SUMMARY
The mechanisms by which Mycobacterium tuberculosis (Mtb) maintains metabolic equilibrium to survive during infection and upon exposure to antimycobacterial drugs are poorly characterized. Ergothioneine (EGT) and mycothiol (MSH) are the major redox buffers present in Mtb, but the contribution of EGT to Mtb redox homeostasis and virulence remains unknown. We report that Mtb WhiB3, a 4Fe-4S redox sensor protein, regulates EGT production and maintains bioenergetic homeostasis. We show that central carbon metabolism and lipid precursors regulate EGT production and that EGT modulates drug sensitivity. Notably, EGT and MSH are both essential for redox and bioenergetic homeostasis. Transcriptomic analyses of EGT and MSH mutants indicate overlapping, but distinct functions of EGT and MSH. Lastly, we show that EGT is critical for Mtb survival in both macrophages and mice. This study has uncovered a dynamic balance between Mtb redox and bioenergetic homeostasis, which critically influences Mtb drug susceptibility and pathogenicity.
INTRODUCTION
Tuberculosis (TB) is the second most common cause of death from an infectious agent after HIV. This is largely due to the ability of Mtb to remain in a dormant, drug-tolerant state for decades in humans before emerging to cause active disease in ~10% of those infected. Mtb is exposed to environments with a wide range of available carbon sources, reactive oxygen intermediates (ROIs) and reactive nitrogen intermediates (RNIs) inside the host that may cause cell death. Therefore, it is strongly anticipated that the ability of Mtb to maintain redox balance and metabolic homeostasis is critical to its pathogenicity and virulence (Kumar et al., 2011). In addition, some front-line TB drugs such as isoniazid are prodrugs that require bioreduction by Mtb for anti-mycobacterial activity (Lei et al., 2000). Thus, a fundamental challenge to global TB control is to understand the mechanisms by which Mtb adapts to diverse carbon sources and redox environments encountered in the host.
Mtb produces mycothiol (MSH; Figure 1A) which acts as a major redox couple to protect against various redox stressors and anti-TB drugs (Buchmeier et al., 2003; Rawat et al., 2007). Mtb also produces a second thiol couple, ergothioneine (EGT; Figure 1B), a sulfur-containing histidine derivative with potent antioxidant properties (Genghof, 1970; Hand and Honek, 2005). However, despite considerable effort, roles for EGT in Mtb and its potential involvement in redox homeostasis and pathogenesis remain unknown. Recently we have shown that EGT levels in Mtb are modulated by protein phosphorylation during transition into late states of growth (Richard-Greenblatt et al., 2015), yet it is still unclear why mycobacteria produce both EGT and MSH to maintain redox homeostasis.
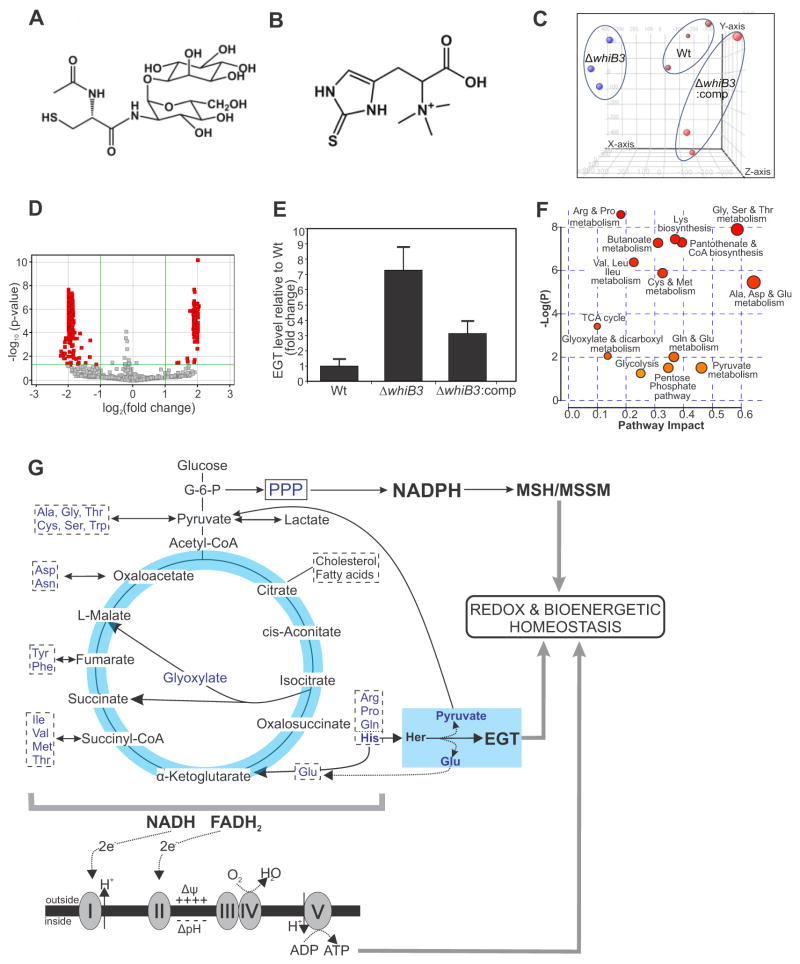
Figure 1. Metabolomic analysis of MtbΔwhiB3 demonstrates increased levels of EGT.
Chemical structures of (A) MSH and (B) EGT. (C) Principal component analysis shows that Wt Mtb, ΔwhiB3 and ΔwhiB3:comp strains can be distinguished based on their overall metabolomic profiles. For metabolomic analysis, each strain was independently examined at least twice in triplicate. (D) Volcano plot of metabolites in Wt Mtb and ΔwhiB3 plotted as fold change versus significance. The green lines demarcate metabolites with a greater than two-fold change on the x-axis and a corrected p value <0.05 (289/3327 molecules). Metabolites with less than a 2-fold change and/or p > 0.05 are shown in gray. See also Table 1. (E) Independent validation of EGT production in Wt Mtb, ΔwhiB3 and ΔwhiB3:comp based on LC/MS analysis after growth on medium containing 5 mM acetate. Error bars denote SEM. (F) Untargeted metabolite profiling assessed by MetPa enrichment analysis, based on changes in the relative abundance of metabolites (see also Figure S1 and Table S1) indicate that WhiB3 has a profound effect on Mtb metabolism, especially carbon catabolism, bioenergetic metabolism, amino acid metabolism, the pentose phosphate pathway (PPP) and glycolysis. (G) EGT is linked to the metabolism of histidine (His) as it is derived from hercynine, (Her), and MSH biosynthesis is regulated by NADPH produced by the PPP. Glycolysis and the TCA cycle both feed into the electron transport chain to regulate the electrical (Δψ) and concentration (ΔpH) gradients across the membrane, which maintain bioenergetic homeostasis. Amino acids acting as anaplerotic and cataplerotic substrates are boxed. See also Figure S1 and Table S1.
Redox balance is essential for energy metabolism including glycolysis, the TCA cycle and oxidative phosphorylation (OXPHOS). Despite this strong interdependence between redox homeostasis and energy metabolism, very few tools are available to investigate mycobacterial bioenergetics in real-time and in a noninvasive manner. Since cellular respiration involves a complex interplay of biological factors, including the availability, nature and concentration of oxidizable substrates as well as energy demand, methods for detecting such bioenergetic perturbations in Mtb will be of great value.
We previously demonstrated that WhiB3, an Mtb 4Fe-4S cluster redox sensor and virulence protein, maintains intracellular redox homeostasis of the mycobacterial cell to provide metabolic and cellular integrity (Muthukumaraswamy et al., 2009; Singh et al., 2007; Steyn et al., 2002). In this study, we examined how WhiB3 controls redox and bioenergetic homeostasis in Mtb to moderate virulence. We used a combination of metabolomic, bioenergetic and transcriptomic approaches and established links between WhiB3 and bioenergetic homeostasis and EGT, a major redox buffer. We characterized the genetic architecture of the EGT biosynthesis operon in Mtb and assessed the contribution of EGT in protection against oxidative stress, antimycobacterial drug susceptibility and in bioenergetic homeostasis. Further, we examined a link between Mtb central carbon catabolism and EGT production and the relationship between EGT and MSH biosynthesis. Using genome-wide expression analysis of genetically defined mutants of MSH and EGT biosynthesis, we identified differentially regulated genes common to all Mtb mutants. Finally, using macrophages and a mouse model of infection, we establish that maintaining redox balance and bioenergetic homeostasis is essential for Mtb virulence.
