SUMMARY
Cyclic dinucleotides are second messengers that target the adaptor STING and stimulate the innate immune response in mammals. Besides protein receptors, there are bacterial riboswitches that selectively recognize cyclic dinucleotides. We recently discovered a natural riboswitch that targets 3’,3’-cGAMP, which is distinguished from the endogenous mammalian signal 2’,3’-cGAMP by its backbone connectivity. Here we report on structures of the aptamer domain of the 3’,3’-cGAMP riboswitch from Geobacter in the 3’,3’-cGAMP and c-di-GMP bound states. The riboswitch adopts a tuning fork-like architecture with a junctional ligand-binding pocket and different orientations of the arms are correlated with the identity of the bound cyclic dinucleotide. Subsequent biochemical experiments revealed that specificity of ligand recognition can be affected by point mutations outside of the binding pocket, which has implications for both the assignment and reengineering of riboswitches in this structural class.
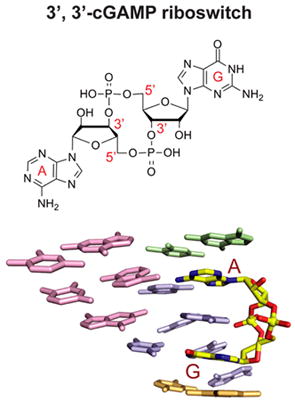
Graphical abstract
INTRODUCTION
Cyclic dinucleotides such as c-di-GMP (reviewed in Hengge, 2009) and c-di-AMP (Witte et al 2013) containing both 3’,5’, 3’,5’ (designated 3’,3’) linkages have established roles as bacterial second messengers, with c-di-GMP contributing to motility, biofilm formation, and virulence, while c-di-AMP regulates sporulation, cell wall metabolism and osmotic stress responses (reviewed in Romling et al. 2013). The mixed purine cyclic dinucleotide cyclic GMP-AMP (cGAMP) has also come into prominence, initially for the role of 3’,3’-cGAMP in intestinal colonization by bacteria (Davies et al. 2012), and more recently, 2’,3’-cGAMP {c[G(2’,5’)pA(3’,5’)p} has been found to serve as an endogenous second messenger in stimulating the innate immune response in mammalian cells (Sun et a. 2013; Wu et al. 2013). There is also now a wealth of evidence that the protein STING (stimulator of interferon genes), an endoplasmic reticulum adaptor that facilitates innate immune signaling (Ishikawa and Barber, 2008), is a direct innate immune sensor of c-di-GMP (Burdette et al. 2011) and both 2’,3’ and 3’,3’ linkage isomers of cGAMP (Ablasser et al. 2013; Diner et al. 2013; Gao et al. 2013; Zhang et al. 2013). This in turn has raised interest in the potential of cyclic dinucleotides as adjuvants for vaccine development (Gray et al. 2012; Kim et al. 2014).
To date, gene regulatory RNA elements called riboswitches (reviewed in Serganov and Nudler, 2013) have been identified that bind and respond to c-di-GMP and c-di-AMP with high specificity and affinity. Two distinct classes of riboswitches target c-di-GMP (Sudarsan et al. 2008; Lee et al. 2010), while one class targets c-di-AMP (Nelson et al. 2013). Structures of c-di-GMP bound to class-I (Smith et al. 2009; Kulshina et al. 2009) and class-II (Smith et al. 2011) GEMM riboswitches and for c-di-AMP bound to the ydaO family of riboswitches (Ren and Patel, 2014; Gao and Serganov, 2014; Jones and Ferre-D’Amare, 2014) have been determined. These structures reveal that the RNAs use different scaffolds and recognition principles for targeting a specific cyclic dinucleotide.
One of our groups has focused on development of RNA-based fluorescent biosensors for live cell imaging of cyclic dinucleotides (Kellenberger et al. 2013), and to this end discovered natural 3’,3’-cGAMP (see formula in Figure S1A, Supplementary Materials) riboswitches (GEMM-Ib), as well as identified a 3’,3’-cGAMP-mediated signaling pathway in Geobacter (Kellenberger et al. 2015). These 3’,3’-cGAMP riboswitches also have been implicated as regulators of genes involved in exoelectrogenesis in Deltaproteobacter (Nelson et al. 2015).
Given the current interest on the role of linkage isomers of cGAMP (Figure S1A) as second messengers modulating the innate immune response (reviewed in Cai et al. 2014; Danlichanka and Mekalanos, 2013; Hornung et al. 2014), we undertook a systematic structural investigation of the 3’,3’-cGAMP riboswitch from Geobacter in the bound state in order to understand the molecular basis for its ligand discrimination and to facilitate the forward engineering of a 2’,3’-cGAMP riboswitch, as natural examples are yet to be identified. The structure of the complex has defined the overall scaffold of the bound RNA, the binding pocket architecture, and intermolecular contacts, whereby the 3’,3’-cGAMP riboswitch targets 3’,3’-cGAMP and discriminates against its 2’,2’ and 2’,3’ linkage counterparts. In addition, biochemical evidence indicates that non-contacting nucleotides affect ligand specificity and enable the 3’,3’-cGAMP riboswitch to preferentially bind 3’,3’-cGAMP over c-di-GMP. The structural basis of this effect is examined and further led to prediction of additional 3’,3’-cGAMP riboswitches from Pelobacter propionicus, which were subsequently validated. Finally, the observation of an unanticipated enol alignment of Gα, or alternately protonation of the N1 of its pairing partner A, for c-di-GMP bound to the 3’,3’-cGAMP riboswitch, suggests an intrinsic mechanism for promiscuous binding that must be overcome to distinguish between these two bacterial second messengers. Taken together, our findings reveal that specificity of ligand recognition by riboswitches can be markedly influenced by point mutations outside of the binding pocket, which has implications for both assigning and reengineering of natural riboswitches.
RESULTS
Our structural efforts have focused on the aptamer domains of 3’,3’-cGAMP riboswitches from Geobacter species, with initial crystallization trials undertaken on two RNAs called Gm0970 and Gs1761. The Gm0970 riboswitch resides upstream of the XRE-PilMNOPQ operon in Geobacter metallireducens, whereas the Gs1761 riboswitch resides upstream of the pgcA gene in Geobacter sulfurreducens (Kellenberger et. al. 2014). Both RNAs yielded crystals, but based on the size and the resolution of their diffraction patterns, we focused our efforts on Gs1761, whose secondary structure and numbering system is shown in Figure 1A. The crystallography construct includes the following modifications to the wild-type aptamer sequence: A2G (to facilitate in vitro transcription), U72C and C73U (these changes reflect an earlier sequencing error that was only corrected on completion of structure determination; residues at positions 72 and 73 are not highly conserved, see Figure S2C). The 84-mer RNA corresponding to the aptamer domain of the 3’,3’-cGAMP riboswitch adopts a three-helical junctional scaffold (Figure 1A), with bulges observed in all three stems (P1, P2 and P3).
Figure 1. Sequence, Structure and Pairing Alignments of the 3’,3’-cGAMP Riboswitch with bound 3’,3’-cGAMP.
