ABSTRACT
Appropriate diversification of cellular lineages from multi-potent progenitors is essential for normal development and homeostasis. The specification of erythroid and megakaryocytic lineages represents an especially vital developmental event whose molecular regulation remains incompletely defined. We now demonstrate the role of Rgs18, a GTPase-activating protein and transcriptional target of the repressor Gfi1b, in regulating these processes in mouse and human cells. Gfi1b stringently represses Rgs18 expression in erythroid cells, whereas, during megakaryocytic differentiation, declining Gfi1b levels facilitate a robust induction of Rgs18. Concordantly, alterations in Rgs18 expression produce disparate outcomes by augmenting megakaryocytic and potently suppressing erythroid differentiation and vice versa. These phenotypes reflect the differential impact of Rgs18 on signaling through p38 MAPK family proteins, and ERK1 and ERK2 (also known as MAPK3 and MAPK1, respectively) in the two lineages, which in turn alter the balance between the mutually antagonistic transcription factors Fli1 and Klf1. Overall, these results identify Rgs18 as a new and crucial effector of Gfi1b that regulates downstream signaling and gene expression programs to orchestrate erythro-megakaryocytic lineage choices. This dual role of Rgs18 in reciprocally regulating divergent lineages could exemplify generic mechanisms characteristic of multiple family members in different contexts.
KEY WORDS: Signaling, Transcription, Differentiation
Summary: This study demonstrates the reciprocal roles of the transcriptional repressor Gfi1b and its gene target and signaling factor Rgs18 in arbitrating the generation of the erythroid and megakaryocytic lineages.
INTRODUCTION
Growth factor independence 1b (Gfi1b) is a zinc-finger transcriptional repressor that is essential for the generation of the erythroid and megakaryocytic lineages (Foudi et al., 2014; Saleque et al., 2002), and is also required for lymphoid development (Doan et al., 2004; Schulz et al., 2012; van der Meer et al., 2010). Although Gfi1b is also moderately expressed in several other tissues, including non-hematopoietic ones (Tong et al., 1998; Vassen et al., 2007), the embryonic lethality resulting from the severe anemia following its germline deletion (Saleque et al., 2002) has, to date, precluded the analysis of its function in other developmental contexts. Dysregulation of Gfi1b and its paralog Gfi1 are also observed in various hematopoietic and non-hematopoietic malignancies (Elmaagacli et al., 2007; Khandanpour et al., 2012; Northcott et al., 2014; Vassen et al., 2009; Wallis et al., 2003).
Gfi1 and Gfi1b mediate transcriptional repression of cognate gene targets by binding to the DNA motif TAAATCAC(A/T)GCA (Grimes et al., 1996; Tong et al., 1998) and recruiting chromatin modifiers and co-repressors such as lysine-specific demethylase 1 (LSD1, also known as KDM1A), the REST corepressors 1 and 2 (Rcor1 and Rcor2), histone deacetylase 1 and 2 (HDAC1 and HDAC2) and histone methyl transferases, such as G9a (also known as EHMT2) and SUV39H1 (Duan et al., 2005; Saleque et al., 2007; Upadhyay et al., 2014; Vassen et al., 2006). The various histone modifications catalyzed by these factors – deacetylation by HDACs, H3K4 demethylation by LSD1 and H3K9 methylation by G9a and SUV39H1 – then lead to stable silencing of the Gfi1 and Gfi1b transcriptional targets in cells (Saleque et al., 2002; van der Meer et al., 2010). Of these co-factors, LSD1 is especially important for Gfi1b function, as shown by the fact that the majority of Gfi1b transcriptional targets (∼80%) are also common to LSD1 in erythroid cells (Saleque et al., 2007). This indicates that LSD1 likely acts as an obligate cofactor of Gfi1b at most targets.
Despite recognition of the essential mechanism of Gfi1b-mediated transcriptional repression, the repertoire of its major gene targets, and their contributions to the processes mediated by this factor and to hematopoietic or other developmental processes are largely unknown. In order to identify relevant gene targets that are major effectors of Gfi1b function, chromatin immunoprecipitation screens (ChIP-on-chip) were previously performed with antibodies against Gfi1b, LSD1 and Rcor1–CoREST in erythroid cells, leading to the identification of >600 putative common ChIP targets (Saleque et al., 2007). To further determine the regulation of these targets by Gfi1b, LSD1 or Rcor1, expression profiling was performed in control versus LSD1-inhibited erythroid cells, given that depletion of LSD1 had previously been shown to upregulate known Gfi1b gene targets (including itself) in erythroid cells (Saleque et al., 2007). This combination of ChIP and microarray profiling screens uncovered the relative hierarchy of Gfi1b–LSD1 targets according to their extent of deregulation upon LSD1 depletion. One such gene highly upregulated in LSD1-knockdown erythroid cells was the signaling molecule regulator of G protein signaling 18 (Rgs18).
Rgs18 is a member of a large family (>30 members) of GTPase-activating proteins (GAPs) that are known to accelerate the rate of GTP hydrolysis by α subunits of heterotrimeric G proteins, thereby controlling the duration and intensity of G-protein-coupled receptor (GPCR) activation and downstream signaling (Bansal et al., 2007; Kach et al., 2012). This signaling molecule is highly expressed in megakaryocytes, platelets and in hematopoietic progenitors of the myeloerythroid lineage (Gagnon et al., 2002; Louwette et al., 2012; Park et al., 2001; Yowe et al., 2001). Although Rgs18 is the most abundantly expressed Rgs protein in platelets, other family members, such as Rgs6, Rgs10 and Rgs16, are also known to be expressed in this lineage (Kim et al., 2006). Rgs18 has been previously shown to bind both Gαq and Gαi proteins in megakaryocytes (Nagata et al., 2001), and to act as a positive regulator of megakaryopoiesis (Delesque-Touchard et al., 2014; Louwette et al., 2012). In contrast, it serves as a negative regulator of platelet function, and limits platelet aggregation and activation (Alshbool et al., 2015; Brass and Ma, 2012; Gegenbauer et al., 2012).
Unlike its known functions in megakaryopoiesis and platelet activation, neither the expression nor the role of Rgs18 in erythroid cells (if any) has to date been documented. Therefore, following the identification of the Rgs18 promoter as a potential target of the Gfi1b–LSD1–Rcor1 transcriptional repressor triad in erythroid cells, we interrogated the expression and regulation of this factor by Gfi1b and LSD1 in both erythroid and megakaryocytic cells. Subsequently, we altered Rgs18 levels in these cells and determined the phenotypic consequences and molecular changes associated with these manipulations. Surprisingly, these experiments demonstrated that, in addition to enhancing megakaryopoiesis, Rgs18 expression potently suppressed erythroid differentiation and that these phenotypes resulted from differential regulation of mitogen-activated protein kinase (MAPK) signaling by Rgs18 in the two lineages. These signaling pathways, in turn, modulated relative expression of the mutually antagonistic cell-fate-specifying transcription factors, erythroid Kruppel-like factor (Eklf) or Kruppel-like factor 1 (Klf1), and friend leukemia integration [site] 1 (Fli1), thus determining phenotypic outcomes. Moreover, the reduced differentiation produced upon Rgs18 inhibition could be reversed by ectopic expression of Fli1 demonstrating its role downstream of Rgs18. Therefore, these results establish Rgs18 as a new and crucial transcriptional target of Gfi1b, which, together with its repressor, regulates the reciprocal regulation of erythro-megakaryocytic lineage specification.
