Abstract
Reverse phase protein array (RPPA) technology evolved from the advent of miniaturized immunoassays and gene microarray technology. Reverse phase protein arrays provide either a low throughput or high throughput methodology for quantifying proteins and their post-translationally modified forms in both cellular and non-cellular samples.
As the demand for patient tailored therapies increases so does the need for precise and sensitive technology to accurately profile the molecular circuitry driving an individual patient’s disease. RPPAs are currently utilized in clinical trials for profiling and comparing the functional state of protein signaling pathways, either temporally within tumors, between patients, or within the same patients before/after treatment. RPPAs are generally employed for quantifying large numbers of samples on one array, under identical experimental conditions. However, the goal of personalized cancer medicine is to design therapies based on the molecular portrait of a patient’s tumor, which in turn result in more efficacious treatments with less toxicity. Therefore, RPPAs are also being validated for low throughput assays of individual patient samples. This review explores reverse phase protein array technology in the cancer research field, concentrating on its role as a fundamental tool for deciphering protein signaling networks and its emerging role in personalized medicine.
1. Introduction
In the last decade, the field of molecular medicine has seen new technological advances in proteomics and genomics, which are rapidly designating molecular profiling as a necessary tool for translational research. Deciphering the molecular pathogenesis of deadly diseases, such as cancer, is fundamental for understanding disease mechanisms and for the rational design of targeted therapy regimens [1, 2]. This new diagnosis and treatment paradigm has several designations - individualized therapy, molecular medicine, or personalized medicine – all of which indicate the need to design therapies based on known/predictive biomarkers, prognostic factors, and a patient’s genomic and/or proteomic disease profile [3]. Pharmacogenetics, assessing the impact of an individual’s genes on drug metabolism/response, is an example of personalized medicine in which genetic information guides specific drug treatment decisions [4]. In contrast, population-based epidemiological approaches for designing therapy rely on analysis of large cohorts of patients with efficacy defined by the outcome of the majority, rather than individuals. Personalized medicine, the term used herein, aims to improve disease detection, predict treatment response, and reduce adverse therapy events by combining common prognostic criteria such as tumor stage, grade, age, etc. with an individual patient’s genomic/proteomic profile [1–3]. The ability to quantify phosphoprotein levels in small amounts of human biopsy material provides a new class of analytes that factor into treatment decisions [5].
While cancer is characterized by accumulation of genomic alterations, it is the proteomic-driven cellular functions and interactions that have a profound effect on the information flow within the cell. The cellular proteome is a complex and dynamic entity, whose fluctuating minute by minute state reflects the in vivo status of the cell. The nucleic acid content (DNA, mRNA, siRNA, ncRNA, etc) cannot provide direct information regarding the state of protein signaling pathways within a cell. Multiple genetic and genomic alterations are currently accepted as the source of malignant transformations; however, the resulting encoded proteins are the actual defective piece of machinery leading to alterations in cellular growth, survival or apoptosis [6, 7]. The faulty protein products of oncogenes and tumor suppressor genes may include protein kinases, growth factors, growth factor receptors, DNA repair enzymes, and growth inhibitors. Protein kinases, however, are often the key molecules in the cellular circuitry, and their aberrant function is frequently at the center of many diseases, including cancer. Although considerable progress has been made in the use of genetics and cancer genomic profiling, molecular therapies, such as tyrosine receptor kinase inhibitors that target specific proteins or protein networks, have rendered a more suitable dynamic approach for cancer treatment [8–11].
Cellular homeostasis is vigilantly safeguarded by continuous rearrangements of proteins through several kinases and phosphatases. The phosphorylation or activation state of kinase-driven signaling networks provides essential information regarding the underlying driving force of an individual’s disease. Characterization of such detailed protein interactions, taking place both inside and outside of the cell even for only a subset of key physiological processes influencing tumor growth, such as survival, proliferation, migration, and apoptosis, could have a profound effect in understanding disease mechanisms. Furthermore, protein-protein and protein–nucleic acid interactions are revealing new potential drug targets. Consequently, mapping the dynamic molecular circuitry, by quantifying post-translationally modified cell signaling proteins, and/or sequencing tumors for known mutations, is now becoming a vital element in designing clinical trials and individualized patient therapies [3–5]. The published treatment response rates for various diseases/conditions vary e.g. 77% response to Cox-2 inhibitors for analgesia of post-operative pain [12, 13], or ~25% response of colorectal cancer to chemotherapy (5–flurouracil plus leucovorin) [14–17]. This wide variation in treatment effectiveness highlights the need for more accurate a priori determinations of treatment efficacy [4]. For example, Epidermal Growth Factor Receptor (EGFR) is a validated therapeutic target in colon cancer [18, 19]. However, EGFR expression levels fail to predict clinical efficacy. 35–40% of patients with KRAS mutations in exon 2 fail to respond to EGFR inhibitors [19]. These examples underscore the importance of deciphering the proteomic and/or genomic underpinnings of individual patient’s tissue to achieve more durable treatment response rates.
Individualizing treatment for cancer patients has always been an aim of molecular medicine, but only recently is this goal being realized and supported by biomedical research [1,2]. The complexity of cancer has proven very challenging; its heterogeneous nature produces unpredictable responses to current drug treatments. Nonetheless, new molecular targets and combination therapies are actively being evaluated in clinical trials, expediting the translation of basic research into clinical applications (Table 1). RPPA analysis includes the following functional information about the state of actionable drug targets: a) protein signal pathway network analysis, b) upstream/downstream linkage analysis, c) protein signaling across classes of samples/treatments, d) predictive treatment efficacy and patient stratification, and e) post-translational proteomic data [5, 20–22]. This data is unattainable by genomic and transcriptomic analyses. RPPA provide post-translational molecular data which facilitates deciphering the underlying cellular biology.
Table 1.
Clinical trials incorporating reverse phase protein microarray analysis.
| Trial identifiera | Acronym | Condition/Tumor type | Study design | Phase | Status |
|---|---|---|---|---|---|
| NCT01563874 | N/A | Lymphoma | Observational | N/A | Completed |
| NCT00407810 | N/A | Metastatic head and neck cancer | Open-label; interventional | II | Completed |
| NCT00867334 | NITMEC | Colorectal cancer | Open-label; interventional | II | Completed |
| NCT01074814 | Side-Out | Metastatic breast cancer | Open-label; interventional | II/III | Completed |
| NCT01919749 | Side-Out 2 | Metastatic breast cancer | Open-label; interventional | II/III | Recruiting |
| NCT01042379 | I-SPY 2 | Breast cancer | Open-label; interventional | II | Recruiting |
| NCT01023477 | PINC | Breast-ductal carcinoma in situ | Open-label; interventional | I/II | Recruiting |
| NCT00798655 | N/A | Head and neck cancer | Open-label; interventional | II | Recruiting |
| NCT02159989 | N/A | Metastatic solid tumors | Open-label; interventional | I | Recruiting |
| NCT00897650 | N/A | Lung cancer | Observational | N/A | Recruiting |
| NCT02133183 | N/A | Adult Glioblastoma | Open-label; interventional | N/A | Recruiting |
Clinicaltrials.gov identifier accessed from http://clinicaltrials.gov, provided by the U.S. National Library of Medicine, as of July 30, 2014.
The tools and information required to truly and proficiently implement personalized medicine are currently being validated in clinical trials and accredited laboratories. Validation entails assessing RPPA performance following strict standard operating procedures with on-going documentation of performance parameters such as inter-assay precision, quality control ranges, and adequacy and performance of calibrators over time [5, 22]. High throughput quantitative proteomic techniques, such as reverse-phase protein arrays in which 80 or more samples can be assessed on the same array under the same experimental conditions, have made the analysis of more complex biological systems achievable. The RPPA platform allows the profiling and comparison of the functional state of protein signaling pathways, over time, to follow the course of disease as well as its response to therapy. Characterization of novel drug targets, and the discovery of potential diagnostic and prognostic biomarkers are placing reverse phase protein array technology as the preferred platform for proteomic cancer studies [1, 2, 10, 22–27]. However, RPPA have been utilized to assess the state of signaling kinases in non-neoplastic conditions, such as during Rift Valley Fever (RFV) infection and in ophthalmologic conditions such as macular degeneration [28, 29]. The focus of this review is to highlight the contributions of reverse phase protein array technology in the cancer research field, concentrating on its role as a fundamental tool for deciphering protein signaling networks and its emerging role in personalized medicine.