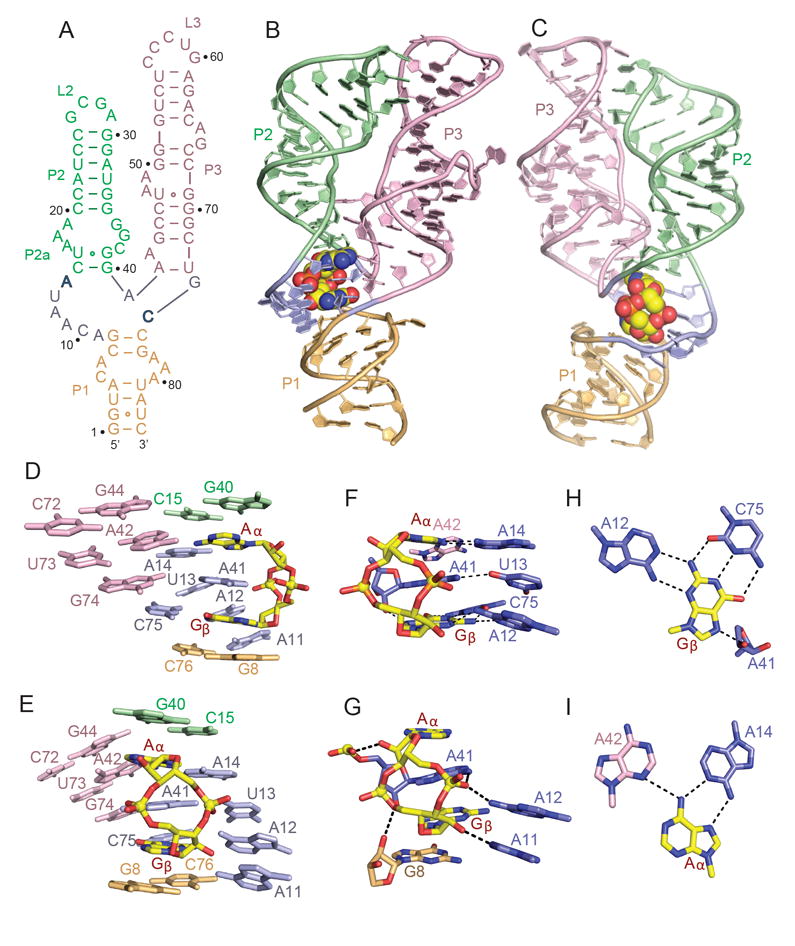
(A) Schematic of the secondary structure of the Gs1761 3’,3’-cGAMP riboswitch. This sequence differs from the natural riboswitch by containing A2G, U72C and C73U substitutions.
(B, C) Two alternate views of the 2.05 A structure of the 3’,3’-cGAMP riboswitch with bound 3’,3’-cGAMP. The riboswitch RNA is shown in a ribbon representation and color coded by segments, while the bound 3’,3’-cGAMP is shown in a space-filling representation. Residues A14 and C74 that interact through base pairing with the bound cGAMP are shown in bold.
(D, E) Two alternate views showing base stacking alignments within the 3’,3’-c-GAMP riboswitch (color-coded) centered about the bound 3’,3’-cGAMP (in yellow).
(F, G) Two alternate views showing intermolecular base-base (panel F) and base-sugar-phosphate (panel G) hydrogen bonding interactions between 3’,3’-cGAMP (in yellow) and the riboswitch RNA residues (color-coded) centered about the binding site in the complex.
(H) Intermolecular recognition of Gβ of 3’,3’-cGAMP (in yellow) by base-base hydrogen bonding with riboswitch RNA bases (in blue) in the complex.
(I) Intermolecular recognition of Aα of 3’,3’-cGAMP (in yellow) by base-base hydrogen bonding with riboswitch RNA bases (color-coded) in the complex. See also Table S1 and Figure S1.
Structure of the 3’,3’-cGAMP Riboswitch Bound to 3’,3’-cGAMP
We have successfully grown crystals of the aptamer domain of the 3’,3’-cGAMP riboswitch (Figure 1A) in the 3’,3’-cGAMP-bound state that diffract to 2.05 Å resolution and determined the structure using the molecular replacement method (PDB code: 3IRW; structure of c-di-GMP bound to GEMM-I class c-di-GMP riboswitch) to solve the phase problem (x-ray statistics listed in Table S1). The aptamer domain of the riboswitch adopts a tuning fork architecture with P1 as the handle and P2 and P3 as the prongs, with the bound cGAMP adopting a single orientation within the zippered up junction (Figure 1B, C). Thus, the global architecture is similar to the Vc2 GEMM-I class riboswitch aptamer domain bound to c-di-GMP (Smith et al. 2009; Kulshina et al. 2009).
Three-way Junction Zippers Up on Complex Formation
The three-way junction involving residues A9-C10-A11-A12-U13-A14, A41, and G74-C75 is zippered up by base stacking and hydrogen bond formation in the structure of the complex (Figure 1D and 1E). The sugar pucker geometries of A41, A42, and A43 in the vicinity of three-way junction are C2’-endo. The bound 3’,3’-cGAMP is an integral part of this junction in the complex, with terminal pair G40-C15 from stem P2 and terminal pair G8-C76 from stem P1 bracketing the bound cyclic dinucleotide (Figure 1D and E). An omit map highlighting the electron density of the bound 3’,3’-cGAMP is shown in Figure S1B. The base A41 is sandwiched between the guanine (labeled Gβ) and adenine (labeled Aα) of the bound cGAMP (Figure 1D, E), making observed intermolecular contacts with the Aα and Gβ bases (Figure 1F) and sugar-phosphate (Figure 1G) of cGAMP. The Gβ base edges of bound cGAMP are all involved in intermolecular hydrogen bond formation, with recognition of its Watson-Crick edge by the Watson-Crick edge of C75, its sugar edge by the Watson-Crick edge of A12, and its Hoogsteen edge by the sugar 2’-OH of A41 (Figure 1H). By contrast, only the Hoogsteen edge of Aα is recognized through intermolecular hydrogen bond formation by the Watson-Crick edges of A14 (trans Hoogsteen-Watson-Crick Aα•A14 non-canonical pair) and A42 (weak hydrogen bond) (Figure 1I).
Parallel Alignment of Stems P2 and P3 in the Complex
Stems P2 and P3 are aligned in a parallel orientation in the structure of the complex (Figure 1B, C) due to formation of loop (L2) - receptor (L3/P3) interactions. The interaction is shown schematically by a red box labeled 1 in Figure 2A with the stabilizing hydrogen bond network depicted in Figure 2B. The base edges of the stacked C27-G28-A29 segment (stable GNRA loop in green) within L2 form hydrogen bonds with the sugar-phosphate backbone (in pink) of the stacked C54-U55-C56 segment emanating from stem P3 (C54-U55) and loop L3 (C56) (Figure 2B). In addition, the parallel alignment is stapled by formation of stacked adjacent minor groove triples between looped out purines on stem P3 (in pink) and receptor pairs (in green) on stem P2 (Figure 2C).
Figure 2. Long-range Loop-Loop Interactions and Topology of an Internal Bulge in Stem P3, P2, and P1 in the Complex.
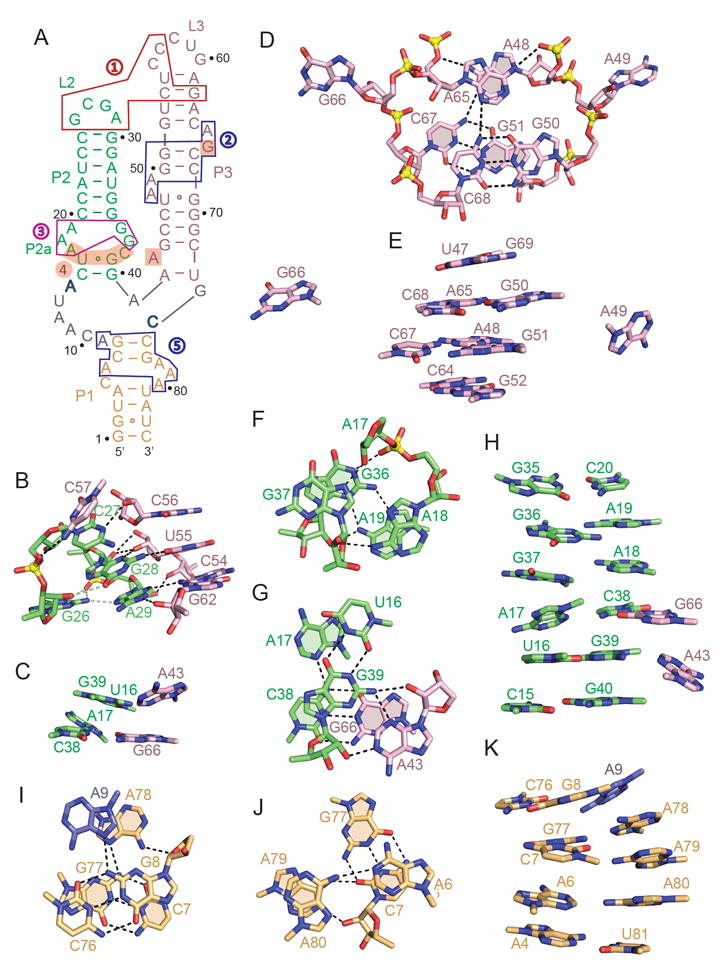
(A) Schematic of the secondary structure of the 3’,3’-cGAMP riboswitch highlighting loop-loop receptor interactions (boxed segment labeled 1 in red) and internal bulges in stem P3 (boxed segment labeled 2 in blue), stem P2 (boxed segments labeled 3 in magenta and labeled 4 with shaded background) and stem P1 (boxed segment labeled 5 in blue) in the complex.
