Abstract
Several candidate drugs for acute radiation syndrome (ARS) have been identified that have low toxicity and significant radioprotective and radiomitigative efficacy. Inasmuch as exposing healthy human volunteers to injurious levels of radiation is unethical, development and approval of new radiation countermeasures for ARS are therefore presently based on animal studies and Phase I safety study in healthy volunteers. The Animal Efficacy Rule, which underlies the Food and Drug Administration approval pathway, requires a sound understanding of the mechanisms of injury, drug efficacy, and efficacy biomarkers. In this context, it is important to identify biomarkers for radiation injury and drug efficacy that can extrapolate animal efficacy results, and can be used to convert drug doses deduced from animal studies to those that can be efficacious when used in humans. Here, we summarize the progress of studies to identify candidate biomarkers for the extent of radiation injury and for evaluation of countermeasure efficacy.
Keywords: Acute radiation syndrome, Animal Efficacy Rule, biomarkers, chromosomal aberration, irradiation, metabolomics, microRNA, radiation countermeasures
Terrorist attacks, with their goals to maximize psychological and economical damage, and mortality to their victims, are an ever-growing worldwide concern in government and public sectors as they become more violent and more sensational. If given the chance, it is likely that terrorists will use chemical, biological, radiological, or nuclear (CBRN) weapons of mass destruction. To thwart these efforts, countermeasures against these weapons of mass destruction are being developed that can be utilized by the military, first responders, medical providers, and exposed victims.[1,2] In the recent past, several drugs have been approved for chemical attacks and biological agents.[1] Developments of radiation protection/mitigation agents are less advanced, and are the subject of this review.
The development of pharmacological radiation countermeasures to prevent, mitigate, and treat acute radiation syndrome (ARS) victims has been a goal since the end of World War II. At the Walter Reed Institute of Research, over 4000 compounds were synthesized and evaluated by the US Army. Out of all these efforts, amifostine (WR2721; 2-(3-aminopropyl) aminoethylphosphorothioate) has been the only systemically effective radioprotective agent to be fully approved for human use by the US Food and Drug Administration (FDA).[3,4] Though fully approved, amifostine is associated with severe side effects and has a very narrow therapeutic time window. Therefore, amifostine has only been authorized for use in a very few narrowly defined medical indications, which include: the reduction of xerostomia (dry mouth) resulting from radiotherapy for head and neck cancers, and the reduction of cumulative renal toxicity associated with repeated administration of cisplatin in patients with advanced ovarian cancer.[1] A second agent, granulocyte colony-stimulating factor (G-CSF), has recently been approved by the US FDA for mitigating hematopoietic ARS (H-ARS), caused by acute lethal doses of radiation.[5] However, the adverse consequences of G-CSF administration need to be taken into consideration.[6] Finally, there are many other radiation countermeasures currently at different stages of development.[1,2,7,8]
One of the difficulties encountered when developing such agents is the inability to obtain human efficacy data. It is, of course, unethical to knowingly expose human volunteers to potentially lethal doses of ionizing radiation. However, in order to obtain US FDA approval and market a drug, pharmaceutical companies must demonstrate potential therapeutic or prophylactic efficacy. Therefore, where typical human clinical trials are not possible, a different approach is required. This approach is embodied in the FDA’s Animal Efficacy Rule, which provides an alternate method for obtaining the necessary efficacy data to facilitate the development of new countermeasures and achieve US FDA approval.[9] To be successful, this strategy needs identified biomarkers in animals that faithfully reflect mitigation effects as they might be found in humans. Presumably, specifically purposed biomarkers will (1) assess the dose of radiation to which a victim is exposed and (2) be reporters of therapeutic success when such countermeasure agents are applied.
FDA’s Animal Efficacy Rule for the development of radiation countermeasures for ARS
The ‘Animal Efficacy Rule’ (21 CFR Parts 314.600-650 for drugs and 21 CFR 601.90 for biological products) was issued by the US FDA in 2002 and was intended to expedite the development of new drugs and biologics that might act as countermeasures against CBRN threats. The rule applies only to new agents for which definitive human efficacy studies cannot be conducted, because it would be unethical to knowingly expose humans to lethal doses of radiation.[9,10] The US FDA may grant marketing approval to new products for which both safety and the likelihood that the drug will produce clinical benefit in humans have been established using adequate and well-controlled animal studies. The criteria of the FDA’s Animal Efficacy Rule relevant to development using animal models are as follows:
There is a reasonably well-understood pathophysiological mechanism for toxicity of the agent (radiation), and its prevention, or substantial reduction by the drug.
The effect of the drug or biologic is demonstrated in more than one animal species, for which the expected effect is predictive for humans. The multiple animal requirement can be abrogated if the effect can be demonstrated in a single well-characterized animal model which sufficiently predicts the response in humans.
The animal study’s endpoint is related to a desired benefit in humans, generally involving the enhancement of survival or prevention of major morbidity.
Pharmacokinetics and pharmacodynamics, or other relevant data or information of the product, in animals and humans, allows selection of an effective human dose.
Because animal models rarely reflect the human disease precisely, the data from animal efficacy studies will never be as convincing as the human efficacy data. To provide confidence that such an agent will be effective in humans, it is especially important to understand how and why a countermeasure may work. Biomarkers for radiation injury and countermeasure efficacy are particularly important in this context.
Biomarkers for radiation injury
According to the FDA, a biomarker, biological marker, is an objective feature that can be measured and can indicate a specific biological, pathological, or therapeutic process.[11] The biomarker may reflect biological processes closely related to the mechanism of disease, or a process that is substantially downstream of the initial deficit. Biomarkers can be used to assess different types of biological characteristics or parameters. These include genetic sequence, receptor expression patterns, radiographic or other imaging-based measurements, blood composition, electrocardiographic parameters, or organ function. Since biomarkers are quantifiable, they can be used to characterize indirect or direct drug performance, dose selection, and potential safety issues related to candidate drug administration. A composite biomarker consists of several individual molecules or cellular changes that are combined in a specified algorithm to reach a single output. The FDA has further defined its qualifications to validate a biomarker by being able to reliably measure it and having a plethora of research and scientific convergence confirming its biological significance.[12]
Biomarkers can be used in the diagnostic, prognostic, predictive, and pharmacodynamic processes of drug development. This categorization is not exclusive, however, and one biomarker may play a role in more than one step of the drug developmental process. A diagnostic biomarker is a disease characteristic that categorizes a person by the presence or absence of a specific physiological or pathophysiological state or disease. A prognostic biomarker is a baseline attribute that categorizes patients by degree of risk for disease occurrence or progression of a specific aspect of a disease. A prognostic biomarker is informative about the natural history of the disorder in that particular patient in the absence of a therapeutic intervention. A predictive biomarker is a baseline characteristic that categorizes individuals by their likelihood of response to a particular treatment relative to no treatment. It may predict a favorable response or an adverse effect. A change in a pharmacodynamic biomarker indicates that a biological response has occurred in an individual who has received a drug; the magnitude of the change is considered pertinent to the response. The decision to use an inappropriate biomarker carries with it the substantial risk of adversely affecting human health, if a biomarker is inappropriately accepted as a surrogate endpoint. Therefore, robust and compelling scientific evidence is needed to validate a biomarker. A passive approach to validation includes extensive peer-reviewed scientific literature and consensus whereas an active approach would utilize in vitro, animal toxicology, genomics, proteomics, metabolomics, and clinical testing (if possible). From a regulation viewpoint, biomarkers have been accepted through several ad hoc pathways in drug regulatory agencies. At the US FDA, the European Medicines Agency (EMEA), and the Pharmaceuticals and Medical Devices Agency (PMDA, Japan), biomarkers have been qualified in recent years on a case-by-case basis. Currently, several biomarkers are approved for specific individual injuries; the US FDA has biomarkers for about 150 drug interactions validated, the EMEA has biomarkers for four injuries approved, and the PMDA has biomarkers for one injury accepted.[13–15] None of these are biomarkers for radiation injury. Multiple potential biomarkers are in the process of being confirmed, including some with radiation applications.
Biomarkers are an important aspect of radiation countermeasure development and can be used as a trigger for intervention as well as in selecting a drug dose and treatment regimen in humans. Biomarkers may also correlate with the mechanism by which the treatment/drug reduces the injury inflicted or used to correlate the desired clinical outcome (i.e. reduction in mortality or major morbidity). Additionally, the human dose of the drug should closely correlate with efficacious doses from well-controlled animal studies.
Biomarkers to assess absorbed radiation dose
In addition to needing biomarkers to determine drug efficacy and dose conversion from animal to humans, biomarkers are needed to assess the dose of absorbed radiation. This is known as biodosimetry. There are large numbers of studies using various strategies to identify the biomarkers for absorbed radiation dose. A brief account is presented below.
Ionizing radiation damages cells and tissues at all levels. Though radiation itself can directly damage DNA, radiation also damages DNA and other biochemical molecules indirectly by forming reactive oxygen species (ROS). ROS are the main contributors to DNA damage and are also responsible for significant macromolecule damage (i.e. cell membranes). The resulting damage can lead to altered cell function and apoptosis. ROS can also stimulate autophagy, the self-digestive process that degrades cellular components.[16] This can result in a positive outcome due to the removal of damage, therefore protecting cells from further serious DNA damage by free cellular components. The sensitivity of any particular cell type to radiation is directly proportional to its mitotic rate and indirectly proportional to the extent of its differentiation. Thus, cells of the lymphatic system, gametocytes, and the lining of the gastrointestinal (GI) tract are particularly vulnerable to the effects of ionizing radiation. By contrast, cells of the central nervous system are relatively resistant.
Depending on the dose of radiation delivered to the subject over a short period of time, there are several distinct sub-syndromes. Exposure to moderate doses (2–6 Gy) of radiation leads to a hematopoietic sub-syndrome (H-ARS), in which bone marrow is severely compromised and severe hemorrhage and infection is common. Much higher doses of radiation lead to discrete sub-syndromes in the GI tract (6–10 Gy; destruction of the intestinal tissue, dehydration, electrolyte imbalance) and neurovascular system (8 Gy). Even with supportive care, the neurovascular sub-syndrome is incurable. Additional sub-syndromes include cutaneous and pulmonary.
