Abstract
To contribute to devise successful beta-cell differentiation strategies for the cure of Type 1 diabetes we sought to uncover barriers that restrict endocrine fate acquisition by studying the role of the transcriptional repressor REST in the developing pancreas. Rest expression is prevented in neurons and in endocrine cells, which is necessary for their normal function. During development, REST represses a subset of genes in the neuronal differentiation program and Rest is down-regulated as neurons differentiate. Here, we investigate the role of REST in the differentiation of pancreatic endocrine cells, which are molecularly close to neurons. We show that Rest is widely expressed in pancreas progenitors and that it is down-regulated in differentiated endocrine cells. Sustained expression of REST in Pdx1+ progenitors impairs the differentiation of endocrine-committed Neurog3+ progenitors, decreases beta and alpha cell mass by E18.5, and triggers diabetes in adulthood. Conditional inactivation of Rest in Pdx1+ progenitors is not sufficient to trigger endocrine differentiation but up-regulates a subset of differentiation genes. Our results show that the transcriptional repressor REST is active in pancreas progenitors where it gates the activation of part of the beta cell differentiation program.
Keywords: NRSF, Beta cells, Diabetes, Islets, Repressor, Pancreas
1. Introduction
Type 1 diabetes (T1D) is caused by an absolute defficiency of insulin secretion due to autoimmune destruction of insulin-secreting beta cells and can be treated by islet cell replacement therapy. However, the lack of high quality donor cells has prompted the search for alternative sources of beta cells to try generating an unlimited supply of beta cells for transplantation. There are promising findings demonstrating in vivo transdifferentiation of acinar, alpha or delta cells into beta cells (Chera et al., 2014; Collombat et al., 2009; Zhou et al., 2008) or differentiation of human embryonic stem cells (ESC) to islet cells (Kroon et al., 2008; Pagliuca et al., 2014; Rezania et al., 2014). However, the differentiation of these cells into beta cells is often partial, a problem which can potentially be solved by better understanding how beta cells differentiate during pancreas embryogenesis.
During development, endocrine cells originate from progenitors in two differentiation waves. Between E8.5 and E12.5, pancreas progenitors are multipotent and can give rise to acinar, ductal and endocrine cells, most of which express glucagon (Kopp et al., 2011; Pan et al., 2013; Solar et al., 2009). After E13.5, the progenitors have become polarized, part of their transcriptional program has changed and they become bi-potent, giving rise to ductal and endocrine cells, while the acinar compartment becomes segregated. Beta cells are essentially born at these stages and until shortly after birth (Johansson et al., 2007; Kopp et al., 2011). We know that a transient Neurog3 expression in progenitors is necessary to direct endocrine differentiation (Gradwohl et al., 2000; Gu et al., 2002) and that the pro-endocrine commitment only proceeds when Neurog3 expression levels reach a threshold (Wang et al., 2008, 2010), triggering direct activation by NEUROG3 of several pro-endocrine transcription factors (Gittes, 2009; Pan and Wright, 2011).
Cell fate decision is, however, the result of inputs from positive as well as negative signals. It is therefore essential to take into account the interplay between the positive drive established by pro-endocrine genes such as Neurog3 and restrictive signals. One such antagonistic signal comes from the Notch effector HES1 which constrains endocrine cell formation by negatively regulating Neurog3 expression (Apelqvist et al., 1999; Jensen et al., 2000). Another factor that might be considered as an attractive new player in this repressive function is the RE-1 Silencing Transcription Factor (REST). This zinc finger transcription factor binds to a 21 bp motif called Repressor Element-1 (RE-1) and recruits several chromatin modiffers to block the expression of its target genes (Ooi and Wood, 2007). Given that the first identified REST targets were associated to terminal function of neurons and because REST is mainly absent from mature neuronal cells, REST has originally been considered as a master repressor of neuronal traits outside of the central nervous system (Chong et al., 1995; Schoenherr and Anderson, 1995). However, a number of new findings have challenged this assertion. First, genome wide analyses of the REST regulon have revealed the existence of a wider than originally thought set of RE-1 containing genes, some of them bearing a non-canonical motif (Otto et al., 2007). Hundreds of new RE-1-bearing genes have been identified and shown to be bound by REST in diverse cell types and contexts (Johnson et al., 2007, 2008; Otto et al., 2007). Importantly, these reports also emphasized that several subsets of REST target genes were associated to non-neuronal functions, showing that REST is not merely a repressor of neuronal traits (Bruce et al., 2004; Johnson et al., 2007, 2008; Mortazavi et al., 2006; Otto et al., 2007; Wu and Xie, 2006). Second, many studies have linked modulations of REST levels in non-neuronal cells to non-neuronal pathologies like colon cancer (Westbrook et al., 2005), cardiac hypertrophy (Kuwahara et al., 2003) or smooth muscle cell neointimal hyperplasia (Cheong et al., 2005); for reviews, see Coulson (2005), Majumder (2006), Thiel et al. (2014).
In the context of pancreatic endocrine cells, in which REST is excluded (Atouf et al., 1997; Martin et al., 2008), we have previously shown using RIP-REST transgenic animals, that RE-1-containing genes are essential for glucose homeostasis. Indeed, we have demonstrated that Rest ectopic expression in pancreatic insulin-producing cells impairs their function and survival by specifically down-regulating the expression of important exocytotic members as well as pro-survival genes (Martin et al., 2008, 2012). As specified for a subset of other genes (Pullen et al., 2010; Quintens et al., 2008), REST is thus “disallowed” in beta cells, as it is in neurons (Atouf et al., 1997). The observation made by ChIP seq analysis that REST binds to the chromatin of drivers of islet cell development (Johnson et al., 2007), together with the fact that REST clearance in neural progenitors has been evoked as a trigger for neural differentiation (Ballas et al., 2005), prompted us to assess the role of REST in the developing pancreas.
In the present study, we show that Rest is expressed in progenitors, and is down-regulated in differentiating endocrine cells during development. We have also generated a transgenic model of REST gain-of-function in pancreatic progenitors to show that REST is sufficient to impair the formation of NEUROG3+ precursors and of differentiated endocrine cells. Finally, using a model of Rest loss-of-function specifically directed to pancreatic progenitors we show that REST inhibits the expression of important factors of endocrine differentiation.
2. Materials and methods
2.1. Mouse strains
The mouse lines (Pdx1-tTA, Pdx1-Cre and REST Fl/Fl) were previously described and are referenced in the result section. The Swiss Veterinary Office and competent Danish authorities approved all animal experiments. Embryos were collected at indicated times; midday on the day of vaginal plug appearance was considered E0.5.
To generate mice with inducible REST expression (TetO-REST mice), an XbaI fragment containing the rabbit β-globin intronic sequence and human Rest cDNA, which served for the generation of RIP-REST mice (Martin et al., 2008), was sub-cloned into a pUHD 10-3 plasmid downstream of a tetracycline operator (TetO) minimal promoter sequence. The resulting XhoI/KasI fragment was used for oocytes injection. These mice were used in the absence of Doxycycline or tetracycline and thus had continuous expression of the transgene in Pdx1-expressing cells, comprising the epithelial cells of the pancreas during development as well as beta cells and a subset of delta cells in the adult.
2.2. Embryonic pancreas preparation
Pancreatic buds were fixed overnight at 4 °C in 4% paraformaldehyde (PFA). Samples were then washed, and immersed in phosphate buffer containing 15% sucrose for 24 h for cryo-preservation. Pancreas were finally incubated in phosphate buffer containing 7% gelatin and 15% sucrose at 37 °C for 1 h, solidified at RT and quickly frozen in methylbutane kept at −65 °C by immersion in dry ice-/EtOH 100%. Blocks were kept at −80 °C until sectioning.
2.3. Mouse islet isolation and cell lines
The rat insulinoma cell lines INS-1E (Merglen et al., 2004) were maintained in complete RPMI 1640 medium as previously described (Martin et al., 2003). Human carcinoma HeLa cells were grown in RPMI 1640 supplemented with 10% fetal bovine serum (FBS), 2 mmol/l L-glutamine, 1 mmol/l sodium pyruvate, 50 U/ml penicillin and 50 μg/ml streptomycin.
Islets of Langerhans were isolated from adult mice bearing REST gain-of-function, Rest loss-of-function and control litter-mates that were anaesthetized by inhalation of 5% halothane (Arovet) and sacrificed. The pancreas was perfused by collagenase injection into the common bile duct. After excision, the pancreas was digested at 37 °C. After filtration on a 100 μm cell strainer (BD Biosciences), the islets were washed in a Hank’s balanced saline solution (HBSS) and cultured in RPMI 1640 containing 11.1 mmol/l glucose supplemented with 10% FCS, 10 mmol/l HEPES, 2 mmol/l L-glutamine, 1 mmol/l sodium pyruvate, 50 μmol/l β-mercaptoethanol, 50 U/ml penicillin and 50 μg/ml streptomycin.
