ABSTRACT
The six serine/threonine kinases in the p21-activated kinase (PAK) family are important regulators of cell adhesion, motility and survival. PAK6, which is overexpressed in prostate cancer, was recently reported to localize to cell–cell adhesions and to drive epithelial cell colony escape. Here we report that PAK6 targeting to cell–cell adhesions occurs through its N-terminus, requiring both its Cdc42/Rac interactive binding (CRIB) domain and an adjacent polybasic region for maximal targeting efficiency. We find PAK6 localization to cell–cell adhesions is Cdc42-dependent, as Cdc42 knockdown inhibits PAK6 targeting to cell–cell adhesions. We further find the ability of PAK6 to drive epithelial cell colony escape requires kinase activity and is disrupted by mutations that perturb PAK6 cell–cell adhesion targeting. Finally, we demonstrate that all type II PAKs (PAK4, PAK5 and PAK6) target to cell–cell adhesions, albeit to differing extents, but PAK1 (a type I PAK) does not. Notably, the ability of a PAK isoform to drive epithelial colony escape correlates with its targeting to cell–cell adhesions. We conclude that PAKs have a broader role in the regulation of cell–cell adhesions than previously appreciated.
KEY WORDS: p21-activated kinase, PAK6, Cdc42, Cell–cell adhesions
Summary: PAK6 targeting to cell–cell contacts is dependent on both a polybasic motif and binding to Cdc42. Disruption of PAK6 targeting to cell–cell contacts inhibits epithelial cell colony escape.
INTRODUCTION
The p21-activated kinases (PAKs) are a family of 6 serine/threonine kinases that have fundamental roles in cellular processes such as adhesion, motility and survival, as well as in cancer progression (Field and Manser, 2012; King et al., 2014). PAKs are effector molecules of the small GTPases Cdc42 and Rac1, and thus participate in a variety of signaling pathways involved in cytoskeletal remodeling and cell adhesion dynamics. As such, all PAKs contain an N-terminal CRIB (Cdc42/Rac interactive binding) domain, capable of interacting with upstream GTPases, and a well-conserved C-terminal catalytic domain, shown to phosphorylate diverse downstream signaling molecules, such as β-catenin, LIM kinases, BAD, GEF-H1 (also known as ARHGEF2), pacsin and paxillin, with varying degrees of PAK isoform substrate specificity (Radu et al., 2014; Ha et al., 2015).
Based on sequence homology, the PAK family is divided into two groups: type I PAKs (PAK1, PAK2, PAK3) and type II PAKs (PAK4, PAK5, PAK6) (Jaffer and Chernoff, 2002), which differ in their modes of activation. GTPase binding to the CRIB domain of type I PAKs triggers release of active PAK monomers through dissociation of an inactive PAK dimer, in which the catalytic domain is inhibited by the autoinhibitory domain (AID) of its dimeric partner (Lei et al., 2000; Parrini et al., 2002; Rane and Minden, 2014). By contrast, type II PAKs are constitutively auto-phosphorylated on the activation loop and are generally not believed to be activated by GTPase binding (Ha et al., 2012, 2015). Cdc42 binding has instead been proposed to determine cellular localization of type II PAKs (Abo et al., 1998; Callow et al., 2002), although Cdc42 might also play a role in conformational activation of PAK4 (Baskaran et al., 2012).
Amplification or overexpression of each of the type II PAK isoforms has been linked to cancer development. The overexpression of PAK4, which is typically ubiquitously expressed at low levels, has been linked to pancreatic, gastric, liver and ovarian cancers, among others (Callow et al., 2002; Rane and Minden, 2014). PAK5 and PAK6 are more tissue-specific and less well-studied than PAK4. Both are predominantly expressed in the brain, and PAK6 is also normally found in the testes (Minden, 2012). Overexpression of PAK5 has been linked to colon cancer (Wen et al., 2014) whereas overexpression of PAK6 has been linked to prostate cancer (Kaur et al., 2008). How type II PAK isoforms contribute to cancer progression at the cellular level is still incompletely understood. Until recently, little was known about the basic cellular role of PAK6 other than its interaction with and phosphorylation of androgen receptor (Yang et al., 2001; Jaffer and Chernoff, 2002; Schrantz et al., 2004; Liu et al., 2013). However, last year, PAK6 was shown to localize to cell–cell adhesions in DU145 prostate cancer cells (Fram et al., 2014). The mechanism of targeting and its isoform specificity were not explored, but PAK6 was shown to promote the disassembly of cell–cell adhesions (Fram et al., 2014), a crucial step in epithelial-to-mesenchymal transition, one of the hallmarks of cancer.
Here, we have mapped the regions of PAK6 necessary and sufficient for cell–cell adhesion targeting. We demonstrate that an intact Cdc42-binding domain is necessary for cell–cell adhesion targeting of PAK6 and that, consistent with this, knockdown of Cdc42 inhibits PAK6 localization at cell–cell adhesions. However, Cdc42 binding is not sufficient for cell–cell adhesion targeting and we find that a polybasic region, just N-terminal to the Cdc42-binding region, is also required. This polybasic region has previously been characterized as a membrane-binding domain in yeast PAK (Takahashi and Pryciak, 2007) and a nuclear localization signal in mammalian PAK4 (Li et al., 2012). Notably, we demonstrate that disruption of cell–cell adhesion targeting impairs the ability of PAK6 to drive epithelial cell colony escape. We further find that PAK6 requires an active kinase domain in order to drive colony escape, and we propose that localization of an active kinase to cell–cell adhesions is a key step in disassembling the adhesions and permitting cell escape. Finally, we expand our study to demonstrate that different PAK isoforms target differentially to cell–cell adhesions, and that this differential targeting correlates with the ability of PAKs to drive epithelial colony escape.
RESULTS
PAK6 targets to cell–cell adhesions
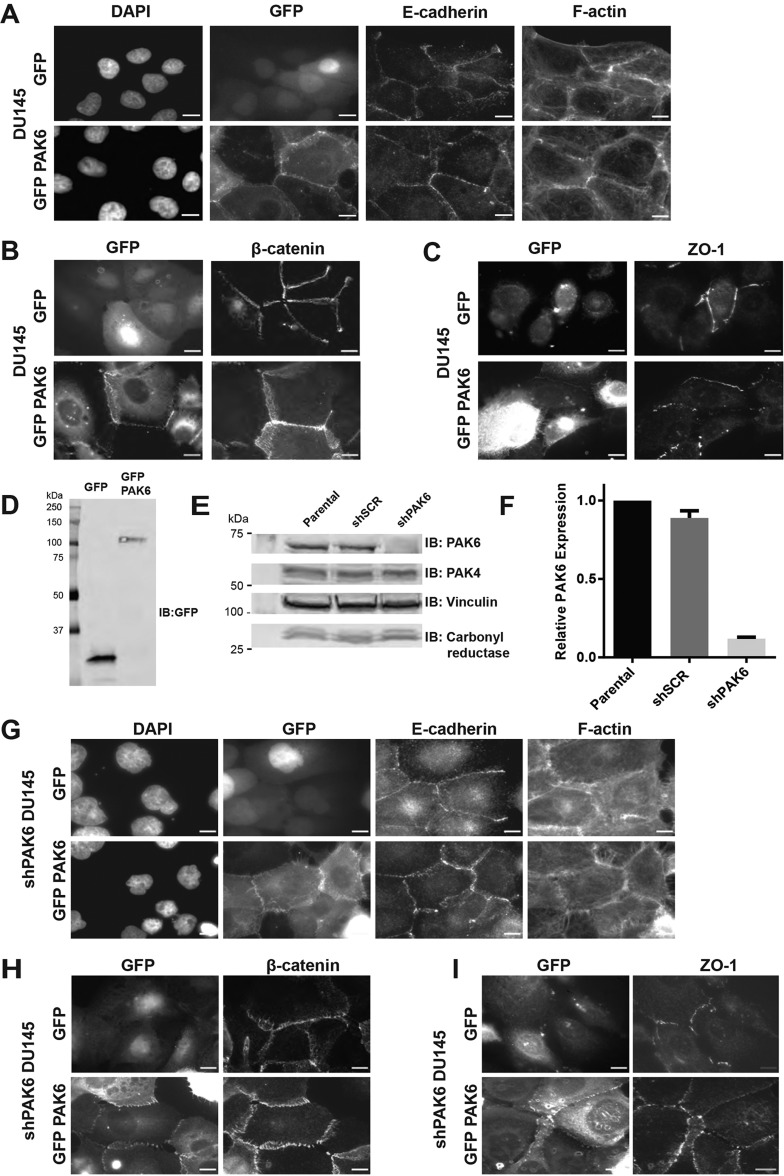
To investigate how PAK6 targets to cell–cell adhesions, we first confirmed that we could observe PAK6 at E-cadherin-rich cell–cell contacts as previously reported (Fram et al., 2014). Using lentiviral infection, we generated stable DU145 prostate cancer cell lines expressing GFP or GFP–PAK6. DU145 prostate cancer cells were chosen because they are known to express endogenous PAK6 (Wen et al., 2009; Zhang et al., 2010) and because they have previously been used to investigate PAK6 localization (Fram et al., 2014). Using these cells we found that GFP–PAK6 localizes to cell–cell adhesions as defined by E-cadherin and F-actin staining, whereas the control GFP construct does not (Fig. 1A). Similar results were obtained using β-catenin or ZO-1 staining to mark cell–cell contacts (Fig. 1B,C). We further validated that the GFP and GFP–PAK6 proteins were full-length and have the expected molecular weights (GFP, ∼27 kDa; GFP–PAK6, ∼102 kDa) by immunoblotting (Fig. 1D). These data indicate that lentiviral delivery of GFP-tagged PAK6 constructs into DU145 cells provides an effective system with which to investigate determinants of PAK6 subcellular localization.
Fig. 1.
