Abstract
The structural diversity and biological activities of fungal indole diterpenes (IDTs) are generated in large part by the IDT cyclases (IDTCs). Identifying different IDTCs from IDT biosynthetic pathway is therefore important towards understanding how these enzymes introduce chemical diversity from a common linear precursor. However, IDTCs involved in the cyclization of the well-known aflavinine subgroup of IDTs have not been discovered. Here using Saccharomyces cerevisiae as a heterologous host and a phylogenetically-guided enzyme mining approach, we combinatorially assembled IDT biosynthetic pathways using IDTCs homologs identified from different fungal hosts. We identified the genetically standalone IDTCs involved in the cyclization of aflavine and anominine, and produced new IDTs not previously isolated. The cyclization mechanisms of the new IDTCs were proposed based on the yeast reconstitution results. Our studies demonstrate heterologous pathway assembly is a useful tool in the reconstitution of unclustered biosynthetic pathways.
The remarkable structural variations observed among natural products of the same biosynthetic origin arise from the catalytic activities of diversity-generating enzymes.1 These enzymes typically function downstream of conserved core enzymes that provide a common starting point for structural decoration. Identifying and reconstituting the activities of the diversity-generating enzymes can enable engineering of pathway towards production of compounds that are not observed in nature. Current approaches for natural product enzyme discovery rely heavily on the clustering of biosynthetic genes in the host genome.2 This feature enables the systematic screening for the desired enzyme using genetic and biochemical methods. However, such strategy is ineffective when the key enzymes are encoded separately in the genome.3 As a result, unclustered enzyme candidates are frequently overlooked and remain difficult to identify. One emerging strategy to overcome this limitation is to perform comprehensive and unbiased genome-wide searches for genes through