SUMMARY
Skeletal muscle growth immediately following birth is a critical for proper body posture and locomotion. However, compared to embryogenesis and adulthood, the processes regulating the maturation of neonatal muscles is considerably less clear. Studies in the 1960s predicted that neonatal muscle growth results from nuclear accretion of myoblasts preferentially at the tips of myofibers. Remarkably, little information has been added since then to resolve how myoblasts migrate to the ends of fibers. Here, we provide insight to this process by revealing a unique NF-κB-dependent communication between NG2+ interstitial cells and myoblasts. NF-κB in NG2+ cells promotes myoblast migration to the tips of myofibers through cell-cell contact. This occurs through expression of ephrinA5 from NG2+ cells, which we further deduce is an NF-κB target gene. Together, results suggest that NF-κB plays an important role in the development of newborn muscles to ensure proper myoblast migration for fiber growth.
Keywords: NF-κB, ephrinA5, skeletal muscle, myoblast, migration, development
INTRODUCTION
In humans, the growth of an infant in the first 1000 days of life is considered vital for maximizing cognitive and behavioral function, as well as minimizing the risk of chronic diseases later in life (Britto and Perez-Escamilla, 2013). The rate at which neonates grow is higher than in any other stage of postnatal development, and the majority of this growth, as measured in mass, derives from skeletal muscle (Davis and Fiorotto, 2009). Surprisingly, in contrast to the wealth of studies that have been devoted to elucidating the development of skeletal muscle during embryogenesis, and the response to injury in adults, little attention has been given to the maturation of newborn muscles.
Following birth, the total number of myofibers are thought to remain fixed throughout adulthood (White et al., 2010). Rather than adding myofibers, muscles grow longitudinally through the accretion of myoblasts, resulting in an increasing pool of myonuclei that regulate the synthesis of contractile proteins and the addition of sarcomeric units (Braun and Gautel, 2011; Ehler and Gautel, 2008). The growth of these fibers is important to support a musculoskeletal apparatus needed for locomotion and posture. In mice, this post-embryonic growth phase is complete by the third week of life (White et al., 2010). Further growth of muscle fibers results from an increase in protein synthesis due to the uptake of nutrients (Davis and Fiorotto, 2009). Early tracer studies postulated that the location where myoblasts fuse along the length of a myofiber is not a random process, but rather a coordinated event occurring at the tip of the fiber (Kitiyakara and Angevine, 1963; Williams and Goldspink, 1971). Nevertheless, how myoblasts from the interstitium receive their cues to migrate to the correct position on the fiber has yet to be identified. In this study we describe a mechanism involving the NF-κB signaling pathway and an intimate communication between an interstitial cell population and myoblasts that together coordinate neonatal muscle growth.
RESULTS
NF-κB is Activated in NG2+ Cells during Neonatal Muscle Development
We previously showed that limb muscles from newborn mice contain a considerably high level of NF-κB activity, composed of the classical p50 and p65 subunits in the signaling pathway (Dahlman et al., 2010; Hayden and Ghosh, 2004, 2012). To identify a cellular source for this activity, we used reporter mice whose EGFP expression in the cytoplasm is under the control of an NF-κB regulated promoter (NFκB-EGFP) (Magness et al., 2004). Results showed that NF-κB activity was localized to the muscle interstitium and declined rapidly throughout the first 4 weeks of life, coinciding with the growth plateau phase of neonatal limb muscles (Figure 1A). Reporter mice also revealed that a significant portion of NF-κB activity in the extra laminar space was localized to a population distinct from Pax7+ or MyoD+ myogenic cells (Figure 1B).
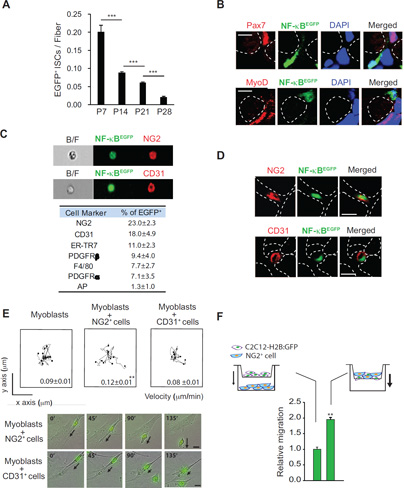
Figure 1. NF-κB Localizes to NG2+ Cells during Neonatal Skeletal Muscle Development.