PAK6 targets to cell–cell adhesions in DU145 prostate cancer cells. (A-C) Stable DU145 cell lines expressing GFP or GFP–PAK6 were plated on glass coverslips, fixed after 24 h and stained for DNA (DAPI), F-actin (phalloidin), or with antibodies to E-cadherin, β-catenin, or ZO-1 as indicated. Scale bars: 10 µm. (D) 5.0×105 cells from A were lysed directly in sample buffer and immunoblotted for GFP to indicate exogenous constructs are the expected molecular weights. (E) DU145 parental cells were infected with scrambled (SCR) or PAK6 shRNA, lysed in RIPA buffer and immunoblotted for endogenous PAK6, PAK4, vinculin and carbonyl reductase. (F) Quantification of protein expression of PAK6 in shSCR and shPAK6 cell lines as compared with parental DU145 cells, mean±s.e.m. of three independent experiments. (G-I) shPAK6 cells expressing GFP or GFP–PAK6 were plated on glass coverslips, fixed after 24 h and stained for DNA (DAPI), F-actin (phalloidin), E-cadherin, β-catenin, or ZO-1 as indicated. Scale bars: 10 µm.
To minimize potential competition between exogenous and endogenous PAK6, we next generated PAK6 knockdown cells. Following lentiviral delivery of shRNA constructs into DU145 cells, we observed a significant (∼90%) reduction in PAK6 protein expression by immunoblotting and confirmed that levels of vinculin and carbonyl reductase loading controls remained unchanged (Fig. 1E,F). The only other type II PAK isoform expressed in DU145 cells, PAK4 (Wells et al., 2010), also remained unchanged upon PAK6 knockdown (Fig. 1E). Fluorescence microscopy shows that the PAK6 knockdown cells retain their ability to form cell–cell adhesions, identified by F-actin, β-catenin or ZO-1 staining, and that GFP–PAK6 targets to cell–cell adhesions in these cells (Fig. 1G-I).
PAK6 localizes at cell–cell adhesions independent of catalytic activity through its N-terminus
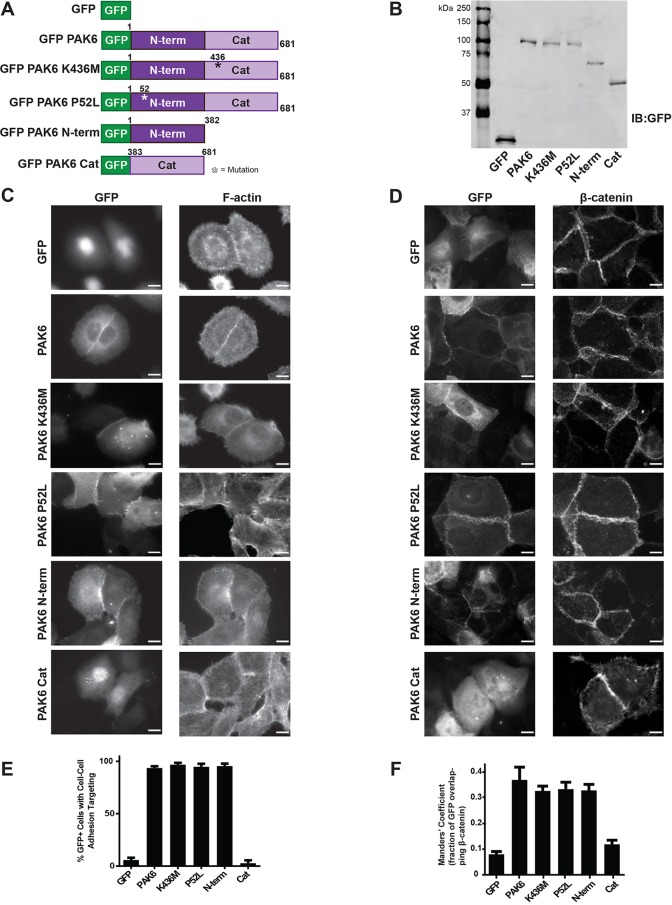
Whereas previous work has established PAK6 localizes to cell–cell adhesions (Fram et al., 2014), it has not been determined how PAK6 is targeted to these sites. Given that PAK6 has been proposed to phosphorylate cell–cell adhesion components such as β-catenin (Fram et al., 2014), we first used GFP–PAK6 point mutants (Fig. 2A,B) to test whether PAK6 catalytic activity could affect its ability to localize to cell–cell adhesions. We found that kinase-dead PAK6 K436M (Schrantz et al., 2004) and hyperactive PAK6 P52L (Gao et al., 2013) both localize to cell–cell adhesions, as identified either by F-actin staining (Fig. 2C) or β-catenin staining (Fig. 2D). Two independent methods were used to quantify GFP localization to cell–cell adhesions (Fig. 2E,F); first, the percentage of GFP-positive cells exhibiting GFP localization in cell–cell contacts was scored manually (Fig. 2E), and second, the percentage of GFP signal colocalizing with β-catenin staining was assessed by calculating the Manders' coefficient using the JACoP plugin on ImageJ (Bolte and Cordelieres, 2006) (Fig. 2F). Together these data indicate that PAK6 localizes to cell–cell adhesions independent of catalytic activity.
Fig. 2.
PAK6 targets to cell–cell adhesions independent of catalytic activity through its N-terminus. (A) Schematic representing generated GFP-tagged PAK6 constructs. (B) 5.0×105 cells expressing the GFP-tagged PAK6 constructs were lysed directly in sample buffer and immunoblotted for GFP to indicate exogenous constructs are the expected molecular weights. (C,D) Stable shPAK6 DU145 cell lines expressing the indicated constructs were plated on glass coverslips, fixed after 24 h and stained for F-actin (phalloidin) or β-catenin. (E) Mean percentage (±s.e.m.) of cells in which GFP-tagged constructs targeted to cell–cell adhesions. n=3 with ≥40 cells per experiment. (F) Mean Manders' coefficient (±s.d.) of cells with GFP-tagged constructs targeted to β-catenin positive cell–cell adhesions (from at least 10 fields, >60 cells per condition). Scale bars: 10 µm.
To determine which region of PAK6 is important for targeting, we generated an N-terminal construct (N-term, residues 1–382) and a C-terminal construct containing the well-conserved PAK catalytic domain (Cat, residues 383–681). We confirmed all exogenous constructs were full-length and of the expected molecular weights by GFP immunoblotting (Fig. 2A,B). Whereas PAK6 Cat is diffuse throughout the cytoplasm and nucleus, PAK6 N-term targets to cell–cell adhesions at the same level as full-length PAK6, indicating that the ability of PAK6 to target to cell–cell adhesions is controlled by the N-terminus (Fig. 2C-F).
PAK6 requires a functional CRIB motif to target to cell–cell adhesions
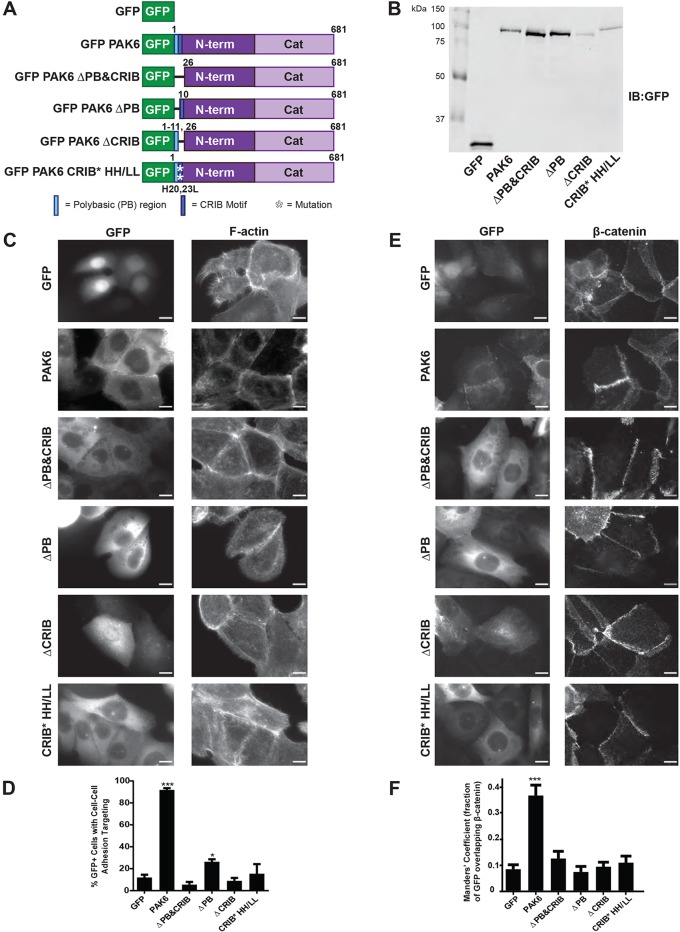
Few protein domains or motifs are ascribed to the PAK6 N-terminus (residues 1–382) with the exceptions of a polybasic (PB) region (residues 3–9), a well-conserved CRIB motif (residues 12–25), and a pseudosubstrate region that inhibits catalytic activity (residues 48–56) (Ha et al., 2012; Gao et al., 2013; Wang et al., 2013). Having found PAK6 cell–cell adhesion targeting is not kinase activity dependent, we focused our attention on the polybasic region and CRIB motif as potential requirements for targeting. We generated constructs lacking the polybasic region and/or the CRIB motif (ΔPB, ΔCRIB, ΔPB&CRIB) as well as a CRIB motif mutant, H20,23L (HH/LL), previously shown to prevent type II PAK binding to Cdc42 (Fig. 3A,B) (Ching et al., 2003; Menzel et al., 2007). We found that the PAK6 CRIB motif is required for localization to cell–cell adhesions, given that constructs lacking the CRIB motif (ΔCRIB and ΔPB&CRIB) or with a mutated CRIB motif (HH/LL) are diffusely cytoplasmic and no longer target to cell–cell adhesions identified by F-actin or β-catenin staining (Fig. 3C,D). Interestingly, the construct lacking the polybasic region (ΔPB) is significantly impaired in its ability to target to cell–cell adhesions, exhibiting only occasional weak targeting to these sites (Fig. 3C-F).
Fig. 3.
PAK6 requires a functional CRIB motif to target to cell–cell adhesions. (A) Schematic representing generated GFP-tagged PAK6 constructs. (B) 5.0×105 cells expressing the GFP-tagged PAK6 constructs were lysed directly in sample buffer and immunoblotted for GFP to indicate exogenous constructs are the expected molecular weights. (C,D) Stable shPAK6 DU145 cell lines expressing the indicated constructs were plated on glass coverslips, fixed after 24 h and stained for F-actin (phalloidin) or β-catenin. (E) Mean percentage (±s.e.m.) of cells in which GFP-tagged constructs targeted to cell–cell adhesions. n=3 with ≥40 cells per experiment; *P<0.05, ***P<0.001. (F) Mean Manders' coefficient (±s.d.) of cells with GFP-tagged constructs targeted to β-catenin positive cell–cell adhesions (from at least 10 fields, >60 cells per condition). Scale bars: 10 µm.
