SUMMARY
The eukaryotic initiation factor eIF5A is a translation factor that, unusually, has been assigned functions in both initiation and elongation. Additionally, it is implicated in transcription, mRNA turnover and nucleocytoplasmic transport. Two eIF5A isoforms are generated from distinct but related genes. The major isoform, eIF5A1, is considered constitutive, is abundantly expressed in most cells, and is essential for cell proliferation. The second isoform, eIF5A2, is expressed in few normal tissues but is highly expressed in many cancers and has been designated a candidate oncogene. Elevated expression of either isoform carries unfavorable prognostic implications for several cancers, and both have been advanced as cancer biomarkers. The amino acid hypusine, a presumptively unique eIF5A post-translational modification, is required for most known eIF5A functions and it renders eIF5A susceptible to inhibitors of the modification pathway as therapeutic targets. eIF5A has been shown to regulate a number of gene products specifically, termed the eIF5A regulon, and its role in translating proline-rich sequences has recently been identified. A model is advanced that accommodates eIF5A in both the initiation and elongation phases of translation. We review here the biochemical functions of eIF5A, the relationship of its isoforms with human cancer, and evolving clinical applications.
Keywords: protein synthesis, tumorigenesis, eukaryotic initiation factor eIF5A, hypusine modification, translational control, cancer therapeutics
Introduction
Eukaryotic translation initiation factor 5A (eIF5A) is as enigmatic as it is unique. Known for some 40 years, its functions remain an active topic of research in diverse arenas encompassing such fields as biochemistry, cell biology, development and oncology. In humans, it is involved in the cell cycle, apoptosis, and viral replication – most notably of HIV-1. eIF5A is present in both the eukaryotic and archaeal domains of life, but its bacterial homolog is an elongation factor known as EF-P [1], and eIF5A has also been implicated in polypeptide chain elongation.
eIF5A is an abundant, small, highly conserved, RNA-binding protein. It associates with ribosomes and with the cytoskeleton, and shuttles between the nucleus and cytoplasm. Remarkably, it contains the amino acid hypusine which has not been detected in any other protein. Hypusine (a portmanteau word merging hydroxyputrescine with lysine) is formed by post-translational modification of a specific lysine residue of eIF5A. Sequential reactions attach and hydroxylate an aminobutyl moiety derived from spermidine to the lysine side-chain (Figure 1). Hypusine appears to be essential to most, if not all, of eIF5A's functions, and the two enzymes responsible for eIF5A modification appear to be dedicated to this pathway, underlining its importance and providing targets for inhibitors and therapeutic intervention. In addition, eIF5A is subject to other modifications which are less well studied and whose implications are less well understood.
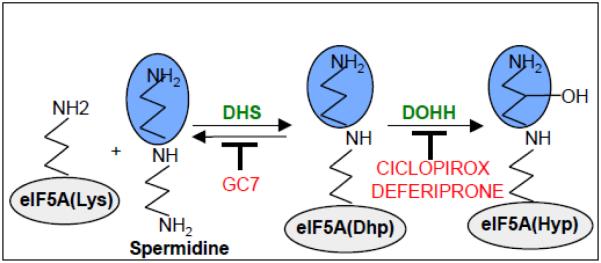
Figure 1. eIF5A modification pathway and inhibitors.
Deoxyhypusine synthase (DHS) catalyzes aminobutyl (blue ) transfer from spermidine to the ε-amino group of lysine-50 of human eIF5A (gray ) using NAD+ as cofactor, yielding deoxyhypusine (Dhp). Deoxyhypusine hydroxylase (DOHH) hydroxylates Dhp to hypusine (Hpu) in an Fe(II)-dependent reaction using molecular oxygen. The spermidine analog GC7 (N1-guanyl-1,7-diaminoheptane) inhibits DHS. The drugs ciclopirox (6-cyclohexyl-1-hydroxy-4-methyl-(1H)-pyrid-2-one) and deferiprone (1,2-dimethyl-3-hydroxypyridin-4-one) inhibit DOHH. (Modified from Hoque et al. [34]; reproduction permitted by BioMed Central.)
Eukaryotes typically harbor two paralogous genes encoding distinct eIF5A isoforms, eIF5A1 and eIF5A2, both of which are hypusinated (see [2] and [3] for reviews). The major form, eIF5A1, is very abundant in most cells and tissues, and essentially all biochemical studies of eIF5A have been carried out with this form. eIF5A2 is rare in most normal tissues but conspicuous in many malignancies. Both isoforms are up-regulated in some tumors, and the association of eIF5A2 with cancer has received considerable attention [4]. Whether and how their differential distribution and sequence differences relate to the physiological and pathophysiological roles of the two isoforms are important questions.
In this article, we review the discovery and functions of eIF5A, with particular emphasis on malignancy, its role as a biomarker, and the potential for cancer therapy. An underlying theme is that the normal functions of the two eIF5A isoforms in proliferation, growth and development have been usurped during the selection and expansion of cancer cells.
Isolation and properties of eIF5A
eIF5A was first identified in 1976 as a mammalian protein that stimulates the translation of poly-uridine mRNA in vitro [5]. The factor was then named IF-M2Bα, later eIF4D, and subsequently eIF5A or eIF5A1 as used here. With a similar translation initiation assay comprising an 80S initiation complex formed with Met-tRNAi, AUG and purified initiation factors, the formation of methionyl-puromycin (analogous to making the first peptide bond) was promoted by eIF5A [6, 7]. While named as an initiation factor, its actual function in these assays is the stimulation of the peptidyl transferase reaction. It was speculated that the requirement for eIF5A was due to the unusual nature of the ribosomal complex involved in formation of the first peptide bond: a positive charge on the aminoacyl-tRNA in the ribosomal P site; and the absence of tRNA in the ribosomal E site.
Mammalian eIF5A exhibits a molecular mass of 16.7 kDa, is acidic (pl = 5.4) and is one of the most abundant of the initiation factors [7, 8]. The human gene encoding eIF5A1 (EIF5A1) was cloned and sequenced [9, 10] and the second human eIF5A gene (EIF5A2) was characterized several years later [11, 12]. In contrast to eIF5A1, which is ubiquitously expressed, eIF5A2 is rare apart from in testis and parts of the brain, and in malignancy [11-13]. The two human eIF5A forms share 84% sequence identity and are 94% similar [13]. eIF5A is found in all eukaryotic species examined and is conserved in sequence from yeast to humans [14]. Two genes from the yeast Saccharomyces cerevisiae have been identified and sequenced as well [15]. The two yeast eIF5A proteins share 90% sequence identify, but differ in their sequences near the C-terminus and in their interactions with other proteins. Both human eIF5A isoforms can individually support the growth of yeast lacking its own eIF5A genes, and the yeast protein functions in vitro in the mammalian eIF5A assay system [16], suggesting that eIF5A activities are functionally interchangeable – to a degree, at least – within and across species.
Formation and role of hypusine
A single lysine residue of eIF5A is modified to form hypusine [1], [17] (Figure 1). This entails two reactions: the transfer of an aminobutyl group from spermidine to the ε-amino group of lysine-50 (in humans) to form deoxyhypusine, catalyzed by deoxyhypusine synthase (DHS; EC 2.5.1.46); and subsequent hydroxylation of the aminobutyl group, catalyzed by deoxyhypusine hydroxylase (DOHH; EC 1.14.99.29). Thus eIF5A(Lys) is converted stepwise to eIF5A(Dhp) and then to mature eIF5A, sometimes called eIF5A(Hyp). These modifications appear to be unique to eIF5A (both isoforms) as no other similarly modified protein has been detected in any organism. However, caution is needed, as it is conceivable that a low-abundant protein might be similarly modified whose detection relative to the highly abundant eIF5A could be missed.
All of the eukaryotic species examined show the ability to synthesize eIF5A(Hyp); similarly, archaea possess aIF5A(Hyp), but bacteria lack eIF5A and fail to generate hypusine [1]. The hypusine modification is required for human eIF5A activity in vitro, although eIF5A(Dhp) is partially active [9]. Deletion of the eIF5A gene or DHS gene is lethal in S. cerevisiae, Caenorhabditis elegans, and mice, consistent with the in vitro results. DOHH deletion is embryonically lethal in C. elegans, Drosophila melanogaster, and mice [18], yet the protein is not essential in yeasts although their growth is impaired [19, 20]. Correspondingly, hydroxylation of EF-P, discussed below, is not essential for peptide bond formation [21, 22]. This raises the question why hydroxylation occurs. Because eIF5A(Dhp) can be converted back to eIF5A(Lys), but eIF5A(Hyp) cannot, this suggests that hydroxylation is needed to preserve the modification [18]. However, inhibition of DOHH leads to the accumulation of eIF5A(Dhp), implying that reversal of the DHS-catalyzed reaction, converting eIF5A(Dhp) back to eIF5A(Lys), is not always rapid [23].
In addition, eIF5A can be modified by transglutaminylation [24], acetylation [25, 26] and phosphorylation [27, 28], but clear effects on its activity have not been detected.
