Abstract
In Escherichia coli cell division is driven by the tubulin-like GTPase, FtsZ, which forms the cytokinetic Z-ring. The Z-ring serves as a dynamic platform for the assembly of the multiprotein divisome, which catalyzes membrane cleavage to create equal daughter cells. Several proteins effect FtsZ assembly, thereby providing spatiotemporal control over cell division. One important class of FtsZ interacting/regulatory proteins is the Z-ring-associated proteins, Zaps, which typically modulate Z-ring formation by increasing lateral interactions between FtsZ protofilaments. Strikingly, these Zap proteins show no discernable sequence similarity, suggesting that they likely harbor distinct structures and mechanisms. The 19.8-kDa ZapC in particular shows no homology to any known protein. To gain insight into ZapC function, we determined its structure to 2.15 Å and performed genetic and biochemical studies. ZapC is a monomer composed of two domains, an N-terminal α/β region and a C-terminal twisted β barrel-like domain. The structure contains two pockets, one on each domain. The N-domain pocket is lined with residues previously implicated to be important for ZapC function as an FtsZ bundler. The adjacent C-domain pocket contains a hydrophobic center surrounded by conserved basic residues. Mutagenesis analyses indicate that this pocket is critical for FtsZ binding. An extensive FtsZ binding surface is consistent with the fact that, unlike many FtsZ regulators, ZapC binds the large FtsZ globular core rather than C-terminal tail, and the presence of two adjacent pockets suggests possible mechanisms for ZapC-mediated FtsZ bundling.
Keywords: cell division, electron microscopy (EM), protein self-assembly, protein-protein interaction, x-ray crystallography, FtsZ Z-ring, ZapC, electron microscopy (EM), polymerization
Introduction
In Escherichia coli, the bacterial tubulin-like GTPase, FtsZ, drives cell division. Unlike the microtubule structures formed by tubulin, FtsZ forms protofilaments that combine to create a ring-like structure at midcell called the Z-ring, which mediates cell division (1–5). FtsZ is an ancient and highly conserved protein that is responsible for cell division in most bacteria as well as many archaea, all chloroplasts, and the mitochondria of primitive eukaryotes (4). FtsZ is made up of five main domains: an N-terminal region, which is largely disordered, a globular core that contains the nucleotide binding site and the T7 synergy loop needed for nucleotide hydrolysis, a long linker of variable sequence and length (∼40–257 residues), a short ∼15-residue C-terminal tail (CTT)3 that contains a highly conserved set of residues critical for the docking of several FtsZ interacting proteins, and a more recently defined C-terminal variable (CTV) region (4, 6). Linear protofilaments are created by GTP binding to the globular domains of FtsZ between different protomers (1, 7, 8). The dynamic assembly and disassembly of protofilaments resulting from cycles of GTP binding and hydrolysis leads to Z-ring remodeling that is thought to contribute to the constrictive force required for cell division (4, 9, 10).
In E. coli the intracellular levels of FtsZ remain essentially the same throughout the cell cycle and exceed the critical concentration required for creation of the Z-ring (11). Therefore, the regulation of Z-ring formation occurs at the level of FtsZ filament assembly. FtsZ interacting proteins modulate FtsZ polymerization and thus play important roles in this process (4, 5, 13–27). In E. coli at least 24 proteins have been identified that promote the assembly/disassembly processes of FtsZ at midcell (28, 29). Cells lacking the division proteins FtsZ and FtsA fail to constrict and exhibit a lethal division phenotype (1, 5). FtsZ regulatory division proteins such as the Zap proteins have functionally redundant roles making them individually nonessential (1, 12, 29). Nevertheless, these regulators have important roles in cell viability as assessed by the resultant inhibition of cell division or synthetic lethal phenotypes resulting when two or more of them is deleted (1, 12). The nature of the selective pressure that ensures the presence of several FtsZ regulatory proteins with redundant functions in Z-ring stability in E. coli is not clearly understood. But it is thought that each of these proteins may perform a beneficial function in FtsZ polymerization and stability under specific environmental conditions. Thus, probing the precise molecular interactions of each FtsZ-associated protein with FtsZ may begin to provide clues into the biology of Z-ring maintenance in E. coli and related species.
Among the many FtsZ regulatory proteins are the FtsZ ring-associated (Zap) proteins, ZapA–E (13–27). With the exception of ZapE, these proteins are recruited early during cytokinesis and have overlapping functions in stabilizing Z-ring formation at the midcell (13–17, 20, 21). ZapE was only recently discovered and represents an FtsZ regulator that functions under specific environmental stress. In particular, it is active during high temperatures (>37 °C) or oxygen depletion (18). ZapE is a Walker ATPase, and by some unknown mechanism it reduces the stability of FtsZ polymers in the presence of ATP (18). By contrast, the remaining Zaps, ZapA–D, act under normal cellular conditions and either directly or indirectly stabilize the Z-ring by increasing lateral interactions and bundling of FtsZ protofilaments. Such bundling is assumed to provide stability to the Z-ring in vivo (12–17, 20, 21). ZapA and ZapB are coiled coil proteins, and studies have shown that ZapA promotes the assembly and stability of the Z-ring by cross-linking adjacent FtsZ protofilaments (14–17, 19, 30–33). The absence of ZapA leads to cell elongation (14). ZapB does not bind FtsZ directly but is recruited to the divisome via its interaction with ZapA where it is thought to enhance the role of ZapA in FtsZ bundling (16, 32). In addition, ZapB plays a role in chromosome segregation through its interaction with MatP (34). ZapC and ZapD interact directly with FtsZ and also promote bundling of FtsZ protofilaments (13, 20, 21). Although ZapA is widely conserved, ZapB–D are mostly restricted to the γ-proteobacteria.
ZapC is arguably the least understood Zap from a molecular standpoint, with no known structures or structures of homologs available. The studies that have been carried out on ZapC show that it is an early assembly protein that co-localizes with FtsZ at midcell and promotes interactions between FtsZ protofilaments in the Z-ring (20, 21). These analyses also revealed that cells lacking ZapC show marginal increases in cell length. However, a zapA zapC double mutant displayed measurably longer cells and a greater variability in the bacterial population compared with cells with zapA or zapC single mutants, thus underscoring the overlapping functions of these proteins in Z-ring stability (13, 21). Consistent with its role in FtsZ bundling, overexpression of ZapC leads to aberrant hyperstable Z-ring structures and lethal filamentation. ZapC binds FtsZ directly in vitro and enhances FtsZ assembly into protofilament bundles. ZapC also suppresses the GTPase activity of FtsZ (20–21).
To gain insight into the function of E. coli ZapC at the molecular level we determined its crystal structure to 2.15 Å resolution. Structural and biochemical studies indicate that ZapC is a monomer with a previously unobserved overall fold. Two pockets were identified on the structure; one in each of its two domains. Genetic and biochemical studies showed that residues in both domains are involved in FtsZ binding and bundling. Additional data demonstrated that ZapC interacts with the globular core of FtsZ, not its C-terminal tail. Thus, the combined findings suggest that ZapC utilizes a novel mechanism in which a monomeric ZapC facilitates FtsZ bundling via interactions within its GTPase globular region.
Experimental Procedures
Purification and Crystallization of E. coli ZapC
An artificial gene encoding zapC that was codon-optimized for expression in E. coli was purchased from Genscript Corp. (Piscataway, NJ). The gene was subcloned into the pET15b vector such that it expressed an N-terminal hexahistidine tag (His tag) for purification. The zapC expressing pET15b vector was transformed into E. coli C41(DE3) cells. For protein expression, the zapC-expressing cells were grown to an A600 of 0.6–0.8 and induced with 1 mm isopropyl β-d-1-thio-galactopyranoside (IPTG) for 4 h at 37 °C. Cells were lysed in buffer A (25 mm Tris, pH 7.5, 300 mm NaCl, 5% glycerol, 10 mm β-mercaptoethanol) using a microfluidizer. Cell debris was removed by centrifugation at 17,000 rpm, and the resultant lysate was loaded onto a Ni-NTA column. The column was washed with increasing concentrations of imidazole in buffer A and ZapC eluted with 100–500 mm imidazole. The protein was >90% pure after Ni-NTA chromatography as assessed by SDS-PAGE analysis. However, the ZapC(K94D) mutant was only ∼75% pure after the Ni-NTA step, and size exclusion chromatography was necessary to obtain >90% pure protein for analyses.
For crystallization, the purified WT protein was buffer-exchanged into 50 mm Tris, pH 7.5, 300 mm NaCl, 5% glycerol, and 10 mm dl-dithiothreitol (DTT). ZapC contains five cysteines. High levels of reducing agent (either 10 mm β-mercaptoethanol or 10 mm DTT) were required for protein stability, likely due to the otherwise rapid oxidation of the cysteine residues.
Selenomethionine-substituted ZapC protein was obtained by using the methionine inhibitory pathway. Briefly, an overnight culture harboring pET15b-zapC was centrifuged, and the cell pellet was thoroughly washed with M9 media to remove any methionine. Washed cells were then resuspended in M9 media and used to inoculate 18 liters of M9 media. The cells were grown to an A600 of ∼0.5 at which point 50 mg/liter selenomethionine and 40 mg/liter amino acids that inhibit selenomethionine biosynthesis were added. The cells were grown for an additional 15 min and induced with 1 mm IPTG for 4 h. The purification of selenomethionine ZapC was performed as for the wild type (WT) protein with the exception that 20 mm reducing agent (β-mercaptoethanol or DTT) was used in all buffers. Crystals of both the native and selenomethionine-substituted protein were grown via hanging drop vapor diffusion by mixing protein (at 20–40 mg/ml) 1:1 with a reservoir of 1.5 m sodium citrate, 0.1 m cacodylate, pH 6.5. Crystals took 2 weeks to grow to maximum size and were cryo-preserved by dipping them in a solution supplemented with 20% glycerol for 4 s before placement in the cryo-stream.