In order to treat ARS, it is important to have an estimate of radiation exposure. This can be determined based on symptoms present in victims, of which the METREPOL system provides a comprehensive triage scheme. Although commercially available dosimeters can provide an accurate dose to the surrounding area, they do not provide information for the absorbed dose, which can be quite different. Furthermore, unless the exposure incident has occurred in a clinic or radiation facility, it is unlikely that a dosimeter would be present. However, to properly treat radiation exposure victims before symptoms become observable, it is necessary to provide medical responders with the best assessment of absorbed dose. This has led to the entirely new field of biodosimetry, which is intrinsically dependent on biomarkers and depends on study and development of methods to more accurately assess the absorbed radiation dose. The principle of biodosimetry is to utilize the changes in biomarkers induced by ionizing radiation, which lead directly to estimates of the dose received, which directly predict the biological consequences of the dose. There are two types of biodosimetry: (1) based on changes in biological parameters (gene activation/chromosomal aberrations) and (2) based on physical changes in tissue detected by electron paramagnetic resonance (EPR; also known as electron spin resonance (ESR)) spectroscopy.
Cytokines, chemokines, and other proteins
G-CSF has been shown to be upregulated in irradiated mouse and nonhuman primates (NHPs) and plays an important role in mediating radiation injury.[17–22] Additionally, several cytokines, chemokines, and other proteins have been identified as candidate protein biomarkers of radiation injury over the last few years using total-body and partial-body exposure in murine and NHP models.[23–26] Among these proteins are interleukin-6 (IL-6), C-reactive protein (CRP), serum amyloid A (SAA), growth arrest and DNA damage-inducible 45 (GADD45) proteins, FMS-like tyrosine kinase 3 ligand (flt3L), and salivary α-amylase.
Previously, interleukin-18 (IL-18) has been shown to be upregulated in several organs collected from CD2F1 mice exposed to 5–10 Gy 60Co γ-radiation, and IL-18 serum concentrations showed a direct correlation to radiation dose in minipigs, NHPs, and mice.[27] Though multiple species were used, only a limited number of samples and time points were analyzed and additional studies are needed to expand and validate such findings. In our attempts to do so, we collected serum samples from a large number of NHPs exposed to three different doses of radiation (5.8, 6.5, and 7.2 Gy) at different time points after irradiation and analyzed them for IL-18 expression. Although serum from irradiated animals had higher levels of IL-18, there was no specific correlation with radiation dose (Figure 1).
Figure 1.
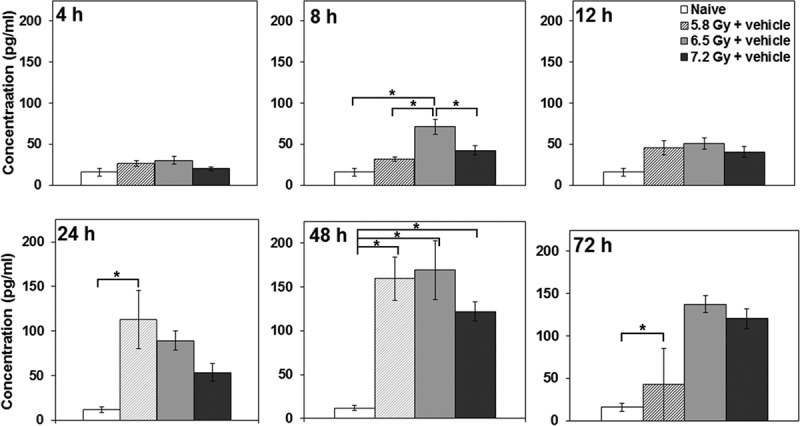
Effects of radiation exposure on NHP plasma IL-18 concentrations at various time points postirradiation. NHPs were irradiated with 5.8, 6.5, or 7.2 Gy (0.6 Gy/min;60Co γ-radiation); blood samples were collected at various time points (4, 8, 12, 24, 48, and 72 h postirradiation); and serum samples were analyzed for IL-18 using the Luminex platform. Data are presented as the mean with error bars representing standard error of the mean. Experimental groups from left to right are Naïve Control, 5.8 Gy + vehicle, 6.5 Gy + vehicle, and 7.2 Gy + vehicle. *Indicates that the difference between groups is significant when equal variance between groups was assumed (p < 0.05).
Peripheral blood counts
Peripheral blood cell counts, collected either at single time points or serially postirradiation, can be robust indicators of absorbed radiation dose. The cells analyzed include granulocytes, lymphocytes, leukocytes, and platelets. Importantly, the correlation exists not only in the early time window (1 or 2 d) but also in the late phase (up to 4 wk) after exposure.[28] However, for an optimal absorbed dose assessment, a complete blood cell count should be taken immediately following exposure. Extensive studies with multiple animal models, and the fact that the peripheral blood cell count has been used to monitor the health of the victims of many radiation-related accidents, support the use of this parameter as a diagnostic tool.[10] Furthermore, the blood cell assay is not only standard for investigating many other clinical indications, but is readily available, automated, and inexpensive. In Figures 2 and 3, we present the data from an experiment where NHPs were exposed to various doses of radiation to investigate the effect of irradiation on neutropenia and thrombocytopenia, respectively. Our results indicate that increasing radiation exposure increases the extent and severity of neutropenia and thrombocytopenia. Furthermore, our results indicate that deploying γ-tocotrienol (GT3) as a candidate countermeasure results in mitigating the radiation-induced effects of neutropenia and thrombocytopenia. Thus, biomarkers of radiation absorption and injury can be used as targets for the development of new radiation countermeasures.
Figure 2.
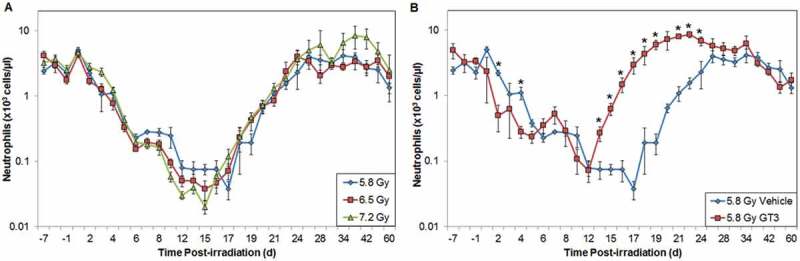
Effects of radiation exposure and GT3 treatment on levels of neutrophils in peripheral blood. (A) NHPs were irradiated with 5.8 (blue), 6.5 (red), or 7.2 Gy (green, 0.6 Gy/min, 60Co γ-radiation) and samples were collected at various time points in relation to irradiation. Neutrophils were counted using an Advia 120 cell counter. Mean neutrophil and standard error of the mean are displayed. (B) Animal exposed to 5.8 Gy radiation had received GT3 (75 mg/kg, red) 24 h prior to irradiation. Vehicle control is colored blue. *The difference between GT3-treated and vehicle-treated groups was significant when equal variance between groups was assumed (p < 0.05).
Figure 3.
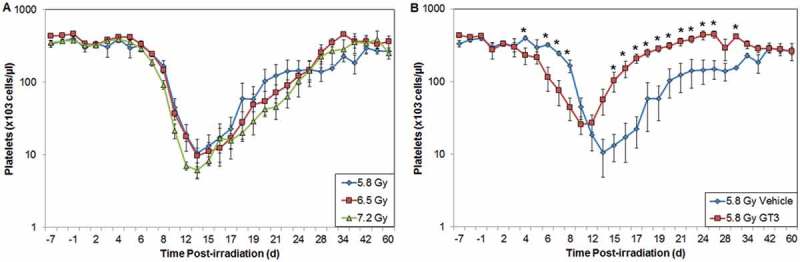
Effects of radiation exposure and GT3 treatment on levels of platelets in peripheral blood. (A) NHPs were irradiated with 5.8 (blue), 6.5 (red), or 7.2 Gy (green, 0.6 Gy/min, 60Co γ-radiation) and samples were collected at various time points in relation to irradiation. Platelets were counted using an Advia 120 cell counter. Mean platelet and standard error of the mean are displayed. (B) Animal exposed to 5.8 Gy radiation had received GT3 (75 mg/kg, red) 24 h prior to irradiation. Vehicle control is colored blue. *The difference between GT3-treated and vehicle-treated groups was significant when equal variance between groups was assumed (p < 0.05).
Chromosomal aberrations
Exposure to ionizing radiation causes both single- and double-stranded DNA breaks. During the subsequent repair process, a dicentric chromosome (DC, viz., an unstable chromosome with two centromeres) may be formed due to a mistake in the repair process and abnormal chromosome replication.[29] The dicentric cytogenetic assay has become the gold standard for cytogenetic biodosimetry after radiation exposure and has been recommended as an indicator of radiation injury by the International Atomic Energy Agency in the event of a radiation emergency. However, in a mass-casualty situation, this may not be the most practical diagnostic approach, as it requires highly trained technical staff and sophisticated equipment. Furthermore, to obtain an accurate dose from a metaphase spread, a dose-response calibration curve needs to be established. Based on reliability and accuracy, ring formation, an unstable chromosomal aberration that occurs less frequently than the DC, and a micronucleus assay, another well-known cytogenetic technique for dosimetry, are regarded as the most appropriate cytogenetic assays for diagnostics in the case of a mass-casualty scenario.[30] Finally, in a study yet to be validated for dosimetry, the premature chromosome condensation (PCC) assay, which identifies interphase cells with radiation-induced chromosomal aberrations, has been shown to accurately discriminate between total- and partial-body exposures. The PCC can be induced upon fusion of mitotic cells, or by treatment with chemicals such as calyculin A or okadaic acid.[31]
Citrulline
Citrulline is a nitrogen end product of glutamine metabolism in small-bowel enterocytes. It has been identified as a potential circulating biomarker for radiation-induced GI damage and epithelial cell loss; in other words, citrulline levels reflect the enterocyte mass. Citrulline is tissue-specific for small intestinal epithelium and its plasma concentration has been inversely correlated to gross histological GI tissue damage. The decrease of intestinal absorptive function following irradiation has been due to the loss of functionally active enterocytes this constitute the absorptive mucosal surface. The correlation between radiation-induced epithelial cell loss and plasma citrulline level has been well validated in mice, [32,33] and several investigators are working with other animal models to validate this biomarker for radiation injury.[34] In NHPs exposed to lower doses of radiation (5.8 and 6.5 Gy), we did not observe reduction in citrulline levels. In this case, enterocyte damage may not have been substantial enough to significantly lower citrulline. However, as presented in Figure 4, exposure to 7.2 Gy of radiation reduced circulating citrulline levels in NHPs.
Figure 4.