RESULTS
WhiB3 Regulates EGT Production in Mtb
Since Mtb WhiB3 is an intracellular redox sensor (Singh et al., 2009), we sought to identify redox-responsive metabolites regulated by WhiB3. We analyzed the metabolomes of Mtb (H37Rv), ΔwhiB3 and the corresponding whiB3-complemented strain (ΔwhiB3:comp) under rigorously controlled conditions (de Carvalho et al., 2010). Metabolites were separated by HPLC and detected by electrospray ionization coupled to mass time-of-flight mass spectrometry (LC-MS/MS) as previously described (Rhee et al., 2011). Among the statistically significant differences detected was the increased production of EGT in ΔwhiB3 (Figure 1C, D and Figure S1). Independent validation showed a 7.3-fold increase in EGT levels in ΔwhiB3 (Figure 1E) and complementation of whiB3 restored the EGT content to near wild-type levels (Figure 1E).
Next, we performed Metabolic Pathway Analysis, (MetPa), which combines pathway enrichment analysis with pathway topology, to detect metabolic differences between Mtb and ΔwhiB3 (Everts et al., 2014; Nandakumar et al., 2014). This analysis highlighted changes in the abundance of metabolites of biochemical pathways in ΔwhiB3 including glycolysis, the pentose phosphate pathway, the tricarboxylic acid (TCA) cycle and several amino acid biosynthesis pathways (Figure 1F, Tables 1 and S1). The pyruvate node is of particular significance as it is a key intermediate of many amino acid metabolism pathways, glycolysis and the TCA cycle (Figure 1G, Table S1).
Table 1.
MetPa analysis of pathways altered in ΔwhiB3 compared to Wt Mtba.
| KEGG Pathway Name | Metabolites | p-value | Impact | |
|---|---|---|---|---|
| Total | Hits | |||
| Arginine and proline metabolism | 41 | 12 | 0.000189 | 0.19 |
| Glycine, serine and threonine metabolism | 32 | 10 | 0.000378 | 0.60 |
| Lysine biosynthesis | 13 | 6 | 0.000588 | 0.38 |
| Pantothenate and CoA biosynthesis | 23 | 8 | 0.000681 | 0.41 |
| Butanoate metabolism | 18 | 7 | 0.000699 | 0.32 |
| Valine, leucine and isoleucine biosynthesis | 26 | 8 | 0.00171 | 0.24 |
| Cysteine and methionine metabolism | 34 | 9 | 0.00282 | 0.34 |
| Alanine, aspartate and glutamine metabolism | 18 | 6 | 0.0043 | 0.66 |
| Citrate cycle (TCA cycle) | 20 | 5 | 0.0327 | 0.11 |
Metabolomic Pathway Analysis (MetPA) combines pathway enrichment analysis with pathway topology analysis to identify the most relevant pathways under the specified conditions. MetPA uses the KEGG metabolic pathways as the backend knowledge base and integrates univariate analysis, over-representation analysis, Global Test, GlobalAncova and network topology analysis into pathway analysis. The total/maximum importance of each pathway = 1; pathway impact value = cumulative % from the matched metabolic nodes. Pathways with the most significant change (p ≤ 0.05) are listed. KEGG; Kyoto Encyclopedia of Genes and Genomes, TCA; tricarboxylic acid. See also Table S1.
Overall, these metabolomic data demonstrate that WhiB3 modulates the production of EGT. Further analyses revealed that WhiB3 also modulates the production of several amino acids, which act as anaplerotic and cataplerotic substrates and converge on glycolysis and the TCA cycle to affect energy metabolism (Figure 1G).
WhiB3 Maintains Bioenergetic Homeostasis in Mtb
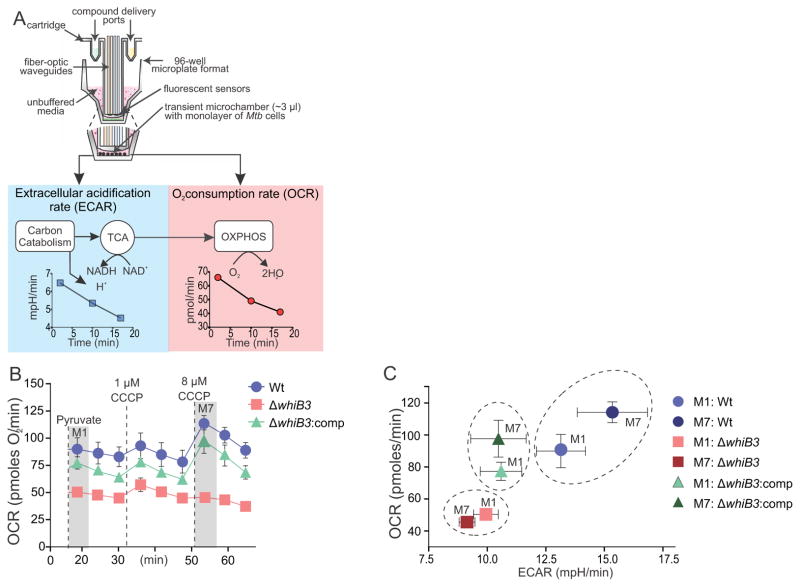
Next, we optimized extracellular flux analysis previously applied only to eukaryotic cells, for the study of Mtb. In this method, fluorescence sensors measure dissolved extracellular oxygen and proton concentration in real time in a microtransient chamber formed when the probes are in the measurement position. Inhibitors, modulators or substrates can be added during the run through delivery ports in the cartridge. This technology allowed us to accurately measure the oxygen consumption rate (OCR) and the extracellular acidification rate (ECAR) of Mtb cells in real time in a noninvasive manner (Figure 2A). The OCR and ECAR are direct and highly sensitive measures of cellular metabolic activity (Ferrick et al., 2008). ECAR gives a measure of H+ extrusion associated with carbon catabolism via glycolysis and the TCA cycle, whereas OCR is indicative of cellular respiration due to OXPHOS. The cell energy phenotype can be deduced from a phenogram (plotting of OCR versus ECAR) that illustrates metabolic shifts between substrate oxidation and glycolysis. We used pyruvate, identified as a key glycolytic intermediate (Figure 1G, Table S1), as the sole carbon source in XF bioenergetic assays. We used carbonyl cyanide m-chlorophenylhydrazone (CCCP), a membrane uncoupler, to examine the capacity of WhiB3 to maintain bioenergetic homeostasis upon uncoupling of OXPHOS from ATP production. In the presence of pyruvate, ΔwhiB3 had a significantly lower OCR than Wt and ΔwhiB3:comp (Figure 2B). After addition of CCCP (8 μM, M7), both the Wt and ΔwhiB3:comp strains responded to the uncoupling stress by increasing OCR (Figure 2B). The OCR and ECAR data from these experiments were plotted to generate a phenogram, which provides a visualization of the overall metabolic profiles of the different strains (Figure 2C). Our data show that ΔwhiB3, in contrast to Wt Mtb, did not respond with an increase in OCR or ECAR to re-establish membrane potential (M7, Figure 2C), indicating that ΔwhiB3 is bioenergetically deficient.
Figure 2. Extracellular flux analysis demonstrates the bioenergetic deficiency of ΔwhiB3.
(A) Diagram of the probe of an XF96 cartridge and a well of the cell culture microplate. During the assay, ECAR is measured as an indication of H+ production from carbon catabolism and OCR as an indication of OXPHOS. (B) OCR profiles of Wt Mtb H37Rv (blue), ΔwhiB3 (red), ΔwhiB3:comp (green) in the presence of pyruvate (50 μM, M1) and after addition of two aliquots of CCCP to a final concentration of 1 μM and 8 μM (M7) indicated by the vertical dotted lines. (C) Phenogram of all three strains at M1 (after pyruvate addition) and M7 (after the second CCCP addition) demonstrating increased OCR and ECAR of Wt Mtb H37Rv and ΔwhiB3:comp after CCCP addition in contrast to the inability of ΔwhiB3 to respond to the uncoupling stress.
In sum, we optimized extracellular flux analysis for the study of Mtb bioenergetics. Adapting this technology toward Mtb has broad implications for studying bioenergetics of microbial pathogens in general. The results demonstrate that in the absence of WhiB3, Mtb cannot respire efficiently when the glycolytic intermediate pyruvate is the sole carbon source. This suggests that the absence of WhiB3 leads to dysfunctional respiration and confirms its involvement in maintaining bioenergetic homeostasis.