Structure Determination and Refinement of the ZapC Structure
The ZapC crystals take the space group C2, with a = 66.0 Å, b = 46.7 Å, c = 62.9 Å, and β = 105.7°. The structure was solved by multiple wavelength anomalous diffraction (MAD) using data obtained to 2.40 Å of crystals grown with selenomethionine substituted ZapC protein (35, 36). Inverse beam data collection was employed at three wavelengths, corresponding to the selenomethionine peak and inflection and a remote wavelength. The heavy atom substructure was obtained via SOLVE and resulted in a figure of merit of 0.53 (35). Density modification (RESOLVE) was then used to generate an improved electron density map, which was readily traced. The map revealed one ZapC subunit in the crystallographic asymmetric unit. Subsequent to model construction into the MAD electron density map and initial refinement of the structure, a native x-ray intensity data set was collected of a wild type (non-selenomethionine substituted) crystal to 2.15 Å at Advanced Light Source beamline 8.3.1 for final refinement, which was performed using Phenix (37). All x-ray intensity data were processed with MOSFLM (38). The final model includes ZapC residues 2–168 (residues 169–180 were disordered) and 109 solvent molecules. ZapC model refinement statistics are presented in Table 1.
TABLE 1.
Data collection and refinement statistics for E. coli ZapC structure
| Data collection | |
| Space group | C2 |
| Cell constants (Å) | a = 66.0, b = 46.7, and c = 62.9; β = 105.7 |
| Resolution (Å) | 60.5 − 2.15 |
| Overall Rsym (%)a | 5.7 (23.4)b |
| Overall I/σ(I) | 11.5 (4.6) |
| No. unique reflections | 9,961 |
| No. total reflections | 28,475 |
| % Complete | 98.0 (98.0) |
| Refinement statistics | |
| Rwork/Rfree(%)c | 19.7/23.1 |
| Root mean square deviation | |
| Bond angles (°) | 1.46 |
| Bond lengths (Å) | 0.009 |
| Ramachandran analysis | |
| Most favored (%) | 96.4 |
| Additional allowed (%) | 3.6 |
| Disallowed (%) | 0.0 |
a Rsym = ΣΣ|Ihkl − Ihkl(j)|/ΣIhkl, where Ihkl(j) is the observed intensity, and Ihkl is the final average value of intensity.
b Values in parentheses are for the highest resolution shell.
c Rwork = Σ‖Fobs| − |Fcalc‖/Σ|Fobs| and Rfree = Σ‖Fobs| − |Fcalc‖/Σ|Fobs|, where all reflections belong to a test set of 5% randomly selected data.
Size Exclusion Chromatography (SEC)
SEC was used to probe the molecular weights of ZapC and ZapC mutants. All SEC experiments were performed using a HiLoad 26/600 Superdex 75 prep grade column. Experiments were performed in a buffer containing 300 mm NaCl, 5% glycerol, and 20 mm Tris HCl, pH 7.5, 10 mm β-mercaptoethanol.
Glutaraldehyde Cross-linking
Glutaraldehyde cross-linking was performed as another method to assess the molecular weight of ZapC. For these experiments, purified ZapC at 3 mg/ml in 50 mm Hepes, pH 7.0, 100 mm NaCl was cross-linked with 0.2% glutaraldehyde (final concentration) at various time points. The reaction was quenched by the addition of 100 mm (final concentration) Tris HCl, pH 8.0. At concentrations >0.3% glutaraldehyde the protein precipitated within minutes. Hence, 0.2% glutaraldehyde was used.
Strains, Plasmids, and Growth Conditions
All strains and plasmids used in the study are listed in Tables 2 and 3. E. coli strains were grown in LB (0.5% NaCl) with appropriate antibiotics (ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; or spectinomycin 100 μg/ml) at 37 °C unless stated otherwise. Yeast strains were maintained in yeast peptone dextrose media or in selective yeast nitrogen broth (YNB) complete media lacking leucine (−L), uracil (−U), or tryptophan (−T) at 30 °C.
TABLE 2.
Bacterial and yeast strains used in the study
| Strain | Genotype | Source or Referencea |
|---|---|---|
| E. coli | ||
| MG1655 | F− λ− ilvG rfb50 rph1 | Laboratory collection |
| BL21 (λ DE3) pLysS | F− ompT hsdSB (rB− mB−) dcm gal λ (DE3) pLysS | 20 |
| C41 (DE3) | F− ompT hsdSB (rB−mB−) gal dcm (DE3) | 6 |
| JD160 | BL21 ( λ DE3) pLysS pET28b-his10-smt3-zapD | 13 |
| JD 161 | BL21 ( λ DE3) pLysS pET28b-his10-smt3-zapC | |
| KHH210 | MG1655 pNG162-zapC | |
| KHH211 | MG1655 pNG162-zapC K37D | |
| KHH212 | MG1655 pNG162-zapC K89D | |
| KHH213 | MG1655 pNG162-zapC K94D | |
| KHH214 | MG1655 pNG162-zapC R150E | |
| KHH216 | MG1655 pNG162 | |
| KHH241 | C41 (DE3) pET28b-his10-smt3-zapC K94D | |
| KHH236 | MG1655 pDSW208-zapC-gfp | |
| KHH237 | MG1655 pDSW208-zapC K89D-gfp | |
| KHH239 | MG1655 pDSW208-zapC K94D-gfp | |
| KHH240 | MG1655 pDSW208-gfp | |
| Saccharomyces cerevisiae | ||
| PJ69-4A | MATa trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2:: GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ | Gift from Beate Schwer |
| SL3004 | MATα ura3 leu2 his3 ade2-101 lys2-801 trp1-901 gal4 gal80 | Gift from Sandra Lemmon |
| KHH-Y3 | SL3004 pGAD-C1 | |
| KHH-Y4 | PJ69–4A pGBDU-C1 | |
a This work, unless otherwise stated.
TABLE 3.
Plasmids used in this study
ori, origin of replication; MCS, multiple cloning site; BD, DNA binding domain; AD, activation domain.
| Plasmid | Genotype/Description | Source or referencea |
|---|---|---|
| pCH320 | bla ura3 Padh1::gal4 BD-zapC | 21 |
| pCH374 | bla ura3 Padh1::gal4 BD-zapC (L22P) | 21 |
| pDSW208 | pDSW204-MCS-gfp | 49 |
| pERV-D8 | pET11b-ftsZ1–372 | |
| pET11b | pBR322 ori pT7, AmpR | Thermo Fisher Scientific |
| pET15b | pBR322 ori pT7-his6, AmpR | |
| pET15b | pET15b-zapC (A54Q F58E R65E) | |
| pET28b-His10-Smt3 | pBR322 ori pT7- his10-smt3, KanR | Gift from S. Shuman |
| CD3–763 | pDEST-GADT7,AmpR | ABRCb |
| CD3–764 | pDEST-GBKT7, KanR | ABRCb |
| pGAD-C1 | bla leu2 Padh1::gal4AD | 21 |
| pGBDU-C1 | bla ura3 Padh1::gal4BD | 21 |
| pKHH18 | pGAD-C1-ftsZ | |
| pKHH19 | pGBDU-C1-zapC (R150A) | |
| pKHH20 | pGBDU-C1-zapC (K94A) | |
| pKHH21 | pGBDU-C1-zapC (K89D) | |
| pKHH22 | pGBDU-C1-zapC (K37D) | |
| pKHH23 | pGBDU-C1-zapC (M38A) | |
| pKHH24 | pET28b-his10-smt3-zapC (K94D) | |
| pKHH25 | pDSW208-zapC-gfp | |
| pKHH26 | pDSW208-zapC K89D-gfp | |
| pKHH27 | pDSW208-zapC K94D-gfp | |
| pLT2 | pNG162-zapC | |
| pLT3 | pNG162-zapC (K37D) | |
| pLT4 | pNG162-zapC (K89D) | |
| pLT5 | pNG162-zapC (K94D) | |
| pLT6 | pNG162-zapC (R150E) | |
| pLT7 | pGBDUC1-zapC (R150E) | |
| pLT8 | pGBDUC1-zapC (K89A) | |
| pLT9 | pGBDUC1-zapC (K94D) | |
| pNG162 | pSC101 ori, SpecR | 31 |
| pPJ2 | pET21b (+)-ftsZ stop | 6 |
| pSC2 | pEXP-GBKT7-zapC | |
| pSC3 | pEXP-GADT7-ftsZ | |
| pSC8 | pEXP-GADT7-ftsZ1–372 | |
| pSC15 | pEXP-GBKT7-zapD | |
| pSC18 | pEXP-GBKT7-zipA | |
| pSC23 | pEXP-GADT7-ftsZ315–383 | |
| pZM25 | pGBDU-C1-zapC (K37A) |
a This work unless otherwise stated.
b Plasmids were obtained from the Arabidopsis Genetic Resource Center (ABRC).
Plasmid Construction
Plasmids expressing WT ZapC or mutant ZapC proteins in the pNG162 vector under the control of an IPTG-inducible promoter were constructed by amplifying zapC or zapC mutants using ZapC BamHI forward and pNG162 ZapC HindIII reverse primers from the pGBDU-C1-zapC or zapC mutant constructs. The PCR products were digested by BamHI and HindIII and ligated into the pNG162 vector at the same sites. To create GFP fusions, zapC or zapC mutants were amplified with ZapC BamHI forward and ZapC no stop SalI reverse primers, and the products were digested with BamHI and SalI and ligated into the pDSW208 vector treated with the same enzymes. The resulting clones were verified by Sanger sequencing (Genewiz).