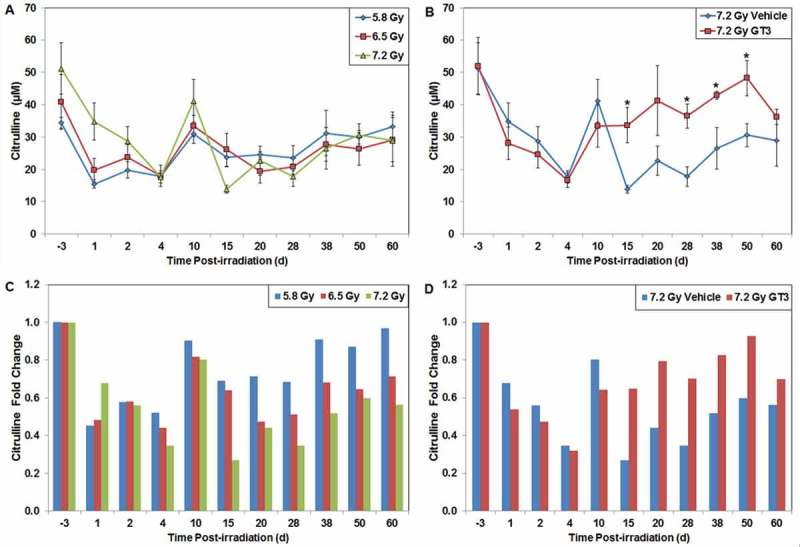
Effects of radiation exposure and GT3 treatment on the levels of citrulline in peripheral blood. (A) NHPs were irradiated with 5.8 (blue), 6.5 (red), or 7.2 Gy (green, 0.6 Gy/min, 60Co γ-radiation) and blood samples were collected at various time points in relation to irradiation. Plasma citrulline concentrations were quantified using liquid chromatography tandem mass spectrometry. Mean citrulline concentration and standard deviation are displayed. (B) Animal exposed to 7.2 Gy radiation had received GT3 (37.5 mg/kg, red) 24 h prior to irradiation. Vehicle control is colored blue. *The difference between GT3-treated and vehicle-treated groups was significant when equal variance between groups was assumed (p < 0.05). (C) Radiation (5.8 – blue, 6.5 – red or 7.2 – green Gy) induced fold change of citrulline concentrations in relation to pre-irradiation samples. (D) GT3 (37.5 mg/kg, red) induced fold change of citrulline concentrations in relation to pre-irradiation samples after exposure to 7.2 Gy radiation compared to vehicle (blue). Values less than 1.0 indicate a decreased citrulline concentration compared to its respective baseline value; values more than 1.0 would signify a citrulline concentration increase compared to baseline.
Tooth enamel- and fingernail-based biomarkers
Exposure to radiation results in the generation of free radicals that induce oxidative stress and cause damage to proteins, lipids, and DNA. The life span of these unpaired electrons is on the scale of nanoseconds in most biological tissues. However, such radiation-induced signals can be fixed for a much longer time in calcified tissues such as tooth, bone, and fingernail, and can be detected by EPR. Although they require high spectrometer frequency and low temperature for optimal performance, EPR-based radiation dosimetry techniques are non- or minimally invasive and do not require the typical processing of other biological-sample based techniques. Tooth enamel consists of 97% hydroxyapatite, and after radiation exposure the radiation-induced free radicals are entrapped in the hydroxyapatite lattice crystalline structure, where they can remain for months, even years. Similarly, free radicals can be incorporated into the α-keratin of fingernails and hair.[35] The output from the EPR dosimetry measurement yields the concentration of the free radicals that have integrated into these tissues and compares them to the established linear dose-response relation. Although the EPR spectroscopy is well established as a viable technique for measuring free radicals in biological samples, its clinical use for humans is challenged with various factors, including the need for customized instrumentation.[36] However, recent developments in EPR probes and measurement techniques offer potential opportunities for clinical dosimetry that could be deployed for triage in a large-scale radiation event.[37]
Recent developments for identifying biomarkers
There have also been many serious attempts to estimate radiation dose exposure using hematological, biochemical, and cytogenetic parameters. Several proteins such as CRP, amylase, cytokines, and growth factors have been investigated for their possible contributions. However, these biological agents have large inter-individual variations and fluctuate as a result of common variables such as inflammation and infection.[38]
Lymphocyte depletion kinetics, clinical observation, and the DC assays are used for postexposure dose assessment. However, these too are not ideal for use in mass-casualty scenarios. The reasons include the facts that lymphocyte depletion analysis requires repeated measurements over a prolonged period of time, and the DC assay is highly technical and labor-intensive.[39] Therefore, the need remains for identification of robust, stable, and novel biomarkers that not only are sensitive to radiation, but also can be repeatedly assayed in a noninvasive or minimally invasive manner. Given these problems of finding a radiation-dependent, practical biomarker for radiation injury, radiation scientists have redirected their efforts to investigate chemical alternatives in the metabolome in biological fluids, biological alternatives in the intestinal microbiome, and biochemical alternatives in the set of microRNAs (miRNAs, miRs) found in blood and elsewhere.
Metabolites as radiation biomarkers
Radiation exposure triggers a complex network of molecular and cellular responses in cells and tissues, which result in widespread changes in circulating metabolites and other types of molecules. In aggregate, these molecular variants are referred to as the metabolome, and components of the metabolome may therefore have potential as candidate biomarkers for radiation dosimetry. Subcategories of the metabolome include nucleotides, lipids, carbon skeletons characteristic of intermediary metabolism, ions, and agents acquired from the environment. Recent developments in fast, high-resolution chromatography, mass spectrometry, and data analysis methods have permitted massively parallel, quantitative assessment of thousands of metabolites found in biofluids that can be obtained in a noninvasive manner. These technological advancements have led to studies of the complex changes of several metabolites affected by radiation exposure, ultimately resulting in the identification of groups of small-molecule biomarkers.[40]
Metabolomic biomarkers in urine
Metabolomics can provide a rapid method for monitoring and assessing an individual’s exposure in a radiological scenario and thus would be useful for triage and injury assessment.[41–46] Metabolomic researchers often times use proteomic techniques and protein targets.
In one study, C57BL/6 mice were exposed to protons, and the levels of metabolites were evaluated in urine at 4 h postexposure. Significant differences were observed in metabolites of the tricarboxylic acid (TCA) cycle and those of fatty acids.[41] The presence of products of purine and pyrimidine metabolism is consistent with DNA damage and/or increased apoptosis. Metabolites of tryptophan, tyrosine, arginine, proline, and phenylalanine were also affected, with possible implications for DNA damage repair. A comparison of these metabolomic data with previously published data from γ- and X-ray strongly suggests a more pronounced effect on metabolism after irradiation with protons.[42–46]
One of the most feared fission radionuclides, 137Cs, can easily spread in water and air and decays by high-energy pathways. The ease of contamination spreading and exposure necessitates identifying 137Cs-induced metabolic pathway perturbations in easily accessible biofluids. A recent study with C57BL/6 mice demonstrated altered fatty acid metabolism, amino acid metabolism, and the TCA cycle.[47] There were similarities between the urinary metabolomic profiles of 137Cs-exposed mice and the γ-irradiated mice. However, the individual metabolites and their abundances differ between the two types of exposure. The 137Cs exposure caused unique effects on the levels of isovalerylglycine and tiglylglycine. Changes in excretion levels of taurine and citrate in urine were also observed after γ-radiation, but caused no attenuation in the levels of hexanoylglycine and N-acetylspermidine. Low- and high-dose rate exposures change many of the same pathways, including the TCA cycle and fatty acid metabolism in urine,[48] suggesting that dose rate does affect the levels of particular metabolites.
Internal emitters such as 90Sr would pose a substantial health risk during and immediately after a nuclear disaster or detonation of an improvised device. Exposure to 90Sr alters the urinary metabolome of mice, particularly the abundances of metabolites pertaining to butanoate metabolism, vitamin B metabolism, glutamate, and fatty acid oxidation, which are either directly or indirectly connected to the TCA cycle, which is central to energy production.[49]
In the event of a radiological incident, individuals with underlying medical conditions could present similar symptoms to radiation injury (nausea, diarrhea, vomiting, and elevated body temperature). Metabolomics has the potential to distinguish between stressors and inflammatory states using easily accessible biofluids such as proinflammatory oxygenated metabolites of arachidonic acid and anti-inflammatory metabolites of omega-3 polysaturated fatty acids (PUFAs).[50] Laiakis et al. studied biomarkers of lipopolysaccharide (LPS) exposure and compared the effects with those of ionizing radiation (3, 8 and 15 Gy of γ-rays). LPS treatment leads to a severe inflammatory response and a cytokine storm, events which are similar to radiation exposure. Cytosine, adenine, and O-propanoylcarnitine showed specificity to LPS at 24 h after treatment when compared to irradiation, further suggesting that metabolomics has the potential to distinguish between indications of radiation exposure and symptoms from underlying medical conditions and even other stressors.[43]
Metabolomics has also been used to identify biomarkers in the urine of total-body irradiated (TBI) humans (1.25 Gy) undergoing hematopoietic stem cell transplantation as part of cancer treatment. Seven markers showed distinct differences between pre- and postexposure samples; however, these levels were found to be gender-dependent. Therefore, separate biomarker signatures might be needed for males and females to distinguish radiation exposure victims.[51]
In aggregate, these studies suggest that differences in urinary excretion levels of certain metabolites could be used to assess an individual’s exposure in a radiobiological event. Further, such agents might have utility for both triage and injury assessment.
Metabolomic biomarkers in blood
Global molecular profiling reveals that serum components undergo a series of significant molecular alterations following radiation exposure. Recent studies suggest that plasma profiling of specific metabolites related to the pyrimidine and tryptophan pathways, as well as spermidine, can be used as candidate biomarkers of radiation exposure.[52,53] Profiling the metabolites and lipids extracted from murine plasma and liver 24 h after total-body γ-irradiation has been reported to yield 37 candidate compounds. These compounds are dominated by pyrimidine and gut microbiome-associated tryptophan metabolism, whose concentrations are correlated with radiation dose.[52] Additional studies suggest that serum spermidine might be used as a biomarker of biological response to 137Cs whole-body sublethal radiation exposure. In addition, changes in serum metabolites associated with amino acid metabolism, fatty acid metabolism, and TCA cycle change significantly after 137Cs internal emitter exposure.[47,53,54] 3-indoxyl sulfate, indole-3-lactic acid, phenyllactic acid, pipecolic acid, hippuric acid, and markers of DNA damage have also been shown to have differential plasma levels 24 h after exposure to 10 Gy radiation.[55]
The levels of methionine, Cys-Gly, and taurocholic acid were affected differently at 1 and 4 d postirradiation (60Co γ, 4 and 8 Gy) in CD2F1 mice. These altered levels varied in GI tissue as well, implicating overall energy metabolism.[56] Additionally, the lipid profile was dysregulated in irradiated mice. The results of the above studies have generated a panel of metabolites that may serve as candidate serum biomarkers for irradiation.
Exposure to γ-radiation significantly induced serum ether phosphatidylcholines (PCs) and decreased diacyl PCs carrying PUFAs.[50] In radiation-exposed mice, levels of pro-inflammatory, oxygenated metabolites of arachidonic acid increased, whereas levels of anti-inflammatory metabolites of omega-3 PUFAs decreased. These data suggest that a specific serum lipidomic biosignature could be used not only to indicate radiation exposure, but also as a potential target for therapeutic intervention.