Cdc42 colocalizes with PAK6 at cell–cell adhesions
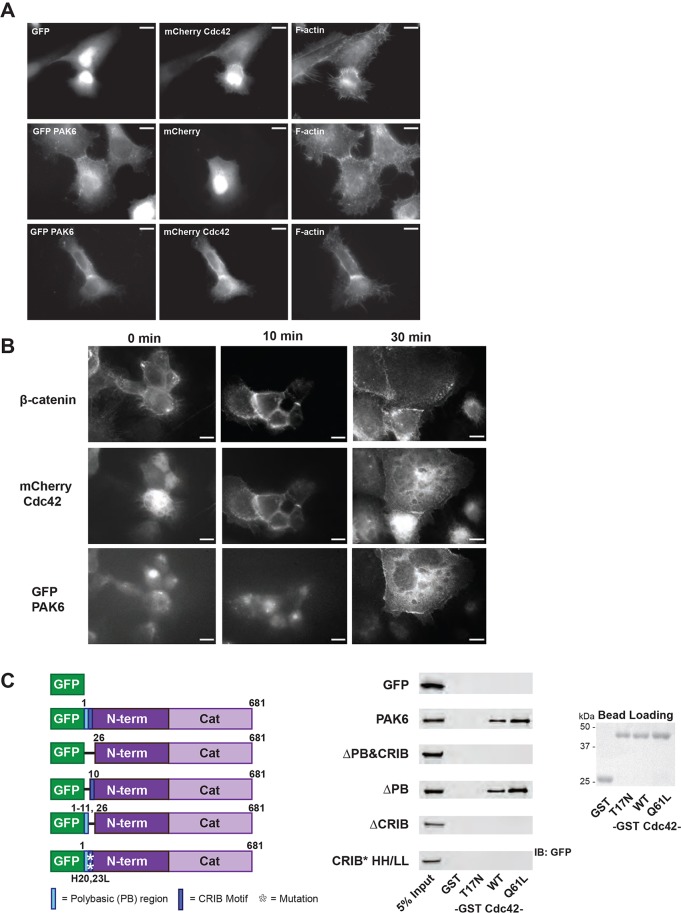
Given that the CRIB motif is required for PAK6 targeting to cell–cell adhesions, we thought it likely that a GTPase that interacts with this motif could contribute to PAK6 localization. Type II PAKs (PAK4, PAK5 and PAK6) have been shown to preferentially bind the GTPase Cdc42 as compared with Rac1 or RhoA (Abo et al., 1998; Lee et al., 2002; Pandey et al., 2002), thus making Cdc42 a potential recruitment candidate. We thus first examined whether Cdc42 localized to cell–cell adhesions. We generated mCherry-tagged Cdc42 and transiently transfected stable DU145 cell lines expressing GFP or GFP–PAK6. We found Cdc42 does indeed target to cell–cell adhesions whereas the control mCherry construct does not. Additionally, we found that GFP–PAK6 and mCherry–Cdc42 colocalize at cell–cell adhesions, further suggesting that Cdc42 is involved in PAK6 targeting (Fig. 4A).
Fig. 4.
PAK6 colocalizes with Cdc42 at cell–cell adhesions and Cdc42 binding is required but not sufficient for maximal targeting efficiency. (A) Stable DU145 cell lines expressing GFP or GFP–PAK6 were plated on glass coverslips, transiently transfected 24 h later with mCherry or mCherry–Cdc42, fixed after 48 h and stained for F-actin (phalloidin). (B) Stable DU145 cells expressing mCherry–Cdc42 were plated on glass coverslips and transfected with GFP–PAK6. 24 h post transfection, cells were cultured in the absence of Ca2+ overnight. Ca2+-containing medium was re-applied and coverslips were fixed at different time points following Ca2+ re-addition and stained for β-catenin. Scale bars: 10 µm. (C) Pull-down of GFP-tagged proteins from stable cell line lysates by bacterially purified GST or GST–Cdc42 loaded on glutathione beads. Samples were immunoblotted for GFP (left) and a 5% input is shown. Bead loading was assessed by Ponceau staining (right).
To investigate the temporal recruitment of Cdc42 and PAK6 to cell–cell adhesions, we transfected DU145 cells stably expressing mCherry–Cdc42 with GFP–PAK6. We cultured these cells overnight in the absence of Ca2+ to disassemble cell–cell contacts and then assessed Cdc42 and PAK6 localization to reforming adhesions upon re-addition of Ca2+-containing medium. As shown in Fig. 4B, although mCherry–Cdc42 was evident in adhesions 10 min after Ca2+ addition, PAK6 was only consistently observed at later time-points. These data are consistent with a model in which Cdc42 recruits PAK6 to adhesions.
Cdc42 binding is required but not sufficient for maximal cell–cell adhesion targeting efficiency
We next confirmed that our GFP–PAK6 constructs bind Cdc42 in the expected CRIB-motif-dependent manner. We bacterially purified and produced GST and three GST-tagged Cdc42 proteins for pull-down assays: wild-type (WT), a nucleotide-binding defective mutant that should not interact with effector proteins (T17N), and an activated mutant that should constitutively interact with effector proteins (Q61L). As expected, GFP–PAK6 pulls down only with Cdc42 WT or Cdc42 Q61L, but not with Cdc42 T17N or GST alone (Fig. 4C). Further, GFP and GFP–PAK6 proteins lacking a functional CRIB domain (ΔPB&CRIB, ΔCRIB and HH/LL) do not bind any of the GST–Cdc42 proteins or GST alone, validating the specificity of our assay. Notably, although PAK6 ΔPB is significantly impaired in its ability to target to cell–cell adhesions (Fig. 3C-F), it binds Cdc42 to the same level as wild-type PAK6 (Fig. 4C), suggesting that Cdc42 binding is required but not sufficient for maximal cell–cell adhesion target efficiency.
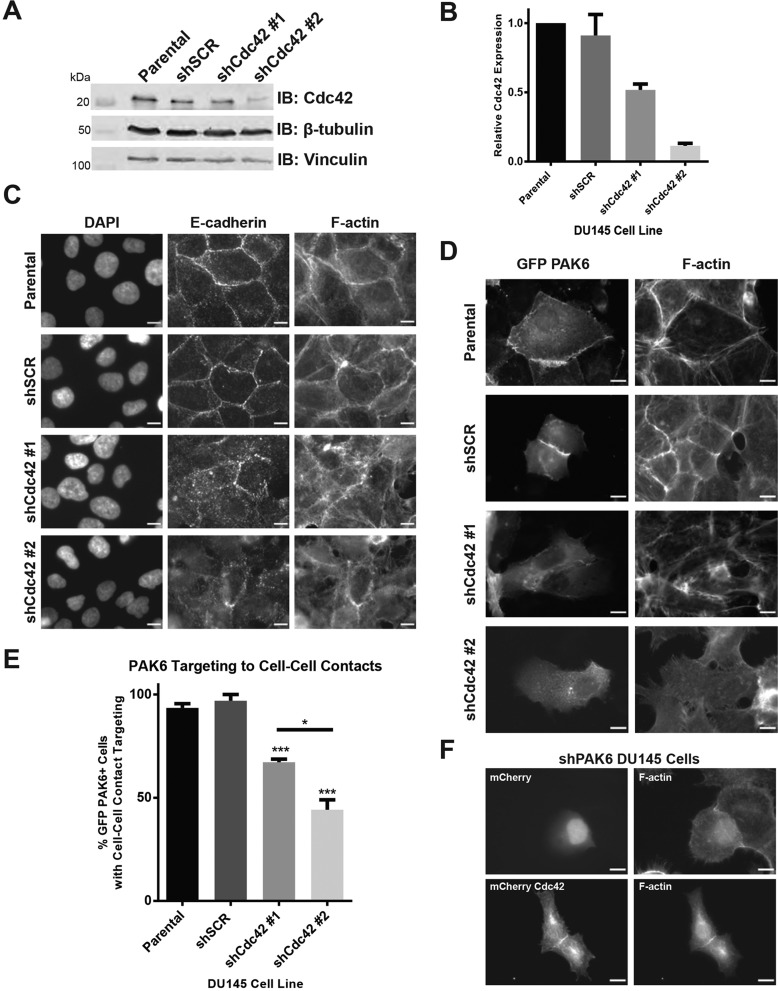
Cdc42 is required for localization of PAK6 to cell–cell contacts
Having found that PAK6 targeting to cell–cell adhesions is CRIB motif dependent (Fig. 3C-F) and that Cdc42 colocalizes with PAK6 at cell–cell adhesions (Fig. 4A), we asked whether Cdc42 is required for PAK6 targeting to these sites. Using lentiviral delivery of shRNA constructs, we established Cdc42 knockdown cell lines with two different hairpins, achieving ∼50% knockdown (shRNA #1) and ∼90% knockdown (shRNA #2) (Fig. 5A,B). In Cdc42 knockdown cells grown on glass coverslips for 24 h, E-cadherin localizes much less efficiently to cell–cell contacts in shCdc42 #1 cells and barely at all in shCdc42 #2 cells as compared with parental and shSCR controls (Fig. 5C). This indicates cell–cell junctions are fewer and/or have been perturbed following reduction in Cdc42 levels, as previously reported (Wallace et al., 2010). In regions where cells are still in contact with each other, as delineated by F-actin staining, transiently transfected GFP–PAK6 targets much less efficiently in Cdc42 knockdown cells and in a manner that correlates with the degree of knockdown (Fig. 5B,D,E). This is consistent with a requirement of Cdc42 for PAK6 localization to cell–cell adhesions and is in keeping with the lack of cell–cell adhesion targeting of Cdc42-binding defective PAK6 mutants (Fig. 3C-F, Fig. 4C).
Fig. 5.