High-resolution structures of eIF5A have been solved for the human factor [29] as well as from numerous other species (reviewed in [3]). The N-terminal domain is positively charged and contains 6 β-strands that form a β-barrel. The domain also contains a flexible loop with the hypusine residue surrounded by a highly conserved sequence. The less-conserved C-terminal domain contains an OB fold β-barrel domain and is negatively charged. Although no high-resolution structure is available for eIF5A bound to the 80S ribosome, hydroxyl radical probing indicates that the protein is bound near the ribosomal E site. Further insight is obtained from studies of EF-P, a bacterial ortholog of eIF5A that lacks the hypusine modification but is β-lysinylated and hydroxylated instead [1], [30]. EF-P stimulates fMet-puromycin synthesis and is thought to promote early peptide bond formation events [31]. A high-resolution structure for EF-P bound to ribosomes indicates that the protein is located in the E site [32]. Based on this structure, together with hydroxyl radical probing, eIF5A was modeled onto the ribosome in the E site, with its flexible hypusine loop positioned near the peptidyl transferase center and the P site tRNA [33] (Figure 2). These structural studies are reviewed in detail [3] and provide key insights into eIF5A function.
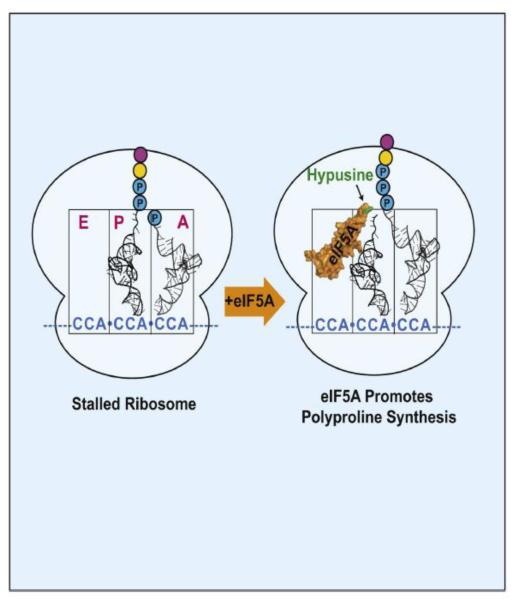
Figure 2. Model of eIF5A action during polypeptide chain elongation.
In S. cerevisiae, eIF5A relieves ribosomal stalling at sequences containing consecutive proline residues (blue-circled P). The model depicts a ribosome stalled at a run of proline codons after condensing 2 proline residues in a nascent polypeptide chain and before the addition of a third proline. The peptidyl-tRNA is in the ribosomal P site and the next residue, on Pro-tRNAPro, is in the A site (left). eIF5A (brown) binds near the ribosomal E site with its hypusine side chain adjacent to the peptidyl-tRNA in the peptidyl transferase center of the ribosome where it can facilitate peptide bond formation (right). (Reprinted from Guttierez et al. [33] with permission from Elsevier.)
Functions of eIF5A
eIF5A clearly is a factor that promotes protein synthesis, but its detailed molecular mechanism is not yet rigorously elucidated. Genetic and reverse genetic studies indicate that eIF5A also participates in other mRNA-related functions – transcription [34, 35], mRNA turnover [36, 37] and nucleocytoplasmic transport [38] – although these are less well defined. The extent to which these processes are affected through eIF5A action during protein synthesis, as opposed to entirely different mechanisms, remains to be elucidated. Based on the methionyl-puromycin assay used in its isolation, eIF5A was called an initiation factor. However, in this assay it appears not to affect the formation of the 80S initiation complex, but rather stimulates peptidyl transferase activity. When yeast eIF5A is partially depleted in vivo, ribosomes shift into lighter polysomes, a phenomenon largely dependent on growth in rich medium where initiation is thought to be more robust [39]. When lysates from such depleted cells are tested in vitro, addition of purified eIF5A stimulates protein synthesis dependent on new initiation [39]. These findings support the view that eIF5A acts at or near the initiation phase of protein synthesis.
Recent studies suggest that eIF5A functions during the elongation phase of protein synthesis. Depletion of yeast eIF5A or partial inactivation of temperature-sensitive mutants results in inhibition of protein synthesis but retention of polysomes [33, 40]. Furthermore, depletion prevents the disassembly of polysomes that normally result from inhibition of initiation by arsenite [41]. In vitro studies with purified components find that tripeptide synthesis is stimulated by eIF5A [40]. Finally, genetic studies show an interaction between eIF5A and eEF2 [42], consistent with involvement during elongation.
Prolines and the ribosomal P site
Fresh insight into how eIF5A might affect elongation was obtained from studies of bacterial EF-P [21, 43]. This factor promotes the synthesis of proline- and glycine- containing peptides by ribosomes, and is all but essential for them to translate oligo-proline regions in proteins, specifically Pro-Pro-Pro and Pro-Pro-Gly. Yeast eIF5A also plays a role in the synthesis of oligo-proline motifs, both in vivo and in vitro [33], as illustrated in Figure 2. Toe-printing experiments show that ribosomes stall when peptidyl-tRNAPro resides in the P site and Pro-tRNAPro resides in the A site [44]. Examination of the yeast proteome for Pro-Pro-Pro/Gly motifs showed about 12.7% of proteins would be expected to have their synthesis stimulated by eIF5A [3]. The frequency of such motifs appears greater in humans, where about 10,000 motifs are found in 18,000 proteins [45]. In a genome-wide search for proteins with Pro-Pro-Pro and Pro-Pro-Gly sequences, the frequency of such motifs was found to be progressively higher as the organisms increased in complexity, suggesting a role for eIF5A in eukaryotic evolution [46].
Numerous stalling sites have been characterized in mouse embryonic stem cell mRNAs [47]. Many of these are proline-rich although they do not routinely correspond to the canonical Pro-Pro-Pro/Gly signals. It is not known if the murine sites are eIF5A-related. However, EF-P-dependent stalling sites in bacteria can be more complex than the canonical signal [48-51]. For example, distinct sequences and context effects are being uncovered. Whether the same is true for mammalian eIF5A is an open question. If stimulation of stalled ribosomes is the major function of eIF5A, this suggests that eIF5A will affect the translation of only a subset of mRNAs, those susceptible to stalling. Partial depletion of yeast eIF5A has only a moderate effect on overall protein synthesis, yet affects cell proliferation strongly, suggesting that proteins required for proliferation may be disproportionally affected [52]. Similarly, experiments using the toxin mimosine or the drugs ciclopirox and deferiprone as inhibitors of DOHH (Figure 1) support the idea that hypusinated eIF5A is required for the translation of specific mRNAs in human cells [23, 53]. Although selective binding of human mRNAs to eIF5A has been documented [54], connections between these mechanistic findings and cellular behavior still need to be solidified.
The claims that eIF5A functions either in initiation or elongation may be reconciled by postulating that it promotes both phases of protein synthesis. In the two sets of claims, the role played by eIF5A appears to be to insert its hypusine loop into the ribosome in order to “correct” a conformation caused by abnormal occupants in the P site, namely non-acylated aminoacyl-tRNA during initiation, and distortions caused by peptidyl-prolyl-tRNA. Since the docking models indicate that the binding of eIF5A on the 80S ribosome is expected to be inhibited by tRNA in the E site, this problem is not present during initiation. During elongation, the tRNA would presumably have to dissociate before eIF5A could bind. Neither the rates of tRNA dissociation from the E site, nor the rates of eIF5A binding to ribosomes, are known. However, it is tempting to speculate that when stalling occurs during elongation, as caused by oligo-prolines, there would be sufficient time for the tRNA to dissociate, thereby enabling eIF5A to bind and “correct” the ribosome conformation to stimulate peptide bond formation. Since ribosome stalling appears to be critical for correct nascent protein folding, it likely occurs during the translation of many mRNAs. Sophisticated kinetic experiments are required to establish if any of these ideas is correct.
Over-expression of eIF5A in cancer
A strong correlation exists between increased levels of eIF5A – often referred to as over-expression – and cancer. This holds for both the eIF5A1 and eIF5A2 isoforms, each of which is associated with several malignancies (Table 1), but the relationship is especially striking in the case of eIF5A2. In contrast to eIF5A1, which is widely expressed at a high level, eIF5A2 is undetectable or only weakly expressed in most normal cells and tissues [13]. Consequently, eIF5A2 over-expression is readily manifest in many tumors, to the extent that it has been deemed a candidate oncogene [12, 13, 55]. eIF5A1 is also often associated with cell proliferation and it can serve as a marker for malignant growth.
Table 1.