RESULTS
Lineage-specific regulation of Rgs18 by Gfi1b and LSD1
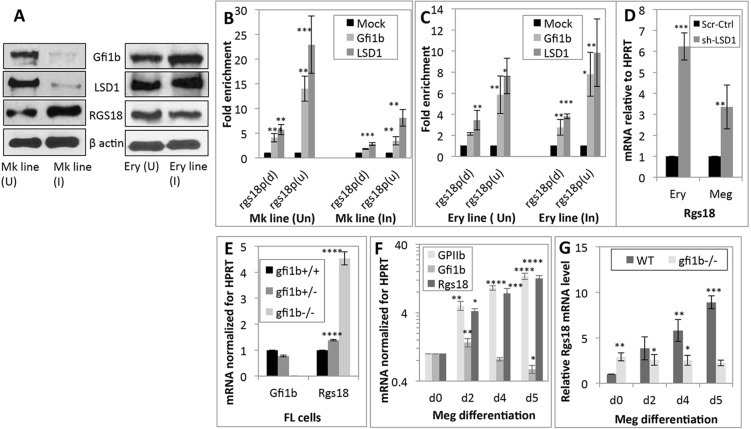
A combination of ChIP-on-chip and expression profiling screens (Saleque et al., 2007) has revealed that the Rgs18 promoter is a prominent target locus for Gfi1b–LSD1–Rcor1. Examination of Rgs18 promoter sequences obtained from the mouse promoter arrays of these ChIP-on-chip screens (Chowdhury et al., 2013; Saleque et al., 2007) revealed the presence of quasi-consensus (AAATCT) and consensus (AAATCA) Gfi1- and Gfi1b-binding sites (Tong et al., 1998) in the 5′ untranslated region (UTR) and proximal protein coding regions of Rgs18, respectively (Fig. S1). Given that Gfi1b and LSD1 generally repress their gene targets (Saleque et al., 2007; Upadhyay et al., 2014), we first monitored expression of Rgs18, Gfi1b, LSD1 and β-actin (as loading control) in immature and mature erythro-megakaryocytic cells. Curiously, Rgs18 was found to be low in the megakaryoblastic cell line L8057 (uninduced), in keeping with strong Gfi1b and LSD1 expression in these cells, but was strongly upregulated upon phorbol-ester-induced differentiation of these cells to megakaryocytes (Ishida et al., 1993) owing to a sharp decline in Gfi1b and LSD1 levels in the induced cells (Fig. 1A). Expression of Gfi1b was strong in uninduced proerythroblast-like murine erythroleukemia (MEL) cells and increased further in DMSO-induced MEL cells, which resemble orthochromatic erythroblasts (Friend et al., 1971) (Fig. 1A). By contrast, Rgs18 expression, which was modest in immature erythroid cells to begin with, exhibited a further reciprocal decline during maturation. As reported previously (Upadhyay et al., 2014), LSD1 levels remained approximately uniform during maturation of these cells.
Fig. 1.
Regulation of Rgs18 by Gfi1b and LSD1. (A) Western blot documenting Gfi1b, LSD1, Rgs18 and β-actin (as loading control) protein levels in uninduced (U) and induced (I) L8057 [megakaryocytic (Mk) line] and MEL [erythroid (Ery) line] cells. 60 µg protein was loaded in each lane. (B,C) Enrichment of Gfi1b and its cofactor LSD1 on upstream (u) and downstream (d) Rgs18 promoter segments (Rgs18p) in uninduced and induced erythroid and megakaryocytic cell lines relative to a mock (IgH switch Sµ) locus. The mean±s.d. from three independent ChIP experiments are shown. (D) Relative Rgs18 mRNA levels in control (Scr-Ctrl; scrambled) and LSD1 inhibited (shRNA against LSD1, sh-LSD1) erythroid (ery; MEL) and megakaryoblastic (meg; L8057) cells. (E) Gfi1b and Rgs18 mRNA levels in wild-type, Gfi1b+/− and Gfi1b−/− e12.5 total fetal liver (FL cells). (F) Time course (from day 0 to day 5) of GPIIb (CD41), Gfi1b and Rgs18 mRNA levels in wild-type e12.5 fetal liver cells differentiated along the megakaryocyte lineage relative to day 0 (d0), following normalization for HPRT. (G) Rgs18 mRNA levels in control (Gfi1b+/−) and mutant (Gfi1b−/−) fetal liver cells cultured along the megakaryocytic lineage for the indicated periods. ****P<0.0001, ***P<0.001, **P<0.01, *P<0.05.
To confirm association of Gfi1b and LSD1 with the above-mentioned Gfi1- and Gfi1b-binding sites, ChIP followed by quantitative real-time PCR (ChIP-qPCR) was performed in both uninduced and induced erythroid (MEL) and megakaryocytic (L8057) cells (Fig. 1B,C). The observed enrichment of Gfi1b and LSD1 on Rgs18 promoter segments corresponded entirely with their total protein levels in these cells. Gfi1b and LSD1 showed substantial enrichment on both Gfi1b-binding elements in uninduced L8057 cells but greatly reduced enrichment in the megakaryocyte-like induced L8057 cells. In contrast, marginally higher enrichment of these proteins was observed in mature (induced) versus immature (uninduced) erythroid (MEL) cells (Fig. 1B,C). Surprisingly, the upstream ‘semi-consensus’ Gfi1b-binding element consistently showed greater enrichment for Gfi1b and LSD1 relative to the downstream ‘consensus’ element defined by previous reports (Tong et al., 1998) in ChIP assays in both lineages, demonstrating the relatively greater affinity of the upstream site for these proteins.
The repression of the Rgs18 promoter following its recruitment of LSD1 was then demonstrated by upregulation of Rgs18 expression in immature erythroid and megakaryocytic cells upon LSD1 inhibition (Fig. S1B), although this occurred to differing extents in the two cell types (Fig. 1D). Overall, these experiments demonstrated inverse expression patterns of Rgs18 and Gfi1b–LSD1 in erythro-megakaryocytic cells resulting from differential repression of Rgs18 by Gfi1b and LSD1 in the two lines.
Given that auto-repression of the Gfi1b promoter by itself (Saleque et al., 2007; Vassen et al., 2005) prevents effective inhibition of this factor by short hairpin RNA (shRNA)-mediated mechanisms, we were unable to replicate these results for Gfi1b. Instead, to confirm analogous repression of Rgs18 by Gfi1b in primary cells, their mRNA levels were monitored in control (wild-type and heterozygous) versus Gfi1b mutant cells. First, we determined Gfi1b and Rgs18 expression in total embryonic day 12.5 (e12.5) fetal liver cells from all three genotypes, given that wild-type e12.5 fetal livers are predominantly erythroid in composition, with ∼80% of cells having cell-surface glycophorin A (i.e. Ter119 antigen) a marker of mature erythroid cells (Fig. S1C), in addition to having limited numbers of hematopoietic stem cells (HSCs), megakaryocyte-erythroid progenitors (MEPs) and megakaryocyte progenitors. Consistent with the indistinguishable phenotypes of wild-type versus Gfi1b+/− fetal livers, as reported previously (Saleque et al., 2002), we observed a marginal decrease in Gfi1b and a similarly modest (∼20%) increase in Rgs18 mRNA levels in the heterozygous cells relative to wild-type controls (Fig. 1E). In contrast, complete absence of Gfi1b in the homozygotes produced a substantial elevation of Rgs18 mRNA relative to wild-type cells in this primarily erythroid tissue (Fig. 1E).
Next, to investigate the expression of Gfi1b and Rgs18 in primary megakaryocytes, we cultured wild-type e12.5 fetal liver cells with megakaryocytic cytokines to preferentially expand this lineage, which is under-represented at this stage of development (Zhang et al., 2003). Differentiation of fetal liver progenitors to megakaryocytes was monitored by visual inspection and profiling for the megakaryocytic differentiation marker glycoprotein IIb (GPIIb, also known as CD41 or ITGA2B) (Fig. 1F). Differentiation of wild-type progenitors into megakaryocytes was accompanied by a gradual decline in Gfi1b, and a concomitant and reciprocal increase in Rgs18 expression (Fig. 1F), demonstrating progressively attenuated repression of the Rgs18 promoter upon decreasing Gfi1b expression. In contrast, although Rgs18 levels were higher in Gfi1b−/− fresh (day 0) total fetal liver cells relative to controls (which are largely erythroid in nature), it remained unchanged upon differentiation of mutant cells along the megakaryocyte lineage relative to that observed on day 0 (Fig. 1G). This lack of a net increase in Rgs18 message levels in Gfi1b−/− cells relative to the day 0 control confirmed minimal or no repression of Rgs18 by Gfi1b during megakaryocytic differentiation and therefore no noticeable rise in Rgs18 expression in the absence of Gfi1b, unlike the increase seen in wild-type cells. The results depicted in Fig. 1 collectively demonstrate dose-dependent and differential repression of Rgs18 by Gfi1b and its cofactor LSD1 in erythroid versus megakaryocytic cells resulting in their reciprocal expression in the two lineages.