(B) Topology and intermolecular hydrogen bonding between residues in stem-loops P2 (in green) and P3 (in pink) in the complex represented by the boxed region (in red) labeled 1 in panel A.
(C) Stacked triples between looped out bases from stem P3 (in pink) and the minor groove of base pairs in stem P2 (in green) that contribute to parallel alignment of stems P2 and P3 in the complex.
(D) Topology and intermolecular hydrogen bonding between residues in the internal bubble in the complex represented by the boxed region (in blue) labeled 2 in panel A.
(E) Stacking alignments spanning the bulged bases in stem P3 of the complex.
(F) Topology and intermolecular hydrogen bonding between residues in the internal bulge in the complex represented by the boxed region (in magenta) labeled 3 in panel A.
(G) Topology and intermolecular hydrogen bonding between residues in the internal bulge in the complex represented by the shaded region background labeled 4 in panel A.
(H) Stacking alignments spanning the bulged bases in stem P2 of the complex.
(I, J) Topology and intermolecular hydrogen bonding between residues in the internal bulge in stem P1 in the complex represented by the boxed region (in blue) labeled 5 in panel A.
(K) Stacking alignments spanning the bulged bases in stem P1 of the complex.
See also Table S1.
Conformation of Bulge in Stem P3 in the Complex
Stem P3 contains a pair of two-residue purine bulges on partner strands that are separated by two base pairs as shown schematically by a blue box labeled 2 in Figure 2A. We observe continuous stacking across both bulges, with bulge bases A48 and A65 stacking on top of each other, while their neighbor bulge bases A49 and G66 are extruded out of the duplex (Figure 2D). Stacked bases A48 and A65 are further anchored in place through the formation of A65•(G50-C68) and A48•(G51-C67) base triples involving pairing of the Watson-Crick edges of the adenines with the major grooves of the G-C base pairs (Figure 2D). Notably, none of the four-bulge bases insert between stacked base pairs in stem P3, so there is continuous stacking across both bulges on partner strands (Figure 2E).
Conformation of Bulge in Stem P2 in the Complex
Stem P2 contains a bulge in the vicinity of the three-way junction associated with three bulged bases positioned opposite each other on partner strands, as shown schematically by a magenta box labeled 3 in Figure 2A that spans residues A17-A18-A19 and G36-G37 on partner strands. We note that A18 and A19 are stacked on each other, as are G36 and G37, with G37 adopting an unanticipated syn alignment (Figure 2F). A hydrogen bond network is observed between A18-A19 and G36-G37 on partner strands, with the base of A18 paired with the sugar of G37, while G36 and A19 form a sheared cis minor-major groove G36•A19 non-canonical pair (Figure 2F).
We also observe medium/long-range interactions between the segments U16-A17 and C38-G39 in green (bulge bases A17 and C38 and flanking cis wobble G39•U16 pair) and residues A43 and G66 in pink, depicted schematically by the shaded segment labeled 4 in Figure 3A. Thus, the Watson-Crick edge of A43 forms an A-minor triple with the cis wobble G39•U16 pair, while C38 forms a long-range Watson-Crick G-C pair with G66 (Figure 2G).
Figure 3. ITC-based Binding Curves of Cyclic Dinucleotides to the 3’,3’-cGAMP Riboswitch containing A2G, U72C and C73U substitutions.
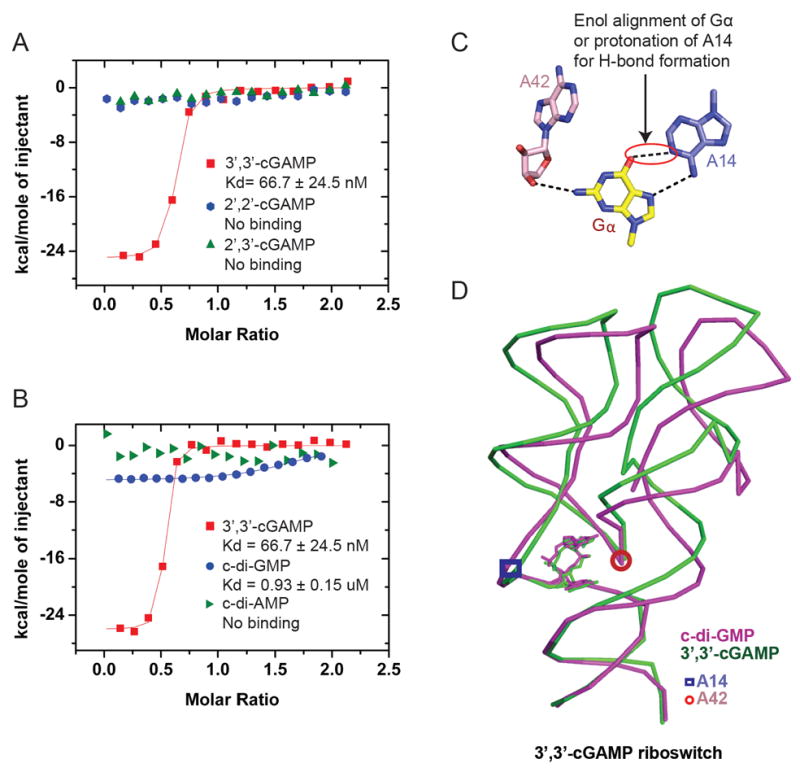
(A) ITC-based studies of the binding of 3’,3’-cGAMP, 2’,2’-cGAMP and 2’,2’-cGAMP to the 3’,3’-cGAMP riboswitch containing A2G, U72C and C73U substitutions.
(B) ITC-based studies of the binding of 3’,3’-cGAMP, c-di-GMP and c-di-AMP to the 3’,3’-cGAMP riboswitch containing A2G, U72C and C73U substitutions.
(C) Pairing of Gα with A14 in the structure of the complex of c-di-GMP with the 3’,3’-cGAMP riboswitch.
(D) Superposition of the structures of the complexes of 3’,3’-cGAMP riboswitch with bound 3’,3’-cGAMP (in green) and c-di-GMP (in magenta). The positions of G42 and A14 are indicated by a closed circles and squares in the two complexes.
See also Table S2.
Notably, all bulge bases stack into the helix, so that three non-canonical pairs separate the flanking Watson-Crick G35-C20 and cis wobble G39•U16 pairs, thereby maintaining a stacking alignment within stem P2 (Figure 2H).
Conformation of Bulge in Stem P1 in the Complex
Stem P1 contains a bulge in the vicinity of the three-way junction associated with bases C5-A6 on one strand positioned opposite A78-A79-A80 on the partner strand, with this bulge and adjacent stem segment shown schematically by a blue box labeled 5 in Figure 2A. We note that A9 and A78 on partner strands stack on each other, and are also involved in Hoogsteen-edge A-minor A9•(G8-C76) and A78•(G77-C7) base triples (Figure 2I). Further, A6 and A80 form a cis Watson-Crick-Hoogsteen A6•A80 non-canonical pair that stacks over the Hoogsteen-edge A-minor groove A79•(G77-C7) triple (Figure 2J). In essence, bulge bases A6 and A80 are inserted opposite each other between G77-C7 and A4-U81 Watson-Crick pairs, allowing retention of continuous stacking in stem P1 (Figure 2K).