Metabolomic and proteomic biomarkers in GI tissue
Though metabolomic biomarkers of radiation from urine and serum have been reported, they have not appeared to be informative or specific for organ-specific changes. A recent study using CD2F1 mice identified a metabolomic profile specific to GI tissue injury by 60Co γ-radiation exposure. In this study, tryptophan was upregulated and glutamic acid and Cys-Gly methionine were downregulated at 24 h post exposure.[56] The endogenous levels of spermidine, taurocholic acid, and eicosenoic acid were also elevated at 4 d after irradiation. Furthermore, the lipid panels were dysregulated at 1 and 4 d after irradiation. Metabolites associated with tryptophan and indoles in plasma reflect radiation-induced gut microbiome effects in the murine model 24 h after 10 Gy whole-body irradiation.[55] These details point toward the possibility that organ-specific biomarkers of radiation injury may yet be found. One such organ-specific biomarker may be doublecortin-like kinase 1 (Dclk1), found in the Notch signaling pathway.[57] This protein can be used to evaluate and correlated to intestinal stem cell survival shortly after radiation injury. When the Notch signaling pathway was inhibited after 40 Gy 137Cs radiation; there was an increase in reduction of regenerative crypts and Dclk1 expression than with radiation alone. Dclk1 was found to be a predictive biomarker for the survival of GI tissue. Several other potential GI biomarkers are Msi1, Lgr5, Bmi1, and Notch 1; these are also directly correlated with crypt and overall survival and can be used as prognostic GI tissue biomarkers.[58] However, more studies in other animal strains and species are needed for confirmation and expansion. Organ-specific biomarkers, such as Dclk1, would be important since such biomarkers might reflect the functional status of the organ system.
Microbiota as novel biomarkers
Humans and animals are hosts to highly complex ecosystems of colonizing microbes, a vast majority of which (10–100 trillion) live in the intestines and are excreted with the feces after irradiation. Several studies using mice, rats, pigs, canine, and human have shown altered abundance of specific intestinal microbiota in the feces, indicating that these bacteria are sensitive to whole- and partial-body irradiation of the host.[59,60] Intestinal microbiota excreted in the feces can act as a biomarker of radiation exposure, and the abundance of specific intestinal microbiota may serve as a diagnostic parameter of radiation exposure. However, these observed changes in specific intestinal microbiota were transient and short-lived. For a biomarker to have utility as a radiation biodosimeter, a biologically significant and sustained signal needs to be present.
Earlier studies on the radiation-sensitive microbiome had relied on the cultivation of fecal bacteria, followed by manual identification and classification. However, the majority of bacteria colonizing the intestines are not readily cultivatable. The modern alternative has been to use a high-throughput molecular approach based on species-specific gene sequences, in hope that the global analysis would provide a more comprehensive view of the ecosystem in response to radiation. Since each bacterial species has a fixed number of genomic copies for the 16S rRNA gene, the quantitative abundance of 16S copies in a fecal sample can provide an index of the relative quantity of that microbial species.
In a recent study using Wistar rats, 16S rRNA levels were shown to respond to single- and multiple-fraction TBI (10 or 18 Gy, X-ray). The levels of 16S rRNA corresponding to 12 members of Bacteroidales, Lactobacillaceae, and Streptococcaceae increased after radiation exposure; 47 Clostridaceae members decreased; and 98 Clostridaceae and Peptostreptococcaceae members remained unchanged.[60] The microbiota that are unaffected by radiation thus serve as internal controls as these are also present in human and rat feces. Using such intestinal microbiota as radiation exposure biomarkers represents a novel approach that can complement conventional chromosome aberrational analysis and may significantly enhance the range and accuracy of biological dose assessments.
Intestinal microbiota-derived metabolomics species have also been recently identified in plasma markers. The profiling of liver and plasma metabolites and lipids, extracted 24 h after total-body irradiation (0, 2, 4, 6, 8 or 10.4 Gy) from C57BL/6 mice, identified 37 compounds whose concentrations correlated with the radiation dose. Pyrimidine levels were positively correlated, and gut microbiome-associated tryptophan metabolism were negatively correlated with radiation dose, suggesting a potential new radiation injury biomarker.[52]
Messenger RNAs as biomarkers
Messenger RNAs (mRNAs) have a pivotal role in gene expression. By transmitting genetic information from DNA, mRNAs are vital for quality and quantity of protein expression. A large set of mRNAs were measurably induced in human cells 24 h postirradiation, and this induction was significant through 72 h postexposure.[61] Several murine cytokine mRNAs have been upregulated in response to irradiation and some specific mRNAs show upregulation after low doses of radiation (2–50 cGy), which indicate a physiological response to nonlethal stress via ionizing radiation.[8,62] More so, radiation-induced mRNA elevations have been shown to be linearly dose-dependent.[62,63] Therefore, mRNA expression may provide estimates of exposure and received radiation doses.[63] Continuing research with identifying mRNAs as biomarkers is promising, as there are ample targets and a wide range of situations that cause signaling available.
miRNA as biomarkers
miRNA are a conserved class of short (typically 19–22 nucleotides), noncoding, regulatory RNAs that generally control gene expression by inducing mRNA cleavage or inhibiting translation by base pairing to partially complementary sequences at the posttranscriptional level. A single miRNA can interact with hundreds of mRNAs, ultimately regulating diverse cellular processes, including development, proliferation, and differentiation in addition to various disease progressions. Several studies have detected and identified circulating miRNAs in a number of body fluids, such as serum, plasma, and urine, [64] as candidate biomarkers for prostate cancer, pancreatic cancer, liver damage, and pregnancy.[65] There are several advantages of using miRNA as biomarkers: (1) they are relatively stable due to their small size and location in the exosome (those found in the serum are inherently stable); (2) their expression alters in response to disease or organ damage; (3) many miRNAs are tissue-specific and can easily be used in high-throughput analysis; and (4) miRNA levels are reproducible in individuals of the same species. It is due to these characteristics that miRNAs have been investigated in detail as candidate biomarkers for radiation exposure.
Based on fold change (≥1.2) and significance criteria (p < 0.05), 70 miRNA sequences were found to be differentially expressed in irradiated (60Co γ, 9.5 Gy, 0.4 Gy/min) B6D2F1/J mice compared to sham irradiated mice. Fourteen of these miRNAs were found to be specific to irradiated mice. From these, nine major miRNA sequences were selected for validation using real-time quantitative polymerase chain reaction (Table 1).[66] Several serum miRNAs had differential expressions at 24 and 48 h post exposure to 0.5, 2, or 10 Gy 137Cs radiation. Of these, miR-150 demonstrated both dose- and time-dependent (24 and 48 h postirradiation) decreases after exposure to 1–8 Gy radiation, thus indicating that it may serve as a candidate biomarker.[67] Additional miRNAs that exhibited serum level increases after irradiation include miR-200b and miR-762. The differential expressions of these miRNAs were more pronounced in animals that received higher doses of radiation. It has also been shown that murine miR-30b and miR-30c are upregulated after 7 and 10 Gy whole-body γ-irradiation (60Co, 0.6 Gy/min).[68] Consistent with a single acute exposure, murine serum miR-150 showed a 50% reduction at 24 h post-fractionated radiation exposure (4 Gy total), and further reduction at higher doses (8 and 12 Gy) at later time points (48 and 72 h). miR-200b and miR-762 were also similar, in that each exhibited an increase after fractionated radiation up to 48 h. However, miRNA-762 decreased at 72 h after 12 Gy fractionated exposure.
Table 1.
miRNA as a biomarker of radiation injury and countermeasure efficacy.
| miRNA | Biomarker for | Effects | Experimental Details | References |
|---|---|---|---|---|
| miR-34b-3p | TBI | Fold change 2.0 ⇑ | B6D2F1/J mice, 60Co, 9.5 Gy, 0.4 Gy/min |
[55] |
| miR-3082-5p | Fold change 2.0 ⇑ | |||
| miR-142-5p | Fold change 1.5 ⇓ | |||
| miR-31 | Fold change 1.5 ⇓ | |||
| miR-185 | Fold change 1.6 ⇓ | |||
| miR-130b | Fold change 1.7 ⇓ | |||
| miR-216b | Fold change 1.7 ⇓ | |||
| miR-130a | Fold change 1.9 ⇓ | |||
| miR-1912 | Fold change 2.1 ⇓ | |||
| miR-150 | TBI and PBI | Down regulated | CBA/J and C57BL/6 mice, 137Cs, 1–12 Gy, 1.1 Gy/min |
[56] |
| miR-200b | Upregulated | |||
| miR-762 | Upregulated | |||
| miR-30b | TBI | Upregulated | CD2F1 mice, 60Co, 7 or 10 Gy, 0.6 Gy/min |
[57] |
| miR-30c | Upregulated | |||
| miR-30a-3p | TBI | Upregulated | C57BL/6 J mice, 137Cs, 8 Gy, 1.1 Gy/min | [58] |
| miR-30c-5p | Upregulated | |||
| miR-187-3p | Downregulated | |||
| miR-194-5p | Downregulated | |||
| miR-27a-3p | Downregulated | |||
| miR-667 | TBI | Differentially | C57BL6 mice, 137Cs, 0.5, 2 and 10 Gy, 0.52 Gy/min | [59] |
| miR-877 | Changed at 6 and 24 h postirradiation with 0, 2, and 10 Gy | |||
| miR-24–2 | ||||
| miR-434-5p | ||||
| miR-501-3p | ||||
| miR-592 | ||||
| miR-148a | ||||
| miR-30a | ||||
| miR-30e | ||||
| miR-690 | CI: TBI and burn | Fold change 1.8 ⇑ | B6D2F1/J mice, 60Co, 9.5 Gy, 0.4 Gy/min | [55] |
| miR-223 | Fold change 1.2 ⇑ | |||
| miR-30b | δ-tocotrienol (75 mg/kg) in TBI model | Downregulated radiation-induced miRNA | CD2F1 mice, 60Co, 7 or 10 Gy, 0.6 Gy/min | [57] |
| miR-30c | ||||
| miR-30a-3p | Amifostine, TBI | Downregulated | C57BL/6 J mice, 137Cs, 8 Gy, 1.1 Gy/min | [58] |
| miR-30c-5p | Downregulated | |||
| miR-187-3p | Upregulated | |||
| miR-194-5p | Upregulated | |||
| miR-27a-3p | Upregulated | |||
| miR-30a-3p | Bone marrow stromal cells, TBI | Downregulated | C57BL/6 J mice, 137Cs, 10.4 Gy, 1.1 Gy/min | [58] |
| miR-30c-5p | Downregulated | |||
| miR-187-3p | Upregulated | |||
| miR-27a-3p | Upregulated |
Only in vivo models have been reviewed. CI, combined injury; PBI, partial-body irradiation; TBI, total-body irradiation.