2.4. Western blotting
Western blots were performed as previously described (Martin et al., 2012). Specific protein levels were revealed with polyclonal rabbit antibodies against human REST (Martin et al., 2003), or with polyclonal rabbit antibodies against murine REST (07-579, Merck Millipore). Monoclonal antibodies against α-tubulin (Sigma-Al-drich) were used to normalize the signals.
2.5. Immunofluorescence and EdU staining
Frozen sections (8 μm) were rehydrated in PBS before 30 min blocking in phosphate buffer saline (PBS), Bovine serum albumin (BSA) 1%, Triton 0.2%. Primary antibodies, anti-Glucagon (rabbit; 1:100; Zymed and guinea pig; 1/400; Linco), anti-Insulin (guinea pig; 1/100; Dako), anti-NEUROG3 (rabbit; 1/500; Beta Cell Biology Consortium), anti-SOX9 (rabbit; 1/500; Chemicon), anti-PDX1 (goat; 1/2000; Beta Cell Biology Consortium), and anti-Amylase (rabbit; 1/500; Calbiochem) were incubated overnight at 4 °C.
Secondary antibodies were incubated 1 h at room temperature. Alexa Fluor secondary antibodies (all from Molecular Probes-Invitrogen) were used for multicolor detection. Cell proliferation was assessed using the EdU Click-iT kit, according to the manufacturer’s instructions (Life technologies). EdU was injected intraperitoneally in pregnant mothers 2 h before dissection. Nuclei were stained with DAPI (50 ng/ml; Sigma). The sections were mounted in 50% PBS:50% glycerol and viewed under a Leica DM5500 or a Zeiss LSM700 confocal microscope.
2.6. In situ hybridization and co-immunohistochemistry
Tissues were cryo-sectionned and post-fixed 10 min with 4% PFA. Slides were washed twice in RNase free PBS, incubated 30 min in 0.3% H2O2 in PBS to inhibit endogenous peroxidase, and treated with 0.2 N HcL for 15 min before incubation 10 min with 2 μg/ml proteinase K in PBS at 37 °C. The reaction was stopped with 5 min incubation in fresh 0.2% glycine in PBS. Slides were washed twice in PBS, post-fixed 10 min in 4% PFA, and washed twice in PBS. Slides were then treated twice with freshly prepared acetic anhydride in triethanolamine at pH8, and finally washed twice in PBS and twice in 5X SSC before pre-hybridization. A solution of hybridization, containing 5X saline sodium citrate (SSC) pH4.5, 50% deionized formamide, blocking reagent (Roche) and yeast total RNA was added onto slides that were placed at 68 °C in a humidified box during 1 h. The buffer was replaced with hybridization buffer containing 1 mu;g/ml of denatured sense or anti-sense full length digoxigenin (DIG)-labeled Rest RNA probe, and the slides, protected with a plastic coverslip, were placed for more than 24 h at 68 °C for probe hybridization. Slides were then washed once with 2XSSC pH4.5, three times with 2XSSC pH4.5, 50% formamide at 60 °C, and finally once in Tris-buffered saline (TBS) tween 0.1% (TBST). For probe detection, slides were blocked 30 min with 0.5% blocking reagent in TBST before incubation overnight at 4 °C with anti-DIG-peroxydase antibodies diluted 1/5000 in TBST. For tyramide signal amplification, a biotinylated tyramide (Perkin-Elmer) was used diluted 1/500 in TBST for 20 min, and after washing, the complex was recognized by adding streptavidin-AlexaFluor 594 (Life Technologies) diluted 1/500 in TBST for 30 min.
To perform immunological detection on the same slides, primary antibodies diluted in PBS were added onto slides over night at 4 °C, and after washing, replaced by diluted secondary antibodies conjugated to appropriate fluorophore for 1 h before washing, DAPI staining and mounting.
2.7. Quantitative real-time PCR
Tissues were dissected in PBS, frozen and kept at −80 °C. Samples were then thawed and mechanically disrupted on ice. Total RNA was extracted using the RNeasy Protect Minikit (Qiagen). Reverse transcription of cDNA was carried out using the ImPromII Reverse transcription kit (Promega). For each tissue analyzed, 1 μg of total RNA was used. The qPCR was carried out using the Fast SYBR Green master mix (Applied Biosystems) and the Viia7 real time PCR system from Applied Biosystems. A list of primer pairs is provided in Supplemental Table 1.
2.8. Plasma glucose levels
Blood samples were obtained from the tail vein of mice to measure glucose levels using a freestyle pen (Abbott, USA).
2.9. Single-cell gene expression analysis
Single cell qPCR was performed as previously described (Greggio et al., 2013). Ninety five single cells were isolated from E12.5 control (36 cells from 2 pancreata) and REST Fl/Fl-Cre (59 cells from 2 pancreata) embryos, and gene-specific cDNA samples were prepared and pre-amplified using 48 gene primers (Supplemental Table 2). qPCR was performed using 48.48 Dynamic Array chips (Fluidigm).
Gene expression data were prepared for analysis using Fluidigm real-time PCR analysis software (v. 4.1.3) with linear derivative baseline correction, a quality correction set to 0.65, and a melting curve analysis with specific temperature range of individual gene amplicons as well as settings of peak sensitivity to 7 and peak ratio threshold to 0.7. Limit of detection for analysis was set to Ct value 24, and samples (cells) with low house-keeping gene expression (Actin and Ubc) and poor overall gene expression were eliminated using automatic outlier identification by principle component analysis (PCA). As a result, 34 primer sets passed the quality controls of adequate melting curves and limit of detection, and 49 cells were selected for statistical analyzes. All statistical analyzes (Outlier identification, ANOVA, hierarchical clustering (HC) analysis, and PCA) and generation of plots (HC, PCA, and Violin plots) were performed by autoAnalysis function, using SINGuLAR Analysis Toolset (Fluidigm; v. 3.5.2) in R (The R Foundation for Statistical Computing).
2.10. Genomic PCR
DNA from pancreas and Langerhans islets of adult control and mutant animals was isolated using the PureLink Genomic DNA kit (Life Technologies) according to the manufacturer’s instructions. 20 ng of each sample was used to perform a 28 cycles PCR program, using primers specified in Supplemental Table 3 and annealing temperature set to 63 °C. The efficiency of Cre-mediated recombination of the fioxed REST allele was evaluated in pancreas and islets of Langerhans from adult REST Fl/+-Cre and REST Fl/Fl-Cre animals, as specified in the legend of Supplemental Fig. 4B.
2.11. Quantification and statistical analysis
For quantification, the entire pancreas was serially sectioned. The relative area occupied by cells per pancreas was obtained by counting immunopositive cells on every 4th (E12.5), 6th (E14.5) or 8th (E18.5) section, normalized to the total epithelium area. NEUROG3 positive cells (matching a DAPI+ nucleus) were counted manually. For Insulin, Glucagon, PDX1, SOX9, and DAPI, the positive pixels (surface of section) were quantified using ImageJ software. All values are shown as mean±SD with n numbers and internal replicates indicated in the figure captions; p values were calculated using the nonparametric unpaired Mann–Whitney test unless otherwise indicated; p < 0.05 was considered significant.
3. Results
3.1. Rest is expressed in pancreas progenitors but not in endocrine cells
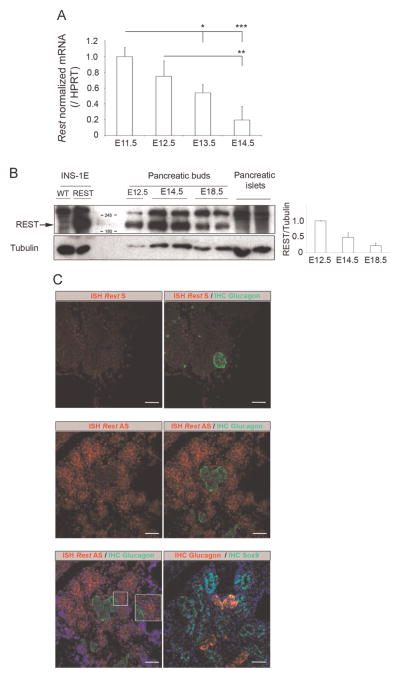
To understand the role of REST during pancreas development, we investigated the spatial and temporal expression patterns of Rest transcripts. We first performed qPCR analysis for Rest mRNA using pancreatic buds at E11.5, 12.5, 13.5 and 14.5 (Fig. 1A). The relative levels of Rest mRNA decreased with time, suggesting that REST levels or the proportion of cells expressing this transcription factor decreases along the differentiation of pancreas. Accordingly, the decreasing abundance of REST protein in developing pancreas was also evidenced by western blot experiments (Fig. 1B). As compared with extracts from native INS-1E cells lacking REST or INS-1E cells transduced with pcDNA encoding murine REST protein, quantification from extracts of WT pancreatic buds at E12.5, E14.5 and E18.5 revealed a decreasing level of REST during development and undetectable repressor in isolated islets (Fig. 1B).
Fig. 1.