Antagonistic functions of Rgs18 in erythroid and megakaryocytic development
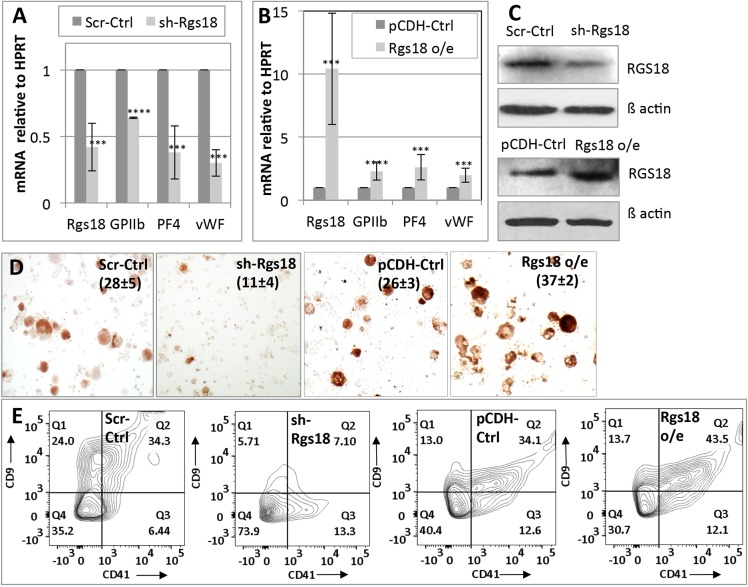
Given that Rgs18 is expressed in both erythroid and megakaryocytic cells, we next interrogated its role in their differentiation. Rgs18 expression was manipulated in fetal liver progenitors and cell lines by shRNA-mediated inhibition and cDNA transfection (i.e. protein overexpression). As per previous reports (Delesque-Touchard et al., 2014; Louwette et al., 2012), inhibition of Rgs18 similarly retarded megakaryocytic differentiation of fetal liver progenitors cultured in vitro (Fig. 2) and differentiation of the murine cell line L8057 induced with 12-O-tetradecanoylphorbol-13-acetate (TPA) (Fig. S2). This effect was demonstrated by a decrease in the expression of differentiation markers relative to a housekeeping gene (HPRT) (Fig. 2A), a reduction in the number of cells positive for acetylcholineesterase (a mouse megakaryocyte marker) (Fig. 2D) and a diminution in the number of cells displaying the mature megakaryocytic cell surface markers CD9 and CD41 (Fig. 2E). The reverse result, namely augmentation of differentiation, was obtained upon overexpression of Rgs18 in this lineage (Fig. 2B,D,E; Fig. S2). These results confirmed previous reports documenting the stimulatory role of Rgs18 in megakaryocytic differentiation (Delesque-Touchard et al., 2014; Louwette et al., 2012).
Fig. 2.
Augmentation of megakaryocytic differentiation by Rgs18. (A) qPCR analysis of Rgs18 mRNA levels and differentiation markers GPIIb (CD41), platelet factor 4 (PF4) and von Willebrand factor (vWF) upon shRNA-mediated Rgs18 inhibition (sh-Rgs18) relative to scrambled shRNA controls (Scr-Ctrl) in e12.5 fetal liver cells differentiated into megakaryocytes. (B) qPCR analysis of analogous markers upon Rgs18 cDNA or protein overexpression (Rgs18 o/e) relative to vector control (pCDH-Ctrl) in the same cells. ****P<0.0001, ***P<0.001. (C) Western blot of total Rgs18 protein levels following inhibition and overexpression as indicated, in primary (fetal-liver-derived) megakaryocytes relative to β-actin. 35 µg of protein was loaded in each lane. (D) Acetylcholineesterase staining of Rgs18-manipulated cells as indicated. The mean±s.d. of acetylcholine-positive cells as a percentage of the total population from three independent experiments is indicated in parentheses. (E) FACS analysis of CD9 and CD41 expression in fetal liver cells cultured into megakaryocytes following the indicated manipulations. One of three representative experiments is shown.
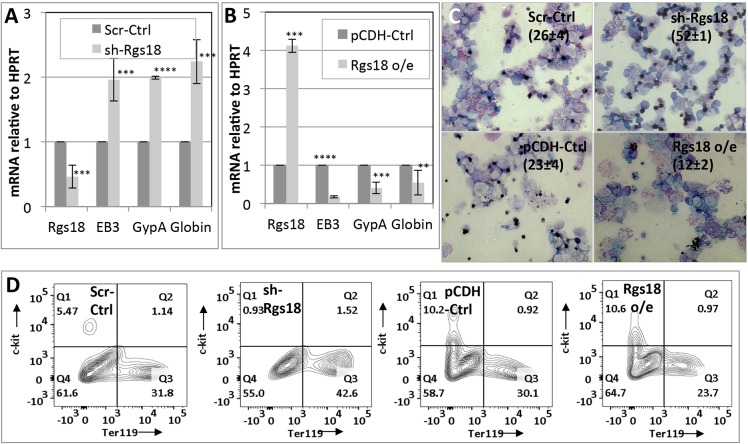
In sharp contrast to the above phenotype, manipulation of Rgs18 produced exactly the opposite outcome in both primary erythroid cells and the murine MEL cell line (Fig. 3; Fig. S2). Rgs18 inhibition produced a marked increase (Fig. 3A,C,D; Fig. S2F–I), and overexpression a commensurate decrease, in differentiation (Fig. 3B–D) as judged from differentiation marker analysis, benzidine staining (for heme positivity) and relative expression of the surface markers Ter119 versus c-Kit (a common marker of uncommitted progenitors). These results demonstrate a hitherto unknown function of Rgs18 in effectively inhibiting erythroid differentiation.
Fig. 3.
Suppression of erythroid differentiation by Rgs18. (A) qPCR analysis of Rgs18 mRNA levels and differentiation markers erythrocyte band 3 (EB3), glycophorin A (GypA, also known as Ter119) and globin (β major globin) upon shRNA-mediated Rgs18 inhibition (sh-Rgs18) relative to controls (Scr-Ctrl) in e12.5 fetal liver cells differentiated along the erythroid lineage. (B) qPCR analysis of the same erythroid markers and cells upon Rgs18 overexpression (Rgs18 o/e) relative to vector control (pCDH-Ctrl). ****P<0.0001, ***P<0.001, **P<0.01. (C) Benzidine staining of Rgs18-manipulated erythroid cells as indicated. The mean±s.d. of acetylcholine-positive cells as a percentage of the total population from three independent experiments is indicated in parentheses. (D) FACS analysis of the surface markers Ter119 and c-Kit in control and Rgs18-manipulated fetal liver cells cultured into erythroid cells as indicated. The mean±s.d. from three independent experiments are shown in A–C; results from one of three representative experiments is shown in D.
To confirm the validity of the Rgs18-knockdown phenotypes and rule out shRNA-mediated off-target effects, we overexpressed the protein-coding segment of Rgs18 cDNA in L8057 and MEL cells depleted for endogenous Rgs18 with shRNAs targeting the 3′UTR of the native transcript but that do not target the recombinant transcript. Following induction with TPA and DMSO, respectively, the cells expressing Rgs18 shRNAs and cDNAs were ‘rescued’ of their Rgs18 depletion phenotypes and reverted to the control state (Fig. S2E,I), confirming the fidelity of the Rgs18-knockdown phenotypes.
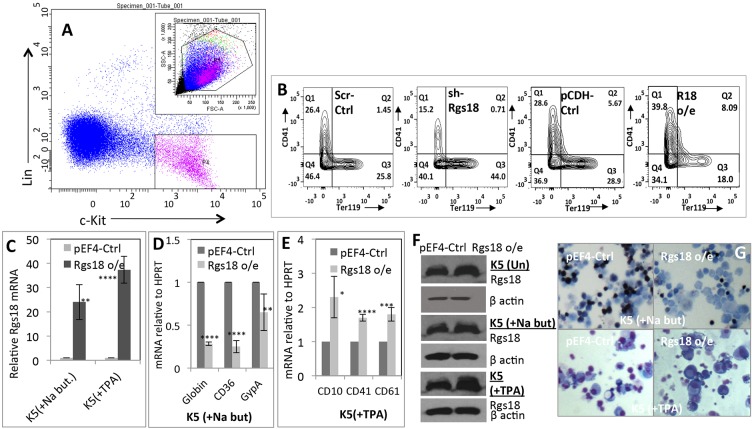
To reiterate the analogous effect of Rgs18 on fetal liver hematopoietic progenitors, lineage-negative (Lin−) c-Kit+ cells comprising HSCs, MEPs and other undifferentiated progenitors were sorted out from e12.5 fetal livers (Fig. 4A), transduced with Rgs18 shRNAs and cDNAs, and simultaneously co-cultured with cytokines promoting both lineages. These results further confirmed the stimulatory effect of Rgs18 expression on megakaryocytic, and suppressive effect on erythroid, differentiation in a progenitor population subjected to mixed culture conditions (Fig. 4B).