Another study found that a set of five miRNAs were capable of distinguishing between mice cohorts irradiated with 0 or 2 Gy and another set of three miRNAs were capable of distinguishing between 2 and 6.5 Gy (low versus high sublethal doses) at both 24 h and 7 d after radiation exposure.[69] A third set of five miRNAs were capable of distinguishing between 6.5 and 8 Gy (high sublethal and lethal doses, respectively) at 24 h. Of these latter five, two could differentiate between cohorts at 3 and 7 d postradiation as well. It is important to note that none of the miRNAs in each set were capable of distinguishing 0 from 2 Gy, 2 from 6.5 Gy, or 6.5 from 8 Gy overlap. This may be due to the fact that the objective of this study was to identify distinct sets of miRNAs with the highest differences between specific doses of radiation. There may therefore be additional miRNAs with the ability to distinguish between larger ranges of radiation doses. Cui et al. were able to quantitate the accuracy, sensitivity, and specificity of murine plasma miRNA that were capable of distinguishing between mice receiving 0.5, 2, or 10 Gy (at 6 or 24 h post exposure), thus further supporting why researchers are focusing on miRNA as potential biomarkers of radiation injury.[70]
Exposure to very high-dose radiation, in combination with burn or wound trauma, can produce more deleterious effects requiring specialist interventions and a separate set of biomarkers. However, precise biomarkers needed to accurately assess and treat such conditions are still undetermined. In a recent study, an attempt was made to identify and explore the possible role of serum miRNA signatures as potential biomarkers for radiation and combined burn injury, in B6D2F1/J mice.[66] Out of 890 differentially expressed miRNAs, microarray analysis showed 47 distinct miRNA sequences significantly associated with combined injury (60Co γ-radiation, 9.5 Gy, 0.4 Gy/min, and 15% total-body-surface-area skin burn) in mice compared to control mice (fold change ≥ 1.2, p < 0.05). Only two major miRNA sequences (miR-690 and miR-223) were validated and shown to be differentially and significantly expressed for combined injury in mice (fold change ≥ 1.5, p < 0.05).
The studies described above demonstrate that miRNA can be predictive of various doses of radiation exposure and can be used to assess radiation exposure in the context of a mass-casualty scenario (radiation exposure or combined injury). miRNA analysis and biomarkers, based on miRNA measurements, may provide valuable tools in developing and implementing effective biodosimetry.
Long noncoding RNA as biomarkers
Long noncoding RNAs (lncRNAs) are a class of mRNA-like transcripts longer than 200 nucleotides that do not code for protein but are present in about 80% of transcriptions.[71] They have many functions including regulating chromatin modification, gene transcription, and posttranslational processes; however, most functions have not yet been discovered. Based on their proximal adjacent protein-coding genes, lncRNAs are generally divided into five classes: sense, antisense, bidirectional, intronic, and intergenic. lncRNAs show cell type-specific expression and respond to various stimuli, suggesting that their expression is largely influenced by transcriptional activity. Though many lncRNA are poorly conserved, several have recently been identified as biomarkers for DNA damage, environmental stressors, cancer, and radiation injury. With regard to radiation injury, several lncRNAs respond to irradiation in a time- and dose-dependent manner.[72,73] This field may hold promise as more lncRNAs are explored as radiation biomarkers or therapeutic targets.
Biomarkers for radiation countermeasure efficacy
As summarized above, the FDA’s Animal Efficacy Rule for the development of radiation countermeasures requires the establishment of human drug doses based on biomarker-guided bioequivalence of effective animal doses. This approach depends on a calculated animal-to-human dose conversion step, yielding a dose range that is tolerable to humans.[9] To estimate the doses of radiation countermeasures likely to produce therapeutic benefit in humans, one needs to use drug-induced biomarker responses as proxies for drug efficacy. Hence, the human efficacious dose can be defined as the drug dose that activates the same biomarker responses to at least the same degree as those that accompany and mediate the drug’s therapeutic efficacy in animals.
Ideal biomarkers for the development of radiation countermeasures.
Biomarker induction should depend on the mechanism of action of countermeasures.
Biomarkers should be induced by countermeasures in unirradiated and irradiated animals.
Biomarker expression should be induced by the full range of efficacious doses of the countermeasure.
Biomarker response should directly correspond with the countermeasure’s dose-dependent efficacy.
The biological nature of the biomarker should be relevant to its effects on reducing the risk of mortality following irradiation.
Biomarkers should be quantifiable using available assays from samples obtained by simple and relatively noninvasive procedures.
Biomarkers should be responsive across multiple species of animals and humans.
Cytokines
Potential protein efficacy biomarkers for the drug CBLB502 (truncated flagellin, Entolimod) in mice, canines, and NHPs have been published, along with a description of the advantages of biomarker-based versus pharmacokinetics-based assessments.[21,74] The biomarkers for CBLB502 were selected based on the fact that its radiomitigative efficacy was mediated via similar sets of cytokines in different animal species. Two of these cytokines, G-CSF and IL-6, were selected as primary biomarkers after being shown to be essential for CBLB502’s radiomitigative activity in a mouse model of ARS. These cytokines were not only correlated with the effect of radiation, but also with the efficacy of CBLB502. In addition, CBLB502-dependent survival was closely associated with the levels of induction of the cytokines G-CSF and IL-6 in mice, canines, and NHPs, as well as with fold change of neutrophil counts at 24 h after drug administration.
In addition, several radiation countermeasures (CBLB502, CBLB612, CBLB613, 5-androstenediol (5-AED), GT3, δ-tocotrienol, and tocopherol succinate (TS)) have been shown to induce G-CSF at the protein level in a dose-dependent manner in mice.[18,20,21,75–82] Two of these agents, CBLB502 [21] and GT3 (unpublished observation), have also been shown to induce G-CSF in NHPs. G-CSF antibody administration abrogated the radioprotective efficacy of CBLB502, 5-AED, GT3, δ-tocotrienol, and TS (an ester of α-tocopherol) in mice, suggesting that G-CSF plays an important role in the radioprotective efficacy of these countermeasures.[21,76–79,83] Increased doses of TS and the associated increased protein levels of G-CSF have been correlated with increased survival in lethally irradiated CD2F1 mice.[78]
In our attempt to expand upon IL-18’s potential as a biomarker for radiation countermeasure efficacy, we found that there were no differences in IL-18 protein expression in irradiated control NHPs treated with vehicle and GT3, except at 72 h postirradiation, when GT3-treated animals had significantly higher levels of IL-18 compared to irradiated animals treated with vehicle (Figure 5). These results, paired with similar findings of nonspecific correlation after irradiation in NHPs described above, suggest that IL-18 may not be used as a biomarker for either radiation injury or countermeasure efficacy.
Figure 5.
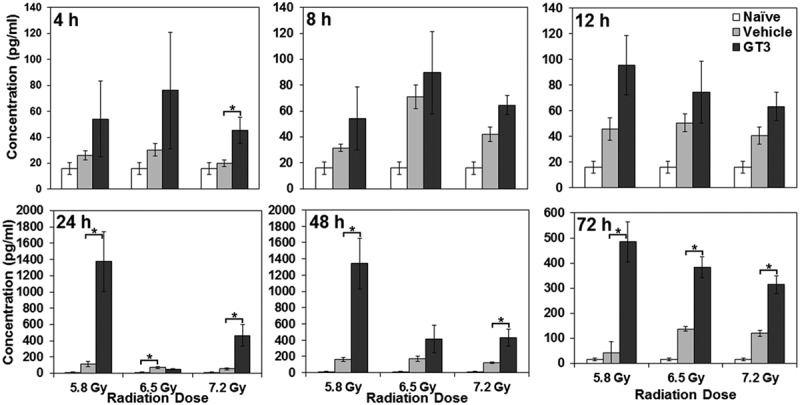
Effects of GT3 on plasma IL-18 concentrations in irradiated NHPs at various time points postirradiation. NHPs received GT3 (75 mg/kg) 24 h prior to 60Co γ-irradiation (5.8, 6.5, or 7.2 Gy at 0.6 Gy/min). Blood samples were collected at various time points (4, 8, 12, 24, 48, and 72 h postirradiation), and serum samples were analyzed for IL-18 using the Luminex platform. Data are presented as means, with error bars representing standard error of the mean. Experimental groups from left to right are naïve control, vehicle, and GT3. *Indicates that the difference between groups is significant when equal variance between groups was assumed (p < 0.05).
In a recent proteomic analysis of 725 proteins in irradiated human CD34+ stem cells, significant changes in expression level of 50 proteins could be protected from radiation-induced reduction by pretreatment with TS.[84] Ingenuity pathway analysis revealed that the modified proteins fell into categories dominated by epigenetic regulation, DNA repair, and inflammation.
Peripheral blood counts
In addition to neutrophil counts acting as a biomarker for radiation injury, the severity and duration of neutropenia have been accepted as biomarkers to assess drug efficacy, most prominently in the case of CBLB502 in conjunction with IL-6 and G-CSF.[21] Similarly, neutropenia may be used as a biomarker for GT3 efficacy, as shown in Figure 2.
Citrulline
As stated above, the small intestine is the principal source of circulating citrulline, and the serum citrulline level is a strong indicator of small intestine health. The effect of recombinant IL-11 (250 μg/day oral beginning 24 h after TBI) on intestinal mucosal injury was assessed at different times after 9.0 Gy 137Cs TBI by measuring plasma citrulline levels in CD2F1 mice.[32] Serum citrulline levels dropped significantly in both vehicle- and IL-11-treated groups at 3.5 d after irradiation (expected nadir). However, in IL-11-treated animals, citrulline levels recovered by 7 d compared to 14 d in the control group, indicating faster recovery of small intestine health in animals treated with IL-11. Similarly, as presented in Figure 4, 7.2 Gy TBI reduced circulating citrulline levels in NHPs, and GT3 treatment was capable of reversing this radiation-induced reduction.
miRNA
In addition to cytokines, there are reports that miRNA may also serve as candidate efficacy biomarkers for radiation countermeasures. Studies indicate that δ-tocotrienol (75 mg/kg) downregulates radiation-induced miR-30b and miR-30c expression in mouse tissues and serum after exposure to either 7 or 10 Gy (0.6 Gy/min) of 60Co γ-radiation.[68]
miRNA levels have also been studied in mice treated with amifostine before radiation exposure to 8 Gy 137Cs radiation (1.1 Gy/min), and compared with those in untreated irradiated mice (Table 1). Five serum miRNAs showed differential expression between amifostine-treated and untreated irradiated mice in blood samples collected 24 h after irradiation.[69] The serum miRNA levels were found to be correlated with the radioprotective efficacy of amifostine. Furthermore, the amifostine-treated lethally irradiated mice (8 Gy) samples resembled mice receiving the sublethal radiation dose (6.5 Gy). Based on Pearson’s correlation, the amifostine-treated lethally-irradiated mice were not significantly different from the sublethally irradiated mice (6.5 Gy). These data thus suggest that the five miRNAs detected in this system (viz., miR-30a-3p, miR-30c-5p, miR-187-3p, miR-194-5p, and miR-27a-3p) may serve as markers of radiation-induced mortality in mice. Four of the five miRNAs above (excluding miR-194-5p) also reported positively on radiomitigation by transplantations of bone marrow stromal cells 5 d after irradiation (10.4 Gy). By contrast, although the decrease in miR-150-5p has emerged as a consistent marker of irradiation in mice,[67,69,70] it did not differ between irradiated and stromal cell-treated irradiated mice in this study.