Preferential Binding of 3’,3’-cGAMP Riboswitch to 3’,3’-cGAMP and Discrimination Against Linkage Isomers, c-di-AMP, and to a Lesser Extent, c-di-GMP
Through ITC binding studies on complex formation between the 3’,3’ c-GAMP riboswitch and linkage isomers of c-GAMP, we observe a robust binding curve for complex formation to c[G(3’,5’)pA(3’,5’)p] (3’,3’-cGAMP) with a Kd = 0.07 μM and a ΔH = -25.1 kcal/mol (Figure 3A and S1C and Table S2). This value likely reflects the effect of the U72C/C73U mutation on the dissociation constant, which has been previously measured as much lower (~0.00053 μM) for the native Gs1761 riboswitch aptamer bound to 3’,3’-cGAMP using the in-line probing assay (Kellenberger et. al. 2015). By contrast, no binding is observed to the 2’,5’-containing linkage isomers c[G(2’,5’)pA(2’,5’)p] (2’,2’-cGAMP) and c[G(2’,5’)pA(3’,5’)p] (2’,3’-cGAMP) (Figure 3A), thereby establishing an exquisite linkage-dependent selectivity of cGAMP for recognition by the riboswitch.
We have measured the ITC-based binding curves for complex formation between the 3’,3’-cGAMP riboswitch and the second messengers 3’,3’-cGAMP, c-di-GMP and c-di-AMP as shown in Figure 4B. Compared to the 3’,3’-cGAMP riboswitch binding to 3’,3’-cGAMP, the binding is measurable, but much weaker compared to c-di-GMP, as reflected in a Kd = 0.93 μM and a ΔH = -5.0 kcal/mol (Figure 3B and S1D and Table S2), with no measurable binding to c-di-AMP (Figure 3B). This value is consistent with the dissociation constant for the native Gs1761 riboswitch bound to c-di-GMP using the inline probing assay (~0.66 μM) (Kellenberger et. al. 2015). This result and data from other binding assays we performed outlined in a later Figure show that the Gs1761 riboswitch binds 3’,3’-cGAMP selectively over other cyclic dinucleotides that act as bacterial second messengers.
Figure 4. A Single Mutation in the c-di-GMP Vc2 Riboswitch Facilitates Binding by 3’,3’-cGAM.
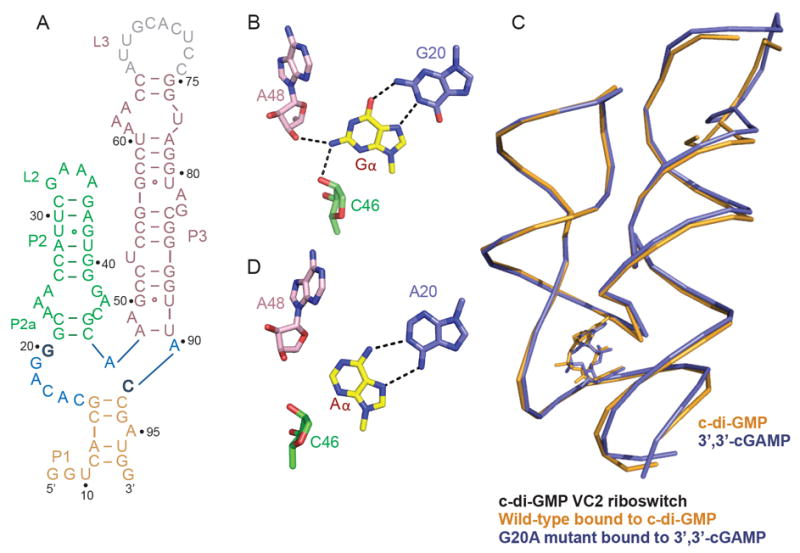
(A) Sequence of the aptamer domain of the c-di-GMP Vc2 riboswitch (Smith et al. 2009). The color-coding is the same as in Figure 1A.
(B) The Gα•G20 non-canonical pairing alignment in the structure of c-di-GMP bound to the c-di-GMP Vc2 riboswitch (PDB: 3IRW ; Smith et al. 2009).
(C) Superposition of the structures of the c-di-GMP Vc2 riboswitch bound to c-di-GMP (in orange; PDB: 3IRW ; Smith et al. 2009) and G20A mutant c-di-GMP Vc2 riboswitch bound to 3’,3’-cGAMP (in blue).
(D) The Aα•A20 non-canonical pairing alignment in the structure of 3’,3’-cGAMP bound to the c-di-GMP riboswitch.
See also Table S3
Unlike Gs1761, the majority of GEMM-I class riboswitches that harbor an A at the position that recognizes the Aα of the ligand (A14 in Gs1761) appear to have similar binding affinities to 3’,3’-cGAMP and c-di-GMP (Kellenberger et. al. 2015). For example, the G20A Vc2 riboswitch binds with comparable affinity to both cyclic dinucleotides (Kellenberger et. al. 2013). To elucidate how non-cognate ligands are differentially accommodated in selective or promiscuous GEMM-I riboswitches, we grew crystals of the 3’,3’-cGAMP riboswitch bound to c-di-GMP and the G20A Vc2 riboswitch bound to 3’,3’-cGAMP, and solved the structure of these complexes to 2.1 Å resolution (x-ray statistics in Table S1 and S3).
Pairing Alignments of c-di-GMP bound to the 3’,3’-cGAMP Riboswitch
The pairing alignment of Gα in c-di-GMP bound to the 3’,3’-cGAMP riboswitch is shown in Figure 3C. An omit map highlighting the electron density of A14 and bound c-di-GMP is shown in Figure S1E. We observe a short distance of 3.0 Å between the O6 oxygen of Gα and the N1 nitrogen of A14, requiring a proton to bridge these two acceptor heteroatoms. This suggests that that Gα binds as either the rare enol tautomer or alternately protonation of the N1 of its pairing partner A (Westhof, 2014; Smith et al. 2010; reviewed in Singh et al. 2015; Kimsey and Al-Hashimi, 2014), thereby accommodating a hydrogen bond between Gα and A14. We do not observe base pairing between Gα and A42, but instead a hydrogen bond is formed between the N2 nitrogen of Gα and the ribose hydroxyl of A42. The result is that A42 twists out of the plane in the c-di-GMP complex (Figure 3C) compared to its planar alignment in the 3’,3’-cGAMP complex (Figure 1I).
Overlay of the RNA folds in the c-di-GMP (in pink) and 3’,3’-cGAMP (in green) complexes with the 3’,3’-cGAMP riboswitch (note that the complexes were crystallized in different space groups, thus implying different packing interactions) indicates that the twist at A42 in the c-di-GMP complex (whose position is shown by a circle) results in perturbations that extend out to the hairpin loops of stems P2 and P3 (Figure 3D and S1F). Thus, binding of the non-cognate ligand, c-di-GMP, to the Gs1761 3’,3’-cGAMP riboswitch leads to a different orientation for the two arms (P2 and P3) of the tuning fork [in the 15-20° range, with a smaller change for P2 relative to P3] versus binding of the cognate ligand, 3’,3’-cGAMP. In the vicinity of the binding pocket, the conformational changes between complexes occur 2-3 nucleotides above A42 in P3 and 1 nucleotide above S42 in P2. Nevertheless, it remains unclear whether c-di-GMP binding causes a change in P2 and P3 or whether the flexibility of these two stems allows binding of c-di-GMP.