Broad Rest expression in early pancreatic epithelium. (A) qPCR analysis shows declining levels of murine Rest transcripts in WT E11.5, 12.5, 13.5 and 14.5 pancreatic buds. Results are mean ± SD. Difference between groups were determined using non parametric one-way ANOVA test. *p < 0.05; **p < 0.01; ***p < 0.001. (B) Left: western blot experiment showing REST production in developing WT pancreas at E12.5, E14.5, and E18.5. Compared with total extracts from wild type INS-1E cells (WT) or INS-1E cells transduced with pcDNA encoding murine Rest (REST), REST antibody revealed a specific band around 200 kDa in total extracts from wild type E12.5, E14.5, and E18.5 pancreatic buds, while this band was absent in islets of Langerhans. Tubulin immunodetection was added as internal control. Right, the quantification of REST signal normalized to tubulin revealed that the levels of REST decreased during pancreas development. (C) Fluorescent in situ hybridization (ISH) for Rest transcript (red) was performed with sense (S) (upper panel) or antisense (AS) probe (middle and lower panels) on E14.5 WT pancreas. The inset shows cytoplasmic signal for Rest transcripts (lower panel). Co-immunostaining performed for Glucagon specifies a region devoid of Rest transcripts (green, upper and middle panel and lower panel, left picture) (red, lower panel, right picture) and staining for SOX9 delineates the domain of bipotent progenitors (green, lower panel, right picture). Blue staining is the DAPI labeling of nuclei. Scale bars, 50 μm.
We next performed in situ hybridization (ISH) to detect Rest mRNA and obtained a specific signal with the antisense probe at E14.5 (Fig. 1C, middle panel). Co-immunostaining on the same section for glucagon (Fig. 1C, middle panel and Supplemental Fig. 1) or insulin (data not shown), together with Rest in situ hybridization, clearly demonstrated that Rest is excluded from differentiated endocrine cells. Although antibodies marking progenitors did not function after in situ hybridization, parallel staining for Rest transcripts and SOX9 protein on two consecutives sections led to the conclusion that Rest and SOX9 expression are overlapping, suggesting that Rest is expressed in the progenitor cells of the developing pancreas (Fig. 1C, lower panel). Rest was widely distributed in the pancreas, including at the periphery, suggestive of expression in exocrine cells, in agreement with previously reported expression in exocrine cell lines (Atouf et al., 1997). Since antibodies against NEUROG3 did not function after in situ hybridization and NEUROG3+ cells are scattered throughout the pancreatic epithelium, it was impossible to draw clear conclusions from staining on adjacent sections. However, several lines of evidence suggest that Rest is repressed in NEUROG3+ endocrine progenitors. First, Rest transcript is decreased 5.8 times in Neu-rog3-YFP+ cells as compared to YFP- cells (http://www.betacell.org/resources/data/studies/view/study_id/3100 and ArrayExpress E-CBIL-48) (Soyer et al., 2010). Second, our transcriptome analysis after NEUROG3 in vivo gain-of-function at E11.25 (Johansson et al., 2007) shows a 1.9 fold decrease of Rest in the pancreatic bud after Neurog3 overexpression (http://www.betacell.org/resources/data/studies/view/study_id/3733) (Cortijo et al., 2012). Altogether, these observations suggest that Rest is widely expressed in the early developing pancreas in multipotent pancreatic progenitors before E12.5 and in bipotent progenitors expressing Sox9 at later developmental stages, and that Rest is down-regulated in differentiated endocrine cells.
3.2. Generation of transgenic mice bearing a conditional, Pdx1-dependent REST expression
To challenge the hypothesis that REST may influence endocrine pancreatic development, we first adopted a gain-of-function strategy to express Rest in all the cells of the epithelium, from early stages on, and to assess whether this mis-expression is sufficient to impair endocrine differentiation. We sub-cloned a fragment containing the human REST cDNA, which served for the generation of RIP-REST mice (Martin et al., 2008), downstream of a tetra-cycline operator (TetO) minimal promoter sequence (Fig. 2A). The construct was then used to generate mice expressing tetracycline-inducible Rest transgene (TetO-REST) by pronuclear injection in oocytes. The four founder lines that we obtained were bred separately with transgenic mice driving the expression of a tetra-cycline-repressible transactivator (tTA) under the control of the endogenous Pdx1 transcriptional regulatory sequence (Pdx1-tTA) (Hale et al., 2005; Holland et al., 2002). Bigenic Pdx1-tTA/TetO-REST mice bear a Tet-off regulatory cassette allowing Rest expression in the absence of Doxycycline (Dox) treatment.
Fig. 2.
Transgenic mice with conditional Rest transgene expression using a Tet-off system, driven by the endogenous Pdx1 promoter. (A) schematic organization of the construct used to generate the transgenic mice for tetracycline-controlled REST activation (TetO-REST mice). The XbaI fragment encompassing β-globin intron and human REST cDNA (Martin et al., 2008) was subcloned into XbaI sites of pUHD10-3. The XhoI/KasI fragment was used for oocytes injection. (B) qPCR analysis shows specific activation of human REST transgene in bigenic Pdx1-tTA/TetO-REST animals. Islets were isolated from adult Pdx1-tTA/TetO-REST (n = 3) from different lines of founders mice that have been treated with Doxycycline (Dox) in utero and during life. Islets were left in culture 24 h with (+) or without (−) Dox and tested for Dox-dependent transgene activation. The Tet-off system allows for REST transgene activation without Dox. The levels of transgene activation were compared to the levels achieved in RIP-REST transgenic mice. Results are mean ± SD. (C) Western blot analysis shows specific production of human REST in islets isolated from bigenic Pdx1-tTA/TetO-REST as compared to control TetO-REST animals. Islets were isolated from adult bigenic Pdx1-tTA/TetO-REST mice that have been treated with Dox in utero and during life. Islets were left 24 h in culture without Dox for REST transgene induction. Nuclear extracts from INS1-E or HeLa cells were used as negative and positive controls, respectively. (D) Immunolabelling for REST (green) and PDX1 (red) shows the pattern of expression of the human REST transgene in comparison with its driver, Pdx1. Staining were performed on E12.5 and E14.5 pancreas from bigenic Pdx1-tTA/TetO-REST mice grown without Dox. Insets show in higher magnification partial co-localization at both stage, demonstrating the mosaic expression of human REST, as compared with the driver, PDX1. Scale bars, 50 μm.
To check for proper transgene activation with the Tet-off system in vivo, we quantified the level of human Rest expression by qPCR, using islets isolated from adult bigenic Pdx1-tTA/TetO-REST. To avoid REST induction in utero and potential associated defects in beta cell formation, the mother and the pups were fed with Dox-containing diet until sacrifice for islet isolation. Then, the islets from each animal were split into two groups and cultured for 24 h with or without Dox. qPCR revealed the absence of Rest in the islets from bigenic mice cultured with Dox (Fig. 2B), thereby attesting for the absence of leakiness of the Tet-off system. Western blot analysis performed with islets of bigenic and control mice grown with Dox and cultured for 24 h without Dox revealed the presence of REST protein in islets of bigenic animals, as compared to negative (INS1-E extract) and positive (HeLa extract) controls (Fig. 2C). The observation of specific signals in bigenic Pdx1-tTA/TetO-REST demonstrated that the Rest transgene is expressed in an inducible manner, in vivo, and leads to the production of REST protein at the expected size. The line 1666 with highest expression levels was selected to pursue the experiments.