Cdc42 knockdown disrupts cell–cell junction formation and inhibits localization of PAK6 to cell–cell contacts. (A) DU145 parental cells were infected with scrambled (SCR) or Cdc42 shRNA, lysed in RIPA buffer and immunoblotted for endogenous Cdc42, β-tubulin and vinculin. (B) Quantification of protein expression of Cdc42 in shSCR and shCdc42 cell lines as compared with parental DU145 cells, mean±s.e.m. of three independent experiments. (C) Control and shCdc42 knockdown lines were plated on glass coverslips, fixed 24 h later, and stained for DNA (DAPI), E-cadherin (DECMA-1) and F-actin (phalloidin). (D) Control and shCdc42 knockdown lines were transiently transfected with GFP–PAK6, fixed and stained for F-actin (phalloidin). (E) Mean percentage (±s.e.m.) of cells in which GFP–PAK6 targeted to cell–cell contacts. n=3 with ≥40 cells per experiment; *P<0.05, ***P<0.001. (F) PAK6 knockdown cells were transiently transfected with mCherry or mCherry–Cdc42, fixed 24 h later and stained for F-actin (phalloidin). Scale bars: 10 µm.
PAK6 is not required for localization of Cdc42 to cell–cell adhesions
To confirm that PAK6 was not required for Cdc42 targeting to cell–cell adhesions we transiently transfected PAK6 knockdown cells (Fig. 1C,D) with mCherry–Cdc42 and mCherry. We found that mCherry–Cdc42 localizes to cell–cell adhesions in PAK6 knockdown cells, indicating that PAK6 is not required for localization of Cdc42 to cell–cell adhesions (Fig. 5F), but rather Cdc42 is required for the recruitment of PAK6.
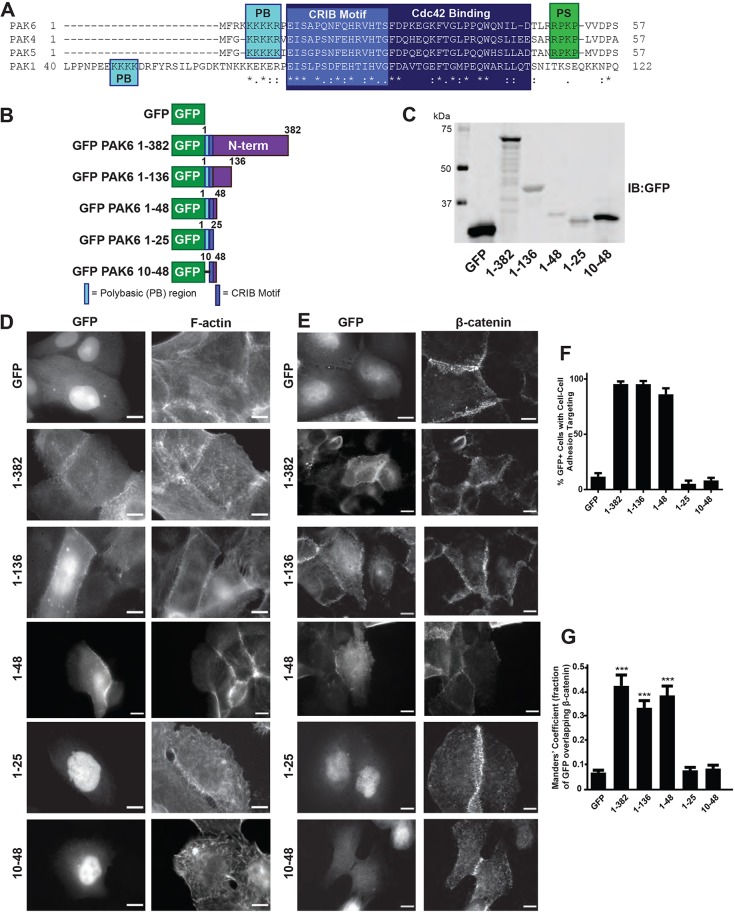
PAK6 1–48 is sufficient for targeting to cell–cell adhesions
Having established that Cdc42 and a functional CRIB motif are required for PAK6 targeting to cell–cell adhesions, we sought to determine the minimal fragment of PAK6 that is capable of targeting to cell–cell adhesions. We generated a series of N-terminal truncation constructs terminating at residues 136, the most similar region of the N-terminus among all type II PAK isoforms and variants; 48, up to the start of the pseudosubstrate region and including the full structurally defined Cdc42-binding portion of PAK6 [Protein Data Bank (PDB): 2ODB]; or 25, up to the end of the CRIB motif (Fig. 6A-C). Analysis of subcellular localization revealed that PAK6 1–382, 1–136 and 1–48 all localize at cell–cell adhesions, but PAK6 1–25 fails to target and is primarily nuclear (Fig. 6D-G). These data suggest that in addition to an intact CRIB motif, residues 25–48 contribute to cell–cell adhesion targeting. This is consistent with our conclusion that targeting to cell–cell contacts requires Cdc42 binding, as the region immediately C-terminal to the CRIB motif has been shown to be important for high-affinity Cdc42 binding to a range of Cdc42 effectors, including a variety of PAKs and WASP (also known as WAS) (Rudolph et al., 1998; Thompson et al., 1998; Su et al., 2005). Furthermore, X-ray crystallography of the PAK6–Cdc42 complex (PDB: 2ODB) supports a role for residues 25–45 in Cdc42 interaction.
Fig. 6.
PAK6 1–48 is sufficient for targeting to cell–cell adhesions. (A) Alignment of Cdc42-interacting region of PAK6 with PAK1, PAK4 and PAK5 completed with Clustal Omega with polybasic (PB), CRIB motif, Cdc42-binding and type II PAK pseudosubstrate (PS) regions indicated. (B) Schematic representing generated GFP-tagged PAK6 constructs. (C) Cells expressing the GFP-tagged PAK6 constructs were lysed and immunoblotted for GFP to indicate exogenous constructs are the expected molecular weights. (D,E) Stable shPAK6 DU145 cell lines expressing the indicated constructs were plated on glass coverslips, fixed after 24 h and stained for F-actin (phalloidin) or β-catenin (D10A8). Scale bars: 10 µm. (F) Mean percentage (±s.e.m.) of cells in which GFP-tagged constructs targeted to cell–cell adhesions. n=3 with ≥40 cells per experiment. (G) Mean Manders' coefficient (±s.d.) of cells with GFP-tagged constructs targeted to β-catenin positive cell–cell adhesions (from at least 10 fields, >60 cells per condition). ***P<0.001.
In addition to Cdc42 binding, our analysis of full-length PAK6 mutants highlighted a role for the polybasic region in targeting PAK6 to cell–cell contacts (Fig. 3C-F). We therefore tested whether removal of the polybasic region from residues 1–48 also impacts localization. As before, all constructs expressed proteins of the expected sizes as assessed by anti-GFP immunoblotting (GFP, ∼27 kDa; GFP–PAK6 1–382, ∼68 kDa; GFP–PAK6 1–136, ∼42 kDa; GFP–PAK6 1–48, ∼33 kDa; GFP–PAK6 10–48, ∼32 kDa; GFP–PAK6 1–25, ∼30 kDa) (Fig. 6C). Indeed, removal of the polybasic region prevents targeting of PAK6 10–48 to cell–cell adhesions, indicating that of the constructs tested, PAK6 1–48 is the minimally sufficient fragment for cell–cell adhesion targeting (Fig. 6D-G). In summary, our analysis of PAK6 mutants and fragments reveals a requirement for the Cdc42-binding extended CRIB domain and an N-terminal polybasic motif for localization at cell–cell adhesions.
PAK6 requires a functional CRIB motif and kinase activity to drive epithelial colony escape
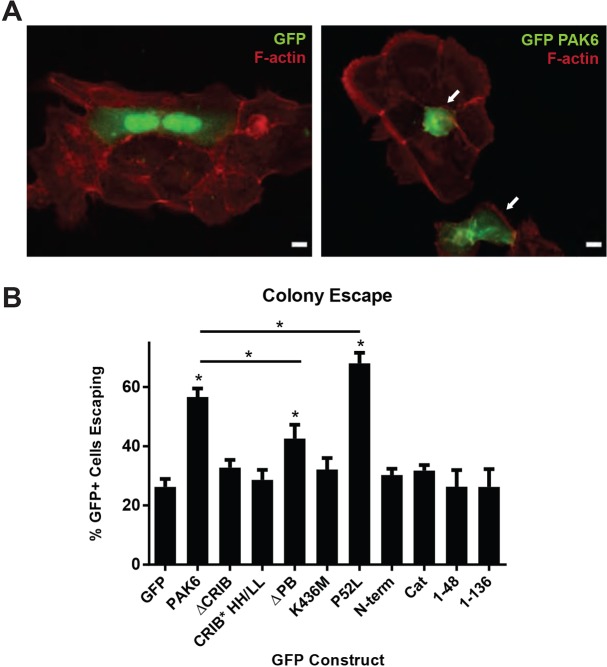
In culture, DU145 cells predominantly grow in monolayer epithelial cell colonies. It has previously been shown that PAK6 expression is able to drive escape or extrusion from epithelial cell colonies, a process thought to be driven, at least in part, by disassembly of cell–cell adhesions (Fram et al., 2014). Having identified the necessary and sufficient regions of PAK6 required for targeting to cell–cell adhesions, we sought to determine whether the ability of PAK6 to target to cell–cell adhesions correlated with its ability to drive epithelial colony escape. To test this, we sparsely plated DU145 cells on glass coverslips and allowed them to form small colonies for 48 h. We then transiently transfected the cells with GFP–PAK6 constructs and fixed the cells 24 h later to assess whether expression of PAK6 drove epithelial colony escape. Using this approach, we confirmed that most cells expressing GFP remained firmly within epithelial colonies (Fig. 7A, left; Fig. 7B), whereas there was a significant increase in the number of GFP–PAK6 expressing cells that were escaping from colonies or had already been extruded (Fig. 7A, right; Fig. 7B).
Fig. 7.
PAK6 requires a functional CRIB motif and kinase activity to drive epithelial colony escape. (A) DU145 cells were seeded in 6-well dishes (1.0×105/well) and allowed to form small colonies for 48 h before transient transfection with the indicated GFP-tagged constructs. Cells were fixed and stained for F-actin (phalloidin) 24 h after transfection and scored. Escaping cells were defined as cells with greater than half of their perimeter detached from neighboring cells (lower arrow) and/or in a different plane than the surrounding cell colony (upper arrow). Scale bars: 10 µm. (B) Quantification of colony escape assay, showing the mean percentage (±s.e.m.) of GFP+ cells escaping colonies. n=3 with ≥50 cells per experiment; *P<0.05.
