ABSTRACT
Cardiolipin (also known as PDL6) is an indispensable lipid required for mitochondrial respiration that is generated through de novo synthesis and remodeling. Here, the cardiolipin remodeling enzyme, acyl-CoA:lysocardiolipin-acyltransferase-1 (Alcat1; SwissProt ID, Q6UWP7) is destabilized in epithelia by lipopolysaccharide (LPS) impairing mitochondrial function. Exposure to LPS selectively decreased levels of carbon 20 (C20)-containing cardiolipin molecular species, whereas the content of C18 or C16 species was not significantly altered, consistent with decreased levels of Alcat1. Alcat1 is a labile protein that is lysosomally degraded by the ubiquitin E3 ligase Skp–Cullin–F-box containing the Fbxo28 subunit (SCF-Fbxo28) that targets Alcat1 for monoubiquitylation at residue K183. Interestingly, K183 is also an acetylation-acceptor site, and acetylation conferred stability to the enzyme. Histone deacetylase 2 (HDAC2) interacted with Alcat1, and expression of a plasmid encoding HDAC2 or treatment of cells with LPS deacetylated and destabilized Alcat1, whereas treatment of cells with a pan-HDAC inhibitor increased Alcat1 levels. Alcat1 degradation was partially abrogated in LPS-treated cells that had been silenced for HDAC2 or treated with MLN4924, an inhibitor of Cullin–RING E3 ubiquitin ligases. Thus, LPS increases HDAC2-mediated Alcat1 deacetylation and facilitates SCF-Fbxo28-mediated disposal of Alcat1, thus impairing mitochondrial integrity.
KEY WORDS: Degradation, Mitochondria, Ubiquitin
Highlighted Article: This study provides new insight into how endotoxin triggers depletion of a key mitochondrial enzyme, Alcat1, by activating an SCF ubiquitin E3 ligase complex that contains Fbxo28.
INTRODUCTION
Gram-negative bacterial infections are major causes of morbidity and mortality in critically ill people, and many of the adverse effects of these infections are related to the pro-inflammatory effects of lipopolysaccharide (LPS) (Force et al., 2012). In the lung, highly virulent pathogenic strains of LPS-containing bacteria trigger strong pro-inflammatory responses leading to disruption of the alveolar capillary barrier, neutrophil infiltration and cytokine secretion, which results in severe ventilatory and gas-exchange abnormalities. At the cellular level, systemic endotoxemia can impair cellular oxygen consumption by adversely affecting mitochondrial function, a process termed cytopathic dysoxia (Creery and Fraser, 2002; Loiacono and Shapiro, 2010). Despite years of intensive study, the molecular mechanisms underlying cytopathic dysoxia are not yet fully understood (Chen et al., 2014a; Huang et al., 2014). Given the mounting evidence that LPS adversely affects mitochondrial function in several systems (Islam et al., 2012; Jeger et al., 2015; Nakahira et al., 2011), we hypothesized that defects within pathways that are vital to mitochondrial structure or function might lead to cytopathic dysoxia.
In eukaryotes, cardiolipin is a mitochondrial-specific phospholipid found almost exclusively in the inner-mitochondrial membrane. The unique dimeric structure of cardiolipin, which has two phosphatidyl residues combining two negative charges with four hydrophobic acyls, makes it indispensable for mitochondrial architecture, and metabolic and signaling functions (Ren et al., 2014). Deficiency of cardiolipin is lethal, resulting in apoptosis, impaired proliferation and reduced energy stores (Kagan et al., 2009; Kirkland et al., 2002; Martens et al., 2014; Sorice et al., 2004). Extracellular cardiolipin levels, such as in lung fluid, are very low, but release of cardiolipin from damaged mitochondria or bacterial membranes into the lung can serve as a damage-associated molecular pattern, causing host cell death under some pathophysiological conditions (Ray et al., 2010). Therefore, tight maintenance of cardiolipin homeostasis is crucial. Cardiolipin generation is governed both by its de novo synthesis and remodeling of existing molecules. The cardiolipin biosynthetic machinery displays limited specificity with regards to its fatty acyl composition (Lu and Claypool, 2015), yet different tissues exhibit characteristic molecular speciation of cardiolipins (Kagan et al., 2014, 2015). This is achieved through processes of its remodeling, during which the fatty acid residues of the nascent cardiolipins are removed, yielding monolysocardiolipins, and are replaced by the typical mature acyls forms, frequently C18:2 (Ye et al., 2014). This reacylation of monolysocardiolipins can be accomplished by one of three enzymes: tafazzin, a phospholipid-lysophospholipid transacylase (Schlame, 2013); monolysocardiolipin acyltransferase 1 (Mlclat1) or acyl-coenzyme A (acyl-CoA):lysocardiolipin-acyltransferase-1 (Alcat1; SwissProt ID, Q6UWP7) (Schlame, 2013; Lu and Claypool, 2015; Shen et al., 2015). Alcat1 is a 414-amino-acid membrane protein that is localized to the endoplasmic reticulum (ER). Alcat1 specifically catalyzes the acyl-CoA-dependent conversion of substrates and recognizes both monolysocardiolipins and dilysocardiolipins as substrates. Recent studies suggest that Alcat1 regulates cardiolipin remodeling in response to oxidative stress and that it might also play a role in pulmonary fibrosis; however, the molecular regulation of Alcat1 remains largely unknown (Cao et al., 2004; Huang et al., 2014). Thus, potentially, many processes that result in impaired remodeling of cardiolipin through dysregulation of mitochondrial acyltransferases could result in decreased cardiolipin generation, mitochondrial structural anomalies and impairment in cellular bioenergetics. One such example is Barth syndrome, in which a mutation causes decreased tafazzin function, leading to a multitude of clinical effects, coupled with impaired cellular bioenergetics due to reduced biologically active cardiolipin (Claypool et al., 2008, 2006, 2011; Whited et al., 2013).
Most cellular proteins are eliminated by ubiquitin-mediated degradation in response to environmental cues, during native cellular homeostatic control or in disease states (Chen et al., 2014a; Popovic et al., 2014; Zou et al., 2011a). Protein ubiquitylation involves an enzymatic cascade that covalently adds a ubiquitin to an acceptor lysine residue within a substrate (Sadowski and Sarcevic, 2010). The terminal step of substrate ubiquitylation involves a large family of enzymes – the ubiquitin E3 ligases that recognize a substrate to ensure a relatively selective mode of removal (Chen and Mallampalli, 2013; Jackson et al., 2000; Xu et al., 2007). Once ubiquitylated, substrates are degraded through the lysosomal or proteasomal degradation pathways. Two major families of E3 ubiquitin ligases are the homologous to the E6-AP C-terminus (HECT)- and Cullin–RING ligase (CRL)-family complexes. One subgroup of CRLs, termed Skp–Cullin–F-box proteins (SCF) has emerged as an important group of E3 complexes that control several fundamental processes, such as cell cycle progression, circadian rhythm and inflammation (Cardozo and Pagano, 2004; Kipreos and Pagano, 2000). Within SCF complexes, there resides a receptor component, the F-box protein, that engages substrates to facilitate their ubiquitylation. One F-box protein, Fbxo28 acts as an SCF subunit to mediate ubiquitin proteasomal degradation of Myc (Cepeda et al., 2013). Fbxo28 might also regulate cognitive function and impact the seizure phenotype observed in a rare microdeletion syndrome (Au et al., 2014). The repertoire of Fbxo28 targets still requires further investigation.
Protein ubiquitylation also can compete with other post-translational modifications on proteins, such as acetylation or methylation. Several families of acetyltransferases catalyze addition of acetyl groups onto acceptor lysine residues within a protein (Roth et al., 2001). Proteomic studies reveal that thousands of cellular proteins are acetylated, particularly those functioning in gene transcription and metabolism (Choudhary et al., 2009, 2014; Henriksen et al., 2012). Deacetylases, such as histone deacetyltransferases (HDACs), enzymatically remove acetyl groups from modified lysine residues, often resulting in gene transcriptional repression (Aapola et al., 2002; Taunton et al., 1996; Wolffe, 1996). Similar to ubiquitylation, reversible acetylation of proteins controls diverse cellular processes, such as the cell cycle (Cai et al., 2000), cell proliferation and tumorigenesis (Drummond et al., 2005).
In the present study we demonstrate that LPS reduces the cellular abundance of Alcat1 protein through actions mediated by an SCF complex comprising the Fbxo28 subunit, which then impairs cardiolipin homeostasis, and mitochondrial structural and functional integrity. Alcat1 is an acetylated enzyme, a modification that enhances its stability. LPS decreases Alcat1 protein levels by enhancing its HDAC2-mediated deacetylation, leading to monoubiquitylation of the enzyme and degradation through the lysosomal pathway.
RESULTS
LPS impairs mitochondrial function and depletes Alcat1
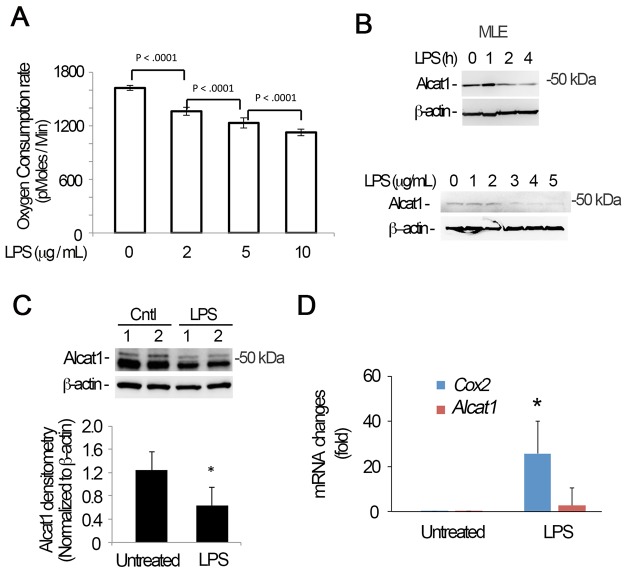
Exposure of MLE cells to LPS (2–10 μg/ml) results in a gradual, modest and yet physiologically important decrease in oxygen consumption, suggestive of a mitochondrial functional defect (Fig. 1A). After 4 h exposure of cells or with >2 μg/ml of LPS, we observed a decrease in the protein levels of Alcat1 (Fig. 1B, upper panels and lower panels). Similar to MLE cells, we observed that LPS reduced Alcat1 protein levels in primary mouse type 2 epithelial cells (Fig. S1). Further, LPS-treated mice exhibited lower levels of immunoreactive Alcat1 in lung tissues (Fig. 1C). Of note, in these experiments we detected at times a doublet representing bands migrating at approximately 50 kDa, perhaps representing the post-translationally modified enzyme. Alcat1 mRNA levels were not significantly altered in MLE cells upon LPS treatment (Fig. 1D). These results suggest that LPS might modulate Alcat1at the protein level, which leads to decreased cellular oxygen consumption.
Fig. 1.
LPS triggers Alcat1 degradation. (A–C) MLE cells were treated with LPS at varying concentrations (B, lower panels) or times (B, upper panel; fixed LPS concentration of 4 µg/ml). The oxygen consumption rate of cells was analyzed with a seahorse XF analyzer (A), or cell lysates were immunoblotted for Alcat1 and β-actin (B). (C) C57BL6 mice were treated with LPS (5 mg/kg) intratracheally overnight, and whole-lung tissue lysates were analyzed by immunoblotting for Alcat1 and β-actin. (D) Total cellular RNA was isolated from untreated or LPS-treated MLE cells in B (4 µg/ml for 16 h), and quantitative PCR analysis was conducted to determine steady-state mRNA levels using Alcat1-specific primers. Cox2 was used as a positive control. *P<0.05, Cox2 mRNA changes in untreated versus LPS-treated cells (Student's t-test). Data in each panel represents n=3 separate experiments. Means±s.e.m. are shown.