EGT Detection Using TLC
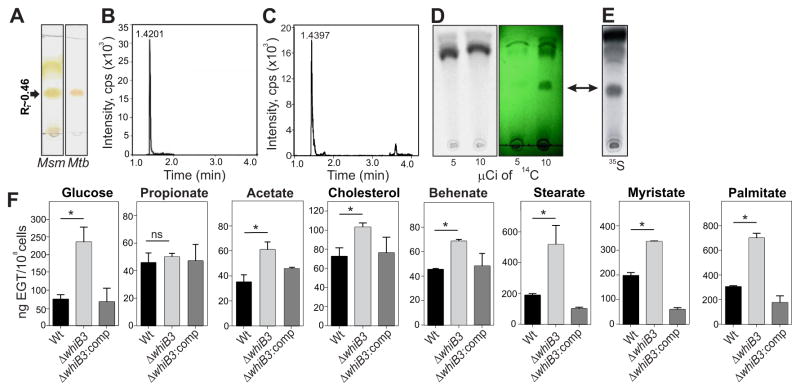
The study of EGT biosynthesis in bacteria has been hampered by the fact that EGT quenches the fluorescence of bimane derivatives, thereby making routine detection of EGT by HPLC analysis difficult. We therefore developed a rapid thin-layer chromatography (TLC)-based radioactive labeling assay and showed that pure EGT has an Rf value of ~ 0.46 (Figure S2A). TLC analysis of concentrated M. smegmatis (Msm) and Mtb extracts revealed bands with Rf values consistent with that of pure EGT (Figure 3A and Figure S2A). LC-MS/MS analysis of these bands and an EGT standard revealed virtually identical retention times, confirmed them to be EGT (Figure 3B,C and Figure S2B). An earlier study in Claviceps purpurea showed that [2-14C]-acetate could be used to label EGT (Heath and Wildy, 1956). However, we could not detect labeled EGT after TLC analysis of extracts from Msm cultured in the presence of [2-14C]-acetate even though UV exposure revealed a band at the same position as pure EGT, indicating that EGT was present (Figure 3D). However, by using [35S]-Cys as a precursor we observed a labeled spot after TLC analysis with the same Rf value as the EGT identified by LC-MS/MS analysis (Figure 3E).
Figure 3. Detection of EGT by TLC analysis and WhiB3 regulation of EGT in response to carbon catabolism.
(A) TLC analysis of Mtb and M. smegmatis (Msm) extracts followed by application of 0.2% ethanolic Gibb’s reagent. EGT appears as a brick-red spot with Rf values near 0.46. (B) This band on the TLC plate was extracted and analyzed by LC-MS/MRM to confirm its identity as EGT. The main elution peak at a retention time of 1.4201 minutes represents EGT (C) LC-MS/MRM analysis of an EGT standard with a retention time of 1.4397 minutes confirmed the band in (B) is EGT. (D) Mtb cells were grown in the presence of 5 or 10 μCi of [2-14C]-acetate followed by extraction in water and TLC analysis using 50,000 counts. Exposure of the TLC plate to a phosphor storage screen and viewing by phosphoimaging (left) or a UV lamp at 256 nm revealed the EGT band (green image; EGT bands; arrow). (E) EGT was detected in extracts of 35S-cysteine-fed cultures followed by TLC and autoradiography and was confirmed by LC-MS/MS. (F) LC-MS/MS quantitation of EGT in Mtb, ΔwhiB3 and ΔwhiB3:comp strains that were cultured in glucose, various fatty acids, or the breakdown products of β-oxidation as the sole carbon sources. Error bars represent SEM, n=3, *p<0.05. See also Figure S2.
Altogether, we have developed a simple radioactive TLC-based assay that facilitates the rapid, routine examination of EGT in mycobacteria and potentially other EGT-producing bacteria. Further, this method revealed differences in the biosynthetic pathways of either EGT or EGT precursors between fungi and mycobacteria.
WhiB3-mediated regulation of EGT production is dependent on central carbon metabolism
Since WhiB3 initiates the metabolic switchover to the preferred in vivo carbon source (fatty acids) and integrates redox signals with core intermediary metabolism (Singh et al., 2009; Singh et al., 2007), we sought to establish whether the type of carbon source influences WhiB3-mediated regulation of EGT production. LC-MS/MS was used to quantify intracellular EGT in Mtb, ΔwhiB3 and ΔwhiB3:comp grown in physiologically relevant concentrations [50 μM (Lee et al., 2013)] of carbon sources, including glucose, lipids and lipid precursors. As shown in Figure 3F, ΔwhiB3 had significantly increased levels of EGT compared to Wt or ΔwhiB3:comp when grown in glucose, acetate, cholesterol, behenate, stearate, myristate or palmitate and no difference when grown in propionate. EGT levels in Wt Mtb ranged from 40 to over 300 ng/108 cells with higher (>200 ng/108 cells) levels occurring in cells grown in myristate (C-14), palmitate (C-16) and stearate (C-18). ΔwhiB3 grown in the same C-14, C-16, C-18 fatty acids produced even higher levels of EGT (≥400 ng/108 cells). We observed EGT levels < 120 ng/108 cells in Wt Mtb grown on glucose, propionate (C-3), acetate (C-2), behenate (C-22) and cholesterol (C-27). ΔwhiB3 grown in these substrates had similarly lower EGT levels albeit significantly higher than the Wt, with the exception of glucose, where levels were >250 ng EGT/108 cells. In sum, these data demonstrate that WhiB3 regulates EGT production in response to catabolism of a diverse set of fatty acid precursors and cholesterol.
The Mtb EGT Operon
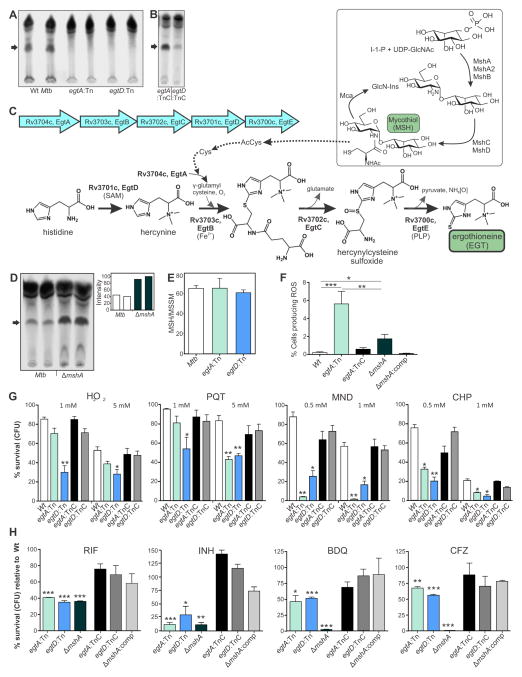
In vitro reconstitution studies using protein preparations of Erwinia tasmaniensis and Msm (Seebeck, 2010), as well as a recent genetic study in Msm (Sao Emani C, 2013) have provided biochemical and genetic evidence for EGT production in Msm. Analysis of the Mtb genome led us to predict that locus Rv3704c-Rv3700c constitutes the EGT biosynthetic operon. Figure 4C shows the genetic organization and biochemical pathway involved in EGT production and the link between EGT and MSH biosynthesis in Mtb. Examination of EGT production in Mtb CDC1551 and corresponding transposon mutants that disrupt egtA or egtD (hereafter referred to as egtA:Tn and egtD:Tn) revealed that Mtb egtA:Tn and egtD:Tn do not produce EGT (Figure 4A). Production of EGT in egtA:Tn was successfully complemented with the addition of the complete egt gene cluster (Rv3704c-Rv3700c, egtA:TnC), and partially complemented in egtD:Tn with the addition of egtD (egtD:TnC, Figure 4B). The arrangement of the egt genes, with egtA/egtB and egtC/egtD having overlapping stop and start codons (GTGA and ATGA, respectively), suggested that they may be operonic. Reverse transcriptase PCR analysis established that egtA through egtD are indeed operonic, while egtE is not (Figure S3). In sum, we have confirmed the genetic components and operonic organization (Rv3704c-3700c) responsible for EGT biosynthesis in Mtb.
Figure 4. Genetic characterization of the EGT biosynthetic operon in Mtb.
(A) TLC and autoradiographic analyses of 35S-cysteine-fed egtA:Tn, egtD:Tn and the corresponding Wt strain (Mtb CDC1551) indicates that disruption of egtA or egtD abrogates EGT production. (B) Genetically complemented strains egtA:TnC and egtD:TnC produce EGT. Complementation in egtD:TnC appeared to be partial. (C) Model depicts a putative link between the MSH and EGT pathways via Acetyl-Cys and Cys (indicated by dotted arrows). (D) TLC and autoradiographic analyses of 35S-cysteine-fed Mtb CDC1551 and its corresponding ΔmshA strain (duplicate samples) shows increased EGT production in the mutant lacking MSH. (E) HPLC measurement of reduced (MSH) and oxidized (MSSM) mycothiol. MSH/MSSM ratio is presented as the mean ± standard deviation (SD) of three independent cultures. (F) Flow cytometric measurement of endogenous ROS with cellROX Green dye in Mtb revealed that the egtA:Tn and ΔmshA mutant had significantly more ROS-producing cells relative to Wt (***p<0.001, **p < 0.005 and *p < 0.05, respectively) or corresponding complemented strains. (G) EGT protects Mtb against oxidative stress. Following exposure to H2O2, PQT, MND and CHP, cells were plated and CFU enumerated. Percentage survival was determined relative to untreated (UT) cells for each strain. Each column represents the mean ± SD (n = 4, *p<0.05, **p<0.01) compared to the Wt strain. (H) Lack of EGT increases susceptibility of Mtb to RIF, INH, BDQ and CFZ. Graphs show, at MIC values obtained for Wt Mtb, the percent survival (CFU) of egtA:Tn, egtD:Tn, ΔmshA relative to the Wt strain (see also Table S2). Complementation restored drug sensitivities to near Wt levels. Each column represents the mean ± SD (n = 4, *p<0.05, **p<0.01, ***p<0.001) compared to Wt strain. See also Figure S3 and Table S2.