Fig. 4.
Reciprocal regulation of erythro-megakaryocytic lineage divergence by Rgs18. (A) Isolation of hematopoietic (lin− c-Kit+) progenitors from e12.5 fetal liver cells for transduction with Rgs18 cDNAs (Rgs18 o/e) and shRNAs (sh-Rgs18) in B. (B) Expression of erythroid and megakaryocytic surface markers Ter119 and CD41, respectively, in lin− c-Kit+ progenitors co-cultured with a mixture of erythroid and megakaryocytic cytokines following the indicated manipulations. sh-Rgs18, shRNA-mediated Rgs18 inhibition; Scr-Ctrl, scrambled shRNA control; Rgs18 o/e, Rgs18 overexpression; pCDH-Ctrl, vector control. (C–E) Expression of Rgs18 (C) and other vectors as indicated in the human hematopoietic cell line K562 transduced with Rgs18 cDNA and differentiated along the erythroid (+Na but) (D) or megakaryocytic (+TPA) (E) lineages. pEF4-Ctrl, vector control. ****P<0.0001, ***P<0.001, **P<0.01, *P<0.05. The mean±s.d. from three independent experiments are shown. (F) Total Rgs18 and β-actin protein levels in uninduced K562 cells (Un; top two panels) and upon differentiation into erythroid (+Na-but; middle two panels) and megakaryocytic cells (+TPA; bottom two panels), respectively. (G) Histochemical staining of K562 cells differentiated along the erythroid (top panels) or megakaryocytic (bottom panels) lineages following transduction with empty vector (pEF4-Ctrl) or Rgs18 cDNA (Rgs18 o/e). Erythroid cells were stained for benzidine (top panels); megakaryocytic cells were visualized by generic May–Grunwald Giemsa staining (bottom panels).
Finally, to delineate the effects of Rgs18 expression in human cells, Rgs18 cDNA was expressed in the human multipotent hematopoietic cell line K562 (Horton et al., 1983) and the cells were induced to differentiate along the erythroid and megakaryocytic lineages (Alitalo, 1990; Andersson et al., 1979). Expression of murine Rgs18 cDNA, which is ∼90% identical to its human counterpart (NP_570138) resulted in suppression of erythroid, and stimulation of megakaryocytic, differentiation as evidenced by differentiation marker mRNA analysis and histochemical staining (Fig. 4D–G).
The cumulative results of these multiple approaches unequivocally demonstrate the dual role of Rgs18 in actively promoting megakaryopoiesis and in restricting erythropoiesis in both mouse and human contexts. This, in conjunction with the reciprocal expression of Rgs18 and its repressor Gfi1b, suggests that the high level of expression of Gfi1b in MEPs and erythroid cells promotes this fate by repressing Rgs18 and keeping megakaryocytic differentiation in check. However, in cells adopting a megakaryocytic fate, declining Gfi1b levels upregulate Rgs18 expression and drive differentiation of these cells.
Differential regulation of MAPK signaling by Rgs18 in erythroid and megakaryocytic cells
To delineate the underlying mechanistic alterations responsible for the phenotypes ensuing from Rgs18 manipulation, we interrogated the status of two branches of MAPK signaling in these cells. The pathways mediated by the p38 MAPK family, and the ERK1 and ERK2 (ERK1/2, also known as MAPK3 and MAPK1, respectively) are known to be impacted by G protein signaling (Goldsmith and Dhanasekaran, 2007), and have also been implicated in erythro-megakaryocytic differentiation (Geest and Coffer, 2009). Specifically, ERK1/2 and p38 MAPK family proteins oppositely regulate these lineages by promoting proliferation of hematopoietic (megakaryocytic) cells and differentiation of erythroid cells, respectively (Geest and Coffer, 2009). Given that Rgs proteins attenuate G protein activation, they are known to negatively impact upon MAPK signaling (Berthebaud et al., 2005; Goldsmith and Dhanasekaran, 2007). Accordingly, Rgs18 levels and activity were expected to correlate inversely with MAPK signaling.
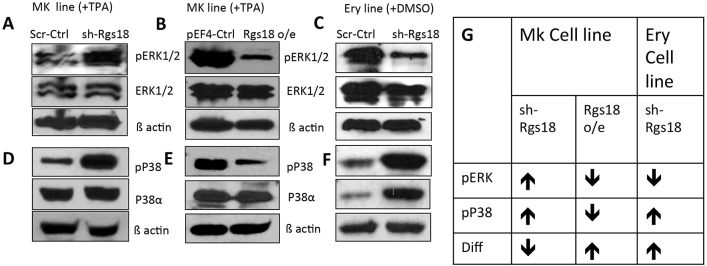
This was indeed found to be the case for ERK1/2 signaling in megakaryocytes (L8057 cells), where Rgs18 inhibition resulted in enhanced, and overexpression in diminished, levels of phosphorylated ERK1/2 (pERK1/2) (Fig. 5A,B), indicating an inverse correlation between Rgs18 expression and the activity of this MAPK pathway. Likewise, p38 MAPK phosphorylation was also sharply attenuated upon Rgs18 overexpression and enhanced upon inhibition (Fig. 5D,E). These observations indicated that both the p38 MAPK and ERK1/2 pathways are negatively impacted by Rgs18 activity in megakaryocytes. These results were confirmed by timecourse induction experiments in L8057 cells that clearly demonstrated attenuated decay of ERK1/2 and p38 MAPK phosphorylation during differentiation in Rgs18-inhibited cells (Fig. S3).
Fig. 5.
Differential regulation of MAPK pathways by Rgs18. (A,B) Effect of Rgs18 inhibition (A) and overexpression (o/e) (B) relative to controls on pERK1/2 levels in induced L8057 cells [megakaryocytic (MK) line +TPA]. Top panel, pERK1/2; middle panel, total ERK1/2; bottom panel, β-actin. (C) Effect of Rgs18 inhibition on the MAPK ERK1/2 pathway in induced MEL cells [erythroid (Ery) line +DMSO]. (D,E) Effect of Rgs18 inhibition (D) and overexpression (E) on p38 MAPK protein (p38α) and phosphorylation (pP38) levels in megakaryocytes. Top panel, phosphorylated p38 MAPK; middle panel, -38α; bottom panel, β-actin. (F) Effect of Rgs18 inhibition on the p38 MAPK pathway in erythroid cells. 50 µg of protein was loaded in panels A, D, E and F and 80 µg was loaded in B and C. One of three representative experiments is shown. (G) Table summarizing the effects of Rgs18 manipulation on the ERK1/2 and p38 pathways versus differentiation of erythro-megakaryocyte cells. Arrows indicate increase (↑) or decrease (↓) in activity and differentiation (Diff), respectively.
Although the effect of ERK1/2 signaling on the proliferation versus differentiation of megakaryoblasts is controversial, with different reports supporting its stimulation of one or the other process (Geest and Coffer, 2009; Whalen et al., 1997), our observation of the heightened differentiation of L8057 cells following downregulation of this pathway (by Rgs18) clearly demonstrates an inverse relationship between ERK1/2 signaling and the differentiation of these cells. By contrast, reduced p38 MAPK signaling upon Rgs18 overexpression likely contributes further to the attenuation of erythroid characteristics and gene expression in these cells (Fig. 5G).
In contrast, Rgs18 depletion in erythroid (MEL) cells, diminished pERK levels (Fig. 5C), whereas it augmented both p38 MAPK protein and phosphorylation levels (Fig. 5F) to co-operatively increase differentiation. Thus, the phosphorylation of p38 MAPK family proteins was similar in both lineages and correlated inversely with Rgs18 activity, whereas the pERK1/2 response was diametrically opposite in erythroid and megakaryocytic cells. Moreover, timecourse induction experiments showed that a decrease in Rgs18 levels disrupted the normal decay of these signaling cascades (Fig. S3), likely by ectopically sustaining G protein signaling. Overall, the alterations in MAPK signaling upon Rgs18 manipulation illustrate the molecular mechanisms responsible for the corresponding phenotypic alterations in differentiation in each lineage (Fig. 5G).