2. Reverse-phase protein arrays
Reverse-phase protein arrays are classified as a subcategory of protein arrays. In contrast to forward phase arrays in which individual proteins, such as antibodies, are immobilized on a substratum, RPPA are comprised of an immobilized cellular or protein-based lysate, or known quantities of a peptide, protein, or recombinant protein [9]. These cellular lysates represent the state of individual tissue cell populations, which can be any type of cell populations including normal, malignant, or the surrounding stroma. Samples, controls and calibrators are printed as individual spots onto a nitrocellulose-coated slide using a robotic arrayer. The high protein binding capacity of the RPPA substratum (nitrocellulose) allows immobilization of protein from dilute cell lysates (e.g. 0.25mg/mL total protein). Using automated staining systems, each array slide is probed with a specific primary antibody, and a corresponding secondary antibody to detect expression of the target epitope [9, 30]. Signal amplification is independent of the immobilized protein, permitting coupling of detection strategies with highly sensitive amplification chemistries [31–34]. Multiplexing is achieved by batching samples, printing them on arrays, and analyzing tens to hundreds of samples under the same experimental conditions for any given analyte on a single array, thus providing direct quantifiable information on post-translational modifications across all samples [8, 9, 35]. However, multiplexing in this context does not lend itself to personalized medicine because an array is constructed only after many different samples have been collected. To address the need for individualized assessment of a patient’s tumor, RPPAs can also be constructed in multi-pad or sector formats [36]. A sector array is comprised of numerous “pads” of nitrocellulose on one glass slide, or in a 96-well format. One patient sample is printed on each of the different nitrocellulose pads, with controls and calibrators in each sector. In a sector format, the nitrocellulose pads can be physically separated from each other by a gasket, thus allowing the same sample to be probed with a different antibody in each sector. The low throughput sector RPPA offers a methodology for assessing several different proteins in an individual sample.
Protein network post-translational modifications can be accurately profiled with RPPAs by probing the arrays using two different antibodies - one antibody is directed against the unmodified form of the protein, for example AKT, while a second antibody is directed against a specific phosphorylation site, for example phospho-AKT Ser473. By comparing the levels of non-phosphorylated protein, phosphorylated protein, and the proportion of phosphorylated protein to the non-phosphorylated form, one can infer the activation status of the protein [8, 24]. The RPPA platform embodies an essential research tool by providing superior sensitivity, combined with broad-scale analysis of phosphoproteins, and the detection of low-abundance proteins from very small amounts of starting material.
Personalized medicine often requires analysis of small numbers of cells, or small volumes of sample, from a single patient (Figure 1). Most proteomics technologies have significant technical limitations mainly due to analytical sensitivity, and these limitations are particularly noticeable during the analysis of very small tissue samples. Most tissue biopsy specimens contain only a few thousand cells and other proteomic methods, such as mass spectrometry or Enzyme-linked Immuno-sorbent Assay (ELISA), usually require relatively large numbers of cells for adequate sensitivity [37]. RPPA combined with laser capture microdissection (LCM) for cell procurement has efficiently crossed that hurdle allowing for the relative quantification a of numerous proteins obtained from a small number of cells directly procured from tissue samples [21, 38–40]. The activation state of proteins in signaling networks fluctuate constantly depending on their cellular microenvironment, which in vitro cell culture models and animal models might not be able to accurately portray [24, 41, 42].
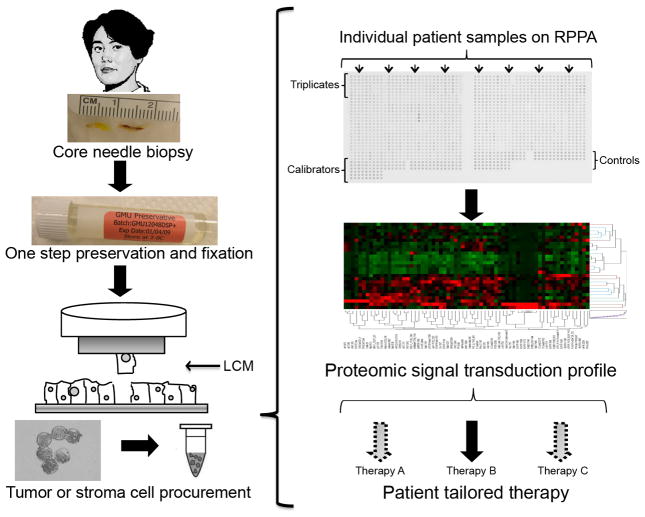
Fig. 1.
Reverse phase protein microarray (RPPA) workflow for clinical samples. Biopsy or surgical samples are immediately stabilized following procurement. Enriched cell populations are procured by Laser Capture Microdissection, followed by cell lysis to produce whole cell lysates that are representative of the microdissected cell populations. RPPAs are constructed using robotic printers. Proteins of interest are detected by immunological staining methods, using validated primary antibodies and signal amplification chemistries to detect low abundance, post-translationally modified proteins. Data analysis encompasses determining spot intensity values, local area background values, normalization, and annotation of the data. Bioinformatic analyses, such as unsupervised hierarchical clustering, provide insights into similarities/dissimilarities between samples and sample groups.
2.1 RPPA data analysis workflow
RPPA data provides relative quantification of phosphorylated, glycosylated, acetylated, cleaved, and total cellular proteins in diseased and non-diseased tissue from multiple samples printed on a series of identical arrays. These arrays can be measured and compared in parallel using commercially available anti-phosphoprotein or other specific antibodies. Antibody specificity is evaluated and validated beforehand using conventional immunoblotting techniques, such as western blotting, on a broad panel of cell lines and human tissues. A single or dominant band at the appropriate molecular weight validates the specificity of the antibody (Figure 2). The relative binding affinity of the antibody determines the optimal antibody dilution for use on the RPPA [43, 44]. However, it is unnecessary to know or calculate the analyte concentration and antibody affinity prior to using an antibody on a RPPA. Samples are printed on each array in a dilution series (e.g. undiluted, 1:2, 1:4, 1:8). The sample dilution series provides an array-specific means of determining both the antibody affinity and analyte concentration because the optimal antibody affinity and protein concentration represent the linear dynamic portion of the sample dilution curve (Figure 1).
Fig. 2.
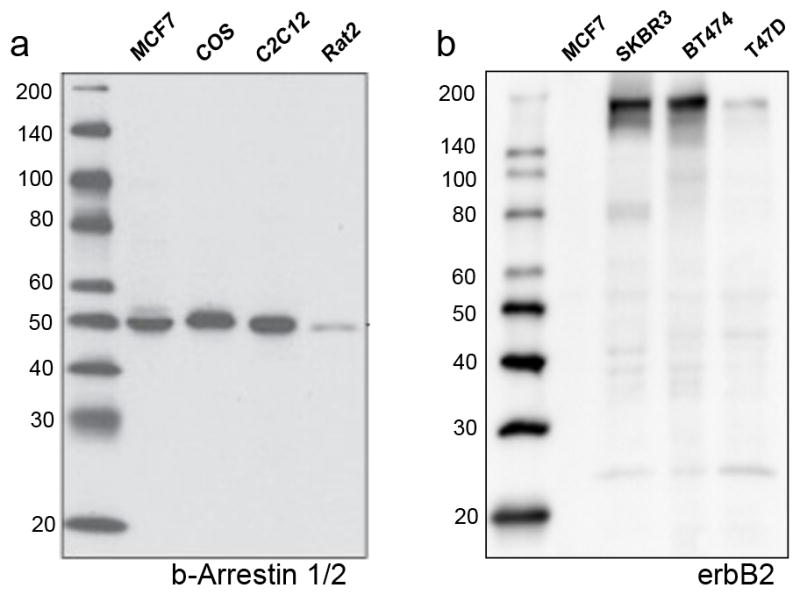
Validation of antibody specificity. (a) Validated antibody: a western blot probed with anti-β-Arrestin 1/2 shows a dominant band at the expected molecular weight using a variety of commercially available cell lysates. (b) Non-validated antibody: a western blot probed with anti-erbB2 shows multiple bands at various molecular weights.