Cancer associations of the two eIF5A isoforms.
| Cancer type | eIF5A isoform |
Clinical, pathological and biological correlates |
Molecular correlates |
Refs. |
|---|---|---|---|---|
| Bladder urothelial carcinoma |
eIF5A2 | Stage, grade, recurrence, progression, EMT, metastasis |
TGFβ1, STAT3 |
59, 76, 77 |
| Cervical carcinoma/ adenocarcinoma |
eIF5A1 | Cell proliferation | Hsp27, NM23, DJ-1, PRDX2, TrpRS, IκB |
23 |
| Chronic myeloid leukemia |
eIF5A1 | DHS, DOHH |
67, 68 |
|
| Colorectal adenoma/carcinoma |
eIF5A1 | Prognosis | AnnexinA3, S100A11, S100P, galectin-1, S100A9, FABPL, IMPDH2, CK20, MAP3K8, CHIP/Stub1 |
62, 63, 64 |
| Colorectal carcinoma |
eIF5A2 | Metastasis, EMT, motility, progression |
MTA1, c-myc |
75, 83 |
| Esophageal squamous cell carcinoma |
eIF5A2 | Metastasis, EMT, hypoxia |
HIF1α, VEGF | 35 |
| Gastric cancer | eIF5A2 | Metastasis, EMT, prognosis |
MTA1 | 82 |
| Glioblastoma | eIF5A1* | Grade-independent | DHS, DOHH | 65 |
| Hepatocellular carcinoma |
eIF5A1 | Tumor nodules | 57 | |
| Hepatocellular carcinoma |
eIF5A2 | Venous infiltration, EMT metastasis, angiogenesis |
DHS, DOHH, RhoA/Rac1, MMP-2 |
57, 78, 79 |
| Lung adenocarcinoma |
eIF5A1 | Poor differentiation, prognosis |
K-ras, p53, AOE372, ATP5D, B4GALT, PPase, GRP58, GSTM4, P4HB, TPI, UCHL1 |
56, 60 |
| Non-small cell lung cancer |
eIF5A2 | Prognosis, EMT |
80, 81 |
|
| Ovarian carcinoma | eIF5A2 | Stage, grade, transformation, tumorigenicity, prognosis |
Ki-67 |
12, 55, 74 |
| Pancreatic ductal adenocarcinoma |
eIF5A1 eIF5A2 |
Tumor growth | PEAK1 | 66 |
| Vulvar intraepithelial neoplasia |
eIF5A1 | Cell proliferation | Ki-67 | 61 |
For each cancer type, the eIF5A isoform(s) reported to be over-expressed are listed together with associated clinical, pathological and biological characteristics and with observed molecular correlations. Italicized proteins are down-regulated.
eIF5A2 was detected in 1 sample.
Expression of the eIF5A genes has been examined in a number of cancers and cancer cell lines, both at the protein and RNA levels, using a range of techniques. For protein analysis, methods include immunohistochemistry (IHC), western blotting (WB), two-dimensional gel electrophoresis (2DG) and mass spectrometry (MS). For RNA, reverse transcription/polymerase chain reaction (RT-PCR) and RNA microarray analysis (RMA) are methods of choice. Often, multiple techniques have been applied in parallel but some studies did not include protein assays, relying on surrogate RNA measures. This introduces an element of uncertainty in view of the imperfect correlation between RNA and protein levels that has been documented for eIF5A1 [56, 57] and eIF5A2 [58, 59]. It also may imply that translational controls, or other post-transcriptional regulatory mechanisms, are in play. Furthermore, interpretation of protein analyses may be complicated by antibody specificity and protein modification issues. Nonetheless, a considerable body of information has been accumulated about eIF5A expression in cancer which we summarize for eIF5A1 and eIF5A2 in turn, considering clinical associations, regulation, and the consequences of over-expression.
eIF5A1 association with cancer
eIF5A1 is up-regulated in several malignancies, as listed in Table 1 together with clinicopathological and molecular correlates that have been reported. These malignancies include lung adenocarcinoma [56, 60], vulvar intraepithelial neoplasia [61], colorectal carcinoma [62-64], glioblastoma [65], pancreatic ductal adenocarcinoma [66], and cervical cancer (both carcinoma and adenocarcinoma) [23]. Increased expression of eIF5A1 was not observed in hepatocellular carcinoma at either the RNA or protein level when tumor tissue was compared to non-malignant tissue. There was, however, a correlation between eIF5A1 RNA – but not protein – and liver tumor nodule number [57].
In addition to solid tumors, eIF5A1 mRNA is also expressed at elevated levels in peripheral blood cells from patients with chronic myeloid leukemia (CML) [67, 68]. Paradoxically, eIF5A1 and other genes in the hypusine pathway were identified as tumor suppressors in lymphoma [69]. While this conclusion is difficult to square with the protein's essential role in cell proliferation in several organisms [58], and its otherwise strong association with malignancy (Table 1), resolution of the discrepancy will undoubtedly be enlightening.
Histopathological observations support the involvement of eIF5A1 in tumor cell proliferation. In vulvar and cervical cancers, the pattern of IHC staining of malignant regions for eIF5A1 closely matches that seen with Ki-67, a standard marker of cell proliferation [23, 61]. Accordingly, eIF5A1 has been proposed as a tumor marker in vulvar and colorectal cancer [61, 62]. In lung adenocarcinoma, higher eIF5A1 protein expression is associated with a poorer prognosis and diminished cellular differentiation [56]. Similarly, in colorectal cancer, greater eIF5A1 RNA expression is also associated with less favorable outcomes – increased metastasis and rate of recurrence, and decreased survival and progression-free survival times [63]. Pathology testing for eIF5A1 protein and RNA may therefore have prognostic value in the clinic.
Regulation of eIF5A1 expression in cancer
The mechanisms whereby eIF5A1 gene expression is regulated are not well established, but relationships with oncogenes have been reported. In tissue culture, the gene has been shown to be myc-regulated [70, 71]. In lung carcinoma, increased eIF5A1 protein expression correlates with oncogenic mutations in K-ras at codons 12 and 13 [56]. Together with the increased expression of eIF5A1 in mouse NIH-3T3 cells brought about by v-HA-Ras [72], these observations point to induction of eIF5A via the K-ras signaling pathway. Treatment of cultured Bcr-Abl– positive K562 cells with imatinib, a drug that inhibits Abl tyrosine kinase, reduces the level of eIF5A1 and its mRNA [67]. This finding, and the increased level of eIF5A1 RNA in CML mentioned above, suggest that eIF5A1 may also be induced by the Bcr-Abl oncogene. On the other hand, comparison of RNA and protein expression levels in lung adenocarcinomas argues for post-transcriptional regulation of eIF5A protein expression [56]. A mechanism based on protein degradation induced by the E3 ubiquitin ligase CHIP/Stub1 has been reported, which can account for the inverse relationship between CHIP/Stub1 and eIF5A1 protein expression observed in colorectal cancers [64]. Evidently eIF5A1 is regulated by oncogene-driven transcription as well as post-transcriptionally. Additional work is needed to augment our understanding of the mechanisms that govern eIF5A1 expression in cancer.
Molecular basis of eIF5A1 action in cancer
In tumor tissue, the eIF5A1 protein has been observed in the cytoplasm of proliferating cells in vulvar neoplasia [61], but in lung tumors it is also distributed in the nuclear compartment [56]. Thus it may be involved in events taking place in the nucleus as well as in the cytoplasm. Proteins and pathways affected by eIF5A1 have been identified using techniques including 2DG, MS and RT-PCR, with results that are summarized in Table 1. For example, depletion of eIF5A1 or DOHH in cervical cancer or osteosarcoma cells and drug-mediated DOHH inhibition by deferiprone or ciclopirox lead to reduced proliferation [23]. Protein and RNA analysis identified three HeLa cell proteins that are down-regulated at the translational level: Hsp27, DJ-1 (PARK7) and NM23, all of which are cancer-associated. The heat shock chaperone Hsp27 regulates IκB and hence NF-κB activity, providing a link between eIF5A and transcriptional effects, apoptosis and HIV-1 inhibition [34, 73]. Another protein, the nonreceptor tyrosine kinase PEAK1, is also down-regulated by suppression of eIF5A synthesis (either isoform) or inhibition of hypusine formation. Concomitantly, the growth and tumorigenicity of pancreatic ductal adenocarcinoma cells is reduced [66].
While Hsp27 and PEAK1 both contain oligo-proline sequences of the type reported to cause eIF5A-conditional stalling, DJ-1 and NM23 do not. It remains to be determined whether this reflects the existence of additional, as-yet uncharacterized determinants of eIF5A dependence. Alternatively, it could be due to secondary effects of the drugs, or of interference with eIF5A function, as suggested by the finding that the synthesis of two proteins – PRDX2 and TrpRS – is up-regulated when DOHH is inhibited [23]. This correlates with increased mRNA levels, presumably resulting from diminished mRNA stability in the absence of mature eIF5A1. We have proposed that these up-regulated and down-regulated proteins are the founder members of an 'eIF5A regulon' (Figure 3) comprising possibly some hundreds of proteins that are subject to control by eIF5A at different levels [23].
Figure 3. Putative eIF5A regulon.