Rgs18 determines the equilibrium between Klf1 and Fli1
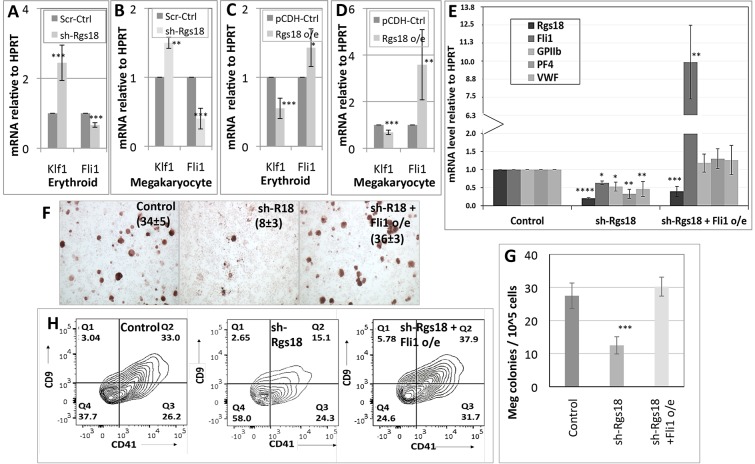
To investigate gene expression changes downstream of Rgs18, we interrogated expression of the mutually antagonistic transcription factors Eklf or Klf1, and Fli1 in Rgs18-manipulated cells. Klf1 and Fli1 are essential for the generation of the erythroid and megakaryocytic lineages, respectively (Drissen et al., 2005; Hart et al., 2000; Parkins et al., 1995), and they also actively suppress alternative lineage fates downstream of MEPs (Starck et al., 2003). Likewise, the relative abundance of these factors determines the lineage choices adopted by the progeny of these bipotent progenitors. However, how one factor gains dominance over the other and vice versa during lineage specification remains unclear. Therefore, to determine whether these factors act downstream of Rgs18, we assessed their levels in cells manipulated for the latter. In both lineages, Rgs18 levels correlated directly with Fli1, and inversely with Klf1, mRNA levels, indicating that Rgs18 arbitrates alternative fates by ultimately regulating relative Klf1 and Fli1 stoichiometries (Fig. 6A–D). Additionally, ectopic expression of Fli1 in fetal liver cells depleted of Rgs18 ‘rescued’ their megakaryocytic potential as evidenced by the recovery of differentiation marker expression (Fig. 6E), histochemical staining of the doubly manipulated cells relative to controls (Fig. 6F), recovery of megakaryocyte colony counts in methylcellulose cultures (Fig. 6G; Fig. S4A) and surface expression of the megakaryocytic markers CD9 and CD41 (Fig. 6H), and confirmed the role of Fli1 downstream of Rgs18 in promoting megakaryocyte differentiation. Overall, these results establish new and important functional connections between Gfi1b, Rgs18, MAPK cascades, and relative Klf1 and Fli1 levels in charting distinct cell fates downstream of bipotential progenitors (Fig. S4C).
Fig. 6.
Transcriptional effectors of Rgs18 and regulation of lineage divergence. (A–D) KLF1 and Fli1 mRNA levels (normalized for HPRT) in control versus Rgs18 inhibited (sh-Rgs18; A,B) or overexpressing (o/e, C,D), fetal-liver-derived erythroid (A,C) and megakaryocytic (B,D) cells. (E,F) Rescue of megakaryocytic differentiation in Rgs18-inhibited primary megakaryocytic cells (sh-Rgs18) upon Fli1 overexpression (sh-Rgs18+Fli1 o/e) as assessed by marker analysis (E) and acetylcholine esterase staining (F). The mean±s.d. of acetylcholine-positive cells as a percentage of the total population from three independent experiments is indicated in parentheses. (G) Megakaryocyte colony numbers obtained from fetal liver cells cultured for 5 days in methyl cellulose supplemented with IL-3 and thrombopoietin following Rgs18 inhibition (sh-Rgs18) and Rgs18 inhibition and Fli1 overexpression (shRgs18; Fli1 o/e) relative to scrambled controls (control). (H) Megakaryocyte marker CD9 and CD41 expression in fetal liver cells cultured for 5 days along the megakaryocytic lineage following the indicated manipulations. ****P<0.0001, ***P<0.001, **P<0.01, *P<0.05.
DISCUSSION
Rgs factors and their regulators in cellular differentiation and divergence
In this report, we have identified Rgs18 as a prominent target of Gfi1b and its co-repressor LSD1 in erythro-megakaryocytic cells. Rgs18 is stringently repressed by Gfi1b and LSD1 in erythroid cells but robustly expressed in megakaryocytes due to reduced expression of these repressors. These expression patterns then support stimulation of megakaryocytic, and suppression of erythroid, differentiation by Rgs18. These results thus introduce Rgs18 as a new arbitrator of erythro-megakaryocytic differentiation and provide a novel perspective on the regulation of their differentiation by Rgs18 and its transcriptional repressor Gfi1b. These results might appear paradoxical in light of the phenotypes of germline and conditional Gfi1b deletions, which lead to the complete abrogation of both lineages in embryos and adult mice, respectively (Saleque et al., 2002; Foudi et al., 2014), and reports of platelet disorders in patients harboring GFI1B mutations (Aminkeng, 2014; Monteferrario et al., 2014). However, although these reports confirm the role of Gfi1b in erythro-megakaryocytic commitment and specification, what our current observations highlight is the differential expression of, and requirement for, Gfi1b subsequent to the initial emergence of these lineages from common progenitors.
Given that Gfi1b performs similar functions in fetal liver and bone marrow hematopoiesis (Foudi et al., 2014; Saleque et al., 2002), this paradigm might also hold true for the differentiation of bone marrow progenitors. Whether Rgs18 arbitrates similar lineage divergence at other cellular branch points and/or is prototypic of the function of other Rgs proteins widely expressed in multiple tissues, hematopoietic and otherwise, are fascinating and germane questions that need to be investigated in the light of the above observations.
Robust and sustained expression of Gfi1b through most of erythroid differentiation (Vassen et al., 2007) is likely required for maintaining erythroid identity and differentiation, in part by suppressing megakaryocytic gene expression, including that of Rgs18, until the erythroblast stage. In contrast, the observed decline in Gfi1b levels subsequent to this stage (Vassen et al., 2007) might be equally important for terminal maturation once the cells have progressed irreversibly along the erythroid program. By contrast, downregulation of Gfi1b is necessary earlier in megakaryopoiesis, and soon after commitment to megakaryoblasts, to enable derepression of genes like Rgs18 and perhaps others that promote differentiation of this lineage.
An earlier report has shown that Rgs18 is repressed in erythroid cells by GATA1 (Johnson et al., 2007), although it was not determined whether this repression was maintained in megakaryocytes given that GATA1 expression is also known to be downregulated during differentiation of these cells (Dai and Murphy, 1993). If this GATA1 repression of Rgs18 is indeed operational in erythro-megakaryocytic cells, then the Rgs18 promoter could potentially be co-operatively repressed by Gfi1b and GATA1 in erythroid cells and potently derepressed by the downregulation of both factors in megakaryocytes.
In addition to Rgs18, several other Rgs factors are derepressed to varying degrees upon loss of Gfi1b (Fig. S4B); of these, Rgs14 and Rgs2 are also chromatin targets of Gfi1b and its cofactors (LSD1 and Rcor1) in erythroid cells (S.S., unpublished data), whereas the derepression of Rgs16 and Rgs10 in the absence of Gfi1b might be indirect. Therefore, several Rgs factors might cooperate with Rgs18 in mediating multi-factorial stimulation of megakaryocytic over erythroid differentiation, whereas their direct or indirect repression by Gfi1b would promote the latter lineage and maintain homeostasis.
Although Rgs18 is also prominently expressed in platelets (Kim et al., 2006), it potently inhibits platelet functions, like activation and aggregation, in these megakaryocyte derivatives by inhibiting G protein signaling (Alshbool et al., 2015; Gegenbauer et al., 2012). Therefore, this GAP factor performs pleiotropic and even opposite functions in megakaryopoiesis versus thrombopoiesis.