RPPA data, while similar to gene microarray data, has a unique set of features that influence the data analysis pipeline. The first unique feature is the presence of total and post-translationally modified proteins within the same sample spot on the array. Unmodified and post-translationally modified proteins can be quantified between multiple sample groups, between replicate sample groups (disease/control), between paired samples (treated/untreated or tumor/stroma), and between time series measurements or drug concentrations. However, RPPAs require an epitope-specific antibody for each available analyte. Therefore the data analysis method must have adequate sensitivity to distinguish total and modified forms of the same protein. The second unique feature is the presence of contaminating proteins, either from blood, the extra-cellular matrix, of buffer components. To normalize spot intensities across all samples on the array, a common analyte, that is unaffected or uniformly affected by the disease, treatment, or sample processing, must be identified. The third unique feature of RPPAs is the presence of controls and calibrators on each array. These features supply a built-in analytical measurement range for each analyte and can furnish low/high cut-off values for assessing activation states of the proteins.
These unique RPPA features have led to the development of several different algorithms for spot intensity analysis. Despite the algorithm selected for data reduction, the typical data analysis workflow follows the same general processes: a) Determine spot intensity values (pixel intensity/fluorescence intensity) [45, 46] and local spot area background; b) Select data analysis methods (e.g. MicroVigene [47], RPPA Analysis Suite [48], NormaCurve/SuperCurve [49], RPPApipe [50]; modified PSCAN Dose Interpolation Algorithm (DI25) [51], RPPAnalyzer Toolbox [52]), c) Annotate data; d) Calculate differential expression; e) Generate graphical representations of the data; and f) Perform bioinformatics such as unsupervised two-way hierarchical clustering to identify clusters of samples and endpoints. A key concept in molecular data analysis is consistency of analysis across samples and sample sets. Intensity values alone cannot be compared across the current RPPA data analysis methods because each method uses different algorithms for generating the spot intensity value. Comparative analysis between and within RPPA data sets requires consistent data analysis methods because each method has slightly different features for normalizing, local spot background correction, and management of non-specific signal.
3. Integration of Laser Capture Microdissection in RPPA based studies
Procurement of enriched cell populations in a vastly heterogeneous tissue sample is required for accurate identification of protein network profiles [40, 53, 54]. In past years, the problem of tissue heterogeneity presented a significant barrier to the molecular analysis of normal and diseased tissue. Initial attempts at protein pathway profiling consisted of whole tissue analysis making it difficult to determine the origin of a given protein signal or the specific cell type responsible for an activated pathway. In many cases, the cell subpopulation of interest in a tissue may constitute only a small fraction of the total tumor mass. In any given tumor sample one may find a combination of several cell types, such as normal and tumor epithelium, stromal cells, nerve cells, immune cells, and vascular cells, which are able to contribute independently to the growth, invasion, and metastasis of the patient’s tumor. Molecular data from heterogeneous samples could be severely compromised if undesired cells contaminate the starting research material, even in samples with >80% tumor [53]. Furthermore, a lack of correlation in protein expression patterns between microdissected and non-microdissected material has been previously reported [40, 54]. Consequently, the isolation of pure individual tissue cell subtypes within the tumor microenvironment is a crucial step in sample preparation for protein pathway profiling based on RPPA analysis and for any other proteomic approach [55]. This issue has been addressed with tissue microdissection. Microdissection, whether manually with a needle/blade or with a laser-guided instrument (Laser Capture Microdissection (LCM)), is considered a well-established technique that allows the procurement of enriched cell populations from a heterogeneous tissue under direct microscopic visualization, thus, enhancing the approach to molecular analysis of pathologic processes. LCM is capable of harvesting target cells for the assessment of molecular changes in diseased and non-diseased tissues and for identification of biomarker content in individual cell populations through RPPA studies [44, 56–61]. Furthermore, analytes present in tumor and host cells may be drug targets. Thus accurately identifying and analyzing the cell populations that harbor the actionable drug target will further support the goals of personalized medicine.
One concern about incorporating LCM into the RPPA workflow has been the additional labor and time required to procure cells. Jameson et al recently reported a turn-around-time of 12–23 days (median 15.5 days) in a multi-site, multi-omic clinical trial (Side-Out, Table 1) in which fresh biopsies were subjected to immunohistochemistry, cDNA microarray analysis, and LCM prior to RPPA [5]. The turnaround-time for LCM-RPPA of clinical biopsy samples in our clinical trials ranges from 3 – 10 business days from biopsy to report [5, 22] [Dr. M. Pierobon, 8 Oct 2014, written personal communication]. Microdissection of heterogeneous tissue samples is required to obtain meaningful information that can reveal unique protein pathway signatures of a patient’s tumor, which in turn can be exploited as therapeutic targets.
4. Tissue Microenvironment: a true representation of disease
Identification of new protein targets is steadily steering away from immortalized cultured cell lines to the discovery of proteins that are modified in diseased human tissue. A patient’s cellular proteome is constantly changing depending on the cellular microenvironment. Therefore, protein signaling events in immortalized cultured cell lines may not accurately reflect the physiological state of the patient’s tissue microenvironment. Although, genomic and proteomic analysis of cultured cell lines are still contributing important information to basic research, the application of molecular profiling to provide individually tailored therapeutics should include direct proteomic pathway analysis of patient material [5, 9, 21, 22, 25, 44, 47, 53, 54, 62–66].
In the era of protein biomarker discoveries, tissue instability presents a real problem to the field of molecular medicine. Tissue collection, handling, storage, and processing can significantly introduce bias and have a major impact in the final data generated from molecular profiling. Following procurement, the tissue is immediately subjected to induced stress and hypoxia and surges of wound repair related signal pathway proteins and transcription factors [67, 68]. Excised tissue is still considered a living cellular entity, and as such, it reacts to ex vivo trauma beginning with oxidative, hypoxic and metabolic stress, nutrient deprivation, wounding, finally resulting in cell death [41, 69, 70]. Consequently, phosphorylation activity of certain kinase substrates may increase due to the persistence of functional signaling, or activation by other stress-response signals [41, 69, 71]. Without tissue stabilization, interpretation of biomarker data might be significantly compromised. Although, chemical and protein-based phosphatase inhibitors are used to prevent substrate dephosphorylation by phosphatases in the absence of kinase activity, there is still room for any remaining active kinases to be affected [72, 73]. The balance between kinases and phosphatases is a major source of false positives and false negatives in the field of biomarker discovery [74]. Tissue samples for RPPA protein pathway profiling require stabilization, or preservation, of the kinases and phophoproteins immediately post tissue procurement to maintain the fidelity of data generated by protein analysis.
Currently, there are new molecular fixatives that are being evaluated in clinical trials to eliminate or greatly minimize sources of pre-analytical variables. These new tissue fixatives are being incorporated and evaluated in large national biobanking efforts and specifically provide the molecular preservation equivalent to snap-frozen tissue, while retaining formalin-like histomorphologic details [41, 70]. There is a broad range of clinical assay variability, including pre-analytical and post-analytical events, which could potentially influence the final data output. The promise of tissue protein biomarkers to provide revolutionary diagnostic and therapeutic information will only be possible with the specific and rapid preservation of phosphoproteins at the time of excision, providing true information representative of the in vivo state of the signaling network within the tissue [70].
5. RPPA and Cancer Profiling
Reverse-phase protein arrays were first described in 2001 by Paweletz et al. [30]in a paper depicting its application to cell signaling analysis of pre-malignant prostate lesions compared to normal epithelium and invasive carcinoma. Signaling pathway profiling of laser-capture microdissected prostate tissue revealed that members of the PI3 kinase/pro-survival protein cascades were activated at the tumor invasion front during prostate cancer progression. Early phosphorylation and activation of AKT was reported to occur as a key step in the progression of cancer, altering cellular turnover due to a decrease in the cell death rate, not by induction of the growth rate [30].
