimates of Diabetes and Its Burden in the United States, 2014. Atlanta, GA: US Department of Health and Human Services; 2014 [Google Scholar]
- 2. Fanali G., di Masi A., Trezza V., Marino M., Fasano M., and Ascenzi P. (2012) Human serum albumin: from bench to bedside. Mol. Aspects Med. 33, 209–290 [DOI] [PubMed] [Google Scholar]
- 3. Sakuma K., Ohyama T., Sogawa K., Fujii-Kuriyama Y., and Matsumura Y. (1987) Low protein-high energy diet induces repressed transcription of albumin mRNA in rat liver. J. Nutr. 117, 1141–1148 [DOI] [PubMed] [Google Scholar]
- 4. Pietrangelo A., Panduro A., Chowdhury J. R., and Shafritz D. A. (1992) Albumin gene expression is down-regulated by albumin or macromolecule infusion in the rat. J. Clin. Invest. 89, 1755–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kimball S. R., Horetsky R. L., and Jefferson L. S. (1995) Hormonal regulation of albumin gene expression in primary cultures of rat hepatocytes. Am. J. Physiol. 268, E6–14 [DOI] [PubMed] [Google Scholar]
- 6. De Feo P., Gaisano M. G., and Haymond M. W. (1991) Differential effects of insulin deficiency on albumin and fibrinogen synthesis in humans. J. Clin. Invest. 88, 833–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jefferson L. S., Liao W. S., Peavy D. E., Miller T. B., Appel M. C., and Taylor J. M. (1983) Diabetes-induced alterations in liver protein synthesis: changes in the relative abundance of mRNAs for albumin and other plasma proteins. J. Biol. Chem. 258, 1369–1375 [PubMed] [Google Scholar]
- 8. Peavy D. E., Taylor J. M., and Jefferson L. S. (1978) Correlation of albumin production rates and albumin mRNA levels in livers of normal, diabetic, and insulin-treated diabetic rats. Proc. Natl. Acad. Sci. U.S.A. 75, 5879–5883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Peavy D. E., Taylor J. M., and Jefferson L. S. (1985) Time course of changes in albumin synthesis and mRNA in diabetic and insulin-treated diabetic rats. Am. J. Physiol. 248, E656–63 [DOI] [PubMed] [Google Scholar]
- 10. Lloyd C. E., Kalinyak J. E., Hutson S. M., and Jefferson L. S. (1987) Stimulation of albumin gene transcription by insulin in primary cultures of rat hepatocytes. Am. J. Physiol. 252, C205–C214 [DOI] [PubMed] [Google Scholar]
- 11. Hutson S. M., Stinson-Fisher C., Shiman R., and Jefferson L. S. (1987) Regulation of albumin synthesis by hormones and amino acids in primary cultures of rat hepatocytes. Am. J. Physiol. 252, E291–8 [DOI] [PubMed] [Google Scholar]
- 12. Flaim K. E., Hutson S. M., Lloyd C. E., Taylor J. M., Shiman R., and Jefferson L. S. (1985) Direct effect of insulin on albumin gene expression in primary cultures of rat hepatocytes. Am. J. Physiol. 249, E447–53 [DOI] [PubMed] [Google Scholar]
- 13. Leavens K. F., and Birnbaum M. J. (2011) Insulin signaling to hepatic lipid metabolism in health and disease. Crit. Rev. Biochem. Mol. Biol. 46, 200–215 [DOI] [PubMed] [Google Scholar]
- 14. Saltiel A. R., and Kahn C. R. (2001) Insulin signalling and the regulation of glucose and lipid metabolism. Nature 414, 799–806 [DOI] [PubMed] [Google Scholar]
- 15. Brown M. S., and Goldstein J. L. (2008) Selective versus total insulin resistance: a pathogenic paradox. Cell Metab. 7, 95–96 [DOI] [PubMed] [Google Scholar]
- 16. Sengupta S., Peterson T. R., and Sabatini D. M. (2010) Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol. Cell 40, 310–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Inoki K., Li Y., Zhu T., Wu J., and Guan K. L. (2002) TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 4, 648–657 [DOI] [PubMed] [Google Scholar]
- 18. Wang X., and Proud C. G. (2006) The mTOR pathway in the control of protein synthesis. Physiology 21, 362–369 [DOI] [PubMed] [Google Scholar]