Site-directed Mutagenesis
Appropriate primer pairs containing the desired mutations were used to amplify zapC using the QuikChange site-directed mutagenesis kit (Agilent). The amplified DNA was incubated with DpnI, which digests the methylated parental DNA strands. The DNA was then transformed into DH5α cells and plated onto Luria broth-agar plates. Plasmids were transformed into C41(DE3) cells, and the mutations were confirmed by sequencing. The resulting transformed cells were used for protein expression and purification as described for the WT protein. For yeast two-hybrid studies, sequence-verified mutant clones were digested with BamHI and SalI restriction enzymes and ligated into the pGBDU-C1 vector.
Protein Expression
Plasmids pET21b-ftsZ and pET28b-His10-Smt3 carrying zapC or zapD were expressed and purified as described (6, 13, 20). Removal of this tag leaves only one non-native residue (serine) at the protein's N terminus. This ZapC protein was used in all functional assays. Plasmid pET11b carrying ftsZ1–372 was created by amplifying an ftsZ fragment with FtsZ372-F1 and FtsZ372R1 primers. The product was digested with KpnI and BamH1 and used to replace the KpnI/BamH1 fragment in the pET11b-ftsZ plasmid. FtsZ1–372 protein was purified essentially as described (20). Plasmids pET28b-His10-Smt3 carrying zapC mutants were constructed by amplifying with SUMO-5 ZapC-BamHI forward primer and SUMO-3 ZapC-HindIII reverse primer from the appropriate pNG162-zapC mutant clones, and the PCR products were digested with BamHI and HindIII and ligated into the same sites in the vector. Sequence verified clones were transformed into C41(DE3) cells for protein expression.
Yeast Two-hybrid (Y2H) Assays
pGBDU-C1- or pGAD-C1-based zapC or ftsZ plasmid clones were chemically transformed into PJ69-4A or SL3004 yeast strains. Appropriate pairs of transformants from −leucine or −uracil plates were mated on yeast extract/peptone/dextrose media at room temperature. Diploid yeast strains were grown selectively in YNB (−leucine/uracil) broth at 30 °C for ∼20 h until A660 = 0.5–1.0, at which point β-galactosidase measurements were conducted using a Y2H assay kit essentially as described by the manufacturer (Thermo Fisher Scientific).
Histidine Growth Assays
The following primer pairs were used to amplify the full-length gene products: ftsZ 5P gateway (GW) and ftsZ 3P GW for ftsZ, ycbW 5P GW and ycbW 3P GW for zapC; yacF 5P GW and yacF 3P GW for zapD; and zipA 5P GW and zipA 3P GW for zipA. Gene deletions in ftsZ were amplified using primer pairs ftsZ315 5P GW and ftsZ 3P GW for ftsZ315–383, and ftsZ 5P GW and ftsZ-372 3P GW. Amplified products were recombined into pDEST-GBKT7 and pDEST-GADT7 vectors using Gateway (Thermo Fisher Scientific). Appropriate pairs of expression plasmids were co-transformed into yeast PJ69-4A strain and plated selectively on YNB (−LT) at 30 °C. Interactions between full-length ftsZ and ftsZ deletions with WT zapC, zapC mutants, zapD, and zipA were examined by patching 4–6 colonies on YNB (−LTH) media lacking histidine with or without the addition of 25 mm 3-amino-1,2,4-triazole supplement. Plates were incubated at 30 °C for 2 days, and then growth was examined, and the plates were imaged (Syngene).
Spot Viability Assays
Overnight cultures of WT (MG1655) strains bearing ZapC or ZapC mutant (native or GFP fusion) plasmids were grown in LB with 0.2% glucose containing appropriate antibiotics at 37 °C. Cells were normalized to A600 =1.0 and were serially diluted from 10−1 to 10−5. 3 μl from each dilution was spotted on LB plates with appropriate antibiotics without or with IPTG at 25, 50, or 100 μm final concentrations and grown for ∼16 h at 37 °C at which point plates were imaged (Syngene).
Fluorescence Microscopy
Overnight cultures of WT (MG1655) cells bearing ZapC-GFP or mutant ZapC-GFP fusion plasmids were grown in LB containing appropriate antibiotics at 37 °C. Cells were subcultured in the same medium at 37 °C until A600 = 0.2–0.3, at which point a range of IPTG at 0–100 μm final concentration was added to the cultures, and growth was continued to A600 = ∼0.6–0.8. Cells were then immobilized on 1.5% agarose pads and imaged using a Nikon TiE microscope essentially as described (13).
Western Analysis
WT cells bearing ZapC- or mutant ZapC-GFP fusion plasmids were grown as described above and prepared for Western analysis. Samples were normalized to A600, and proteins were separated on a 12.5% SDS-PAGE gel. Levels of ZapC, FtsZ, and RpoD (loading control) were probed using a rabbit polyclonal anti-GFP (Thermo Fisher Scientific), a rabbit polyclonal anti-FtsZ (Genscript), or a mouse monoclonal anti-RpoD primary antibody (BioLegend). Appropriate IRDye secondary antibodies were used for detection (LI-COR). Band intensities were measured with an Odyssey® CLx Infrared Imaging System (LI-COR).
Sedimentation Assay
FtsZ sedimentation assays were conducted essentially as described (13, 21). Briefly, 5 μm of purified FtsZ or FtsZ1–372 was combined with purified 1 or 5 μm ZapC, mutant ZapC, or ZapD proteins in a reaction buffer (50 mm K-MOPS, pH 6.5, 50 mm KCl, 2.5 mm MgCl2) containing 3 μm BSA. Polymerization was initiated by the addition of 1 mm GTP to the assembled reaction. Reaction mixtures (100 μl) were processed at room temperature, and products were recovered by centrifugation using a TLA100.3 rotor at 80,000 rpm for 10 min. Equivalent amounts of supernatant and pellet fractions were separated on 15% SDS-PAGE gels and visualized by staining with Simply Blue Safe Stain (Thermo Fisher Scientific).
Electron Microscopy
FtsZ polymerization reactions were performed essentially as described above but without the addition of BSA. After incubating for 5 min at room temperature, a 10-μl aliquot was placed on a carbon-coated copper grid and negatively stained with 2% uranyl acetate for ∼5 min and wicked dry. Images were collected on a JEOL 2100 Lab6 transmission electron microscope operated at 200 kV with 30 pA/cm2 current density and recorded on a US1000 XP1 camera at a nominal magnification of ×30,000.
Results
Structure Determination of E. coli ZapC
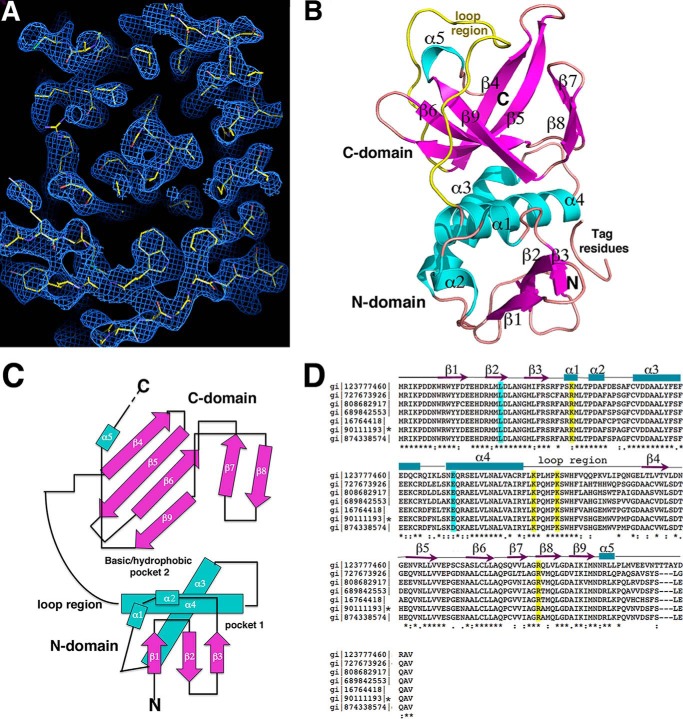
To gain insight into the molecular function of ZapC we determined its structure by selenomethionine MAD (“Experimental Procedures”). The quality of the MAD electron density map was excellent and permitted facile tracing of the structure (Fig. 1A). After initial model construction, a native x-ray intensity data set was obtained to 2.15 Å resolution and used for final refinement. The ZapC model had Rwork/Rfree values of 19.7%/23.1% (Table 1).
FIGURE 1.
Crystal structure of E. coli ZapC. A, experimental MAD electron density map (blue mesh) contoured at 1.0 σ and calculated to 2.4 Å with the final model included (yellow sticks). B, ribbon diagram of the ZapC structure. Secondary structural elements are labeled and colored magenta for β-strands and cyan for α-helices. Also labeled are the domains and the long loop region (yellow). This figure and Fig. 2A, Fig. 3, A–D, and Fig. 8 were generated using PyMOL. C, topology diagram of ZapC with β-strands shown as arrows and colored magenta, as in B, α-helices are colored cyan and shown as rods. D, sequence alignment of selected ZapC proteins. Highlighted in cyan are residues previously shown to be important in ZapC function (21) and in yellow, the basic residues that line the C-domain pocket that were analyzed for their affects on FtsZ binding and bundling. Identical/always conserved, highly similar/conserved, and semi-conserved residues are indicated by asterisks, double dots, and dots, respectively, under the alignment. Also shown over the sequence are the secondary structural elements obtained from the structure. The sequences denote the following bacteria: gi 123777460 sp Q7CHK5.1 Yersinia pestis; gi 727673926 gb KHF69007.1 Klebsiella pneumonia; gi 808682917 emb CQR76953.1 Enterobacter cloacae; gi 689842553 dbj GAL42017.1 Citrobacter freundii NBRC; gi 16764418 ref NP_460033.1 Salmonella enterica subsp. enterica serovar typhimurium str. LT2; gi 90111193 ref NP_415466.4 E. coli str. K-12 substr. MG1655, gi 874338574 emb CEP58367.1 Shigella flexneri 2a. An asterisk denotes the E. coli sequence.