RPPA has been utilized to address inherent differences in tumor biology of primary and metastatic lesions. Ovarian carcinoma often disseminates to the peritoneum beyond the pelvis. Prognosis of metastatic ovarian cancer is often governed by the metastatic cells, rather than the primary tumor. In a study performed by Sheehan et al. [65] six primary ovarian tumors and patient-matched omental metastases were procured simultaneously during surgery. RPPA phosphoproteomic data was evaluated with unsupervised hierarchical clustering and principle component analysis to assess cell signaling events in patient matched primary and metastatic tumors and to determine if there were common dysregulated proteins that develop in the metastatic tumors. Hierarchical clustering revealed two data clusters: activated and non-activated protein, with considerable heterogeneity in protein levels between patients. The primary peritoneal carcinomas phosphoproteomic levels were not significantly different compared to the primary ovarian carcinomas. However, variation in phosphoproteomic levels were noted in the metastatic tumor group compared to the patient matched primary carcinoma group, without a common dominant protein pathway between these groups. Phosphorylation of c-Kit was dramatically elevated in the majority of metastatic tumors compared to the primary lesions. Principle component analysis identified phospho-c-kit, phospho-Ask, phospho-Ikβα and phospho-Ras-Grf as highly variable phosphoproteins between the primary and metastatic tumors [65]. Each patient’s proteomic signature appeared to have evolved as the tumor spread to a metastatic site.
RPPA proteomic signal pathway patterns can complement mRNA pattern analysis of clinical response and histopathologic subtypes for a more complete understanding of the true drivers of the malignant process [75]. To address the lack of response to chemotherapy in children with rhabdomyosarcoma, Petricoin et al. [21]utilized RPPA to provide a phosphoproteomic network analysis of microdissected tumor cells. Frozen tissue samples were obtained from children with Rhabdomyosarcoma enrolled in the Children’s Oncology Group Intergroup Rhabdomyosarcoma Study (IRS) IV, D9502 and D9803. Rhabdomyosarcoma exists in three histomorphological subtypes: alveolar, embryonal and botyroid, each one of which is associated with the presence/absence of genetic transformations and prognostic indicators. To assess the state of cell signaling networks, samples representing each of the tumor subtypes were procured prior to treatment. The tissue samples were microdissected, and profiled by RPPA to investigate potential differences in pathway networks reflective of the tumor histomorphology. Surprisingly, the histological subtype was not correlated with protein signaling network data. An altered interconnection was found however, between phosphorylated forms of mTOR, IRS-1 and AKT/mTOR pathway proteins for tumors from patients unresponsive to therapy compared to therapy responders. The authors functionally demonstrated that the negative feedback loop between AKT-IRS-1-mTOR was disrupted in the non-responder group (p<0.001). Elevated levels of phospho-AKT Ser473, phospho-mTOR Ser2448, and phospho-p70S6K ser389 were found to be linked to elevated levels of phospho-IRS-1 Ser612 in the responder group, indicating that IRS-1 negative feedback loop was intact (Spearman rho non-parametric correlations). However, in the therapy non-responder group these correlations were not statistically significant. The significance of the AKT/mTOR pathway was further confirmed in a mouse xenograft model treated with CCI-779, an mTOR inhibitor. CCI-779 significantly reduced tumor growth in the mouse xenograft model of Rhabdomyosarcoma [21]. Thus, the authors concluded that combination therapies targeting both AKT/mTOR and IGF-IRS pathways prior to standard of care chemotherapy may impart substantial cellular stress on the tumor cells, effectively enhancing the chemotherapy.
RPPA was also used to address the epithelial-stromal signaling pathway crosstalk of colorectal carcinoma and matched normal tissue samples. The ‘seed versus soil hypothesis” of tumor metastasis postulates that metastasis requires cross-talk between tumor (seed) and the distant tissue (soil) [76]. Sheehan et al evaluated phosphoproteomic levels of colon tumor and stroma, both biochemically via RPPA, and histologically with hematoxylin stained tissue sections during laser capture microdissection of distinct tumor and stromal cell compartments [66]. Stromal and epithelium similarities were more evident in colon tumors than in normal tissue from the same patient, suggesting activation of epithelial-mesenchymal transition (EMT). Activation of Ras-GRF, phosphop38 and phospho-IkB proteins suggested that EMT is driven by cell proliferation pathways, specifically the mitogen-activated protein kinase pathway [66]. In this case, therapies targeting active molecules in both stroma and cancer cells may be more advantageous for the treatment of colorectal cancer.
In a recent lung cancer study consisting of 101 case-matched normal and tumor tissue samples, RPPA technology was utilized to investigate differences in the activation state of 126 proteins and their phosphorylation sites involved in cell proliferation, DNA repair, signal transduction and lipid metabolism [77]. Ku80 levels, a tumor suppressor gene, were reported as significantly higher in tumors of non-smokers compared to smokers. Furthermore, overexpression of Cyclin B1 was detected in poorly differentiated tumors and significantly higher levels of Cox2 were observed in neuroendocrine tumors. An important finding was the noticeable increased expression of Stat5. Given its association with favorable clinical outcomes Stat5 has been proposed as a prognostic biomarker for lung cancer [77]. Coupling RPPA with bioinformatics has revealed protein networks/co-regulations that were not discernable from single probability data distribution of sample data in a breast tissue cohort [63]. These network interactions have the potential to elucidate protein crosstalk, which can then be studied mechanistically in cell culture models of protein over-expression and/or knock-down. Using hierarchical clustering from RPPA data of 56 patient matched tumor and normal adjacent breast tissue samples, Gujral et al showed phospho-cMet Tyr1349 was not clustered near common prognostic proteins Her2, Estrogen Receptor (ER) and Progesterone Receptor (PR) [63]. However, phospho-cMet Tyr1349 levels were statistically associated with Axl receptor tyrosine kinase (Axl) and Stat3. Structured Bayesian inference network analysis, using class variables cancer versus normal, revealed a likely network in which phospho-cMet interacted with Axl, and with phospho-Raf, and phospho-Raf interacted with phospho-Akt. While cMet overexpression is associated with poor clinical outcome, Axl receptor tyrosine kinase (Axl) and phosphorylation of the cMet were not known to be functionally linked in a cell signaling kinase pathway. Mechanistic studies with the triple negative cell line MDA-MB-231 (ER, PR, Her2 negative) that overexpress both Axl and cMet supported the Axl-cMet receptor cross-talk finding in the human RPPA breast tissue samples [63]. This study highlights the application of RPPA for discovering kinase combinations that may be susceptible to novel bi-specific inhibitors.
6. Application of Personalized Medicine and RPPA
The growing awareness that every cancer develops as a unique molecular entity depending on the patient’s individual genetic background is leading the development of personalized therapy. Although two tumors could appear morphologically similar under the microscope, they could be driven by different aberrant signaling pathways. For this reason, a population of cancer patients treated with the same drug may result in only a few responders and many others could suffer from unnecessary toxic side effects. RPPA analysis quantifies specific phosphorylation events and offers an approach to profile the activity state of protein cascades that contain potential drug targets to segregate responders from non-responders [10, 21, 24, 27, 40, 44]. Personalized medicine based on an individual patient’s molecular profile will provide clinicians with the required information to efficiently treat individual cancers and lessen toxicity from generalized treatments by determining a priori which treatment has a high likelihood of efficacy [1–3, 6, 18].
The concept that each patient’s cancer has a unique set of pathogenic molecular derangements continues to be supported by emerging molecular information derived from RPPAs [23,30,36]. RPPA are capable of monitoring changes in protein phosphorylation over time, before and after treatment, between disease and non-disease states, and between responders and non-responders. Identification of critical nodes or intersections within protein networks is a potential starting point for drug development and the design of individual therapeutic regimens [9, 21, 22, 30, 65, 66, 78]. The ideal clinical scenario would be to present a menu of treatment choices, or treatment combinations specifically targeted to a patient’s molecular profile of his/her tumor. The following examples of ongoing clinical trials illustrate the emerging use of RPPA within clinical research.