EGT and Mycobacterial Redox Balance
Next, to examine the biological significance of EGT, and whether EGT and MSH are linked metabolically, we measured EGT levels in Mtb defective in MSH production. We noticed substantially increased EGT levels in ΔmshA compared to Wt (Figure 4D). HPLC analyses of reduced MSH (2MSH) and oxidized (disulfide) MSH (MSSM) levels in Mtb CDC1551, egtA:Tn and egtD:Tn indicated that the MSH/MSSM ratio does not differ in the Mtb EGT mutants (Figure 4E).
We then examined the endogenous ROS levels in Mtb strains defective in EGT or MSH production. Flow cytometry analysis of Wt, egtA:Tn and ΔmshA following exposure to the ROS-responsive dye CellROX Green revealed that egtA and mshA mutants have a significantly increased percentage of cells that produce ROS under normal growth conditions (Figure 4F). Compared to Wt, egtA:Tn had 25-fold and ΔmshA had 8-fold more ROS-producing cells. Further, the egtA:Tn strain had 3-fold more ROS-producing cells compared to ΔmshA. The increased EGT levels observed in ΔmshA (Figure 4D) likely mitigates the endogenous ROS in this mutant. We conclude that while a lack of MSH substantially increases levels of EGT, loss of EGT does not lead to a corresponding increase in MSH/MSSM, which may be the cause of increased endogenous ROS levels observed in the egtA mutant. These data establish a physiological link between MSH and EGT production in Mtb and confirm their role in controlling endogenous ROS.
Role of EGT in Protection Against Oxidative Stress
The role of MSH in protecting mycobacteria from oxidative stress is well documented (Buchmeier et al., 2003; Miller et al., 2007). However, we know little about the role of EGT in combating oxidative stress in Mtb. We investigated the ability of EGT to protect Mtb under various conditions of oxidative stress. We observed that both egtA:Tn and egtD:Tn were significantly more sensitive than Wt upon exposure to hydrogen peroxide (H2O2), paraquat (PQT), menadione (MND) and cumene hydroperoxide (CHP) (Figure 4G). Survival of egtA:Tn and egtD:Tn was reduced most significantly by CHP and MND. Compared to egtA:Tn, viability of egtD:Tn was significantly decreased following exposure to H2O2 or PQT (Figure 4G). We noted that egtA:Tn is susceptible only at the higher concentration of PQT. Complementation of the mutants restored protection against the oxidants and viability was similar to that of Wt Mtb. Altogether, these findings provide evidence that EGT protects Mtb from a diverse set of oxidative stressors.
EGT and Antimycobacterial Drug Susceptibility
The redox status and metabolic state of Mtb can affect its sensitivity to anti-TB drugs (Baek et al., 2011; Kumar et al., 2011; Nandakumar et al., 2014). Here we tested the hypothesis that lack of EGT in Mtb modulates susceptibility to front line anti-TB drugs. To address this, we used a colony-forming unit (CFU)-based viability assay to determine drug susceptibilities. We observed that, compared to Wt Mtb, the minimal inhibitory concentration (MIC) of rifampicin (RIF), isoniazid (INH), bedaquiline (BDQ) and clofazimine (CFZ) were significantly reduced for the egtA:Tn, egtD:Tn and ΔmshA strains (Table S2). At MIC values obtained for Wt Mtb, the percent survival of egtA:Tn, egtD:Tn and ΔmshA was <40% and <25% of Wt Mtb in the presence of RIF and INH, respectively (Figure 4H). Similarly, against CFZ and the recently developed drug BDQ, survival of egtA:Tn and egtD:Tn strains was reduced to <70% and <50% of Wt Mtb, respectively. Genetic complementation of the egtA/D and mshA mutants reduced drug susceptibility to near wild type levels. As shown in Figure 4H, drug susceptibilities of the egt mutants against INH and RIF were comparable to that of ΔmshA (MIC= 0.015 and 0.006 μg/ml, respectively). Of note, ΔmshA was significantly more susceptible to BDQ and CFZ compared to Wt Mtb and the egtA/D mutants, revealing a greater role for MSH in protecting against these drugs. These data demonstrate that the lack of EGT results in increased susceptibility to anti-TB drugs and indicate that EGT and MSH are not fully redundant and have overlapping, but distinct functional roles in Mtb physiology.
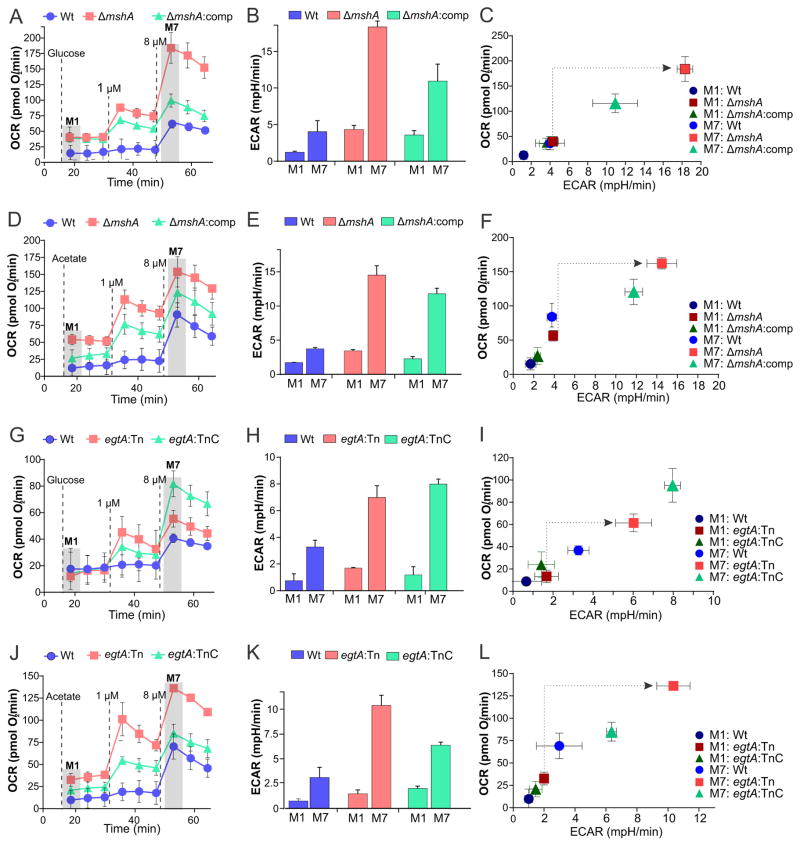
EGT and MSH are Critical for Bioenergetic Homeostasis in Mtb
Since oxidation/reduction reactions redox and energy metabolism are intricately linked, we tested the hypothesis that redox buffers MSH and EGT are essential for maintaining bioenergetic homeostasis. Extracellular flux analysis showed alterations in the bioenergetic profiles (OCR and ECAR) of mshA and egtA mutant strains when using glucose or acetate as the sole carbon source. Genetic complementation of these mutants restored the bioenergetic phenotype comparable to Wt, suggesting that lack of either EGT or MSH alters respiration in Mtb (Figure 5A–L). The OCR (M1) of egtA:Tn and ΔmshA grown in acetate was higher than that of their respective Wt strains (Figure 5D and J). However, addition of CCCP (M7) resulted in an increase in OCR (Figure 5A, D and J) and ECAR (Figure 5B, E, and K) of egtA and mshA mutants that was significantly greater than that of Wt and complemented strains, with the exception of the egtA complemented strain in glucose (M7, Figure 5G, H). This is also highlighted in the phenograms in Figures 5C, F, I and L where clear shifts from low OCR and ECAR in the mutants at M1 to higher OCR and ECAR than the Wt and complemented strains at M7. Importantly, the substantially different bioenergetic profiles upon addition of an uncoupler suggest electron transport chain dysfunction in the mutants resulting in greater OCR and ECAR in order to reestablish their membrane potentials. The unexpectedly high OCR observed in the egtA complemented strain at the higher concentration of CCCP (M7) (Figure 5G) could be due to aberrant behavior of constitutive hsp60 promoter in the episomal plasmid of the complement and has been observed previously in metabolic studies (Hartman et al., 2014). In sum, our data support the paradigm that alterations in redox status can modulate bioenergetic functions in Mtb cells.