Overall Structure of E. coli ZapC
Database analyses show that there are currently no proteins in the database that display significant overall structural similarity to ZapC (39). The ZapC structure has the following topology, β1(residues 9–15)-β2(18–23)-β3(29–31)-α1(36–38)-α2(41–44)-α3(51–65)-α4(71–87)-β4(112–116)-β5(122–128)-β6(135–140)-β7(144–147)-β8(150–154)-β9(155–161)-α5(162–164) (Fig. 1, B and C). Only C-terminal residues 169–180 are disordered in the structure, consistent with the fact that although the ZapC C-terminal region is relatively conserved among enteric bacteria, this region shows significant diversity in both length and sequence among ZapC homologs in other bacteria (Fig. 1D).
The ZapC structure is composed of two domains, an N-domain from residues 1–89, and a C-domain, from residues 90–168. The region linking α4 in the N-domain to β4 is composed of a long loop (Fig. 1B). The ZapC N-domain consists of two long helices that pack nearly perpendicular to a three-stranded antiparallel β-sheet. This domain also contains two short helices, α1 and α2. The C-domain is composed almost entirely of β-strands and has a β-barrel-like fold with a central, four-stranded twisted antiparallel β-sheet. This twisted β-sheet is located in the center of the domain and is surrounded on one side by the β7-β8 hairpin and the other by α5 and the long loop region (Fig. 1B).
ZapC Is a Monomer
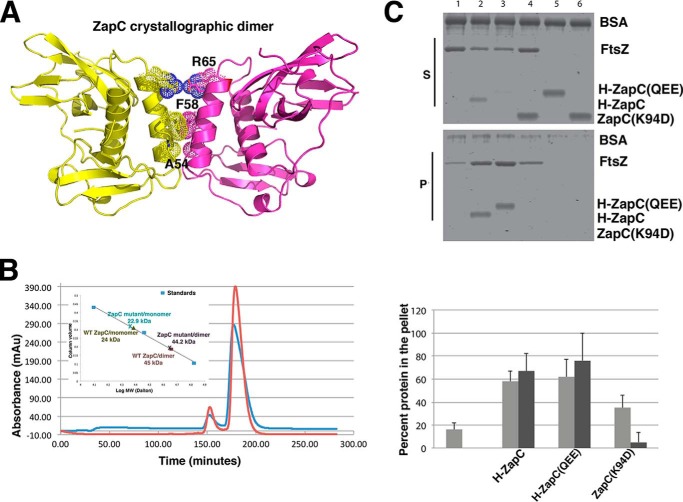
ZapC plays a similar role as ZapA in promoting the bundling of FtsZ protofilaments (20, 21). Recent studies have indicated that ZapA carries out this role by functioning as a cross-linking agent to bridge FtsZ protofilaments between subunits of its dimer (33). The oligomeric state of ZapC, however, has been unclear. But prior genetic and biochemical data suggested that unlike ZapA, this protein may be a monomer (20). Consistent with this, analysis of the ZapC crystal packing revealed no oligomers with significant buried surface areas. Only one possible dimer interface was revealed by PISA as being somewhat energetically favorable as its interface is largely hydrophobic with a ΔiG, p value = 0.187, where ΔiG p values <0.5 are suggestive of possible dimers (Fig. 2A) (40). However, the buried surface area of this interface is only 550 Å2 (for the dimer), which is much lower than known stable dimers.
FIGURE 2.
Analysis of ZapC oligomeric state. A, ribbon diagram of possible ZapC dimer revealed by crystal packing. Shown as dotted surfaces are the three residues, Ala-54–Phe-58–Arg-65, that participate in this dimer and that were mutated to assess dimer formation. The small size of the Ala-54 side chain is essential for dimer packing, the two Phe-58 side chains stack together, and Arg-65 makes a salt bridge with Glu-64′. The buried surface area in this interface is 550 Å2, which is significantly lower than known stable dimers; however, the interface is largely hydrophobic with a (ΔiG, p value = 0.187), where ΔiG p value indicates the p value of the observed solvation free energy gain (40). The p value measures the probability of getting a lower than observed ΔiG when the interface atoms are picked randomly from the protein surface. p < 0.5 designates interfaces with higher than would-be average for given structures due to the hydrophobicity of the interface, suggesting that the interface could be interaction-specific. B, SEC analyses of WT ZapC and ZapC(A54Q/F58E/R65E). Both proteins eluted primarily as monomers (≥90%). The inset is a plot used to deduce the Mr. Proteins used as standards to generate the curve were cytochrome c, carbonic anhydrase, and albumin. mAu, milliabsorbance units. C, sedimentation analyses of the ZapC(A54Q/F58E/R65E) triple mutant (QEE) showing properties essentially identical to WT ZapC, His-tagged ZapC, and ZapC(QEE) were used in these assays and were essentially similar to tag-free ZapC/FtsZ interactions. Equivalent aliquots of supernatants (S) and pellets (P) from co-sedimentation reactions in the presence of GTP were separated by SDS-PAGE and visualized. Lanes: 1, FtsZ alone; 2, FtsZ with WT ZapC; 3, FtsZ with ZapC(QEE); 4, FtsZ with ZapC(K94D); 5, ZapC(QEE) alone; 6, ZapC(K94D) alone. The bar graph represents the percent FtsZ (gray) and His-ZapC or His-ZapC(QEE)/ZapC(K94D) (black) amounts found in the pellet. Band intensities were measured using Image J (National Institutes of Health). Each bar shows the average amounts with S.D. of three independent trials. The first bar represents the reaction carried out with FtsZ alone and no ZapC variant.
As the oligomeric state of ZapC has key implications for its function, we performed SEC experiments to probe its molecular weight. SEC of WT ZapC revealed that it was primarily monomeric even at high concentrations (∼5 mg/ml); 92% of the protein eluted as a monomer with 8% running at the molecular weight corresponding to a dimer (Fig. 2B). To ascertain if the “dimer” population was due to the partially hydrophobic dimer observed in the crystals, a ZapC triple mutant, ZapC(A54Q/F58E/R65E), was constructed to disrupt the dimer. Each of the three mutations alone would be predicted to strongly disfavor the dimer. The side chain of Ala-54 is 4.0 Å from that of Ala-54′ (where the ′ indicates other subunit in the dimer), and hence any substitution other than glycine would lead to steric clash. Phe-58 stacks closely with Phe-58′ so that a F58E substitution would be electrostatically unfavorable, and finally Arg-65 makes salt bridges with Glu-64′, and therefore a R65E substitution would also cause strong charge repulsion (Fig. 2A). SEC analyses of the triple mutant revealed essentially the same monomer/dimer ratio (90%/10%). Thus, the data indicate that the dimer observed in the ZapC crystal is not found in solution, and WT ZapC appears to be a functional monomer. The small dimer peak appears to be due to the noted aggregation that takes place even in the presence of high levels of reducing agents. Moreover, glutaraldehyde cross-linking showed that ZapC remained almost entirely monomeric under conditions when a control oligomeric protein readily dimerized (data not shown). Thus, our experiments indicate that ZapC is monomeric and that the Ala-54–Phe-58–Arg-65 region is not involved in oligomerization. However, to assess if the hydrophobic surface region containing Ala-54–Phe-58–Arg-65 is involved in the FtsZ binding/bundling function of ZapC, we performed sedimentation studies with the triple mutant. The results revealed that ZapC(A54Q/F58E/R65E) displayed sedimentation properties that were essentially indistinguishable from the WT protein (Fig. 2C). Thus, these data indicate that the ZapC surface encompassing residues Ala-54–Phe-58–Arg-65 (Fig. 2A) does not contribute to its FtsZ bundling function.
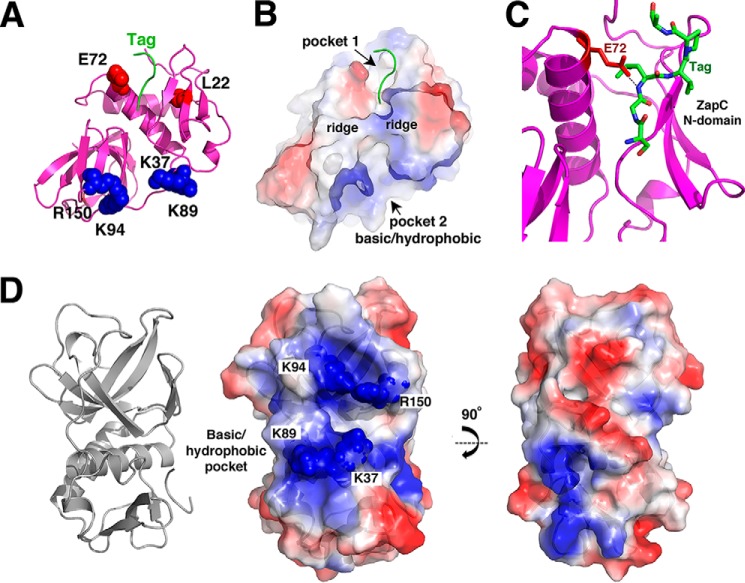
The ZapC Structure Revealed Pockets Lined by Residues Involved in FtsZ Binding
The finding that ZapC does not form a dimer or other oligomer indicates that it mediates FtsZ bundling in a manner distinct from the other Zaps. Closer examination of the ZapC structure revealed the presence of two adjacent pockets, one on the N-domain and one on the C-domain (Figs. 1C and 3, A and B). The pockets are separated by ridges formed by two loops; one composed of β3-α1 on the N-domain and the other, the β8-β9 connection in the C-domain (Fig. 3B). Notably, the N-domain contains two residues previously implicated as important for ZapC function, Leu-22 and Glu-72 (21). Substitution of Leu-22 to a proline led to an inactive ZapC that could not bundle FtsZ. Interestingly, the ZapC(L22P) was found to have increased self-association, possibly through alterations in the overall structure. If this pocket is involved in FtsZ binding, such structural changes would be predicted to impede the ZapC-FtsZ interaction. Residue Glu-72, also in this N-domain pocket, was identified as part of a double ZapC mutant, E72G/R164A, that was nonfunctional (21). In the ZapC structure, Arg-164 is located on the C-domain, and its side chain participates in hydrogen bonds with the C-terminal carbonyl groups of residues on the long interdomain loop and the side chain of Asp-163. As a result, Arg-164 is not exposed and thus not free to mediate contacts with outside proteins. Because Glu-72 is exposed to the surface, it seems a likely candidate for participating in protein-protein interactions, such as with FtsZ. Strikingly, tag residues (from the His tag attached to the N terminus of the protein and used for purification) were found to bind the N-domain and interact directly with the Glu-72 side chain. Specifically, six of the tag residues are docked within the pocket and form a distorted β-strand that hydrogen bonds with Glu-72 as well as β3 in the N-domain (Fig. 3C). The finding that Glu-72 contacts a docked “peptide” lends support to the notion that it may be a residue involved in substrate (FtsZ) binding.