Alcat1 depletion impairs mitochondrial morphology and function
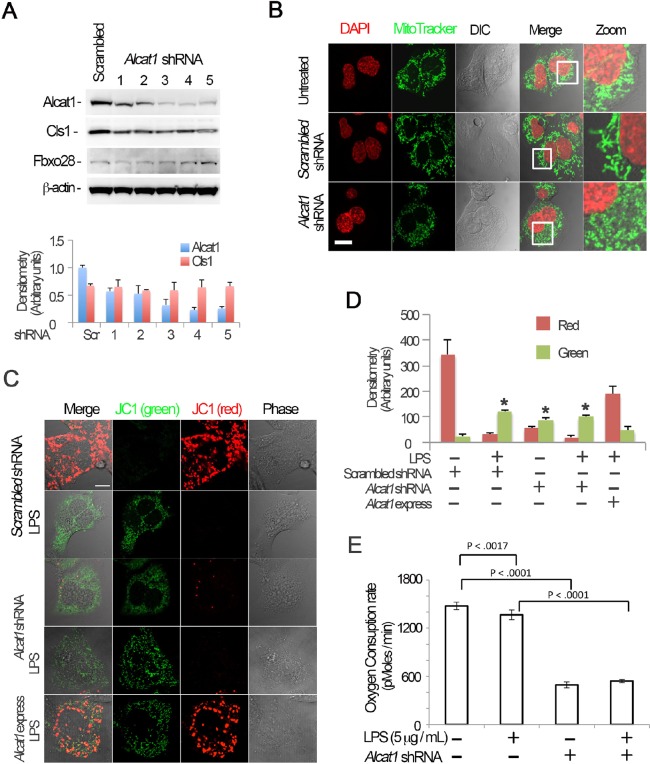
A recent study has shown that Alcat1 affects mitochondrial morphology, bioenergetics and biogenesis (Li et al., 2012). Hence, we depleted Alcat1 to further assess the requirement for the enzyme in lung epithelia. Small hairpin (sh)RNA constructs targeting Alcat1 (Alcat1 shRNA) effectively reduced the levels of immunoreactive Alcat1 compared to a scrambled control without affecting the amount of cardiolipin synthase1 (Cls1), β-actin or Fbxo28 (Fig. 2A, top panel and lower panels). Silencing of Alcat1 using these shRNA constructs in the cells was followed up with analysis of mitochondrial morphology. Results from fluorescent immunostaining showed that knockdown of Alcat1 substantially triggered mitochondrial fragmentation (Fig. 2B, lower panels) compared to untreated or to scrambled shRNA groups (Fig. 2B, upper and middle panels, respectively). To further link these LPS-induced changes in Alcat1 with a physiological readout, we measured the membrane potential (ΔΨ) in MLE cells using JC-1 staining. JC-1 dye displays potential-dependent accumulation in mitochondria, indicated by a fluorescence-emission shift from green to red. Thus, mitochondrial depolarization is indicated by a decrease in the red:green fluorescence ratio. Either treatment of cells with LPS or Alcat1 depletion using shRNAs resulted in impaired membrane potential, indicative of bioenergetic changes compared with control cells (Fig. 2C,D). In addition, overexpression of Alcat1 in MLE cells partially rescued the LPS-induced mitochondrial dysfunction (Fig. 2C, bottom row, Fig. 2D). To determine the role of Alcat1 in oxygen consumption, we depleted cells of the enzyme and observed a >60% reduction in oxygen consumption versus LPS alone, the latter producing more modest effects; LPS in combination with Alcat1 depletion did not further reduce oxygen uptake in an additive or synergistic manner (Fig. 2E). Overall, these data indicate that Alcat1 availability in lung epithelial cells is crucial because its depletion impairs mitochondrial morphology and function.
Fig. 2.
Alcat1 is required for maintenance of mitochondrial function and morphology. (A) Alcat1 was depleted using one of several (numbered 1–5) candidate lentiviral shRNA constructs for 48 h in MLE cells. Cell lysates were analyzed by immunoblotting for Alcat1 or for one of several additional control proteins (Cls1, cardiolipin synthase1; Fbxo28, F-box only protein 28; β-actin). The lower panel shows plots of the densitometry analysis of blots in A. The densitometry analysis values were normalized to those of β-actin. Scr, scrambled. (B) shRNA against Alcat1 (Alcat1 shRNA, shRNA construct 3 shown in Fig. 2A) or scrambled RNA constructs were introduced into cells for 48 h, and the cells were subjected to MitoTracker staining, or DAPI in order to visualize the nucleus. Scale bar: 10 μm. (C) JC-1 staining of Alcat1-shRNA- and LPS-treated MLE cells. Cells were electroporated with scrambled RNA or Alcat1 shRNA plasmid for 48 h with or without LPS. A plasmid encoding Alcat1 was also overexpressed in cells (Alcat1 express). Cells were treated with LPS for 5 min before staining with JC-1 (2 µM) for 20 min. Cells were washed with warm PBS five times before analysis by using confocal microscopy. Scale bar: 10 μm. (D) Densitometry analysis of images detailed in C. *P<0.05, green versus red staining (Student's t-test). (E) Cells were infected with lentiviral Alcat1 shRNA or scrambled RNA with or without LPS treatment, and the oxygen consumption rate was measured using a seahorse XF analyzer. Data in each panel represents n=3 separate experiments. Means±s.e.m. are shown.
LPS modulates cardiolipin molecular species
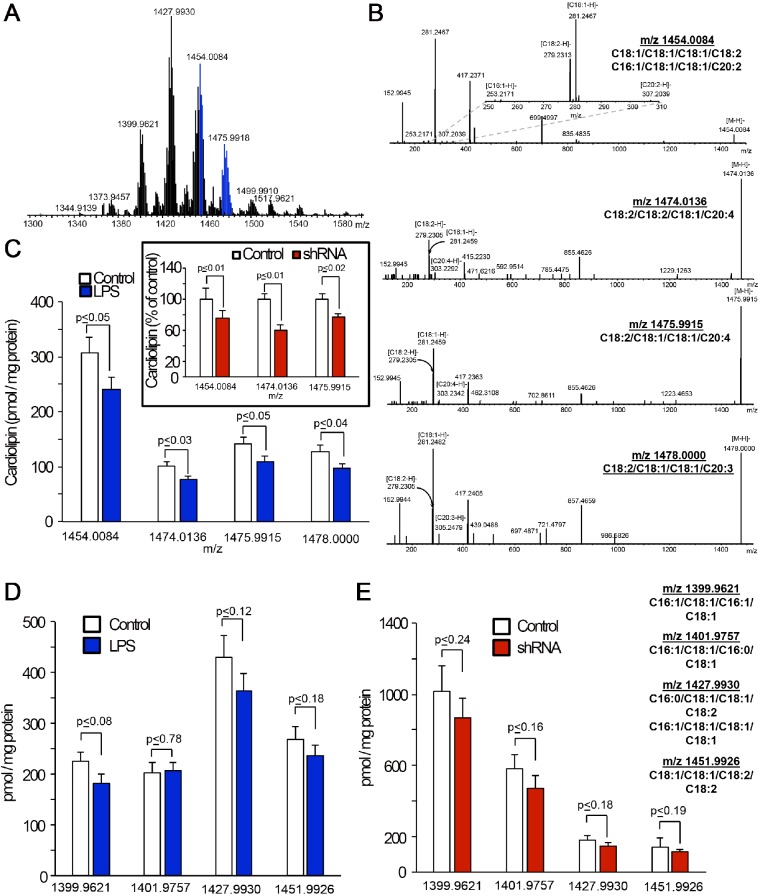
Mammalian cells utilize three different proteins to catalyze the remodeling reactions of cardiolipins: tafazzin, Alcat1 and Mlclat1 (Ren et al., 2014). Because the two latter enzymes require acyl-CoA species as donors for remodeling of monolysocardiolipin, the entire process might display specificity with regards to the reaction substrates and products (Claypool and Koehler, 2012). Of the three enzymes, Mlclat1 is the only one that is cardiolipin-specific, whereas the two other proteins can catalyze the reacylation of different lyso-phospholipids (Cao et al., 2009). Moreover, Mlclat1 seems to prefer C18:2 as the acyl-donor, whereas Alcat1 might utilize longer, particularly C20- and C22-based polyunsaturated acyls (Taylor and Hatch, 2003, 2009). Overall, the deficiency of Alcat1 could lead to the decreased content of cardiolipins and accumulation of monolysocardiolipins (in the absence of alternative reacylation reactions and catabolic degradation of monolysocardiolipin). With this in mind, we performed comparative liquid chromatography and mass spectrometry (LC-MS)-based analysis of cardiolipins and monolysocardiolipins in control versus LPS-treated or Alcat1-shRNA-transfected MLE cells (Fig. 3). We found that exposure to LPS resulted in selectively decreased levels of C20-containing cardiolipin molecular species (Fig. 3A–C), whereas the content of C18- or C16-containing species was not significantly changed (Fig. 3D). The total content of monolysocardiolipins was significantly lower in LPS-treated cells as compared to that in controls (data not shown), with a notable exception of monolysocardiolipin with m/z 1165.7671, the content of which increased from 33.9±9.8 to 67.9±11.4 pmol/mg of protein (P≤0.03, n=3; mean±s.e.m.). Knockdown of Alcat1 with shRNAs caused similar effects – a selective decrease of C20-containing molecular species of cardiolipins (Fig. 3C, insert) without significantly affecting the levels of C18- or C16-containing species (Fig. 3E), and selective accumulation of one monolysocardiolipin species with m/z 1165.7671, the content of which increased from 9.9±3.5 to 21.7±2.2 pmol/mg of protein (P≤0.02, n=3). This particular monolysocardiolipin is the only one that can be definitively associated with the parental C20-containing cardiolipin species with m/z 1454.0084, containing different combinations of other fatty acid residues (such as C16:1, C18:1, C18:1, C20:2). Because genetic manipulations of Alcat1 expression have been associated with the accumulation of oxidation-sensitive C20:4- and C22:6-containing species of cardiolipins, we also assessed the content of oxidation products in C20:4-containing cardiolipins (of note, C22:6-containing cardiolipin species were essentially undetectable in MLE cells). Very low levels of di-oxygenated derivatives of cardiolipin species with m/z 1505.9704 and 1507.9861 originating from the species containing C20:4 with m/z 1474.0136 and 1475.9915, respectively, were detectable in control and LPS-treated MLE cells. These levels of cardiolipin oxidation products in the cells were not different between the control and exposed samples [1.5±0.3 and 0.02±0.01 pmol/mg protein in control samples (m/z 1505.9704 and 1507.9861, respectively); 2.3±0.6 and 1.1±0.3 pmol/mg protein in LPS-treated samples (m/z 1505.9704 and 1507.9861, respectively)].
Fig. 3.
Silencing of Alcat1 or treatment with LPS decreases C20-containing cardiolipin molecular species in MLE cells. (A) Typical mass spectrum of cardiolipins obtained from MLE cells. Signals from C20-containing molecular species of cardiolipins are highlighted in blue. (B) Fragmentation patterns from tandem mass spectrometry analyses of C20-containing molecular species of cardiolipin [m/z 1454.0084, 1474.0136, 1475.9915 and 1478.0000 (top to bottom)] in MLE cells. Upper panel inset, a zoomed segment of a tandem spectrometry spectrum of cardiolipin molecular species with m/z 1454.0084 in the m/z range 250 to 310 showing the signals from [C18:2-H]− and [C18:1-H]− [C16:1-H]− and [C20:2-H]− ions. (C) Quantitative assessments of C20-containing molecular species of cardiolipin in MLE cells that had been treated with LPS (4 µg/ml, 16 h, 37°C) or shRNA (inset). Quantitative LC-MS analysis of the cardiolipin species with the indicated m/z (x-axis) in MLE cells that had been treated with LPS (D) or Alcat1 shRNA (E). Data are mean±s.d., n=3.
To identify the intracellular compartment where cardiolipin changes take place, we performed assessments of cardiolipins in mitochondrial and ER fractions isolated from wild-type (controls) and Alcat1-knockdown MLE cells, and cells that had been exposed to LPS. We found that, overall, cardiolipins, particularly the molecular species containing fatty acids with 20 carbon atoms (with signals at m/z of 1454.0084, 1474.0136, 1475.9915 and 1478.0000), were predominantly localized to mitochondria in control and treated MLE cells. Only very low levels of these C20-containing cardiolipin molecular species were detected in ER. Thus, mitochondrial cardiolipins, rather than ER cardiolipins, are the major contributors to the observed changes in the cardiolipin species that are affected by manipulations of Alcat1 (Fig. S2). Overall, our data on LPS- and shRNA-induced changes in cardiolipins are compatible with the decreased levels of Alcat1 that we detected. They also indicate that alternative pathways for cardiolipin reacylation, as well as catabolic reactions of monolysocardiolipin degradation, might contribute the overall changes in mitochondrial cardiolipins.