Figure 5. MSH and EGT are critical for maintaining bioenergetic homeostasis in Mtb.
(A, D) OCR profiles of Wt Mtb (blue), ΔmshA (red) and ΔmshA:comp (green). (B, E) corresponding ECAR measurements and (C, F) phenograms of the three strains at M1 and M7, with 50 μM glucose (A, B, C) or 50 μM acetate (D, E, F) as carbon sources. (G, J) OCR profiles of Wt Mtb (blue), egtA:Tn (red) and egtA:TnC (green); (H, K) corresponding ECAR measurements. (I, L) phenograms of the three strains at M1 and M7, with 50 μM glucose (G, H, I) or 50 μM acetate (J, K, L) as a carbon source. Error bars represent SEM, n≥16.
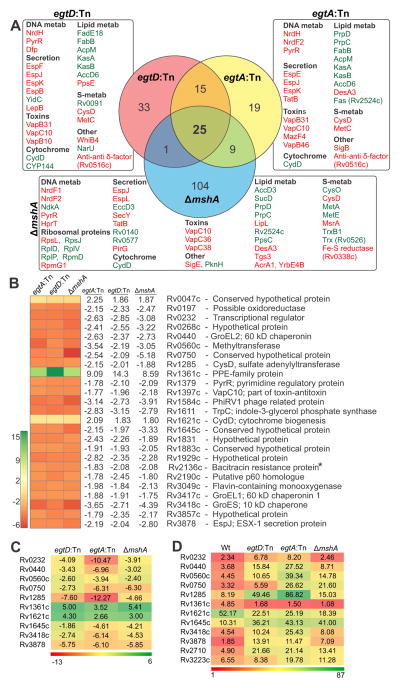
Identification of Differentially Regulated Genes in EGT- and MSH-deficient Mtb
Genes that contribute toward maintaining endogenous redox balance in Mtb are not well defined. To identify this core subset of genes, we carried out whole-genome transcriptomic profiling and compared the gene expression profiles of Mtb strains deficient in egtA (Figure S4), egtD (Figure S5) and mshA (Figure S6). Compared to Wt Mtb, 139 genes are differentially expressed in ΔmshA, and 68 and 74 genes in the egtA and egtD mutant strains, respectively (Figure 6A). Notably, a common set of 25 genes was differentially regulated across all three strains (Figure 6B). The changes in gene expression observed in the microarray analysis were validated independently in egtA:Tn, egtD:Tn and ΔmshA by quantitative RT-PCR analysis on a select subset of 10 genes (Figure 6C). Quantitative RT-PCR analysis of egtA:TnC, egtD:TnC and ΔmshA:comp yielded gene expression values comparable to Wt (Table S3).
Figure 6. Whole-genome transcriptional profiling of Wt Mtb, egtA:Tn, egtD:Tn and ΔmshA.
(A) Illustration of the overlap of the transcriptomic response of Mtb mutants defective in EGT and MSH production (Venn diagram). Also shown are selected sub-sets of differentially regulated genes in each strain classified according to function. Green: up-regulation, red: down-regulation See Figs. S4, S5 and S6 for a detailed list of differentially regulated genes. (B) Microarray analysis of egtA;Tn, egtD:Tn and ΔmshA grown under normal conditions identified a common set of 25 genes, 22 of which were downregulated. Genes displaying at least 1.75-fold variation in expression, with a q value of 1% were considered to be differentially regulated. The q value is the equivalent of the p value after multiple-testing correction (see Supplemental Experimental Procedures). (C) Quantitative RT-PCR analysis of a subset (10 of 25) of genes under normal growth conditions to validate microarray profiles. Expression of sigA was used as an internal control and gene expression values were normalized to Wt (Manganelli, et al., 1999). (D) Fold change in gene expression upon exposure to oxidative stress (CHP) relative to expression in unexposed cells. Expression of sigB (Rv2710) and sigH (Rv3223c) was used as a positive control to evaluate oxidative stress response. *Sole gene (Rv2136c) unaffected by any uncoupler, respiratory inhibitor, membrane potential or ΔpH modulator (Boshoff et al., 2004). See also Figures S4–S6 and Tables S3 and S4.
Importantly, 24 of the 25 differentially regulated genes shared by the EGT- and MSH-deficient strains are differentially regulated by inhibitors of respiration (KCN, thioridazine, 2,4-dinitrophenol), uncouplers (CCCP and N,N′-dicyclohexylcarbodiimide) or compounds that target an energized membrane (valinomycin) or ΔpH (nigericin) (Boshoff et al., 2004). Further, exposure of Wt and all three mutant strains to oxidative stress (CHP) resulted in substantially increased expression of the same subset of 10 genes used in the microarray validation (Figure 6D). This observation underscores the importance of this core set of genes in redox regulation in Mtb in the absence of EGT and MSH under conditions of increased oxidative stress.
These data demonstrate that a lack of EGT or MSH results in differential expression of genes involved in nucleotide and sulfur metabolism, secretion, toxin/anti-toxins, lipid metabolism and cytochrome biogenesis, likely as a compensatory response to the loss of redox couples. The regulation of trx genes by MSH, but not by EGT, is possibly an indicator of distinct chemical and functional requirements of mycobacterial redox buffers.
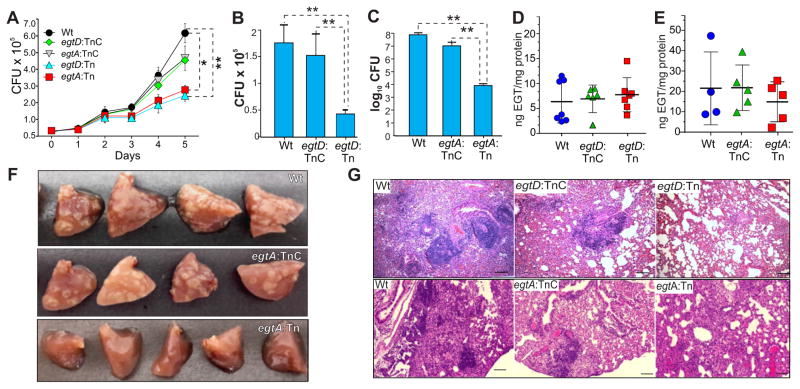
EGT Biosynthesis by Mtb is Required for Survival in Macrophages and Mice
We next examined the role of EGT in intracellular Mtb survival and virulence. Compared to Wt Mtb, we observed significantly reduced survival of egtA:Tn and egtD:Tn in RAW264.7 macrophages at 5 days post-infection (Figure 7A). Complementation of the egtA/D mutants restored intracellular survival to near Wt levels. To determine the role of EGT in Mtb pathogenicity, mice were infected with Wt, egtD:Tn, egtA:Tn or the complemented strains in two independent experiments. We observed significantly reduced bacillary burdens in the lungs of mice infected with egtD:Tn or egtA:Tn compared to mice infected with Wt Mtb. Complementation of egtD:Tn and egtA:Tn successfully restored bacillary load to Wt levels. The egtD:Tn showed ~4-fold decrease in lung bacillary burden, whereas the egtA:Tn showed a 4-log10 reduction (Figure 7B, C). Multiple lesions were observed in the lungs of mice infected with Wt and egtA:TnC strains compared to lungs of mice infected with egtA:Tn (Figure 7F). Histopathological analysis of lung sections from mice infected with Wt Mtb, egtA:TnC and egtD:TnC strains revealed large areas of alveolar consolidation due to lymphocyte-rich granulomatous infiltration (Figure 7G). In addition, multinucleated giant cells were observed in and around granulomas in the lungs of these mice. In contrast, egtA:Tn- and egtD:Tn-infected lungs showed diffuse areas of lymphocyte infiltration and exhibited visible alveolar spaces with partial lung parenchyma.
Figure 7. Survival and Pathogenicity of Mtb egt mutants in macrophages and mice.