sequence similarity, prioritize the candidates through phylogenetic analysis, and screen for activities via combinatorial assembly of pathways in genetically robust heterologous hosts.4 With the advent of powerful synthetic biology tools and vast genome resources, this strategy can accelerate the mining of new diversity-generating, biosynthetic enzymes.
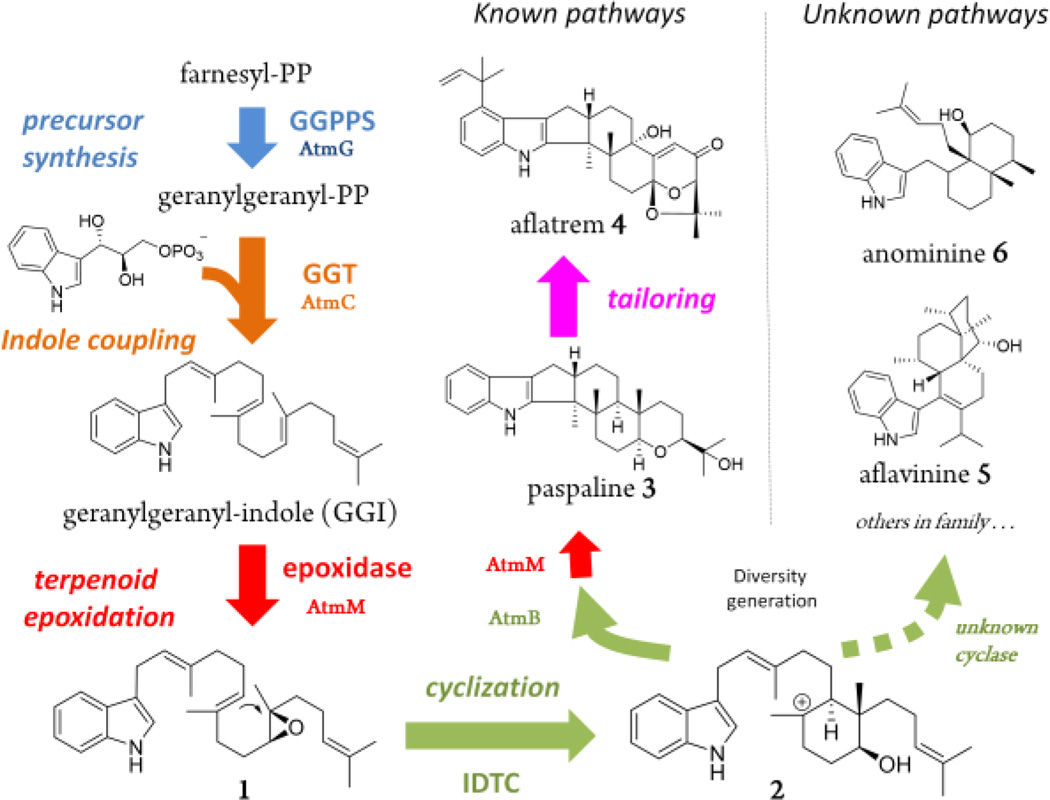
To apply the heterologous pathway assembly strategy, we focused on the fungal indole diterpenes (IDTs) and the associated diversity generating enzyme IDT cyclases (IDTC). IDTs form a structurally diverse family of natural products with a wide spectrum of biological activities, including insecticidal, anti-MRSA, and blocking of ion channels, etc.5 The structural complexity of IDTs has generated considerable total synthetic effort in recent years.6 Core enzymes in the pathway include GGPP synthase (GGPPS) that synthesize geranylgeranyl-PP (GGPP); geranylgeranyl transferase (GGT) that couples GGPP with indole-3-glycerol-phosphate to yield 3-geranylgeranyl-indole (GGI); and a regioselective flavin-dependent epoxidase that can produce the epoxide 1 (Figure 1).7 IDTCs from different pathways are proposed to cyclize 1 into at least two known large subgroups of IDTs via the common intermediate carbocation 2.7 IDTCs responsible for the formation of one subgroup of IDTs, which include paxilline,8 terpendole,9 lolitrem,10 penitrem,11 and aflatrem 4,12 have been discovered to be clustered with the core enzymes. On the other hand, IDTCs responsible for generating the other large subgroup with different diterpene cyclization and insecticidal profiles, as exemplified by aflavinine 513 and anominine 6,14 have remained undiscovered to date. Moreover, since the cyclization mechanisms of forming the aflavinine family of compounds are significantly different from those in the paspaline family, it remains unclear whether additional enzymes may be involved to generate the cyclized diterpenoids. As a result, no mechanistic proposal for the formation of these compounds is available. Therefore, uncovering these cyclizations steps by heterologously reassembled pathways can afford insights into the origin of structural diversity among IDTs and opportunities for engineering additional derivatives.