(A) Muscle sections were prepared from NFκB-EGFP mice at P7, P14, P21, and P28 for laminin staining. EGFP+ interstitial cells (ISCs) located outside of laminin were counted per cross sectional area and normalized to fiber numbers. Error bars indicate SEM. ***p < 0.001. (B) Muscle sections from P7 NFκB-EGFP mice were analyzed for Pax7 (top, red) or MyoD (bottom, red) and NFκB (green) positive cells. Dotted white cells represent myofiber boundaries. DAPI was used for nuclear staining. (C) Representative images of NG2+ EGFP+ (top) and CD31+ EGFP+ cells (bottom) from the FACS analysis. Table generated from FACS analysis summarizing the percentage of mononuclear cells from P7 NFκB-EGFP mice expressing EGFP and indicated cell surface markers used to identify cell types. (D) Muscle sections from P7 NFκB-EGFP mice were analyzed for cell expressing both EGFP and NG2 (top) or CD31 (bottom). Dotted white cells represent myofiber boundaries. (E) C2C12-H2B:GFP myoblasts were co-cultured with NG2 or CD31 positive cells isolated from P2 – P5 wild type mice, and then recorded for 10h using time lapse fluorescence microscopy (top). The migration of C2C12-H2B:GFP myoblasts was measured after contacting with CD31+ or NG2+ cells and the data are presented as mean ± s.e.m. Representative still shots from the time-lapse fluorescence microscopy are shown (bottom). Arrows indicate the movements of C2C12-H2B:GFP myoblasts. (F) Migration of C2C12-H2B:GFP myoblasts was monitored using a Boyden chamber system where NG2+ cells were plated either in the lower chamber alone (left) or added to the upper chamber (right) at a 1:1 ratio with C2C12-H2B:GFP myoblasts. Relative migration was normalized to C2C12-H2B:GFP myoblasts migration by themselves. B/F: bright field image. Scale bars = 5 µm (B) and 10 µm (D) and (E). **p < 0.01 by the student’s t-test. See accompanying Figure S1 and Movie S1. Error bars, SEM
Fluorescence-activated cell sorting (FACS) analysis was used to characterize those populations positive for NF-κB within neonatal muscle. Results showed that a major fraction of activity resided in both NG2+ (23.0 ± 2.3%) and CD31+ (18.0 ± 4.9%) cells, with less activity present in cells positive for F4/80, PDGFRα, or ER-TR7 markers (Figure 1C). While CD31 is present on endothelial cells, NG2 is expressed on the surface of pericytes and other cell types (Cappellari and Cossu, 2013). Immunohistochemical staining of P7 hindlimb muscles verified that cells positive for NF-κB co-localized with cells expressing NG2 and CD31 (Figure 1D). In neonatal muscle, alkaline phosphatase (AP) also serves as a marker for pericytes (Dellavalle et al., 2011). However, only a minor portion of GFP+ cells was positive for AP (<1.5%), and rarely did AP+ cells appear in the same population as NG2+ cells (Figure S1A). In addition, and consistent with FACS results, rarely did we observe cells positive for both NF-κB and AP (Figure S1B). Similar results were obtained for another pericyte marker, PDGFRβ (Figure 1C). These data demonstrate that, in newborn muscle, NF-κB predominantly resides in both endothelial cells and an interstitial NG2+ population that appears distinct from pure pericytes.
NF-κB Functions in NG2+ Cells to Promote Myoblast Migration Through Cell-Cell Contact
To investigate the functional role of CD31+ and NG2+ cells during neonatal muscle growth, we established co-cultures of these cells with C2C12 myoblasts stably expressing a H2B-GFP reporter gene. This system allowed us to monitor myoblasts over time using fluorescent, live cell imaging. A clear difference was observed in the motility of myoblasts when co-cultured with NG2+ but not CD31+ cells, which could not be accounted for by differences in proliferation rates, as monitored by Ki67 staining (movie S1, and Figure S1C). Co-culture studies also revealed that NG2+ cells and myoblasts underwent cell-cell contact. Tracking of cell movements showed that myoblasts were 33% (p = 0.0023) more mobile in the presence of NG2+ compared to CD31+ cells (Figure 1E), suggesting that cell-cell contact likely contributed to the increased motility of myoblasts. Further tracking examination revealed that following cell contact, myoblasts tended to migrate away from NG2+ cells (Figure 1F) indicating that the movement of myoblasts results from contact-mediated repulsion. To further assess the migration stimulating activity of NG2+ cells, Boyden chambers were used so that C2C12-H2B:GFP myoblasts could be grown either in the same well as NG2+ cells, or separated by a transwell filter (Figure 1F). Compared to the migration of C2C12-H2B:GFP myoblasts when plated by themselves (set to a value of 1), the addition of NG2+ cells in a separate well had no effect on myoblast migration. In contrast, combining the two cell types in the same well promoted C2C12-H2B:GFP myoblasts migration through the filter (Figure 1F). This strongly suggested that NG2+ cells function to stimulate myoblast motility through cell-cell contact rather than through a diffusible factor.