Single Mutation Enables the c-di-GMP Vc2 Riboswitch to Bind 3’,3’-cGAMP
The 3’,3’-cGAMP (Figure 1A) and 3’,3’-c-di-GMP (Figure 4A) riboswitches adopt similar secondary folds but contain distinct patterns of bulged bases. The structures of the 3’,3’-cGAMP riboswitch with bound c-GAMP and the c-di-GMP Vc2 riboswitch with bound c-di-GMP are superimposed in Figure S1G.
We reasoned that it should be possible to convert a previously studied c-di-GMP GEMM-I class Vc2 riboswitch (Smith et al. 2009) (Figure 4A) from one that bound c-di-GMP to one that also bound 3’,3’-c-GAMP by a single mutation. In the previously reported structure of c-di-GMP bound to the c-di-GMP Vc2 riboswitch, the Gα base of the ligand was paired with G20 through a trans Gα•G20 non-canonical pair involving the Hoogsteen edge of Gα and the Watson-Crick edge of G20 (Figure 4B). We therefore set out to replace the Gα•G20 pair by its isosteric Aα•A20 pair (Kellenberger et al. 2013), such that 3’,3’ c-GAMP could potentially target the c-di-GMP Vc2 riboswitch containing a G20A mutant. ITC binding studies established that 3’,3’-cGAMP bound to the c-di-GMP Vc2 riboswitch containing a G20A mutant with a Kd = 1.5 μM and a ΔH = -22.3 kcal/mol (Table S2) and we grew crystals and solved the structure of this complex to 2.1 Å resolution (x-ray statistics in Table S3). The structures of c-di-GMP bound to the c-di-GMP Vc2 riboswitch (in orange) and 3’,3’-cGAMP bound to the c-di-GMP Vc2 riboswitch containing a G20A mutant (in blue) superposition very well (Figure 4C), with Aα of cGAMP forming an Hoogsteen-Watson-Crick Aα•A20 non-canonical pair (Figure 4D) that is isosteric to the Gα•G20 pair (Figure 4B).
Analysis of the Role of the P2 Stem in Ligand Discrimination
In the 3’,3’-cGAMP riboswitch, a subtle shift in the A42 contact with the ligand (compare Figure 1I with bound cGAMP and Figure 3C with bound c-di-GMP) propagates to a detectable change in orientation of P2 and P3 stems, which may be correlated with its ability to distinguish between 3’,3’-cGAMP and c-di-GMP. We have obtained a second line of evidence for the P2 stem being involved in ligand discrimination. Using riboswitch-Spinach aptamer fusion constructs (Kellenberger et. al. 2013), we interrogated a variety of natural riboswitch sequences harboring an A at the position that recognizes the Aα of the ligand (A14 in Gs1761, G20A in Vc2) (Figure S2A). A trend was revealed upon grouping the sequences based on functional assessment of selectivity or promiscuity for binding 3’,3’-cGAMP versus c-di-GMP. Namely, selective sequences exhibit conservation of nucleotide identity in the two base pairs of the P2 stem that stack directly on top of Aα (Figure 5A, B). In contrast, promiscuous sequences exhibit different types of base pairs at those positions.
Figure 5. The P2a Region Affects Ligand Selectivity of the Gm0970 3’,3’-cGAMP Riboswitch.
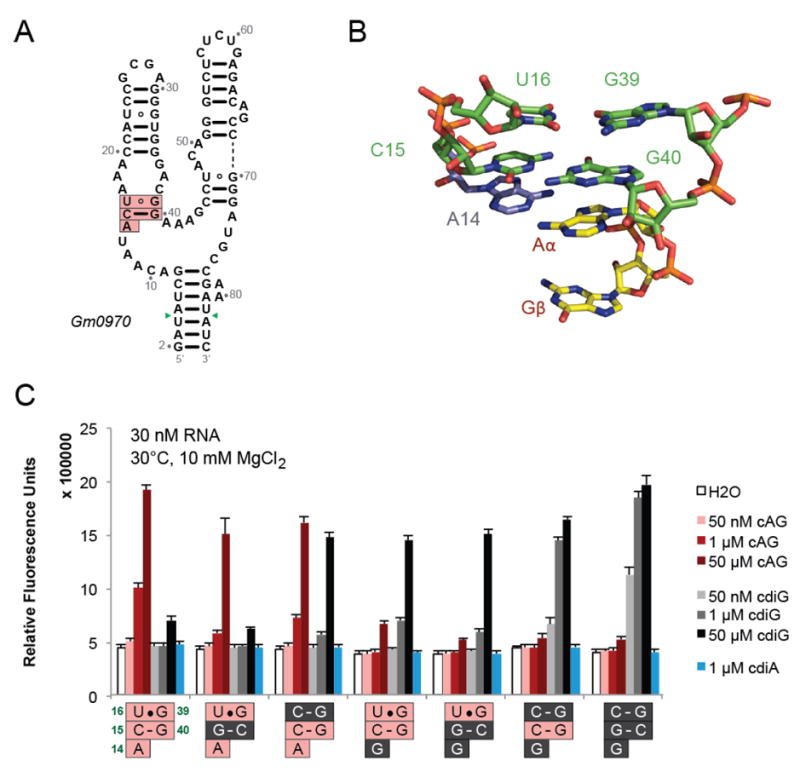
(A) Secondary structure of Gm0970, another 3’,3’-cGAMP-selective riboswitch. The nucleotide numbering is set to match that of Gs1761 in the P2a region (pink boxes). Green arrows indicate the positions in the P1 stem to which the Spinach aptamer was fused.
(B) Structure of the P2a region for Gs1761 with bound 3’,3’-cGAMP.
(C) Spinach-based selectivity screen of Gm0970 riboswitch constructs with mutations to the P2a region shown. Fluorescence activation was measured in the presence of no ligand or different cyclic dinucleotides at the indicated concentrations. The nucleotides from Gm0970 (pink) were changed to those from the c-di-GMP-selective riboswitch, Vc2 (gray).
A series of mutants of the 3’,3’-cGAMP riboswitch focused on this conserved P2a region were evaluated for their ability to bind either 3’,3’-cGAMP or c-di-GMP. The mutations were chosen to swap the nucleotide or base pair with the corresponding sequence from the Vc2 riboswitch, which is selective for c-di-GMP. The mutations show the same effects for both Gm0970 (Figure 5C) and Gs1761 (Figure S3B) riboswitches, but the former exhibits higher fluorescence signal and binding affinity when fused to the Spinach aptamer.
It was found that changing only two nucleotides in the Gm0970/Gs1761 riboswitch aptamers was sufficient to switch ligand recognition from 3’,3’-cGAMP to c-di-GMP (Figure 5C and S3B). Specifically, U16C makes these aptamers more promiscuous for binding c-di-GMP and is further additive to the effects of A14G (nucleotides are numbered in correspondence to the Gs1761 structure). Interestingly, U16C has a more pronounced effect than switching the orientation of the C-G base pair that directly stacks with either Aα or Gα (Figure 5C). Based on the structure of Gs1761, this may be due to its proximity to the two stapling interactions at the base of the P2 and P3 stems. Additional mutations focused on the U16•G39 wobble pair show that replacement with Watson-Crick pairs generally increases recognition of c-di-GMP (Figure S4A). We found that either moving or reversing the wobble pair, or replacing it with unpaired bases, also makes the riboswitch more promiscuous and leads to further loss of fluorescence signal, suggesting that these mutations may destabilize the RNA fold.
In contrast, performing the reverse set of mutations to the c-di-GMP selective Vc2 riboswitch did not result in a full switch of ligand specificity (Figure S4B). At most, mutations led to promiscuous binding of 3’,3’-cGAMP and c-di-GMP. Complete loss of fluorescence signal was observed upon full conversion of the conserved nucleotides to match the Gm0970/Gs1761 sequence, likely due to misfolding of the RNA. These results show that the extent of ligand discrimination is dependent on the overall sequence context as well, and that the conserved nucleotides in the P2a region are not sufficient to define whether a GEMM-I riboswitch discriminates between 3’,3’-cGAMP and c-di-GMP. In a related example, we found that both wild-type Gs1761 and the U72C/C73U construct exhibit selectivity for 3’,3’-cGAMP, but mutations of the P2a region showed different effects depending on the identity of those nucleotides (Figure S3C).