Figure 1.
The modular biosynthetic pathway of indole diterpenes. In the aflatrem biosynthetic gene cluster, AtmG, AtmC, AtmM and AtmB serve as GGPPS, GGT, epoxidase and IDTC, respectively.
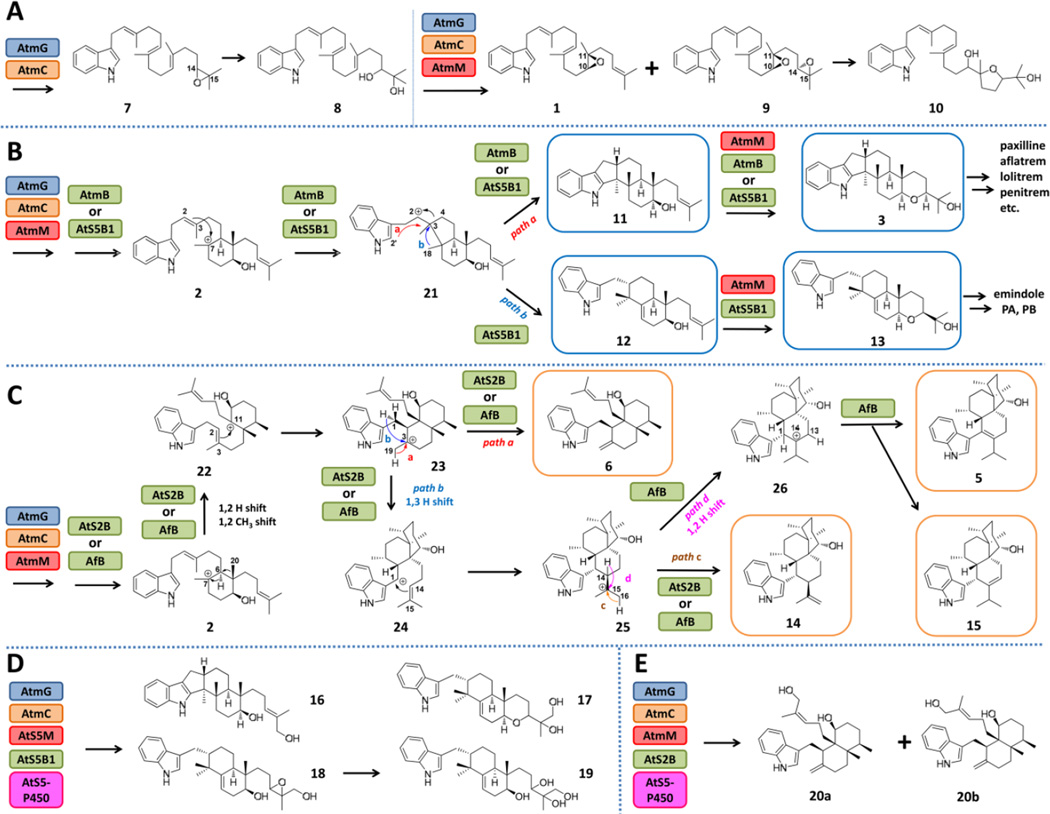
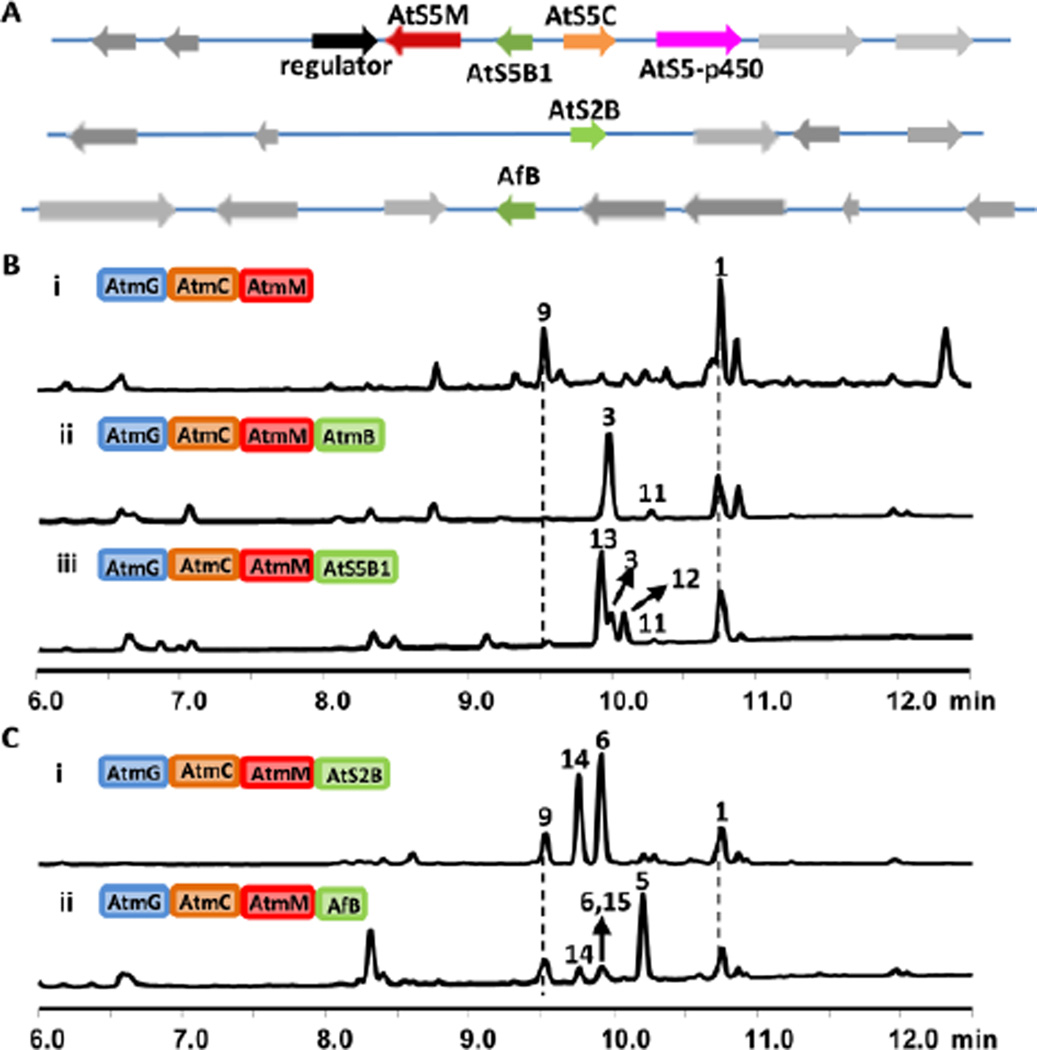
We chose to first reassemble the core biosynthetic steps leading to 1 in Saccharomyces cerevisiae, a host that has been widely used for producing and engineering fungal nature products.15 Genes from the aflatrem biosynthetic pathway (Figure 1),12 including atmG, atmC, and atmM were each inserted into 2µ vectors under the control of ADH2 promoters and introduced into yeast. When AtmG (GGPPS) and AtmC (GGT) were coexpressed, instead of the expected product GGI, a new compound 7 (MW=405) was formed (Figure S1, trace i). During the isolation process, 7 rapidly converted to the diol 8 (0.2 mg/L, all values represent purified titers) (Figures S28–S32, Table S7), which suggested that 7 is GGI containing a C14-C15 epoxide (Figure 4A). Upon additional coexpresion of the epoxidase AtmM, we isolated two indole containing compounds (Figure 2B, trace i). The MW 405 compound was verified to be the expected 1 (~ 1 mg/L) (Figures S4–S9, Table S3), while the MW 421 compound 9 was readily converted to the tetrahydrofuran-containing 10 (0.3 mg/L) (Figures S33–S37, Table S8), which can be formed through the hydrolytic epoxide opening of bis-epoxide 9. These results suggest that while yeast is able to produce 1, the terminal olefin is prone to oxidation by an endogenous yeast oxidase as seen in 7 and 9 in the absence of a dedicated downstream IDTC. Indeed, when AtmB was introduced into the above strain, the major product detected from the extract was paspaline 3 (1.5 mg/L) (Figures S10–S15, Table S4), while 9 was attenuated (Figure 2B, trace ii). We also detected small amounts of the precursor 11 (Figures S38–S43, Table S9), which is cyclized from the