Since we found that NF-κB is active in NG2+ cells within newborn muscles, we next asked whether this activity is required for myoblast migration. Thus, co-culture assays were repeated with C2C12-H2B:GFP myoblasts and NG2+ cells genetically ablated for the p65 subunit of NF-κB. Results showed that myoblasts favored contacting p65+/+ NG2+ cells, which associated with a 47% (p = 0.0352) increase in myoblast motility compared to co-cultures with p65−/− NG2+ cells (movie S2 and Figure 2A). To substantiate these results, we performed a separate migration assay using a two-chamber system, which, as illustrated in Figure 2B, allows the simultaneous culture of two distinct cell populations in the same culture dish. Unlabeled C2C12 myoblasts were plated in one chamber and subsequently differentiated into myotubes prior to plating a mixture of C2C12-H2B:GFP myoblasts with p65+/+ or p65−/− NG2+ cells in the opposite chamber. Following the removal of a gasket separating the chambers, we quantitated the number of C2C12-H2B:GFP myoblasts that migrated across the gap in the direction of differentiated myotubes. Results showed that myoblast migration was considerably increased by 209% (p = 0.00683) in the presence of p65+/+ NG2+ cells compared to p65−/− NG2+ cells (Figure 2B and movie S3). This regulation was selective to the p65 subunit, since similar differences in myoblast migration were not observed between p50+/+ and p50−/− NG2+ cultures (Figure S2A). We also found that migration was more efficient when myoblasts were exposed to myotubes rather than an empty chamber (Figure S2B), consistent with the notion that myotubes secrete chemotactic factors to stimulate myoblast motility (Griffin et al., 2010). We previously demonstrated that p65 regulation of the inducible nitric oxide synthase gene (iNOS) and secretion of nitric oxide (NO) from stromal fibroblasts promotes myoblast nuclear accretion to myofibers (Dahlman et al., 2010). To determine whether this regulation was also relevant in myoblast migration, we repeated migration assays in the presence of NO. However, neither the NO donor, sodium nitroprusside (SNP), or iNOS−/− NG2+ cells had a significant effect on the migration of C2C12-H2B:GFP myoblasts, suggesting that p65 regulation of iNOS is not involved in mobilizing skeletal muscle cells (Figure S2C and S2D).
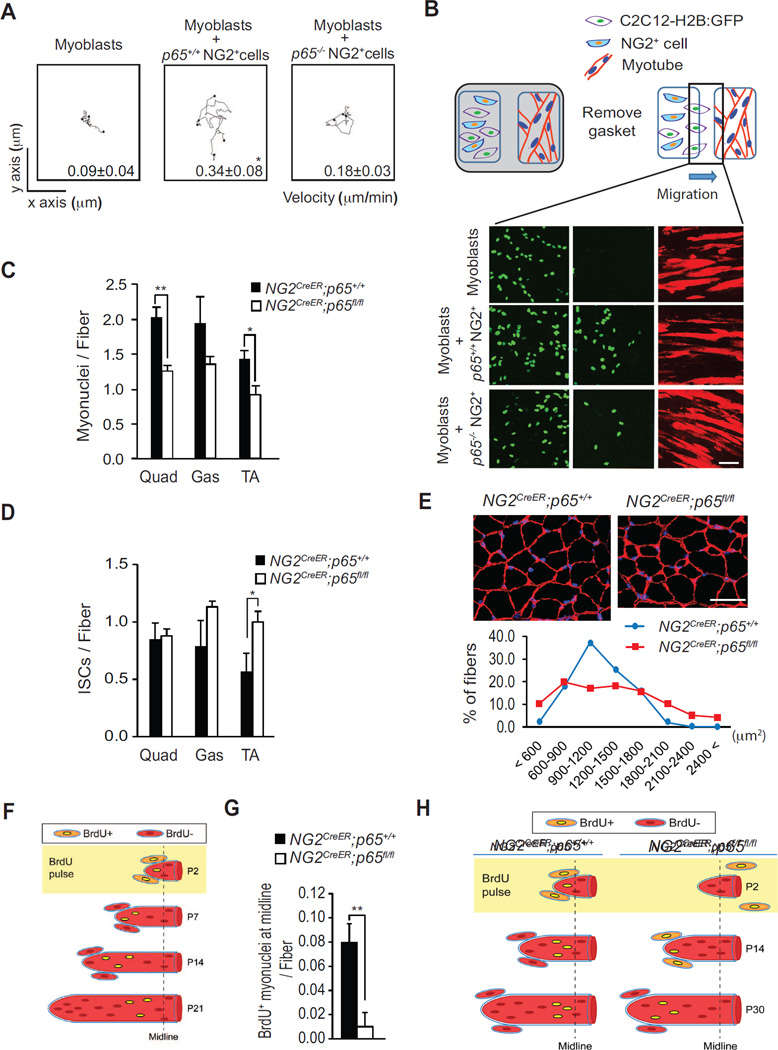
Figure 2. The p65 Activity of NF-κB Functions in NG2 Cells to Promote Myoblast Migration in vitro and in vivo.