Alcat1 is labile and degraded within the lysosome
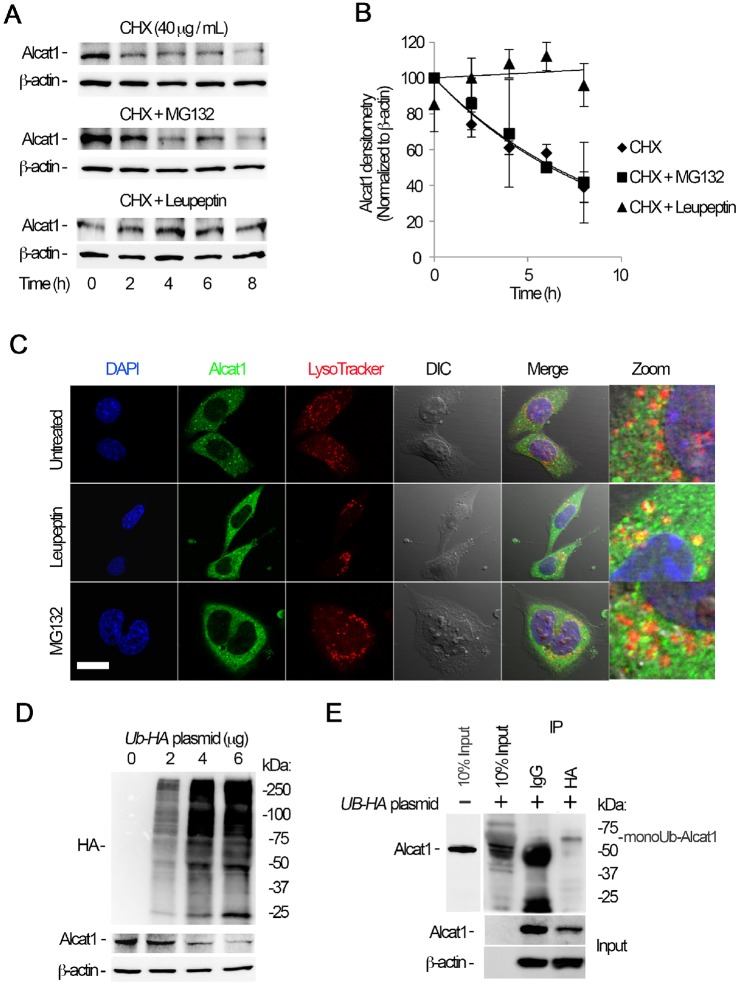
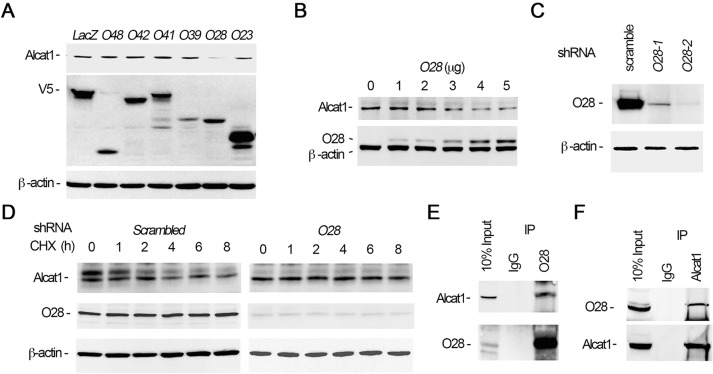
The observation that LPS reduces Alcat1 levels led us to investigate the molecular regulation of Alcat1, and specifically its turnover in cells, of which little is known. To evaluate this, we first examined Alcat1 degradation in MLE cells. In the presence of the protein synthesis inhibitor cycloheximide (CHX), we found that Alcat1 was unstable with a t½ of approximately 3 h (Fig. 4A, upper panels). The majority of the proteins are degraded through the proteasomal degradation pathway, yet some proteins are also sorted to the lysosome for elimination in cells. Hence, we treated cells with the proteasome inhibitor MG132 or a lysosome inhibitor, leupeptin, along with CHX. Unexpectedly, the addition of MG132 did not affect Alcat1 degradation, but leupeptin effectively abrogated degradation of Alcat1, indicating that Alcat1 is degraded through a lysosomal pathway (Fig. 4A, middle panel and lower panels, Fig. 4B). To confirm this observation, we performed fluorescent immunostaining of cells to identify localization of Alcat1 within these organelles. The results showed that Alcat1 proteins are mainly compartmentalized in the cytoplasm of untreated cells (Fig. 4C, upper panels). In the presence of leupeptin, Alcat1 was sorted and colocalized with LysoTracker, indicating the existence of Alcat1 trafficking within the lysosome (Fig. 4C, middle panels). However, this phenomenon was not observed in MG132-treated cells (Fig. 4C, lower panels). These data indicate that Alcat1 is unstable and is degraded through a lysosomal degradation pathway. Proteins that are destined for degradation in general are ubiquitylated. To determine if ubiquitin is sufficient to trigger Alcat1 degradation, we overexpressed a plasmid encoding hemagglutinin (HA)-tagged ubiquitin in cells and assayed endogenous Alcat1 protein. Immunoblotting results showed that ubiquitin had been successfully expressed in the cells and that ectopically expressed HA–ubiquitin promotes Alcat1 instability (Fig. 4D). We then examined whether Alcat1 is ubiquitylated. HA–ubiquitin was expressed in MLE cells, and the cell lysates were subjected to immunoprecipitation of HA followed by immunoblotting of Alcat1. As expected, results from immunoprecipitation studies showed a slower migrating band on SDS-PAGE gels, the size of which was approximately 8 kDa larger than that of the native Alcat1 band detected in the input (Fig. 4E). Given the mass of one ubiquitin moiety (∼8.5 kDa), these data strongly suggest that Alcat1 is monoubiquitylated. Taken together, these data demonstrate that Alcat1 is a short-lived protein that is monoubiquitylated and degraded through the lysosomal degradation pathway.
Fig. 4.
Alcat1 is monoubiquitylated and degraded through a lysosomal pathway. (A) MLE cells were treated with cycloheximide (CHX), CHX with MG132, or CHX with leupeptin, and cells were harvested at a variety of time points; lysates were analyzed by immunoblotting for Alcat1 and β-actin. (B) The bands shown in A were quantified, and the densitometry results are shown graphically. (C) Alcat1 colocalizes with lysosomes. Cells were treated with leupeptin or MG132 for 6 h and then immunostained for Alcat1 or with LysoTracker. The nucleus was visualized with DAPI staining. Untreated cells were used as a control. Scale bar: 10 μm. (D) Ectopically expressed HA–ubiquitin stimulates Alcat1 degradation. Various amounts of HA–ubiquitin (UB-HA) plasmids were introduced into MLE cells for 48 h. Cell lysates were processed for immunostaining of HA, Alcat1 and β-actin. (E) Cell expressing HA–ubiquitin were subjected to immunoprecipitation of HA, followed by immunoblotting for Alcat1 and β-actin. Data in each panel represent n=3 separate experiments. IP, immunoprecipitation; monoUb-Alact1, monoubiquitylated Alcat1. Means±s.e.m. are shown.
Alcat1 is selectively targeted for degradation by SCF containing Fbxo28
The final step of substrate ubiquitylation involves an E3 ubiquitin ligase that specifically recognizes the protein substrate. In the process of screening effects of ectopically expressed E3 ligase plasmids in cells, we have previously observed that SCF subunits modulate cardiolipin generation and mitochondrial function (Chen et al., 2014a; data not shown). By screening using a biobank of SCF E3 ubiquitin ligase F-box plasmids, we observed that overexpression of the SCF F-box protein Fbxo28 efficiently mediated Alcat1 degradation in MLE cells (Fig. 5A). Ectopic expression of Fbxo28 generally triggered Alcat1 degradation in a plasmid-dose-dependent manner (Fig. 5B). We next determined the effects of Fbxo28 knockdown with shRNA on Alcat1 stability. Immunoblotting analysis results showed that Fbxo28 was successfully silenced at the protein level with shRNAs o28-1 and o28-2, with a pronounced effect of o28-2 (Fig. 5C). Cellular depletion of Fbxo28 with shRNA o28-2 showed that Alcat1 levels exhibited prolonged stability in CHX half-life studies compared to that upon transfection of cells with scrambled shRNA (Fig. 5D). Further, co-immunoprecipitation studies demonstrated that Fbxo28 and Alcat1 interacted in lung epithelia (Fig. 5E,F). These data demonstrate that Fbxo28 is an authentic component of the E3 ubiquitin ligase complex and that Alcat1 is selectively targeted by this F-box protein in lung epithelial cells.
Fig. 5.
Alcat1 is selectively targeted by the ubiquitin E3 ligase SCF-Fbxo28. (A) Overexpression of Fbxo28 degrades Alcat1. Various V5-tagged F-box-encoding plasmids were expressed in MLE cells for 48 h (O48, Fbxo48; O42, Fbxo42; O41, Fbxo41; O39, Fbxo39; O28, Fbxo28; O23, Fbxo23). Cell lysates were analyzed by immunoblotting for Alcat1, V5 or β-actin. LacZ was used as a negative control. (B) Fbxo28 mediates Alcat1 degradation. Different amounts of V5–Fbxo28 expression plasmid were introduced into cells for 24 h. Cell lysates were analyzed by immunoblotting for Alcat1, V5 or β-actin. (C) Fbxo28 was knocked down by using shRNA (Fbxo28 shRNA). Lentiviral shRNA constructs against Fbxo28 (O28-1 and O28-2) were introduced into cells for 48 h. Cell lysates were analyzed by immunoblotting for Fbxo28 and β-actin. (D) Depletion of Fbxo28 by using shRNA stabilizes Alcat1 levels. An Fbxo28 shRNA construct (O28-2 in C) was introduced into cells for 48 h. Cells were treated with CHX for various time points. Cell lysates were analyzed by immunoblotting for Alcat1, Fbxo28 or β-actin. A scrambled lentiviral shRNA construct was used as a control. (E,F) Fbxo28 associates with Alcat1. MLE cell lysates were immunoprecipitated (IP) with antibodies against Fbxo28 (E) or Alcat1 (F), and the precipitates were subjected to immunoblotting as indicated. Data in each panel represents n=3 separate experiments.
Identification of a ubiquitin acceptor within Alcat1
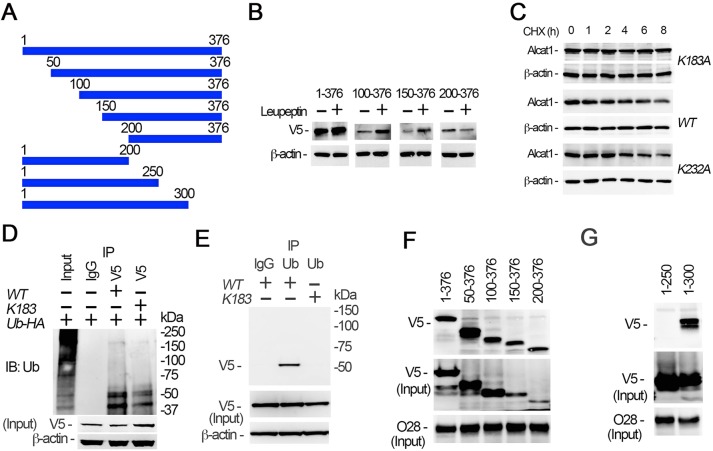
We next investigated the ubiquitin-acceptor site within Alcat1 that is targeted by an SCF complex containing Fbxo28 (SCF-Fbxo28). To map the ubiquitylation site, we constructed a series of truncation deletion Alcat1 mutants (Fig. 6A). We expressed wild-type Alcat1 and the truncated mutants in MLE cells, and assessed their relative accumulation in the presence of leupeptin, with the assumption being that if a fragment harbors the lysine acceptor, the ubiquitylated fragment would accumulate in the presence of lysosomal inhibition. The results showed that the full-length protein and fragments containing residues 100–376 and residues 150–376 accumulated upon lysosomal blockade, but not a fragment containing residues 200–376, suggesting that a putative ubiquitin-acceptor site resides between residues 150–200 of the primary sequence (Fig. 6B). Analysis of the primary sequence of this fragment revealed that this region contains one lysine residue at position 183. We conducted site-directed mutagenesis by replacing K183 with an alanine residue and ectopically expressed this plasmid (Alcat1K183A), the wild-type protein or a control plasmid (Alcat1K232A) in MLE cells and examined protein half-life (Fig. 6C). The degradation assays indicated that the K183A mutation within Alcat1 conferred an extended half-life (Fig. 6C, upper panels). As a control, the lifespan of the K232A mutant was comparable to that of wild-type Alcat1 (Fig. 6C, middle and lower panels). To further support the notion that K183 is a ubiquitin acceptor, we conducted an immunoprecipitation study to assay the ubiquitylation of wild-type and K183A mutant Alcat1. The V5-tagged wild-type and K183A mutant Alcat1 proteins were co-expressed with HA–ubiquitin, and the cell lysates were used for immunoprecipitation of the V5 tag. Immunoblotting of the precipitates for HA demonstrated that the K183A mutant showed reduced monoubiquitylation as compared with that of the wild type, suggesting that K183 is the major ubiquitylation site within Alcat1 (Fig. 6D). In reciprocal co-immunoprecipitation studies, immunoblotting the ubiquitin precipitates for V5 indicated that wild-type Alcat1, but not the K183A mutant was monoubiquitylated (Fig. 6E). Because Fbxo28 binds to Alcat1, we next assessed the docking site of Fbxo28 within Alcat1. We synthesized the V5-tagged recombinant deletion mutants in vitro using a rabbit reticulocyte transcription and translation system. The synthesized truncated mutants were successfully expressed. Endogenous Fbxo28 was immunoprecipitated using an antibody. Pull-down assays indicated that the stretch of amino acids spanning 200–250 within Alcat1 were crucial for binding of Fbxo28 (Fig. 6F,G). Taken together, these data indicate that Fbxo28 docks within a central domain of Alcat1 and catalyzes the monoubiquitylation of the remodeling enzyme at K183 to promote lysosomal degradation.