anti-Markovnikov addition product 21 to yield the 6-5-5-6-6 ring system (Figure 4B, path a). Oikawa et al previously showed homologs of AtmM and AtmB in the paxilline pathway can function iteratively to first form 11 from GGI, followed by additional epoxidation and cyclization to yield 3 (Figure 4B).7 Formation of 3 by the four Atm gene cassette in yeast demonstrates this host is suitable for producing IDTs and assaying IDTC functions.
Figure 4.
IDTs produced from the engineered yeast host and the proposed mechanisms of cyclization by the cloned IDTCs
Figure 2.
Reassemble IDT biosynthesis in yeast. A) The genetic loci containing IDTCs found in the genomes of A. tubingensis (At) and A. flavus (Af). atS2B and afB are standalone IDTCs. Annotation of genes can be found in Figure S2. B–C) HPLC analysis (λ=280 nm) of the extracts of yeast strains expressing different combinations of IDT biosynthetic enzymes from the B) paspaline-like and C) aflavinine–like pathways. For detailed MS and UV spectra of each peak, see Figure S3.
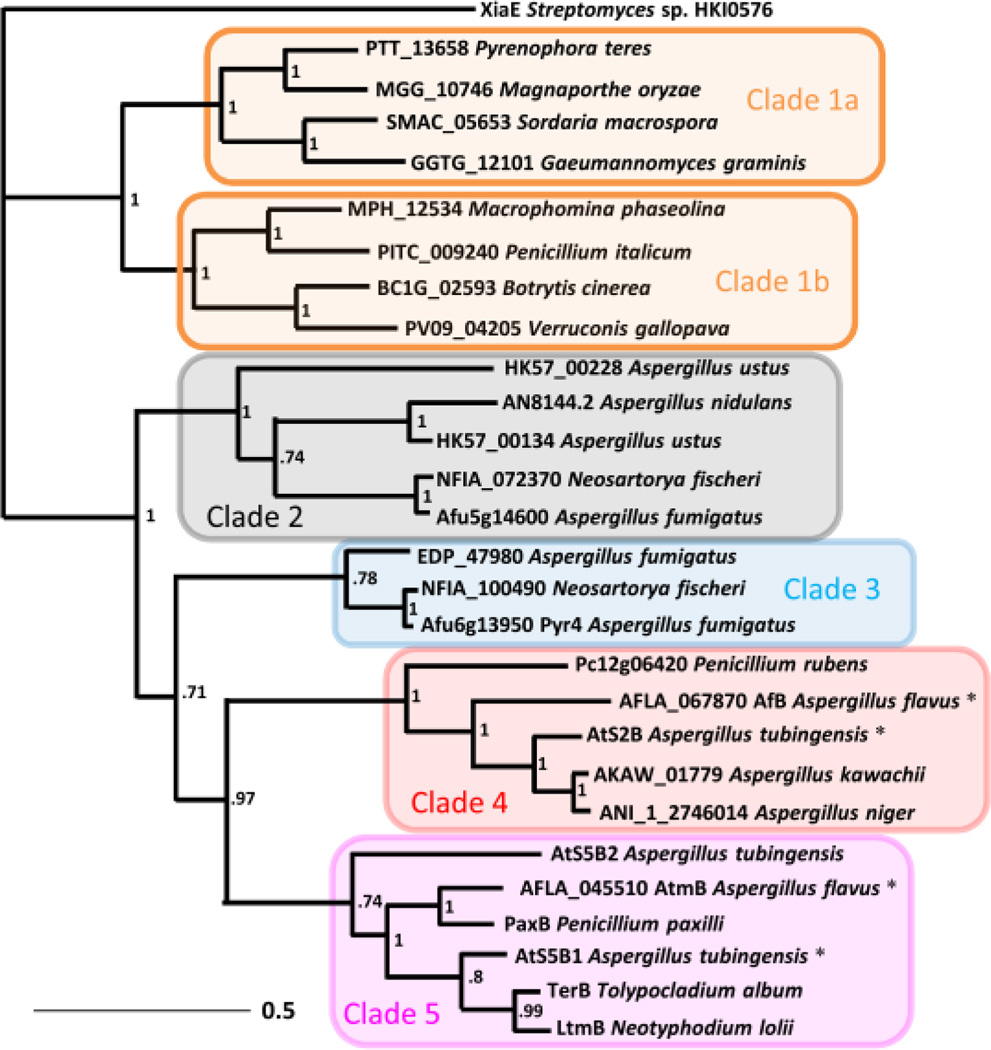
To facilitate search for IDTCs that may be responsible for the cyclization of 5 and 6, as well as to assess the sequence space of this structure-diversifying enzyme, we constructed a phylogenetic tree using fungal AtmB homologs identified through Protein BLAST (Supporting Information). AtmB is an integral membrane enzyme with seven transmembrane helices as predicted by the TMHMM server. More than 140 homologous enzymes were identified which can be loosely grouped into five clades (Figure 3 and Figure S90). Clade 5 includes known IDTC AtmB,12 PaxB,7 and TerB,9 which are all involved in the formation of paspaline family of compounds. Clade 3 is represented by Pyr4,17 which is responsible for the cyclization of sesquiterpene units of meroterpenoids such as pyripyropene. None of the AtmB homologs found in the other clades have known functions. Some of the Clade 1 and Clade 2 homologs are neighbored by PKS genes and are also likely involved in cyclization of polyketide-terpene hybrid compounds. Numerous members of the large Clade 1 and the entire Clade 4 are standalone in their respective fungal genomes, and are not flanked by clear secondary metabolite biosynthetic genes.