Nevertheless, we used the five conserved nucleotides to search for additional GEMM-I sequences that are selective for 3’,3’-cGAMP. Most representatives were found in the Geobacter genus, which have been validated as harboring many 3’,3’-cGAMP riboswitches (Kellenberger et. al. 2015). However, four matching sequences were found in the genome of Pelobacter propionicus. Through the Spinach fusion assay, three of these riboswitch candidates were confirmed to be selective for 3’,3’-cGAMP, while the fourth did not show signal in the assay (Figure S2B).
DISCUSSION
Tuning-Fork Architecture
The 3’,3’-cGAMP riboswitch adopts a tuning-fork architecture in the 3’,3’-cGAMP bound state. Such tuning-fork-like architectures have previously been reported for purine (Batey et al. 2004; Serganov et al. 2004), thiamine pyrophosphate (Serganov et al. 2006; Thore et al. 2006), and class-I c-di-GMP (Smith et al. 2009; Kulshina et al. 2009) riboswitches in the ligand-bound state. In each case, loop-loop (purine riboswitches) or loop-receptor (thiamine pyrophosphate and c-di-GMP riboswitches) interactions contribute to parallel alignments of two of the stems that constitute the prongs of the tuning fork scaffold. Such loop-loop receptor interactions are also observed in the current structure of the 3’,3’-cGAMP riboswitch in the bound state (Figure 1B, C), except that the parallel alignment of stems P2 and P3 are further restricted by inter-stem interactions involving formation of a pair of stacked minor groove-aligned base triples (Figure 2C and 3C). Indeed, formation of a long-range Watson-Crick G66-C38 pair as part of the base triple (Figure 3C) identifies a ‘linchpin’ interaction, which, if disrupted, could contribute to a realignment of stem P2 and P3, thereby acting as an RNA switch (Ganser et al. 2014). According to previously published SAXS data, the corresponding ‘linchpin’ G-C base pair is unlikely to form in the free state of the c-di-GMP riboswitch (Kulshina et al., 2009; see also Smith et al., 2010), and the same is likely to hold for the c-GAMP riboswitch in the free state.
Topology of Bulges Within Helical Stems of the 3’,3’-cGAMP Riboswitch in the Bound State
The bulges in stems P1 (labeled 5 in Figure 3A), stem P2 (labeled 3 and 4 in Figure 3A) and P3 (labeled 2 in Figure 2A) are distinctly different, being composed of different sequences and lengths on partner strands. Thus, individual bulges adopt different topologies but retain continuous stacking across the bulge, either through looping the bases out of the stem (A49 and G66 in Figure 2D), stacking of the bulge bases within the grooves (A48 and A65 in Figure 2D), or insertion of one (Figure 2K) or more (Figure 2H) bulge bases/pairs into the duplex. A looped out base (G66 in Figure 2D) can in turn be further involved in long-range pairing (Watson-Crick G66-C38 ‘linchpin’ pair, Figure 2G). Interestingly, whereas the stem P2 bulges appears conserved between all 3’,3’-cGAMP riboswitches, the stem P3 bulges are more variable (Fig. S2C). In particular, the 5’ bulge on stem P3 can vary from one to three nucleotides in length, with some differences in sequence composition. The conservation of the stem P1 bulges is difficult to determine because of the variability of the P1 stem sequence.
Consequence of Two Accidental Mutations in the Aptamer Domain of 3’,3’-cGAMP Riboswitch Used in the Current Study
The positions of the two accidental mutations at nucleotides 72 and 73 are not conserved amongst 3’,3’-cGAMP riboswitches in this class. It does not appear that these two mutations impact on our conclusions based on their interactions in the structure of the complex. Thus, C72 is paired with G44 in a Watson-Crick alignment in our structure. Replacement by the wild-type residue U72 could be accommodated with a small change associated with a wobble alignment. U73 is paired with A42 in a trans Hoogsteen-Watson-Crick alignment in our structure. Replacement by the wild-type residue C73 would require a small adjustment to generate an alignment stabilized by a A42 N7 to C73 NH2-6 hydrogen bond.
Comparison of the 3’,3’-cGAMP Riboswitch with the c-di-GMP Vc2 Riboswitch
Inspection of the secondary structures of the 3’,3’-cGAMP (Figure 1A) and c-di-GMP Vc2 (Figure 4A) riboswitches establish striking similarities in the topology of the three-way junction. Thus, the number but not the sequence of nucleotides that constitute the three-way junction are the same in the two riboswitches. Notably, an important difference occurs between junctional bases at identical positions involved in base specific recognition of the cyclic dinucleotides in the two complexes (in bold in Figures 1A and 4A). Thus, a junctional C involved in pairing with Gβ (Figure 1H) is common to both riboswitches (Figure 1A and 4A), while a junctional A (Figure 1A) involved in pairing with Aα in the cGAMP riboswitch (Figure 1I) is replaced by G (Figure 4A) necessary for isosteric pairing with Gα in the c-di-GMP Vc2 riboswitch (Smith et al. 2009; Kulshina et al. 2009).
Smith et al. 2009 were able to modulate the specificity of the c-di-GMP Vc2 riboswitch from c-di-GMP to favor c-di-AMP by replacement of two junctional nucleotides (C in bold to U and G in bold to A in Figure 4A) involved in base specific recognition of the second messengers. The corresponding mutation to the 3’,3’-cGAMP riboswitch only requires replacement of the single nucleotide that recognizes Gβ from C to U; however we did not see binding of c-di-AMP to this variant in the Spinach fusion (data not shown). A very large (105-fold) decrease in binding affinity was observed by Smith et. al. (2010) for the double mutant as well, which underscores the contribution of other differential factors (e.g. stacking interactions) in ligand recognition. Instead, we were able to fully switch the specificity of the 3’,3’-cGAMP riboswitch from 3’,3’-cGAMP to c-di-GMP by replacement of two nucleotides (A14G and U16C) (Figure 6C). The latter nucleotide does not make direct contact with the ligand, but clearly modulates specificity of the riboswitch (Figure S4A).
The bulges are very different between the 3’,3’-cGAMP (Figure 1A) and c-di-GMP Vc2 (Figure 5A) riboswitches. Hence, an important aspect of the current study that is distinct from the earlier structures of the c-di-GMP Vc2 riboswitch (Smith et al. 2009; Kulshina et al. 2009) is the topology adopted by the bulges in stems P1 (Figure 3a), P2 (Figure 3A) and P3 (Figure 2A) in our structure of the 3’,3’-cGAMP riboswitch.
Structural Basis of Specific Ligand Recognition by the 3’,3’-cGAMP Riboswitch
The 3’,3’-cGAMP riboswitch binds its cognate ligand in a single orientation with asymmetric recognition through base pairing in the complex. Thus, base-specific recognition involves the Watson-Crick and minor groove edges of Gβ (Figure 1H) and the major groove edge of Aα (Figure 1I) in the complex. Importantly, the 3’,3’-cGAMP riboswitch binds neither 2’,2’-cGAMP nor 2’,3’-cGAMP (Figure 3A). Thus, the RNA receptor does not tolerate these changes in the cyclic dinucleotide backbone that may misalign the Gβ and Aα bases for pairing interactions. Ligand backbone changes also may impair interactions with the highly conserved nucleotide A41 that stacks in between Gβ and Aα and hydrogen bonds with the G(3’,5’)pA linkage that is alternately a G(2’,5’)pA linkage in the mammalian second messenger.