3.3. Forced expression of REST in PDX1+ progenitor cells impairs the formation of NEUROG3+ endocrine-committed precursors and subsequent differentiation of endocrine cells
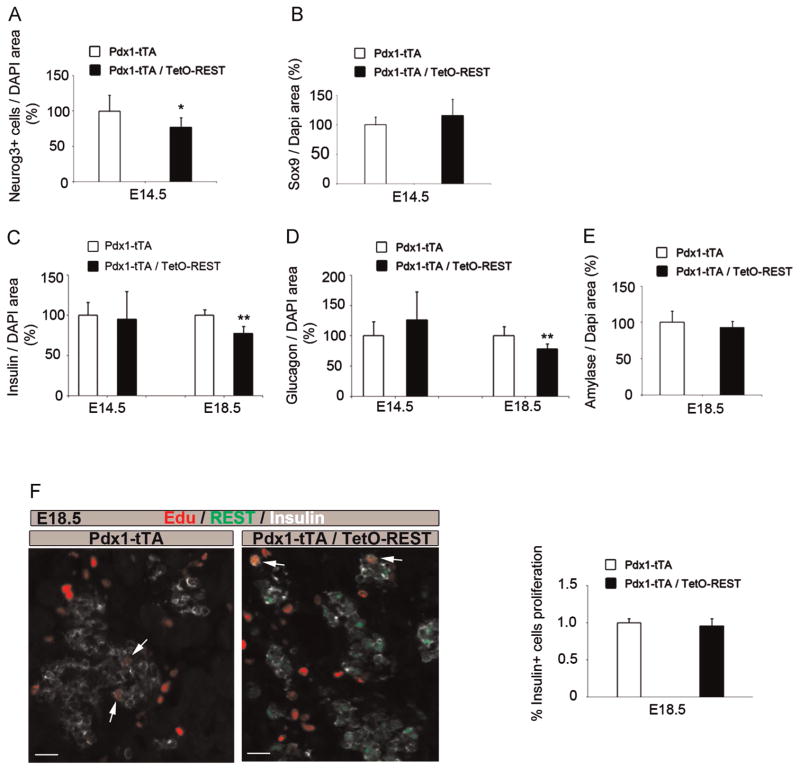
Since Pdx1 gene dosage is important for pancreas formation and islet architecture (Fujitani et al., 2006) and that Pdx1-tTA mice are similar to a Pdx1+/− genotype, we used these as controls rather than the TetO-REST mice. REST was widely detected in the epithelium of the pancreas at E12.5 and E14.5 (Fig. 2D), although the antibody has a low affinity and does not detect endogenous levels (data not shown). The transgene was expressed at higher levels in PDX1-high cells (Fig. 2D and Supplemental Fig. 2). Despite this heterogeneity, we observed a significant reduction by 20% in the number of NEUROG3+ endocrine progenitors in E14.5 pancreas of Pdx1-tTA/TetO-REST as compared to the Pdx1-tTA control mice (Fig. 3A), while there was no detectable alteration in the area of SOX9+ pancreas progenitors (Fig. 3B). Moreover, we observed 20% and 30% decrease in beta- and alpha-cell area, respectively, at E18.5, whereas there was no difference in beta- and alpha-cell area at E14.5 (Fig. 3C and D). We did not detect any change in the exocrine compartment at E18.5 (Fig. 3E). To further distinguish whether the decrease in beta cell formation arises from a defect in differentiation or proliferation, we measured EdU incorporation in insulin+ cells of E18.5 pancreas from control and bigenic embryos (Fig. 3F). No difference was observed in beta cell proliferation at this stage. Altogether, our observations strongly suggest that overexpression of REST in progenitor cells of the developing pancreas from E8.5 (beginning of Pdx1 expression) is sufficient to impair the differentiation of endocrine cells observed at late stages.
Fig. 3.
Quantification of endocrine progenitor and differentiated cell populations in bigenic Pdx1-tTA/TetO-REST. (A) NEUROG3+ cell number normalized to DAPI area was quantified in E14.5 pancreas from Pdx1-tTA control (n = 6) and bigenic Pdx1-tTA/TetO-REST (n = 8) mice grown without Dox. NEUROG3+ cell number is reduced in REST-overexpressing pancreas. *p < 0.05 versus values of control littermates. (B) SOX9+/DAPI area was quantified in E14.5 pancreas from Pdx1-tTA control (n = 6) and bigenic Pdx1-tTA/TetO-REST (n = 8) mice grown without Dox. (C) Insulin+/DAPI area was quantified in E14.5 and E18.5 pancreas from Pdx1-tTA control (n = 5) and bigenic Pdx1-tTA/TetO-REST (n = 5) mice grown without Dox. Insulin+ cell number is reduced in REST-overexpressing pancreas at E18.5. **p < 0.01 versus values of control littermates. (D) Glucagon+/DAPI area was quantified in E14.5 and E18.5 pancreas from Pdx1-tTA control (n = 5) and bigenic Pdx1-tTA/TetO-REST (n = 5) mice grown without Dox. Glucagon+ cell number is reduced in REST-overexpressing pancreas at E18.5. **p < 0.01 versus values of control littermates. (E) Amylase+/DAPI area was quantified in E18.5 pancreas from Pdx1-tTA control (n = 5) and bigenic Pdx1-tTA/TetO-REST (n = 5) mice grown without Dox. (F) Left, representative image of EdU (red), REST (green) and Insulin (white) staining in E18.5 pancreas from Pdx1-tTA control and bigenic Pdx1-tTA/TetO-REST. Arrows indicate proliferative Insulin+/EdU+ double positive cells. Scale bars, 25 μm. Right, quantification of the % of Insulin+/EdU+ in E18.5 pancreas from Pdx1-tTA control and bigenic Pdx1-tTA/TetO-REST.
3.4. Pdx1-tTA/TetO-REST adult mice are diabetic
Our previous study has shown that ectopic expression of REST in mature beta cells under the control of the insulin promoter (RIP-REST mice) impairs both beta cell secretory function as well as survival (Martin et al., 2008). By analyzing a new RIP-REST founder line, we further showed that higher levels of REST trans-gene expression drastically compromised beta cell survival, leading to diabetes (Martin et al., 2012). Here, we report the same impairment in glucose homeostasis with another transgenic system driving REST expression in adult mature beta cells (under the control of Pdx1 regulatory region) (Fig. 4A). Indeed, we observed hyperglycemia in adult bigenic Pdx1-tTA/TetO-REST animals (Fig. 4B). This is likely to be due to REST interfering with beta-cell function, since a 20% reduction in beta-cell mass would not be expected to have such strong physiological outcomes.
Fig. 4.
Pdx1-tTA/TetO-REST adult mice are diabetic. (A) Representative immunostaining of pancreas from adult bigenic Pdx1-tTA/TetO-REST for PDX1 (red) and REST (green) (left panel) or Glucagon (red), REST (green) and insulin (white) (right panel). Left panel shows partial overlap of REST and PDX1 stainings, indicative of a mosaic expression of REST. Right panel shows Insulin+/REST+ double positive cells, while none of the glucagon+ cells are labeled for REST. Scale bars, 25 μm. (B) Blood glucose measurements in adult animals show hyperglycemia in bigenic Pdx1-tTA/TetO-REST (n = 6), as compared to Pdx1-tTA animals (n = 6). ***p < 0.001 versus values of control littermates.
3.5. REST blocks a subset of the endocrine differentiation program
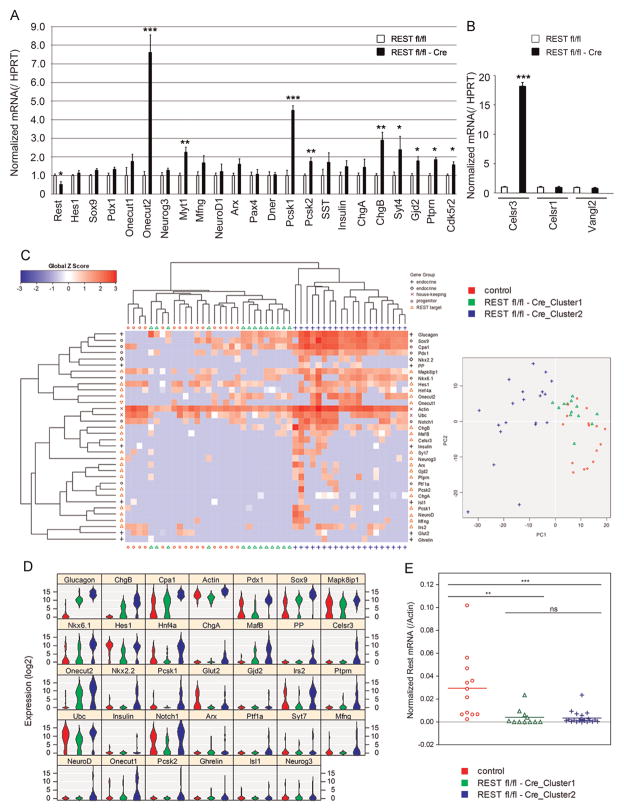
After showing that REST overexpression is sufficient to impair endocrine differentiation, we assessed whether REST loss-of-function in pancreatic progenitors promotes endocrine differentiation. To address this question, we specifically deleted the first coding exon of REST in epithelial pancreatic progenitors by crossing REST Fl/Fl mice (Mao et al., 2011) with mice expressing Cre recombinase under the control of the Pdx1 promoter (Pdx1-Cre) (Hingorani et al., 2003). Rest mRNA was decreased by 50% in REST Fl/Fl-Cre pancreata at E12.5, arguing for important but incomplete inactivation at this stage (Fig. 6A). Genomic PCR in the adult pancreas also confirmed efficient but incomplete inactivation (Supplemental Fig. 4).
Fig. 6.