Figure 3.
Inferred phylogeny of selected IDTC and homologous proteins. Node support is shown as Bayesian posterior probabilities. The bacterial terpene cyclase XiaE16 is used as an outgroup. A comprehensive tree including all homologs from fungi is shown in Figure S90.
Multiple AtmB homologs are found in Clades 4 and 5 from A. tubingenesis, A. niger, A. flavus, which have all been noted to produce IDTs belonging to both subgroups.5, 18, 19 For example, A. tubingenesis has been shown to produce the cis-decalin containing 6, while A. flavus has been noted to produce both 6 and the 3-vinyl indole 5.5, 19 The three AtmB homologs from A. tubingenesis (AtS2B and AtS5B1) and A. flavus (AfB), and their neighboring regions are shown in Figures 2A and S2. Both AtS2B and AfB are Clade 4, standalone IDTC homologs, while AtS5B1 is grouped in Clade 5 with AtmB. We reason that phylogenetically grouped IDTC should have similar cyclization regioselectivity, hence AtS5B1 is unlikely to be responsible for formation of 5 and 6. To test this hypothesis, we cloned atS5B1 into the yeast strain expressing AtmGCM. Both 3 (0.3 mg/L) and 11 were present in the extract, confirming that AtS5B1 functions similarly as AtmB (Figure 2B, trace iii). In addition, two new IDTs 12 (0.3 mg/L) (Figures S44–S49, Table S10) and 13 (1 mg/L) (Figures S50–S55, Table S11) were detected and characterized. The ring-system present in 12 and 13, of which the indole ring is not fused to the cyclized terpene portion, is different from that observed in 11 and 3. Compound 13 serves as the precursor to several IDTs, such as emindole PA and emindole PB.20 Formation of the 3-alkyl indole 12 and 13 requires AtS5B1 to also catalyze a different mode of cyclization from 21, which involves 1,2-methyl migration and concomitant ring expansion to yield the 6-6 bicyclic terpene moiety (Figure 4B, path b). Therefore, AtS5B1 can catalyze the formation of two different IDTs (3 and 13) from the shared 21.