Fig. 6.
SCF-Fbxo28 selectively ubiquitylates Alcat1 at residue K183 and docks within the protein. (A) Schematic presentation of Alcat1 truncation mutants, which were tagged with V5. Amino acid positions are numbered. (B) The wild type (WT) (residues 1–376) and Alcat1 truncated mutants were expressed in MLE cells for 48 h. The cells were treated with leupeptin for 6 h. Cell lysates were analyzed by immunoblotting for V5 or β-actin. (C) WT V5–Alcat1, V5–K183A or V5-K232A Alcat1 mutants were expressed in cells for 48 h, and cells were then treated with CHX for various times. Cell lysates were subjected to immunoblotting for V5 and β-actin. (D) WT Alcat1 or the K183A mutant were co-expressed with HA–ubiquitin (Ub-HA) in cells for 48 h. Equal amounts of cell lysate (1 mg total protein) were subjected to immunoprecipitation with an antibody against V5; the precipitates were analyzed by immunoblotting for ubiquitin (Ub). The input is 5% of the total cell lysate. (E) WT V5–Alcat1 or a V5–K183A mutant was expressed in cells for 48 h. Equal amounts of cell lysate (1 mg total protein) were subjected to immunoprecipitated with an antibody against ubiquitin; the precipitates were analyzed by immunoblotting for V5. (F,G) Pull-down assays were conducted with truncated mutants to map the region within Alcat1 with which Fbxo28 interacts. Precipitates were then immunoblotted for V5 or Fbxo28 (O28).
HDAC2 deacetylates K183 within Alcat1 to enhance its degradation
Lysine sites are highly prone to a variety of post-translational modifications aside from ubiquitylation. Further, competition occurs in some systems between ubiquitylation and acetylation (Grönroos et al., 2002). Our data showed that monoubiquitylation at residue K183 within Alcat1 contributes to its instability. This clue prompted us to study Alcat1 acetylation status, which might also act to modulate enzyme stability. V5-tagged wild-type Alcat1, and the K183A and K232A mutants were expressed in cells, and cell lysates were assayed by immunoprecipitating V5 and immunoblotting for acetyl-lysine. Interestingly, the results showed that wild-type Alcat1 was highly acetylated under these culture conditions (Fig. 7A). Alcat1 acetylation levels were drastically reduced when the K183A mutant was assessed, indicating that residue K183 is a major acetylation site within Alcat1. Because K183 is acetylated and is also prone to Fbxo28-mediated monoubiquitylation, acetylation of this residue must be reversed for subsequent ubiquitylation to occur. Therefore, we hypothesized that Alcat1 undergoes deacetylation. Indeed, treatment of the cells with an acetyltransferase inhibitor, anacardic acid, modestly decreased steady-state Alcat1 protein mass, but treatment with a deacetylase inhibitor, trichostatin A, increased immunoreactive Alcat1 levels in the cells (Fig. 7B). Hence, the acetylation–deacetylation state of Alcat1 is crucial for its stability. Because inhibition of a deacetylase leads to accumulation of Alcat1, we next investigated potential deacetylases that might modulate Alcat1 cellular concentrations. Using a candidate plasmid screen (data not shown), we identified that HDAC2 and Alcat1 interact in co-immunoprecipitation studies (Fig. 7C,D). These results suggest that HDAC2-catalyzed deacetylation could contribute to Alcat1 stability. Because LPS decreases Alcat1 levels, we next examined whether inhibition of the Cullin-1 E3 ubiquitin ligase complexes with the inhibitor MLN4924 could restore acetylated Alcat1 levels in the presence of LPS. LPS reduced acetylated Alcat1 levels, an effect that was partly rescued by treatment with MLN4924 (Fig. 7E). We next tested whether LPS-induced Alcat1 degradation was HDAC2 dependent. We treated cells with LPS in the presence or absence of trichostatin A. Immunoblotting results showed that LPS-mediated Alcat1 degradation was attenuated by the HDAC2 inhibitor (Fig. 7F) in MLE cells. We next tested Alcat1 stability upon overexpression or knockdown of HDAC2 in cells (Fig. 7G). Immunoblotting results showed that HDAC2 protein was successfully overexpressed or silenced in MLE cells (Fig. 7G). Compared to expression of a vector control (Fig. 7G, left panel), overexpression of HDAC2 accelerated Alcat1 degradation (Fig. 7G, middle panel), whereas knockdown of HDAC2 stabilized Alcat1 levels (Fig. 7G, right panel). HDAC2 protein is believed to be a nuclear protein that might relocate into the cytoplasm in order to deacetylate cytosolic proteins. To test this possibility, we treated the MLE cells with LPS overnight and monitored the levels of HDAC2 in subcellular fractions. Interestingly, HDAC2 under native unstimulated conditions was consistently detected in the nuclear fraction but was also present at modest levels in the cytoplasmic (soluble) fraction (Fig. S3). Unexpectedly, HDAC2 was upregulated at the protein level only in the cytoplasmic fraction after LPS stimulation and not in the nuclear fraction. In addition, LPS did not reduce total HDAC2 protein levels in MLE cells (Fig. S3). Thus, deacetylation of Alcat1 contributes to its instability. Collectively, these data indicate that LPS downregulates Alcat1 concentrations in cells through HDAC2-mediated deacetylation of the cardiolipin remodeling enzyme.
Fig. 7.
HDAC2 promotes Alcat1 degradation. (A) V5-tagged WT Alcat1, or the K183A or K232A mutants were separately expressed in MLE cells for 48 h. Cell lysates were subjected to immunoprecipitation of V5 followed by immunoblotting for acetyl-lysine, V5 or Alcat1. (B) Cells were treated with an acetylation inhibitor, anacardic acid (AA), or a deacetylation inhibitor, trichostatin A (TSA), overnight. Cell lysates were immunoblotted for Alcat1 and β-actin. (C,D) Alcat1 interacts with HDAC2. Cell lysates were subjected to immunoprecipitation of HDAC2 (C) or Alcat1 (D), and precipitates were immunoblotted for the indicated proteins. (E) MLE cells or Alcat1-silenced cells (Alcat1 sh) were treated with LPS (4 μg/ml) in the presence of the Cullin-1 inhibitor MLN. Cell lysates were subjected to immunoprecipitation with an antibody against Alcat1; the precipitates were analyzed by immunoblotting for acetyl lysine or Alcat1. (F) Cells were treated with different concentrations of LPS with or without the deacetylase inhibitor TSA. Cell lysates were analyzed by immunoblotting for Alcat1 and β-actin. (G) A plasmid encoding HDAC2 was expressed (HDAC2 express), or HDAC2 was silenced by using retroviral shRNA constructs (HDAC2 shRNA) (48 h). Alcat1 degradation was assayed by using CHX, and lysates were immunoblotted for Alcat1, HDAC2 and β-actin. Vector-only-transfected cells were used as control. Data in each panel represent n=2 separate experiments.
DISCUSSION
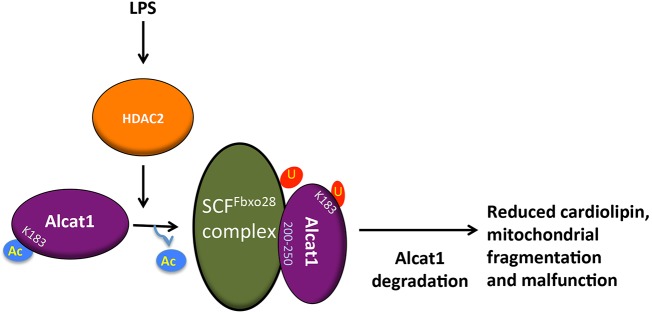
There is limited data on the molecular regulation of Alcat1. The primary findings in this study are: (i) Alcat1 is an unstable lysosomally degraded enzyme, (ii) SCF-Fbxo28 mediates degradation of Alcat1 through site-specific docking and monoubiquitylation, (iii) Alcat1 is an acetylated enzyme and is subject to HDAC2-mediated deacetylation that contributes to Alcat1 instability, and (iv) the adverse effects of LPS on cardiolipin synthesis and mitochondrial cellular bioenergetics could be mediated by coordinate actions of HDAC2 and SCF-Fbxo28 that reduce Alcat1 lifespan in lung epithelia (Fig. 8). Interestingly, mitochondrial defects that result in an inability to utilize oxygen at the cellular level (cytopathic dysoxia) is a hallmark of fulminant systemic infection (e.g. septic shock), the molecular basis of which remains unclear. There is ample evidence that LPS incites abnormalities in mitochondrial function (Jeger et al., 2015), and that in hepatocytes LPS interacts with cell membranes and disperses within the cytoplasm where it can modulate organelle activity (Díaz-Laviada et al., 1991). The results here support a unique model whereby LPS directly or indirectly depletes levels of a crucial lipogenic remodeling enzyme by regulating the deacetylation and ubiquitylation of the enzyme, which ultimately modifies cardiolipin molecular species, mitochondrial structure and energetics. The demonstration that LPS-induced impairment of mitochondrial function can be protected through mitochondrial transfer using bone-marrow-derived stromal cells provides an intriguing translational opportunity to reconstitute Alcat1, especially given that cell-based therapies are currently under investigation for disorders that are linked to severe pulmonary cell injury (Islam et al., 2012; Liu et al., 2014).
Fig. 8.
LPS impacts mitochondrial morphology and function by destabilizing Alcat1. Treatment with LPS activates the HDAC2 deacetylase, which targets Alcat1 at K183. SCF-Fbxo28 docks with deacetylated Alcat1 (in the region of Alcat1 comprising residues 200–250) and catalyzes monoubiquitylation of K183 within Alcat1. Monoubiquitylated Alcat1 is degraded through a lysosomal pathway. Reduction in the cellular concentration of Alcat1 leads to altered cardiolipin content, and impaired mitochondrial structure and bioenergetics. Ac, acetylation; U, ubiquitin.
Alcat1 is one of the enzymes involved in the regulation of mitochondrial cardiolipins and functions by controlling the remodeling of existing cardiolipins (Lu and Claypool, 2015). The Alcat1-driven reacylation of monolysocardiolipin, as a part of the cardiolipin remodeling process, occurs in a specialized compartment of the ER, the mitochondrial associated membrane (MAM) (Cao et al., 2004; Cao et al., 2009; Li et al., 2010). Alcat1-mediated reacylation preferentially incorporates coenzyme A that is loaded with long-chain polyunsaturated fatty acids (Cao et al., 2004). The remodeling process is initiated in mitochondria, by a Ca2+-independent phospholipase PLA2 (iPLA2), which removes an acyl chain from cardiolipin, thus generating the remodeling substrate monolysocardiolipin. By contrast, the product of Alcat1-mediated remodeling, mature cardiolipin, is generated in the ER, albeit in close proximity to mitochondria. Given that the preponderance of mature cardiolipin is found in mitochondria, more specifically in the inner mitochondrial membrane, transmembrane redistribution of both the substrates and the products between mitochondria and ER membranes seems to be required for the effective completion of the entire remodeling process (Claypool and Koehler, 2012). However, the details of this complex multi-stage process are just beginning to emerge. Externalization of cardiolipin to the mitochondrial surface, including to locales in the proximity of MAMs, has been documented (Chu et al., 2013; Kagan et al., 2015). Speculatively, at least two translocases – nucleoside diphosphate kinase isoform D (NDPKD) (Schlattner et al., 2013) and phospholipid scramblase 3 (PLS3) (Zhou et al., 1998) are candidate proteins that could be involved in cardiolipin redistribution and Alcat1-based cardiolipin remodeling. In accordance with these concepts on Alcat1 involvement in cardiolipin remodeling, we found that mitochondrial cardiolipins, rather than ER cardiolipins, are predominantly affected by manipulations of Alcat1 and are the major contributors to the observed changes in the cardiolipin species.