Rest knock-out in multipotent progenitors relieves expression of key regulatory elements. (A) qPCR analysis on E12.5 pancreatic buds from controls (REST Fl/Fl; n = 5) and Rest KO (REST Fl/Fl-Cre; n = 5) embryos shows a decrease of 50% in the relative expression of Rest. We observed a relieved expression for several REST target genes, including Onecut2, Pcsk1, Myt1, Pcsk2, Chgb, Syt4, Gjd2, Ptprn, and Cdk5r2. Results are mean ± SD. *p < 0.05; **p < 0.01; ***p < 0.001. (B) qPCR analysis on E12.5 pancreatic buds from controls (n = 5) and Rest KO (n = 5) embryos shows a drastic increase in the levels of expression of Celsr3, but not of the other family member Celsr1 or Vangl2. Results are mean ± SD. ***p < 0.001. (C) Left, heatmap of unsupervised hierarchical clustering analysis of gene expression from single cells isolated from E12.5 pancreatic buds of control or Rest KO embryos. Each column represents a single cell from either genotype, for which qPCR analysis yielded clustering of different cell populations (control, REST Fl/Fl-Cre_Cluster 1 and 2), and rows represent clustering of the 34 genes analyzed, according to global Z score. REST Fl/Fl-Cre_Cluster 2 cells differ from control cells, as they express higher levels of endocrine and REST target genes. Right, PCA plot shows clustering of the REST Fl/Fl-Cre_Cluster 1 cells with control cells, while REST Fl/Fl-Cre_Cluster 2 cells differs from both control cells. (D) Violin plot of gene expression ranked according to the order of PCA gene score. REST Fl/Fl-Cre_Cluster 2 cells express higher levels of endocrine genes (including Glucagon, Nkx6.1, PP, and Nkx2.2), as well as REST target genes (including Chgb, Mapk8ip1, Hnf4a, Chga, Mafb, Celsr3, Onecut2, Pcsk1, Gjd2, Ptprn, Mfng, Neurod1, Onecut1 and Pcsk2). E. qPCR analysis of Rest mRNA from single cells used in panels C and D. While control cells express various levels of Rest, both REST Fl/Fl-Cre_Cluster 1 and 2 cells do not express Rest. Differences between groups were determined using non-parametric two-way Mann–Whitney test. **p < 0.01; ***p < 0.001; ns, not significant.
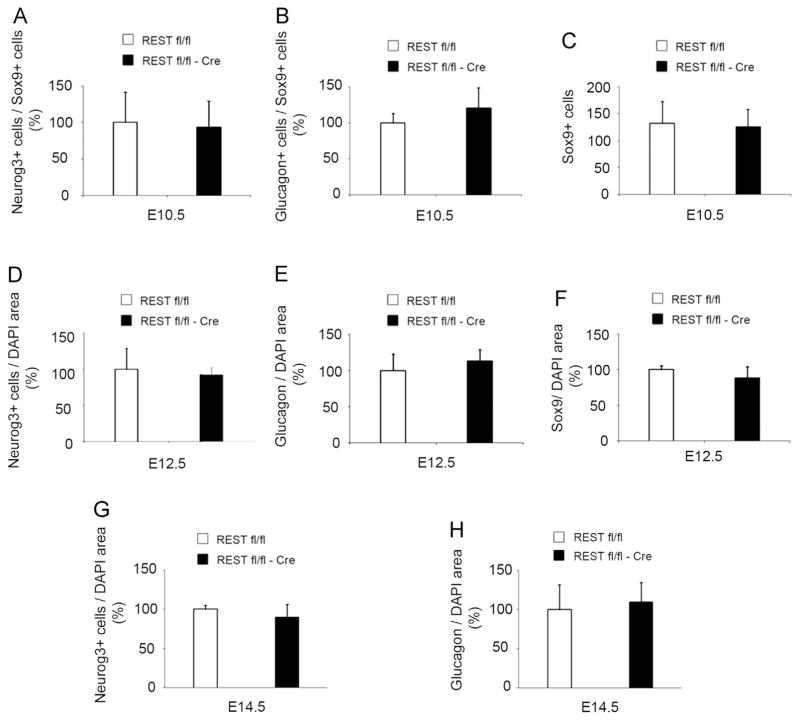
If REST imposes a brake on endocrine differentiation, its inactivation from E8.5 on, might result in ectopic or accelerated pancreatic endocrine differentiation. However, no change in the proportion of NEUROG3+ cell number was detected in REST Fl/Fl; Pdx1-Cre embryos (abbreviated REST Fl/Fl-Cre) as compared to controls (REST Fl/Fl), at E10.5 (Fig. 5A), E12.5 (Fig. 5D) and E14.5 (Fig. 5G). Neurog3 mRNA levels were also unchanged at E12.5 (Fig. 6A). The same observation was made for Glucagon+ cells (Fig. 5B, E and H), and we detected no impairment in the fraction of SOX9+ progenitors at E10.5 or E12.5 (Fig. 5C and F). Since in the nervous system REST represses only part of the neuronal differentiation program (Mao et al., 2011), we investigated whether endocrine differentiation markers were perturbed after REST inactivation.
Fig. 5.
Rest knock-out in multipotent progenitors does not alter pancreas differentiation. (A–H) Cell quantification in E10.5 (A–C), E12.5 (D–F) or E14.5 (G, H) pancreas from controls (REST Fl/Fl; n = 5) and Rest KO (REST Fl/Fl-Cre; n = 5). (A) NEUROG3+ cell number normalized to SOX9+ cell number. (B) Glucagon+ cell number normalized to SOX9+ cell number. (C) SOX9+ cell number. (D and G) NEUROG3+ cell number normalized to DAPI area. (E and H) Glucagon+ cell number normalized to DAPI area. (F) SOX9+ cell number normalized to DAPI area. There was no difference in cell populations at each stage.
Using ChIPSeq analysis in a T cell line, the study of Johnson et al., has identified RE-1 motifs in the chromatin of genes coding for known crucial transcription factors acting during pancreas development, although in the lowest range of intensity, indicative of a weak binding site (Johnson et al., 2007). We investigated the expression of these putative targets of pancreatic relevance based on two databases (Bruce et al., 2004; Johnson et al., 2007). We provide their RE-1 motifs as Supplemental Fig. 3. We also added some key functional genes of endocrine function and tested by qPCR in E12.5 pancreata to investigate whether Rest deletion unlocks their expression. Several important genes involved in endocrine cell differentiation, including several genes with RE-1 motifs were increased in REST Fl/Fl-Cre pancreata including Celsr3, Onecut2, Myt1, Pcsk1 and 2, Chgb, Syt4, Gjd2, Ptprn and Cdk5r2 (Fig. 6A and B). The most striking activation of expression that we observed among RE-1-containing genes in E12.5 Rest mutants was nevertheless that of Celsr3 (Fig. 6B), a key component of the planar cell polarity pathway (PCP) expressed in progenitors and enriched as they differentiate (Cortijo et al., 2012). This did not correspond to a general activation of PCP components since expression of Celsr1 and Vangl2 were not modified in Rest mutant animals (Fig. 6B). Some endocrine genes bearing a previously reported RE-1 motif did not increase, as for example Neurog3, Mfng, Neurod1, Arx, Pax4, Sst or Chga. Progenitor genes were not modified (Hes-1, Sox9, Pdx1, Onecut1, Dner), even those with previously reported RE-1 motifs (Hes-1, Onecut1, Dner).
Because the inactivation of REST was partial, we resorted to single-cell PCR to assess the effect of Rest inactivation on individual cells. The transcripts tested above on whole buds, as well as additional known REST targets and differentiation genes were assessed. We found that 60% of the knock-out cells (cluster 2) were detected as different from control cells by non-supervised clustering and principal component analysis (Fig. 6C). Because the sensitivity of our initial Fluidigm assays was not sufficient to measure Rest transcripts, we performed subsequent nested qPCR on the same samples and found that this population had decreased Rest expression (Fig. 6E). These inactivated cells exhibit an up-regulation of several REST target genes (Chgb, Mapk8ip1, Hnf4a, Chga, Mafb, Celsr3, Onecut2, Pcsk1, Gjd2, Ptprn, Mfng, Neurod1, Onecut1 and Pcsk2) shown in violin plot (Fig. 6D). In addition, endocrine markers such as glucagon, Chgb, Chga, Mafb, and others were up-regulated together with progenitor markers Sox9, Pdx1, Cpa1, Ptf1a and Nkx6.1 suggesting that REST inactivation leads to a confused cell state between a progenitor and an endocrine cell. Among the other 40% cells that were more similar to control cells, a partial response was seen in 10 of 13 cells (cluster 1), which also exhibit a more moderate increase of glucagon, Sox9 and Cpa1. These cells may be recently or heterozygously recombined. Rest expression was also largely absent in cluster 1. Control cells exhibited heterogeneous expression levels of Rest which may be due to stochasticity of detection at low expression levels (Fig. 6E). From our observations, we conclude that even though REST down-regulation is not able to promote precocious or increased differentiation of endocrine cells at E12.5 or E14.5, it normally acts in progenitors to block the expression of several important endocrine differentiation genes.