FIGURE 3.
Pockets on the ZapC structure. A, ribbon diagram of ZapC showing relative location of pockets in the N-domain and C-domain. Residues previously identified as important in ZapC function/FtsZ binding, Glu-72 and Leu-22 (colored red), that line the N-domain pocket are shown as spheres. Basic residues that surround the larger C-domain pocket are shown as spheres and are colored blue. Also shown is the N-terminal tag that was found docked into the N-domain pocket (green). B, surface representation of ZapC shown in the same orientation as A and colored according to electrostatic potential, with blue and red representing electropositive and electronegative regions, respectively. This figure underscores the strong electropositive nature of the sides of the C-domain pocket. This figure also underscores the hydrophobic nature of the pocket cavities (indicated as white surfaces). C, close up of the interaction between the tag residues docked in the N-domain pocket and the side chain of Glu-72. D, focus on the C-domain pocket and the location of the basic residues that line the pocket, which were mutated to assess their roles in ZapC function. Left is the ribbon diagram to orient the location of the pocket, and center is the electrostatic surface. Right is the same electrostatic figure but rotated by 90° to underscore that this surface has neither strong charge characteristics nor does it contain a deep pocket.
The N-domain pocket containing Leu-22 and Glu-72 is directly adjacent to a larger pocket located on the C-domain (Fig. 3, A, B, and D). Electrostatic surface analysis of ZapC shows that this large, exposed pocket has an exposed hydrophobic center with sides lined by multiple basic residues (Fig. 3, A and B). Residues that contribute to the electropositive nature of the outside of the pocket include Lys-37, Lys-89, and Lys-94, and Arg-150 (Fig. 3D). Lys-37 is located on the ledge formed by β3 and α1, and its side chain points into the C-domain pocket. Surface-exposed charged residues are typically not retained between protein homologs unless they are of functional importance. Therefore, it is notable that these basic residues which encircle the ZapC C-domain pocket are strongly conserved among ZapC proteins as either arginines or lysines (Fig. 1D).
The presence of the large hydrophobic/basic pocket in the ZapC C-domain was intriguing because it lies adjacent to the N-domain pocket and also because it harbors a strongly electropositive surface. The latter is of note because its interacting partner, FtsZ, is electronegative, and FtsZ polymerization properties are known to be affected by charge. Indeed, the importance of charge in FtsZ function was underscored by recent studies on the Bacillus subtilis FtsZ protein, which demonstrated that not only cations, but the electropositive residues located on its own CTT, facilitated its ability to polymerize and bundle (6, 41). Although E. coli FtsZ filamentation does not appear to be similarly affected by its own CTT, it is also electronegative, and cations have been shown to impact its polymerization properties (4–6). Thus, these combined data suggested the basic/hydrophobic C-domain pocket of ZapC as a possible FtsZ interaction site.
Residues in ZapC Pockets Contributed to Interaction with FtsZ in a Y2H System
To probe the ZapC-FtsZ interaction and test structure based hypotheses, we employed a Y2H assay. The WT ZapC protein showed robust binding to full-length (FL) FtsZ, whereas ZapC(L22P) showed no interaction with FL FtsZ (21). We next examined whether mutations of charged and hydrophobic residues (Lys-37, Met-38, Lys-89, Lys-94, and Arg-150) within the C-domain pocket of ZapC affected FtsZ binding. Mutations in each of these residues to alanines resulted in interactions with FtsZ comparable with that of WT ZapC (Table 4). On the other hand, substitutions to the opposite charge at each of the lysine or arginine residues significantly reduced interactions with FtsZ (Table 4). ZapC(K89D) and ZapC(K94D) substitutions failed to interact with FtsZ similar to the ZapC(L22P) mutant (Table 4). These results suggest that the C-domain pocket residues in ZapC may serve as an interaction site with FtsZ. To further test this hypothesis we carried out additional genetic and cytological studies.
TABLE 4.
Residues in the C-domain basic pocket of ZapC are important for interaction with FtsZ in a yeast two-hybrid assay
| BD-fused proteina | AD-fused proteina | β-Galactosidase activityb | S.D. |
|---|---|---|---|
| –c | FtsZ | 20.2 | 4.2 |
| ZapC | –c | 16.5 | 4 |
| ZapC | FtsZ | 100 | 6 |
| ZapC L22P | FtsZ | 17.9 | 1.1 |
| ZapC K37A | FtsZ | 78.8 | 11.7 |
| ZapC M38A | FtsZ | 116.1 | 19.1 |
| ZapC K89A | FtsZ | 75.1 | 15.7 |
| ZapC K94A | FtsZ | 65.4 | 6.1 |
| ZapC R150A | FtsZ | 77.8 | 7.7 |
| ZapC K37D | FtsZ | 36.1 | 12.6 |
| ZapC K89D | FtsZ | 25.4 | 9.8 |
| ZapC K94D | FtsZ | 23.9 | 1.2 |
| ZapC R150E | FtsZ | 43.5 | 5.1 |
a WT ZapC and various ZapC mutants fused to the DNA binding domain (BD) were co-expressed with FtsZ fused to activation domain (AD) in yeast cells.
b β-Galactosidase activity is provided in Miller units ± S.D. from at least three independent experiments. Miller units were calculated based on the formula (1000 × A420)/(time × volume × A660) where time and volume relate to reaction time and volumes. All interactions are reported relative to the full-length ZapC/FtsZ interaction, which is normalized to 100.
c Dash indicates unfused binding or activation domains.
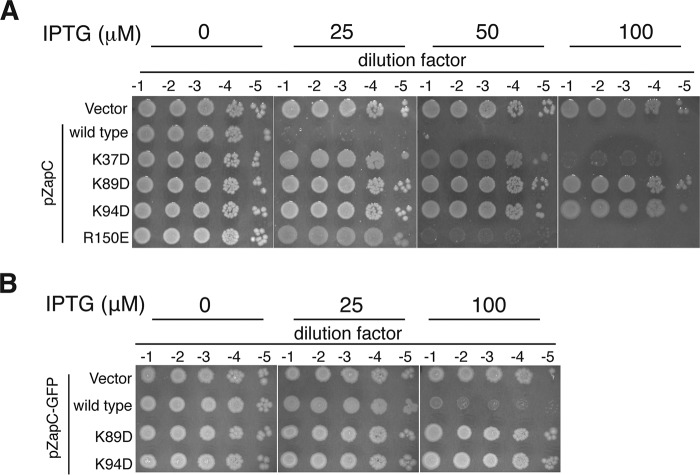
Overexpression of ZapC(K89D) or ZapC(K94D) Did Not Lead to Lethal Filamentation
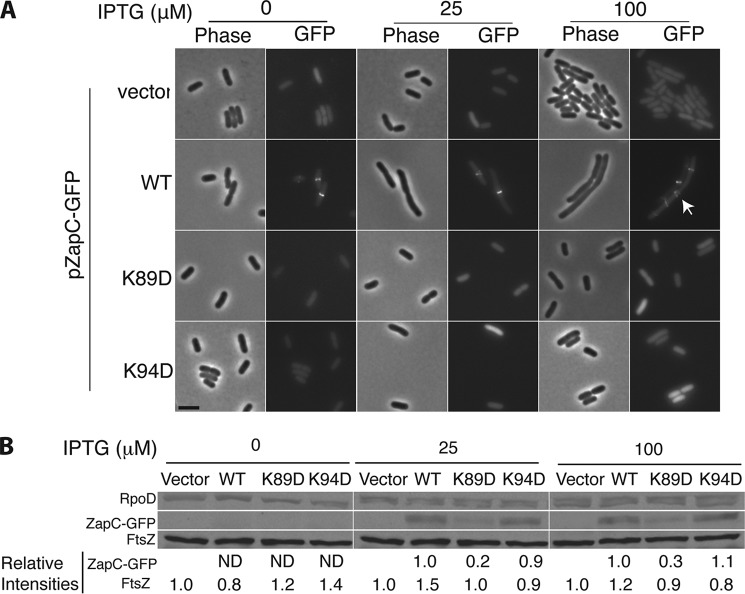
Increased expression of WT ZapC interferes with cell division by hyperstabilizing Z-rings and leading to lethal filamentation (20, 21). We tested the ZapC C-domain pocket mutants for their ability to interfere with cell division upon overexpression by monitoring their viability. WT ZapC expression under leaky or at low inducer concentrations led to a severe plating defect. These viability defects were exacerbated at higher inducer concentrations (Fig. 4A). In contrast, overexpression of ZapC single mutants showed varying degrees of survival with ZapC(K89D) and ZapC(K94D) displaying growth even at the highest level of inducer concentration tested (Fig. 4A). Consistent with these data, overexpression of a ZapC-GFP fusion was toxic to the cell, but multicopy expression of ZapC(K89D)-GFP or ZapC(K94D)-GFP did not display any obvious plating deficiencies (Fig. 4B).