The RPPA platform has been recently used in the I-SPY 1 TRIAL (Investigation of Serial Studies to Predict Your Therapeutic Response with Imaging and Molecular Analysis) sponsored by the Foundation for the National Institutes of Health (USA). This multicenter clinical study was designed to identify novel tumor biomarkers and establish standards for collecting molecular and imaging data of neoadjuvant breast cancer samples over the course of patient care. Magnetic resonance imaging (MRI) imaging changes were evaluated as potential predictors to patient therapy response and recurrence free/overall survival after neoadjuvant treatment. Additionally, it was the intention of this clinical trial to produce clinical, proteomic, genomic, and imaging biomarker data in order to generate molecular portraits of breast malignancies [79]. As part of this study, RPPA coupled with LCM was specifically utilized to investigate the signaling profile of HER family members in 127 fine-needle biopsies collected before neoadjuvant treatment. Wulfkuhle et al. [47] reported high levels of phospho-HER2 Y1248 in a small set of patients showing no signs of HER2 over-expression. Furthermore, associated increases in activation of HER3, EGFR, and downstream substrates suggested that HER-mediated signaling might be determined by heterodimerization of HER receptor family members. This subgroup of patients may benefit from receiving HER-targeted therapies even though they did not demonstrate amplification of membrane bound HER2 [47].
I-SPY 2 is utilizing an adaptive trial design to correlate response and outcome with molecular targeted inhibitors. Association between activated biomarkers in protein cascades and the response to investigational targeted therapeutic agents is being evaluated in patients newly diagnosed with invasive breast cancer. Tumor biopsies are currently collected before treatment begins and patients are placed in different treatment regimens determined only by the molecular characteristics of their tumor. Thus, the main purpose of this clinical trial is to determine if the addition of a therapeutic agent to neoadjuvant treatment is superior to standard of care alone [62].
The PINC (Prevention of Invasive breast Neoplasia by Chloroquine) trial is a neoadjuvant therapy for patients with breast ductal carcinoma in situ (DCIS). Analysis of the mechanisms used by DCIS cells to survive in the nutrient deprived intraductal environment has revealed autophagy as a therapeutic target and chloroquine phosphate is being evaluated as an anti-proliferative agent [41,42]. This clinical trial is examining the safety and effectiveness of chloroquine administration for a 4-week period for patients with low-grade, intermediate-grade or high-grade DCIS, regardless of hormone receptor status. Magnetic resonance imaging is conducted on each patient before enrollment and just before the designated standard-of-care surgical therapy. Effectiveness in this DCIS trial design will be uniquely measured directly at the molecular level in the DCIS tissue utilizing RPPA before and after treatment. The activated state of signaling pathway proteins associated with autophagy, hypoxia, cell-adhesion, apoptosis, and p53 mediated cell survival in microdissected epithelial and stroma cells will be measured before and after the 4 week therapy. Furthermore, DCIS living organoids and DCIS progenitor cells are harvested and characterized by organ culture, xenograft transplantation, and molecular cytogenetics [20, 80].
7. Concluding Remarks
RPPA technology is emerging as an indispensable tool in the development of patient tailored therapies. Broad-scale pathway activation mapping using techniques such as the RPPA provide a means to quantify the current state of hundreds of key signaling proteins and cascades at once from clinical samples. RPPA quantitatively analyses phosphorylated, glycosylated, acetylated, cleaved, or total cellular proteins from multiple samples simultaneously [9, 30]. Microdissection of the heterogeneous tissue sample is required to obtain meaningful information that can reveal unique protein pathway signatures in a patient’s tumor which in turn can be exploited as therapeutic targets. The cellular kinome represents a rich source of new targets for molecular therapeutics, and technologies such as RPPA have greatly facilitated the assessment of cellular activity of these molecules for the realization of patient-tailored therapy [81, 82].
However it is imperative to mechanistically link the discovery of a biomarker to the biology of the patient’s tumor. Generating a list of activated proteins and pathways found in a diseased sample may not have any clinical impact if the biology of the tumor is not known or ignored. Confidence in biomarker applications for early detection, high-risk screening, recurrence monitoring, or individualized therapy could significantly increase if there is an understanding of the biological and biochemical effects of existing overexpressing and/or under-expressing proteins in an patient’s tumor [74].
8. Expert opinion
The technology for printing, staining and analyzing reverse phase protein arrays has evolved into sophisticated, commercially available systems, thus facilitating the adoption of this technology by many laboratories. However, the cost and labor to produce and analyze a RPPA is the main limiting factor to widespread clinical adoption of this technology. Nevertheless, improvements in the technology are either currently available or under development to address these current technological shortcomings. Examples of improvements are: a) pre-printed arrays with known samples and controls, b) commercial RPPA printing services, c) a variety of commercially available nitrocellulose substrata and formats, d) fluorescent, colorimetric, and chemiluminescent protein multiplexing strategies, and e) development of multi-use scanners that allow for alternative RPPA detection methods.
As reverse phase protein arrays continue to evolve as a clinical research assay, the RPPA community is striving to establish guidelines for published RPPA data, as suggested by guidelines such as minimum information for a proteomic experiment (MIAPE) [23, 83]. Performance characteristics such as precision, accuracy, analytical measurement range, and interfering substance are currently being evaluated in accredited labs within the USA in an effort to validate RPPA as a laboratory developed test. Establishing a world-wide RPPA society and user group meetings is the first step in sharing knowledge essential for transforming RPPA into a clinical assay for personalized medicine [23].
A critical unmet need in the field of proteomics is the development of reference intervals (also known as normal ranges) for the proteins, including signaling kinases and tyrosine receptor kinases, to which molecular targeted inhibitors exist. Although RPPAs incorporate controls and calibrators on each array which can be utilized to establish high and low protein levels, we currently don’t know the true range of “normal” values for many proteins and transcription factors. Analogous to normal ranges for serum electrolytes or glucose, we foresee a future in which large cohorts of tissue samples, or individual patient tissues, from normal and diseased tissue are analyzed by RPPA to establish reference ranges for these emerging molecular targets. Despite the current lack of knowledge regarding normal levels of cell signaling proteins, reverse phase protein arrays provide an unrivalled technology for deciphering and monitoring aberrant proteins and their post-translationally modified states from a small amount of cellular material.
Key Points.
Reverse phase protein arrays allow a multiplex, broad-scale characterization of activated/deactivated proteins and their post-translationally modified forms. Specific cell signaling pathways can be interrogated, which can elucidate therapeutic protein targets in the context of individualized treatment.
The use of reverse phase protein arrays as a tool for mapping protein signaling networks of tumor tissue and the tumor microenvironment has advanced from a novel research technique to clinical trials.
Further refinements of the reverse phase protein array technology as an in vitro diagnostic test are on-going. Validation of clinical assay performance, such as precision, accuracy, linearity, and analytical measurement range, will further enhance the utility and application of this technology in the clinical setting.
Acknowledgments
This work was supported in part by George Mason University, the Department of Defense Breast Cancer Research Program through a grant to L. Liotta and V. Espina (W81XVVH-10-1-0781), and the National Institutes of Health Innovative Molecular Analysis Technologies program through a grant to L. Liotta and V. Espina (1R33CA157403-01). Lance Liotta is Principle Investigator for the PINC trial and kindly provided editorial advice for this manuscript. The funding sources did not have any role in the study design; the collection, analysis and interpretation of data; manuscript preparation; or the decision to submit the paper for publication.
Footnotes
Conflict of interest statement: VE is an inventor of technologies discussed in this article and, as a university employee, may receive patent royalties per university policies. VE is entitled to stock options from Theranostics Health, Inc.
Author contributions: RG and VE wrote the manuscript. RG contributed the figures and tables.
