Abstract
Purpose
To illustrate how conditional growth percentiles can be adapted for use to systematically identify implausible measurements in growth trajectory data.
Methods
The use of conditional growth percentiles as a tool to assess serial weight data was reviewed. The approach was applied to 86,427 weight measurements (kg) taken between birth and age 6.5 years in 8217 girls participating in the Promotion of Breast Feeding Intervention Trial in Belarus. A conditional mean and variance was calculated for each weight measurement, which reflects the expected weight at a current visit given the girl’s previous weights. Measurements were flagged as outliers if they were more than 4 standard deviation (SD) above or below the expected (conditional) weight.
Results
The method identified 234 weight measurements (0.3%) from 216 girls as potential outliers. Review of these trajectories confirmed the implausibility of the flagged measurements, and that the approach identified observations that would not have been identified using a conventional cross-sectional approach (−4 SD of the population mean) for identifying implausible values. Stata code to implement the approach is provided.
Conclusions
Conditional growth percentiles can be used to systematically identify implausible values in growth trajectory data and may be particularly useful for large data sets where the high number of trajectories makes ad hoc approaches unfeasible.
Keywords: Longitudinal growth data, Data cleaning, Outliers identification
Introduction
Patterns of growth during pregnancy, infancy, and childhood have important consequences for long-term health. A growing number of epidemiologic studies are collecting serial length, height, and body weight measurements with the aims of identifying determinants of growth trajectories and establishing the consequences of different growth patterns on future health risk [1–3]. Collecting serial anthropometric measurements adds an additional layer of complexity to the process of identifying outliers, and data errors that may be present in the research data set. In studies where there is only one measurement per individual, outliers and implausible values are often identified by comparing a given observation to that of the population distribution (e.g., a child’s height measurement 4 standard deviation (SD) below or above the population average). With longitudinal growth data, however, the plausibility of a given observation depends not only on its absolute value at each time point but also on the individual’s previous and subsequent measurements. Despite the large number of studies analyzing longitudinal measurements, the approaches used to ensure that the data are free from errors are often not reported. Approaches that are reported are highly subjective (i.e., “eyeballing” growth plots for unusual patterns), not feasible for large sample sizes (where the number of trajectories for visual inspection becomes extremely cumbersome), or do not take into account the plausibility of the observation in relation to the other measurements in the trajectory [4–6].
In this study, our goal was to demonstrate how conditional growth percentiles, percentiles that are established conditional on an individual’s previous weight measurements, can be adapted for use as a systematic, reproducible approach for cleaning longitudinal data in epidemiologic studies of human growth. We enclose annotated Stata code to enable epidemiologists to apply the method to their own data.
Methods
Conditional growth percentiles
Conditional growth percentiles were initially developed as a method to identify fetuses with intrauterine growth restriction [7]. Conditional fetal growth percentiles are weight percentiles that are calculated given (conditional on) a fetus’ weight earlier in pregnancy. The fetus’ ultrasound estimated weight from a previous visit and information on population fetal growth patterns are used to calculate its expected weight at a current visit. If the fetus’ current weight is below an 80% coverage limit of its expected weight (i.e., below −1.28 SD or the 10th percentile of its expected weight), the infant is classified as “small for gestational age” and considered to be at increased risk of complications due to intrauterine growth restriction. Reference values for two populations [8,9] and evaluations of the clinical utility of conditional percentiles [9,10] have been published.
We propose that conditional growth percentiles can be adapted for use as a tool to flag outliers and implausible values in growth trajectory data. Instead of calculating an 80% coverage limit to identify the range of “normal” growth, we calculate a much broader range (i.e., a 4 SD coverage limit), which allows us to identify weight measurements that are extremely unlikely given the previous measurements of an individual’s growth trajectory.