- 19. Thoreen C. C., Chantranupong L., Keys H. R., Wang T., Gray N. S., and Sabatini D. M. (2012) A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485, 109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li S., Brown M. S., and Goldstein J. L. (2010) Bifurcation of insulin signaling pathway in rat liver: mTORC1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 3441–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Laplante M., and Sabatini D. M. (2010) mTORC1 activates SREBP-1c and uncouples lipogenesis from gluconeogenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 3281–3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Düvel K., Yecies J. L., Menon S., Raman P., Lipovsky A. I., Souza A. L., Triantafellow E., Ma Q., Gorski R., Cleaver S., Vander Heiden M. G., MacKeigan J. P., Finan P. M., Clish C. B., Murphy L. O., and Manning B. D. (2010) Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol. Cell 39, 171–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Biggs W. H. 3rd, Meisenhelder J., Hunter T., Cavenee W. K., and Arden K. C. (1999) Protein kinase B/Akt-mediated phosphorylation promotes nuclear exclusion of the winged helix transcription factor FKHR1. Proc. Natl. Acad. Sci. U.S.A. 96, 7421–7426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rena G., Guo S., Cichy S. C., Unterman T. G., and Cohen P. (1999) Phosphorylation of the transcription factor forkhead family member FKHR by protein kinase B. J. Biol. Chem. 274, 17179–17183 [DOI] [PubMed] [Google Scholar]
- 25. Rena G., Woods Y. L., Prescott A. R., Peggie M., Unterman T. G., Williams M. R., and Cohen P. (2002) Two novel phosphorylation sites on FKHR that are critical for its nuclear exclusion. EMBO J. 21, 2263–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nakae J., Park B. C., and Accili D. (1999) Insulin stimulates phosphorylation of the forkhead transcription factor FKHR on serine 253 through a wortmannin-sensitive pathway. J. Biol. Chem. 274, 15982–15985 [DOI] [PubMed] [Google Scholar]
- 27. Hall R. K., Yamasaki T., Kucera T., Waltner-Law M., O'Brien R., and Granner D. K. (2000) Regulation of phosphoenolpyruvate carboxykinase and insulin-like growth factor-binding protein-1 gene expression by insulin: the role of winged helix/forkhead proteins. J. Biol. Chem. 275, 30169–30175 [DOI] [PubMed] [Google Scholar]
- 28. Schmoll D., Walker K. S., Alessi D. R., Grempler R., Burchell A., Guo S., Walther R., and Unterman T. G. (2000) Regulation of glucose-6-phosphatase gene expression by protein kinase B and the forkhead transcription factor FKHR: evidence for insulin response unit-dependent and -independent effects of insulin on promoter activity. J. Biol. Chem. 275, 36324–36333 [DOI] [PubMed] [Google Scholar]
- 29. Michael M. D., Kulkarni R. N., Postic C., Previs S. F., Shulman G. I., Magnuson M. A., and Kahn C. R. (2000) Loss of insulin signaling in hepatocytes leads to severe insulin resistance and progressive hepatic dysfunction. Mol. Cell 6, 87–97 [PubMed] [Google Scholar]
- 30. Matsumoto M., Pocai A., Rossetti L., Depinho R. A., and Accili D. (2007) Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor Foxo1 in liver. Cell Metab. 6, 208–216 [DOI] [PubMed] [Google Scholar]
- 31. Miller R. A., Chu Q., Xie J., Foretz M., Viollet B., and Birnbaum M. J. (2013) Biguanides suppress hepatic glucagon signalling by decreasing production of cyclic AMP. Nature 494, 256–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Deng X., Zhang W., O-Sullivan I., Williams J. B., Dong Q., Park E. A., Raghow R., Unterman T. G., and Elam M. B. (2012) FoxO1 inhibits sterol regulatory element-binding protein-1c (SREBP-1c) gene expression via transcription factors Sp1 and SREBP-1c. J. Biol. Chem. 287, 20132–20143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tuteja G., Jensen S. T., White P., and Kaestner K. H. (2008) Cis-regulatory modules in the mammalian liver: composition depends on strength of Foxa2 consensus site. Nucleic Acids Res. 36, 4149–4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cho H., Mu J., Kim J. K., Thorvaldsen J. L., Chu Q., Crenshaw E. B. 3rd, Kaestner K. H., Bartolomei M. S., Shulman G. I., and Birnbaum M. J. (2001) Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKB β). Science 292, 1728–1731 [DOI] [PubMed] [Google Scholar]