(A) Survival of Wt Mtb, egtA:Tn, egtD:Tn and the corresponding complemented strains following macrophage infection. At the indicated time points, infected cells were lysed (n = 3) and lysates plated to enumerate CFU. Each data point represents the mean ± SEM (*p = 0.0142, **p = 0.0093). (B–E) Mice were infected with Wt Mtb, egtA:Tn, egtD:Tn or the complemented strains. At five weeks post infection, lungs were processed and bacillary burden (B, C) was determined. Columns represent the mean and error bars represent the standard deviation of the mean of 4–7 mice per group (**p<0.01). Corresponding EGT concentrations (D, E) were determined. Horizontal lines show the mean and error bars represent the standard deviation of the mean of 4–7 mice per group. (F) Gross pathology of mouse lungs infected with Wt Mtb, egtA:Tn and egtA:TnC. (G) H&E staining of lung tissue from infected mice (10x magnification). Scale bar = 100 μm.
Although it is known that EGT accumulates in organs that encounter oxidative stress (Cheah and Halliwell, 2012), it is not known whether infection by Mtb promotes accumulation of host-derived EGT and/or whether Mtb itself contributes significantly to the host EGT pool. Infection of macrophages with Wt Mtb, egtD:Tn, egtA:Tn and complemented strains (egtD:TnC and egtA:TnC) showed no significant difference in intracellular EGT concentrations (data not shown). Also, in mice infected with Wt Mtb, egtD:Tn, egtA:Tn or the complemented strains, we observed no statistically significant differences in lung EGT concentrations (Figure 7D, E). Altogether, our findings suggest that Mtb is not a significant contributor to host EGT during infection. In sum, independent disruption of two different genes within the EGT biosynthetic pathway results in attenuation in vivo and provides strong evidence for a role EGT in Mtb intracellular survival and virulence.
DISCUSSION
The aim of this study was to examine how Mtb WhiB3 maintains intracellular redox homeostasis and bioenergetics, to regulate virulence. Here we discovered that the virulence factor WhiB3 is a regulator of bioenergetic homeostasis, and controls the production of a major redox buffer EGT in a carbon source-dependent manner to maintain redox balance. Furthermore, by successfully adapted extracellular flux analysis technology, we have shown that a lack of EGT or MSH significantly alters Mtb respiratory activity, which establishes a direct link between redox homeostasis and bioenergetic metabolism. Importantly, we provide evidence that redox and bioenergetic homeostasis contribute to Mtb survival in macrophages and in mice, and are critical components in drug susceptibility and sensitivity to oxidative stress. The identification of bioenergetic deficiencies in Mtb mutants that display no apparent phenotypic defects when grown in vitro, but demonstrate attenuation upon mouse infection, points to bioenergetic homeostasis as a largely overlooked, but important factor in Mtb pathogenicity. We anticipate that bioenergetic homeostasis will broadly influence the study of microbial bioenergetics and pathogenesis. In sum, we have uncovered a dynamic balance between Mtb redox and bioenergetic homeostasis, which critically influence Mtb drug susceptibility and pathogenicity.
Our data indicate interdependency among central carbon metabolism, bioenergetic homeostasis and redox balance in Mtb that ultimately affects drug susceptibility. This is in line with increasing evidence of the link between bacterial metabolism and drug susceptibility. For example, Lobritz et al. (2015) demonstrated a link between cellular respiration in Escherichia coli and Staphylococcus aureus and antibiotic efficacy. Peng et al. (2015) showed that promoting TCA flux to increase NADH and consequently proton motive force in multidrug-resistant Edwardsiella tarda restored susceptibility to kanamycin. In the case of Mtb, metabolic profiling revealed that exposure to sub-lethal concentrations INH, RIF and streptomycin remodeled Mtb central carbon metabolism to enable antibiotic tolerance (Nandakumar et al., 2014). In addition, growth-limiting stresses that redirect Mtb carbon flux away from central carbon metabolism towards triglyceride synthesis have been shown to diminish antibiotic sensitivity (Baek et al., 2011). Our data affirm this link between metabolism and antibiotic efficacy and show how the bioenergetic and redox status of Mtb, in which EGT and MSH play a role, affect drug susceptibility. Likewise, our study indicates that the roles of EGT and MSH in bioenergetic and redox homeostasis are essential for the resilience of Mtb against oxidative stress, which is necessary for full virulence of Mtb.
What is the biological significance of having two redox buffers, EGT and MSH, to protect Mtb against oxidative environments? One explanation rests in the inherent physicochemical properties of these redox intermediaries. For example, the reduction potential (E0′) of EGT is −60 mV, and although the E0′ of MSH has not been determined, it is speculated to be in the range of −200 to −320 mV, as is typical for other naturally occurring thiols (Hartman, 1990). This would enable EGT and MSH to respond differently to a spectrum of oxido-reductive stresses to facilitate a successful infection. This redox hierarchy may be critical to infection as EGT has been shown to function as a scavenger of singlet oxygen, OH•, ONOO−, and HOCl radicals (Akanmu et al., 1991). Conversely, MSH has been shown to play a protective role against •NO, and CHP (alkyl peroxy radicals) (Miller et al., 2007). Notably, EGT does not readily undergo auto-oxidation and exists predominantly in the thione form rather than the thiol form, which confers greater stability compared to other thiols such as glutathione and MSH (Heath and Wildy, 1956). The reduction of MSSM to MSH requires an enzymatic system comprised of a mycoredoxin, mycothiol disulfide reductase (Mtr) and NADPH (Patel and Blanchard, 1999) making this a highly complex system compared to EGT (Cheah and Halliwell, 2012). Altogether, these physicochemical differences likely explain the overlapping but distinct functions of EGT and MSH and the need of redundancy in redox buffers in this highly evolved pathogen.
This EGT/MSH redox hierarchy is also a critical parameter in drug susceptibility and therefore has strong clinical relevance. For example, we found that egtA/D and mshA mutants are more susceptible than Wt Mtb to RIF, INH, CFZ and the new anti-TB drug BDQ. The egtA/D mutants are less sensitive than the mshA mutant to BDQ and CFZ, both of which target the Mtb electron transport chain (Complex V and Complex I, respectively), thus implicating redox balance and bioenergetic homeostasis in modulating drug susceptibility. The dissimilarities in drug susceptibility phenotypes between EGT and MSH mutants may be due to the increased levels of EGT observed in the MSH mutant. MSH deficiency has been reported to lead to ethionamide resistance (Vilcheze et al., 2008) and rifampin sensitivity (Buchmeier et al., 2003). Altogether, our results highlight similar yet distinct roles for EGT and MSH in maintaining redox balance and conferring drug susceptibility through different redox or energy-mediated mechanisms.
The ability of EGT to protect against oxidative stress caused by CHP, PQT, H2O2 and MND confirms that Mtb produces EGT to maintain redox balance. Not all oxidative stresses are equal as evidenced by differences in Mtb survival in the presence of various oxidants and the fact that a broad spectrum of free radical reactions exist, ranging from very reducing (e.g., Eo′ = −1800 mV for CO2/CO2•−) to very oxidizing (e.g., Eo′ = 2310 mV for HO•, H+/H2O) (Buettner, 1993). The production of both EGT and MSH to counter free radicals may partially explain the remarkable success of Mtb as an intracellular pathogen that persists for decades in the oxidative environment of the lung. This work also establishes a relationship between EGT and MSH production. Notably, the increased EGT levels and reduced endogenous ROS (Figure 4F) in ΔmshA reveal the presence of a compensatory mechanism wherein EGT production is increased to mitigate endogenous oxidative stress in the absence of MSH. Collectively, the data advocate that the level of EGT in the cell, which is modulated by WhiB3 and the availability of carbon sources, serves as a buffer against redox stress such as O2•− associated with cellular metabolic activity. The implication of this finding is significant as carbon sources such as fatty acids or oxidation products can modulate redox balance and thus drug susceptibility during infection by varying EGT levels.
To understand the effects of EGT or MSH deficiency on global gene expression in Mtb, we performed transcriptomic analysis of egtA, egtD and mshA mutants and identified 25 differentially regulated gene common to all three mutants. Our quantitative RT-PCR validation underscored the accuracy of our microarray-based gene expression analysis. Of these 25 genes, 24 are also modulated by respiration inhibitors, uncouplers or compounds that target an energized membrane (Boshoff et al., 2004), implicating EGT and MSH in maintenance of bioenergetic homeostasis. Our observations elucidate a bioenergetics-centered model of WhiB3-mediated redox balance through EGT and represent a paradigm for redox homeostasis in prokaryotes.