(A) Plots of time-lapse microscopy, recording the migration of C2C12-H2B:GFP myoblasts cultured alone, or with either p65+/+ NG2+, or with p65−/− NG2+ cells. The migration of C2C12-H2B:GFP myoblasts was measured following there contact with p65+/+ or p65−/− NG2+ cells. Values are presented as mean ± s.e.m. (B) Two-chamber migration system showing differentiated C2C12 myotubes (red) on the right side of the chamber and C2C12-H2B:GFP myoblasts (green) on the left side cultured either alone, with p65+/+ NG2+ cells, or with p65−/− NG2+ cells. Upon removal of the gasket, C2C12-H2B:GFP myoblasts migrate towards myotubes and cell number was scored across the gap at 24h. Relative migration was expressed as mean ± s.e.m. Myotubes were visualized by immunostaining for Troponin T. (C) Myonuclei number per fiber was measured from quadriceps (Quad), gastrocnemius muscles (Gas) and tibialis anterior (TA) muscles from NG2CreER;p65+/+ and NG2CreER;p65fl/fl mice. Approximately, 800 myonuclei for each genotypes were analyzed. (D) DAPI+ interstitial cells (ISCs) were measured from Quad, Gas, and TA muscles from P30 NG2CreER;p65+/+ and NG2CreER;p65fl/fl mice. (E) (top) Representative cross sections of Gas from P30 NG2CreER;p65+/+ and NG2CreER;p65fl/fl mice immunostained for laminin and counterstained with DAPI. (bottom) Quantitative distribution of fiber size from NG2CreER;p65+/+ and NG2CreER;p65fl/fl mice recorded from a minimum of 1000 fibers. (F) Schematic of BrdU injection experiment where BrdU+ cells are depicted with yellow nuclei. (G) BrdU was injected in P2 NG2CreER;p65+/+ and NG2CreER;p65fl/fl mice, and at P30 EDL muscle sections were analyzed for the sublaminar localization of BrdU+ cells per myofiber, measured at the midline of the muscle. (H) A model for BrdU+ cell accretion in EDL muscles from NG2CreER;p65+/+ and NG2CreER;p65fl/fl mice. Scale bars = 100 µm for (B) and (C). *p < 0.05, **p < 0.01 by the student’s t-test. See accompanying Figure S2; Movie S2, and Movie S3. Error bars, SEM
NF-κB Functions in NG2+ Cells to Promote Myoblasts Migration in vivo
Next, we asked if NF-κB controls myoblast migration in vivo. Mice containing an CreERT2 allele under the control of the NG2 promoter were crossed with p65flox/flox (p65fl/fl) mice (Steinbrecher et al., 2008). Resulting progenies with genotypes NG2CreER;p65+/+ and NG2CreER;p65fl/fl were administered tamoxifen at P2 to induce Cre-mediated deletion of p65 in NG2 cells. Hindlimb muscles were subsequently analyzed at P30, when postnatal muscle development is stabilized. No differences were observed in myofiber number per area (Figure S2E). However, we noted that the number of myonuclei per fiber were significantly reduced in NG2CreER;p65fl/fl mice, coinciding with an increase in the number of cells residing in the muscle interstitium (Figure 2C and 2D). In addition, we observed an unusual heterogeneity in myofiber size in NG2CreER;p65fl/fl mice compared to NG2CreER;p65+/+ mice (Figure 2E).