To calculate conditional percentiles [7], a random-effects (hierarchical) model is first built to describe the repeated weight measurements as a function of age. This model provides estimates of the population average weights across time (age), as well as estimates of the between-individual and within-individual variation in growth. These estimates are used to calculate a conditional mean weight and 4 SD range for an individual’s weight at time t, given their weight at time t – 1. Weight measurements which are below −4 SD or above +4 SD are classified as outliers. We use a limit of 4 SD based on the statistical convention that observations 4 or more SD from the expected mean can be considered to be “far outliers”[11], We also conducted sensitivity analyses using 3 SD as a cutoff to evaluate the extent to which the choice of cutoff alters results. We illustrate our results of identification of outlying values based on the 4 SD of the conditional mean by contrasting the results based on a conventional cross-sectional approach of using 4 SD of the population average at each time point. The specific formulae used to calculate the conditional percentiles and exemplary plots of outliers identified shown in our results are provided in Appendix A along with annotated Stata code.
Data
We used growth data from children who participated in the Promotion of Breastfeeding Intervention Trial (PROBIT). A full description of PROBIT has been published elsewhere [12]. In brief, PROBIT is a cluster-randomized controlled trial of a breastfeeding promotion intervention modeled on the World Health Organization and/or United Nations International Children Emergency Fund Baby-Friendly Hospital Initiative in the Republic of Belarus. A total of 17,046 healthy full-term (≥37 completed weeks of gestation) infants who weighed 2500 g or more were recruited from 31 maternity hospitals and affiliated polyclinics and followed-up at 1, 2, 3, 6, 9, and 12 months and at age 6.5 years during which study pediatricians measured weight and length and/or height. Weights (kg) and lengths and/or heights (cm) between 12 months and the 6.5-year follow-up visit were abstracted from the polyclinic records of routine checkups. For simple illustration, we focused on weight trajectory among girls in this study. There were 8217 girls with a median of 11 measures of weight (interquartile range [IQR] = 8–13; range, 1–14) from birth to the median age of 78 months (IQR = 77–79 months), yielding a total of 86,427 weight measurements.
To apply the conditional percentiles approach, we built a random-effects model with weight as a function of age. Because early childhood growth trajectory is not linear, we modeled growth using a restricted cubic spline with five knots at 0, 3, 9, 24, and 78 months. This model showed the best fit according to the Akaike Information Criterion [13] among models we compared using different number of knots and knots at fixed ages. Estimates from this model were then used to calculate conditional percentiles for each weight observation for each girl.
Results
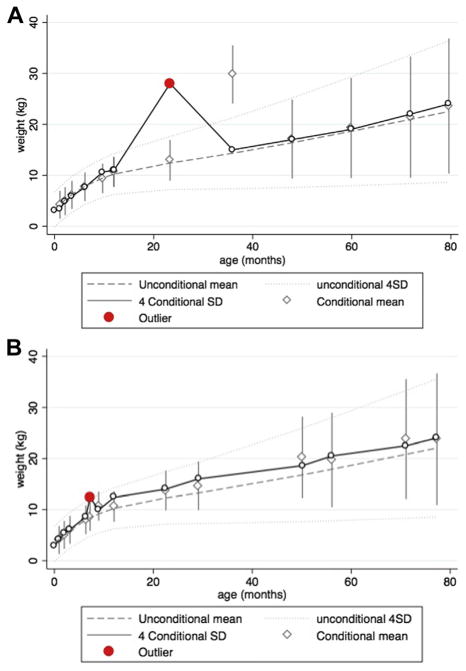
Using 4 SD from the conditional mean as a criterion (as specified in Appendix A, equations 2–5) identified 234 (0.27%) outliers of 86,427 weight measurements. These outliers were from 216 individuals: 201 girls with one outlier, 12 girls with two outliers, and three girls with three outliers. Figure 1 presents examples of the weight trajectories of two girls with identified outliers.
Fig. 1.
Examples of outliers identified by conditional percentiles on prior weight.