A substantial body of the published data in the literature indicates that the absence of taffazin, rather than the loss of Alcat1 or Mlclat1, causes alterations in cardiolipins and their molecular speciation (Vreken et al., 2000; Baile et al., 2014; Bissler et al., 2002; Acehan et al., 2011, 2009; Dudek et al., 2013; Gonzalvez et al., 2013; Gu et al., 2004; Houtkooper et al., 2009a,b; Valianpour et al., 2005, 2002; Schlame et al., 2003, 2005; Xu et al., 2006a,b, 2005, 2009; Wang et al., 2014); there are also indications that Alcat1 is involved in ‘pathological’ remodeling of cardiolipins, leading to mitochondrial dysfunction and oxidative stress (Li et al., 2010; Liu et al., 2012). It has been demonstrated that targeted deletion of Alcat1 restores effective mitophagy and removal of damaged mitochondria, thus contributing to the maintenance of functional mitochondrial architecture, mitochondrial DNA (mtDNA) fidelity and oxidative phosphorylation (Wang et al., 2015).
These conclusions, however, are based on experiments with limited lipidomics analysis. Our LC-MS data included detailed characterization of >35 individual molecular species of cardiolipin. These data indicate that both treatment with LPS and shRNA induces Alcat1 deficiency that is associated with the uniform and selective loss of predominantly mitochondrial C20-polyunsaturated fatty acid (PUFA) species of cardiolipins in MLE cells and that such deficiency impairs mitochondrial function in these cells. These data are in sharp contrast to those reported by others (Li et al., 2010; Liu et al., 2012) in which expression, rather than depletion, of Alcat1 is associated with the production of ‘pathological’ highly oxidizable PUFA-containing (mostly C22:6) cardiolipin species, leading to mitochondrial dysfunction. A thorough lipidomics analysis did not reveal any significant changes in cardiolipin oxidation that is related to LPS treatment or Alcat1 deficiency (shRNA).
Cardiolipin deficiency is lethal and results in decreased cell growth, apoptosis and reduced energy stores (Choi et al., 2007; Kagan et al., 2009; Kirkland et al., 2002). In eukaryotic cells, cardiolipin is synthesized by two pathways (de novo synthesis and remodeling of the pre-existing protein) that involve enzymatic activities that, under certain scenarios, are indispensable for cell viability. For example, mitochondrial dysfunction ensues in Barth syndrome – where tafazzin is mutated (Claypool et al., 2008, 2006, 2011) – and when Cls1 is defective (Pineau et al., 2013). In our related studies, bacterial infection with Staphylococcus aureas downregulates Cls1 levels through activation of another E3 ubiquitin ligase, SCF-Fbxo15, which decreases cardiolipin availability and disrupts mitochondrial function in lung epithelial cells (Chen et al., 2014a). Here, endotoxin treatment triggered E3-ligase-dependent degradation of Alcat1, which produced a similar mitochondrion phenotype. Interestingly, Alcat1-knockout mice are not prone to embryonic lethality but have variable alterations in fatty acyl composition of phospholipids depending on the nature of the diet (Imae et al., 2012; Li et al., 2010). The vulnerability of these mice to endotoxin challenge awaits further study, but the lack of an overt phenotype in Alcat1-knockout mice suggests the presence of compensatory activities of other pathways. Of note, forced expression of Alcat1 triggers mitochondrial fragmentation through oxidative stress and impairs processes important for mitochondrial fusion (Li et al., 2012). Thus, these observations collectively suggest a tight physiological margin of Alcat1 concentration in cells whereby the enzyme maintains the mitochondrial balance of cardiolipins. Our results suggest that these concentrations are diametrically governed by post-translational processing through ubiquitylation and deacetylation.
The stability of key proteins has emerged as an interesting area of study in disease. Recent studies report that the stability of many molecules at the protein level in the lung are dysregulated during tissue injury. In the pathogenesis of bacterial infection, functional proteins are impaired by pathogens that might directly or indirectly modulate protein stability, or through mRNA transcriptional repression. The ability of pathogens to impair host protein stability could represent a more energy-efficient mechanism, whereby these microbial factors undermine key pathways instead of silencing enzyme de novo synthesis. Our observations here are consistent with the ability of pathogens to modulate the stability of key biomolecules at the protein level, such as that of the surfactant biosynthetic enzymes CCT (Chen et al., 2011) and Lpcat1 (Zou et al., 2011a).
Post-translational modification appears to be a prerequisite for SCF E3 ligases to be recruited to substrates. A well-studied paradigm is the ability of phosphorylated targets (through phosphodegrons) to recruit F-box proteins that regulate ubiquitin–proteasomal degradation or sorting of proteins. However, increasing evidence indicates that the prototypical post-translational modification acetylation also plays an important role in regulation of ubiquitin–proteasomal or lysosomal degradation. For example, site-specific acetylation of the multifunctional enzyme nucleoside diphosphate kinase A impairs its interaction with an SCF containing the Fbxo24 subunit, and the stability of the protein Ras association domain family member 5 is enhanced through reduced binding to the HECT-family E3 ligase Itch in cells after treatment with a deacetylation inhibitor (Chen et al., 2015b; Suryaraja et al., 2013). Further, p300 acetylates Skp2 to increase its stability through impairment of Cdh1-mediated proteolysis in cancer cells (Inuzuka et al., 2012). In addition to these proteins, mitochondrial enzymes are highly modified by acetylation. For example, ATP synthase, enzymes within the tricarboxylic acid cycle and those involved in mitochondrial β-oxidation exhibit high levels of acetylation, raising the possibility of roles for this modification in regulating the half-lives of these proteins (Kerner et al., 2015). At times, the processes of ubiquitylation and acetylation can compete for access to a common molecular site. Our data suggest intermolecular competition at a single residue, K183, between acetylation – which stabilizes Alcat1 – and destabilizing monoubiquitylation that is mediated by SCF-Fbxo28, although confirmation of this would require additional studies. Whether acetylation serves other roles in addition to stabilizing protein levels, such as preserving catalytic activity or modulating other aspects of enzymatic behavior (protein sorting), is unclear.
There is mounting evidence that deacetylases mediate responses to endotoxin. For example, inhibition of the deacetylase sirtuin 2 lessens the severity of inflammation in response to LPS in microglia (Chen et al., 2015a). Further, inhibition of histone deacetylases improves survival in a murine model of LPS-induced shock and hepatotoxicity (Zhao et al., 2015). Other studies show that inhibition of histone deacetylase 8 reduces synthesis of pro-inflammatory cytokines (Li et al., 2015). Here, endotoxin increased soluble HDAC2 content in lung epithelia, HDAC2 associated with Alcat1 and deacetylase inhibition attenuated SCF-Fbxo28-mediated degradation of Alcat1 after exposure to LPS. Thus, acetylation and deacetylation might regulate protein stability by facilitating or impairing the E3 ubiquitin ligase recognition of this substrate, thereby modulating mitochondrial function in sepsis.
MATERIALS AND METHODS
Cell line and reagents
Murine lung epithelial (MLE) cells were cultured in HITES medium (500 ml of DMEM/F12, 2.5 mg of insulin, transferrin, sodium selenite, 2.5 mg of transferrin, 10 μM hydrocortisone, 10 μM β-estradiol, 10 mM Hepes, and 2 mM l-glutamine) and supplemented with 10% fetal bovine serum, as described previously (Zou et al., 2011b). MG132 (20 µM final concentration), leupeptin (20 µM final) and cycloheximide (10 µM) were purchased from Calbiochem. The antibody against Alcat1 (1:1000, catalog number MFG40984, lot number QC14266) was purchased from Aviva Systems Biology. Antibodies against HA (1:1000, catalog number 3724S, lot number 5), HDAC2 (1:1000, catalog number 5113P, lot number 1), laminin A/C (1:1000, catalog number 4777S, lot number 1), ubiquitin (1:1000, catalog number 3933S, lot number 4) and acetyl lysine (1:1000, catalog number 9441L, lot number 11) were purchased from Cell Signaling Technology. Lentiviral shRNA plasmids against Alcat1 (LCLAT1) were purchased from ThermoScientific. shRNA constructs against Fbxo28 and HDAC2 were purchased from Origene. Immobilized protein A/G beads were purchased from Pierce. In vitro TnT kits were purchased from Promega. Trypan Blue and a cell viability counter were purchased from Bio-Rad. Complete protease inhibitor cocktail was from Roche. LPS was purchased from Sigma. The antibody against Fbxo28 (1:1000, catalog number SC-134723, lot number C0120) was from Santa Cruz Biotechnology. The antibody against V5 (1:5000, catalog number 46-1157, lot number 1653719) and the pcDNA3.1D-V5-His plasmid were purchased from Invitrogen.
Immunostaining
Fluorescent immunostaining was conducted as previously described (Zou et al., 2011b). Briefly, cells (2×105) were plated at 70% confluence onto 35-mm MatTek glass-bottomed culture dishes. Immunofluorescent cell imaging was performed with a Nikon A1 confocal microscope. Cells were washed with PBS and fixed with 4% paraformaldehyde for 20 min, then exposed to 15% BSA, 1:500 dilutions of primary antibodies, and 1:1000 dilutions of Alexa-488- or Alexa-647-labeled goat anti-mouse or anti-rabbit secondary antibodies sequentially for immunostaining.
Construction of plasmids and cell transfection
The series truncation mutations of Alcat1 were constructed by performing PCR-based mutagenesis, with Alcat1 cDNA (Origene) as the template and the following primers: Alcat1 forward, 5′-ATGGTGTCATGGAAGGGGATTTAC-3′; Alcat1 N50 forward, 5′-ATGCTCACACTTCCTGTGGCATTG -3′; Alcat1 N100 forward, 5′-ATGTACAGCTACCTCAGGGTGGAG-3′; Alcat1 N150 forward, 5′-ATGTTTTGTGCCATCCATGAACCACTACAG-3′; Alcat1 N200 forward, 5′-ATGACCTTTGTGGTGGACCGCCTAAGAG-3′; Alcat1 C200 reverse, 5′-AAAGCCAGTGGTTCTTGGGTG-3′; Alcat1 C250 reverse, 5′-AGTCAGCTGGATACCGCTGGACGTG-3′; Alcat1 C300 reverse, 5′-AGACTTGCAAGGTGGAACTGTACTCTG-3′; Alcat1 C345 reverse, 5′-CTGCAGCACACGAAGAACAATGCTG-3′; and Alcat1 reverse, 5′-CTCATTTTTCTTTGAATTTAAATGTGG-3′. The resulting PCR products were purified and cloned into a pCDNA3.1D-V5-His plasmid. The accuracy of the cloned genes were verified by DNA sequencing. Quickchange II XL site-directed mutagenesis kits (Clontech) were used to introduce mutagenesis following the manufacturer's instructions. Primers used for mutagenesis were as follows: Alcat1K183R forward, 5′-GTAATGATTTTGCTGAGAGGAACGGACTTCAGAAATATG-3′; and Alcat1K183R reverse, 5′-CATATTTCTGAAGTCCGTTCCTCTCAGCAAAATCATTAC-3′. Plasmids were introduced into MLE cells by using nucleofection as previously described (Zou et al., 2013). Briefly, 1×106 MLE cells that had been suspended in 100 μl of nucleofection buffer (20 mM of Hepes in PBS buffer) were mixed with 3 μg of plasmid DNA (including the expression and shRNA plasmids) in an electroporation cuvette. Electroporation was conducted using the Nucleofection™ II system (Amaxa Biosystems).