Lineage-specific regulation of MAPK signaling by Rgs18
Several studies have demonstrated the role of p38 MAPK family proteins and ERK1/2 signaling in the regulation of hematopoietic stem and progenitor cell expansion and differentiation (Geest and Coffer, 2009; Whalen et al., 1997). However, their relationship with upstream or downstream transcription factors, and the processes regulated by them, continue to remain nebulous. The observations reported here now establish coherent connections between the erythro-megakaryocytic transcription factor Gfi1b and MAPK pathways through regulation of G protein signaling mechanisms. They further extend the signaling chain to reveal regulation of downstream factors Klf1 and Fli1 by these pathways. Previously, manipulation of Rgs16, but not Rgs18, has been found to impact on MAPK signaling in megakaryocytes (Berthebaud et al., 2005), and our results now clearly illustrate the effect of Rgs18 manipulation on p38 MAPK and ERK1/2 signaling in both erythroid and megakaryocytic cells. This apparent discrepancy between the earlier results and our current results might be a consequence of the different cell types (MO7e versus L8057 and MEL) or the assays employed in each study. Our results also provide unprecedented insights into the regulation of signal transduction, particularly MAPK pathways, by Gfi1b through its transcriptional target Rgs18. Although the mechanism(s) responsible for the differential, or even opposite effects, exerted by Rgs18 on ERK1/2 signaling in erythroid versus megakaryocytic cells remains unknown, it is likely to be a major determinant in specifying distinct outcomes in the two lineages.
Relevance of Rgs18 function to hematopoietic diseases and their control
Various lines of evidence demonstrate that Rgs18 stimulates megakaryocytic differentiation (Delesque-Touchard et al., 2014; Louwette et al., 2012), and limits platelet aggregation and activation by turning off Gq signaling (Alshbool et al., 2015; Brass and Ma, 2012; Gegenbauer et al., 2012). We now demonstrate the key role played by this factor in ensuring erythro-megakaryocytic homeostasis by actively suppressing the former and promoting the latter. These insights could be utilized to rectify imbalances between them that lead to hematopoietic abnormalities. Accordingly, a deficit in erythropoiesis could conceivably be compensated for by inhibiting Rgs18 (or other Rgs proteins that work similarly and co-operatively in these cells) and/or by indirectly stimulating G protein signaling. Conversely, thrombocytopenias resulting from ectopic and excessive G protein signaling could be offset by stimulating Rgs18 expression or activity thereby rolling back G protein signaling.
In conclusion, our study presents Rgs18 as a dual regulator of erythro-megakaryocytic differentiation, downstream of the transcription factor Gfi1b. These effects are mediated by differential MAPK signaling and alterations in the Klf1 to Fli1 ratio. As mentioned above, these molecular insights on the generation of erythroid and megakaryocytic cells from MEPs could also provide rational platforms for developing strategies for stimulating one or the other lineage when depleted by diseases or environmental assaults.
MATERIALS AND METHODS
ChIP
ChIP experiments were performed in MEL and L8057 cells as previously described (Chowdhury et al., 2013; Saleque et al., 2007) with anti-Gfi1b (Sc8559, Santa Cruz Biotechnology) and anti-LSD1 (ab17721, Abcam) antibodies. Primers used for qPCR amplification of ChIP DNA were: Rgs18 promoter (upstream), 5′-TCATTTCCTTCAACAATTCAGTACA-3′ and 5′-CGAATCTTTCCTCAGATTTTTCTTA-3′; Rgs18 promoter (downstream), 5′-ATGTGTGAATCAAAAGAGAAAACTTT-3′ and 5′-CACAGATATTCATCAATCATGCTACTT-3′; and Sµ, 5′-CTTGAGCCAAAATGAAGTAGACTGT-3′ and 5′-ACAGTCCAGTGTAGGCAGTAGAGTT-3′.
Plasmid construction and expression
Murine Rgs18 cDNA (accession number NM_022881) was PCR amplified with the primers: 5′-ATGTCACTGGTTTTCTTCTCTCAATT-3′ and 5′-TAACCAAATGGCAACATCTGACTTTACAT-3′ from total RNA from L8057 cells and sub-cloned into pEF4/myc-His vector (Invitrogen) and pCDH-MSCV™ vector (Systems Biosciences). Fli1 (accession number NM_008026) cDNA was also PCR amplified from L8057 RNA with the primers 5′-ATGGACGGGACTATTAAGGAGGCT-3′ and 5′-GTATGGGTAGTAGCTGCCTAAGTGTGAAGG-3′ and subcloned into pCDH. Inserted cDNAs were verified by sequencing and protein expression was confirmed in 293T cells. Commercially available RGS18 shRNAs were purchased from the Mission™ collection (Sigma-Aldrich) and were as follows: RGS18 shRNA1 (coding), 5′-CCGGCTCCTGAAGAAGCAGTGAAATCTCGAGATTTCACTGCTTCTTCAGGAGTTTTTG-3′; RGS18 shRNA2 (3′UTR), 5′-CCGGAGTAATGTCACATCTAGTTTGCTCGAGCAAACTAGATGTGACATTACTTTTTTG-3′; and RGS18 shRNA3 (3′UTR), 5′-CCGGCATCATCTATCTTCCGAAATACTCGAGTATTTCGGAAGATAGATGATGTTTTTTG-3′.
Cell culture and cell line production
Stable overexpression lines were generated by nucleofection (Amaxa) of Rgs18 pEF4/myc-His vector into L8057 (megakaryoblastic cell line) (Ishida et al., 1993) and MEL (murine erythroleukemia) (Friend et al., 1971) followed by zeocin (0.5 mg/ml) selection. Stable Rgs18-shRNA-expressing (knockdown) cell lines were created as previously described (Upadhyay et al., 2014). MEL cells were induced to differentiate with 1.5% dimethylsulfoxide (DMSO) for 4 days and L8057 cells were induced with 50 nM 12-O-tetradecanoyl phorbol-13-acetate (TPA) for 5 days. K562 cells (ATCC CCL243) (Horton et al., 1983) were cultured in Iscove's modified Dulbecco's medium (IMDM) supplemented with 10% fetal bovine serum and antibiotics and induced to differentiate into erythroid and megakaryocytic cells by treatment with 2 mM sodium butyrate and 50 nM TPA, respectively (Alitalo, 1990; Andersson et al., 1979). Uninduced and induced cells were harvested as needed for protein and RNA collection or histological staining.
Culture and manipulation of fetal liver cells
Total or sorted fetal liver cells (∼105) from embryonic day 12.5 (e12.5) embryos were harvested and cultured directly or transduced with lentiviruses carrying Rgs18 cDNA, Rgs18 shRNA and/or Fli1 cDNA. Cells were differentiated either in liquid culture or in a semi-solid methylcellulose medium (M3234; Stem Cell Technologies) along the erythroid or megakaryocytic lineages by culturing in erythropoietin (2 U/ml) and stem cell factor (SCF, 25 ng/ml), or thrombopoietin (20 ng/ml), IL-3 (10 ng/ml) and SCF (10 ng/ml), respectively; and selected with puromycin (0.5–1 µg/ml). Cells were harvested from liquid culture for various assays and megakaryocyte colonies counted in methylcellulose cultures.
Animal husbandry and manipulation
All mice were maintained, manipulated and euthanized in the CCNY vivarium according to the approved CCNY IACUC (Institutional Animal Care and Use Committee) protocol. 6–8-week-old wild-type or mutant mice were subject to timed matings to obtain staged embryos for tissue (blood, yolk sac and fetal liver) collection as discussed above.
Preparation of cell lysates and western blotting
Harvested cells were lysed in whole-cell lysis buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA and 1% Triton X-100) supplemented with 1 mM PMSF, protease inhibitors (Sigma) and phosphatase inhibitors (eBioscience) as needed. Lysates were resolved on SDS-PAGE, and western blotted with specific antibodies. Antibodies used were anti-Gfi1b (Sc8559, Santa Cruz Biotechnology), anti-LSD1 (ab17721, Abcam) and anti-Rgs18 (ab25917, Abcam) antibodies, and anti-ERK1/2 (4695), anti-pERK1/2 (4370S), anti-p38 MAPK (2371) and anti-phosphorylated-p38 MAPK (4511S) antibodies from Cell Signaling Technologies.
qPCR, histological assays and flow cytometry
Total RNA was isolated and quantified by qPCR in an ABI 7500 machine (Applied Biosciences). Marker expressions were normalized to that for hypoxanthine phosphoribosyl transferase (HPRT). The data for all qPCR reactions represents the mean±s.d. from three independent experiments. P-values were calculated by one-way analysis of variance (ANOVA) (for comparison of three or more datasets) or by multiple t-test (for two datasets) followed by Holm-Sidak post-hoc test as applicable, for comparisons. P<0.05 was considered statistically significant and has, therefore, not been indicated in any Figs. Murine and human qPCR primer sequences have either been reported previously (Saleque et al., 2007) or are listed in Table S1.