We next tested whether PAK6 mutants defective in targeting to cell–cell contacts could drive colony escape. We found that GFP–PAK6 constructs lacking the CRIB motif (ΔCRIB), or containing a mutated CRIB motif (HH/LL), are unable to drive epithelial colony escape above GFP levels, indicating that PAK6 requires a functional CRIB motif to drive colony escape (Fig. 7B). We also tested PAK6 lacking the polybasic region (PAK6 ΔPB), which has significantly diminished cell–cell adhesion targeting (Fig. 3C,D). Consistent with this, PAK6 ΔPB has a significantly inhibited ability to drive epithelial colony escape compared with wild-type PAK6 (Fig. 7B). Thus, the ability of PAK6 to drive colony escape correlates with its ability to target to cell–cell adhesions.
It has recently been proposed that type II PAKs might phosphorylate cell–cell adhesion components such as β-catenin in order to disassemble cell–cell adhesions (Fram et al., 2014; Selamat et al., 2015). We thus asked whether PAK6 catalytic activity is required to drive colony escape. Though kinase-dead PAK6 (K436M) targets to cell–cell adhesions (Fig. 2B,C), it is unable to drive colony escape (Fig. 7B). Interestingly, hyperactive PAK6 (P52L), which targets well to cell–cell adhesions (Fig. 2C,D), drives a greater percentage of cells to escape colonies than PAK6 wild-type (Fig. 7B). Thus, PAK6 requires not only the ability to target to cell–cell adhesions but also catalytic activity to drive colony escape. This is further confirmed by tests of the PAK6 N-terminus (N-term, 1–382) or smaller N-terminal fragments (1–48 and 1–136), which, despite efficient targeting to cell–cell adhesions, lack catalytic activity and cannot drive colony escape (Fig. 7B). Furthermore, the isolated catalytic domain (Cat), which retains kinase activity but cannot target to cell–cell adhesions, is also incapable of driving colony escape (Fig. 7B). Thus, the ability of PAK6 to drive colony escape requires targeting of active kinase to cell–cell adhesions.
PAK isoforms target differentially to cell–cell adhesions
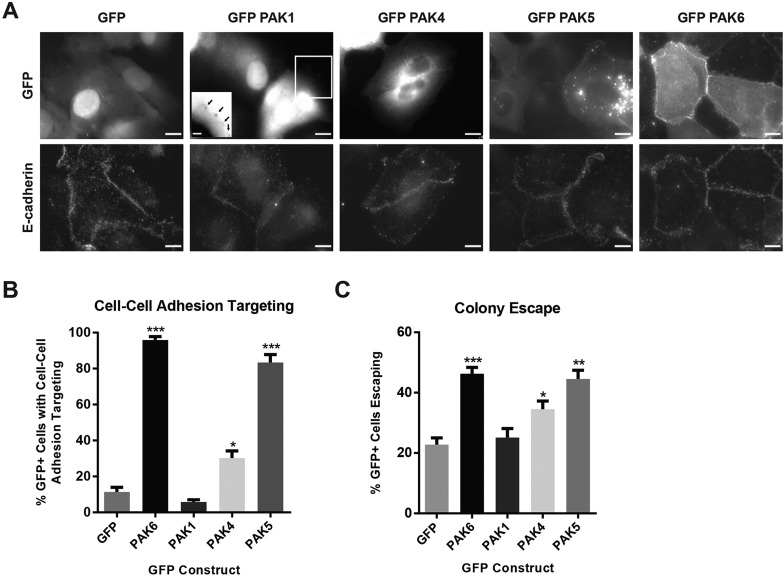
PAK6 is one of six serine/threonine kinases that belong to the PAK family (Ha et al., 2015). In addition to a well-conserved C-terminal kinase domain, all PAK family kinases contain an N-terminal CRIB motif capable of binding Cdc42. To test whether other PAK isoforms could also target to cell–cell adhesions, we generated stable DU145 cell lines expressing PAK4 and PAK5, the other two type II PAKs most closely related to PAK6, and a representative type I PAK, PAK1. We found that all type II PAKs target to cell–cell adhesions, although to varying degrees. PAK5 and PAK6 target in ∼85% and 95% of cells, respectively, whereas PAK4 targets more weakly to cell–cell adhesions in ∼30% of cells (Fig. 8A,B). We observed that PAK5 forms aggregates in DU145 cells and targets with less intensity to cell–cell adhesions, perhaps because PAK5 is not endogenous in DU145 cells. We did not observe PAK1 in cell–cell adhesions, though PAK1 is present in focal adhesions (Fig. 8A, inset; Fig. 8B), as previously reported for PAK1 (Brown et al., 2002; Field and Manser, 2012).
Fig. 8.
PAK isoform differential targeting correlates with ability to drive epithelial colony escape. (A) Stable DU145 cell lines expressing the indicated PAK isoforms were plated on glass coverslips, fixed after 24 h and stained for E-cadherin (DECMA-1). Inset depicts color-inverted, enlarged image of GFP–PAK1 targeted to focal adhesions, marked by arrows. Scale bars: 10 µm. (B) Mean percentage (±s.e.m.) of cells in which GFP-tagged constructs targeted to cell–cell adhesions; n=3 with ≥40 cells per experiment. (C) Quantification of colony escape assay, showing the mean percentage (±s.e.m.) of GFP+ cells escaping colonies. n=3 with ≥50 cells per experiment. *P<0.05, **P<0.01, ***P<0.001.
PAK isoform differential targeting correlates with ability to drive epithelial colony escape
Having found that PAK6 targeting correlated with its ability to drive colony escape, we tested whether the differential targeting of PAK isoforms also correlates with their ability to drive colony escape. Indeed, as we had observed for PAK6, the better PAK isoforms targeted to cell–cell adhesions, the greater the degree of colony escape we observed (Fig. 8C). Taken together, PAK ability to drive epithelial colony escape correlates with its ability to target to cell–cell adhesions, which is dependent on Cdc42, and with its catalytic activity.
DISCUSSION
The PAKs are a family of serine/threonine kinases with important roles in a variety of fundamental cell processes, including cell adhesion, motility and survival (Wells and Jones, 2010). It was previously reported that PAK6 localizes at cell–cell adhesions where it is proposed to phosphorylate β-catenin to promote dissociation of cell–cell junctions, a process crucial to basic cellular functioning as well as cancer progression (Fram et al., 2014). However, prior to our current study, it was unknown how PAK6 targets to cell–cell adhesions, what protein(s) are required for recruitment to these sites, and if localization at cell–cell adhesions is indeed required to drive cell–cell dissociation. Further, it had not been explored whether other members of the PAK family could target to cell–cell adhesions and contribute to cell–cell dissociation. Here, using a series of knockdown studies, fluorescence microscopy and biochemical analyses, we establish that PAK6 targets to cell–cell adhesions through its N-terminus in a Cdc42-dependent manner, requiring both a functional CRIB motif and a polybasic region for maximal targeting efficiency, and that this targeting is required, in addition to kinase activity, to functionally drive epithelial cell colony escape. We further found that the other type II PAK isoforms, PAK4 and PAK5, also drive epithelial colony escape to varying extents that correlate with their ability to target to cell–cell adhesions.
PAK6 has been reported to directly bind IQGAP1 (IQ motif containing GTPase activating protein 1) (Kaur et al., 2008; Fram et al., 2014), which binds cell–cell junction proteins β-catenin and E-cadherin, among many other partners (Abel et al., 2015). At first an attractive candidate for the recruitment of PAK6 to cell–cell adhesions, IQGAP1 binds the catalytic domain of PAK6 (Fram et al., 2014), which we have found does not localize at cell–cell adhesions. Instead, we found that the N-terminus of PAK6 is necessary and sufficient for targeting to cell–cell adhesions, indicating that another direct binding partner must be involved in its recruitment. In keeping with this, we found that PAK6 constructs with altered catalytic activity (kinase-dead K436M, hyperactive P52L) but with intact N-termini still localize at cell–cell adhesions, indicating that PAK6 targeting to cell–cell adhesions is independent of catalytic activity. In agreement with our findings, the Xenopus type II PAK, PAK5 (also known as PAK7), targets to cell–cell junctions independent of catalytic activity (Faure et al., 2005).
Through biochemical mapping and immunofluorescence studies, we show that a functional CRIB motif is required for PAK6 targeting to cell–cell adhesions and its ability to promote cell–cell dissociation as observed through epithelial cell colony escape assays. We found that PAK6 constructs lacking a CRIB motif, or with a mutated CRIB motif (HH/LL), do not localize at cell–cell adhesions, suggesting a requirement of GTPase binding for PAK6 localization to cell–cell adhesions. Type II PAKs, including PAK6, directly interact with small GTPases and have been shown to preferentially bind Cdc42 compared with other widely studied GTPases, Rac and RhoA (Abo et al., 1998; Lee et al., 2002; Pandey et al., 2002). Having found through fluorescence microscopy that Cdc42 and PAK6 colocalize at cell–cell adhesions, we thus identified Cdc42 as a candidate for PAK6 recruitment to cell–cell adhesions.
Our knockdown studies indicate that Cdc42 is required for PAK6 localization at cell–cell adhesions, and not vice versa. We found that Cdc42 knockdown cells are impaired in their ability to form cell–cell adhesions, as previously reported by others (Wallace et al., 2010; Selamat et al., 2015). Even still, these cells maintain some cell–cell contacts, as is delineated by F-actin staining, and PAK6 is significantly impaired in its ability to target to these regions. Interestingly, this is in keeping with a previous finding that the Drosophila type II PAK homolog, Mbt, requires Cdc42 for recruitment to adherens junctions and proper photoreceptor cell morphogenesis (Schneeberger and Raabe, 2003). It is also important to note that Cdc42 targets to cell–cell adhesions in PAK6 knockdown cells, indicating, albeit unsurprisingly given the tissue-specific expression of PAK6 and ubiquitous nature of Cdc42, that Cdc42 is not dependent on PAK6 to localize at cell–cell adhesions. Further, in synchronized time-course studies of PAK6 and Cdc42 recruitment to cell–cell adhesions, we observe Cdc42 localizing to cell–cell adhesions prior to PAK6 accumulation, again in keeping with a model in which Cdc42 recruits PAK6 and not vice versa.