Immunoprecipitation and immunoblotting
Immunoprecipitation and immunoblotting analyses were conducted as previously described (Chen et al., 2015b). Briefly, Alcat1 was immunoprecipitated from 1 mg of MLE cell lysate through incubation with the anti-Alcat1 antibody (2 μg) for 2 h; coupling to protein A/G beads was performed by incubation for an additional hour in 1 ml of immunoprecipitation buffer [150 mM NaCl, 50 mM Tris-HCl, 1 mM EDTA, 2 mM DTT, 0.5% Triton X-100 (v/v)] at room temperature. After washing, the precipitates were processed for immunoblotting of Alcat1. For immunoblotting analysis, cell lysates were prepared by briefly sonicating them in Buffer A (150 mM NaCl, 50 mM Tris, 1.0 mM EDTA, 2 mM dithiothreitol, 0.025% sodium azide and 1 mM phenylmethylsulfonyl fluoride. pH 7.4) at 4°C.
Pull-down assays
We performed in vitro pull-down assays to identify the Fbxo28-binding domain within Alcat1, as described previously (Chen et al., 2014b). The Alcat1 truncated mutant proteins that had been tagged with V5 were expressed in vitro using a TnT-coupled reticulocyte lysate system (Promega). Endogenous Fbxo28 protein was obtained by immunoprecipitating it from MLE cell lysate (1 mg protein) using an antibody against Fbxo28 and protein A/G agarose beads (ThermoScientific). Fbxo28-bound precipitated beads were incubated with various Alcat1 truncation mutants in 0.5 ml of binding buffer [150 mM NaCl, 50 mM Tris-HCl, 0.3% Tween20 (v/v) and 1:1000 protease inhibitor mixture (pH 7.4)] for 2 h at room temperature. The beads were subsequently washed with the binding buffer three times and analyzed by V5 immunoblotting.
Isolation of mitochondria and ER
Cells were harvested by trypsinization and re-suspended in 4 ml PBS. Cells were re-suspended in 500 µl of mitochondria isolation buffer, transferred to a Dounce homogenizer and ruptured with 90 strokes at 1600 rpm on ice. The homogenized cells were centrifuged at 700 g for 5 min at 4°C to pellet unbroken cells, nuclei and chromatin. The resulting supernatants were then centrifuged at 12,000 g for 15 min at 4°C. The pellets were collected as mitochondrial fractions and the supernatants were collected as ER fractions.
Analysis of cardiolipin
Lipids were extracted from MLE cells by using the Folch procedure (Folch et al., 1957). LC/MS analysis of cardiolipin molecular species was performed in negative mode using a Dionex Ultimate™ 3000 RSLCnano system coupled online with a Q-Exactive hybrid quadrupole-orbitrap mass spectrometer (ThermoScientific) (Tyurina et al., 2014). Total lipids were separated on a normal phase column [Silica Luna 3 µm, 100A, 150×2 mm, (Phenomenex, Torrance, CA)] with a flow rate of 0.2 ml/min using gradient solvents containing 5 mM CH3COONH4 [solvent A – n-hexane:2-propanol:water, 43:57:1 (v/v/v) and solvent B – n-hexane:2-propanol:water, 43:57:10 (v/v/v)]. The resolution was set up at 140,000, which corresponds to 5 ppm in m/z measurement error. Thus, m/z values for cardiolipins and their oxidation species were presented to four decimal places. Tetra-myristoyl-cardiolipin (Avanti polar lipids, Alabaster, AL) was used as an internal standard for mass spectrometry.
Oxygen consumption rate
The oxygen consumption rate (OCR) was measured in MLE cells using a XF24 Extracellular Flux Analyzer (Seahorse Bioscience). MLE cells (2×105 cells/well) were plated and transfected in a XF24 cell culture microplate (Seahorse Bioscience). Medium was changed to unbuffered DMEM 48 h after transfection, and the plate was incubated in a non-CO2 incubator at 37°C for 60 min before the assay was performed. The OCR measurements were taken every 2 min after a 2 min mixing period and a 3 min wait.
Isolation and culture of mouse lung type II cells
Lung alveolar type II cells were isolated as per the following protocol. Lungs of C57BL/6 male mice (8–12 weeks old) were perfused with cold PBS. Lungs were harvested after instillation of 2 ml of dispase (20 U/ml) into the lungs through a catheter. Lungs were then removed quickly, rinsed in cold PBS, placed in 3 ml PBS in Miltenyi GentleMACS C tubes containing 0.1 mg/ml collagenase-dispase and 0.1 mg/ml DNase I, and the tubes were incubated at 37°C for 45 min with continuous rotation to allow digestion. The digested tissue was carefully teased away from the airways, and then cells from the tissue were dissociated using a Miltenyi GentleMACS dissociator. Total cells were isolated after passing them through a 100-μm cell strainer and washing twice with 2% FBS–PBS followed by red blood cell lysis. The isolated cells were then stained with a biotin-labeled rat antibody against mouse CD45 (1.5 μg/million cells) and a rat antibody against mouse CD32 (0.65 μg/million cells), and incubated for 15 min at 4°C. Unbound antibodies were removed by centrifugation of the tubes at 159 g for 5 min at 4°C, and the supernatants were removed. The stained cells were then incubated with anti-biotin microbeads (20 μl per 10 million cells) for 10 min at 4°C in the dark and then centrifuged. The cell pellet was resuspended in Automacs running buffer, and the negative fraction containing epithelial cells was collected using an equilibrated LS column. The cells were then centrifuged and resuspended in BEBM medium supplemented with 5% FBS and 1% penicillin–streptomycin and gentamycin and plated onto a collagen-coated 24-well plate at a density of 0.2×106–0.5×106 cells/well. After 24 h, three-quarters of the medium was aspirated off gently, and the culture was replenished with fresh medium.
Animal studies
Ten-week-old male C57BL/6 mice from Jackson Laboratories were acclimated and maintained at the Animal Care Facility at University of Pittsburgh. Mice were anesthetized, and LPS at a dose of 5 mg/kg was intratracheally administered. After 24 h, the animals were euthanized, and the lung tissues were collected for immunoblotting analysis. All of the procedures followed all federal and institutional animal guidelines and were approved by the University of Pittsburgh Institutional Animal Care and Use Committee.
Statistical analysis
Statistical comparisons were performed using the Prism program version 4.03 (GraphPad Software, San Diego, CA) using an ANOVA or an unpaired two-sample t-test with P<0.05 indicative of significance.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Experiments were conducted by C.Z., M.J.S., J.L., S.X., M.M., Y.L., B.B.C., Y.Z., S.S., Y.Y.T., J.J., J.S.L, S.D., A.R. and P.R. The manuscript was written by C.Z., V.E.K. and R.K.M.
Funding
This material is based upon work supported, in part, by the US Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development, Biomedical Laboratory Research and Development. This work was supported by a Merit Review Award from the US Department of Veterans Affairs; National Institutes of Health [R01 grant numbers HL096376, HL097376, HL098174, HL081784, 1UH2HL123502 and P01HL114453 to R.K.M.; and ES020693, U19AIO68021, CA 165065, OH008282 (to V.E.K.)]; and Human Frontier Science Program [grant numbers HFSPRGP0013/2014 (to V.E.K.); HL116472 (to B.B.C.); HL01916 and HL112791 (to Y.Z.); and HL125435 and 12SDG12040330 (to C.Z.)]. S.X. was supported from the MOST of China [grant number 2012ZX09103-301-033 and 2012ZX09202-301-001]. The contents presented do not represent the views of the Department of Veterans Affairs or the United States Government. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.176701/-/DC1
References
- Aapola U., Liiv I. and Peterson P. (2002). Imprinting regulator DNMT3L is a transcriptional repressor associated with histone deacetylase activity. Nucleic Acids Res. 30, 3602-3608. 10.1093/nar/gkf474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acehan D., Khuchua Z., Houtkooper R. H., Malhotra A., Kaufman J., Vaz F. M., Ren M., Rockman H. A., Stokes D. L. and Schlame M. (2009). Distinct effects of tafazzin deletion in differentiated and undifferentiated mitochondria. Mitochondrion 9, 86-95. 10.1016/j.mito.2008.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acehan D., Vaz F., Houtkooper R. H., James J., Moore V., Tokunaga C., Kulik W., Wansapura J., Toth M. J., Strauss A. et al. (2011). Cardiac and skeletal muscle defects in a mouse model of human Barth syndrome. J. Biol. Chem. 286, 899-908. 10.1074/jbc.M110.171439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au P. Y., Argiropoulos B., Parboosingh J. S. and Micheil Innes A. (2014). Refinement of the critical region of 1q41q42 microdeletion syndrome identifies FBXO28 as a candidate causative gene for intellectual disability and seizures. Am. J. Med. Genet. A 164A, 441-448. 10.1002/ajmg.a.36320 [DOI] [PubMed] [Google Scholar]
- Baile M. G., Sathappa M., Lu Y.-W., Pryce E., Whited K., McCaffery J. M., Han X., Alder N. N. and Claypool S. M. (2014). Unremodeled and remodeled cardiolipin are functionally indistinguishable in yeast. J. Biol. Chem. 289, 1768-1778. 10.1074/jbc.M113.525733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissler J. J., Tsoras M., Göring H. H. H., Hug P., Chuck G., Tombragel E., McGraw C., Schlotman J., Ralston M. A. and Hug G. (2002). Infantile dilated X-linked cardiomyopathy, G4.5 mutations, altered lipids, and ultrastructural malformations of mitochondria in heart, liver, and skeletal muscle. Lab. Invest. 82, 335-344. 10.1038/labinvest.3780427 [DOI] [PubMed] [Google Scholar]
- Cai R. L., Yan-Neale Y., Cueto M. A., Xu H. and Cohen D. (2000). HDAC1, a histone deacetylase, forms a complex with Hus1 and Rad9, two G2/M checkpoint Rad proteins. J. Biol. Chem. 275, 27909-27916. [DOI] [PubMed] [Google Scholar]
- Cao J., Liu Y., Lockwood J., Burn P. and Shi Y. (2004). A novel cardiolipin-remodeling pathway revealed by a gene encoding an endoplasmic reticulum-associated acyl-CoA:lysocardiolipin acyltransferase (ALCAT1) in mouse. J. Biol. Chem. 279, 31727-31734. 10.1074/jbc.M402930200 [DOI] [PubMed] [Google Scholar]
- Cao J., Shen W., Chang Z. and Shi Y. (2009). ALCAT1 is a polyglycerophospholipid acyltransferase potently regulated by adenine nucleotide and thyroid status. Am. J. Physiol. Endocrinol. Metab. 296, E647-E653. 10.1152/ajpendo.90761.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo T. and Pagano M. (2004). The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 5, 739-751. 10.1038/nrm1471 [DOI] [PubMed] [Google Scholar]
- Cepeda D., Ng H.-F., Sharifi H. R., Mahmoudi S., Cerrato V. S., Fredlund E., Magnusson K., Nilsson H., Malyukova A., Rantala J. et al. (2013). CDK-mediated activation of the SCF(FBXO) (28) ubiquitin ligase promotes MYC-driven transcription and tumourigenesis and predicts poor survival in breast cancer. EMBO Mol. Med. 5, 1067-1086. 10.1002/emmm.201202341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B. B. and Mallampalli R. K. (2013). F-box protein substrate recognition: a new insight. Cell Cycle 12, 1009-1010. 10.4161/cc.23071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B. B., Coon T. A., Glasser J. R. and Mallampalli R. K. (2011). Calmodulin antagonizes a calcium-activated SCF ubiquitin E3 ligase subunit, FBXL2, to regulate surfactant homeostasis. Mol. Cell. Biol. 31, 1905-1920. 10.1128/MCB.00723-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B. B., Coon T. A., Glasser J. R., Zou C., Ellis B., Das T., McKelvey A. C., Rajbhandari S., Lear T., Kamga C. et al. (2014a). E3 ligase subunit Fbxo15 and PINK1 kinase regulate cardiolipin synthase 1 stability and mitochondrial function in pneumonia. Cell Rep. 7, 476-487. 10.1016/j.celrep.2014.02.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Li J., Dunn S., Xiong S., Chen W., Zhao Y., Chen B. B., Mallampalli R. K. and Zou C. (2014b). Histone deacetylase 2 (HDAC2) protein-dependent deacetylation of mortality factor 4-like 1 (MORF4L1) protein enhances its homodimerization. J. Biol. Chem. 289, 7092-7098. 10.1074/jbc.M113.527507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Wu D., Ding X. and Ying W. (2015a). SIRT2 is required for lipopolysaccharide-induced activation of BV2 microglia. Neuroreport 26, 88-93. 10.1097/WNR.0000000000000305 [DOI] [PubMed] [Google Scholar]