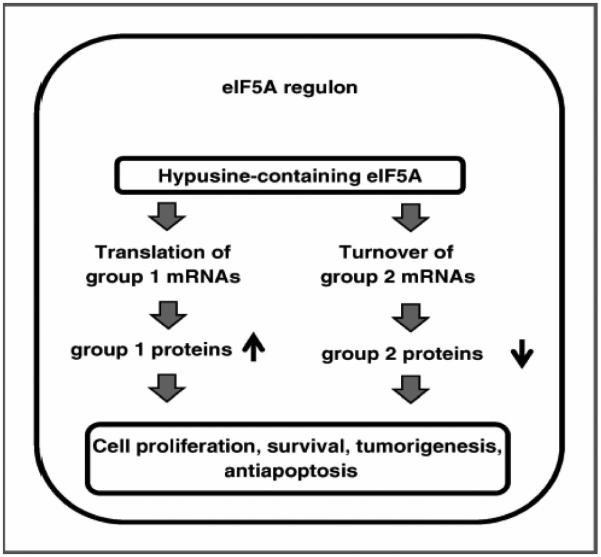
Hypusine-containing eIF5A is proposed to affect protein expression by enhancing the translation of Group 1 mRNAs (resulting in up-regulation), and the turnover of Group 2 mRNAs (resulting in down-regulation). The targets may be direct or secondary to eIF5A's biochemical action, are collectively considered the eIF5A regulon, and influence tumorigenesis and related cellular processes. (Reproduced from Mémin et al. [23] as permitted by AACR.)
eIF5A2 association with cancer
The association and functions of eIF5A2 have been studied in several tumors. eIF5A2 transcripts were initially detected by analysis of an amplified chromosomal region from ovarian cancer [12] and database searches [11]. The eIF5A2 protein was first detected by WB in an ovarian cancer cell line, where it occurs at a high level, and to a lesser extent in an adenocarcinoma cell line [13]. eIF5A2 mRNA exists in several forms and can be detected in a number of cell lines; it appears to be inefficiently translated compared to eIF5A1 [58].
eIF5A2 is weakly detectable, if at all, in normal tissues but is up-regulated in a number of malignancies (Table 1). Its over-expression, usually visualized by IHC, has been observed in cancer of the ovary [55, 74], colorectal carcinoma [75], bladder cancer [59, 76, 77], hepatocellular carcinoma [78, 79], non-small cell lung cancer [80, 81], pancreatic cancer [66], esophageal squamous cell carcinoma [35], and gastric cancer [82]. Both eIF5A1 and eIF5A2 isoforms are present in hepatocellular [57] and pancreatic [66] cancer. On the other hand, eIF5A2 is not generally overexpressed in CML [68] or glioblastoma [65], and it is poorly expressed in some lymphoma and leukemia cell lines [11, 13]. These observations argue that eIF5A2 over-expression is not an invariable hallmark of cancer and that either isoform may be sufficient for establishment of the malignant phenotype.
As with eIF5A1, higher expression of eIF5A2 is associated with less favorable clinical features and outcomes. In cancers that have been studied, elevated eIF5A2 often correlates with a more advanced and aggressive disease state, increased likelihood of recurrence after surgery, metastasis, and diminished survival (Table 1). For example, correlation with more advanced stage and/or grade was observed for ovarian, colorectal, gastric and non-small cell lung cancer [55, 74, 80, 82, 83], although not bladder cancer [76]. Correlation with decreased survival has been reported for many malignancies, including cancer of the ovary, lower intestine, bladder, liver, lung, stomach and esophagus [35, 74, 76, 79, 80, 82, 83]. These data support consideration of eIF5A2 as a prognostic marker in a variety of malignant conditions.
Metastasis and lymph node invasion are eIF5A2-related pathological features that have attracted attention from a mechanistic point of view. eIF5A2 over-expression is visualized at hepatocellular carcinoma margins [78]. Interestingly, eIF5A2 is one of only three genes found to be predictive of lymph node metastasis in gastric cancer [84]. Association of eIF5A2 with metastasis is also seen in colorectal cancer [75] and esophageal squamous cell carcinoma [35]. How the eIF5A2-associated malignant phenotype relates to biochemical and molecular characteristics of the tumors is discussed below.
Regulation of eIF5A2 expression in cancer
As with eIF5A1, the regulation of eIF5A2 expression is poorly understood although advances are being made. The EIF5A2 gene is often, but not invariably, amplified in cancers and cancer cell lines [12, 35, 59, 74, 75, 80]. While tumors exhibiting gene amplification generally exhibit high eIF5A2 expression, many have high eIF5A2 levels without gene amplification so additional mechanisms of up-regulation must exist. No doubt these include transcriptional activation by oncogenes such as K-ras, as in pancreatic cancer [66]. Recently, the involvement of hypoxia and the hypoxia- inducible factor HIF1α has been described [35]. In esophageal squamous cell carcinoma cells, hypoxia increases eIF5A2 RNA levels, at least in part via HIF1α. (A reciprocal action of eIF5A2 on HIF1α transcription is mentioned below.)
Post-transcriptional regulatory mechanisms are likely to contribute to eIF5A2 expression. As a result of differential polyadenylation site usage, and possibly alternative splicing, there are at least 5 size classes of eIF5A2 mRNA [58]. The various mRNA forms have distinct stabilities presumably due to differences in their untranslated regions. Translation of eIF5A2 mRNA is relatively inefficient compared to eIF5A1 [58], and this has been attributed to ribosome stalling, presumptively mediated by determinants located in untranslated regions of the mRNA. It is likely, therefore, that eIF5A2 is subject to controls acting at multiple levels, perhaps reflecting its roles in normal processes, such as development or differentiation, that await discovery.
Molecular and cell biological basis of eIF5A2 action in cancer
eIF5A2 depletion by antisense DNA or RNA interference slows the growth of ovarian cancer cells [55] and several other types of tumor cells in culture and in vivo, including cells from liver [57], pancreatic [66], esophageal [35], and gastric [82] cancers. eIF5A2 knockdown also reduces cell migration in tissue culture, in wound healing and matrigel invasiveness assays, as well as metastatic properties in vivo in xenograft experiments with a range of cancer types [35, 57, 77, 82, 83]. Small molecule inhibitors of DHS and DOHH, such as the spermidine analog GC7 and the drug ciclopirox (Figure 1), mimic many of these effects; conversely, eIF5A2 overexpression increases these mobility and metastatic properties in many of the same cell types. Correspondingly, eIF5A2 silencing and overexpression experiments demonstrated that the protein favors the metastasis-related epithelial-mesenchymal transition (EMT), as reflected in changes in the levels of epithelial cell markers such as E-cadherin and β-catenin and mesenchymal markers such as fibronectin and vimentin [35, 78, 81-83].
A variety of downstream events has been reported (Table 1). In colorectal and gastric cancer, metastasis-related protein MTA1, a key regulator of metastasis and invasiveness is up-regulated by eIF5A2, mediated by c-myc [82, 83]. In bladder cancer, TGFβ1 is responsible for eIF5A-inducd EMT and invasiveness, acting through STAT3 protein stabilization [77]. Inhibition of eIF5A2 expression or of hypusination reduces the level of the PEAK1 protein which is required for pancreatic cancer cell growth and metastasis [66]. In esophageal squamous cell carcinoma cell lines, hypoxia leads to an increase in eIF5A2 which engages in a complex interplay with HIF1α and angiogenesis [35]. One notable finding is the binding of eIF5A2 protein to a region upstream of the HIF1α promoter in chromatin immunoprecipitation assays, supporting a direct role for eIF5A2 in transcriptional regulation. Further, the vascular endothelial growth factor VEGF is also increased when eIF5A2 is overexpressed, consistent with a role in angiogenesis [35]. eIF5A is also associated with angiogenesis via another mechanism: in hepatocellular carcinoma, eIF5A overexpression increases matrix metalloproteinase MMP-2 RNA and activity via p38 MAPK and JNK/c-Jun transcriptional pathways [79].
Overall, the picture that emerges is of a broad up-regulation of tumorigenic parameters triggered, directly or indirectly, by eIF5A2 over-expression.
Therapeutic developments
The involvement of eIF5A isoforms in cancer, and the singularity of their post-translational modification pathway, has led to considerable attention from a therapeutic perspective. Both eIF5A isoforms and both of the enzymes required for hypusine formation have been exploited as targets for drug development and cancer therapy. Modalities explored include suppression of eIF5A production, over-expression of an eIF5A1 mutant, drugs and chemical inhibitors of DHS and DOHH, and microRNAs directed against DOHH. Combination therapy with drugs such as imatinib, 5-fluorouracil and cisplatin have also been explored [67, 68], [79], [81].
At the nucleic acid level, strategies to limit eIF5A-dependent cell proliferation in culture have included decreasing the eIF5A2 gene copy number by treatment with hydroxyurea to eliminate double minute chromosomes in ovarian cancer [55]. A more generally applicable approach is to attenuate eIF5A1 or eIF5A2 gene expression with antisense DNA or interfering RNA, as in several studies mentioned above. In a related approach, siRNA-mediated down-regulation of eIF5A1 is coupled with the over-expression of a mutant form of the protein that cannot be hypusinated as a result of a substitution at lysine-50 [85]. The siRNA and expression vector are introduced together in a particle called SNS01-T that is in early stage clinical trial (Phase 1b/2a) for treatment of multiple myeloma and lymphoma. In mouse studies, SNS01-T inhibited cancer cell growth of xenografts and extended the animals' survival, and it synergized with standard-of-care drugs bortezomib and lenalidomide. Apoptosis appears to contribute to its mechanism of action [86], consistent with previous studies in which an adenovirus vector expressing eIF5A1 reduced tumor growth, induced apoptosis and improved mouse survival [87]. An adenoviral vector expressing eIF5A2 was also effective, albeit to a somewhat lesser degree. In view of the preponderance of evidence that eIF5A promotes cell proliferation, it is germane to consider why over-expression of either isoform would be pro-apoptotic. Remarkably, the induction of apoptosis may be due to the accumulation of incompletely hypusinated forms of eIF5A, carrying either lysine or deoxyhypusine at position 50, as a result of DHS and DOHH limitations [58].