Results above are consistent with our in vitro data supporting the hypothesis that p65 functions in NG2+ cells to regulate migration of myoblasts to their target myofibers. We then performed an in vivo myoblast migration assay adopted from earlier studies. This technique depends on DNA analogues to track dividing cells in newborn muscles, and was originally used to demonstrate that the accretion of myoblasts to growing myofibers preferentially occurs at the ends of the fibers (Kitiyakara and Angevine, 1963; Williams and Goldspink, 1971). As illustrated in Figure 2F, when this approach was performed by pulsing hindlimbs of P2 mice with the thymidine analogue, 5-bromo-2-deoxyuridine (BrdU), the position of BrdU+ nuclei in extensor digitorum longus (EDL) muscles could be tracked over time due to the sequential accretion of unlabeled myoblasts. For example, by serial cross section and whole-mount analysis of EDL muscles, BrdU+ myonuclei were positioned closer to the midline at P21 compared to P14 (Figure S2F), which supports the model that the fusion of myoblasts to a growing myofiber is coordinated in a spatial rather than in a random process. We proceeded to use the same labeling strategy with P2 NG2CreER;p65+/+ and NG2CreER;p65fl/fl mice injected with BrdU and tamoxifen. BrdU+ myonuclei were subsequently scored from EDL muscle sections. Results showed that the number of BrdU+ myonuclei localized at the midline of EDL muscles was significantly reduced by 87% (p = 0.00533) in NG2CreER;p65fl/fl compared to NG2CreER;p65+/+ mice (Figure 2G, 2H, and S2G). Whole-mount muscle analysis showed that this difference was not due to any changes to the position of NG2+ cells along the length of the myofiber resulting from p65 deletion (Figure S2H). Taking into account both in vitro and in vivo results, these findings strongly support the notion that in newborn muscles, NF-κB functions in NG2+ cells to stimulate the migration of myoblasts to the tips of myofibers for accretion to occur.
EFNA5 is Directly Regulated by NF-κB Signaling
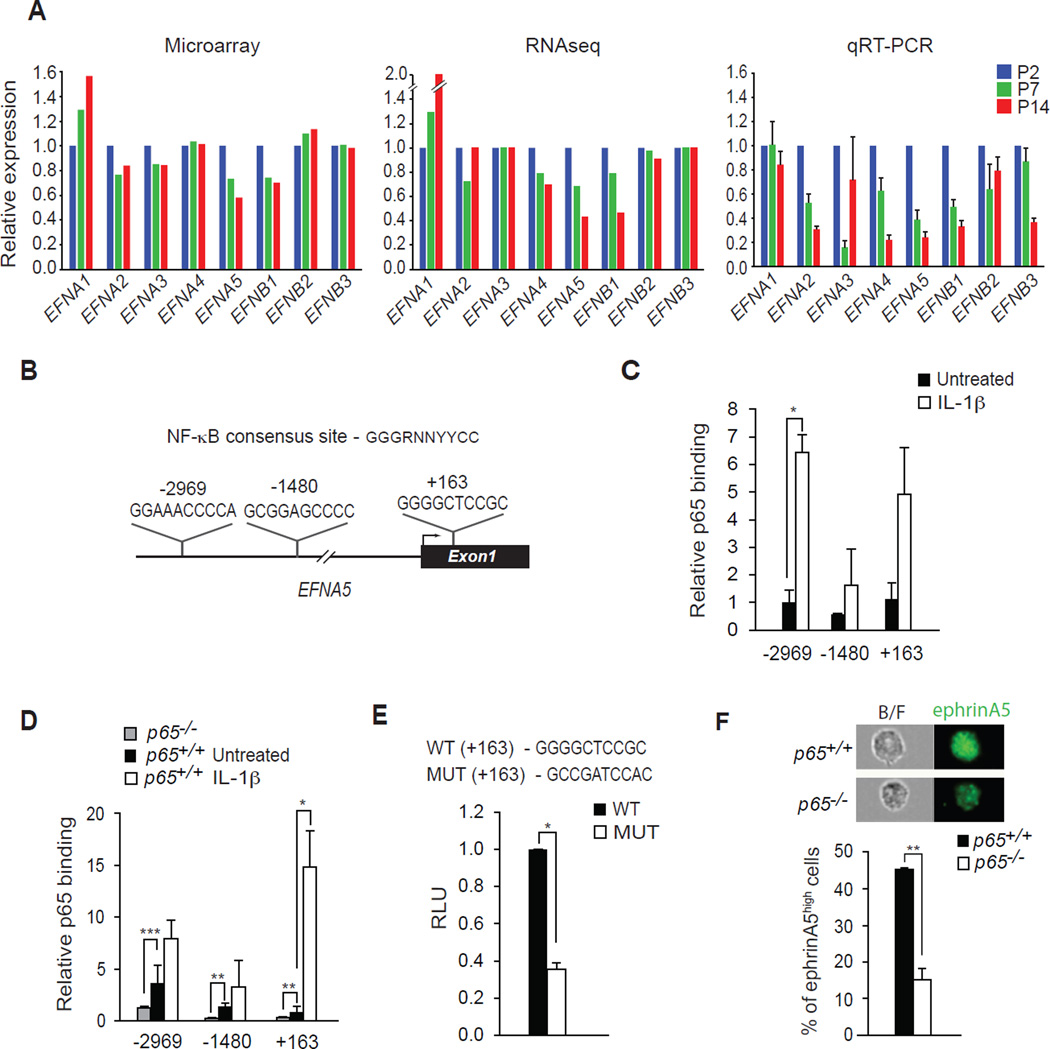
To gain insight into the mechanism by which NF-κB functions in this capacity, we performed genome wide expression profiling using microarray and RNA Seq analyses from early (P2), mid (P7), and late (P14) time points of neonatal muscle development. Annotation from both methods revealed the predicted regulatory expression changes in fetal and neonatal myosin genes (Figure S3A and