The biomarkers above are popular and are commonly used for drug efficacy, though there are several others with which various investigators are working and many more may be identified in the future for such use.[24,44,85–89]
Expert commentary
Only one agent, G-CSF, has received approval from the US FDA as a radiomitigator for H-ARS; however, several other agents are currently under development. To develop radiation countermeasures, radiation exposure dose assessment, identification of biomarkers for radiation injury, and efficacy biomarkers for countermeasures are important milestones.
It is not easy to identify universal biomarkers for radiation exposure that would be useful under a variety of scenarios. Combined injury has a higher mortality rate than radiation exposure alone in both, human and animal models.[90–92] The wound or burn effect complicates and alters biodosimetric assessments, and reliable biochemical and molecular biomarkers are critical for future emergency planning and national preparedness to accurately assess and implement effective countermeasures for such an incident. The increase in radiation-induced mortality by wounding is triggered by sustained activation of (1) nitric oxide synthase pathways; (2) persistent alteration of cytokine homeostasis; and (3) increased susceptibility to bacterial infection. Therefore, cytokine measurements have been widely adopted for biodosimetric evaluation and assessment of radiation dose and injury. Recent results suggest that agents inhibiting wound responses may prove therapeutic responses for radiation combined injury and reduce related mortality.[93] Serum miRNA signatures of mice, following combined burn injury, provide new insights into the molecular and biochemical pathways associated with radiation combined skin-burn trauma in vivo.
It is a general consensus in the radiation dosimetry field that there is no perfect biomarker to assess the absorbed radiation dose, and that complimentary methods must be used. Measuring chromosomal aberrations is best suited for triage in the event of acute whole-body irradiation, but this technique requires highly trained personnel and relatively longer time, during a period where time is of the essence.
Rapid screening of a large population for radiation-exposed victims presents new challenges to which several approaches are being developed.[40] Protein and gene expression biomarkers are commonly used for assessing the absorbed radiation dose, and gene transcription biomarkers (lncRNA) may soon be incorporated for radiation temporal and dose dependence parameters. Metabolomic studies show promise for detecting responses to irradiation as well as variation in metabolic products as a result of a chain of amplifying proteomic and transcriptomic events, indicating their sensitivity.
Metabolomics is a powerful tool to study postirradiation changes even before the onset of clinical symptoms and can augment population screening in real-life scenarios. Improvements in metabolite identification databases are likely to facilitate the development of robust, multi-metabolite panels for potential clinical use. Urine metabolomics are especially promising due to thoroughly completed recent studies involving radiation dose and time dependence in rat and NHP models.[94,95]
Five-year view
There are several major difficulties that need to be faced when defining biomarkers, few of which have clear defined solutions. Though several biomarkers have been validated by the US FDA, none have been validated for radiation injury. This is due to several reasons; the first of which is the unethicality of irradiating humans; another being the slow process of determining biomarkers through the Animal Efficacy Rule first by investigating in a small animal model and then in a large animal model. Furthermore, if a radiological attack or accident were to occur, medical personnel would be focused on triaging victims instead of collecting and processing samples for biomarker identification and validation, even though biomarkers would need to be assessed over a realistic time frame post-incident. Likewise, much of the radiation exposure research conducted focuses more on whole-/total-body exposure and less with partial-body irradiations and combined injuries (wounding plus irradiation, burn plus irradiation), which, in a radiological incident, will both be more likely than homogenous whole-body irradiation. Therefore, biomarker assessment in partial-body exposure and combination injury needs to be explored to a greater extent, as separate biomarkers for each for these situations (whole, partial, and combined injury) will be needed. Some solutions to these issues would be to expand the scope of radiobiology research and make it a more attractive field for future researchers.
Analysis of intestinal microbiota from feces by microarray and polymerase chain reaction (evaluation of 16S rRNA) is a noninvasive technique and provides a sustained level of reporting signals that are increased several folds following exposure to radiation. Intestinal microbiota may therefore serve as novel biomarkers of radiation exposure, and may complement conventional chromosome aberrational analysis to significantly enhance biological dose assessments.
Radiation metabolomics is a highly promising area that involves noninvasive measurements of radiation-induced changes in the body. Several promising biomarkers have been identified for radiation injury as well as the efficacy of radiation countermeasures under advanced stages of development; however, these biomarkers need further investigation and validation using different animal models and human volunteers. Recent data demonstrate that plasma cytokines, chemokines, citrulline, apoptotic pathway molecules, lncRNA, and miRNA profiles can be highly predictive of different levels of radiation exposure.
Key issues.
Only one radiation countermeasure, G-CSF, has received the US FDA approval as a radiomitigator for H-ARS.
To develop radiation countermeasures, important milestones include: radiation exposure dose assessment and biomarker identification of radiation injury and efficacy of radiation countermeasures.
Quantification of chromosomal aberrations, the gold standard for assessing absorbed dose, will not be practically possible in a mass-casualty scenario due to the time and types of equipment needed.
During the last decade, several cytokines/growth factors and proteins have been identified as biomarkers for assessing radiation exposure dose. Cytokines, such as G-CSF and IL-6, have also been identified as efficacy biomarkers for radiation countermeasures such as CBLB502 and tocols.
Plasma citrulline concentration is inversely proportional to gross histological GI tissue damage and the dose of radiation exposure, making it a promising biomarker for radiation-induced GI damage and epithelial cell loss.
miRNAs can be predictive of radiation exposure and extent of countermeasure efficacy.
Recent studies suggest that metabolomic markers, which are easily accessible in biofluids, may prove useful biomarkers for radiation exposure and provide a rapid method for monitoring radiation exposure.
Using microbiota as biomarkers of radiation exposure represents a novel approach to significantly enhance radiation dose assessments.
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Acknowledgements
The opinions or assertions contained herein are the professional views of the authors and do not necessarily represent the Uniformed Services University of the Health Sciences, or the Department of Defense, USA. The authors gratefully acknowledge the research support from Defense Medical Research and Development Program (project # D_I_14_J7_735 to VKS) and other funding agencies of US Department of Defense. Mention of specific therapeutic agents does not constitute endorsement by the U.S. Department of Defense, and trade names are used only for the purpose of clarification. We apologize to those who have contributed substantially to the topics discussed herein that we were unable to cite due to space constraints.
References
- Papers of special note have been highlighted as:
- • of interest
- •• of considerable interest
- Singh VK, Romaine PL, Seed TM. Medical countermeasures for radiation exposure and related injuries: characterization of medicines, FDA-approval status and inclusion into the strategic national stockpile. Health Phys. 2015;108:607–630. doi: 10.1097/HP.0000000000000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh VK, Newman VL, Romaine PL. Radiation countermeasure agents: an update (2011 - 2014) Expert Opin Ther Pat. 2014;24:1229–1255. doi: 10.1517/13543776.2014.964684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MedImmune [[cited 2013 Sep 30;]];Available from: 2013 http://www.medimmune.com/docs/default-source/pdfs/prescribing-information-for-amifostine.pdf
- Seed TM, Inal CE, Singh VK. Radioprotection of hematopoietic progenitors by low dose amifostine prophylaxis. Int J Radiat Biol. 2014;90:594–604. doi: 10.3109/09553002.2014.899450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Food and Drug Administration [[cited 2015 Apr 17;]];FDA approves Neupogen® for treatment of patients with radiation-induced myelosuppression following a radiological/nuclear incident. 2015 http://www.fda.gov/EmergencyPreparedness/Counterterrorism/MedicalCountermeasures/AboutMCMi/ucm443245.htm Available from:
- Singh VK, Newman VL, Seed TM. Colony-stimulating factors for the treatment of the hematopoietic component of the acute radiation syndrome (H-ARS): a review. Cytokine. 2015;71:22–37. doi: 10.1016/j.cyto.2014.08.003. [DOI] [PubMed] [Google Scholar]
- Singh VK, Romaine PL, Newman VL. Biologics as countermeasures for acute radiation syndrome: where are we now? Expert Opin Biol Ther. 2015;15:465–471. doi: 10.1517/14712598.2015.986453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh VK, Ducey EJ, Brown DS. A review of radiation countermeasure work ongoing at the Armed Forces Radiobiology Research Institute. Int J Radiat Biol. 2012;88:296–310. doi: 10.3109/09553002.2012.652726. [DOI] [PubMed] [Google Scholar]
- U.S. Food and Drug Administration [[cited 2014 Jul 18;]];Guidance for industry: product developoment under the animal rule. 2014 http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM399217.pdf Available from:
•• This is the US FDA draft guidance document for the drug development under the Animal Efficacy Rule and demonstrates the relevance of biomarkers for the development of radiation countermeasures.