References
- 1.Liotta LA, Kohn EC, Petricoin EF. Clinical proteomics: personalized molecular medicine. JAMA. 2001;286(18):2211–4. doi: 10.1001/jama.286.18.2211. [DOI] [PubMed] [Google Scholar]
- 2.Petricoin EF, Zoon KC, Kohn EC, Barrett JC, Liotta LA. Clinical proteomics: translating benchside promise into bedside reality. Nat Rev Drug Discov. 2002;1(9):683–95. doi: 10.1038/nrd891. [DOI] [PubMed] [Google Scholar]
- 3.de Bono JS, Ashworth A. Translating cancer research into targeted therapeutics. Nature. 2010;467(7315):543–9. doi: 10.1038/nature09339. [DOI] [PubMed] [Google Scholar]
- 4.Spear BB, Heath-Chiozzi M, Huff J. Clinical application of pharmacogenetics. Trends Mol Med. 2001;7(5):201–4. doi: 10.1016/s1471-4914(01)01986-4. [DOI] [PubMed] [Google Scholar]
- 5.Jameson GS, Petricoin EF, Sachdev J, Liotta LA, Loesch DM, Anthony SP, et al. A pilot study utilizing multi-omic molecular profiling to find potential targets and select individualized treatments for patients with previously treated metastatic breast cancer. Breast Cancer Res Treat. 2014;147(3):579–88. doi: 10.1007/s10549-014-3117-1. [DOI] [PubMed] [Google Scholar]
- 6.Boja ES, Rodriguez H. Proteogenomic convergence for understanding cancer pathways and networks. Clin Proteomics. 2014;11(1):22. doi: 10.1186/1559-0275-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patel LR, Nykter M, Chen K, Zhang W. Cancer genome sequencing: understanding malignancy as a disease of the genome, its conformation, and its evolution. Cancer Lett. 2013;340(2):152–60. doi: 10.1016/j.canlet.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Espina V, Mehta AI, Winters ME, Calvert V, Wulfkuhle J, Petricoin EF, 3rd, et al. Protein microarrays: molecular profiling technologies for clinical specimens. Proteomics. 2003;3(11):2091–100. doi: 10.1002/pmic.200300592. [DOI] [PubMed] [Google Scholar]
- 9.Liotta LA, Espina V, Mehta AI, Calvert V, Rosenblatt K, Geho D, et al. Protein microarrays: meeting analytical challenges for clinical applications. Cancer Cell. 2003;3(4):317–25. doi: 10.1016/s1535-6108(03)00086-2. [DOI] [PubMed] [Google Scholar]
- 10.Petricoin EF, 3rd, Bichsel VE, Calvert VS, Espina V, Winters M, Young L, et al. Mapping molecular networks using proteomics: a vision for patient-tailored combination therapy. J Clin Oncol. 2005;23(15):3614–21. doi: 10.1200/JCO.2005.02.509. [DOI] [PubMed] [Google Scholar]
- 11.Wulfkuhle J, Espina V, Liotta L, Petricoin E. Genomic and proteomic technologies for individualisation and improvement of cancer treatment. Eur J Cancer. 2004;40(17):2623–32. doi: 10.1016/j.ejca.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 12.Chen LC, Elliott RA, Ashcroft DM. Systematic review of the analgesic efficacy and tolerability of COX-2 inhibitors in post-operative pain control. J Clin Pharm Ther. 2004;29(3):215–29. doi: 10.1111/j.1365-2710.2004.00558.x. [DOI] [PubMed] [Google Scholar]
- 13.Desjardins PJ, Mehlisch DR, Chang DJ, Krupa D, Polis AB, Petruschke RA, et al. The time to onset and overall analgesic efficacy of rofecoxib 50 mg: a meta-analysis of 13 randomized clinical trials. Clin J Pain. 2005;21(3):241–50. doi: 10.1097/00002508-200505000-00007. [DOI] [PubMed] [Google Scholar]
- 14.de Gramont A, Figer A, Seymour M, Homerin M, Hmissi A, Cassidy J, et al. Leucovorin and fluorouracil with or without oxaliplatin as first-line treatment in advanced colorectal cancer. J Clin Oncol. 2000;18(16):2938–47. doi: 10.1200/JCO.2000.18.16.2938. [DOI] [PubMed] [Google Scholar]
- 15.Labianca R, Pancera G, Luporini G. Factors influencing response rates for advanced colorectal cancer chemotherapy. Ann Oncol. 1996;7(9):901–6. doi: 10.1093/oxfordjournals.annonc.a010791. [DOI] [PubMed] [Google Scholar]
- 16.Moreau LC, Rajan R, Thirlwell MP, Alcindor T. Response to chemotherapy in metastatic colorectal cancer after exposure to oxaliplatin in the adjuvant setting. Anticancer Res. 2013;33(4):1765–8. [PubMed] [Google Scholar]
- 17.Advanced Colorectal Cancer Meta-Analysis Project. Modulation of fluorouracil by leucovorin in patients with advanced colorectal cancer: evidence in terms of response rate. J Clin Oncol. 1992;10(6):896–903. doi: 10.1200/JCO.1992.10.6.896. [DOI] [PubMed] [Google Scholar]
- 18.Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26(10):1626–34. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 19.Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369(11):1023–34. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 20.Espina V, Mariani BD, Gallagher RI, Tran K, Banks S, Wiedemann J, et al. Malignant precursor cells pre-exist in human breast DCIS and require autophagy for survival. PLoS One. 2010;5(4):e10240. doi: 10.1371/journal.pone.0010240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petricoin EF, 3rd, Espina V, Araujo RP, Midura B, Yeung C, Wan X, et al. Phosphoprotein pathway mapping: Akt/mammalian target of rapamycin activation is negatively associated with childhood rhabdomyosarcoma survival. Cancer Res. 2007;67(7):3431–40. doi: 10.1158/0008-5472.CAN-06-1344. [DOI] [PubMed] [Google Scholar]
- 22.Pierobon M, Silvestri A, Spira A, Reeder A, Pin E, Banks S, et al. Pilot phase I/II personalized therapy trial for metastatic colorectal cancer: evaluating the feasibility of protein pathway activation mapping for stratifying patients to therapy with imatinib and panitumumab. J Proteome Res. 2014;13(6):2846–55. doi: 10.1021/pr401267m. [DOI] [PubMed] [Google Scholar]
- 23.Akbani R, Becker KF, Carragher N, Goldstein T, de Koning L, Korf U, et al. Realizing the promise of reverse phase protein arrays for clinical, translational, and basic research: a workshop report: the RPPA (Reverse Phase Protein Array) society. Mol Cell Proteomics. 2014;13(7):1625–43. doi: 10.1074/mcp.O113.034918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Espina V, Wulfkuhle JD, Calvert VS, Petricoin EF, 3rd, Liotta LA. Reverse phase protein microarrays for monitoring biological responses. Methods Mol Biol. 2007;383:321–36. doi: 10.1007/978-1-59745-335-6_21. [DOI] [PubMed] [Google Scholar]
- 25.Mueller C, Liotta LA, Espina V. Reverse phase protein microarrays advance to use in clinical trials. Mol Oncol. 2010;4(6):461–81. doi: 10.1016/j.molonc.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishizuka S, Chen ST, Gwadry FG, Alexander J, Major SM, Scherf U, et al. Diagnostic markers that distinguish colon and ovarian adenocarcinomas: identification by genomic, proteomic, and tissue array profiling. Cancer Res. 2003;63(17):5243–50. [PubMed] [Google Scholar]
- 27.Speer R, Wulfkuhle J, Espina V, Aurajo R, Edmiston KH, Liotta LA, et al. Development of reverse phase protein microarrays for clinical applications and patient-tailored therapy. Cancer Genomics Proteomics. 2007;4(3):157–64. [PubMed] [Google Scholar]