Inhibitors of these enzymes have been studied extensively in tissue culture. DHS is a metastasis signature gene [88] and its inhibitor GC7 has been used in many studies. Further small-molecule inhibitors have also been synthesized and tested [19, 68]. They inhibit cell growth in tissue culture, and GC7 has been reported to reduce tumorigenicity, suppress cell migration and reverse the EMT in vivo in liver, lung and pancreatic cancer models [78], [81], [66] and in a murine melanoma model [89].
Ablation of DOHH reduces cell proliferation and transformation in culture [18]. The drugs ciclopirox (an antifungal) and deferiprone (an iron chelator used to relieve iron overload in thalassemia) are DOHH inhibitors that have been studied in culture and in vivo [23], [34, 73]. Ciclopirox inhibits the growth of pancreatic tumor cells in culture [66]. Administered orally, it is effective against human breast cancer and leukemia xenografts in mice [90, 91], and improvement in clinical parameters was observed in some acute myeloid leukemia patients during a phase 1 trial [92]. Mimosine, a natural compound that also inhibits DOHH activity, acts synergistically with two different microRNAs (miR-331-3p and miR-642-5p) that reduce DOHH mRNA and protein expression to suppress the proliferation of prostate cancer cells in culture [93]. This raises the prospect of tumor chemotherapy with combinations of agents that target the eIF5A pathway.
Perspectives
Appreciation of the pivotal role of translational control in cancer is steadily increasing. In regard to eIF5A, the field has arrived at an interesting juncture, where biochemical and clinical approaches to function intersect and inform each other. Biochemical and genetic studies have defined protein sequences that impose a requirement for this translation factor, while processes leading to cell transformation and tumorigenicity have been described through clinical observations and related cell biological and animal model experimentation. Within the common ground salient topics are beginning to come into focus, including the regulation of eIF5A gene expression, the identification of eIF5A's molecular targets, and the definition of the pathways and mechanisms affected by these targets. Progress has been made on each of these topics as outlined above, but much still remains to be learned. For example, the 'eIF5A regulon' (Figure 3) needs to be substantiated, its membership catalogued, and its control deciphered.
One of the most seminal unanswered questions relates to the functions of the two eIF5A isoforms. Both eIF5A genes are implicated in cancer, as documented in Table 1, yet their sequences and gene expression patterns are distinct and it is not clear whether their biochemical activities differ. Evidence from yeast [16] and mammalian cell experiments [66, 87] is consistent with functional redundancy of eIF5A1 and eIF5A2, with the caveat that these systems may not model cancer closely enough to be definitive. Further work is required to uncover more subtle functional differences if they exist. An alternative interpretation would posit that the isoforms differ in the conditions and circumstances of their expression, i.e., the regulation of their gene expression, rather than in their protein functions. Taking the two genes together, controls have been reported that act at various levels, including translation, transcription, RNA processing, and protein modification, localization and turnover. Fuller comprehension of eIF5A's functions may allow differences between the isoforms, either in their expression or their function, to be exploited therapeutically.
Another set of issues concerns the hypusine modification, which is generally held to be specific for eIF5A (both isoforms) because no other hypusine-containing protein has been characterized. Likewise, the modifying enzymes DHS and DOHH are thought to be dedicated to eIF5A. A variety of experimental and therapeutic interventions has been devised and exploited based on these premises, thereby enabling a great deal of progress. Yet the premises are subject to limitations and some of the inhibitors employed – particularly drugs and other small molecules – can have additional or alternative targets. Furthermore, when such interventions bring about changes in gene expression and/or biological activities, it is not always obvious which changes are primary (i.e., direct actions on the target proteins) as opposed to secondary (e.g., downstream effects resulting from the failure to produce a regulator such as a transcription factor). In addition, the question arises whether the effects of DHS or DOHH inhibition are due simply to a decrease in mature eIF5A, or do the accumulating lysine- or deoxyhypusine-containing forms of eIF5A exert separate and distinctive functions as proposed in apoptosis [86]?
Building on our current appreciation of eIF5A's function in translation, a crucial next step will be to establish the precise role of eIF5A and its hypusine modification in protein synthesis and translational control. It remains to be determined whether the translational effects are all related to proline-dependent stalling, and more broadly, to evaluate the contribution of eIF5A's actions on translational initiation and other processes such as mRNA production, transport and stability. If eIF5A also affects the initiation stage of protein synthesis, it will be important to determine whether such stimulation affects all mRNAs similarly, or whether only some specific mRNAs are significantly stimulated. Ribosome profiling analysis [47] has the potential to provide decisive information about eIF5A's translational activities. In the context of the malignant phenotype, it will be important to catalog the gene products whose expression is influenced by eIF5A – the eIF5A regulon (or regulons, if the two isoforms prove to regulate distinct sets of genes) – and which play essential roles in tumorigenesis. Identification of these gene products may suggest additional downstream targets for therapeutic intervention. Meanwhile, the canonical components of the pathway – eIF5A1, eIF5A2, DHS and DOHH – offer promise and inspiration for the development of drugs and biologicals to combat cancer.
Acknowledgments
We thank Dr. Augusto Luchessi and Dr. Tsafi Pe'ery for critical reading of the manuscript. MBM received support from NIH grant R21HG006339.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Rossi D, Kuroshu R, Zanelli CF, Valentini SR. eIF5A and EF-P: two unique translation factors are now traveling the same road, Wiley interdisciplinary reviews. RNA. 2014;5:209–222. doi: 10.1002/wrna.1211. [DOI] [PubMed] [Google Scholar]
- [2].Caraglia M, Park MH, Wolff EC, Marra M, Abbruzzese A. eIF5A isoforms and cancer: two brothers for two functions? Amino acids. 2011;44:103–109. doi: 10.1007/s00726-011-1182-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Dever TE, Gutierrez E, Shin BS. The hypusine-containing translation factor eIF5A. Critical reviews in biochemistry and molecular biology. 2014;49:413–425. doi: 10.3109/10409238.2014.939608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wang FW, Guan XY, Xie D. Roles of eukaryotic initiation factor 5A2 in human cancer. International journal of biological sciences. 2013;9:1013–1020. doi: 10.7150/ijbs.7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kemper WM, Berry KW, Merrick WC. Purification and properties of rabbit reticulocyte protein synthesis initiation factors M2Balpha and M2Bbeta. The Journal of biological chemistry. 1976;251:5551–5557. [PubMed] [Google Scholar]
- [6].Schreier MH, Erni B, Staehelin T. Initiation of mammalian protein synthesis. I. Purification and characterization of seven initiation factors. Journal of molecular biology. 1977;116:727–753. doi: 10.1016/0022-2836(77)90268-6. [DOI] [PubMed] [Google Scholar]
- [7].Benne R, Brown-Luedi ML, Hershey JW. Purification and characterization of protein synthesis initiation factors eIF-1, eIF-4C, eIF-4D, and eIF-5 from rabbit reticulocytes. The Journal of biological chemistry. 1978;253:3070–3077. [PubMed] [Google Scholar]
- [8].Duncan RF, Hershey JW. Changes in eIF-4D hypusine modification or abundance are not correlated with translational repression in HeLa cells. The Journal of biological chemistry. 1986;261:12903–12906. [PubMed] [Google Scholar]
- [9].Smit-McBride Z, Schnier J, Kaufman RJ, Hershey JW. Protein synthesis initiation factor eIF-4D. Functional comparison of native and unhypusinated forms of the protein. The Journal of biological chemistry. 1989;264:18527–18530. [PubMed] [Google Scholar]