S3B). We then mined for candidate genes involved in cell migration whose protein function is dependent on cell-cell contact. One class of genes that fit this criterion was the Eph/Ephrin receptor tyrosine kinase family, which function by transmitting their signals between ligand (Ephrin) and receptor (Eph) to promote directional migration (Petrie et al., 2009; Poliakov et al., 2004). Interestingly, several Eph/Ephrin members have been described to regulate skeletal muscle progenitor cell motility (Li and Johnson, 2013; Siegel et al., 2009; Stark et al., 2011; Swartz et al., 2001). Both microarray and RNA Seq revealed that several of the Eph/Ephrin genes (EFNA2, EFNA4, EFNA5, EFNB1) were down regulated in hindlimb muscles during the first two weeks of life, results that we validated by quantitative RT-PCR analysis (Figure 3A). Since this gene expression pattern was consistent with what we had earlier observed for NF-κB activity (Figure 1A), we considered that Eph/Ephrin genes might be transcriptional targets of NF-κB. We therefore used the rVista database to query Eph/Ephrin promoters for potential NF-κB regulatory elements. One gene, EFNA5, contained conserved NF-κB binding sites at positions −2969, −1480, and +163, relative to the transcriptional start site (Figure 3B). Chromatin immunoprecipitation experiments in NG2+ cells confirmed the binding of p65 to each of these sites, which were enriched in response to an NF-κB inducer (IL-1β, Figure 3C). Similar binding of p65 to EFNA5 was observed in p65+/+ mouse embryonic fibroblasts (MEFs), which as expected was pronouncedly reduced in p65−/− cells (Figure 3D). We next tested the functionality of this site by generating a luciferase reporter construct containing the wild type and mutated version of the +163 NF-κB binding site. Results showed that the EFNA5 reporter was active in MEFs, but this activity was significantly diminished when assays were repeated with a mutated NF-κB binding site (Figure 3E). Such results provided evidence that the EFNA5 gene is a direct target of NF-κB. For further validation, we tested the expression of EFNA5 in NG2+ cells by FACS analysis. EFNA5 expression was significantly reduced in p65−/− compared to p65+/+ NG2+ cells (Figure 3F). Similar results were obtained with p65+/+ and p65−/− MEFs (Figure S3C), which together demonstrated that expression of EFNA5 in neonatal muscle is under NF-κB control.
Figure 3. EFNA5 is Transcriptionally Regulated by NF-κB.