FIGURE 4.
Overexpression of ZapC(K89D) and ZapC(K94D) did not interfere with cell division distinct from WT ZapC. A, 10-fold serial dilutions of WT (MG1655) cells expressing vector alone (KHH216), WT ZapC (KHH210), or mutant ZapC (KHH211, KHH212, KHH213, and KHH214) were spotted on LB Spec media without or with IPTG and incubated overnight at 37 °C. B, 10-fold serial dilutions of WT cells expressing GFP fusion plasmid alone (KHH240), WT ZapC-GFP (KHH236), ZapC(K89D)-GFP (KHH237), or ZapC(K94D)-GFP (KHH239) were spotted on LB Amp agar without or with IPTG and incubated at 37 °C.
To examine whether the reduced toxicity upon overexpression of ZapC(K89D) or ZapC(K94D) arose from an inability of these mutant proteins to localize to midcell, we visualized the cell morphology and localization phenotypes of the mutant proteins. WT ZapC localized GFP to midcell even upon leaky expression from a multicopy plasmid (Fig. 5A). As reported previously, with increasing inducer concentrations, filamentation and aberrant ZapC-GFP structures were observed at periodic intervals along the cells (Fig. 5A). In contrast, cells expressing either ZapC(K89D)-GFP or ZapC(K94D)-GFP displayed diffuse localization of GFP even at all inducer concentrations with no obvious division defects (Fig. 5A).
FIGURE 5.
ZapC(K89D) and ZapC(K94D) mutants were unable to localize GFP to midcell. A, representative phase and fluorescence images of WT (MG1655) cells expressing ZapC-GFP (KHH236) or mutant ZapC-GFP fusions (KHH237 and KHH239) were grown and visualized as described in “Materials and Methods.” The arrow in the image panel expressing WT ZapC upon 100 μm IPTG induction indicates aberrant localization of the ZapC-GFP fusion. Bar = 5 μm. B, quantitative immunoblotting of FtsZ, ZapC and mutant ZapC proteins upon overexpression of ZapC- or mutant ZapC-GFP fusions. Whole cell proteins from equal quantities of each strain (KHH236, KHH237, and KHH239) were separated by SDS-PAGE and immunoblotted with anti-GFP, anti-FtsZ, or anti-RpoD antibodies. A representative blot (of five independent trials) with relative intensity values is reported. ND = signal not detectable.
To investigate whether the ZapC(K89D)-GFP and ZapC(K94D)-GFP overexpression phenotypes were simply due to changes in FtsZ levels or the instability of the ZapC-GFP fusion proteins, we probed both ZapC and FtsZ amounts by immunoblotting. FtsZ levels were unperturbed under all conditions tested (Fig. 5B). Although ZapC(K94D) levels were essentially similar to that of WT ZapC overexpression amounts, ZapCK89D levels were significantly lower, suggesting poor expression or excessive processing of this mutant protein (Fig. 5B). Therefore, we chose ZapC(K94D) for further study.
ZapC(K94D) Failed to Interact with FtsZ in Vitro
We examined the effects of the ZapC(K94D) mutant on FtsZ polymerization by using purified proteins in a FtsZ sedimentation assay and by visualizing morphological properties of FtsZ polymers by transmission electron microscopy. Notably, the K94D mutation did not affect folding as assessed by CD analyses. Moreover, ZapC(K94D) eluted with a near identical profile to the WT protein (92% monomeric and 8% dimeric). When incubated in the presence of GTP alone, ∼20% of WT FtsZ was found in the pellet, and this amount increased 2–3-fold in the presence of ZapC at 1:0.2 and 1:1 ratios of the two proteins with a significant amount of ZapC co-sedimenting with FtsZ under these reaction conditions (Fig. 6A). In contrast, the ZapC(K94D) mutant failed to promote an increase in pelletable FtsZ amounts even at 1:1 ratios, and ZapC(K94D) was mostly retained in the supernatant fractions (Fig. 6A).
FIGURE 6.
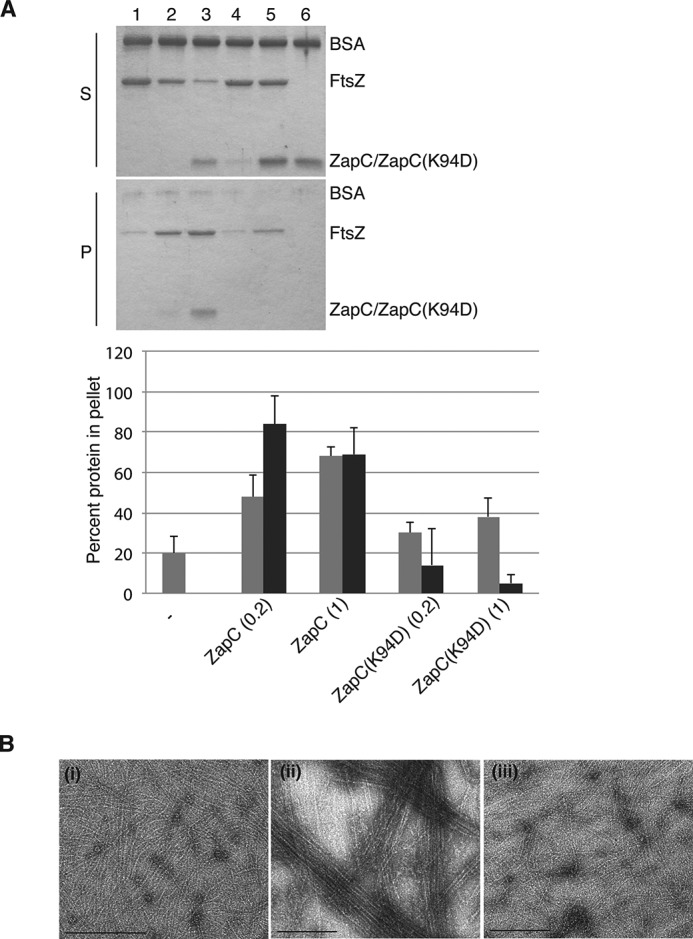
The ZapC(K94D) mutant did not promote FtsZ polymeric assemblies in vitro. A, equivalent aliquots of supernatants (S) and pellets (P) from co-sedimentation reactions in the presence of GTP containing FtsZ with either WT ZapC or ZapC(K94D) were separated by SDS-PAGE and visualized. Lane 1, FtsZ alone; lanes 2 and 3, FtsZ with ZapC at 1 and 5 μm, respectively; lanes 4 and 5, FtsZ with ZapC(K94D) at 1 and 5 μm, respectively; lane 6, ZapC(K94D) alone. The bar graph shows average amounts with S.D. of FtsZ (gray) or ZapC or ZapC(K94D) (black) amounts that were recovered in the pellet in each reaction and was compiled from three independent trials. Band intensities were measured using Image J. B, negative-stained electron microscopy images of FtsZ polymer assemblies in the presence of GTP alone (i), with WT ZapC (ii), and with ZapC(K94D) (iii). Bar = 200 nm.
Upon visualization of the reaction mixtures, FtsZ forms single protofilaments in the presence of GTP and large bundled polymeric assemblies in the presence of GTP and ZapC (Fig. 6B) (20, 21). Unlike WT ZapC, in the presence of ZapC(K94D), FtsZ protofilaments do not associate to form polymeric assemblies (Fig. 6B). These results are consistent with the Lys-94 residue of ZapC being critical in mediating interactions with FtsZ.
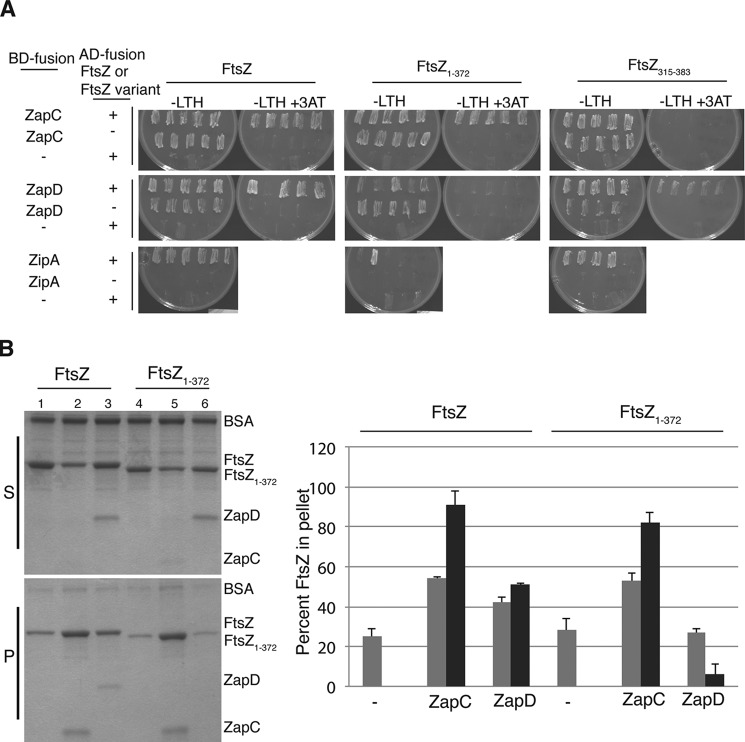
ZapC Did Not Require the FtsZ CTT for Interaction
Unlike a number of FtsZ modulatory proteins, previous studies suggested that ZapC does not require the CTT for interaction with FtsZ (13). However, to analyze this in more detail we carried out Y2H and FtsZ sedimentation assays. These data revealed that ZapC bound to FtsZ fusions lacking the CTT residues (FtsZ1–372)but failed to interact with FtsZ lacking the N-terminal and globular core residues (FtsZ315–383) (Fig. 7A). The essential division protein ZipA, whose interaction with FtsZ CTT has been well characterized, and the FtsZ modulatory protein ZapD, which also interacts with FtsZ CTT, failed to interact with a FtsZ variant lacking the CTT residues (Fig. 7A) (13, 42). To test more directly for interactions between ZapC and FtsZ, we conducted a FtsZ sedimentation assay with purified FL FtsZ and FtsZ1–372. Residues in the E. coli FtsZ CTT are not required for its polymerization (43, 44). Consistent with earlier results, in the presence of GTP, purified ZapD increased pelletable amounts of FL FtsZ but failed to increase sedimentable amounts of FtsZ1–372, indicating the requirement of the CTT residues for the ZapD-FtsZ interaction (Fig. 7B) (13). By contrast, ZapC enhanced the sedimentation (∼2×) of both FL FtsZ and truncated FtsZ1–372 proteins, and appreciable levels of ZapC were present in the pellet in these reactions (Fig. 7B). These results align with the Y2H data and indicate that the FtsZ globular core mediates interactions with ZapC. Recent work by Bhattacharya et al. (45) also showed that the FtsZ CTT is not required for interaction with ZapC.