In Figure 1, the girls’ observed weight measurements (shown by circles) are superimposed on the population unconditional mean (i.e., the 50th percentile of weight for age in the cohort, shown by the dashed line) and 2 SD (i.e., the population 2.5th and 97.5th percentiles, shown by the dotted lines). The conditional means for her individual weight measurements with the 4 SD range are shown by the gray diamonds with the range lines. The weight trajectory of the girl shown in Figure 1a was tracking steadily close to the population average until the clinic visit at age 12 months. Given this initial trajectory, she would be expected to have a weight close to the population average at the next visit (as seen by the conditional mean of 13 kg at 23 months). However, her observed weight was 28 kg, which was more than 4 SD outside her expected weight, so this weight was flagged as an implausible value (red circle). Her observed weight then returned close to the population average weight for subsequent visits. Of note, the conditional mean at her next visit, at 36 months, was much greater than the population average because it was calculated given the previous outlying weight measurement. In our code, we specify that the next observation after an outlier should be evaluated for plausibility based not on the outlier but on an observation before the outlier. In Figure 1b, the girl’s weight increased from 8.6 kg at age 6 months to 12.4 kg at age 7 months (red circle) showing a gain of close to 4 kg at 1 month and returned to 9.9 kg at her next visit at 9 months. The weight at age 7 months was within 4 SD of the population average weight, so would not have been identified as an outlier using a conventional cross-sectional approach to identifying outliers. However, it was more than 4 SD from her expected weight given her previous weight measurements, so was identified as an outlier according to the conditional percentiles. When we used 3 SD from the conditional mean as a threshold, the approach detected 537 measures of weight (0.62%) from 493 girls as outliers.
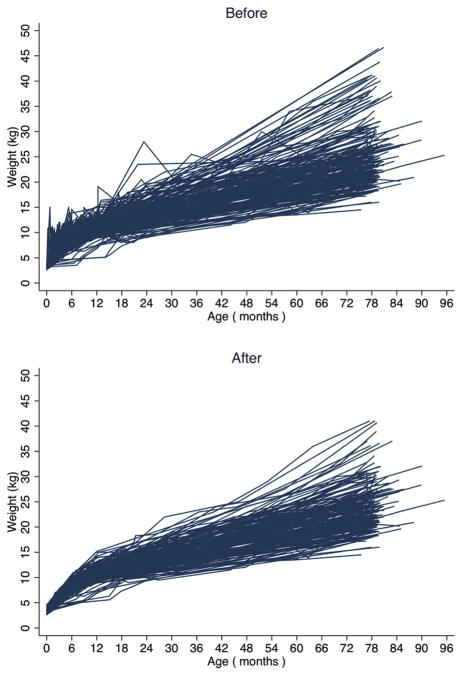
The impact of excluding weight observations identified as outliers using conditional percentiles can be seen in Figure 2. The upper panel shows the trajectories of girls with at least one outlier identified before exclusion of any implausible values. A number of jagged growth patterns are evident. After removing these outliers (bottom panel), the resulting weight gain trajectories appear more biologically plausible and smooth.
Fig. 2.
Weight trajectories of a sample of 216 girls in PROBIT before and after exclusion of implausible values identified by conditional centiles.
Discussion
In this report, we illustrated how conditional growth percentiles can be used to identify implausible values in pregnancy and pediatric growth trajectory data. In our cohort, the approach enabled us to screen a large number (n = 8217) of trajectories in an effficient manner, visually review the trajectories of only a small, manageable subset (n = 216), and ensure that decisions on exclusion of implausible measurements were reproducible and systematic.
As with all data cleaning procedures, identification of extreme observations through this approach does not necessarily imply that the observations should be automatically discarded from analyses. However, those identified implausible values warrant further investigation. This may involve going back to the data collection tool to identify data entry errors or reviewing the individual’s clinical notes to assess plausibility. For data in which verification with source data is not possible, examination of the trajectories in which an outlier was identified is still advisable to gain a subjective understanding of the observations being flagged.
Although we used 4 SD as threshold for identifying outliers based on statistical convention, alternative thresholds could be used as shown in our sensitivity analysis. We present our results with 4 SD as threshold, a rather extreme threshold, because we wanted to ensure our flagged outliers were “true” implausible values in our study that included a very large number of observations (86,427 weight measurements from 8217 girls). Large sample sizes and/or short intervals between measurements will yield tighter confidence intervals around the conditional means. The choice of threshold would also vary across studies according to factors such as the expected accuracy of the study’s measurement (i.e., the larger measurement error expected such as fetal growth estimates [14], the less restrictive thresholds). The time and/or resources required to review measurements flagged as implausible may be important in choosing a threshold as well: our sensitivity analysis using 3 SD as threshold yielded twice as many trajectories to review as those from 4 SD. Thus, individual studies need to take their design, characteristics of their growth measurement, and resources into consideration in determining a threshold to identify implausible and/or outlying values. Regardless, it is important to use an extreme cutoff to avoid removing any true measurements and to identify outliers in the tails of the distribution where there should be no data.