- 35. Lu M., Wan M., Leavens K. F., Chu Q., Monks B. R., Fernandez S., Ahima R. S., Ueki K., Kahn C. R., and Birnbaum M. J. (2012) Insulin regulates liver metabolism in vivo in the absence of hepatic Akt and Foxo1. Nat. Med. 18, 388–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Friedman A. D., Landschulz W. H., and McKnight S. L. (1989) CCAAT/enhancer binding protein activates the promoter of the serum albumin gene in cultured hepatoma cells. Genes Dev. 3, 1314–1322 [DOI] [PubMed] [Google Scholar]
- 37. Sekine K., Chen Y. R., Kojima N., Ogata K., Fukamizu A., and Miyajima A. (2007) Foxo1 links insulin signaling to C/EBPα and regulates gluconeogenesis during liver development. EMBO J. 26, 3607–3615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Qiao L., and Shao J. (2006) SIRT1 regulates adiponectin gene expression through Foxo1-C/enhancer-binding protein transcriptional complex. J. Biol. Chem. 281, 39915–39924 [DOI] [PubMed] [Google Scholar]
- 39. Everett L. J., Le Lay J., Lukovac S., Bernstein D., Steger D. J., Lazar M. A., and Kaestner K. H. (2013) Integrative genomic analysis of CREB defines a critical role for transcription factor networks in mediating the fed/fasted switch in liver. BMC Genomics 14, 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim J. J., Li P., Huntley J., Chang J. P., Arden K. C., and Olefsky J. M. (2009) FoxO1 haploinsufficiency protects against high-fat diet-induced insulin resistance with enhanced peroxisome proliferator-activated receptor activation in adipose tissue. Diabetes 58, 1275–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xiong X., Tao R., DePinho R. A., and Dong X. C. (2013) Deletion of hepatic FoxO1/3/4 genes in mice significantly impacts on glucose metabolism through downregulation of gluconeogenesis and upregulation of glycolysis. PLOS ONE 8, e74340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang W., Patil S., Chauhan B., Guo S., Powell D. R., Le J., Klotsas A., Matika R., Xiao X., Franks R., Heidenreich K. A., Sajan M. P., Farese R. V., Stolz D. B., Tso P., Koo S. H., Montminy M., and Unterman T. G. (2006) FoxO1 regulates multiple metabolic pathways in the liver: effects on gluconeogenic, glycolytic, and lipogenic gene expression. J. Biol. Chem. 281, 10105–10117 [DOI] [PubMed] [Google Scholar]
- 43. Murphy C. T., McCarroll S. A., Bargmann C. I., Fraser A., Kamath R. S., Ahringer J., Li H., and Kenyon C. (2003) Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424, 277–283 [DOI] [PubMed] [Google Scholar]
- 44. Schuster E., McElwee J. J., Tullet J. M., Doonan R., Matthijssens F., Reece-Hoyes J. S., Hope I. A., Vanfleteren J. R., Thornton J. M., and Gems D. (2010) DamID in C. elegans reveals longevity-associated targets of DAF-16/FoxO. Mol. Syst. Biol. 6, 399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tepper R. G., Ashraf J., Kaletsky R., Kleemann G., Murphy C. T., and Bussemaker H. J. (2013) PQM-1 complements DAF-16 as a key transcriptional regulator of DAF-2-mediated development and longevity. Cell 154, 676–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hirota K., Daitoku H., Matsuzaki H., Araya N., Yamagata K., Asada S., Sugaya T., and Fukamizu A. (2003) Hepatocyte nuclear factor-4 is a novel downstream target of insulin via FKHR as a signal-regulated transcriptional inhibitor. J. Biol. Chem. 278, 13056–13060 [DOI] [PubMed] [Google Scholar]
- 47. Dowell P., Otto T. C., Adi S., and Lane M. D. (2003) Convergence of peroxisome proliferator-activated receptor and Foxo1 signaling pathways. J. Biol. Chem. 278, 45485–45491 [DOI] [PubMed] [Google Scholar]
- 48. Hirota K., Sakamaki J., Ishida J., Shimamoto Y., Nishihara S., Kodama N., Ohta K., Yamamoto M., Tanimoto K., and Fukamizu A. (2008) A combination of HNF-4 and Foxo1 is required for reciprocal transcriptional regulation of glucokinase and glucose-6-phosphatase genes in response to fasting and feeding. J. Biol. Chem. 283, 32432–32441 [DOI] [PubMed] [Google Scholar]
- 49. Fan W., Imamura T., Sonoda N., Sears D. D., Patsouris D., Kim J. J., and Olefsky J. M. (2009) FOXO1 transrepresses peroxisome proliferator-activated receptor γ transactivation, coordinating an insulin-induced feed-forward response in adipocytes. J. Biol. Chem. 284, 12188–12197 [DOI] [PMC free article] [PubMed] [Google Scholar]