4. Discussion
In this study, we identified a novel transcription factor in the gene regulatory network that controls endocrine cell fate allocation. As Rest expression was evidenced in neuronal progenitors but not in mature neurons, it was speculated that Rest extinction might play a role in neuronal differentiation (Chong et al., 1995; Schoenherr and Anderson, 1995). Ballas et al. were the first to characterize and describe in vitro the gradual clearance of REST from the chromatin of crucial genes as a trigger for neuronal differentiation to proceed (Ballas et al., 2005). Analysis of inducible knock-out mice for Rest in early progenitors of the developing brain, under the dependence of the Sox1 promoter, supported the conclusion that REST is not required for neurogenesis although inactivation outside of the brain leads to de-repression of REST targets (Aoki et al., 2012). In contrast, inducible deletion of Rest in adult neural stem cells under the dependence of the Nestin promoter triggered a transient increase in neurogenesis and depletion of the hippocampal stem cell pool, indicating a role for REST in maintaining this pool in a quiescent state by restraining the neurogenic program (Gao et al., 2011). Another study, in which Rest was deleted in retinal ganglion progenitor cells under the control of the Six3 promoter, reported that REST controls neurogenesis in the developing retina. Rest deletion led to an increased number of retinal ganglion cells, via the upregulation of crucial retinal ganglion genes, and this occurred independently of the pro-neural gene Atoh7, which is considered as a master promoter of retinal neurogenesis (Mao et al., 2011).
Taking into account that pancreatic beta cells share many similarities with neurons, including the fact that REST is also disallowed in these endocrine cells (Atouf et al., 1997; Martin et al., 2008, 2012; Thiel and Schuit, 2008), we initiated this study to evaluate the role of REST in endocrine differentiation. We report that REST is expressed in pancreas progenitors and that its expression decreases in endocrine progenitors and endocrine cells. This is in agreement with recent findings showing that the Rest gene acquires a Polycomb-mediated H3K27me3 repressive mark after the pancreatic precursor stage which coincides with the activation of a core beta-cell de-repression program (van Arensbergen et al., 2010).
To gain further insights into the role that REST may play in pancreas development, we adopted a gain-of-function strategy, and forced the expression of Rest in pancreas progenitors to assess whether REST clearance is a required event for the pancreas to differentiate. This led to a decrease in the number of endocrine-committed progenitors by E14.5 and ultimately reduced the numbers of glucagon+ and insulin+ cells in E18.5 pancreas. Eventually, adult mice developed diabetes, an observation that we already made in RIP-REST mice overexpressing REST specifically in beta cells (Martin et al., 2012). Diabetes in Pdx1-tTA/TetO-REST mice is most likely caused by an impaired function, rather than being caused by the moderate reduction in insulin-producing cells. A previous report based on REST gain-of-function experiment via electroporation in rat neocortical neurons documented a delayed differentiation and a blocked migration of progenitors upon sustained REST expression (Mandel et al., 2011). Sequence variations in the RE-1 binding motif establish a hierarchy of binding affinity for REST to its target genes, indicating that genes bearing a weakly conserved RE-1 motif have a suboptimal binding affinity for REST (Bruce et al., 2009). We previously noticed a positive correlation between Rest levels of expression and the severity of the phenotype obtained in our RIP-REST animals (Martin et al., 2012). Reaching higher levels of REST expression in pancreatic progenitors of our transgenic mice may have caused a stronger phenotype.
We then used the conditional Rest knock-out line generated to study retinal ganglion cell genesis (Mao et al., 2011) to investigate the requirement for REST in pancreas differentiation. If REST gates the entry of some progenitors into the developmental pathway, by repressing the expression of critical genes of the regulatory network driving differentiation, then its inactivation would be expected to cause premature commitment of these progenitors. However, deleting Rest in early progenitors that express Pdx1 did not result in any increase of endocrine progenitors and endocrine cells. This is surprising as REST was shown to bind the Neurog3 regulatory sequences, and our gain-of-function experiments indeed suggest that Neurog3 is repressed by REST. It is probable that Neurog3 expression and endocrine pathway entry are gated not only by REST but also by other REST-independent repression mechanisms. Other pathways gating endocrine differentiation such as the Notch pathway and its effector HES1 may restrain endocrine differentiation in parallel (Jensen et al., 2000). Different repressive complexes have been involved in development, especially those driving chromatin modifications (Laugesen and Helin, 2014). It was recently shown that REST and Polycomb repressions act in parallel in neuronal (McGann et al., 2014) as well as in pancreatic beta-cells differentiations (van Arensbergen et al., 2010). Indeed, in pancreatic progenitors, PcG marks a wide number of disallowed genes for beta cells, including regulators of alternate developmental fate, while REST binds in ES cells to genes that later selectively lack PcG repression in beta cells. Furthermore, the Rest gene acquires a PcG-mediated H3K27me3 repressive mark after the pancreatic precursor stage, that coincides with the activation of a core beta-cell derepression program (van Arensbergen et al., 2010). CoREST, first identified as a component of the REST complex (Andres et al., 1999), also appears to act independently of REST, by binding to a differential set of genes (Abrajano et al., 2009), and to be sufficient in itself to control cortical neuron differentiation (Ballas et al., 2005; Fuentes et al., 2012; Volvert et al., 2014). It may also control endocrine differentiation with or without REST. Although Rest depletion is not sufficient to initiate endocrine cell formation, an important subset of the endocrine differentiation program is activated and is therefore not under the control of multiple repressors. Indeed, several endocrine markers are up-regulated in the KO. The control over a subprogram is in agreement with observations in Rest-deficient mice (Aoki et al., 2012) or zebrafish (Kok et al., 2012) in the context of neuron specification. The targets appear to affect a variety of functions rather than a specific class. Indeed, genes as diverse as those encoding transcription factors (Onecut1, Onecut2, Hnf4a, Myt1, Arx, Neurod1, Mafb), proteins acting in insulin maturation (Pcsk1, Pcsk2), insulin secretion (Chga, Chgb, Syt4), cell to cell communication (Gjd2, Mfng), anti-apoptotic activity (Ptprn, Cdk5r2) or differentiation functions (Celsr3) were affected upon Rest deletion. Some genes with a RE-1 motif were detected as REST-dependent only in the more sensitive single-cell PCR assay (Hnf4a, Onecut1, Arx, Chga, Mnfg, Neurod1) and some were not de-repressed as for example Neurog3 and Hes1. This was independent of the level of conservation of their RE-1 motif (Supplemental Fig. 3). The expression of unaffected genes may be gated by multiple repressors. Of interest, only a subset of the target genes of REST in the pancreas is also under REST repression in the nervous system. This is for example the case of the gene under strongest repression in pancreas, Celsr3 (Jia et al., 2014) and Syt4, although this has been shown in vitro (Ballas et al., 2005), but not confirmed in vivo (Aoki et al., 2012). In contrast, the fact that other targets regulated in the pancreas are not in the nervous system, as observed for Gjd2 (Hohl and Thiel, 2005) and for Pcsk1 (McClelland et al., 2014), suggests that transcriptional repression by REST may function in a cell type-specific manner. Indeed, the fact that REST represses distinct sets of genes in different cell types has already been noticed and accounted for by the level of REST protein and the sequence variations in the RE-1 motif (Bruce et al., 2009; Johnson et al., 2008). The single-cell PCR also revealed that inactivated cells tend to increase both Glucagon, an endocrine marker, and progenitor genes. The activation of progenitor genes may be due to non-cell autonomous outcomes of partial or non-synchronous REST inactivation inducing feed-back responses. In addition, the induction of Glucagon and to some extent Insulin and other endocrine genes devoid of known RE-1 motifs may be secondary. Primary REST targets inducing this effect remain unknown and may not be detected as robustly as Glucagon, as single cell PCR is noisy for genes expressed at low levels, which is frequent for transcription factors. Interestingly the induction of Glucagon did not result in an increase of cells expressing Glucagon protein, which may result from translational control or too low a level of Glucagon transcript induction.
Taken together our study shows for the first time that endogenous REST plays a repressive role during pancreas development and that multiple repression programs function in parallel, but with likely overlaps, to gate endocrine differentiation. This positions REST as a potential repressor of maturation limiting the production of fully functional beta cells from ES cells in vitro. It will be important to monitor its repression during differentiation protocols producing beta cells, all the more that during the in vitro production the expression levels of some of REST target genes do not reach the expression levels found in beta cells in vivo (Pagliuca et al., 2014; Rezania et al., 2014).
Supplementary Material
Acknowledgments
This project was financed by EPFL and the Novo Nordisk Foundation, Denmark to AGB, as well as by a Swiss National Science Foundation Grant SNSF 31-155897 to JAH. CAM is supported by a National Institutes of Health Grant (NEI, R01EY024376). We would like to thank Dr. Keiichi Katsumoto and Benedikte Traasdahl for help with the knock-out line.
Appendix A. Supplementary material
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.ydbio.2015.07.002.
Footnotes
Author contributions
DM designed and performed the vast majority of experiments, YHK and AGB designed and performed single cell PCR experiments leading to Fig. 6, DS performed adult tissue collection and tests leading to Fig. S4, CAM contributed REST FL animals. AGB contributed to experimental design. DM and AGB wrote the manuscript and all co-authors participated in interpretation and writing.