Upregulation of an operon involved in mycolic acid biosynthesis (Rv2243-2247) in the egtA/D mutants, and of the Rv1130-1131 operon (encoding PrpD and PrpC in the methylcitrate cycle) in the mshA and egtA mutants suggest that perturbations in redox balance due to lack of EGT or MSH affect biosynthesis of different classes of lipids, which may act as sinks for reducing equivalents (Muthukumaraswamy et al., 2009). An important dissimilarity between the expression profiles of egtA/D and mshA mutants is that thioredoxin B (trxB1, Rv1471) and a putative thioredoxin-like gene (trx, Rv0526) are upregulated in the mshA mutant, but not in the egtA/D mutants. This observation is consistent with the requirement of MSH for thioredoxins as reducing partners (Attarian et al., 2009). The majority of the differentially regulated genes (22 of 25) in the egtA/D and mshA mutants are downregulated. Interestingly, 17 of the 22 downregulated genes are upregulated (2 to 5-fold) during Mtb infection of macrophages or under nutrient deprivation conditions in vitro (Schnappinger et al., 2003; Betts et al., 2002). It is therefore reasonable to posit that redox buffers EGT or MSH may also modulate the survival of Mtb inside macrophages and thus influence the outcome of infection.
Our in vitro and in vivo infection studies clearly show that EGT-deficient Mtb strains are attenuated and establish that EGT is critical for the intracellular survival and virulence of Mtb (Figure 7A–C, F). These results provide important insights into the role of EGT in Mtb virulence in addition to its role in drug susceptibility, both of which appear to be distinct from MSH (Vilcheze et al., 2008). The observation that egtA and egtD contribute to different degrees to intracellular survival is supported by the finding that different intermediates of EGT biosynthesis may perform antioxidant functions (Song et al., 2015) and feed into different pathways of central carbon metabolism. Consequently, disruption of different steps in the EGT biosynthetic pathway in the egtA and egtD mutants likely explains the differences in attenuation (Figure 7A–C, F, G).
In summary, the findings in this study reveal new mechanisms by which Mtb perceives and relays metabolic signals to maintain redox balance and bioenergetic homeostasis. These findings establish important roles for EGT in oxidative stress protection, drug susceptibility and virulence in Mtb, which are unlike MSH. The discovery of an EGT/MSH hierarchy of redox defense mechanisms with distinct physicochemical properties to counteract a spectrum of host-generated free radicals and metabolites has significant implications for how Mtb maintains redox and bioenergetic homeostasis during persistence.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Culture Conditions
All Mtb strains used in this study, unless otherwise specified, were grown at 37°C in Gibco Middlebrook (MB)7H9 (broth) or MB7H10/7H11 (agar) media supplemented with 0.2% glycerol, 1x ADST (albumin saline enrichment (albumin-NaCl) with 0.05% tyloxapol) as described in Supplemental Experimental Procedures.
Liquid Chromatography/Mass Spectrometry (LC/MS) for Metabolome Analysis
Metabolites were separated on a Cogent Diamond Hydride Type C column and analyzed by LC/MS as described in Supplemental Experimental Procedures.
Bioenergetic Analysis of Mtb
OCR and ECAR of Mtb strains adhered to Cell-Tak® coated XF cell culture microplates (Seahorse Bioscience, USA) at 2×106 bacilli/well, were measured using a XF96 Extracellular Flux Analyzer as described in Supplemental Experimental Procedures.
TLC and LC/MS/MS Analysis of EGT
Cell extracts containing radiolabeled EGT were concentrated to dryness and resuspended in 50–100 μl of acetonitrile:methanol:50 mM ammonium acetate (40:40:20) [AMAA]. At least 30,000 cpm in a maximum of 15 μl was loaded onto silica gel TLC plates and developed in a 3:1 methanol:water solvent system as described in Supplemental Experimental Procedures. To validate TLC analysis of EGT, concentrated, unlabeled Msm extract was loaded on a silica gel TLC plate and developed as above. The suspected EGT spot was visualized under UV light, scraped from the plate, and extracted four times with 50 μl AMAA. It was then concentrated to dryness, resuspended in 20 μl AMAA and examined by an AB Sciex API-3200 Triple Quadrupole coupled to a Waters Acquity LC system. Data were collected in the positive electrospray mode. EGT levels in Mtb and mouse lungs were also determined using LC-MS/MS as described in Supplemental Experimental Procedures.
Measurement of Endogenous ROS in Mtb
The percentage of ROS-producing Mtb cells was determined using CellROX® Green (Life Technologies) and flow cytometry as described in Supplemental Experimental Procedures.
Determination of Mtb Susceptibility to Oxidants and Anti-TB Drugs
Mtb cells were cultured as described above. Mtb strains were exposed to oxidizing agents (6 hrs) or various concentrations of anti-TB drugs (7 days). Cells were plated on MB7H11 agar plates, colonies were enumerated after 4–5 weeks of incubation at 37°C, and percentage survival was determined. Additional details are provided in Supplemental Experimental Procedures.
Microarray Analysis
Mtb RNA was isolated and prepared as described in Supplemental Experimental Procedures.
Quantitative RT-PCR Analysis
Total RNA was isolated from mycobacterial cells and quantitative real time PCR (qRT-PCR) analysis was performed as described in Supplemental Experimental Procedures.
Mtb Infection Studies and Quantitation of EGT
For in vitro infection studies, RAW264.7 macrophages were infected with Mtb strains at an MOI of 10. Mice were used as our in vivo model of infection. Experimental details are outlined in Supplemental Experimental Procedures.
Statistics
Statistical computations were performed with Prism 6.0 software (GraphPad, La Jolla, California). Pairwise comparison was performed by Students t-test for normally distributed data. Multiple comparisons were performed using One-way ANOVA (Tukey’s test) module of GraphPad.
Supplementary Material
Acknowledgments
This work was supported by NIH grants AI076389, AI058131, DOD Discovery Award PR121320 and pilot funds from the UAB Centers for AIDS Research and Free Radical Biology, and UAB School of Medicine Infectious Diseases and Global Health and Vaccines Initiative to AJCS. AJCS and KR are Burroughs Welcome Investigators in the Pathogenesis of Infectious Disease. VS was supported by Senior Research Training Fellowship RT-232840-N from the American Lung Association. The research from which this publication emanated was co-funded by the South African Medical Research Council. The authors thank Amit Singh for helpful discussions, Nicholas J. Eustace for technical assistance, William R. Jacobs, Jr. (Albert Einstein College of Medicine) for providing ΔmshA and Digby Warner (University of Cape Town) for providing bedaquiline.
Footnotes
Data Submission: Microarray data reported in this study can be accessed from the EMBL ArrayExpress public data repository (accession number E-MTAB-4099).
Supplemental Information includes Supplemental Experimental Procedures, 6 figures and 4 tables and can be found with this article online.