Though PAK6 requires Cdc42 to target to cell–cell adhesions, we found that the ability to interact with Cdc42 is not sufficient for maximal targeting efficiency. Though PAK6 lacking the polybasic region (ΔPB) still binds Cdc42 efficiently, it is significantly impaired in its ability to target to cell–cell adhesions. This is further corroborated by the observation PAK6 10-48 does not localize at cell–cell adhesions whereas its counterpart containing the PB region, PAK6 1-48, does. This indicates the polybasic region is involved in PAK6 localization at cell–cell adhesions, likely as a membrane-targeting or membrane-anchoring sequence, as has been observed for polybasic regions of other cytoplasmic adhesion-related molecules, talin (Goult et al., 2010) and kindlin (Bouaouina et al., 2012). This is further corroborated by reports that the polybasic regions of the yeast ortholog STE20 bind lipids (Takahashi and Pryciak, 2007), which could potentially aid in kinase interactions with the membrane. The PAK4 polybasic region has previously been characterized as a nuclear localization signal (Li et al., 2012), but in the context of cell–cell adhesion localization, we propose that the membrane-binding role is likely to be the most relevant.
In expanding our study to the other type II PAK isoforms, PAK4 and PAK5, and a representative type I PAK isoform, PAK1, we discovered that PAK isoforms exhibit differential targeting to cell–cell adhesions. We found that PAK4, PAK5 and PAK6 all target to cell–cell adhesions to varying degrees, indicating cell–cell adhesion targeting is not unique to PAK6 among the PAK family, but is a characteristic of type II PAKs. This is in agreement with previously published work showing that type II PAKs from other species, Xenopus PAK5 and Drosophila Mbt, also target to cell–cell adhesions (Schneeberger and Raabe, 2003; Faure et al., 2005). Further, while this manuscript was in preparation, a study was published confirming our observations that GFP–PAK4 localizes weakly to cell–cell adhesions (Selamat et al., 2015). By contrast, although we observe the type I PAK isoform, PAK1, in focal adhesions as previously reported (Brown et al., 2002), we do not observe PAK1 localized at cell–cell adhesions.
Given that all PAK isoforms can interact with Cdc42 but PAK1 does not localize to cell–cell adhesions, our isoform findings are in agreement with our PAK6 mapping studies that the ability to bind Cdc42 is not sufficient for targeting to cell–cell adhesions. Thus, there must be additional factors at play in determining type I versus type II PAK localization. We have already found that the polybasic region of PAK6 contributes to cell–cell adhesion targeting, and this region is well-conserved in PAK4 and PAK5 immediately adjacent to the CRIB motif (Fig. 6A). Although a few basic residues are present N-terminal to the CRIB motif in PAK1, they are interrupted by positively charged glutamic acid residues, which might be enough to interrupt any cell–cell targeting functionality. PAK1 does contain a polybasic stretch further upstream, which has been reported to function as a nuclear localization signal (Singh et al., 2005) and to interact with membrane lipids to contribute to PAK1 activation and membrane recruitment (Strochlic et al., 2010), but the presence of this sequence, at least in the context of GFP-tagged PAK1, does not appear to be sufficient for specific cell–cell adhesion targeting. It also remains a possibility that differences in the Cdc42-binding region of the PAK isoforms might contribute to differential targeting, even among type II isoforms, although the CRIB motif and many of the residues C-terminal to the motif shown to be important for Cdc42 binding are well-conserved (PDB: 2ODB) (Thompson et al., 1998). What determines type I versus type II PAK localization thus remains an area for exploration and additional studies will be required to reveal why PAK1 fails to target to cell–cell contacts.
Taken together, we have demonstrated that the ability of PAK6 to target to cell–cell adhesions is correlated with its ability to promote cell–cell dissociation, and that this phenomenon is characteristic of all human type II PAKs. Given that it has been proposed that type II PAKs phosphorylate β-catenin (or its homologs) at cell–cell adhesions to drive disassembly (Menzel et al., 2008; Fram et al., 2014; Selamat et al., 2015), the careful regulation of PAK targeting to and activation at cell–cell adhesions is likely important for the global regulation of cell–cell (dis)association. The proposed role for PAK6 in driving cell–cell contact disassembly and colony escape is, at first glance, apparently at odds with our demonstration of PAK6 recruitment at cell–cell contacts. We propose that this is explained by the kinetics of the process and suggest that Cdc42-mediated PAK6 recruitment to cell–cell contacts is an early step leading to phosphorylation of PAK6 substrates at adhesions and subsequent adhesion disassociation. Consistent with this model, our studies confirm that Cdc42 recruitment to cell–cell contacts precedes PAK6 recruitment, that Cdc42 is important for recruitment, and that the kinase-inactive mutant GFP–PAK6 K436M targets to cell–cell contacts but does not drive colony escape. Furthermore, PAK6 Cat, which is hyperactive in the absence of the regulatory N-terminus but is not locally concentrated at cell–cell adhesions, does not drive epithelial cell colony escape. It is possible that the active kinase is targeted to the membrane to phosphorylate cell–cell adhesion components and drive adhesion disassembly, or that the kinase is targeted to and then activated at the membrane and/or cell–cell adhesions. Given that PAK6 targets to cell–cell adhesions independent of catalytic activity, and that PAK6 readily accumulates at cell–cell adhesions before their disassembly, the latter possibility seems more likely. Indeed, as noted earlier, the polybasic membrane-targeting site of PAK1 has been reported to contribute to PAK1 activation (Strochlic et al., 2010) and the same might be true for type II PAKs. Additional studies will be required to identify potential membrane-associated local activators of type II PAKs, an area already of growing interest given their unique activation mechanisms (Baskaran et al., 2012; Ha et al., 2012).
MATERIALS AND METHODS
Antibodies
Primary antibodies against E-cadherin (mouse monoclonal DECMA-1, ab11512, Abcam), GFP (goat polyclonal 600-101-215, Rockland), PAK6 (AF4265, R&D Systems), vinculin (mouse monoclonal hVIN-1, Sigma), carbonyl reductase (rabbit polyclonal C-18, Santa Cruz), Cdc42 (2462, Cell Signaling Technology), β-catenin (D10A8, Cell Signaling Technology), ZO-1 (1A12, ThermoFisher Scientific) and β-tubulin (mouse monoclonal 2 28 33, Life Technologies) were purchased. A polyclonal antibody raised against PAK4 was kindly provided by Joseph Schlessinger (Yale University). IRDye-800-conjugated donkey anti-mouse (C31024-04, Li-Cor), LiCor IRDye-680 donkey anti-rabbit (C30724-04) and Alexa-Fluor-568-conjugated goat anti-mouse IgG (84E2-1, Invitrogen) secondary antibodies were purchased, and used at 1:2000 dilution for immunoblotting and at 1:500 dilution for immunofluorescence.
DNA constructs
Human PAK constructs were generated in pEGFP by PCR amplification from pCMV6M PAK1 (Uniprot Q13153) kindly provided by Pradeep Uchil (Yale University) and pLX304 PAK4 isoform 1 (Uniprot O96013-1), PAK5 (Uniprot Q9P286) and PAK6 (Uniprot Q9NQU5) constructs kindly provided by Michael Calderwood (Dana-Farber Cancer Institute). Mutations and deletions were introduced by QuikChange (Agilent Technologies) mutagenesis or PCR amplification and subcloning into pEGFP (Takara Bio Inc.) or pLENTI-CMV-Hygro-GFP (Draheim et al., 2015) vectors. Cdc42 DNA was kindly provided by Brian Rosenberg (Yale University) and subcloned into pmCherry (in-house vector derived from pEGFP). Human Cdc42 (UniProt ID: P60953) cDNA (Open Biosystems) encoding residues 1–177 was sub-cloned into modified pET28a vector with an N-terminal 6×His and GST tag. Cdc42 mutations were made by QuikChange mutagenesis.
Cell culture and transfection
DU145 prostate cancer cells were kindly provided by Raymond Baumann (Yale University) and HEK-293T cells were obtained from ATCC. Cells were grown in Dulbecco's modified essential medium (DMEM) (Life Technologies) containing 9% fetal bovine serum (FBS) (Life Technologies) and penicillin/streptomycin (Life Technologies) in a humidified atmosphere at 37°C with 5% CO2. Cells were transfected using PEI (Linear Polyethylenimine, Polysciences, Inc.). All lines were confirmed to be mycoplasma free by testing with MycoAlert Plus (Lonza).
Lentiviral knockdown and overexpression
Lentiviral shRNA pLKO constructs were obtained from a TRC shRNA library (Sigma) targeting PAK6 (TRCN0000196314), Cdc42 #1 (TRCN0000047632), Cdc42 #2 (TRCN0000310772) as well as a scrambled control (SHC002). Viral production occurred in HEK293-T cells following co-transfection with pLKO (knockdown) or pLENTI (overexpression) constructs, pCMV ΔR8.91 packaging vector, and pCMV-VSV-G envelope vector (provided by Soosan Ghazezadeh, SUNY, Stony Brook, NY). Viral supernatant was collected 48 h after transfection, passed through a 0.45 µm filter, and incubated overnight with adherent DU145 cells with 8 µg/ml polybrene (Sigma). Infected cells were selected with puromycin (2 µg/ml, pLKO) or hygromycin (500 µg/ml, pLENTI) 24 h after infection to generate stable lines.