References
- Abrajano JJ, Qureshi IA, Gokhan S, Zheng D, Bergman A, Mehler MF. Differential deployment of REST and CoREST promotes glial subtype specification and oligodendrocyte lineage maturation. PloS One. 2009;4:e7665. doi: 10.1371/journal.pone.0007665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres ME, Burger C, Peral-Rubio MJ, Battaglioli E, Anderson ME, Grimes J, Dallman J, Ballas N, Mandel G. CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc Natl Acad Sci U S A. 1999;96:9873–9878. doi: 10.1073/pnas.96.17.9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki H, Hara A, Era T, Kunisada T, Yamada Y. Genetic ablation of Rest leads to in vitro-specific derepression of neuronal genes during neurogenesis. Development. 2012;139:667–677. doi: 10.1242/dev.072272. [DOI] [PubMed] [Google Scholar]
- Apelqvist A, Li H, Sommer L, Beatus P, Anderson DJ, Honjo T, Hrabe de Angelis M, Lendahl U, Edlund H. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400:877–881. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- Atouf F, Czernichow P, Scharfmann R. Expression of neuronal traits in pancreatic beta cells. Implication of neuron-restrictive silencing factor/re-pressor element silencing transcription factor, a neuron-restrictive silencer. J Biol Chem. 1997;272:1929–1934. doi: 10.1074/jbc.272.3.1929. [DOI] [PubMed] [Google Scholar]
- Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Bruce AW, Donaldson IJ, Wood IC, Yerbury SA, Sadowski MI, Chapman M, Gottgens B, Buckley NJ. Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc Natl Acad Sci U S A. 2004;101:10458–10463. doi: 10.1073/pnas.0401827101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce AW, Lopez-Contreras AJ, Flicek P, Down TA, Dhami P, Dillon SC, Koch CM, Langford CF, Dunham I, Andrews RM, Vetrie D. Functional diversity for REST (NRSF) is defined by in vivo binding affinity hierarchies at the DNA sequence level. Genome Res. 2009;19:994–1005. doi: 10.1101/gr.089086.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong A, Bingham AJ, Li J, Kumar B, Sukumar P, Munsch C, Buckley NJ, Neylon CB, Porter KE, Beech DJ, Wood IC. Downregulated REST transcription factor is a switch enabling critical potassium channel expression and cell proliferation. Mol Cell. 2005;20:45–52. doi: 10.1016/j.molcel.2005.08.030. [DOI] [PubMed] [Google Scholar]
- Chera S, Baronnier D, Ghila L, Cigliola V, Jensen JN, Gu G, Furuyama K, Thorel F, Gribble FM, Reimann F, Herrera PL. Diabetes recovery by age-dependent conversion of pancreatic delta-cells into insulin producers. Nature. 2014;514:503–507. doi: 10.1038/nature13633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong JA, Tapia-Ramirez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, Altshuller YM, Frohman MA, Kraner SD, Mandel G. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- Collombat P, Xu X, Ravassard P, Sosa-Pineda B, Dussaud S, Billestrup N, Madsen OD, Serup P, Heimberg H, Mansouri A. The ectopic expression of Pax4 in the mouse pancreas converts progenitor cells into alpha and subsequently beta cells. Cell. 2009;138:449–462. doi: 10.1016/j.cell.2009.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortijo C, Gouzi M, Tissir F, Grapin-Botton A. Planar cell polarity controls pancreatic beta cell differentiation and glucose homeostasis. Cell Rep. 2012;2:1593–1606. doi: 10.1016/j.celrep.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulson JM. Transcriptional regulation: cancer, neurons and the REST. Curr Biol. 2005;15:R665–R668. doi: 10.1016/j.cub.2005.08.032. [DOI] [PubMed] [Google Scholar]
- Fuentes P, Canovas J, Berndt FA, Noctor SC, Kukuljan M. CoREST/LSD1 control the development of pyramidal cortical neurons. Cereb Cortex. 2012;22:1431–1441. doi: 10.1093/cercor/bhr218. [DOI] [PubMed] [Google Scholar]
- Fujitani Y, Fujitani S, Boyer DF, Gannon M, Kawaguchi Y, Ray M, Shiota M, Stein RW, Magnuson MA, Wright CV. Targeted deletion of a cis-regulatory region reveals differential gene dosage requirements for Pdx1 in foregut organ differentiation and pancreas formation. Genes dev. 2006;20:253–266. doi: 10.1101/gad.1360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Ure K, Ding P, Nashaat M, Yuan L, Ma J, Hammer RE, Hsieh J. The master negative regulator REST/NRSF controls adult neurogenesis by restraining the neurogenic program in quiescent stem cells. J Neurosci: Off J Soc Neurosci. 2011;31:9772–9786. doi: 10.1523/JNEUROSCI.1604-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittes GK. Developmental biology of the pancreas: a comprehensive review. Dev Biol. 2009;326:4–35. doi: 10.1016/j.ydbio.2008.10.024. [DOI] [PubMed] [Google Scholar]
- Gradwohl G, Dierich A, LeMeur M, Guillemot F. neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci U S A. 2000;97:1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greggio C, De Franceschi F, Figueiredo-Larsen M, Gobaa S, Ranga A, Semb H, Lutolf M, Grapin-Botton A. Artificial three-dimensional niches deconstruct pancreas development in vitro. Development. 2013;140:4452–4462. doi: 10.1242/dev.096628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu G, Dubauskaite J, Melton DA. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129:2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- Hale MA, Kagami H, Shi L, Holland AM, Elsasser HP, Hammer RE, MacDonald RJ. The homeodomain protein PDX1 is required at mid-pancreatic development for the formation of the exocrine pancreas. Dev Biol. 2005;286:225–237. doi: 10.1016/j.ydbio.2005.07.026. [DOI] [PubMed] [Google Scholar]
- Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA, Kawaguchi Y, Johann D, Liotta LA, Crawford HC, Putt ME, Jacks T, Wright CV, Hruban RH, Lowy AM, Tuveson DA. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- Hohl M, Thiel G. Cell type-specific regulation of RE-1 silencing transcription factor (REST) target genes. Eur J Neurosci. 2005;22:2216–2230. doi: 10.1111/j.1460-9568.2005.04404.x. [DOI] [PubMed] [Google Scholar]
- Holland AM, Hale MA, Kagami H, Hammer RE, MacDonald RJ. Experimental control of pancreatic development and maintenance. Proc Natl Acad Sci U S A. 2002;99:12236–12241. doi: 10.1073/pnas.192255099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen J, Pedersen EE, Galante P, Hald J, Heller RS, Ishibashi M, Kageyama R, Guillemot F, Serup P, Madsen OD. Control of endodermal endocrine development by Hes-1. Nat Genet. 2000;24:36–44. doi: 10.1038/71657. [DOI] [PubMed] [Google Scholar]
- Jia Z, Guo Y, Tang Y, Xu Q, Li B, Wu Q. Regulation of the protocadherin Celsr3 gene and its role in globus pallidus development and connectivity. Mol Cell Biol. 2014;34:3895–3910. doi: 10.1128/MCB.00760-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson KA, Dursun U, Jordan N, Gu G, Beermann F, Gradwohl G, Grapin-Botton A. Temporal control of neurogenin3 activity in pancreas progenitors reveals competence windows for the generation of different endocrine cell types. Dev Cell. 2007;12:457–465. doi: 10.1016/j.devcel.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Johnson R, Teh CH, Kunarso G, Wong KY, Srinivasan G, Cooper ML, Volta M, Chan SS, Lipovich L, Pollard SM, Karuturi RK, Wei CL, Buckley NJ, Stanton LW. REST regulates distinct transcriptional networks in embryonic and neural stem cells. PLoS Biol. 2008;6:e256. doi: 10.1371/journal.pbio.0060256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok FO, Taibi A, Wanner SJ, Xie X, Moravec CE, Love CE, Prince VE, Mumm JS, Sirotkin HI. Zebrafish rest regulates developmental gene expression but not neurogenesis. Development. 2012;139:3838–3848. doi: 10.1242/dev.080994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp JL, Dubois CL, Schaffer AE, Hao E, Shih HP, Seymour PA, Ma J, Sander M. Sox9+ ductal cells are multipotent progenitors throughout development but do not produce new endocrine cells in the normal or injured adult pancreas. Development. 2011;138:653–665. doi: 10.1242/dev.056499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon E, Martinson LA, Kadoya K, Bang AG, Kelly OG, Eliazer S, Young H, Richardson M, Smart NG, Cunningham J, Agulnick AD, D’Amour KA, Carpenter MK, Baetge EE. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat Biotechnol. 2008;26:443–452. doi: 10.1038/nbt1393. [DOI] [PubMed] [Google Scholar]