- [10].Smit-McBride Z, Dever TE, Hershey JW, Merrick WC. Sequence determination and cDNA cloning of eukaryotic initiation factor 4D, the hypusine-containing protein. The Journal of biological chemistry. 1989;264:1578–1583. [PubMed] [Google Scholar]
- [11].Jenkins ZA, Haag PG, Johansson HE. Human eIF5A2 on chromosome 3q25-q27 is a phylogenetically conserved vertebrate variant of eukaryotic translation initiation factor 5A with tissue-specific expression. Genomics. 2001;71:101–109. doi: 10.1006/geno.2000.6418. [DOI] [PubMed] [Google Scholar]
- [12].Guan XY, Sham JS, Tang TC, Fang Y, Huo KK, Yang JM. Isolation of a novel candidate oncogene within a frequently amplified region at 3q26 in ovarian cancer. Cancer research. 2001;61:3806–3809. [PubMed] [Google Scholar]
- [13].Clement PM, Henderson CA, Jenkins ZA, Smit-McBride Z, Wolff EC, Hershey JW, Park MH, Johansson HE. Identification and characterization of eukaryotic initiation factor 5A-2. Eur J Biochem. 2003;270:4254–4263. doi: 10.1046/j.1432-1033.2003.03806.x. [DOI] [PubMed] [Google Scholar]
- [14].Gordon ED, Mora R, Meredith SC, Lee C, Lindquist SL. Eukaryotic initiation factor 4D, the hypusine-containing protein, is conserved among eukaryotes. The Journal of biological chemistry. 1987;262:16585–16589. [PubMed] [Google Scholar]
- [15].Schnier J, Schwelberger HG, Smit-McBride Z, Kang HA, Hershey JW. Translation initiation factor 5A and its hypusine modification are essential for cell viability in the yeast Saccharomyces cerevisiae. Molecular and cellular biology. 1991;11:3105–3114. doi: 10.1128/mcb.11.6.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Schwelberger HG, Kang HA, Hershey JW. Translation initiation factor eIF-5A expressed from either of two yeast genes or from human cDNA. Functional identity under aerobic and anaerobic conditions. The Journal of biological chemistry. 1993;268:14018–14025. [PubMed] [Google Scholar]
- [17].Cooper HL, Park MH, Folk JE, Safer B, Braverman R. Identification of the hypusine-containing protein hy+ as translation initiation factor eIF-4D. Proceedings of the National Academy of Sciences of the United States of America. 1983;80:1854–1857. doi: 10.1073/pnas.80.7.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Sievert H, Pallmann N, Miller KK, Hermans-Borgmeyer I, Venz S, Sendoel A, Preukschas M, Schweizer M, Boettcher S, Janiesch PC, Streichert T, Walther R, Hengartner MO, Manz MG, Brummendorf TH, Bokemeyer C, Braig M, Hauber J, Duncan KE, Balabanov S. A novel mouse model for inhibition of DOHH-mediated hypusine modification reveals a crucial function in embryonic development, proliferation and oncogenic transformation. Disease models & mechanisms. 2014;7:963–976. doi: 10.1242/dmm.014449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Weir BA, Yaffe MP. Mmd1p, a novel, conserved protein essential for normal mitochondrial morphology and distribution in the fission yeast Schizosaccharomyces pombe. Molecular biology of the cell. 2004;15:1656–1665. doi: 10.1091/mbc.E03-06-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Park JH, Aravind L, Wolff EC, Kaevel J, Kim YS, Park MH. Molecular cloning, expression, and structural prediction of deoxyhypusine hydroxylase: a HEAT-repeat-containing metalloenzyme. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:51–56. doi: 10.1073/pnas.0509348102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Doerfel LK, Wohlgemuth I, Kothe C, Peske F, Urlaub H, Rodnina MV. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science. 2013;339:85–88. doi: 10.1126/science.1229017. [DOI] [PubMed] [Google Scholar]
- [22].Bullwinkle TJ, Zou SB, Rajkovic A, Hersch SJ, Elgamal S, Robinson N, Smil D, Bolshan Y, Navarre WW, Ibba M. (R)-beta-lysine-modified elongation factor P functions in translation elongation. The Journal of biological chemistry. 2013;288:4416–4423. doi: 10.1074/jbc.M112.438879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Mémin E, Hoque M, Jain MR, Heller DS, Li H, Cracchiolo B, Hanauske-Abel HM, Pe'ery T, Mathews MB. Blocking eIF5A modification in cervical cancer cells alters the expression of cancer-related genes and suppresses cell proliferation. Cancer research. 2014;74:552–562. doi: 10.1158/0008-5472.CAN-13-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Beninati S, Nicolini L, Jakus J, Passeggio A, Abbruzzese A. Identification of a substrate site for transglutaminases on the human protein synthesis initiation factor 5A. The Biochemical journal. 1995;305:725–728. doi: 10.1042/bj3050725. Pt 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ishfaq M, Maeta K, Maeda S, Natsume T, Ito A, Yoshida M. The role of acetylation in the subcellular localization of an oncogenic isoform of translation factor eIF5A. Bioscience, biotechnology, and biochemistry. 2012;76:2165–2167. doi: 10.1271/bbb.120620. [DOI] [PubMed] [Google Scholar]
- [26].Ishfaq M, Maeta K, Maeda S, Natsume T, Ito A, Yoshida M. Acetylation regulates subcellular localization of eukaryotic translation initiation factor 5A (eIF5A) FEBS letters. 2012;586:3236–3241. doi: 10.1016/j.febslet.2012.06.042. [DOI] [PubMed] [Google Scholar]
- [27].Kang HA, Schwelberger HG, Hershey JW. Translation initiation factor eIF-5A, the hypusine-containing protein, is phosphorylated on serine in Saccharomyces cerevisiae. The Journal of biological chemistry. 1993;268:14750–14756. [PubMed] [Google Scholar]
- [28].Klier H, Wohl T, Eckerskorn C, Magdolen V, Lottspeich F. Determination and mutational analysis of the phosphorylation site in the hypusine-containing protein Hyp2p. FEBS letters. 1993;334:360–364. doi: 10.1016/0014-5793(93)80712-4. [DOI] [PubMed] [Google Scholar]
- [29].Tong Y, Park I, Hong BS, Nedyalkova L, Tempel W, Park HW. Crystal structure of human eIF5A1: insight into functional similarity of human eIF5A1 and eIF5A2. Proteins. 2009;75:1040–1045. doi: 10.1002/prot.22378. [DOI] [PubMed] [Google Scholar]
- [30].Glick BR, Ganoza MC. Identification of a soluble protein that stimulates peptide bond synthesis. Proceedings of the National Academy of Sciences of the United States of America. 1975;72:4257–4260. doi: 10.1073/pnas.72.11.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Glick BR, Chladek S, Ganoza MC. Peptide bond formation stimulated by protein synthesis factor EF-P depends on the aminoacyl moiety of the acceptor. European journal of biochemistry / FEBS. 1979;97:23–28. doi: 10.1111/j.1432-1033.1979.tb13081.x. [DOI] [PubMed] [Google Scholar]
- [32].Blaha G, Stanley RE, Steitz TA. Formation of the first peptide bond: the structure of EF-P bound to the 70S ribosome. Science. 2009;325:966–970. doi: 10.1126/science.1175800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Gutierrez E, Shin BS, Woolstenhulme CJ, Kim JR, Saini P, Buskirk AR, Dever TE. eIF5A promotes translation of polyproline motifs. Molecular cell. 2013;51:35–45. doi: 10.1016/j.molcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hoque M, Hanauske-Abel HM, Palumbo P, Saxena D, D'Alliessi Gandolfi D, Park MH, Pe'ery T, Mathews MB. Inhibition of HIV-1 gene expression by Ciclopirox and Deferiprone, drugs that prevent hypusination of eukaryotic initiation factor 5A. Retrovirology. 2009;6:90. doi: 10.1186/1742-4690-6-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Li Y, Fu L, Li JB, Qin Y, Zeng TT, Zhou J, Zeng ZL, Chen J, Cao TT, Ban X, Qian C, Cai Z, Xie D, Huang P, Guan XY. Increased expression of EIF5A2, via hypoxia or gene amplification, contributes to metastasis and angiogenesis of esophageal squamous cell carcinoma. Gastroenterology. 2014;146:1701–1713. doi: 10.1053/j.gastro.2014.02.029. e1709. [DOI] [PubMed] [Google Scholar]
- [36].Zuk D, Jacobson A. A single amino acid substitution in yeast eIF-5A results in mRNA stabilization. The EMBO journal. 1998;17:2914–2925. doi: 10.1093/emboj/17.10.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Schrader R, Young C, Kozian D, Hoffmann R, Lottspeich F. Temperature-sensitive eIF5A mutant accumulates transcripts targeted to the nonsense-mediated decay pathway. The Journal of biological chemistry. 2006;281:35336–35346. doi: 10.1074/jbc.M601460200. [DOI] [PubMed] [Google Scholar]