The 3’,3’-cGAMP riboswitch for which we solved the structure was identified in Geobacter sulfurreducens, an organism that was shown to produce all three bacterial cyclic dinucleotides: c-di-GMP, 3’,3’-cGAMP, and c-di-AMP (Kellenberger et. al. 2015). Thus, discrimination between these 3’,3’ cyclic dinucleotides is critical to the regulatory function of this riboswitch, and we have shown that the wild-type Gm0970 and Gs1761 riboswitches from Geobacter are over ~1,600-fold selective for 3’,3’-cGAMP versus other cyclic dinucleotides (Kellenberger et. al. 2015). It is not surprising that c-di-AMP does not bind the wild-type 3’,3’-cGAMP riboswitch (Figure 3A) since replacement of Gβ by Aβ would completely disrupt base pairing with C75 (Figure 1H). However, c-di-GMP has been shown to bind with comparable affinity as 3’,3’-cGAMP to the G20A Vc2 riboswitch (Kellenberger et. al. 2013) and to the U16C Gm0970 riboswitch in this study. The discrepancy in ligand selectivity between these riboswitches and the wild-type 3’,3’-cGAMP riboswitches is remarkable because all of these RNAs harbor identical nucleotides at the positions that base pair with the ligand. This observation raises the important question of what other sequence and structural aspects dictate whether binding is selective or promiscuous for these two cyclic dinucleotides.
Our structural and biochemical experiments revealed that recognition of Aα or Gα is particularly sensitive to changes in the P2a stem (Figure S4A, B). This region is involved directly in stacking and is connected to the critical intercalating nucleotide A41 (Figure 1D). It also is connected to A14, the nucleotide that base pairs with Aα or Gα (Figure 1I and 3C). Finally, it forms an A-minor triple with A43 (Figure 32G), so the P2a stem makes interactions on both sides of the A42 “hinge.” However, we also showed that the sequences of the base pairing nucleotides and the P2a stem are not always sufficient to dictate ligand selectivity, as we were unable to convert c-di-GMP Vc2 riboswitch into a 3’,3’-cGAMP-selective riboswitch. This latter result indicates that there are other important differences between the RNA scaffolds for 3’,3’-cGAMP and class-I c-di-GMP riboswitches.
On the outset, it was surprising that c-di-GMP and 3’,3’-cGAMP could ever bind with similar affinities, given that Aα in the 3’,3’-cGAMP complex should form two hydrogen bonds to A14, whereas Gα in the c-di-GMP complex should form one hydrogen bond to A14. There are several possible explanations for how these riboswitches can be promiscuous. One possibility is that the second hydrogen bond between Aα and A14 is weak and does not make a significant contribution to the stability of the complex. Another possibility is that Gα actually forms two hydrogen bonds to A14, which is effected either by a perturbation of the pKa, such that A14 is protonated at N1 (Smith et. al. 2010), or by perturbation of the keto:enol equilibrium, such that Gα binds as the enol tautomer (Figure S3C) (Singh et al. 2015; Westhof, 2014). Both our structure of the 3’,3’-cGAMP riboswitch bound to c-di-GMP and the published structure of the G20A Vc2 riboswitch bound to c-di-GMP (Smith et. al. 2010) show that the orientation of A14 (or A20, its counterpart in Vc2) is consistent with two hydrogen bonds. A final possibility is that differences in the stacking interactions can compensate for the difference in hydrogen bonds. In fact, each of these factors may be influenced by changes to the proximal P2a stem that instead lead to selective binding of 3’,3’-cGAMP over c-di-GMP.
The three crystal structures solved in this study, along with the published structure of c-di-GMP complexed with G20A Vc2 riboswitch, provide a useful comparison of how a selective versus promiscuous riboswitch binds 3’,3’-cGAMP and c-di-GMP. It is striking that the selective riboswitch (Gs1761) displays different orientations of the P2 and P3 stems depending on the identity of the bound ligand (Figure 3D), whereas the promiscuous riboswitch (G20A Vc2) adopts nearly identical orientations when bound to either ligand (Figure 4C). This correlation does not distinguish between cause and effect, however, as 3’,3’-cGAMP either binds a lower energy conformation or induces a lower energy conformation of the Gs1761 riboswitch.
Future Prospects
Interestingly, the 3’,3’-cGAMP riboswitch is exquisitely sensitive to linkage isomers of cGAMP (Figure 3B) and it remains to be seen what modification(s) would either loosen or alter this specificity. Our current efforts are also aimed towards the identification and subsequent structure determination of a 2’,3’-cGAMP riboswitch that could target the mammalian second messenger 2’,3’-cGAMP and discriminate against other linkage isomers. It remains unclear whether such a 2’,3’-cGAMP riboswitch will adopt a global and binding pocket topology related to or distinct from the 3’,3’-cGAMP riboswitch.
Our structural and biochemical data have also shown how non-contacting nucleotides can impact ligand nucleobase selectivity for the GEMM-I riboswitch scaffold. These insights are necessary to accurately predict whether a given sequence is a c-di-GMP Vc2 riboswitch or a 3’,3’-cGAMP riboswitch, and would assist in ongoing studies of the prevalence of 3’,3’-cGAMP signaling in bacteria. Finally, the observation that subtle mutations outside of the binding pocket can change ligand recognition, as previously observed for the 2’-deoxyguanosine riboswitch (Kim et al. 2007; Edwards and Batey, 2009; Pikovskaya et al. 2011), demonstrates the potential, as well as the challenge, of re-engineering natural riboswitch scaffolds for new sensing functions.
EXPERIMENTAL PROCEDURES
RNA Crystallization, Structure Determination and ITC Experiments
All the RNA samples were transcribed in vitro using T7 RNA polymerase (Pikovskaya et al. 2009). All the crystallization complexes were generated by annealing the purified RNA at 70 C with bound 3’,3’-cGAMP or c-di-GMP in a 1:2 molar ratio in a buffer containing 5 mM MgCl2. The crystals of the aptamer domain of the 3’,3’-cGAMP riboswitch with bound 3’,3’-cGAMP or c-di-GMP, as well as G20A mutant c-di-GMP riboswitch with 3’,3’-cGAMP were grown under specific crystallization buffer and additive conditions and flash-frozen in liquid nitrogen for data collection. All the structures were solved by the molecular replacement method. The initial RNA model was traced and built in COOT (Emsley and Cowtan, 2004) and refined in PHENIX (Adams et al. 2002). The x-ray statistics of the crystals are listed in Table S1 and S3. ITC experiments were performed in an experimental buffer at 35 °C. The thermograms were integrated and analyzed using the model of one set of sites in Origin 7.0 software (Microcal, Inc.). Details are provided under Extended Experimental Procedures in Supplementary Materials.
Spinach-based Fluorescence Assays for Ligand Binding
Riboswitch-Spinach fusion RNAs were transcribed in vitro using T7 RNA polymerase and purified by denaturing (7.5 M urea) 6% PAGE gel. Fluorescence activation assays were performed as previously described (Kellenberger et. al. 2013). Briefly, RNAs were refolded in binding buffer (40 mM HEPES, 125 mM KCl, and 10 mM MgCl2 at pH 7.5) and added to wells on a 96-well-plate containing binding buffer, 10 μM DFHBI, and ligand at given concentrations. The reaction plate was incubated at 30 °C and fluorescence measurements were taken on a SpectraMax Paradigm plate reader (Molecular Devices) at 460 nm excitation / 500 nm emission or 448 nm excitation / 506 nm emission (for Fig. S3 inset). Details are provided under Extended Experimental Procedures in Supplementary Materials.
Supplementary Material
Acknowledgments
We thank the synchrotron beamline staff at the Argonne National laboratory for their assistance. D.J.P. was supported by NIH grant NIH grant 1 U19 CA179564, M.C.H. was supported by NIH grant DP2 OD008677, C.A.K. was supported in part by a DoD NDSEG fellowship. M.C.H. holds a Career Award at the Scientific Interface from the Burroughs Wellcome Fund.