- Chen W., Xiong S., Li J., Li X., Liu Y., Zou C. and Mallampalli R. K. (2015b). The ubiquitin E3 ligase SCF-FBXO24 recognizes deacetylated nucleoside diphosphate kinase A to enhance its degradation. Mol. Cell. Biol. 35, 1001-1013. 10.1128/MCB.01185-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S.-Y., Gonzalvez F., Jenkins G. M., Slomianny C., Chretien D., Arnoult D., Petit P. X. and Frohman M. A. (2007). Cardiolipin deficiency releases cytochrome c from the inner mitochondrial membrane and accelerates stimuli-elicited apoptosis. Cell Death Differ. 14, 597-606. 10.1038/sj.cdd.4402020 [DOI] [PubMed] [Google Scholar]
- Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V. and Mann M. (2009). Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834-840. 10.1126/science.1175371 [DOI] [PubMed] [Google Scholar]
- Choudhary C., Weinert B. T., Nishida Y., Verdin E. and Mann M. (2014). The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol. 15, 536-550. 10.1038/nrm3841 [DOI] [PubMed] [Google Scholar]
- Chu C. T., Ji J., Dagda R. K., Jiang J. F., Tyurina Y. Y., Kapralov A. A., Tyurin V. A., Yanamala N., Shrivastava I. H., Mohammadyani D. et al. (2013). Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat. Cell Biol. 15, 1197-1205. 10.1038/ncb2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claypool S. M. and Koehler C. M. (2012). The complexity of cardiolipin in health and disease. Trends Biochem. Sci. 37, 32-41. 10.1016/j.tibs.2011.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claypool S. M., McCaffery J. M. and Koehler C. M. (2006). Mitochondrial mislocalization and altered assembly of a cluster of Barth syndrome mutant tafazzins. J. Cell Biol. 174, 379-390. 10.1083/jcb.200605043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claypool S. M., Boontheung P., McCaffery J. M., Loo J. A. and Koehler C. M. (2008). The cardiolipin transacylase, tafazzin, associates with two distinct respiratory components providing insight into Barth syndrome. Mol. Biol. Cell 19, 5143-5155. 10.1091/mbc.E08-09-0896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claypool S. M., Whited K., Srijumnong S., Han X. and Koehler C. M. (2011). Barth syndrome mutations that cause tafazzin complex lability. J. Cell Biol. 192, 447-462. 10.1083/jcb.201008177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creery D. and Fraser D. D. (2002). Tissue dysoxia in sepsis: getting to know the mitochondrion. Crit. Care Med. 30, 483-484. [DOI] [PubMed] [Google Scholar]
- Díaz-Laviada I., Ainaga J., Portolés M. T., Carrascosa J. L., Muncio A. M. and Pagani R. (1991). Binding studies and localization of Escherichia coli lipopolysaccharide in cultured hepatocytes by an immunocolloidal-gold technique. Histochem. J. 23, 221-228. 10.1007/BF01462244 [DOI] [PubMed] [Google Scholar]
- Drummond D. C., Noble C. O., Kirpotin D. B., Guo Z., Scott G. K. and Benz C. C. (2005). Clinical development of histone deacetylase inhibitors as anticancer agents. Annu. Rev. Pharmacol. Toxicol. 45, 495-528. 10.1146/annurev.pharmtox.45.120403.095825 [DOI] [PubMed] [Google Scholar]
- Dudek J., Cheng I.-F., Balleininger M., Vaz F. M., Streckfuss-Bömeke K., Hübscher D., Vukotic M., Wanders R. J. A., Rehling P. and Guan K. (2013). Cardiolipin deficiency affects respiratory chain function and organization in an induced pluripotent stem cell model of Barth syndrome. Stem Cell Res. 11, 806-819. 10.1016/j.scr.2013.05.005 [DOI] [PubMed] [Google Scholar]
- Folch J., Lees M. and Sloane Stanley G. H. (1957). A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226, 497-509. [PubMed] [Google Scholar]
- Force A. D. T., Ranieri V. M., Rubenfeld G. D., Thompson B. T., Ferguson N. D., Caldwell E., Fan E., Camporota L. and Slutsky A. S. (2012). Acute respiratory distress syndrome: the Berlin Definition. JAMA 307, 2526-2533. [DOI] [PubMed] [Google Scholar]
- Gonzalvez F., D'Aurelio M., Boutant M., Moustapha A., Puech J.-P., Landes T., Arnauné-Pelloquin L., Vial G., Taleux N., Slomianny C. et al. (2013). Barth syndrome: cellular compensation of mitochondrial dysfunction and apoptosis inhibition due to changes in cardiolipin remodeling linked to tafazzin (TAZ) gene mutation. Biochim. Biophys. Acta 1832, 1194-1206. 10.1016/j.bbadis.2013.03.005 [DOI] [PubMed] [Google Scholar]
- Grönroos E., Hellman U., Heldin C. H. and Ericsson J. (2002). Control of Smad7 stability by competition between acetylation and ubiquitination. Mol. Cell 10, 483-493. 10.1016/S1097-2765(02)00639-1 [DOI] [PubMed] [Google Scholar]
- Gu Z., Valianpour F., Chen S., Vaz F. M., Hakkaart G. A., Wanders R. J. and Greenberg M. L. (2004). Aberrant cardiolipin metabolism in the yeast taz1 mutant: a model for Barth syndrome. Mol. Microbiol. 51, 149-158. 10.1046/j.1365-2958.2003.03802.x [DOI] [PubMed] [Google Scholar]
- Henriksen P., Wagner S. A., Weinert B. T., Sharma S., Bacinskaja G., Rehman M., Juffer A. H., Walther T. C., Lisby M. and Choudhary C. (2012). Proteome-wide analysis of lysine acetylation suggests its broad regulatory scope in Saccharomyces cerevisiae. Mol. Cell. Proteomics 11, 1510-1522. 10.1074/mcp.M112.017251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper R. H., Rodenburg R. J., Thiels C., van Lenthe H., Stet F., Poll-The B. T., Stone J. E., Steward C. G., Wanders R. J., Smeitink J. et al. (2009a). Cardiolipin and monolysocardiolipin analysis in fibroblasts, lymphocytes, and tissues using high-performance liquid chromatography–mass spectrometry as a diagnostic test for Barth syndrome. Anal. Biochem. 387, 230-237. 10.1016/j.ab.2009.01.032 [DOI] [PubMed] [Google Scholar]
- Houtkooper R. H., Turkenburg M., Poll-The B. T., Karall D., Pérez-Cerdá C., Morrone A., Malvagia S., Wanders R. J., Kulik W. and Vaz F. M. (2009b). The enigmatic role of tafazzin in cardiolipin metabolism. Biochim. Biophys. Acta 1788, 2003-2014. 10.1016/j.bbamem.2009.07.009 [DOI] [PubMed] [Google Scholar]
- Huang L. S., Mathew B., Li H., Zhao Y., Ma S.-F., Noth I., Reddy S. P., Harijith A., Usatyuk P. V., Berdyshev E. V. et al. (2014). The mitochondrial cardiolipin remodeling enzyme lysocardiolipin acyltransferase is a novel target in pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 189, 1402-1415. 10.1164/rccm.201310-1917OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imae R., Inoue T., Nakasaki Y., Uchida Y., Ohba Y., Kono N., Nakanishi H., Sasaki T., Mitani S. and Arai H. (2012). LYCAT, a homologue of C. elegans acl-8, acl-9, and acl-10, determines the fatty acid composition of phosphatidylinositol in mice. J. Lipid Res. 53, 335-347. 10.1194/jlr.M018655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inuzuka H., Gao D., Finley L. W. S., Yang W., Wan L., Fukushima H., Chin Y. R., Zhai B., Shaik S., Lau A. W. et al. (2012). Acetylation-dependent regulation of skp2 function. Cell 150, 179-193. 10.1016/j.cell.2012.05.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M. N., Das S. R., Emin M. T., Wei M., Sun L., Westphalen K., Rowlands D. J., Quadri S. K., Bhattacharya S. and Bhattacharya J. (2012). Mitochondrial transfer from bone-marrow–derived stromal cells to pulmonary alveoli protects against acute lung injury. Nat. Med. 18, 759-765. 10.1038/nm.2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson P. K., Eldridge A. G., Freed E., Furstenthal L., Hsu J. Y., Kaiser B. K. and Reimann J. D. (2000). The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 10, 429-439. 10.1016/S0962-8924(00)01834-1 [DOI] [PubMed] [Google Scholar]
- Jeger V., Brandt S., Porta F., Jakob S. M., Takala J. and Djafarzadeh S. (2015). Dose response of endotoxin on hepatocyte and muscle mitochondrial respiration in vitro. Biomed. Res. Int. 2015, 353074 10.1155/2015/353074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan V. E., Bayir H. A., Belikova N. A., Kapralov O., Tyurina Y. Y., Tyurin V. A., Jiang J., Stoyanovsky D. A., Wipf P., Kochanek P. M. et al. (2009). Cytochrome c/cardiolipin relations in mitochondria: a kiss of death. Free Radic. Biol. Med. 46, 1439-1453. 10.1016/j.freeradbiomed.2009.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan V. E., Chu C. T., Tyurina Y. Y., Cheikhi A. and Bayir H. (2014). Cardiolipin asymmetry, oxidation and signaling. Chem. Phys. Lipids 179, 64-69. 10.1016/j.chemphyslip.2013.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan V. E., Tyurina Y. Y., Tyurin V. A., Mohammadyani D., Angeli J. P. F., Baranov S. V., Klein-Seetharaman J., Friedlander R. M., Mallampalli R. K., Conrad M. et al. (2015). Cardiolipin signaling mechanisms: collapse of asymmetry and oxidation. Antioxid. Redox Signal. 22, 1667-1680. 10.1089/ars.2014.6219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerner J., Yohannes E., Lee K., Virmani A., Koverech A., Cavazza C., Chance M. R. and Hoppel C. (2015). Acetyl-l-carnitine increases mitochondrial protein acetylation in the aged rat heart. Mech. Ageing Dev. 145, 39-50. 10.1016/j.mad.2015.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipreos E. T. and Pagano M. (2000). The F-box protein family. Genome Biol. 1, reviews3002-reviews3002.7. 10.1186/gb-2000-1-5-reviews3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkland R. A., Adibhatla R. M., Hatcher J. F. and Franklin J. L. (2002). Loss of cardiolipin and mitochondria during programmed neuronal death: evidence of a role for lipid peroxidation and autophagy. Neuroscience 115, 587-602. 10.1016/S0306-4522(02)00512-2 [DOI] [PubMed] [Google Scholar]
- Li J., Romestaing C., Han X., Li Y., Hao X., Wu Y., Sun C., Liu X., Jefferson L. S., Xiong J. et al. (2010). Cardiolipin remodeling by ALCAT1 links oxidative stress and mitochondrial dysfunction to obesity. Cell Metab. 12, 154-165. 10.1016/j.cmet.2010.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Liu X., Wang H., Zhang W., Chan D. C. and Shi Y. (2012). Lysocardiolipin acyltransferase 1 (ALCAT1) controls mitochondrial DNA fidelity and biogenesis through modulation of MFN2 expression. Proc. Natl. Acad. Sci. USA 109, 6975-6980. 10.1073/pnas.1120043109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Fossati G., Marchetti C., Modena D., Pozzi P., Reznikov L. L., Moras M. L., Azam T., Abbate A., Mascagni P. et al. (2015). Specific inhibition of histone deacetylase 8 reduces gene expression and production of proinflammatory cytokines in vitro and in vivo. J. Biol. Chem. 290, 2368-2378. 10.1074/jbc.M114.618454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Ye B., Miller S., Yuan H., Zhang H., Tian L., Nie J., Imae R., Arai H., Li Y. et al. (2012). Ablation of ALCAT1 mitigates hypertrophic cardiomyopathy through effects on oxidative stress and mitophagy. Mol. Cell. Biol. 32, 4493-4504. 10.1128/MCB.01092-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K. D., Wilson J. G., Zhuo H., Caballero L., McMillan M. L., Fang X., Cosgrove K., Calfee C. S., Lee J.-W., Kangelaris K. N. et al. (2014). Design and implementation of the START (STem cells for ARDS Treatment) trial, a phase 1/2 trial of human mesenchymal stem/stromal cells for the treatment of moderate-severe acute respiratory distress syndrome. Ann. Intensive Care 4, 1334 10.1186/s13613-014-0022-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiacono L. A. and Shapiro D. S. (2010). Detection of hypoxia at the cellular level. Crit. Care Clin. 26, 409-421. 10.1016/j.ccc.2009.12.001 [DOI] [PubMed] [Google Scholar]