We then turned to the closely related, standalone Clade 4 AtmB homologs AtS2B and AfB as IDTC candidates for the formation of 5 and 6. AtS2B and AfB were individually introduced into yeast strains that express AtmGCM. When AtS2B was expressed, two new IDT products with the same MW (405) were produced at equal levels (Figure 2C, trace i). NMR analysis and comparison to a synthetic standard confirmed one of the products to be 6 (~1 mg/L) (Figures S22–S27, Table S6).6d The other compound was solved to be 10,23-dihydro-24,25-dehydroaflavinine 14 (~1 mg/L) (Figures S56–S61, Table S12), which was also isolated from A. tubingenesis along with 6.19a Similarly, when AfB was coexpressed with AtmGCM, the yeast host produced numerous new compounds all exhibiting the same MW (Figure 2C, trace ii). The major product was characterized to be 5 (~1 mg/L) (Figures S16–S21, Table S5), while the minor products corresponded to 6, 14 and 15 (Figures S62–S67, Table S13) (~0.1 mg/L each).
Therefore, phylogeny-guided pathway reassembly uncovered the standalone IDTCs AtS2B and AfB in Clade 4 are solely responsible for the alternative mode of terpene cyclization in the biosynthesis of IDTs. These results also confirmed 1, which is the product of AtmGCM (Figure 1) is also the substrate for the new IDTCs. This allows us to propose the cyclization steps as shown in Figure 4C. Instead of the intermediate 21 required for paspaline like IDTs, IDTCs such as AfB and AtS2B most likely catalyze a concerted 1,2-hydride shift and 1,2-methyl shift of 2 to yield 22, in which the C20 methyl is migrated to C6 (Figure 4C). Attack of C2 on C11 carbocation then forms the cis-decalin carbocation 23. The cyclization steps leading to 5 and 6 then diverges. Deprotonation of the methyl group at C19 forms the exo-methylene containing 6 (Figure 4C, path a), which can be further cyclized into the carbazole containing IDT such as tubingensin A.6d Alternatively, 23 can undergo 1,3-hydride shift to yield the indole-stabilized carbocation at C1 in 24 (Figure 4C, path b), which can be attacked by the terminal olefin to form 25. From there, different paths of hydride shift and deprotonation result in formation of 5 and related congeners such as 14 and 15 (Figure 4C). Based on the product profiles, AtS2B can catalyze both cyclization paths (Figure 4C, paths a and b) from 23, but is unable to promote the additional 1,2-hydride shift to form 26 (Figure 4C, path d), which is required for the formation of 5. This observation is consistent with 5 not being reported to be isolated from A. tubingenesis. In contrast, it appears AfB to be considerably more promiscuous than AtS2B and can singlehandedly catalyze all four paths of cyclization starting from 22 as shown in Figure 4. Interestingly, A. flavus has been reported to be producers of all the products derived from observed in the reconstituted yeast host (5, 6, 14 and 15)5, 19. Therefore, these results validate the reassembled pathways in yeast can capture the differences in cyclization mechanisms of the individual IDTCs, and can generate products that are consistent with chemotypes of the parent fungi.