FIGURE 7.
ZapC did not require the FtsZ CTT for interaction with FtsZ. A, interactions between FL ftsZ (pSC3) and ftsZ lacking the C-terminal tail residues (pSC8) fused to activation domains (AD) with WT zapC (pSC2), zapD (pSC15), or zipA (pSC18) fused to binding domains (BD) were visualized by examining the growth of 4–6 diploid yeast colonies patched selectively on YNB media (−LTH) without or with 25 mm 3-amino-1,2,4-triazole (3AT) at 30 °C for 2 days. Note: BD-ZipA fusion alone displayed no background growth in YNB (−LTH) plates, and therefore, FtsZ/ZipA interactions could be scored under those conditions. In contrast, BD-ZapC and BD-ZapD fusions alone showed considerable growth in YNB (−LTH) plates, and therefore interactions could only be scored on YNB (−LTH) with 3-amino-1,2,4-triazole. The – indicates unfused binding or activation domains. B, equivalent aliquots of co-sedimentation reactions in the presence of GTP containing ZapC or ZapD (1 μm) with either FL FtsZ or FtsZ1–372 (5 μm) were separated by SDS-PAGE and visualized. Lane 1, FL FtsZ alone; lane 2, FL FtsZ with WT ZapC; lane 3, FL FtsZ with WT ZapD; lane 4, FtsZ1–372 alone; lane 5, FtsZ1–372 with ZapC; lane 6, FtsZ1–372 with ZapD. The bar graph represents the amounts of FtsZ (gray), ZapC, or ZapD (black) proteins that were recovered in the pellets. Band intensities were measured using Image J. Average and S.D. values of three independent experiments are reported. S, supernatants; P, pellets; - indicates FtsZ alone.
Discussion
The ZapA-D proteins enhance FtsZ lateral contacts, which promotes the interactions that give rise to Z-ring formation required for cell division. Both ZapA and ZapD are dimeric and hence could act as cross-linking reagents of FtsZ protofilaments. Recent structural and biochemical studies on the E. coli ZapA have provided strong support for the hypothesis that it functions as a dimeric cross-linking reagent, binding FtsZ protofilaments using its globular core regions (33). The molecular mechanism by which ZapC may affect FtsZ polymerization has been less clear as no structure has been obtained for it or for a homolog. Here we show that ZapC contains a previously unobserved fold that is composed of two domains. Furthermore, biochemical analyses showed that, unlike its Zap counterparts, ZapC is monomeric, suggesting that it employs a mechanism distinct from ZapA and ZapD in FtsZ bundling. Insight into ZapC function was provided by examination of the structure, which revealed two pockets that are present on each domain. A fortuitously bound tag pointed to a largely hydrophobic pocket in the N-domain as a potential protein binding site. Strikingly, this pocket is flanked by two residues shown to be important for ZapC's function as a Zap, with one of these residues, Glu-72, directly contacting the bound tag. Thus, this pocket appears to form at least part of the FtsZ interacting surface. Further analysis of the structure revealed that this pocket is directly adjacent to a larger pocket within the C-domain that is hydrophobic and basic in character. Our subsequent analyses of ZapC proteins with charge reversal mutations of residues in this region strongly support this pocket as also being important in FtsZ binding. Thus, the two pockets appear to form the nexus of the FtsZ binding surface of ZapC. Indeed, the mutation of residues, A54Q/F58E/R65E, which lie on the surface that is opposite to these pockets, had no effect on FtsZ binding.
The ZapC N-domain and C-domain pockets provide an extensive surface for protein-protein interactions, which is consistent with analyses showing that the FtsZ globular core binds ZapC and not its small CTT. Recent studies suggested that hydrophobic residues are important for the ZapC-FtsZ interaction (45). Our structure shows that the N-domain and C-domain ZapC pockets that interact with FtsZ both contain exposed hydrophobic cavities, consistent with that supposition (45). However, we also found that basic residues at the sides of the C-domain pocket impacted the interaction of ZapC with FtsZ. This effect was only notable upon substitution of the basic residues to acidic residues; substitution to noncharged alanines had no effect. Thus, these residues may play nonspecific roles in complex formation, such as steering or charge stabilization. Further studies will be needed to dissect the contributions of specific ZapC residues in the ZapC-FtsZ interaction.
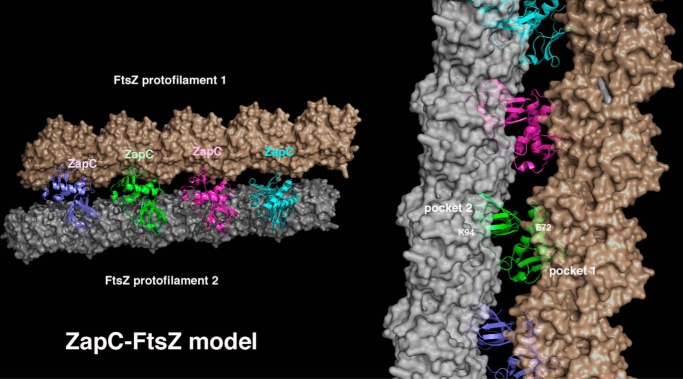
Thus, our combined data point to a general mechanism for ZapC-mediated FtsZ binding/bundling that involves two adjacent pockets on a ZapC monomer (Fig. 8). Strikingly, modeling shows that binding by each of these regions of the ZapC monomer to FtsZ molecules from distinct protofilaments brings the protofilaments within very close proximity to facilitate bundling without leading to steric clash between the protofilaments (Fig. 8). Moreover, the continued addition of ZapC monomers leads to a “zipper” model in which non-interacting ZapC subunits align to fasten together the two protofilaments (Fig. 8). An alternative to this model is that each ZapC pocket region could bind FtsZ molecules and aid in their incorporation into a single protofilament. This might facilitate FtsZ bundling, as previous studies have indicated that the generation of longer, more stable FtsZ protofilaments facilitates the eventual formation lateral contacts (46, 47). Previous data have indicated that ZapC decreases the GTPase activity of FtsZ (20, 21). This could be explained by the zipper model in which multiple ZapC molecules bind close to the GTP binding pocket between subunits. Clearly, further studies will be needed to probe these models and obtain an understanding of the likely complex molecular mechanism by which this novel FtsZ regulatory protein functions.
FIGURE 8.
Molecular model for how ZapC may enhance FtsZ interactions/bundling. In this model the adjacent pockets of ZapC bind adjacent FtsZ protofilaments, effectively zipping them together. FtsZ protofilaments (FtsZ, PDB code; 4DXD) are shown as schematic surfaces as the ZapC interacting residues on FtsZ are currently unknown. In this model the ZapC pockets appear optimally positioned to stimulate protofilament bundling without steric clash. The right shows a close-up of the modeled interaction highlighting how the ZapC pockets (with residues shown to be important for FtsZ contacts labeled) dock onto FtsZ protofilaments.
Author Contributions
M. A. S. and A. J. designed the study and wrote the paper. W. Z. purified the proteins. M. A. S. crystallized ZapC and determined its x-ray structure. K.-H. H. and L. T. performed all functional studies including EM and sedimentation studies. All authors analyzed the results and approved the final version of the manuscript.
Acknowledgments
We thank the Advanced Light Source and their support staff. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, and Material Science Division of the United States Department of Energy at the Lawrence Berkeley National Laboratory. We are grateful to Piet de Boer, Beate Schwer, and Sandra Lemmon for generous gifts of plasmids and strains. We acknowledge Jorge Morales for assistance with transmission electron microscopy and Chase Budell, Sumra Chaudhry, and Eugene Rivkin for technical assistance.
This work was supported by National Institutes of Health Grants GM074815 (to M. A. S.) and 8G12MD007603-29 (NIMHD, for core facility support to CCNY). This work was also supported by National Science Foundation Grant MCB1158059 (to A. J.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The atomic coordinates and structure factors (code 5E1L) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- CTT
- C-terminal tail
- IPTG
- isopropyl β-d-1-thio-galactopyranoside
- Ni-NTA
- nickel-nitrilotriacetic acid
- SEC
- size exclusion chromatography
- YNB
- yeast nitrogen broth
- Y2H
- yeast two-hybrid
- FL
- full-length
- MAD
- multiple wavelength anomalous diffraction.