- 28.Davuluri G, Espina V, Petricoin EF, 3rd, Ross M, Deng J, Liotta LA, et al. Activated VEGF receptor shed into the vitreous in eyes with wet AMD: a new class of biomarkers in the vitreous with potential for predicting the treatment timing and monitoring response. Arch Ophthalmol. 2009;127(5):613–21. doi: 10.1001/archophthalmol.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Popova TG, Turell MJ, Espina V, Kehn-Hall K, Kidd J, Narayanan A, et al. Reverse-phase phosphoproteome analysis of signaling pathways induced by Rift valley fever virus in human small airway epithelial cells. PLoS One. 2010;5(11):e13805. doi: 10.1371/journal.pone.0013805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paweletz CP, Charboneau L, Bichsel VE, Simone NL, Chen T, Gillespie JW, et al. Reverse phase protein microarrays which capture disease progression show activation of pro-survival pathways at the cancer invasion front. Oncogene. 2001;20(16):1981–9. doi: 10.1038/sj.onc.1204265. [DOI] [PubMed] [Google Scholar]
- 31.Bobrow MN, Harris TD, Shaughnessy KJ, Litt GJ. Catalyzed reporter deposition, a novel method of signal amplification. Application to immunoassays. J Immunol Methods. 1989;125(1–2):279–85. doi: 10.1016/0022-1759(89)90104-x. [DOI] [PubMed] [Google Scholar]
- 32.Bobrow MN, Shaughnessy KJ, Litt GJ. Catalyzed reporter deposition, a novel method of signal amplification. II. Application to membrane immunoassays. J Immunol Methods. 1991;137(1):103–12. doi: 10.1016/0022-1759(91)90399-z. [DOI] [PubMed] [Google Scholar]
- 33.King G, Payne S, Walker F, Murray GI. A highly sensitive detection method for immunohistochemistry using biotinylated tyramine. J Pathol. 1997;183(2):237–41. doi: 10.1002/(SICI)1096-9896(199710)183:2<237::AID-PATH893>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 34.Zhu H, Qian J. Applications of functional protein microarrays in basic and clinical research. Adv Genet. 2012;79:123–55. doi: 10.1016/B978-0-12-394395-8.00004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.VanMeter A, Signore M, Pierobon M, Espina V, Liotta LA, Petricoin EF., 3rd Reverse-phase protein microarrays: application to biomarker discovery and translational medicine. Expert Rev Mol Diagn. 2007;7(5):625–33. doi: 10.1586/14737159.7.5.625. [DOI] [PubMed] [Google Scholar]
- 36.Espina V, Petricoin E, Liotta L, Geho D. Application of sector protein microarrays to clinical samples. Clin Proteomics. 2004;1(91):91–99. [Google Scholar]
- 37.Gorg A, Weiss W, Dunn MJ. Current two-dimensional electrophoresis technology for proteomics. Proteomics. 2004;4(12):3665–85. doi: 10.1002/pmic.200401031. [DOI] [PubMed] [Google Scholar]
- 38.Belluco C, Mammano E, Petricoin E, Prevedello L, Calvert V, Liotta L, et al. Kinase substrate protein microarray analysis of human colon cancer and hepatic metastasis. Clin Chim Acta. 2005;357(2):180–3. doi: 10.1016/j.cccn.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 39.Gulmann C, Espina V, Petricoin E, 3rd, Longo DL, Santi M, Knutsen T, et al. Proteomic analysis of apoptotic pathways reveals prognostic factors in follicular lymphoma. Clin Cancer Res. 2005;11(16):5847–55. doi: 10.1158/1078-0432.CCR-05-0637. [DOI] [PubMed] [Google Scholar]
- 40.Wulfkuhle JD, Speer R, Pierobon M, Laird J, Espina V, Deng J, et al. Multiplexed cell signaling analysis of human breast cancer applications for personalized therapy. J Proteome Res. 2008;7(4):1508–17. doi: 10.1021/pr7008127. [DOI] [PubMed] [Google Scholar]
- 41.Espina V, Edmiston KH, Heiby M, Pierobon M, Sciro M, Merritt B, et al. A portrait of tissue phosphoprotein stability in the clinical tissue procurement process. Mol Cell Proteomics. 2008;7(10):1998–2018. doi: 10.1074/mcp.M700596-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ornstein DK, Gillespie JW, Paweletz CP, Duray PH, Herring J, Vocke CD, et al. Proteomic analysis of laser capture microdissected human prostate cancer and in vitro prostate cell lines. Electrophoresis. 2000;21(11):2235–42. doi: 10.1002/1522-2683(20000601)21:11<2235::AID-ELPS2235>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 43.van Oostrum J, Calonder C, Rechsteiner D, Ehrat M, Mestan J, Fabbro D, et al. Tracing pathway activities with kinase inhibitors and reverse phase protein arrays. Proteomics Clin Appl. 2009;3(4):412–22. doi: 10.1002/prca.200800070. [DOI] [PubMed] [Google Scholar]
- 44.VanMeter AJ, Rodriguez AS, Bowman ED, Jen J, Harris CC, Deng J, et al. Laser capture microdissection and protein microarray analysis of human non-small cell lung cancer: differential epidermal growth factor receptor (EGPR) phosphorylation events associated with mutated EGFR compared with wild type. Mol Cell Proteomics. 2008;7(10):1902–24. doi: 10.1074/mcp.M800204-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gallagher RI, Silvestri A, Petricoin EF, 3rd, Liotta LA, Espina V. Reverse phase protein microarrays: fluorometric and colorimetric detection. Methods Mol Biol. 2011;723:275–301. doi: 10.1007/978-1-61779-043-0_18. [DOI] [PubMed] [Google Scholar]
- 46.Pin E, Federici G, Petricoin EF., 3rd Preparation and use of reverse protein microarrays. Curr Protoc Protein Sci. 2014;75(Unit 27):7. doi: 10.1002/0471140864.ps2707s75. [DOI] [PubMed] [Google Scholar]
- 47.Wulfkuhle JD, Berg D, Wolff C, Langer R, Tran K, Illi J, et al. Molecular analysis of HER2 signaling in human breast cancer by functional protein pathway activation mapping. Clin Cancer Res. 2012;18(23):6426–35. doi: 10.1158/1078-0432.CCR-12-0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chiechi A, Mueller C, Boehm KM, Romano A, Benassi MS, Picci P, et al. Improved data normalization methods for reverse phase protein microarray analysis of complex biological samples. Biotechniques. 2012;0(0):1–7. doi: 10.2144/000113926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Troncale S, Barbet A, Coulibaly L, Henry E, He B, Barillot E, et al. NormaCurve: a SuperCurve-based method that simultaneously quantifies and normalizes reverse phase protein array data. PLoS One. 2012;7(6):e38686. doi: 10.1371/journal.pone.0038686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eichner J, Heubach Y, Ruff M, Kohlhof H, Strobl S, Mayer B, et al. RPPApipe: A pipeline for the analysis of reverse-phase protein array data. Biosystems. 2014;122:19–24. doi: 10.1016/j.biosystems.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 51.Nishizuka S, Charboneau L, Young L, Major S, Reinhold WC, Waltham M, et al. Proteomic profiling of the NCI-60 cancer cell lines using new high-density reverse-phase lysate microarrays. Proc Natl Acad Sci U S A. 2003;100(24):14229–34. doi: 10.1073/pnas.2331323100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.von der Heyde S, Sonntag J, Kaschek D, Bender C, Bues J, Wachter A, et al. RPPanalyzer Toolbox: An improved R package for analysis of reverse phase protein array data. Biotechniques. 2014;57(3):125–35. doi: 10.2144/000114205. [DOI] [PubMed] [Google Scholar]
- 53.Mueller C, deCarvalho AC, Mikkelsen T, Lehman NL, Calvert V, Espina V, et al. Glioblastoma cell enrichment is critical for analysis of phosphorylated drug targets and proteomic-genomic correlations. Cancer Res. 2014;74(3):818–28. doi: 10.1158/0008-5472.CAN-13-2172. [DOI] [PubMed] [Google Scholar]
- 54.Silvestri A, Colombatti A, Calvert VS, Deng J, Mammano E, Belluco C, et al. Protein pathway biomarker analysis of human cancer reveals requirement for upfront cellular-enrichment processing. Lab Invest. 2010;90(5):787–96. doi: 10.1038/labinvest.2010.47. [DOI] [PubMed] [Google Scholar]