To further demonstrate the yeast platform can be engineered to include additional downstream diversity generating enzymes, we examined P450 enzymes that may modify the IDTs. Several Clade 5 IDTCs are grouped closely with AtS5B1, such as AKAW_02101 from A. kawachii and ANI_1_1736094 from A. niger. Each of these gene clusters contains an additional P450 that show low sequence homology (<20%) to characterized P450s from the paxilline pathway (PaxP and PaxQ).7 To determine the role of the P450 and the IDTs produced from these clusters, we expressed AtmGC, AtS5M (homolog of AtmM), AtS5B1 and the AtS5-P450 in the yeast strain RC01, which encodes a genome-integrated copy of the A. terreus cytochrome P450 reductase (CPR) gene. Three hydroxylated IDTs (16–18) related to 11–13 were detected and isolated (Figure S1, trace ii). Structural elucidation revealed that these compounds are oxidized at the terminal C17 methyl group (Figure 4D, Figures S68-S78 and Table S14–S15). The structure of 18 was inferred from that of the tetra-ol 19 (Figures S79-S83, Table S16), which readily formed during the purification of 18. Therefore, AtS5-P450 can hydroxylate terminal methyl groups of cyclized IDTs. Unexpectedly, when AtS5-p450 was combinatorially coexpressed with AtmGCM and AtS2B, hydroxylation of the terminal methyl group in 6 led to 20a and 20b (Figure 4E, and Figure S1, trace iii) (Figures S84–S89, Table S17). The formation of these new IDT compounds showcases the potential for combining previously unclustered genes to yield novel products.
The combinatorial assembly of fungal IDT pathways in yeast guided by phylogenetic classification enabled us to identify new cyclases that would have otherwise been overlooked due to its unclustered nature. This approach can be applied towards the reconstruction of other structurally diverse natural product families. There is much to explore with just the IDTC homologs shown in Figure 3 and S90. More than 90% of the identified IDTC homologs have unknown functions. In addition to the possibility of producing novel IDT structures, we speculate that some of these homologs are responsible for the synthesis of indole sesquiterpenes, which has attracted significant attention both synthetically and biosynthetically recently,21 although no biosynthetic pathway is known. In addition, a number of these homologs with polyketide synthases nearby may lead to the synthesis of new meroterpenoid compounds. Each of these pathways generates diversity from a common intermediate, and is therefore amendable to study using the approach described in this work. An exhaustive characterization of the IDTC homologs and reassembly of the corresponding pathways will afford new and engineered natural products.
Supplementary Material
Acknowledgments
This work was supported by NIH 1U01GM110706-01 and 1DP1GM106413. We thank Prof. Li Ang at SIOC for an authentic standard of 6. We thank Prof. James Gloer for insightful discussions. NMR instrumentation was supported by the NSF CHE-1048804. We thank Michael Corsello for assistance with spectra collection.
Footnotes
ASSOCIATED CONTENT
Supporting Information
Experimental details and spectroscopic data. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.(a) Jakubczyk D, Cheng JZ, O’Connor SE. Nat. Prod. Rep. 2014;31:1328. doi: 10.1039/c4np00062e. [DOI] [PubMed] [Google Scholar]; (b) Geris R, Simpson TJ. Nat. Prod. Rep. 2009;26:1063. doi: 10.1039/b820413f. [DOI] [PubMed] [Google Scholar]
- 2.(a) Owen JG, Reddv BV, Ternei MA, Charlop-Powers Z, Calle PY, Kim JH, Brady SF. Proc. Natl. Acad. Sci. U. S. A. 2013;110:11797. doi: 10.1073/pnas.1222159110. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Yamanaka K, Reynolds KA, Kersten RD, Ryan KS, Gonzalez DJ, Nizet V, Dorrestein PC, Moore BS. Proc. Natl. Acad. Sci. U. S. A. 2014;111:1957. doi: 10.1073/pnas.1319584111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Richter TK, Hughes CC, Moore BS. Environ. Microbiol. 2015;17:2158. doi: 10.1111/1462-2920.12669. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Sanchez JF, Entwistle R, Hung JH, Yaegashi J, Jain S, Chiang YM, Wang CC, Oakley BR. J. Am. Chem. Soc. 2011;133:4010. doi: 10.1021/ja1096682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Lau W, Sattely ES. Science. 2015;349:1224. doi: 10.1126/science.aac7202. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Hawkins KM, Smolke CD. Nat. Chem. Biol. 2008;4:564. doi: 10.1038/nchembio.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gloer JB. Acc. Chem. Res. 1995;28:343. [Google Scholar]