- [38].Hofmann W, Reichart B, Ewald A, Muller E, Schmitt I, Stauber RH, Lottspeich F, Jockusch BM, Scheer U, Hauber J, Dabauvalle MC. Cofactor requirements for nuclear export of Rev response element (RRE)-and constitutive transport element (CTE)-containing retroviral RNAs. An unexpected role for actin. The Journal of cell biology. 2001;152:895–910. doi: 10.1083/jcb.152.5.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Henderson A, Hershey JW. Eukaryotic translation initiation factor (eIF) 5A stimulates protein synthesis in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:6415–6419. doi: 10.1073/pnas.1008150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Saini P, Eyler DE, Green R, Dever TE. Hypusine-containing protein eIF5A promotes translation elongation. Nature. 2009;459:118–121. doi: 10.1038/nature08034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Li CH, Ohn T, Ivanov P, Tisdale S, Anderson P. eIF5A promotes translation elongation. polysome disassembly and stress granule assembly, PloS one. 2010;5:e9942. doi: 10.1371/journal.pone.0009942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Dias CA, Gregio AP, Rossi D, Galvao FC, Watanabe TF, Park MH, Valentini SR, Zanelli CF. eIF5A interacts functionally with eEF2. Amino acids. 2012;42:697–702. doi: 10.1007/s00726-011-0985-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ude S, Lassak J, Starosta AL, Kraxenberger T, Wilson DN, Jung K. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science. 2013;339:82–85. doi: 10.1126/science.1228985. [DOI] [PubMed] [Google Scholar]
- [44].Woolstenhulme CJ, Parajuli S, Healey DW, Valverde DP, Petersen EN, Starosta AL, Guydosh NR, Johnson WE, Wilson DN, Buskirk AR. Nascent peptides that block protein synthesis in bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E878–887. doi: 10.1073/pnas.1219536110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Morgan AA, Rubenstein E. Proline: the distribution, frequency, positioning, and common functional roles of proline and polyproline sequences in the human proteome. PloS one. 2013;8:e53785. doi: 10.1371/journal.pone.0053785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Mandal A, Mandal S, Park MH. Genome-wide analyses and functional classification of proline repeat-rich proteins: potential role of eIF5A in eukaryotic evolution. PloS one. 2014;9:e111800. doi: 10.1371/journal.pone.0111800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Elgamal S, Katz A, Hersch SJ, Newsom D, White P, Navarre WW, Ibba M. EF-P dependent pauses integrate proximal and distal signals during translation. PLoS genetics. 2014;10:e1004553. doi: 10.1371/journal.pgen.1004553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Hersch SJ, Wang M, Zou SB, Moon KM, Foster LJ, Ibba M, Navarre WW. Divergent protein motifs direct elongation factor P-mediated translational regulation in Salmonella enterica and Escherichia coli. mBio. 2013;4:e00180–00113. doi: 10.1128/mBio.00180-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Peil L, Starosta AL, Lassak J, Atkinson GC, Virumae K, Spitzer M, Tenson T, Jung K, Remme J, Wilson DN. Distinct XPPX sequence motifs induce ribosome stalling, which is rescued by the translation elongation factor EF-P. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:15265–15270. doi: 10.1073/pnas.1310642110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Starosta AL, Lassak J, Peil L, Atkinson GC, Virumae K, Tenson T, Remme J, Jung K, Wilson DN. Translational stalling at polyproline stretches is modulated by the sequence context upstream of the stall site. Nucleic acids research. 2014;42:10711–10719. doi: 10.1093/nar/gku768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kang HA, Hershey JW. Effect of initiation factor eIF-5A depletion on protein synthesis and proliferation of Saccharomyces cerevisiae. The Journal of biological chemistry. 1994;269:3934–3940. [PubMed] [Google Scholar]
- [53].Hanauske-Abel HM, Slowinska B, Zagulska S, Wilson RC, Staiano-Coico L, Hanauske AR, McCaffrey T, Szabo P. Detection of a sub-set of polysomal mRNAs associated with modulation of hypusine formation at the G1-S boundary. Proposal of a role for eIF-5A in onset of DNA replication. FEBS letters. 1995;366:92–98. doi: 10.1016/0014-5793(95)00493-s. [DOI] [PubMed] [Google Scholar]
- [54].Xu A, Jao DL, Chen KY. Identification of mRNA that binds to eukaryotic initiation factor 5A by affinity co-purification and differential display. The Biochemical journal. 2004;384:585–590. doi: 10.1042/BJ20041232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Guan XY, Fung JM, Ma NF, Lau SH, Tai LS, Xie D, Zhang Y, Hu L, Wu QL, Fang Y, Sham JS. Oncogenic role of eIF-5A2 in the development of ovarian cancer. Cancer research. 2004;64:4197–4200. doi: 10.1158/0008-5472.CAN-03-3747. [DOI] [PubMed] [Google Scholar]
- [56].Chen G, Gharib TG, Thomas DG, Huang CC, Misek DE, Kuick RD, Giordano TJ, Iannettoni MD, Orringer MB, Hanash SM, Beer DG. Proteomic analysis of eIF-5A in lung adenocarcinomas. Proteomics. 2003;3:496–504. doi: 10.1002/pmic.200390063. [DOI] [PubMed] [Google Scholar]
- [57].Lee NP, Tsang FH, Shek FH, Mao M, Dai H, Zhang C, Dong S, Guan XY, Poon RT, Luk JM. Prognostic significance and therapeutic potential of eukaryotic translation initiation factor 5A (eIF5A) in hepatocellular carcinoma. International journal of cancer. Journal international du cancer. 2010;127:968–976. doi: 10.1002/ijc.25100. [DOI] [PubMed] [Google Scholar]
- [58].Clement PM, Johansson HE, Wolff EC, Park MH. Differential expression of eIF5A-1 and eIF5A-2 in human cancer cells. Febs J. 2006;273:1102–1114. doi: 10.1111/j.1742-4658.2006.05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Luo JH, Hua WF, Rao HL, Liao YJ, Kung HF, Zeng YX, Guan XY, Chen W, Xie D. Overexpression of EIF-5A2 predicts tumor recurrence and progression in pTa/pT1 urothelial carcinoma of the bladder. Cancer science. 2009;100:896–902. doi: 10.1111/j.1349-7006.2009.01126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Chen G, Gharib TG, Huang CC, Thomas DG, Shedden KA, Taylor JM, Kardia SL, Misek DE, Giordano TJ, Iannettoni MD, Orringer MB, Hanash SM, Beer DG. Proteomic analysis of lung adenocarcinoma: identification of a highly expressed set of proteins in tumors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2002;8:2298–2305. [PubMed] [Google Scholar]
- [61].Cracchiolo BM, Heller DS, Clement PM, Wolff EC, Park MH, Hanauske-Abel HM. Eukaryotic initiation factor 5A-1 (eIF5A-1) as a diagnostic marker for aberrant proliferation in intraepithelial neoplasia of the vulva. Gynecologic oncology. 2004;94:217–222. doi: 10.1016/j.ygyno.2004.03.018. [DOI] [PubMed] [Google Scholar]
- [62].Lam FF, Jankova L, Dent OF, Molloy MP, Kwun SY, Clarke C, Chapuis P, Robertson G, Beale P, Clarke S, Bokey EL, Chan C. Identification of distinctive protein expression patterns in colorectal adenoma. Proteomics Clin Appl. 2010;4:60–70. doi: 10.1002/prca.200900084. [DOI] [PubMed] [Google Scholar]
- [63].Tunca B, Tezcan G, Cecener G, Egeli U, Zorluoglu A, Yilmazlar T, Ak S, Yerci O, Ozturk E, Umut G, Evrensel T. Overexpression of CK20, MAP3K8 and EIF5A correlates with poor prognosis in early-onset colorectal cancer patients. Journal of cancer research and clinical oncology. 2013;139:691–702. doi: 10.1007/s00432-013-1372-x. [DOI] [PubMed] [Google Scholar]
- [64].Shang Y, Zhao X, Tian B, Wang Y, Ren F, Jia B, Zhai Y, Chen W, He D, Chang Z. CHIP/Stub1 interacts with eIF5A and mediates its degradation. Cellular signalling. 2014;26:1098–1104. doi: 10.1016/j.cellsig.2014.01.030. [DOI] [PubMed] [Google Scholar]
- [65].Preukschas M, Hagel C, Schulte A, Weber K, Lamszus K, Sievert H, Pallmann N, Bokemeyer C, Hauber J, Braig M, Balabanov S. Expression of eukaryotic initiation factor 5A and hypusine forming enzymes in glioblastoma patient samples: implications for new targeted therapies. PloS one. 2012;7:e43468. doi: 10.1371/journal.pone.0043468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Fujimura K, Wright T, Strnadel J, Kaushal S, Metildi C, Lowy AM, Bouvet M, Kelber JA, Klemke RL. A hypusine-eIF5A-PEAK1 switch regulates the pathogenesis of pancreatic cancer. Cancer research. 2014;74:6671–6681. doi: 10.1158/0008-5472.CAN-14-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Balabanov S, Gontarewicz A, Ziegler P, Hartmann U, Kammer W, Copland M, Brassat U, Priemer M, Hauber I, Wilhelm T, Schwarz G, Kanz L, Bokemeyer C, Hauber J, Holyoake TL, Nordheim A, Brummendorf TH. Hypusination of eukaryotic initiation factor 5A (eIF5A): a novel therapeutic target in BCR-ABL-positive leukemias identified by a proteomics approach. Blood. 2007;109:1701–1711. doi: 10.1182/blood-2005-03-037648. [DOI] [PubMed] [Google Scholar]