- 55.Mukherjee S, Rodriguez-Canales J, Hanson J, Emmert-Buck MR, Tangrea MA, Prieto DA, et al. Proteomic analysis of frozen tissue samples using laser capture microdissection. Methods Mol Biol. 2013;1002:71–83. doi: 10.1007/978-1-62703-360-2_6. [DOI] [PubMed] [Google Scholar]
- 56.Bonner RF, Emmert-Buck M, Cole K, Pohida T, Chuaqui R, Goldstein S, et al. Laser capture microdissection: molecular analysis of tissue. Science. 1997;278(5342):1481, 1483. doi: 10.1126/science.278.5342.1481. [DOI] [PubMed] [Google Scholar]
- 57.Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, et al. Laser capture microdissection. Science. 1996;274(5289):998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- 58.Espina V, Wulfkuhle J, Liotta LA. Application of laser microdissection and reverse-phase protein microarrays to the molecular profiling of cancer signal pathway networks in the tissue microenvironment. Clin Lab Med. 2009;29(1):1–13. doi: 10.1016/j.cll.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 59.Espina V, Wulfkuhle JD, Calvert VS, VanMeter A, Zhou W, Coukos G, et al. Laser-capture microdissection. Nat Protoc. 2006;1(2):586–603. doi: 10.1038/nprot.2006.85. [DOI] [PubMed] [Google Scholar]
- 60.Gallagher RI, Blakely SR, Liotta LA, Espina V. Laser capture microdissection: Arcturus(XT) infrared capture and UV cutting methods. Methods Mol Biol. 2012;823:157–78. doi: 10.1007/978-1-60327-216-2_11. [DOI] [PubMed] [Google Scholar]
- 61.Xu BJ. Combining laser capture microdissection and proteomics: methodologies and clinical applications. Proteomics Clin Appl. 2010;4(2):116–23. doi: 10.1002/prca.200900138. [DOI] [PubMed] [Google Scholar]
- 62.Barker AD, Sigman CC, Kelloff GJ, Hylton NM, Berry DA, Esserman LJ. I-SPY 2: an adaptive breast cancer trial design in the setting of neoadjuvant chemotherapy. Clin Pharmacol Ther. 2009;86(1):97–100. doi: 10.1038/clpt.2009.68. [DOI] [PubMed] [Google Scholar]
- 63.Gujral TS, Karp RL, Finski A, Chan M, Schwartz PE, MacBeath G, et al. Profiling phospho-signaling networks in breast cancer using reverse-phase protein arrays. Oncogene. 2013;32(29):3470–6. doi: 10.1038/onc.2012.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rapkiewicz A, Espina V, Zujewski JA, Lebowitz PF, Filie A, Wulfkuhle J, et al. The needle in the haystack: application of breast fine-needle aspirate samples to quantitative protein microarray technology. Cancer. 2007;111(3):173–84. doi: 10.1002/cncr.22686. [DOI] [PubMed] [Google Scholar]
- 65.Sheehan KM, Calvert VS, Kay EW, Lu Y, Fishman D, Espina V, et al. Use of reverse phase protein microarrays and reference standard development for molecular network analysis of metastatic ovarian carcinoma. Mol Cell Proteomics. 2005;4(4):346–55. doi: 10.1074/mcp.T500003-MCP200. [DOI] [PubMed] [Google Scholar]
- 66.Sheehan KM, Gulmann C, Eichler GS, Weinstein JN, Barrett HL, Kay EW, et al. Signal pathway profiling of epithelial and stromal compartments of colonic carcinoma reveals epithelial-mesenchymal transition. Oncogene. 2008;27(3):323–31. doi: 10.1038/sj.onc.1210647. [DOI] [PubMed] [Google Scholar]
- 67.Li J, Gould TD, Yuan P, Manji HK, Chen G. Post-mortem interval effects on the phosphorylation of signaling proteins. Neuropsychopharmacology. 2003;28(6):1017–25. doi: 10.1038/sj.npp.1300112. [DOI] [PubMed] [Google Scholar]
- 68.Li X, Friedman AB, Roh MS, Jope RS. Anesthesia and post-mortem interval profoundly influence the regulatory serine phosphorylation of glycogen synthase kinase-3 in mouse brain. J Neurochem. 2005;92(3):701–4. doi: 10.1111/j.1471-4159.2004.02898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Espina V, Mueller C, Edmiston K, Sciro M, Petricoin EF, Liotta LA. Tissue is alive: New technologies are needed to address the problems of protein biomarker pre-analytical variability. Proteomics Clin Appl. 2009;3(8):874–882. doi: 10.1002/prca.200800001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mueller C, Edmiston KH, Carpenter C, Gaffney E, Ryan C, Ward R, et al. One-step preservation of phosphoproteins and tissue morphology at room temperature for diagnostic and research specimens. PLoS One. 2011;6(8):e23780. doi: 10.1371/journal.pone.0023780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grellner W, Madea B. Demands on scientific studies: vitality of wounds and wound age estimation. Forensic Sci Int. 2007;165(2–3):150–4. doi: 10.1016/j.forsciint.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 72.Goldstein BJ. Protein-tyrosine phosphatases: emerging targets for therapeutic intervention in type 2 diabetes and related states of insulin resistance. J Clin Endocrinol Metab. 2002;87(6):2474–80. doi: 10.1210/jcem.87.6.8641. [DOI] [PubMed] [Google Scholar]
- 73.Neel BG, Tonks NK. Protein tyrosine phosphatases in signal transduction. Curr Opin Cell Biol. 1997;9(2):193–204. doi: 10.1016/s0955-0674(97)80063-4. [DOI] [PubMed] [Google Scholar]
- 74.Liotta LA, Petricoin EF., 3rd -Omics and cancer biomarkers: link to the biological truth or bear the consequences. Cancer Epidemiol Biomarkers Prev. 2012;21(8):1229–35. doi: 10.1158/1055-9965.EPI-12-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tell RW, Horvath CM. Bioinformatic analysis reveals a pattern of STAT3-associated gene expression specific to basal-like breast cancers in human tumors. Proc Natl Acad Sci U S A. 2014;111(35):12787–92. doi: 10.1073/pnas.1404881111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3(6):453–8. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 77.He Y, Zhou Z, Hofstetter WL, Zhou Y, Hu W, Guo C, et al. Aberrant expression of proteins involved in signal transduction and DNA repair pathways in lung cancer and their association with clinical parameters. PLoS One. 2012;7(2):e31087. doi: 10.1371/journal.pone.0031087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Angenendt P. Progress in protein and antibody microarray technology. Drug Discov Today. 2005;10(7):503–11. doi: 10.1016/S1359-6446(05)03392-1. [DOI] [PubMed] [Google Scholar]
- 79.Esserman LJ, Berry DA, DeMichele A, Carey L, Davis SE, Buxton M, et al. Pathologic complete response predicts recurrence-free survival more effectively by cancer subset: results from the I-SPY 1 TRIAL--CALGB 150007/150012, ACRIN 6657. J Clin Oncol. 2012;30(26):3242–9. doi: 10.1200/JCO.2011.39.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Espina V, Liotta LA. What is the malignant nature of human ductal carcinoma in situ? Nat Rev Cancer. 2010;11(1):68–75. doi: 10.1038/nrc2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gulmann C, Sheehan KM, Kay EW, Liotta LA, Petricoin EF., 3rd Array-based proteomics: mapping of protein circuitries for diagnostics, prognostics, and therapy guidance in cancer. J Pathol. 2006;208(5):595–606. doi: 10.1002/path.1958. [DOI] [PubMed] [Google Scholar]
- 82.Wulfkuhle JD, Edmiston KH, Liotta LA, Petricoin EF., 3rd Technology insight: pharmacoproteomics for cancer--promises of patient-tailored medicine using protein microarrays. Nat Clin Pract Oncol. 2006;3(5):256–68. doi: 10.1038/ncponc0485. [DOI] [PubMed] [Google Scholar]
- 83.Taylor CF, Paton NW, Lilley KS, Binz PA, Julian RK, Jr, Jones AR, et al. The minimum information about a proteomics experiment (MIAPE) Nat Biotechnol. 2007;25(8):887–93. doi: 10.1038/nbt1329. [DOI] [PubMed] [Google Scholar]