(A) Microarray, RNAseq, and quantitative RT-PCR were performed probing for the expression of ephrin genes from skeletal muscles at P2, P7, and P14. (B) A schematic map of the 5’ region of the EFNA5 gene and locations of conserved NF-κB DNA binding sites at −2969, −1480, and +163 relative to the transcriptional start site. (C) ChIP assays for p65 binding to the EFNA5 promoter in NG2+ cells untreated and treated with 10 ng/ml of IL-1β. (D) ChIP assays for p65 binding to the EFNA5 promoter in p65+/+ MEFs untreated and treated with 10 ng/ml of IL-1β, compared with untreated p65−/− MEFs *** p < 0.001. (E) A portion of EFNA5 gene containing the +163 binding site of NF-κB was cloned into a luciferase reporter construct. Luciferase activity was measured following the transfection of an EFNA5 promoter reporter construct in MEFs, and compared to activity generated from an EFNA5 promoter reporter construct containing a mutation in the +163 NF-κB binding site. (F) Representative flow images of ephrinA5 from p65+/+ and p65−/− NG2 cells (top). Percentage of ephrinA5high NG2 cells isolated from p65+/+ and p65−/− cells (bottom). RLU: Relative Luciferase Units. B/F: bright field image. *p < 0.05, **p < 0.01 by the student’s t-test. See accompanying Figure S3. Error bars, SEM
EphrinA5 is a Regulator of Myoblast Migration
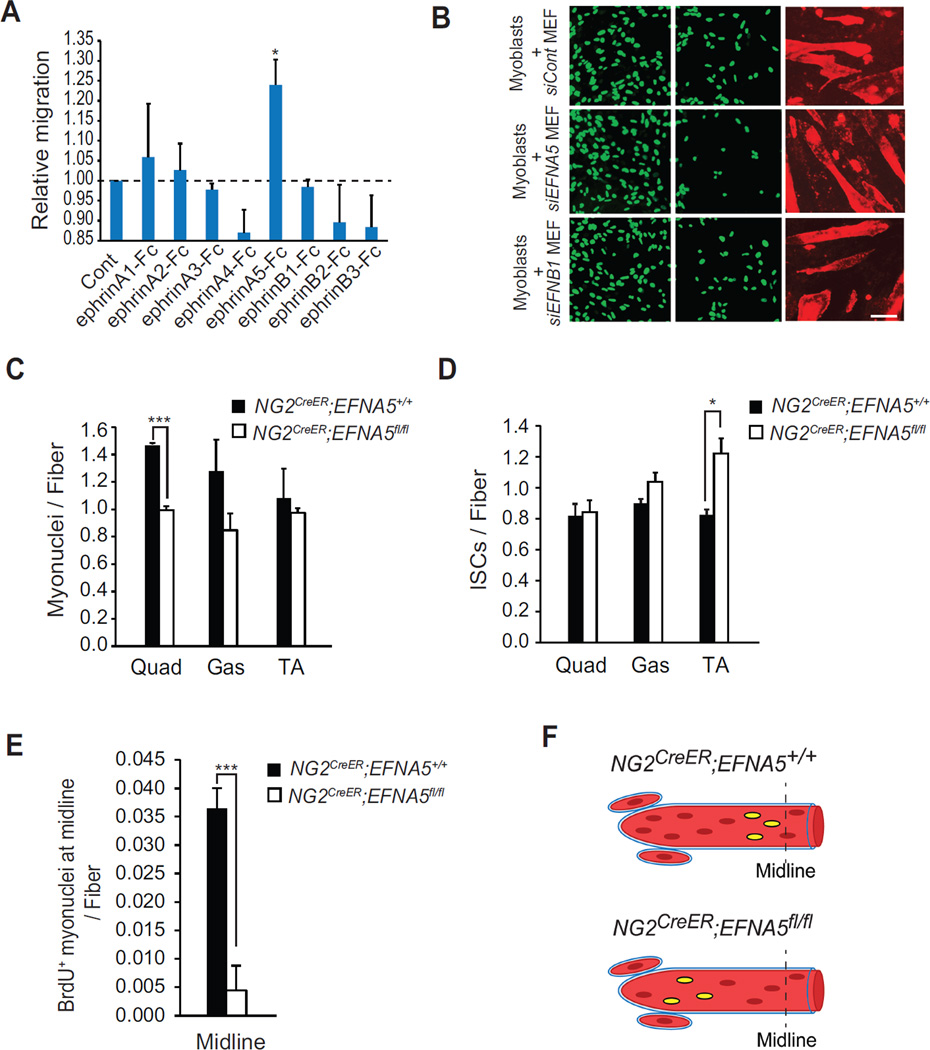
Next, we assessed the functional relevance of EFNA5 in myoblast migration by using a Boyden chamber assay. C2C12 myoblasts were cultured in the upper well and subsequently treated with eight different types of recombinant Fc-clustered ephrin ligands (ephrinA1–A5 and ephrinB1–B3). Whereas some myoblast migratory activity was observed from ephrinA1-Fc and A2-Fc, the greatest stimulation came from ephrinA5-Fc (Figure 4A). To confirm this specificity to EFNA5, C2C12-H2B:GFP myoblasts were co-cultured with MEFs that had been previously transfected with an siRNA against EFNA5 (Figure S4A) and myoblast migration was monitored in a two chamber migration system as described in Figure 2. Knockdown of EFNA5, but not siControl or an siRNA against EFNB1, caused a significant reduction in myoblast motility (Figure 4B and S4B).