For validation by immunoblotting, knockdown cells were lysed in RIPA buffer (50 mM Tris pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) with protease inhibitor tablets (Roche). Protein content was measured by BCA Assay (Thermo Scientific) and 100–150 µg of protein was mixed with sample buffer and loaded per well. Overexpression cells were lysed directly in sample buffer (5.0×105 cells per well) or in cell lysis buffer (10 mM PIPES pH 6.8, 50 mM NaCl, 150 mM sucrose, 1 mM Na3VO4, 50 mM NaF, 40 mM NaPPi, 0.05% Triton X-100, 0.1 mM PMSF) with protease inhibitor tablets (Roche). Samples were run on polyacrylamide gels and transferred to 0.2 µm nitrocellulose or PVDF membranes at 100 V for 2 h on ice. Membranes were blocked with 5% milk in Tris-buffered saline with Tween 20 (TBS-T) for 15–30 min at room temperature. Primary and secondary antibodies were diluted in 5% milk in TBS-T and incubated with membranes for at least 30 min up to overnight. Anti-PAK6 antibodies were used at 1:200 dilution, anti-GFP and anti-carbonyl-reductase antibodies at 1:1000, anti-tubulin antibody at 1:5000 and anti-vinculin antibodies at 1:10000. Membranes were rinsed in TBS-T prior to image acquisition using the Odyssey infrared imaging system (LI-COR) and image analysis with Image Studio (LI-COR).
Immunofluorescence and scoring of cell–cell adhesion targeting
DU145 cells were seeded on glass coverslips in 24-well dishes in complete medium (DMEM, 9% FBS, 1% pen/strep) and fixed with 4% paraformaldehyde containing 0.1% Triton X-100 in PBS for 15 min at room temperature. Coverslips were washed 3 times with 300 µl PBS and then incubated for 30 min with 300 µl blocking buffer (0.2% bovine serum albumin, 0.1% Triton X-100, 50 mM ammonium chloride in PBS). Primary and secondary antibodies were diluted in blocking buffer and incubated with coverslips in a humidity chamber for 1 h at room temperature (37°C for anti-β-catenin antibody). Anti-E-cadherin, anti-ZO-1 and anti-β-catenin antibodies were used at 1:100 dilution, and anti-Cdc42 antibodies at 1:500 dilution. Cells were mounted in FluorSave Reagent (EMD Millipore) containing DAPI and/or fluorophore-conjugated phalloidin (Invitrogen). Images were captured using a Nikon T1 microscope with a 40× or 100× immersion oil objective and analyzed using ImageJ (NIH).
The percentage of GFP-positive cells exhibiting cell–cell adhesion targeting was scored in at least three independent experiments for each construct with at least 40 cells per experiment. Only well-spread cells exhibiting low-to-moderate green fluorescence were scored. Student's t-test was performed using GraphPad Prism.
The fraction of GFP signal colocalizing with β-catenin staining was measured by calculating the Manders' coefficient using JACoP (Just Another Colocalization Plugin) in ImageJ. Prior to analysis, the threshold of β-catenin staining was determined using the RenyiEntropy setting in JACoP. Threshold of GFP–PAK6 was determined manually to incorporate all GFP signals. The Manders' coefficient was calculated and the fraction of GFP signal overlapping with β-catenin stain was recorded for at least 10 independent, non-overlapping frames for each condition.
Ca2+ switch assay
DU145 cells stably expressing mCherry–Cdc42 were generated by infection with retrovirus encoding mCherry–Cdc42 (gift from Brian Rosenberg, Yale University), and were seeded on glass coverslips and transfected with GFP–PAK6 in growth medium. 24 h post transfection, growth medium was replaced with Ca2+-free medium and cells incubated overnight at 37°C. Ca2+-containing growth medium was then re-introduced to the cells. Coverslips were fixed at various time points post Ca2+ addition and imaged by fluorescence microscopy.
Protein production and purification
6×His-GST, 6×His-GST-Cdc42 T17N (loss-of-function), 6×His-GST-Cdc42 WT (wild-type), and 6×His-GST-Cdc42 Q61L (gain-of-function) proteins were produced and purified in BL21 E. coli. Upon reaching OD600=0.6, cultures were induced with 1 mM IPTG and grown for 3 h at 37°C. To prepare GST–Cdc42 beads, bacterial pellets from 250 ml of culture were lysed in 35 ml of PBS with 1 mM DTT, 1% Triton X-100, 1.2 µg/ml lysozyme, and protease inhibitor tablets (Roche), and subsequently sonicated on ice. Lysates were clarified, added to 250 µl of 80% slurry glutathione-sepharose 4B beads (GE Healthcare), and incubated while rolling for 2 h at 4°C. Beads were washed twice in PBS with 1 mM DTT, twice in PBS, and resuspended to a 50% slurry in PBS. Bead loading was quantified by Coomassie staining on a polyacrylamide gel alongside BSA standards.
Cdc42 binding assay
DU145 cells stably expressing GFP-tagged PAK constructs were lysed in cell lysis buffer (5 mM PIPES pH 6.8, 0.5 mM Na3VO4, 25 mM NaF, 20 mM NaPPi, 25 mM NaCl, 75 mM sucrose, 0.05% Triton X-100, 100 µM PMSF, 0.1% deoxycholate) with protease inhibitor tablets (Roche) per confluent plate. The lysates were clarified and balanced with parental lysate to achieve similar inputs among cell lines. Lysate was applied to 5 µg of GST or GST–Cdc42 proteins on glutathione-sepharose beads (as described above) in Buffer X-T (5 mM Pipes pH 6.8, 0.5 mM Na3VO4, 25 mM NaF, 20 mM NaPPi, 25 mM NaCl, 75 mM sucrose, 0.05% Triton X-100). Beads and lysate were rocked for 1 h at 4°C then washed three times in 750 µl Buffer X-T. Samples were run on polyacrylamide gels, transferred to nitrocellulose and immunoblotted for GFP in the presence of 5% input controls.
Epithelial colony escape assay
DU145 cells were seeded on glass coverslips in 6-well dishes (1.0×105 cells/well) and allowed to form small colonies (∼5–10 cells) for 48 h. Cells were then transiently transfected with the indicated pEGFP constructs using PEI. Twenty-four hours after transfection, cells were fixed with 4% paraformaldehyde and stained with phalloidin and imaged as previously described in this section. Escaping cells were scored and defined as cells with greater than half of the cell periphery no longer in contact with neighboring cells, as previously published (Fram et al., 2014). At least 50 cells were scored per experiment in at least three independent experiments. Student's t-test was performed using GraphPad Prism.
Acknowledgements
We would like to thank past and present members of the Calderwood laboratory for helpful insights and discussion and Brain Rosenberg (Yale University) for providing mCherry–Cdc42 expressing retrovirus.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
E.M.M., X.S. and D.A.C. conceived and designed the experiments. E.M.M., X.S. and J.R.O. generated reagents and performed experiments. B.H.H. and T.J.B. edited the paper and provided key reagents and expertise. E.M.M., X.S., J.R.O. and D.A.C. analyzed data and wrote the paper.
Funding
This work was supported by the National Institutes of Health [grant numbers T32GM007223, R01GM068600 to D.A.C., R01GM102262 to T.J.B., P50CA121974 to T.J.B.] and a National Science Foundation Graduate Research Fellowship [grant number DGE-1122492 to E.M.M.]. Deposited in PMC for release after 12 months.
References
- Abel A. M., Schuldt K. M., Rajasekaran K., Hwang D., Riese M. J., Rao S., Thakar M. S. and Malarkannan S. (2015). IQGAP1: insights into the function of a molecular puppeteer. Mol. Immunol. 65, 336-349. 10.1016/j.molimm.2015.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abo A., Qu J., Cammarano M. S., Dan C., Fritsch A., Baud V., Belisle B. and Minden A. (1998). PAK4, a novel effector for Cdc42Hs, is implicated in the reorganization of the actin cytoskeleton and in the formation of filopodia. EMBO J. 17, 6527-6540. 10.1093/emboj/17.22.6527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskaran Y., Ng Y.-W., Selamat W., Ling F. T. P. and Manser E. (2012). Group I and II mammalian PAKs have different modes of activation by Cdc42. EMBO Rep. 13, 653-659. 10.1038/embor.2012.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S. and Cordelieres F. P. (2006). A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213-232. 10.1111/j.1365-2818.2006.01706.x [DOI] [PubMed] [Google Scholar]
- Bouaouina M., Goult B. T., Huet-Calderwood C., Bate N., Brahme N. N., Barsukov I. L., Critchley D. R. and Calderwood D. A. (2012). A conserved lipid-binding loop in the kindlin FERM F1 domain is required for kindlin-mediated alphaIIbbeta3 integrin coactivation. J. Biol. Chem. 287, 6979-6990. 10.1074/jbc.M111.330845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M. C., West K. A. and Turner C. E. (2002). Paxillin-dependent paxillin kinase linker and p21-activated kinase localization to focal adhesions involves a multistep activation pathway. Mol. Biol. Cell 13, 1550-1565. 10.1091/mbc.02-02-0015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callow M. G., Clairvoyant F., Zhu S., Schryver B., Whyte D. B., Bischoff J. R., Jallal B. and Smeal T. (2002). Requirement for PAK4 in the anchorage-independent growth of human cancer cell lines. J. Biol. Chem. 277, 550-558. 10.1074/jbc.M105732200 [DOI] [PubMed] [Google Scholar]
- Ching Y.-P., Leong V. Y. L., Wong C.-M. and Kung H.-F. (2003). Identification of an autoinhibitory domain of p21-activated protein kinase 5. J. Biol. Chem. 278, 33621-33624. 10.1074/jbc.C300234200 [DOI] [PubMed] [Google Scholar]