Footnotes
AUTHOR CONTRIBUTIUONS
X-ray structure determination and ITC binding curves of ligand-riboswitch complexes were performed by A.R. under the supervision of D.J.P. The solution to the phase problem related to structure determination of the ligand-riboswitch complex was provided by K.R.R. c-GAMP linkage isomers were provided by R.J.J. Bioinformatics, design of riboswitch-Spinach constructs, and fluorescence binding assays were performed by X.C.W. and C.A.K. under the supervision of M.C.H.
ACCESSION NUMBERS
The following coordinates have been deposited in the Protein Data Bank: 3’,3’-cGAMP bound to the 3’,3’-cGAMP riboswitch (4YAZ), c-di-GMP bound to the 3’,3’-cGAMP riboswitch (4YB0), and 3’,3’-cGAMP bound to the G20A mutant c-di-GMP Vc2 riboswitch (4YB1).
SUPPLEMENTAL INFORMATION
Supplemental Information includes five Figures and five tables and can be found with this article on line at ………..
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Ming C. Hammond, Email: mingch@berkeley.edu.
Dinshaw J. Patel, Email: pateld@mskcc.org.
References
- Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, Hopfner KP, Ludwig J, Hornung V. cGAS produces a 2’-5’-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Grosse-Kunstleve RW, Hung LW, Loerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Chiu YH, Chen ZJ. The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Mol Cell. 2014;54:289–296. doi: 10.1016/j.molcel.2014.03.040. [DOI] [PubMed] [Google Scholar]
- Danilchanka O, Mekalanos JJ. Cyclic dinucleotides and the innate immune response. Cell. 2013;154:962–970. doi: 10.1016/j.cell.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies BW, Bogard RW, Young TS, Mekalanos JJ. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell. 2012;149:358–370. doi: 10.1016/j.cell.2012.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diner EJ, Burdette DL, Wilson SC, Monroe KM, Kellenberger CA, Hyodo M, Hayakawa Y, Hammond MC, Vance RE. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep. 2013;3:1355–1361. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AL, Batey RT. A structural basis for the recognition of 2’-deoxyguanosine by the purine riboswitch. J Mol Biol. 2009;385:938–948. doi: 10.1016/j.jmb.2008.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Ganser LR, Mustoe AM, Al-Hashimi HM. An RNA tertiary switch by modifying how helices are tethered. Genome Biol. 2014;15:425. doi: 10.1186/s13059-014-0425-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao A, Serganov A. Structural insights into recognition of c-di-AMP by the ydaO riboswitch. Nat Chem Biol. 2014;10:787–792. doi: 10.1038/nchembio.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, Serganov AA, Liu Y, Jones RA, Hartmann G, et al. Cyclic [G(2’,5’)pA(3’,5’)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013;153:1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray PM, Forrest G, Wisniewski T, Porter G, Freed DC, DeMartino JA, Zaller DM, Guo Z, Leone J, Fu TM, et al. Evidence for cyclic diguanylate as a vaccine adjuvant with novel immunostimulatory activities. Cell Immunol. 2012;278:113–119. doi: 10.1016/j.cellimm.2012.07.006. [DOI] [PubMed] [Google Scholar]
- Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- Hornung V, Hartmann R, Ablasser A, Hopfner KP. OAS proteins and cGAS: unifying concepts in sensing and responding to cytosolic nucleic acids. Nat Rev Immunol. 2014;14:521–528. doi: 10.1038/nri3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CP, Ferre-D’Amare AR. Crystal structure of a c-di-AMP riboswitch reveals an internally pseudo-dimeric RNA. Embo J. 2014;33:2692–2703. doi: 10.15252/embj.201489209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellenberger CA, Wilson SC, Sales-Lee J, Hammond MC. RNA-Based Fluorescent Biosensors for Live Cell Imaging of Second Messengers Cyclic di-GMP and Cyclic AMP-GMP. J Am Chem Soc. 2013;135:4906–4909. doi: 10.1021/ja311960g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellenberger CA, Wilson SC, Hickey SF, Gonzalez TL, Su Y, Hallberg ZF, Brewer TF, Iavarone AT, Carlson HK, Hsieh YF, Hammond MC. GEMM-I riboswitches from geobacter sense the bacterial second messenger 3’,3’ c-AMP-GMP. 2015 doi: 10.1073/pnas.1419328112. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JN, et al. Guanine riboswitch variants from Mesoplasma florum selectvely recognize 2’-deoxyguanosine. Proc Natl Acad Scis USA. 2007;104:16092–16097. doi: 10.1073/pnas.0705884104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Li L, Maliga Z, Yin Q, Wu H, Mitchison TJ. Anticancer flavonoids are mouse-selective STING agonists. ACS Chem Biol. 2013;8:1396–1401. doi: 10.1021/cb400264n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimsey I, Al-Hashimi HM. Increasing occurences and functional roles for high energy purine-pyrimidine base pairs in nucleic acids. Curr Opin Struct Biol. 2014;24:72–80. doi: 10.1016/j.sbi.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulshina N, Baird NJ, Ferre-D’Amare AR. Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch. Nat Struct Mol Biol. 2009;16:1212–1217. doi: 10.1038/nsmb.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ER, Baker JL, Weinberg Z, Sudarsan N, Breaker RR. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science. 2010;329:845–848. doi: 10.1126/science.1190713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JW, Sudarsan N, Furukawa K, Weinberg Z, Wang JX, Breaker RR. Riboswitches in eubacteria sense the second messenger c-di-AMP. Nat. Chem. Biol. 2013;9:834–839. doi: 10.1038/nchembio.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JW, et al. Control of Exoelectrogenesis in Geobacter spp. by c-AMP-GMP. Proc Natl Acad Scis USA. 2015 in press. [Google Scholar]
- Pikovskaya O, Polonskaia A, Patel DJ, Serganov A. Structural princoples of nucleosie selectivity in a 2’-deoxyguanosine riboswitch. Nat Chem Biol. 2011;7:748–755. doi: 10.1038/nchembio.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren A, Patel DJ. c-di-AMP binds the ydaO riboswitch in two pseudo-symmetry-related pockets. Nat Chem Biol. 2014;10:780–786. doi: 10.1038/nchembio.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romling U, Galperin MY, Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Nudler E. A decade of riboswitches. Cell. 2013;152:17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Yuan YR, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, Hobartner C, Micura R, Breaker RR, Patel DJ. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V, Fedeles BI, Essigmann JM. Role of tautomerism in RNA biochemistry. Rna. 2015;21:1–13. doi: 10.1261/rna.048371.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KD, Lipchock SV, Ames TD, Wang J, Breaker RR, Strobel SA. Structural basis of ligand binding by a c-di-GMP riboswitch. Nat Struct Mol Biol. 2009;16:1218–1223. doi: 10.1038/nsmb.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KD, Lipchock SV, Livingston AL, Shanahan CA, Strobel SA. Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch. Biochemistry. 2010;49:7351–7359. doi: 10.1021/bi100671e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KD, Shanahan CA, Moore EL, Simon AC, Strobel SA. Structural basis of differential ligand recognition by two classes of bis-(3’-5’)-cyclic dimeric guanosine monophosphate-binding riboswitches. Proc Natl Acad Sci U S A. 2011;108:7757–7762. doi: 10.1073/pnas.1018857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thore S, Leibundgut M, Ban N. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- Westhof E. Isostericity and tautomerism of base pairs in nucleic acids. FEBS Lett. 2014;588:2464–2469. doi: 10.1016/j.febslet.2014.06.031. [DOI] [PubMed] [Google Scholar]
- Wilson SC, Cohen DT, Wang XC, Hammond MC. A neutral pH thermal hydrolysis method for quantification of structured RNAs. RNA. 2014;20:1153–1160. doi: 10.1261/rna.045856.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte CE, Whiteley AT, Burke TP, Sauer JD, Portnoy DA, Woodward JJ. Cyclic di-AMP is critical for Listeria monocytogenes growth, cell wall homeostasis, and establishment of infection. MBio. 2013;4:e00282–00213. doi: 10.1128/mBio.00282-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, Chen ZJ. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol Cell. 2013;51:226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