- Singh VK, Newman VL, Berg AN. Animal models for acute radiation syndrome drug discovery. Expert Opin Drug Discov. 2015;10:497–517. doi: 10.1517/17460441.2015.1023290. [DOI] [PubMed] [Google Scholar]
- U.S. Food and Drug Administration [[cited 2015 Oct 23;]];Guidance for industry and FDA staff: qualification process for drug development tools. 2014 http://www.fda.gov/downloads/drugs/guidancecomplianceregulatoryinformation/guidances/ucm230597.pdf Available from:
- U.S. Food and Drug Administration [[cited 2015 Oct 23;]];Guidance on pharmacogenomic data submissions examples of voluntary submissions or submissions required under 21 CFR 312, 314, or 601. 2005 http://www.fda.gov/downloads/drugs/guidancecomplianceregulatoryinformation/guidances/ucm079851.pdf Available from:
- U.S. Food and Drug Administration [[cited 2015 Oct 25;]];Table of pharmacogenomic biomarkers in drug labeling. 2015 http://www.fda.gov/drugs/scienceresearch/researchareas/pharmacogenetics/ucm083378.htm Available from:
- European Medicines Agency [[cited 2015 Oct 25;]];Qualification of novel methodologies for medicine development. 2015 http://www.ema.europa.eu/ema/index.jsp?curl=pages/regulation/document_listing/document_listing_000319.jsp&mid=WC0b01ac0580022bb0 Available from:
- Pharmaceutical and Medical Devices Agency [[cited 2015 Oct 25;]];Record of consultations on pharmacogenomics/biomarkers. 2010 https://www.pmda.go.jp/english/review-services/consultations/0001.html Available from:
- Wang X, Zhang J, Fu J. Role of ROS-mediated autophagy in radiation-induced bystander effect of hepatoma cells. Int J Radiat Biol. 2015;91:452–458. doi: 10.3109/09553002.2015.1012308. [DOI] [PubMed] [Google Scholar]
- Singh VK, Fatanmi OO, Singh PK. Role of radiation-induced granulocyte colony-stimulating factor in recovery from whole body gamma-irradiation. Cytokine. 2012;58:406–414. doi: 10.1016/j.cyto.2012.03.011. [DOI] [PubMed] [Google Scholar]
- Singh VK, Ducey EJ, Fatanmi OO. CBLB613: a TLR 2/6 agonist, natural lipopeptide of Mycoplasma arginini, as a novel radiation countermeasure. Radiat Res. 2012;177:628–642. doi: 10.1667/rr2657.1. [DOI] [PubMed] [Google Scholar]
- Singh VK, Christensen J, Fatanmi OO. Myeloid progenitors: a radiation countermeasure that is effective when initiated days after irradiation. Radiat Res. 2012;177:781–791. doi: 10.1667/rr2894.1. [DOI] [PubMed] [Google Scholar]
- Singh VK, Brown DS, Kao TC. Alpha-tocopherol succinate protects mice from gamma-radiation by induction of granulocyte-colony stimulating factor. Int J Radiat Biol. 2010;86:12–21. doi: 10.3109/09553000903264515. [DOI] [PubMed] [Google Scholar]
- Krivokrysenko VI, Shakhov AN, Singh VK. Identification of granulocyte colony-stimulating factor and interleukin-6 as candidate biomarkers of CBLB502 efficacy as a medical radiation countermeasure. J Pharmacol Exp Ther. 2012;343:497–508. doi: 10.1124/jpet.112.196071. [DOI] [PMC free article] [PubMed] [Google Scholar]
•• Based on murine, canine, and NHP models, this study suggests that G-CSF and IL-6, along with neutrophil counts, can serve as efficacy biomarkers for CBLB502.
- Singh PK, Wise SY, Ducey EJ. Radioprotective efficacy of tocopherol succinate is mediated through granulocyte-colony stimulating factor. Cytokine. 2011;56:411–421. doi: 10.1016/j.cyto.2011.08.016. [DOI] [PubMed] [Google Scholar]
- Ossetrova NI, Sandgren DJ, Blakely WF. Protein biomarkers for enhancement of radiation dose and injury assessment in nonhuman primate total-body irradiation model. Radiat Prot Dosimetry. 2014;159:61–76. doi: 10.1093/rpd/ncu165. [DOI] [PubMed] [Google Scholar]
- Redon CE, Nakamura AJ, Martin OA. Recent developments in the use of gamma-H2AX as a quantitative DNA double-strand break biomarker. Aging (Albany NY) 2011;3:168–174. doi: 10.18632/aging.100284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossetrova NI, Condliffe DP, Ney PH. Early-response biomarkers for assessment of radiation exposure in a mouse total-body irradiation model. Health Phys. 2014;106:772–786. doi: 10.1097/HP.0000000000000094. [DOI] [PubMed] [Google Scholar]
- Blakely WF, Sandgren DJ, Nagy V. Further biodosimetry investigations using murine partial-body irradiation model. Radiat Prot Dosimetry. 2014;159:46–51. doi: 10.1093/rpd/ncu127. [DOI] [PubMed] [Google Scholar]
- Ha CT, Li XH, Fu D. Circulating interleukin-18 as a biomarker of total-body radiation exposure in mice, minipigs, and nonhuman primates (NHP) PLoS One. 2014;9:e109249. doi: 10.1371/journal.pone.0109249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Blakely WF, Cucinotta FA. HEMODOSE: a biodosimetry tool based on multi-type blood cell counts. Health Phys. 2015;109:54–68. doi: 10.1097/HP.0000000000000295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KF, Siu LL, Ainsbury E. Cytogenetic biodosimetry: what it is and how we do it. Hong Kong Med J. 2013;19:168–173. [PubMed] [Google Scholar]
- Vral A, Fenech M, Thierens H. The micronucleus assay as a biological dosimeter of in vivo ionising radiation exposure. Mutagenesis. 2011;26:11–17. doi: 10.1093/mutage/geq078. [DOI] [PubMed] [Google Scholar]
- Puig R, Barrios L, Pujol M. Suitability of scoring PCC rings and fragments for dose assessment after high-dose exposures to ionizing radiation. Mutat Res. 2013;757:1–7. doi: 10.1016/j.mrgentox.2013.03.013. [DOI] [PubMed] [Google Scholar]
- Burnett AF, Biju PG, Lui H. Oral interleukin 11 as a countermeasure to lethal total-body irradiation in a murine model. Radiat Res. 2013;180:595–602. doi: 10.1667/RR13330.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawar SA, Shao L, Chang J. C/EBPdelta deficiency sensitizes mice to ionizing radiation-induced hematopoietic and intestinal injury. PLoS One. 2014;9:e94967. doi: 10.1371/journal.pone.0094967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herodin F, Richard S, Grenier N. Assessment of total- and partial-body irradiation in a baboon model: preliminary results of a kinetic study including clinical, physical, and biological parameters. Health Phys. 2012;103:143–149. doi: 10.1097/HP.0b013e3182475e54. [DOI] [PubMed] [Google Scholar]
- Rana S, Kumar R, Sultana S. Radiation-induced biomarkers for the detection and assessment of absorbed radiation doses. J Pharm Bioallied Sci. 2010;2:189–196. doi: 10.4103/0975-7406.68500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz HM, Williams BB, Zaki BI. Clinical EPR: unique opportunities and some challenges. Acad Radiol. 2014;21:197–206. doi: 10.1016/j.acra.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz HM, Williams BB, Flood AB. Overview of the principles and practice of biodosimetry. Radiat Environ Biophys. 2014;53:221–232. doi: 10.1007/s00411-014-0522-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakely WF, Ossetrova NI, Whitnall MH. Multiple parameter radiation injury assessment using a nonhuman primate radiation model-biodosimetry applications. Health Phys. 2010;98:153–159. doi: 10.1097/HP.0b013e3181b0306d. [DOI] [PubMed] [Google Scholar]
- Blakely WF, Madrid JP, Sandgren DJ. Biodosimetry medical recording-use of the Biodosimetry Assessment Tool. Health Phys. 2010;99(Suppl 5):S184–S191. doi: 10.1097/HP.0b013e3181f26895. [DOI] [PubMed] [Google Scholar]
- Coy SL, Cheema AK, Tyburski JB. Radiation metabolomics and its potential in biodosimetry. Int J Radiat Biol. 2011;87:802–823. doi: 10.3109/09553002.2011.556177. [DOI] [PMC free article] [PubMed] [Google Scholar]
• This article reviews the developments in the area of metabolomics and suggests that differential mobility mass spectrometry/ion mobility spectrometry appears promising.
- Laiakis EC, Trani D, Moon BH. Metabolomic profiling of urine samples from mice exposed to protons reveals radiation quality and dose specific differences. Radiat Res. 2015;183:382–390. doi: 10.1667/RR3967.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Brenner DJ, Brown TR. Identification of urinary biomarkers from X-irradiated mice using NMR spectroscopy. Radiat Res. 2011;175:622–630. doi: 10.1667/RR2388.1. [DOI] [PubMed] [Google Scholar]
- Laiakis EC, Hyduke DR, Fornace AJ. Comparison of mouse urinary metabolic profiles after exposure to the inflammatory stressors gamma radiation and lipopolysaccharide. Radiat Res. 2012;177:187–199. doi: 10.1667/rr2771.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
•• Metabolomics has the potential to distinguish between different inflammatory responses based on differential ion signatures.
- Manna SK, Krausz KW, Bonzo JA. Metabolomics reveals aging-associated attenuation of noninvasive radiation biomarkers in mice: potential role of polyamine catabolism and incoherent DNA damage-repair. J Proteome Res. 2013;12:2269–2281. doi: 10.1021/pr400161k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyburski JB, Patterson AD, Krausz KW. Radiation metabolomics. 1. Identification of minimally invasive urine biomarkers for gamma-radiation exposure in mice. Radiat Res. 2008;170:1–14. doi: 10.1667/RR1265.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyburski JB, Patterson AD, Krausz KW. Radiation metabolomics. 2. Dose- and time-dependent urinary excretion of deaminated purines and pyrimidines after sublethal gamma-radiation exposure in mice. Radiat Res. 2009;172:42–57. doi: 10.1667/RR1703.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudarzi M, Weber W, Mak TD. Development of urinary biomarkers for internal exposure by cesium-137 using a metabolomics approach in mice. Radiat Res. 2014;181:54–64. doi: 10.1667/RR13479.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudarzi M, Mak TD, Chen C. The effect of low dose rate on metabolomic response to radiation in mice. Radiat Environ Biophys. 2014;53:645–657. doi: 10.1007/s00411-014-0558-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudarzi M, Weber WM, Mak TD. A comprehensive metabolomic investigation in urine of mice exposed to strontium-90. Radiat Res. 2015;183:665–674. doi: 10.1667/RR14011.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
• This is the first in vivo metabolomics study to evaluate the effects of exposure to 90Sr using urine, the most easily accessible biofluid.
- Laiakis EC, Strassburg K, Bogumil R. Metabolic phenotyping reveals a lipid mediator response to ionizing radiation. J Proteome Res. 2014;13:4143–4154. doi: 10.1021/pr5005295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laiakis EC, Mak TD, Anizan S. Development of a metabolomic radiation signature in urine from patients undergoing total body irradiation. Radiat Res. 2014;181:350–361. doi: 10.1667/RR13567.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
• This is the first radiation metabolomics study in human urine for the use of metabolomics in biodosimetry and biomarker identification based on the overlap between animal models and humans.