References
- 1. Adams D. W., and Errington J. (2009) Bacterial cell division, assembly, maintenance and disassembly of the Z ring. Nat. Rev. Microbiol. 7, 642–653 [DOI] [PubMed] [Google Scholar]
- 2. Bi E. F., and Lutkenhaus J. (1991) FtsZ ring structure associated with division in Escherichia coli. Nature 354, 161–164 [DOI] [PubMed] [Google Scholar]
- 3. de Boer P. A. (2010) Advances in understanding E. coli cell fission. Curr. Opin. Microbiol. 13, 730–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Erickson H. P., Anderson D. E., and Osawa M. (2010) FtsZ in bacterial cytokinesis, cytoskeleton and force generator all in one. Microbiol. Mol. Biol. Rev. 74, 504–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lutkenhaus J., Pichoff S., and Du S. (2012) Bacterial cytokinesis, from Z ring to Divisome. Cytoskeleton 69, 778–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Buske P. J., and Levin P. A. (2012) Extreme C terminus of bacterial cytoskeletal protein FtsZ plays fundamental role in assembly independent of modulatory proteins. J. Biol. Chem. 287, 10945–10957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Boer P., Crossley R., and Rothfield L. (1992) The essential bacterial cell-division protein FtsZ is a GTPase. Nature 359, 254–256 [DOI] [PubMed] [Google Scholar]
- 8. RayChaudhuri D., and Park J. T. (1992) Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature 359, 251–254 [DOI] [PubMed] [Google Scholar]
- 9. Mukherjee A., and Lutkenhaus J. (1998) Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J. 17, 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Milam S. L., Osawa M., and Erickson H. P. (2012) Negative-stain electron microscopy of inside-out FtsZ rings reconstituted on artificial membrane tubules show ribbons of protofilaments. Biophys. J. 103, 59–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Weart R. B., and Levin P. A. (2003) Growth rate-dependent regulation of medial FtsZ ring formation. J. Bacteriol. 185, 2826–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang K. H., Durand-Heredia J., and Janakiraman A. (2013) FtsZ ring stability, of bundles, tubules, cross-links, and curves. J. Bacteriol. 195, 1859–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Durand-Heredia J., Rivkin E., Fan G., Morales J., and Janakiraman A. (2012) Identification of ZapD as a cell division factor that promotes the assembly of FtsZ in Escherichia coli. J. Bacteriol. 194, 3189–3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mohammadi T., Ploeger G. E., Verheul J., Comvalius A. D., Martos A., Alfonso C., van Marle J., Rivas G., and den Blaauwen T. (2009) The GTPase activity of Escherichia coli FtsZ determines the magnitude of the FtsZ polymer bundling by ZapA in vitro. Biochemistry 48, 11056–11066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ebersbach G., Galli E., Møller-Jensen J., Löwe J., and Gerdes K. (2008) Novel coiled-coil cell division factor ZapB stimulates Z ring assembly and cell division. Mol. Microbiol. 68, 720–735 [DOI] [PubMed] [Google Scholar]
- 16. Galli E., and Gerdes K. (2012) FtsZ-ZapA-ZapB interactome of Escherichia coli. J. Bacteriol. 194, 292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pacheco-Gómez R., Cheng X., Hicks M. R., Smith C. J., Roper D. I., Addinall S., Rodger A., and Dafforn T. R. (2013) Tetramerisation of ZapA is required for FtsZ bundling. Biochem. J. 449, 795–802 [DOI] [PubMed] [Google Scholar]
- 18. Marteyn B. S., Karimova G., Fenton A. K., Gazi A. D., West N., Touqui L., Prevost M. C., Betton J. M., Poyraz O., Ladant D., Gerdes K., Sansonetti P. J., and Tang C. M. (2014) ZapE is a novel cell division protein interacting with FtsZ and modulating the Z-ring dynamics. MBio 5, e00022–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Low H. H., Moncrieffe M. C., and Löwe J. (2004) The crystal structure of ZapA and its modulation of FtsZ polymerisation. J. Mol. Biol. 341, 839–852 [DOI] [PubMed] [Google Scholar]
- 20. Durand-Heredia J. M., Yu H. H., De Carlo S., Lesser C. F., and Janakiraman A. (2011) Identification and characterization of ZapC, a stabilizer of the FtsZ ring in Escherichia coli. J. Bacteriol. 193, 1405–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hale C. A., Shiomi D., Liu B., Bernhardt T. G., Margolin W., Niki H., and de Boer P. A. (2011) Identification of Escherichia coli ZapC (YcbW) as a component of the division apparatus that binds and bundles FtsZ polymers. J. Bacteriol. 193, 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Flynn J. M., Neher S. B., Kim Y. I., Sauer R. T., and Baker T. A. (2003) Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol. Cell 11, 671–683 [DOI] [PubMed] [Google Scholar]
- 23. Camberg J. L., Hoskins J. R., and Wickner S. (2009) ClpXP protease degrades the cytoskeletal protein, FtsZ, and modulates FtsZ polymer dynamics. Proc. Natl. Acad. Sci. U.S.A. 106, 10614–10619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hamoen L. W., Meile J.-C., de Jong W., Noirot P., and Errington J. (2006) SepF, a novel FtsZ-interacting protein required for a late step in cell division. Mol. Microbiol. 59, 989–999 [DOI] [PubMed] [Google Scholar]
- 25. Król E., van Kessel S. P., van Bezouwen L. S., Kumar N., Boekema E. J., and Scheffers D. J. (2012) Bacillus subtilis SepF binds to the C-terminus of FtsZ. PLoS ONE 7, e43293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Goley E. D., Dye N. A., Werner J. N., Gitai Z., and Shapiro L. (2010) Imaging based identification of a critical regulator of FtsZ protofilament curvature in Caulobacter. Mol. Cell 39, 975–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Potluri L.-P., Kannan S., and Young K. D. (2012) ZipA is required for FtsZ dependent pre-septal peptidoglycan synthesis prior to invagination during cell division. J. Bacteriol. 194, 5334–5342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tsang M. J., and Bernhardt T. G. (2015) Guiding divisome assembly and controlling its activity. Curr. Opin. Microbiol. 24, 60–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ortiz C., Natale P., Cueto L., and Vicente M. (2015) The keepers of the ring: regulators of FtsZ assembly. FEMS Microbiol. Rev., Epub [DOI] [PubMed] [Google Scholar]
- 30. Gueiros-Filho F. J., and Losick R. (2002) A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 16, 2544–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Goehring N. W., Gonzalez M. D., and Beckwith J. (2006) Premature targeting of cell division proteins to midcell reveals hierarchies of protein interactions involved in divisome assembly. Mol. Microbiol. 61, 33–45 [DOI] [PubMed] [Google Scholar]
- 32. Galli E., and Gerdes K. (2010) Spatial resolution of two bacterial cell division proteins; ZapA recruits ZapB to the inner face of the Z-ring. Mol. Microbiol. 76, 1514–1526 [DOI] [PubMed] [Google Scholar]
- 33. Roach E. J., Kimber M. S., and Khursigara C. M. (2014) Crystal structure and site-directed mutagenesis reveals key residues involved in Escherichia coli ZapA function. J. Biol. Chem. 289, 23276–23286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Espéli O., Borne R., Dupaigne P., Thiel A., Gigant E., Mercier R., and Boccard F. (2012) MatP-divisome interaction coordinates chromosome segregation with cell division in E. coli. EMBO J. 31, 3198–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Terwilliger T. C. (2003) SOLVE and RESOLVE, Automated structure solution and density modification. Methods Enzymol. 374, 22–37 [DOI] [PubMed] [Google Scholar]
- 36. Terwilliger T. C., and Berendzen J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., and Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Battye T. G., Kontogiannis L., Johnson O., Powell H. R., and Leslie A. G. (2011) iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Holm L., and Rosenström P. (2010) Dali server, conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Krissinel E., and Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 41. Buske P. J., and Levin P. A. (2013) A flexible C-terminal linker is required for proper FtsZ assembly in vitro and cytokinetic ring formation in vivo. Mol. Microbiol. 89, 249–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Haney S. A., Glasfeld E., Hale C., Keeney D., He Z., and de Boer P. (2001) Genetic analysis of the Escherichia coli FtsZ. ZipA interaction in the yeast two-hybrid system: characterization of FtsZ residues essential for the interactions with ZipA and FtsA. J. Biol. Chem. 276, 11980–11987 [DOI] [PubMed] [Google Scholar]
- 43. Hale C. A., Rhee A. C., and de Boer P. A. (2000) ZipA-induced bunlding of FtsZ polymers mediated by an interaction between C-terminal domains. J. Bacteriol. 182, 5153–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang X., Huang J., Mukherjee A., Cao C., and Lutkenhaus J. (1997) Analysis of the interaction of FtzZ with itself, GTP and FtsA. J. Bacteriol. 179, 5551–5559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bhattacharya A., Ray S., Singh D., Dhaked H. P., and Panda D. (2015) ZapC promotes assembly and stability of FtsZ filaments by binding at a different site on FtsZ than ZipA. Int. J. Biol. Macromol. 81, 435–442 [DOI] [PubMed] [Google Scholar]
- 46. Bisson-Filho A. W., Discola K. F., Castellen P., Blasios V., Martins A, Sforça M. L., Garcia W., Zeri A. C., Erickson H. P., Dessen A., and Gueiros-Filho F. J. (2015) FtsZ filament capping by MciZ, a developmental regulator of bacteria division. Proc. Natl. Acad. Sci. U.S.A. 112, E2130–E2138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lan G., Dajkovic A., Wirtz D., and Sun S. X. (2008) Polymerization and bundling kinetics of FtsZ filaments. Biophys. J. 95, 4045–4056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Delano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA [Google Scholar]
- 49. Chen J. C., and Beckwith J. (2001) FtsQ, FtsL, and FtsI require FtsK, but not FtsN, for co-localization with FtsZ during Escherichia coli cell division. Mol. Microbiol. 42, 395–413 [DOI] [PubMed] [Google Scholar]