Author Contributions
Conceptualization: V.S., L.G. and A.J.C.S.; Investigation: mass spectrometric and metabolomic analyses (J.H.A., H.E., K.R.), quantitation of EGT and MSH (L.G., Y.A-G., H.B.), XF assays (D.L.), ROS determination (B.M.C.), transcriptomic profiling and analysis (V.S.), macrophage assays (K.C.C.), drug and oxidative stress susceptibility assays (V.S. and V.P.R.), animal studies at UAB (V.S., V.P.R., K.C.C., and J.N.G.) and KRITH (J.M. and M.R-G.) and histopathological analysis (V.S. and V.P.R.); Validation: V.S. and K.C.C.; Data curation: V.S., D.L. and K.R.: Writing: V.S., B.M.C., L.G., J.N.G., and A.J.C.S.; Funding acquisition: A.J.C.S. All authors discussed the results and commented on the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akanmu D, Cecchini R, Aruoma OI, Halliwell B. The antioxidant action of ergothioneine. Arch Biochem Biophys. 1991;288:10–16. doi: 10.1016/0003-9861(91)90158-f. [DOI] [PubMed] [Google Scholar]
- Attarian R, Bennie C, Bach H, Av-Gay Y. Glutathione disulfide and S-nitrosoglutathione detoxification by Mycobacterium tuberculosis thioredoxin system. FEBS Lett. 2009;583:3215–3220. doi: 10.1016/j.febslet.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Baek SH, Li AH, Sassetti CM. Metabolic regulation of mycobacterial growth and antibiotic sensitivity. PLoS Biol. 2011;9:e1001065. doi: 10.1371/journal.pbio.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol. 2002;43:717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- Boshoff HI, Myers TG, Copp BR, McNeil MR, Wilson MA, Barry CE., 3rd The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism: novel insights into drug mechanisms of action. J Biol Chem. 2004;279:40174–40184. doi: 10.1074/jbc.M406796200. [DOI] [PubMed] [Google Scholar]
- Buchmeier NA, Newton GL, Koledin T, Fahey RC. Association of mycothiol with protection of Mycobacterium tuberculosis from toxic oxidants and antibiotics. Mol Microbiol. 2003;47:1723–1732. doi: 10.1046/j.1365-2958.2003.03416.x. [DOI] [PubMed] [Google Scholar]
- Buettner GR. The Pecking Order of Free Radicals and Antioxidants: Lipid Peroxidation, -Tocopherol, and Ascorbate. Arch Biochem Biophys. 1993;300:535–535. doi: 10.1006/abbi.1993.1074. [DOI] [PubMed] [Google Scholar]
- Cheah IK, Halliwell B. Ergothioneine; antioxidant potential, physiological function and role in disease. BBA Mol Basis Dis. 2012;1822:784–793. doi: 10.1016/j.bbadis.2011.09.017. [DOI] [PubMed] [Google Scholar]
- de Carvalho LP, Fischer SM, Marrero J, Nathan C, Ehrt S, Rhee KY. Metabolomics of Mycobacterium tuberculosis reveals compartmentalized co-catabolism of carbon substrates. Chem Biol. 2010;17:1122–1131. doi: 10.1016/j.chembiol.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Everts B, Amiel E, Huang SC, Smith AM, Chang CH, Lam WY, Redmann V, Freitas TC, Blagih J, van der Windt GJ, et al. TLR-driven early glycolytic reprogramming via the kinases TBK1-IKKvarepsilon supports the anabolic demands of dendritic cell activation. Nat Immunol. 2014;15:323–332. doi: 10.1038/ni.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrick DA, Neilson A, Beeson C. Advances in measuring cellular bioenergetics using extracellular flux. Drug Discov Today. 2008;13:268–274. doi: 10.1016/j.drudis.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Genghof DS. Biosynthesis of ergothioneine and hercynine by fungi and Actinomycetales. J Bacteriol. 1970;103:475–478. doi: 10.1128/jb.103.2.475-478.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand CE, Honek JF. Biological chemistry of naturally occurring thiols of microbial and marine origin. J Nat Prod. 2005;68:293–308. doi: 10.1021/np049685x. [DOI] [PubMed] [Google Scholar]
- Hartman PE. Ergothioneine as antioxidant. Methods Enzymol. 1990;186:310–318. doi: 10.1016/0076-6879(90)86124-e. [DOI] [PubMed] [Google Scholar]
- Hartman T, Weinrick B, Vilchèze C, Berney M, Tufariello J, Cook GM, Jacobs WR., Jr Succinate Dehydrogenase is the Regulator of Respiration in Mycobacterium tuberculosis. PLoS Pathog. 2014;10:e1004510. doi: 10.1371/journal.ppat.1004510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath H, Wildy J. The biosynthesis of ergothioneine and histidine by Claviceps purpurea. 1. The incorporation of [2-14C]-acetate. Biochem J. 1956;64:612. doi: 10.1042/bj0640612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Farhana A, Guidry L, Saini V, Hondalus M, Steyn AJ. Redox homeostasis in mycobacteria: the key to tuberculosis control? Expert Rev Mol Med. 2011;13:e39. doi: 10.1017/S1462399411002079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, VanderVen BC, Fahey RJ, Russell DG. Intracellular Mycobacterium tuberculosis exploits host-derived fatty acids to limit metabolic stress. J Biol Chem. 2013;288:6788–6800. doi: 10.1074/jbc.M112.445056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei B, Wei CJ, Tu SC. Action mechanism of antitubercular isoniazid activation by Mycobacterium tuberculosis KatG, isolation, and characterization of InhA inhibitor. J Biol Chem. 2000;275:2520–2526. doi: 10.1074/jbc.275.4.2520. [DOI] [PubMed] [Google Scholar]
- Lobritz MA, Belenky P, Porter CB, Gutierrez A, Yang JH, Schwarz EG, Dwyer DJ, Khalil AS, Collins JJ. Antibiotic efficacy is linked to bacterial cellular respiration. Proc Natl Acad Sci U S A. 2015;112:8173–8180. doi: 10.1073/pnas.1509743112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manganelli R, Dubnau E, Tyagi S, Kramer FR, Smith I. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol Microbiol. 1999;31:715–724. doi: 10.1046/j.1365-2958.1999.01212.x. [DOI] [PubMed] [Google Scholar]
- Miller CC, Rawat M, Johnson T, Av-Gay Y. Innate protection of Mycobacterium smegmatis against the antimicrobial activity of nitric oxide is provided by mycothiol. Antimicrob Agents Chemother. 2007;51:3364–3366. doi: 10.1128/AAC.00347-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthukumaraswamy SD, Edden RA, Jones DK, Swettenham JB, Singh KD. Resting GABA concentration predicts peak gamma frequency and fMRI amplitude in response to visual stimulation in humans. Proc Natl Acad Sci U S A. 2009;106:8356–8361. doi: 10.1073/pnas.0900728106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar M, Nathan C, Rhee KY. Isocitrate lyase mediates broad antibiotic tolerance in Mycobacterium tuberculosis. Nat Commun. 2014;5:4306. doi: 10.1038/ncomms5306. [DOI] [PubMed] [Google Scholar]
- Patel MP, Blanchard JS. Expression, purification, and characterization of Mycobacterium tuberculosis mycothione reductase. Biochemistry. 1999;38:11827–11833. doi: 10.1021/bi991025h. [DOI] [PubMed] [Google Scholar]
- Peng B, Su YB, Li H, Han Y, Guo C, Tian YM, Peng XX. Exogenous alanine and/or glucose plus kanamycin kills antibiotic-resistant bacteria. Cell Metab. 2015;21:249–261. doi: 10.1016/j.cmet.2015.01.008. [DOI] [PubMed] [Google Scholar]
- Rawat M, Johnson C, Cadiz V, Av-Gay Y. Comparative analysis of mutants in the mycothiol biosynthesis pathway in Mycobacterium smegmatis. Biochem Biophys Res Commun. 2007;363:71–76. doi: 10.1016/j.bbrc.2007.08.142. [DOI] [PubMed] [Google Scholar]
- Rhee KY, de Carvalho LP, Bryk R, Ehrt S, Marrero J, Park SW, Schnappinger D, Venugopal A, Nathan C. Central carbon metabolism in Mycobacterium tuberculosis: an unexpected frontier. Trends Microbiol. 2011;19:307–314. doi: 10.1016/j.tim.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard-Greenblatt M, Bach H, Adamson J, Peña-Diaz S, Li W, Steyn AJ, Av-Gay Y. Regulation of Ergothioneine Biosynthesis and its effect on Mycobacterium tuberculosis Growth and Infectivity. J Biol Chem. 2015;290:23064–23076. doi: 10.1074/jbc.M115.648642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sao Emani C, Williams MJ, Wiid IJ, Hiten NF, Viljoen AJ, Pietersen RD, van Helden PD, Baker B. Ergothioneine is a secreted antioxidant in Mycobacterium smegmatis. Antimicrob Agents Chemother. 2013;57:3202–3207. doi: 10.1128/AAC.02572-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, Monahan IM, Dolganov G, Efron B, Butcher PD, Nathan C. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages insights into the phagosomal environment. J Exp Med. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seebeck FP. In vitro reconstitution of Mycobacterial ergothioneine biosynthesis. J Am Chem Soc. 2010;132:6632–6633. doi: 10.1021/ja101721e. [DOI] [PubMed] [Google Scholar]
- Singh A, Crossman DK, Mai D, Guidry L, Voskuil MI, Renfrow MB, Steyn A. Mycobacterium tuberculosis WhiB3 maintains redox homeostasis by regulating virulence lipid anabolism to modulate macrophage response. PLoS Pathog. 2009;5:e1000545–e1000545. doi: 10.1371/journal.ppat.1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Guidry L, Narasimhulu KV, Mai D, Trombley J, Redding KE, Giles GI, Lancaster JR, Steyn AJC. Mycobacterium tuberculosis WhiB3 responds to O2 and nitric oxide via its [4Fe-4S] cluster and is essential for nutrient starvation survival. Proc Natl Acad Sci U S A. 2007;104:11562. doi: 10.1073/pnas.0700490104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Hu W, Naowarojna N, Her AS, Wang S, Desai R, Qin L, Chen X, Liu P. Mechanistic studies of a novel C-S lyase in ergothioneine biosynthesis: the involvement of a sulfenic acid intermediate. Sci Rep. 2015;5:11870. doi: 10.1038/srep11870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyn AJC, Collins D, Hondalus M, Jacobs WR, Kawakami RP, Bloom BR. Mycobacterium tuberculosis WhiB3 interacts with RpoV to affect host survival but is dispensable for in vivo growth. Proc Natl Acad Sci U S A. 2002;99:3147–3152. doi: 10.1073/pnas.052705399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilcheze C, Av-Gay Y, Attarian R, Liu Z, Hazbon MH, Colangeli R, Chen B, Liu W, Alland D, Sacchettini JC, et al. Mycothiol biosynthesis is essential for ethionamide susceptibility in Mycobacterium tuberculosis. Mol Microbiol. 2008;69:1316–1329. doi: 10.1111/j.1365-2958.2008.06365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.