- Draheim K. M., Li X., Zhang R., Fisher O. S., Villari G., Boggon T. J. and Calderwood D. A. (2015). CCM2-CCM3 interaction stabilizes their protein expression and permits endothelial network formation. J. Cell Biol. 208, 987-1001. 10.1083/jcb.201407129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure S., Cau J., de Santa Barbara P., Bigou S., Ge Q., Delsert C. and Morin N. (2005). Xenopus p21-activated kinase 5 regulates blastomeres’ adhesive properties during convergent extension movements. Dev. Biol. 277, 472-492. 10.1016/j.ydbio.2004.10.005 [DOI] [PubMed] [Google Scholar]
- Field J. and Manser E. (2012). The PAKs come of age: celebrating 18 years of discovery. Cell. Logist. 2, 54-58. 10.4161/cl.22084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fram S., King H., Sacks D. B. and Wells C. M. (2014). A PAK6-IQGAP1 complex promotes disassembly of cell–cell adhesions. Cell. Mol. Life Sci. 71, 2759-2773. 10.1007/s00018-013-1528-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Ha B. H., Lou H. J., Morse E. M., Zhang R., Calderwood D. A., Turk B. E. and Boggon T. J. (2013). Substrate and inhibitor specificity of the type II p21-Activated Kinase, PAK6. PLoS ONE 8, e77818 10.1371/journal.pone.0077818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goult B. T., Bouaouina M., Elliott P. R., Bate N., Patel B., Gingras A. R., Grossmann J. G., Roberts G. C. K., Calderwood D. A., Critchley D. R. et al. (2010). Structure of a double ubiquitin-like domain in the talin head: a role in integrin activation. EMBO J. 29, 1069-1080. 10.1038/emboj.2010.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha B. H., Davis M. J., Chen C., Lou H. J., Gao J., Zhang R., Krauthammer M., Halaban R., Schlessinger J., Turk B. E. et al. (2012). Type II p21-activated kinases (PAKs) are regulated by an autoinhibitory pseudosubstrate. Proc. Natl. Acad. Sci. USA 109, 16107-16112. 10.1073/pnas.1214447109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha B. H., Morse E. M., Turk B. E. and Boggon T. J. (2015). Signaling, regulation, and specificity of the type II p21-activated kinases. J. Biol. Chem. 290, 12975-12983. 10.1074/jbc.R115.650416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffer Z. M. and Chernoff J. (2002). p21-activated kinases: three more join the Pak. Int. J. Biochem. Cell Biol. 34, 713-717. 10.1016/S1357-2725(01)00158-3 [DOI] [PubMed] [Google Scholar]
- Kaur R., Yuan X., Lu M. L. and Balk S. P. (2008). Increased PAK6 expression in prostate cancer and identification of PAK6 associated proteins. Prostate 68, 1510-1516. 10.1002/pros.20787 [DOI] [PubMed] [Google Scholar]
- King H., Nicholas N. S. and Wells C. M. (2014). Role of p-21-activated kinases in cancer progression. Int. Rev. Cell Mol. Biol. 309, 347-387. 10.1016/B978-0-12-800255-1.00007-7 [DOI] [PubMed] [Google Scholar]
- Lee S. R., Ramos S. M., Ko A., Masiello D., Swanson K. D., Lu M. L. and Balk S. P. (2002). AR and ER interaction with a p21-activated kinase (PAK6). Mol. Endocrinol. 16, 85-99. 10.1210/mend.16.1.0753 [DOI] [PubMed] [Google Scholar]
- Lei M., Lu W., Meng W., Parrini M.-C., Eck M. J., Mayer B. J. and Harrison S. C. (2000). Structure of PAK1 in an autoinhibited conformation reveals a multistage activation switch. Cell 102, 387-397. 10.1016/S0092-8674(00)00043-X [DOI] [PubMed] [Google Scholar]
- Li Y., Shao Y., Tong Y., Shen T., Zhang J., Li Y., Gu H. and Li F. (2012). Nucleo-cytoplasmic shuttling of PAK4 modulates beta-catenin intracellular translocation and signaling. Biochim. Biophys. Acta 1823, 465-475. 10.1016/j.bbamcr.2011.11.013 [DOI] [PubMed] [Google Scholar]
- Liu X., Busby J., John C., Wei J., Yuan X. and Lu M. L. (2013). Direct interaction between AR and PAK6 in androgen-stimulated PAK6 activation. PLoS ONE 8, e77367 10.1371/journal.pone.0077367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel N., Schneeberger D. and Raabe T. (2007). The Drosophila p21 activated kinase Mbt regulates the actin cytoskeleton and adherens junctions to control photoreceptor cell morphogenesis. Mech. Dev. 124, 78-90. 10.1016/j.mod.2006.09.007 [DOI] [PubMed] [Google Scholar]
- Menzel N., Melzer J., Waschke J., Lenz C., Wecklein H., Lochnit G., Drenckhahn D. and Raabe T. (2008). The Drosophila p21-activated kinase Mbt modulates DE-cadherin-mediated cell adhesion by phosphorylation of Armadillo. Biochem. J. 416, 231-241. 10.1042/BJ20080465 [DOI] [PubMed] [Google Scholar]
- Minden A. (2012). PAK4-6 in cancer and neuronal development. Cell. Logist. 2, 95-104. 10.4161/cl.21171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey A., Dan I., Kristiansen T. Z., Watanabe N. M., Voldby J., Kajikawa E., Khosravi-Far R., Blagoev B. and Mann M. (2002). Cloning and characterization of PAK5, a novel member of mammalian p21-activated kinase-II subfamily that is predominantly expressed in brain. Oncogene 21, 3939-3948. 10.1038/sj.onc.1205478 [DOI] [PubMed] [Google Scholar]
- Parrini M. C., Lei M., Harrison S. C. and Mayer B. J. (2002). Pak1 kinase homodimers are autoinhibited in trans and dissociated upon activation by Cdc42 and Rac1. Mol. Cell 9, 73-83. 10.1016/S1097-2765(01)00428-2 [DOI] [PubMed] [Google Scholar]
- Radu M., Semenova G., Kosoff R. and Chernoff J. (2014). PAK signalling during the development and progression of cancer. Nat. Rev. Cancer 14, 13-25. 10.1038/nrc3645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane C. K. and Minden A. (2014). P21 activated kinases: structure, regulation, and functions. Small GTPases 5, e28003 10.4161/sgtp.28003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph M. G., Bayer P., Abo A., Kuhlmann J., Vetter I. R. and Wittinghofer A. (1998). The Cdc42/Rac interactive binding region motif of the Wiskott Aldrich syndrome protein (WASP) is necessary but not sufficient for tight binding to Cdc42 and structure formation. J. Biol. Chem. 273, 18067-18076. 10.1074/jbc.273.29.18067 [DOI] [PubMed] [Google Scholar]
- Schneeberger D. and Raabe T. (2003). Mbt, a Drosophila PAK protein, combines with Cdc42 to regulate photoreceptor cell morphogenesis. Development 130, 427-437. 10.1242/dev.00248 [DOI] [PubMed] [Google Scholar]
- Schrantz N., da Silva Correia J., Fowler B., Ge Q., Sun Z. and Bokoch G. M. (2004). Mechanism of p21-activated kinase 6-mediated inhibition of androgen receptor signaling. J. Biol. Chem. 279, 1922-1931. 10.1074/jbc.M311145200 [DOI] [PubMed] [Google Scholar]
- Selamat W., Tay P.-L. F., Baskaran Y. and Manser E. (2015). The Cdc42 effector kinase PAK4 localizes to cell–cell junctions and contributes to establishing cell polarity. PLoS ONE 10, e0129634 10.1371/journal.pone.0129634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R. R., Song C., Yang Z. and Kumar R. (2005). Nuclear localization and chromatin targets of p21-activated kinase 1. J. Biol. Chem. 280, 18130-18137. 10.1074/jbc.M412607200 [DOI] [PubMed] [Google Scholar]
- Strochlic T. I., Viaud J., Rennefahrt U. E. E., Anastassiadis T. and Peterson J. R. (2010). Phosphoinositides are essential coactivators for p21-activated kinase 1. Mol. Cell 40, 493-500. 10.1016/j.molcel.2010.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z., Osborne M. J., Xu P., Xu X., Li Y. and Ni F. (2005). A bivalent dissectional analysis of the high-affinity interactions between Cdc42 and the Cdc42/Rac interactive binding domains of signaling kinases in Candida albicans. Biochemistry 44, 16461-16474. 10.1021/bi050846l [DOI] [PubMed] [Google Scholar]
- Takahashi S. and Pryciak P. M. (2007). Identification of novel membrane-binding domains in multiple yeast Cdc42 effectors. Mol. Biol. Cell 18, 4945-4956. 10.1091/mbc.E07-07-0676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson G., Owen D., Chalk P. A. and Lowe P. N. (1998). Delineation of the Cdc42/Rac-binding domain of p21-activated kinase. Biochemistry 37, 7885-7891. 10.1021/bi980140+ [DOI] [PubMed] [Google Scholar]
- Wallace S. W., Durgan J., Jin D. and Hall A. (2010). Cdc42 regulates apical junction formation in human bronchial epithelial cells through PAK4 and Par6B. Mol. Biol. Cell 21, 2996-3006. 10.1091/mbc.E10-05-0429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Lim L., Baskaran Y., Manser E. and Song J. (2013). NMR binding and crystal structure reveal that intrinsically-unstructured regulatory domain auto-inhibits PAK4 by a mechanism different from that of PAK1. Biochem. Biophys. Res. Commun. 438, 169-174. 10.1016/j.bbrc.2013.07.047 [DOI] [PubMed] [Google Scholar]
- Wells C. M. and Jones G. E. (2010). The emerging importance of group II PAKs. Biochem. J. 425, 465-473. 10.1042/BJ20091173 [DOI] [PubMed] [Google Scholar]
- Wells C. M., Whale A. D., Parsons M., Masters J. R. W. and Jones G. E. (2010). PAK4: a pluripotent kinase that regulates prostate cancer cell adhesion. J. Cell Sci. 123, 1663-1673. 10.1242/jcs.055707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X., Li X., Liao B., Liu Y., Wu J., Yuan X., Ouyang B., Sun Q. and Gao X. (2009). Knockdown of p21-activated kinase 6 inhibits prostate cancer growth and enhances chemosensitivity to docetaxel. Urology 73, 1407-1411. 10.1016/j.urology.2008.09.041 [DOI] [PubMed] [Google Scholar]
- Wen Y.-Y., Zheng J.-N. and Pei D.-S. (2014). An oncogenic kinase: putting PAK5 forward. Expert Opin. Ther. Targets 18, 807-815. 10.1517/14728222.2014.918103 [DOI] [PubMed] [Google Scholar]
- Yang F., Li X., Sharma M., Zarnegar M., Lim B. and Sun Z. (2001). Androgen receptor specifically interacts with a novel p21-activated kinase, PAK6. J. Biol. Chem. 276, 15345-15353. 10.1074/jbc.M010311200 [DOI] [PubMed] [Google Scholar]
- Zhang M., Siedow M., Saia G. and Chakravarti A. (2010). Inhibition of p21-activated kinase 6 (PAK6) increases radiosensitivity of prostate cancer cells. Prostate 70, 807-816. 10.1002/pros.21114 [DOI] [PMC free article] [PubMed] [Google Scholar]