For histochemical analysis, 105 cells were cyto-centrifuged and stained for benzidine, acetylcholine esterase and with May Grunwald Giemsa as previously described (Saleque et al., 2002). The number of positively staining cells relative to total for a fixed area were determined using the ImageJ™ cell imaging and counting software (Schneider et al., 2012). For flow cytometric analyses of surface markers, ≥105 cells were stained with FITC-conjugated anti-mouse CD9 or CD71 antibodies and APC-conjugated anti-mouse CD41 or Ter119 antibodies (eBioscience), respectively, and analyzed on the BD LSRII Analyzer (Becton Dickinson). For FACS sorting, cells were labeled with lin–FITC and c-Kit–phycoerythrin and the lin− c-Kit+ population collected following elimination of dead cells, doublets and aggregates (gates P1–P3) on a BD FACS Aria sorter (Becton Dickinson).
Acknowledgements
The authors thank Dr Jayanta Chaudhuri and his laboratory for providing crucial reagents for the project and Dr Malcolm Moore for the gift of K562 cells. We thank Mr Jeffrey Walker for help with flow cytometry analysis and sorting.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
The project and experiments were conceived and designed by S.S. The majority of experiments were performed by A.S. G.U. performed ChIP, primary cell manipulations and contributed to data analysis and quantification. S.S. contributed to the construction of expression vectors. The manuscript was written by A.S. and S.S.
Funding
This work was supported by National Institute on Minority Health and Health Disparities Research Centers in Minority Institutions [grant numbers G12RR03060-26A1 and 8G12MD007603-27 to The City University of New York]; and by the National Heart, Lung, and Blood Institute grant [grant number 5SC1HL104638 to S.S.]. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.177519/-/DC1
References
- Alitalo R. (1990). Induced differentiation of K562 leukemia cells: a model for studies of gene expression in early megakaryoblasts. Leuk. Res. 14, 501-514. 10.1016/0145-2126(90)90002-Q [DOI] [PubMed] [Google Scholar]
- Alshbool F. Z., Karim Z. A., Vemana H. P., Conlon C., Lin O. A. and Khasawneh F. T. (2015). The regulator of G-protein signaling 18 regulates platelet aggregation, hemostasis and thrombosis. Biochem. Biophys. Res. Commun. 462, 378-382. 10.1016/j.bbrc.2015.04.143 [DOI] [PubMed] [Google Scholar]
- Aminkeng F. (2014). GFI1B mutation causes autosomal dominant gray platelet syndrome. Clin. Genet. 85, 534-535. 10.1111/cge.12380 [DOI] [PubMed] [Google Scholar]
- Andersson L. C., Jokinen M. and Gahmberg C. G. (1979). Induction of erythroid differentiation in the human leukaemia cell line K562. Nature 278, 364-365. 10.1038/278364a0 [DOI] [PubMed] [Google Scholar]
- Bansal G., Druey K. M. and Xie Z. (2007). R4 RGS proteins: regulation of G-protein signaling and beyond. Pharmacol. Ther. 116, 473-495. 10.1016/j.pharmthera.2007.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthebaud M., Rivière C., Jarrier P., Foudi A., Zhang Y., Compagno D., Galy A., Vainchenker W. and Louache F. (2005). RGS16 is a negative regulator of SDF-1-CXCR4 signaling in megakaryocytes. Blood 106, 2962-2968. 10.1182/blood-2005-02-0526 [DOI] [PubMed] [Google Scholar]
- Brass L. F. and Ma P. (2012). Applying the brakes to platelet activation. Blood 119, 3651-3652. 10.1182/blood-2012-02-406629 [DOI] [PubMed] [Google Scholar]
- Chowdhury A. H., Ramroop J. R., Upadhyay G., Sengupta A., Andrzejczyk A. and Saleque S. (2013). Differential transcriptional regulation of meis1 by Gfi1b and its co-factors LSD1 and CoREST. PLoS ONE 8, e53666 10.1371/journal.pone.0053666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W. and Murphy M. J. Jr. (1993). Downregulation of GATA-1 expression during phorbol myristate acetate-induced megakaryocytic differentiation of human erythroleukemia cells. Blood 81, 1214-1221. [PubMed] [Google Scholar]
- Delesque-Touchard N., Pendaries C., Volle-Challier C., Millet L., Salel V., Hervé C., Pflieger A.-M., Berthou-Soulie L., Prades C., Sorg T. et al. (2014). Regulator of G-protein signaling 18 controls both platelet generation and function. PLoS ONE 9, e113215 10.1371/journal.pone.0113215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan L. L., Porter S. D., Duan Z., Flubacher M. M., Montoya D., Tsichlis P. N., Horwitz M., Gilks C. B. and Grimes H. L. (2004). Targeted transcriptional repression of Gfi1 by GFI1 and GFI1B in lymphoid cells. Nucleic Acids Res. 32, 2508-2519. 10.1093/nar/gkh570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drissen R., von Lindern M., Kolbus A., Driegen S., Steinlein P., Beug H., Grosveld F. and Philipsen S. (2005). The erythroid phenotype of EKLF-null mice: defects in hemoglobin metabolism and membrane stability. Mol. Cell. Biol. 25, 5205-5214. 10.1128/MCB.25.12.5205-5214.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z., Zarebski A., Montoya-Durango D., Grimes H. L. and Horwitz M. (2005). Gfi1 coordinates epigenetic repression of p21Cip/WAF1 by recruitment of histone lysine methyltransferase G9a and histone deacetylase 1. Mol. Cell. Biol. 25, 10338-10351. 10.1128/MCB.25.23.10338-10351.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmaagacli A. H., Koldehoff M., Zakrzewski J. L., Steckel N. K., Ottinger H. and Beelen D. W. (2007). Growth factor-independent 1B gene (GFI1B) is overexpressed in erythropoietic and megakaryocytic malignancies and increases their proliferation rate. Br. J. Haematol. 136, 212-219. 10.1111/j.1365-2141.2006.06407.x [DOI] [PubMed] [Google Scholar]
- Foudi A., Kramer D. J., Qin J., Ye D., Behlich A.-S., Mordecai S., Preffer F. I., Amzallag A., Ramaswamy S., Hochedlinger K. et al. (2014). Distinct, strict requirements for Gfi-1b in adult bone marrow red cell and platelet generation. J. Exp. Med. 211, 909-927. 10.1084/jem.20131065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friend C., Scher W., Holland J. G. and Sato T. (1971). Hemoglobin synthesis in murine virus-induced leukemic cells in vitro: stimulation of erythroid differentiation by dimethyl sulfoxide. Proc. Natl. Acad. Sci. USA 68, 378-382. 10.1073/pnas.68.2.378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon A. W., Murray D. L. and Leadley R. J. (2002). Cloning and characterization of a novel regulator of G protein signalling in human platelets. Cell Signal. 14, 595-606. 10.1016/S0898-6568(02)00012-8 [DOI] [PubMed] [Google Scholar]
- Geest C. R. and Coffer P. J. (2009). MAPK signaling pathways in the regulation of hematopoiesis. J. Leukoc. Biol. 86, 237-250. 10.1189/jlb.0209097 [DOI] [PubMed] [Google Scholar]
- Gegenbauer K., Elia G., Blanco-Fernandez A. and Smolenski A. (2012). Regulator of G-protein signaling 18 integrates activating and inhibitory signaling in platelets. Blood 119, 3799-3807. 10.1182/blood-2011-11-390369 [DOI] [PubMed] [Google Scholar]
- Goldsmith Z. G. and Dhanasekaran D. N. (2007). G protein regulation of MAPK networks. Oncogene 26, 3122-3142. 10.1038/sj.onc.1210407 [DOI] [PubMed] [Google Scholar]
- Grimes H. L., Chan T. O., Zweidler-McKay P. A., Tong B. and Tsichlis P. N. (1996). The Gfi-1 proto-oncoprotein contains a novel transcriptional repressor domain, SNAG, and inhibits G1 arrest induced by interleukin-2 withdrawal. Mol. Cell. Biol. 16, 6263-6272. 10.1128/MCB.16.11.6263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart A., Melet F., Grossfeld P., Chien K., Jones C., Tunnacliffe A., Favier R. and Bernstein A. (2000). Fli-1 is required for murine vascular and megakaryocytic development and is hemizygously deleted in patients with thrombocytopenia. Immunity 13, 167-177. 10.1016/S1074-7613(00)00017-0 [DOI] [PubMed] [Google Scholar]