Conditional growth percentiles approach offers advantages over existing approaches. A common approach is to plot and visually examine the trajectories for unusual-looking patterns. For instance, the superimposition by translation and rotation model [15] allows researchers to graph individuals’ trajectories based on the fitted model and click to highlight trajectories that appear “abnormal”. However, this is subjective and becomes impractical with large data sets. Identifying implausible values based only on absolute values (e.g., observations 3 SD or greater from the population mean value at that age) [5] fails to recognize that the plausibility of growth measurements depends on the individual’s earlier measurements. A more advanced approach is to convert raw growth data to z scores using a standard growth reference such as CDC or World Health Organization growth chart, which will convert the nonlinear growth curves to straight lines. Researchers then can use an extreme change in z scores between visits as a criterion for identifying outliers. However, this method requires an external standard that is not always available for many “growth” data. In addition, the approach does not account for the time interval between measurements, which affects plausibility (i.e., a large change in z scores is more plausible if there were 2 years between measurements than if the measurements were 2 weeks apart).
As the conditional growth percentiles approach requires a prior measurement to “condition on” to evaluate the plausibility of the next measurement, it naturally cannot be applied to an individual’s first measurement of growth. The first measurement of growth should, therefore, be evaluated by a conventional cross-sectional approach. It is also important to note that the approach, like any model-based approach, depends on the accuracy with which the growth model was fit. If the fitted model does not adequately describe the underlying growth pattern, implausible values identified by this approach would be less meaningful.
In conclusion, the conditional growth percentiles approach represents a novel tool to systematically examine and identify implausible values of growth measurements. The approach may be particularly useful in large data sets where the large number of trajectories to be examined makes “eyeballing” trajectories impractical. Reduction of data errors through the application of this approach will help increasing the accuracy and precision of estimates obtained from statistical models of pediatric and pregnancy growth. This, in turn, will improve our understanding of the determinants and consequences of different human growth trajectories.
Acknowledgments
This work was supported by the U.S. National Institute of Child Health and Human Development (R01 HD072008 and R01 NR014245 to J.A.H.) and the Canadian Institutes of Health Research (MOP-53155 to S.Y.) and the Bill and Melinda Gates Foundation (OPP1119659 to S.Y.). J.A.H. holds a Canadian Institutes of Health Research New Investigator Award and is a Career Scholar of the Michael Smith Foundation for Health Research.
Appendix A
Part 1: Conditional percentiles method
The following equation (Equation 1) describes a basic random-effects model whereby both the intercept (β0) and slope (βAge) are allowed to vary by individual:
| (1) |
where, “i” denotes the “ith” individual, “j” denotes the “jth” measurement occasion, Weight denotes the response variable (fetal, infant, or pediatric weight), ε denotes the within-individual variability in weights, X is the independent variable of age, β0i=β0+ui; ui being the random effect at the level of the individual to allow each individual to have its own intercept, βAgei=βAge+μAgei; μAgei again being the random effect at the level of the individual, here, allowing each individual to have its own slope (i.e., growth rate that varies across individuals during pregnancy or childhood), , and cov(β0i,βAgei) = σβ0βAge. For simplicity, the model is described as a linear growth model but should be modified to reflect nonlinear growth patterns as appropriate.