- [68].Ziegler P, Chahoud T, Wilhelm T, Pallman N, Braig M, Wiehle V, Ziegler S, Schroder M, Meier C, Kolodzik A, Rarey M, Panse J, Hauber J, Balabanov S, Brummendorf TH. Evaluation of deoxyhypusine synthase inhibitors targeting BCR-ABL positive leukemias. Investigational new drugs. 2012;30:2274–2283. doi: 10.1007/s10637-012-9810-1. [DOI] [PubMed] [Google Scholar]
- [69].Scuoppo C, Miething C, Lindqvist L, Reyes J, Ruse C, Appelmann I, Yoon S, Krasnitz A, Teruya-Feldstein J, Pappin D, Pelletier J, Lowe SW. A tumour suppressor network relying on the polyamine-hypusine axis. Nature. 2012;487:244–248. doi: 10.1038/nature11126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Coller HA, Grandori C, Tamayo P, Colbert T, Lander ES, Eisenman RN, Golub TR. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:3260–3265. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Boon K, Caron HN, van Asperen R, Valentijn L, Hermus MC, van Sluis P, Roobeek I, Weis I, Voute PA, Schwab M, Versteeg R. N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. The EMBO journal. 2001;20:1383–1393. doi: 10.1093/emboj/20.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Chen ZP, Chen KY. Marked elevation of hypusine formation activity on eukaryotic initiation factor 5A in v-HA-RAS transformed mouse NIH3T3 cells. Cancer letters. 1997;115:235–241. doi: 10.1016/s0304-3835(97)04741-1. [DOI] [PubMed] [Google Scholar]
- [73].Hanauske-Abel HM, Saxena D, Palumbo PE, Hanauske AR, Luchessi AD, Cambiaghi TD, Hoque M, Spino M, D'Alliessi Gandolfi D, Heller DS, Singh S, Park MH, Cracchiolo BM, Tricta F, Connelly J, Popowicz AM, Cone RA, Holland B, Pe'ery T, Mathews MB. Drug-induced reactivation of apoptosis abrogates HIV-1 infection. PloS one. 2013;8:e74414. doi: 10.1371/journal.pone.0074414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Yang GF, Xie D, Liu JH, Luo JH, Li LJ, Hua WF, Wu HM, Kung HF, Zeng YX, Guan XY. Expression and amplification of eIF-5A2 in human epithelial ovarian tumors and overexpression of EIF-5A2 is a new independent predictor of outcome in patients with ovarian carcinoma. Gynecologic oncology. 2009;112:314–318. doi: 10.1016/j.ygyno.2008.10.024. [DOI] [PubMed] [Google Scholar]
- [75].Xie D, Ma NF, Pan ZZ, Wu HX, Liu YD, Wu GQ, Kung HF, Guan XY. Overexpression of EIF-5A2 is associated with metastasis of human colorectal carcinoma. Human pathology. 2008;39:80–86. doi: 10.1016/j.humpath.2007.05.011. [DOI] [PubMed] [Google Scholar]
- [76].Chen W, Luo JH, Hua WF, Zhou FJ, Lin MC, Kung HF, Zeng YX, Guan XY, Xie D. Overexpression of EIF-5A2 is an independent predictor of outcome in patients of urothelial carcinoma of the bladder treated with radical cystectomy. Cancer Epidemiol Biomarkers Prev. 2009;18:400–408. doi: 10.1158/1055-9965.EPI-08-0754. [DOI] [PubMed] [Google Scholar]
- [77].Wei JH, Cao JZ, Zhang D, Liao B, Zhong WM, Lu J, Zhao HW, Zhang JX, Tong ZT, Fan S, Liang CZ, Liao YB, Pang J, Wu RH, Fang Y, Chen ZH, Li B, Xie D, Chen W, Luo JH. EIF5A2 predicts outcome in localised invasive bladder cancer and promotes bladder cancer cell aggressiveness in vitro and in vivo. British journal of cancer. 2014;110:1767–1777. doi: 10.1038/bjc.2014.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Tang DJ, Dong SS, Ma NF, Xie D, Chen L, Fu L, Lau SH, Li Y, Guan XY. Overexpression of eukaryotic initiation factor 5A2 enhances cell motility and promotes tumor metastasis in hepatocellular carcinoma. Hepatology. 2010;51:1255–1263. doi: 10.1002/hep.23451. [DOI] [PubMed] [Google Scholar]
- [79].Wang FW, Cai MY, Mai SJ, Chen JW, Bai HY, Li Y, Liao YJ, Li CP, Tian XP, Kung HF, Guan XY, Xie D. Ablation of EIF5A2 induces tumor vasculature remodeling and improves tumor response to chemotherapy via regulation of matrix metalloproteinase 2 expression. Oncotarget. 2014;5:6716–6733. doi: 10.18632/oncotarget.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].He LR, Zhao HY, Li BK, Liu YH, Liu MZ, Guan XY, Bian XW, Zeng YX, Xie D. Overexpression of eIF5A-2 is an adverse prognostic marker of survival in stage I non-small cell lung cancer patients. International journal of cancer. Journal international du cancer. 2011;129:143–150. doi: 10.1002/ijc.25669. [DOI] [PubMed] [Google Scholar]
- [81].Xu G, Yu H, Shi X, Sun L, Zhou Q, Zheng D, Shi H, Li N, Zhang X, Shao G. Cisplatin sensitivity is enhanced in non-small cell lung cancer cells by regulating epithelial-mesenchymal transition through inhibition of eukaryotic translation initiation factor 5A2. BMC pulmonary medicine. 2014;14:174. doi: 10.1186/1471-2466-14-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Meng QB, Kang WM, Yu JC, Liu YQ, Ma ZQ, Zhou L, Cui QC, Zhou WX. Overexpression of Eukaryotic Translation Initiation Factor 5A2 (EIF5A2) Correlates with Cell Aggressiveness and Poor Survival in Gastric Cancer. PloS one. 2015;10:e0119229. doi: 10.1371/journal.pone.0119229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Zhu W, Cai MY, Tong ZT, Dong SS, Mai SJ, Liao YJ, Bian XW, Lin MC, Kung HF, Zeng YX, Guan XY, Xie D. Overexpression of EIF5A2 promotes colorectal carcinoma cell aggressiveness by upregulating MTA1 through C-myc to induce epithelial-mesenchymaltransition. Gut. 2012;61:562–575. doi: 10.1136/gutjnl-2011-300207. [DOI] [PubMed] [Google Scholar]
- [84].Marchet A, Mocellin S, Belluco C, Ambrosi A, DeMarchi F, Mammano E, Digito M, Leon A, D'Arrigo A, Lise M, Nitti D. Gene expression profile of primary gastric cancer: towards the prediction of lymph node status. Annals of surgical oncology. 2007;14:1058–1064. doi: 10.1245/s10434-006-9090-0. [DOI] [PubMed] [Google Scholar]
- [85].Francis SM, Taylor CA, Tang T, Liu Z, Zheng Q, Dondero R, Thompson JE. SNS01-T modulation of eIF5A inhibits B-cell cancer progression and synergizes with bortezomib and lenalidomide. Molecular therapy : the journal of the American Society of Gene Therapy. 2014;22:1643–1652. doi: 10.1038/mt.2014.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Taylor CA, Sun Z, Cliche DO, Ming H, Eshaque B, Jin S, Hopkins MT, Thai B, Thompson JE. Eukaryotic translation initiation factor 5A induces apoptosis in colon cancer cells and associates with the nucleus in response to tumour necrosis factor alpha signalling. Experimental cell research. 2007;313:437–449. doi: 10.1016/j.yexcr.2006.09.030. [DOI] [PubMed] [Google Scholar]
- [87].Jin S, Taylor CA, Liu Z, Sun BY, Thompson JE. Suppression of primary and disseminated murine tumor growth with eIF5A1 gene therapy. Gene Therapy and Molecular Biology. 2008:207–218. [Google Scholar]
- [88].Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nature genetics. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- [89].Jasiulionis MG, Luchessi AD, Moreira AG, Souza PP, Suenaga AP, Correa M, Costa CA, Curi R, Costa-Neto CM. Inhibition of eukaryotic translation initiation factor 5A (eIF5A) hypusination impairs melanoma growth. Cell Biochem Funct. 2007;25:109–114. doi: 10.1002/cbf.1351. [DOI] [PubMed] [Google Scholar]
- [90].Zhou H, Shen T, Luo Y, Liu L, Chen W, Xu B, Han X, Pang J, Rivera CA, Huang S. The antitumor activity of the fungicide ciclopirox. International journal of cancer. Journal international du cancer. 2010;127:2467–2477. doi: 10.1002/ijc.25255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Eberhard Y, McDermott SP, Wang X, Gronda M, Venugopal A, Wood TE, Hurren R, Datti A, Batey RA, Wrana J, Antholine WE, Dick JE, Schimmer AD. Chelation of intracellular iron with the antifungal agent ciclopirox olamine induces cell death in leukemia and myeloma cells. Blood. 2009;114:3064–3073. doi: 10.1182/blood-2009-03-209965. [DOI] [PubMed] [Google Scholar]
- [92].Minden MD, Hogge DE, Weir SJ, Kasper J, Webster DA, Patton L, Jitkova Y, Hurren R, Gronda M, Goard CA, Rajewski LG, Haslam JL, Heppert KE, Schorno K, Chang H, Brandwein JM, Gupta V, Schuh AC, Trudel S, Yee KW, Reed GA, Schimmer AD. Oral ciclopirox olamine displays biological activity in a phase I study in patients with advanced hematologic malignancies. American journal of hematology. 2014;89:363–368. doi: 10.1002/ajh.23640. [DOI] [PubMed] [Google Scholar]
- [93].Epis MR, Giles KM, Kalinowski FC, Barker A, Cohen RJ, Leedman PJ. Regulation of expression of deoxyhypusine hydroxylase (DOHH), the enzyme that catalyzes the activation of eIF5A, by miR-331-3p and miR-642-5p in prostate cancer cells. The Journal of biological chemistry. 2012;287:35251–35259. doi: 10.1074/jbc.M112.374686. [DOI] [PMC free article] [PubMed] [Google Scholar]