- Broin PÓ, Vaitheesvaran B, Saha S. Intestinal microbiota-derived metabolomic blood plasma markers for prior radiation injury. Int J Radiat Oncol Biol Phys. 2015;91:360–367. doi: 10.1016/j.ijrobp.2014.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh C, Yu DK, Kim I. The biological response of spermidine induced by ionization radiation. Molecules. 2012;17:145–150. doi: 10.3390/molecules17010145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudarzi M, Weber WM, Mak TD. Metabolomic and lipidomic analysis of serum from mice exposed to an internal emitter, cesium-137, using a shotgun LC-MS(E) approach. J Proteome Res. 2015;14:374–384. doi: 10.1021/pr500913n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland IJ, Broin PO, Golden A. Integrative metabolic signatures for hepatic radiation injury. PLoS One. 2015;10:e0124795. doi: 10.1371/journal.pone.0124795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh SP, Singh R, Chakraborty K. Metabolomic changes in gastrointestinal tissues after whole body radiation in a murine model. Mol Biosyst. 2013;9:723–731. doi: 10.1039/c3mb25454b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu D, May R, Sureban SM. Inhibition of Notch signaling reduces the number of surviving Dclk1+ reserve crypt epithelial stem cells following radiation injury. Am J Physiol Gastrointest Liver Physiol. 2014;306:G404–G411. doi: 10.1152/ajpgi.00088.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureban SM, May R, Qu D. Dietary pectin increases intestinal crypt stem cell survival following radiation injury. PLoS One. 2015;10:e0135561. doi: 10.1371/journal.pone.0135561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Kim J, Park SJ. High-throughput 16S rRNA gene sequencing reveals alterations of mouse intestinal microbiota after radiotherapy. Anaerobe. 2015;33:1–7. doi: 10.1016/j.anaerobe.2015.01.004. [DOI] [PubMed] [Google Scholar]
- Lam V, Moulder JE, Salzman NH. Intestinal microbiota as novel biomarkers of prior radiation exposure. Radiat Res. 2012;177:573–583. doi: 10.1667/rr2691.1. [DOI] [PubMed] [Google Scholar]
- Amundson SA, Do KT, Shahab S. Identification of potential mRNA biomarkers in peripheral blood lymphocytes for human exposure to ionizing radiation. Radiat Res. 2000;154:342–346. doi: 10.1667/0033-7587(2000)154[0342:iopmbi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Amundson SA, Do KT, Fornace AJ., Jr Induction of stress genes by low doses of gamma rays. Radiat Res. 1999;152:225–231. [PubMed] [Google Scholar]
- Kang CM, Park KP, Song JE. Possible biomarkers for ionizing radiation exposure in human peripheral blood lymphocytes. Radiat Res. 2003;159:312–319. doi: 10.1667/0033-7587(2003)159[0312:pbfire]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Cortez MA, Bueso-Ramos C, Ferdin J. MicroRNAs in body fluids–the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8:467–477. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chim SS, Shing TK, Hung EC. Detection and characterization of placental microRNAs in maternal plasma. Clin Chem. 2008;54:482–490. doi: 10.1373/clinchem.2007.097972. [DOI] [PubMed] [Google Scholar]
- Islam A, Ghimbovschi S, Zhai M. An exploration of molecular correlates relevant to radiation combined skin-burn trauma. PLoS One. 2015;10:e0134827. doi: 10.1371/journal.pone.0134827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob NK, Cooley JV, Yee TN. Identification of sensitive serum microRNA biomarkers for radiation biodosimetry. PLoS One. 2013;8:e57603. doi: 10.1371/journal.pone.0057603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XH, Ha CT, Fu D. Delta-tocotrienol suppresses radiation-induced microRNA-30 and protects mice and human CD34+ cells from radiation injury. PLoS One. 2015;10:e0122258. doi: 10.1371/journal.pone.0122258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya SS, Fendler W, Watson J. Serum microRNAs are early indicators of survival after radiation-induced hematopoietic injury. Sci Transl Med. 2015;7:287ra69. doi: 10.1126/scitranslmed.aaa6593. [DOI] [PMC free article] [PubMed] [Google Scholar]
•• Serum miRNAs can serve as functional dosimeters for early assessment of radiation-induced hematopoietic damage and timely use of medical countermeasures to mitigate the long-term impact of radiation.
- Cui W, Ma J, Wang Y. Plasma miRNA as biomarkers for assessment of total-body radiation exposure dosimetry. PLoS One. 2011;6:e22988. doi: 10.1371/journal.pone.0022988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Peng G. Non-coding RNAs: an emerging player in DNA damage response. Mutat Res Rev Mutat Res. 2015;763:202–211. doi: 10.1016/j.mrrev.2014.11.003. [DOI] [PubMed] [Google Scholar]
- Kabacik S, Manning G, Raffy C. Time, dose and ataxia telangiectasia mutated (ATM) status dependency of coding and noncoding RNA expression after ionizing radiation exposure. Radiat Res. 2015;183:325–337. doi: 10.1667/RR13876.1. [DOI] [PubMed] [Google Scholar]
- Nie J, Peng C, Pei W. A novel role of long non-coding RNAs in response to X-ray irradiation. Toxicol In Vitro. 2015 doi: 10.1016/j.tiv.2015.09.007. [DOI] [PubMed] [Google Scholar]
- Krivokrysenko VI, Toshkov IA, Gleiberman AS. The Toll-like receptor 5 agonist Entolimod mitigates lethal acute radiation syndrome in non-human primates. PLoS One. 2015;10:e0135388. doi: 10.1371/journal.pone.0135388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni SS, Cary LH, Gambles K. Gamma-tocotrienol, a radiation prophylaxis agent, induces high levels of granulocyte colony-stimulating factor. Int Immunopharmacol. 2012;14:495–503. doi: 10.1016/j.intimp.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Kulkarni S, Singh PK, Ghosh SP. Granulocyte colony-stimulating factor antibody abrogates radioprotective efficacy of gamma-tocotrienol, a promising radiation countermeasure. Cytokine. 2013;62:278–285. doi: 10.1016/j.cyto.2013.03.009. [DOI] [PubMed] [Google Scholar]
- Singh VK, Romaine PL, Newman VL. Tocols induce G-CSF and mobilise progenitors that mitigate radiation injury. Radiat Prot Dosimet. 2014;162:83–87. doi: 10.1093/rpd/ncu223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh VK, Beattie LA, Seed TM. Vitamin E: tocopherols and tocotrienols as potential radiation countermeasures. J Radiat Res. 2013;54:973–988. doi: 10.1093/jrr/rrt048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grace MB, Singh VK, Rhee JG. 5-AED enhances survival of irradiated mice in a G-CSF-dependent manner, stimulates innate immune cell function, reduces radiation-induced DNA damage and induces genes that modulate cell cycle progression and apoptosis. J Radiat Res. 2012;53:840–853. doi: 10.1093/jrr/rrs060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakhov AN, Singh VK, Bone F. Prevention and mitigation of acute radiation syndrome in mice by synthetic lipopeptide agonists of Toll-like receptor 2 (TLR2) PLoS One. 2012;7:e33044. doi: 10.1371/journal.pone.0033044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh VK, Shafran RL, Inal CE. Effects of whole-body gamma irradiation and 5-androstenediol administration on serum G-CSF. Immunopharmacol Immunotoxicol. 2005;27:521–534. doi: 10.1080/08923970500416707. [DOI] [PubMed] [Google Scholar]
- Singh PK, Wise SY, Ducey EJ. Alpha-tocopherol succinate protects mice against radiation-induced gastrointestinal injury. Radiat Res. 2012;177:133–145. doi: 10.1667/rr2627.1. [DOI] [PubMed] [Google Scholar]
- Singh VK, Wise SY, Scott JR. Radioprotective efficacy of delta-tocotrienol, a vitamin E isoform, is mediated through granulocyte colony-stimulating factor. Life Sci. 2014;98:113–122. doi: 10.1016/j.lfs.2014.01.065. [DOI] [PubMed] [Google Scholar]
- Srivastava A, Leighton X, Eidelman O. Personalized radioproteomics: identification of a protein biomarker signature for preemptive rescue by tocopherol succinate in CD34-positive irradiated progenitor cells isolated from a healthy control donor. J Proteomics Bioinform. 2015;8 doi: 10.4172/jpb.1000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCarlo AL, Jackson IL, Shah JR. Development and licensure of medical countermeasures to treat lung damage resulting from a radiological or nuclear incident. Radiat Res. 2012;177:717–721. doi: 10.1667/rr2881.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Narayanan J, Joneikis C. Enalapril mitigates focal alveolar lesions, a histological marker of late pulmonary injury by radiation to the lung. Radiat Res. 2013;179:465–474. doi: 10.1667/RR3127.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JW, Scott AJ, Tudor G. Identification and quantitation of biomarkers for radiation-induced injury via mass spectrometry. Health Phys. 2014;106:106–119. doi: 10.1097/HP.0b013e3182a4ed3b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herodin F, Valente M, Abend M. Useful radiation dose biomarkers for early identification of partial-body exposures. Health Phys. 2014;106:750–754. doi: 10.1097/HP.0000000000000059. [DOI] [PubMed] [Google Scholar]
- Shim S, Jang WS, Lee SJ. Development of a new minipig model to study radiation-induced gastrointestinal syndrome and its application in clinical research. Radiat Res. 2014;181:387–395. doi: 10.1667/RR13207.1. [DOI] [PubMed] [Google Scholar]
•• This paper discusses minipig model for studying GI ARS.
- Kiang JG, Jiao W, Cary LH. Wound trauma increases radiation-induced mortality by activation of iNOS pathway and elevation of cytokine concentrations and bacterial infection. Radiat Res. 2010;173:319–332. doi: 10.1667/RR1892.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCarlo AL, Maher C, Hick JL. Radiation injury after a nuclear detonation: medical consequences and the need for scarce resources allocation. Disaster Med Public Health Prep. 2011;5(Suppl 1):S32–S44. doi: 10.1001/dmp.2011.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCarlo AL, Ramakrishnan N, Hatchett RJ. Radiation combined injury: overview of NIAID research. Health Phys. 2010;98:863–867. doi: 10.1097/HP.0b013e3181a6ee32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiang JG, Garrison BR, Burns TM. Wound trauma alters ionizing radiation dose assessment. Cell Biosci. 2012;2:20. doi: 10.1186/2045-3701-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak TD, Tyburski JB, Krausz KW. Exposing to ionizing radiation reveals global dose and time-dependent changes in the urinary metabolome of rat. Metabolomics. 2015;11:1082–1094. doi: 10.1007/s11306-014-0765-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannkuk EL, Laiakis EC, Authier S. Global metabolomic identification of long-term dose-dependent urinary biomarkers in nonhuman primates exposed to ionizing radiation. Radiat Res. 2015;184:121–133. doi: 10.1667/rr14091.1. [DOI] [PMC free article] [PubMed] [Google Scholar]