The variance of weight for individual i at time j (the unconditional variance) is calculated as
| (2) |
Having estimated the unconditional mean and variance of growth in the population using the random effects model, the conditional mean weight for an individual at time 2, given their weight at time 1 is calculated as
| (3) |
where
The conditional variance of Weight2 given Weight1 is:
| (4) |
With the conditional mean and variance established, the upper and lower limits of a 4 standard deviation range for the conditional mean can be calculated as:
| (5) |
Part 2: Stata code for calculating conditional percentiles
**SETUP: * data should be in long format. * rename the subject’s unique identifier ‘id’ * rename the variable for time (e.g. gestational age, child’s age) ‘age’ ** Note: ‘age’ variable should correspond to the function of time in growth trajectory (assumed ‘linear’ in this code for simplicity) * rename the variable for growth (e.g. weight, weight gain, estimated fetal weight) ‘weight’ * *REMOVING MISSING OBSERVATIONS drop if weight==. drop if age==. sort id age. by id, sort: gen visit=_n * *CREATING A RESTRICTED CUBIC SPLINE FOR AGE (Default knots and positions used here; download ‘rc_spline’ package by William Dupont if needed) rc_spline age, nknots(5) * **RANDOM EFFECTS MODEL DESCRIBING (UNCONDITIONAL) MEAN WEIGHT BY AGE (EQUATION 1) xtmixed weight age || id: age, cov(unstr) mle variance. predict uncond_mean, xb. label var uncond_mean “Unconditional mean” * **UNCONDITIONAL VARIANCE (EQUATION 2): local var_resid= (exp(2 * [lnsig_e]_cons)) estat recov matrix mymatrix=r(cov) local var_slope=mymatrix[1,1] local var_cons=mymatrix[2,2] local cov=mymatrix[2,1] gen uncond_var=‘var_cons’ + (‘var_slope’)*age^2 + 2*age* ‘cov’+‘var_resid’ label var uncond_var “Unconditional variance” * ** CONDITIONAL CENTILES. *Covariance of Y1, Y2: sort id age. gen cov12=”var_cons’ + (age[_n-1] + age)*‘cov’ + (age[_n-1] *age*‘var_slope’) if visit>1. label var cov12 “Covariance” * *Conditional mean of Y2|Y1 (EQUATION 3): gen cond_mean= uncond_mean + (weight[_n-1]-uncond_mean [_n-1])*cov12/uncond_var[_n-1] if visit>1. label var cond_mean “Conditional mean” * *Conditional variance of Y2 | Y1 (EQUATION 4): gen cond_var= uncond_var-(cov12^2/uncond_var[_n-1]) if visit>1 label var cond_var “Conditional variance” * ** IDENTIFYING OUTLIERS USING THE CRITERIA OF > 4SD (EQUATION 5) *+/−4SD .= E(Y2|Y1)+/−4*sqrt(conditional variance) gen ul_cond_centile= cond_mean+4*sqrt(cond_var) gen ll_cond_centile= cond_mean-4*sqrt(cond_var) * gen outlier= 0 replace outlier=1 if weight> ul_cond_centile & ul_cond_centile!=. & visit!=1 replace outlier=1 if weight< ll_cond_centile & ll_cond_centile!=.& visit!=1 replace outlier=0 if outlier==1 & outlier[_n-1]==1 tab outlier *This is the percent of weight observations flagged as outliers * by id, sort: egen outlier_child=max(outlier) gen number=_n if outlier_child==1 tab outlier_child *This is the number of children who have at least one weight measurement flagged as an outlier. * ** GRAPHING THE OUTLIERS sort id age. tab id if outlier==1. local i=[insert here id of a selected outlier] graph twoway (rspike ul_cond_centile ll_cond_centile age if id==‘i’, lcolor(gs8)) (scatter cond_mean age if id==‘i’ , title(ID ‘i’) msymbol(diamond) mfcolor(white) mlcolor(gs8) lpattern(dash)) (connected weight age if id==‘i’, connect(ascending) lcolor(black) mcolor(black) msize(large)) (scatter weight age if id==‘i’ & outlier==1, mcolor(red) msize(large)) (line uncond_mean age, lcolor(gs8) lpattern(dash)), legend(order (5 “Unconditional mean” 1 “4 Conditional SD” 2 “Conditional mean” 4 “Outlier”)) *
References