- Horton M. A., Cedar S. H., Maryanka D., Mills F. C. and Turberville C. (1983). Multiple differentiation programs in K562 erythroleukemia cells and their regulation. Prog. Clin. Biol. Res. 134, 305-322. [PubMed] [Google Scholar]
- Ishida Y., Levin J., Baker G., Stenberg P. E., Yamada Y., Sasaki H. and Inoue T. (1993). Biological and biochemical characteristics of murine megakaryoblastic cell line L8057. Exp. Hematol. 21, 289-298. [PubMed] [Google Scholar]
- Johnson K. D., Boyer M. E., Kang J.-A., Wickrema A., Cantor A. B. and Bresnick E. H. (2007). Friend of GATA-1-independent transcriptional repression: a novel mode of GATA-1 function. Blood 109, 5230-5233. 10.1182/blood-2007-02-072983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kach J., Sethakorn N. and Dulin N. O. (2012). A finer tuning of G-protein signaling through regulated control of RGS proteins. Am. J. Physiol. Heart Circ. Physiol. 303, H19-H35. 10.1152/ajpheart.00764.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandanpour C., Krongold J., Schutte J., Bouwman F., Vassen L., Gaudreau M.-C., Chen R., Calero-Nieto F. J., Diamanti E., Hannah R. et al. (2012). The human GFI136N variant induces epigenetic changes at the Hoxa9 locus and accelerates K-RAS driven myeloproliferative disorder in mice. Blood 120, 4006-4017. 10.1182/blood-2011-02-334722 [DOI] [PubMed] [Google Scholar]
- Kim S. D., Sung H. J., Park S. K., Kim T. W., Park S. C., Kim S. K., Cho J. Y. and Rhee M. H. (2006). The expression patterns of RGS transcripts in platelets. Platelets 17, 493-497. 10.1080/09537100600758123 [DOI] [PubMed] [Google Scholar]
- Louwette S., Labarque V., Wittevrongel C., Thys C., Metz J., Gijsbers R., Debyser Z., Arnout J., Van Geet C. and Freson K. (2012). Regulator of G-protein signaling 18 controls megakaryopoiesis and the cilia-mediated vertebrate mechanosensory system. FASEB J. 26, 2125-2136. 10.1096/fj.11-198739 [DOI] [PubMed] [Google Scholar]
- Monteferrario D., Bolar N. A., Marneth A. E., Hebeda K. M., Bergevoet S. M., Veenstra H., Laros-van Gorkom B. A. P., MacKenzie M. A., Khandanpour C., Botezatu L. et al. (2014). A dominant-negative GFI1B mutation in the gray platelet syndrome. N. Engl. J. Med. 370, 245-253. 10.1056/NEJMoa1308130 [DOI] [PubMed] [Google Scholar]
- Nagata Y., Oda M., Nakata H., Shozaki Y., Kozasa T. and Todokoro K. (2001). A novel regulator of G-protein signaling bearing GAP activity for Galphai and Galphaq in megakaryocytes. Blood 97, 3051-3060. 10.1182/blood.V97.10.3051 [DOI] [PubMed] [Google Scholar]
- Northcott P. A., Lee C., Zichner T., Stütz A. M., Erkek S., Kawauchi D., Shih D. J. H., Hovestadt V., Zapatka M., Sturm D. et al. (2014). Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature 511, 428-434. 10.1038/nature13379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I.-K., Klug C. A., Li K., Jerabek L., Li L., Nanamori M., Neubig R. R., Hood L., Weissman I. L. and Clarke M. F. (2001). Molecular cloning and characterization of a novel regulator of G-protein signaling from mouse hematopoietic stem cells. J. Biol. Chem. 276, 915-923. 10.1074/jbc.M005947200 [DOI] [PubMed] [Google Scholar]
- Parkins A. C., Sharpe A. H. and Orkin S. H. (1995). Lethal beta-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature 375, 318-322. 10.1038/375318a0 [DOI] [PubMed] [Google Scholar]
- Saleque S., Cameron S. and Orkin S. H. (2002). The zinc-finger proto-oncogene Gfi-1b is essential for development of the erythroid and megakaryocytic lineages. Genes Dev. 16, 301-306. 10.1101/gad.959102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleque S., Kim J., Rooke H. M. and Orkin S. H. (2007). Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol. Cell 27, 562-572. 10.1016/j.molcel.2007.06.039 [DOI] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S. and Eliceiri K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671-675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz D., Vassen L., Chow K. T., McWhirter S. M., Amin R. H., Moroy T. and Schlissel M. S. (2012). Gfi1b negatively regulates Rag expression directly and via the repression of FoxO1. J. Exp. Med. 209, 187-199. 10.1084/jem.20110645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck J., Cohet N., Gonnet C., Sarrazin S., Doubeikovskaia Z., Doubeikovski A., Verger A., Duterque-Coquillaud M. and Morle F. (2003). Functional cross-antagonism between transcription factors FLI-1 and EKLF. Mol. Cell. Biol. 23, 1390-1402. 10.1128/MCB.23.4.1390-1402.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong B., Grimes H. L., Yang T.-Y., Bear S. E., Qin Z., Du K., El-Deiry W. S. and Tsichlis P. N. (1998). The Gfi-1B proto-oncoprotein represses p21 WAF1 and inhibits myeloid cell differentiation. Mol. Cell. Biol. 18, 2462-2473. 10.1128/MCB.18.5.2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay G., Chowdhury A. H., Vaidyanathan B., Kim D. and Saleque S. (2014). Antagonistic actions of Rcor proteins regulate LSD1 activity and cellular differentiation. Proc. Natl. Acad. Sci. USA. 111, 8071-8076. 10.1073/pnas.1404292111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer L. T., Jansen J. H. and van der Reijden B. A. (2010). Gfi1 and Gfi1b: key regulators of hematopoiesis. Leukemia 24, 1834-1843. 10.1038/leu.2010.195 [DOI] [PubMed] [Google Scholar]
- Vassen L., Fiolka K., Mahlmann S. and Möröy T. (2005). Direct transcriptional repression of the genes encoding the zinc-finger proteins Gfi1b and Gfi1 by Gfi1b. Nucleic Acids Res. 33, 987-998. 10.1093/nar/gki243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassen L., Fiolka K. and Möröy T. (2006). Gfi1b alters histone methylation at target gene promoters and sites of gamma-satellite containing heterochromatin. EMBO J. 25, 2409-2419. 10.1038/sj.emboj.7601124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassen L., Okayama T. and Moroy T. (2007). Gfi1b:green fluorescent protein knock-in mice reveal a dynamic expression pattern of Gfi1b during hematopoiesis that is largely complementary to Gfi1. Blood 109, 2356-2364. 10.1182/blood-2006-06-030031 [DOI] [PubMed] [Google Scholar]
- Vassen L., Khandanpour C., Ebeling P., van der Reijden B. A., Jansen J. H., Mahlmann S., Dührsen U. and Möröy T. (2009). Growth factor independent 1b (Gfi1b) and a new splice variant of Gfi1b are highly expressed in patients with acute and chronic leukemia. Int. J. Hematol. 89, 422-430. 10.1007/s12185-009-0286-5 [DOI] [PubMed] [Google Scholar]
- Wallis D., Hamblen M., Zhou Y., Venken K. J. T., Schumacher A., Grimes H. L., Zoghbi H. Y., Orkin S. H. and Bellen H. J. (2003). The zinc finger transcription factor Gfi1, implicated in lymphomagenesis, is required for inner ear hair cell differentiation and survival. Development 130, 221-232. 10.1242/dev.00190 [DOI] [PubMed] [Google Scholar]
- Whalen A. M., Galasinski S. C., Shapiro P. S., Nahreini T. S. and Ahn N. G. (1997). Megakaryocytic differentiation induced by constitutive activation of mitogen-activated protein kinase kinase. Mol. Cell. Biol. 17, 1947-1958. 10.1128/MCB.17.4.1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yowe D., Weich N., Prabhudas M., Poisson L., Errada P., Kapeller R., Yu K., Faron L., Shen M., Cleary J. et al. (2001). RGS18 is a myeloerythroid lineage-specific regulator of G-protein-signalling molecule highly expressed in megakaryocytes. Biochem. J. 359, 109-118. 10.1042/bj3590109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Socolovsky M., Gross A. W. and Lodish H. F. (2003). Role of Ras signaling in erythroid differentiation of mouse fetal liver cells: functional analysis by a flow cytometry-based novel culture system. Blood 102, 3938-3946. 10.1182/blood-2003-05-1479 [DOI] [PubMed] [Google Scholar]