- Lu Y.-W. and Claypool S. M. (2015). Disorders of phospholipid metabolism: an emerging class of mitochondrial disease due to defects in nuclear genes. Front. Genet. 6, 3 10.3389/fgene.2015.00003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens J.-C., Keilhoff G., Gardemann A. and Schild L. (2014). Oxidation of cardiolipin is involved in functional impairment and disintegration of liver mitochondria by hypoxia/reoxygenation in the presence of increased Ca(2)(+) concentrations. Mol. Cell Biochem. 394, 119-127. 10.1007/s11010-014-2087-2 [DOI] [PubMed] [Google Scholar]
- Nakahira K., Haspel J. A., Rathinam V. A. K., Lee S.-J., Dolinay T., Lam H. C., Englert J. A., Rabinovitch M., Cernadas M., Kim H. P. et al. (2011). Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 12, 222-230. 10.1038/ni.1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineau B., Bourge M., Marion J., Mauve C., Gilard F., Maneta-Peyret L., Moreau P., Satiat-Jeunemaitre B., Brown S. C., De Paepe R. et al. (2013). The importance of cardiolipin synthase for mitochondrial ultrastructure, respiratory function, plant development, and stress responses in Arabidopsis. Plant Cell 25, 4195-4208. 10.1105/tpc.113.118018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic D., Vucic D. and Dikic I. (2014). Ubiquitination in disease pathogenesis and treatment. Nat. Med. 20, 1242-1253. 10.1038/nm.3739 [DOI] [PubMed] [Google Scholar]
- Ray N. B., Durairaj L., Chen B. B., McVerry B. J., Ryan A. J., Donahoe M., Waltenbaugh A. K., O'Donnell C. P., Henderson F. C., Etscheidt C. A. et al. (2010). Dynamic regulation of cardiolipin by the lipid pump Atp8b1 determines the severity of lung injury in experimental pneumonia. Nat. Med. 16, 1120-1127. 10.1038/nm.2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren M., Phoon C. K. L. and Schlame M. (2014). Metabolism and function of mitochondrial cardiolipin. Prog. Lipid Res. 55, 1-16. 10.1016/j.plipres.2014.04.001 [DOI] [PubMed] [Google Scholar]
- Roth S. Y., Denu J. M. and Allis C. D. (2001). Histone acetyltransferases. Annu. Rev. Biochem. 70, 81-120. 10.1146/annurev.biochem.70.1.81 [DOI] [PubMed] [Google Scholar]
- Sadowski M. and Sarcevic B. (2010). Mechanisms of mono- and poly-ubiquitination: Ubiquitination specificity depends on compatibility between the E2 catalytic core and amino acid residues proximal to the lysine. Cell Div. 5, 19 10.1186/1747-1028-5-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlame M. (2013). Cardiolipin remodeling and the function of tafazzin. Biochim. Biophys. Acta. 1831, 582-588. 10.1016/j.bbalip.2012.11.007 [DOI] [PubMed] [Google Scholar]
- Schlame M., Kelley R. I., Feigenbaum A., Towbin J. A., Heerdt P. M., Schieble T., Wanders R. J. A., DiMauro S. and Blanck T. J. (2003). Phospholipid abnormalities in children with Barth syndrome. J. Am. Coll. Cardiol. 42, 1994-1999. 10.1016/j.jacc.2003.06.015 [DOI] [PubMed] [Google Scholar]
- Schlame M., Ren M., Xu Y., Greenberg M. L. and Haller I. (2005). Molecular symmetry in mitochondrial cardiolipins. Chem. Phys. Lipids 138, 38-49. 10.1016/j.chemphyslip.2005.08.002 [DOI] [PubMed] [Google Scholar]
- Schlattner U., Tokarska-Schlattner M., Ramirez S., Tyurina Y. Y., Amoscato A. A., Mohammadyani D., Huang Z., Jiang J., Yanamala N., Seffouh A. et al. (2013). Dual function of mitochondrial Nm23-H4 protein in phosphotransfer and intermembrane lipid transfer: a cardiolipin-dependent switch. J. Biol. Chem. 288, 111-121. 10.1074/jbc.M112.408633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z., Ye C., McCain K., Greenberg M. L. (2015). The Role of Cardiolipin in Cardiovascular Health. BioMed Res. Int. 2015, 891707 10.1155/2015/891707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorice M., Circella A., Cristea I. M., Garofalo T., Di Renzo L., Alessandri C., Valesini G. and Esposti M. D. (2004). Cardiolipin and its metabolites move from mitochondria to other cellular membranes during death receptor-mediated apoptosis. Cell Death Differ. 11, 1133-1145. 10.1038/sj.cdd.4401457 [DOI] [PubMed] [Google Scholar]
- Suryaraja R., Anitha M., Anbarasu K., Kumari G. and Mahalingam S. (2013). The E3 ubiquitin ligase Itch regulates tumor suppressor protein RASSF5/NORE1 stability in an acetylation-dependent manner. Cell Death Dis. 4, e565 10.1038/cddis.2013.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taunton J., Hassig C. A. and Schreiber S. L. (1996). A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272, 408-411. 10.1126/science.272.5260.408 [DOI] [PubMed] [Google Scholar]
- Taylor W. A. and Hatch G. M. (2003). Purification and characterization of monolysocardiolipin acyltransferase from pig liver mitochondria. J. Biol. Chem. 278, 12716-12721. 10.1074/jbc.M210329200 [DOI] [PubMed] [Google Scholar]
- Taylor W. A. and Hatch G. M. (2009). Identification of the human mitochondrial linoleoyl-coenzyme A monolysocardiolipin acyltransferase (MLCL AT-1). J. Biol. Chem. 284, 30360-30371. 10.1074/jbc.M109.048322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyurina Y. Y., Poloyac S. M., Tyurin V. A., Kapralov A. A., Jiang J., Anthonymuthu T. S., Kapralova V. I., Vikulina A. S., Jung M.-Y., Epperly M. W. et al. (2014). A mitochondrial pathway for biosynthesis of lipid mediators. Nat. Chem. 6, 542-552. 10.1038/nchem.1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valianpour F., Wanders R. J. A., Overmars H., Vreken P., van Gennip A. H., Baas F., Plecko B., Santer R., Becker K. and Barth P. G. (2002). Cardiolipin deficiency in X-linked cardioskeletal myopathy and neutropenia (Barth syndrome, MIM 302060): a study in cultured skin fibroblasts. J. Pediatr. 141, 729-733. 10.1067/mpd.2002.129174 [DOI] [PubMed] [Google Scholar]
- Valianpour F., Mitsakos V., Schlemmer D., Towbin J. A., Taylor J. M., Ekert P. G., Thorburn D. R., Munnich A., Wanders R. J., Barth P. G. et al. (2005). Monolysocardiolipins accumulate in Barth syndrome but do not lead to enhanced apoptosis. J. Lipid Res. 46, 1182-1195. 10.1194/jlr.M500056-JLR200 [DOI] [PubMed] [Google Scholar]
- Vreken P., Valianpour F., Nijtmans L. G., Grivell L. A., Plecko B., Wanders R. J. A. and Barth P. G. (2000). Defective remodeling of cardiolipin and phosphatidylglycerol in Barth syndrome. Biochem. Biophys. Res. Commun. 279, 378-382. 10.1006/bbrc.2000.3952 [DOI] [PubMed] [Google Scholar]
- Wang G., McCain M. L., Yang L., He A., Pasqualini F. S., Agarwal A., Yuan H., Jiang D., Zhang D., Zangi L. et al. (2014). Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat. Med. 20, 616-623. 10.1038/nm.3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Liu X., Nie J., Zhang J., Kimball S. R., Zhang H., Zhang W. J., Jefferson L. S., Cheng Z., Ji Q. et al. (2015). ALCAT1 controls mitochondrial etiology of fatty liver diseases, linking defective mitophagy to steatosis. Hepatology 61, 486-496. 10.1002/hep.27420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whited K., Baile M. G., Currier P. and Claypool S. M. (2013). Seven functional classes of Barth syndrome mutation. Hum. Mol. Genet. 22, 483-492. 10.1093/hmg/dds447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe A. P. (1996). Histone deacetylase--a regulator of transcription. Science 272, 371 10.1126/science.272.5260.371 [DOI] [PubMed] [Google Scholar]
- Xu Y., Sutachan J. J., Plesken H., Kelley R. I. and Schlame M. (2005). Characterization of lymphoblast mitochondria from patients with Barth syndrome. Lab. Invest. 85, 823-830. 10.1038/labinvest.3700274 [DOI] [PubMed] [Google Scholar]
- Xu Y., Condell M., Plesken H., Edelman-Novemsky I., Ma J., Ren M. and Schlame M. (2006a). A Drosophila model of Barth syndrome. Proc. Natl. Acad. Sci. USA 103, 11584-11588. 10.1073/pnas.0603242103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Malhotra A., Ren M. and Schlame M. (2006b). The enzymatic function of tafazzin. J. Biol. Chem. 281, 39217-39224. 10.1074/jbc.M606100200 [DOI] [PubMed] [Google Scholar]
- Xu S., Abbasian M., Patel P., Jensen-Pergakes K., Lombardo C. R., Cathers B. E., Xie W., Mercurio F., Pagano M., Giegel D. et al. (2007). Substrate recognition and ubiquitination of SCFSkp2/Cks1 ubiquitin-protein isopeptide ligase. J. Biol. Chem. 282, 15462-15470. 10.1074/jbc.M610758200 [DOI] [PubMed] [Google Scholar]
- Xu Y., Zhang S., Malhotra A., Edelman-Novemsky I., Ma J., Kruppa A., Cernicica C., Blais S., Neubert T. A., Ren M. et al. (2009). Characterization of tafazzin splice variants from humans and fruit flies. J. Biol. Chem. 284, 29230-29239. 10.1074/jbc.M109.016642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C., Shen Z. and Greenberg M. L. (2014). Cardiolipin remodeling: a regulatory hub for modulating cardiolipin metabolism and function. J. Bioenerg. Biomembr., 1-11. 10.1007/s10863-014-9591-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Zhou P., Liu B., Bambakidis T., Mazitschek R., Alam H. B. and Li Y. (2015). Protective effect of suberoylanilide hydroxamic acid against lipopolysaccharide-induced liver damage in rodents. J. Surg. Res. 194, 544-550. 10.1016/j.jss.2014.10.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Sims P. J. and Wiedmer T. (1998). Identity of a conserved motif in phospholipid scramblase that is required for Ca2+-accelerated transbilayer movement of membrane phospholipids. Biochemistry 37, 2356-2360. 10.1021/bi972625o [DOI] [PubMed] [Google Scholar]
- Zou C., Butler P. L., Coon T. A., Smith R. M., Hammen G., Zhao Y., Chen B. B. and Mallampalli R. K. (2011a). LPS impairs phospholipid synthesis by triggering beta-transducin repeat-containing protein (beta-TrCP)-mediated polyubiquitination and degradation of the surfactant enzyme acyl-CoA:lysophosphatidylcholine acyltransferase I (LPCAT1). J. Biol. Chem. 286, 2719-2727. 10.1074/jbc.M110.192377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou C., Ellis B. M., Smith R. M., Chen B. B., Zhao Y. and Mallampalli R. K. (2011b). Acyl-CoA:lysophosphatidylcholine acyltransferase I (Lpcat1) catalyzes histone protein O-palmitoylation to regulate mRNA synthesis. J. Biol. Chem. 286, 28019-28025. 10.1074/jbc.M111.253385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou C., Chen Y., Smith R. M., Snavely C., Li J., Coon T. A., Chen B. B., Zhao Y. and Mallampalli R. K. (2013). SCF(Fbxw15) mediates histone acetyltransferase binding to origin recognition complex (HBO1) ubiquitin-proteasomal degradation to regulate cell proliferation. J. Biol. Chem. 288, 6306-6316. 10.1074/jbc.M112.426882 [DOI] [PMC free article] [PubMed] [Google Scholar]