- Kuwahara K, Saito Y, Takano M, Arai Y, Yasuno S, Nakagawa Y, Takahashi N, Adachi Y, Takemura G, Horie M, Miyamoto Y, Morisaki T, Kuratomi S, Noma A, Fujiwara H, Yoshimasa Y, Kinoshita H, Kawakami R, Kishimoto I, Nakanishi M, Usami S, Saito Y, Harada M, Nakao K. NRSF regulates the fetal cardiac gene program and maintains normal cardiac structure and function. Embo J. 2003;22:6310–6321. doi: 10.1093/emboj/cdg601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugesen A, Helin K. Chromatin repressive complexes in stem cells, development, and cancer. Cell Stem Cell. 2014;14:735–751. doi: 10.1016/j.stem.2014.05.006. [DOI] [PubMed] [Google Scholar]
- Majumder S. REST in good times and bad: roles in tumor suppressor and oncogenic activities. Cell Cycle. 2006;5:1929–1935. doi: 10.4161/cc.5.17.2982. [DOI] [PubMed] [Google Scholar]
- Mandel G, Fiondella CG, Covey MV, Lu DD, Loturco JJ, Ballas N. Repressor element 1 silencing transcription factor (REST) controls radial migration and temporal neuronal specification during neocortical development. Proc Natl Acad Sci U S A. 2011;108:16789–16794. doi: 10.1073/pnas.1113486108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao CA, Tsai WW, Cho JH, Pan P, Barton MC, Klein WH. Neuronal transcriptional repressor REST suppresses an Atoh7-independent program for initiating retinal ganglion cell development. Dev Biol. 2011;349:90–99. doi: 10.1016/j.ydbio.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D, Allagnat F, Chaffard G, Caille D, Fukuda M, Regazzi R, Abderrahmani A, Waeber G, Meda P, Maechler P, Haefiiger JA. Functional significance of repressor element 1 silencing transcription factor (REST) target genes in pancreatic beta cells. Diabetologia. 2008;51:1429–1439. doi: 10.1007/s00125-008-0984-1. [DOI] [PubMed] [Google Scholar]
- Martin D, Allagnat F, Gesina E, Caille D, Gjinovci A, Waeber G, Meda P, Haefiiger JA. Specific silencing of the REST target genes in insulin-secreting cells uncovers their participation in beta cell survival. PloS One. 2012;7:e45844. doi: 10.1371/journal.pone.0045844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D, Tawadros T, Meylan L, Abderrahmani A, Condorelli DF, Waeber G, Haefiiger JA. Critical role of the transcriptional repressor neuron-restrictive silencer factor in the specific control of connexin36 in insulin-producing cell lines. J Biol Chem. 2003;278:53082–53089. doi: 10.1074/jbc.M306861200. [DOI] [PubMed] [Google Scholar]
- McClelland S, Brennan GP, Dube C, Rajpara S, Iyer S, Richichi C, Bernard C, Baram TZ. The transcription factor NRSF contributes to epileptogenesis by selective repression of a subset of target genes. eLife. 2014;3:e01267. doi: 10.7554/eLife.01267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGann JC, Oyer JA, Garg S, Yao H, Liu J, Feng X, Liao L, Yates JR, 3rd, Mandel G. Polycomb- and REST-associated histone deacetylases are independent pathways toward a mature neuronal phenotype. eLife. 2014;3:e04235. doi: 10.7554/eLife.04235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merglen A, Theander S, Rubi B, Chaffard G, Wollheim CB, Maechler P. Glucose sensitivity and metabolism-secretion coupling studied during two-year continuous culture in INS-1E insulinoma cells. Endocrinology. 2004;145:667–678. doi: 10.1210/en.2003-1099. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Leeper Thompson EC, Garcia ST, Myers RM, Wold B. Comparative genomics modeling of the NRSF/REST repressor network: from single conserved sites to genome-wide repertoire. Genome Res. 2006;16:1208–1221. doi: 10.1101/gr.4997306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi L, Wood IC. Chromatin crosstalk in development and disease: lessons from REST. Nat Rev Genet. 2007;8:544–554. doi: 10.1038/nrg2100. [DOI] [PubMed] [Google Scholar]
- Otto SJ, McCorkle SR, Hover J, Conaco C, Han JJ, Impey S, Yochum GS, Dunn JJ, Goodman RH, Mandel G. A new binding motif for the transcriptional repressor REST uncovers large gene networks devoted to neuronal functions. J Neurosci: Off J Soc Neurosci. 2007;27:6729–6739. doi: 10.1523/JNEUROSCI.0091-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliuca FW, Millman JR, Gurtler M, Segel M, Van Dervort A, Ryu JH, Peterson QP, Greiner D, Melton DA. Generation of functional human pancreatic beta cells in vitro. Cell. 2014;159:428–439. doi: 10.1016/j.cell.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan FC, Bankaitis ED, Boyer D, Xu X, Van de Casteele M, Magnuson MA, Heimberg H, Wright CV. Spatiotemporal patterns of multipotentiality in Ptf1a-expressing cells during pancreas organogenesis and injury-induced facultative restoration. Development. 2013;140:751–764. doi: 10.1242/dev.090159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan FC, Wright C. Pancreas organogenesis: from bud to plexus to gland. Dev Dyn. 2011;240:530–565. doi: 10.1002/dvdy.22584. [DOI] [PubMed] [Google Scholar]
- Pullen TJ, Khan AM, Barton G, Butcher SA, Sun G, Rutter GA. Identification of genes selectively disallowed in the pancreatic islet. Islets. 2010;2:89–95. doi: 10.4161/isl.2.2.11025. [DOI] [PubMed] [Google Scholar]
- Quintens R, Hendrickx N, Lemaire K, Schuit F. Why expression of some genes is disallowed in beta-cells. Biochem Soc Trans. 2008;36:300–305. doi: 10.1042/BST0360300. [DOI] [PubMed] [Google Scholar]
- Rezania A, Bruin JE, Arora P, Rubin A, Batushansky I, Asadi A, O’Dwyer S, Quiskamp N, Mojibian M, Albrecht T, Yang YH, Johnson JD, Kieffer TJ. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat Biotechnol. 2014;32:1121–1133. doi: 10.1038/nbt.3033. [DOI] [PubMed] [Google Scholar]
- Schoenherr CJ, Anderson DJ. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- Solar M, Cardalda C, Houbracken I, Martin M, Maestro MA, De Medts N, Xu X, Grau V, Heimberg H, Bouwens L, Ferrer J. Pancreatic exocrine duct cells give rise to insulin-producing beta cells during embryogenesis but not after birth. Dev Cell. 2009;17:849–860. doi: 10.1016/j.devcel.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Soyer J, Flasse L, Raffelsberger W, Beucher A, Orvain C, Peers B, Ravassard P, Vermot J, Voz ML, Mellitzer G, Gradwohl G. Rfx6 is an Ngn3-dependent winged helix transcription factor required for pancreatic islet cell development. Development. 2010;137:203–212. doi: 10.1242/dev.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel G, Ekici M, Rossler OG. RE-1 silencing transcription factor (REST): a regulator of neuronal development and neuronal/endocrine function. Cell Tissue Res. 2015;359:99–109. doi: 10.1007/s00441-014-1963-0. [DOI] [PubMed] [Google Scholar]
- Thiel G, Schuit F. No REST for healthy beta cells. Diabetologia. 2008;51:1343–1346. doi: 10.1007/s00125-008-1043-7. [DOI] [PubMed] [Google Scholar]
- van Arensbergen J, Garcia-Hurtado J, Moran I, Maestro MA, Xu X, Van de Casteele M, Skoudy AL, Palassini M, Heimberg H, Ferrer J. Dere-pression of Polycomb targets during pancreatic organogenesis allows insulin-producing beta-cells to adopt a neural gene activity program. Genome Res. 2010;20:722–732. doi: 10.1101/gr.101709.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volvert ML, Prevot PP, Close P, Laguesse S, Pirotte S, Hemphill J, Rogister F, Kruzy N, Sacheli R, Moonen G, Deiters A, Merkenschlager M, Chariot A, Malgrange B, Godin JD, Nguyen L. MicroRNA targeting of CoREST controls polarization of migrating cortical neurons. Cell Rep. 2014;7:1168–1183. doi: 10.1016/j.celrep.2014.03.075. [DOI] [PubMed] [Google Scholar]
- Wang S, Hecksher-Sorensen J, Xu Y, Zhao A, Dor Y, Rosenberg L, Serup P, Gu G. Myt1 and Ngn3 form a feed-forward expression loop to promote endocrine islet cell differentiation. Dev Biol. 2008;317:531–540. doi: 10.1016/j.ydbio.2008.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Yan J, Anderson DA, Xu Y, Kanal MC, Cao Z, Wright CV, Gu G. Neurog3 gene dosage regulates allocation of endocrine and exocrine cell fates in the developing mouse pancreas. Dev Biol. 2010;339:26–37. doi: 10.1016/j.ydbio.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbrook TF, Martin ES, Schlabach MR, Leng Y, Liang AC, Feng B, Zhao JJ, Roberts TM, Mandel G, Hannon GJ, Depinho RA, Chin L, Elledge SJ. A genetic screen for candidate tumor suppressors identifies REST. Cell. 2005;121:837–848. doi: 10.1016/j.cell.2005.03.033. [DOI] [PubMed] [Google Scholar]
- Wu J, Xie X. Comparative sequence analysis reveals an intricate network among REST, CREB and miRNA in mediating neuronal gene expression. Genome Biol. 2006;7:R85. doi: 10.1186/gb-2006-7-9-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.