- 6.(a) Zou Y, Melvin JE, Gonzales SS, Spafford MJ, Smith AB., III J. Am. Chem. Soc. 2015;137:7095. doi: 10.1021/jacs.5b04728. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Sharpe RJ, Johnson JS. J. Am. Chem. Soc. 2015;137:4968. doi: 10.1021/jacs.5b02631. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Goetz AE, Silberstein AL, Corsello MA, Garg NK. J. Am. Chem. Soc. 2014;136:3036. doi: 10.1021/ja501142e. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Bian M, Wang Z, Xiong X, Sun Y, Matera C, Nicolaou KC, Li A. J. Am. Chem. Soc. 2012;134:8078. doi: 10.1021/ja302765m. [DOI] [PubMed] [Google Scholar]; (e) Enomoto M, Morita A, Kuwahara S. Angew. Chem. Int. Ed. 2012;51:12833. doi: 10.1002/anie.201206299. [DOI] [PubMed] [Google Scholar]
- 7.Tagami K, Liu C, Minami A, Noike M, Isaka T, Fueki S, Shichijo Y, Toshima H, Gomi K, Dairi T, Oikawa H. J. Am. Chem. Soc. 2013;135:1260. doi: 10.1021/ja3116636. [DOI] [PubMed] [Google Scholar]
- 8.Young C, McMillan L, Telfer E, Scott B. Mol. Microbiol. 2001;39:754. doi: 10.1046/j.1365-2958.2001.02265.x. [DOI] [PubMed] [Google Scholar]
- 9.Motoyama T, Hayashi T, Hirota H, Ueki M, Osada H. Chem. Biol. 2012;19:1611. doi: 10.1016/j.chembiol.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 10.Young CA, Felitti S, Shields K, Spangenberg G, Johnson RD, Bryan GT, Saikia S, Scott B. Fungal Genet. Biol. 2006;43:679. doi: 10.1016/j.fgb.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Liu C, Tagami K, Minami A, Matsumoto T, Frisvad JC, Suzuki H, Ishikawa J, Gomi K, Oikawa H. Angew. Chem. Int. Ed. 2015;54:5748. doi: 10.1002/anie.201501072. [DOI] [PubMed] [Google Scholar]
- 12.Nicholson MJ, Koulman A, Monahan BJ, Pritchard BL, Payne GA, Scott B. Appl. Environ. Microbiol. 2009;75:7469. doi: 10.1128/AEM.02146-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallagher RT, Clardy J, Wilson BJ. Tetrahedron Lett. 1980;21:239. [Google Scholar]
- 14.Rinderknecht BL, Gloer JB. J. Org. Chem. 1989;54:2530. [Google Scholar]
- 15.(a) Tsunematsu Y, Ishiuchi K, Hotta K, Watanabe K. Nat. Prod. Rep. 2013;30:1139. doi: 10.1039/c3np70037b. [DOI] [PubMed] [Google Scholar]; (b) Jacubczyk D, Caputi L, Hatsch A, Nielsen CAF, Diefenbacher M, Klein J, Molt A, Schroder H, Cheng JZ, Naesby M, O’Connor SE. Angew. Chemie. Intl. Ed. 2015;54:5117. doi: 10.1002/anie.201410002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu Z, Baunach M, Ding L, Hertweck C. Angew. Chemie. Intl. Ed. 2012;51:10293. doi: 10.1002/anie.201204087. [DOI] [PubMed] [Google Scholar]
- 17.Itoh T, Tokunaga K, Matsuda Y, Fujii I, Abe I, Ebizuka Y, Kushiro T. Nat. Chem. 2010;2:858. doi: 10.1038/nchem.764. [DOI] [PubMed] [Google Scholar]
- 18.Frisvad JC, Petersen LM, Lyhne EK, Larsen TO. PLoS One. 2014;9:e94857. doi: 10.1371/journal.pone.0094857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.(a) TePaske MR, Gloer JB. Tetrahedron. 1989;45:4961. [Google Scholar]; (b) Gloer JB, TePaske MR, Sima JS. J. Org. Chem. 1988;53:5457. [Google Scholar]; (c) TePaske MR, Gloer JB. J. Nat. Prod. 1992;55:1080. [Google Scholar]
- 20.Hosoe T, Itabashi T, Kobayashi N, Udagawa S, Kawai K. Chem. Pharm. Bull. (Tokyo) 2006;54:185. doi: 10.1248/cpb.54.185. [DOI] [PubMed] [Google Scholar]
- 21.(a) Marcos IS, Moro RF, Costales I, Basabe P, Díez D. Nat. Prod. Rep. 2013;30:1509. doi: 10.1039/c3np70067d. [DOI] [PubMed] [Google Scholar]; (b) Zhou S, Chen H, Luo Y, Zhang W, Li A. Angew. Chem. Int. Ed. 2015;54:6878. doi: 10.1002/anie.201501021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.