- 1.Barker DJP, Osmond C, Forsen TJ, Kajantie E, Eriksson JG. Trajectories of growth among children who have coronary events as adults. N Engl J Med. 2005;353(17):1802–9. doi: 10.1056/NEJMoa044160. [DOI] [PubMed] [Google Scholar]
- 2.Baird J, Fisher D, Lucas P, Kleijnen J, Roberts H, Law C. Being big or growing fast: systematic review of size and growth in infancy and later obesity. BMJ. 2005;331(7522):929–34. doi: 10.1136/bmj.38586.411273.E0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang S, Tilling K, Martin R, Davies N, Ben-Shlomo Y, Kramer MS. Pre-natal and post-natal growth trajectories and childhood cognitive ability and mental health. Int J Epidemiol. 2011;40(5):1215–26. doi: 10.1093/ije/dyr094. [DOI] [PubMed] [Google Scholar]
- 4.Surkan PJ, Ettinger AK, Hock RS, Ahmed S, Strobino DM, Minkovitz CS. Early maternal depressive symptoms and child growth trajectories: a longitudinal analysis of a nationally representative US birth cohort. BMC Pediatr. 2014;14:185. doi: 10.1186/1471-2431-14-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hutcheon JA, Platt RW, Abrams B, Himes KP, Simhan HN, Bodnar LM. A weight-gain-for-gestational-age z score chart for the assessment of maternal weight gain in pregnancy. Am J Clin Nutr. 2013;97(5):1062–7. doi: 10.3945/ajcn.112.051706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richard SA, McCormick BJ, Miller MA, Caulfield LE, Checkley W MALED Network Investigators. Modeling environmental influences on child growth in the MAL-ED cohort study: opportunities and challenges. Clin Infect Dis. 2014;59(Suppl 4):S255–60. doi: 10.1093/cid/ciu436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Royston P. Calculation of unconditional and conditional reference intervals for foetal size and growth from longitudinal measurements. Stat Med. 1995;14(13):1417–36. doi: 10.1002/sim.4780141303. [DOI] [PubMed] [Google Scholar]
- 8.Johnsen SL, Rasmussen S, Wilsgaard T, Sollien R, Kiserud T. Longitudinal reference ranges for estimated fetal weight. Acta Obstet Gynecol Scand. 2006;85(3):286–97. doi: 10.1080/00016340600569133. [DOI] [PubMed] [Google Scholar]
- 9.Owen P, Ogston S. Conditional centiles for the quantification of fetal growth. Ultrasound Obstet Gynecol. 1998;11(2):110–7. doi: 10.1046/j.1469-0705.1998.11020110.x. [DOI] [PubMed] [Google Scholar]
- 10.Hutcheon JA, Egeland GM, Morin L, Meltzer SJ, Jacobsen G, Platt RW. The predictive ability of conditional fetal growth percentiles. Paediatr Perinat Epidemiol. 2010;24(2):131–9. doi: 10.1111/j.1365-3016.2010.01101.x. [DOI] [PubMed] [Google Scholar]
- 11.Tukey JW. Exploratory Data Analysis. Reading, MA: Addison-Wesley; 1977. [Google Scholar]
- 12.Kramer MS, Chalmers B, Hodnett ED, Sevkovskaya Z, Dzikovich I, Shapiro S, et al. Promotion of Breastfeeding Intervention Trial (PROBIT): a randomized trial in the Republic of Belarus. JAMA. 2001;285(4):413–20. doi: 10.1001/jama.285.4.413. [DOI] [PubMed] [Google Scholar]
- 13.Harrell FE. Regression Modeling Strategies: with applications to linear models, logistic regression, and survival analysis. Nashville, TN: Springer; 2001. [Google Scholar]
- 14.Dudley NJ. A systematic review of the ultrasound estimation of fetal weight. Ultrasound Obstet Gynecol. 2005;25(1):80–9. doi: 10.1002/uog.1751. [DOI] [PubMed] [Google Scholar]
- 15.Cole TJ, Donaldson MDC, Ben-Shlomo Y. SITAR-a useful instrument for growth curve analysis. Int J Epidemiol. 2010;39(6):1558–66. doi: 10.1093/ije/dyq115. [DOI] [PMC free article] [PubMed] [Google Scholar]