Figure 4. EFNA5 from NG2+ Cells Mediates Myoblast Migration in vitro and in vivo.
(A) A Boyden chamber was used to measure migration of myoblasts when exposed for 24h to different recombinant Fc-clustered ephrins. (B) A two-chamber assay system was used to measure migration of C2C12-H2B:GFP myoblasts toward myotubes when co-cultured with MEFs transfected with siRNAs against EFNA5 or EFNB1. Relative migration is shown as mean ± s.e.m. Myonuclei number per fiber (C) and DAPI+ ISCs (D) was measured from Quad, Gas and TA muscles from P30 NG2CreER;EFNA5+/+ and NG2CreER;EFNA5fl/fl mice. (E) BrdU was injected in NG2CreER;EFNA5+/+ and NG2CreER;EFNA5fl/fl mice and at P30 muscle sections were analyzed for the sublaminar localization of BrdU+ cells at the midline of EDL muscles. (F) A model for BrdU+ cell accretion in muscles from NG2CreER;EFNA5+/+ and NG2CreER;EFNA5fl/fl mice. Scale bars = 100 µm for (B) *p < 0.05, ***p < 0.001 by the student’s t-test. See accompanying Figure S4. Error bars, SEM
To determine if a similar function for EFNA5 could be recapitulated in vivo, we generated conditional mutant mice using a targeting vector where exon 2 of EFNA5 was flanked by flox alleles (Figure S4C). These mice were crossed with NG2CreERT2 mice to generate NG2CreER;EFNA5fl/fl progeny. P2 neonates were treated with tamoxifen to cause deletion of EFNA5 from NG2+ cells (Figure S4D) and hindlimb muscles were subsequently examined histologically at P30. Similar to the muscle phenotypes of NG2CreER;p65fl/fl, loss of EFNA5 had no effect on myofiber number or the position of NG2+ cells along the myofiber (Figure S4E, S4G), but did lead to a reduction in myonuclei number per fiber from multiple muscles, which was inversely correlated with an increase in interstitial nuclei (Figure 4C, 4D). To examine the ability of EFNA5 to aid in myoblast migration, we simultaneously pulsed hindlimb muscles from NG2CreER;EFNA5fl/fl mice with BrdU and tamoxifen. EDL sections were prepared and the position of BrdU+ myonuclei was then scored along the length of the muscle. The number of BrdU+ myonuclei localized at the midline of EDL muscles was significantly reduced by 88% (p = 0.07 × 10−4) in NG2CreER;EFNA5fl/fl compared to NG2CreER;EFNA5+/+ mice (Figure 4E and 4F). These findings phenocopy results obtained in p65−/− NG2+ cells and suggest that EFNA5 plays an important role in guiding the migration of myoblasts to the ends of growing myofibers. However, unlike NG2CreER;p65fl/fl mice, loss of EFNA5 only trended to showing differences in the distribution of fiber diameter size (Figure S4F). Together, these results suggest that EFNA5 is involved in regulating myoblast migration during neonatal muscle growth.
DISCUSSION
In our current study, we explored the underlying mechanism by which myoblasts migrate to their targeted myofibers. Our results revealed the involvement of an NF-κB signaling pathway functioning in an NG2+ interstitial cell population. NF-κB is activated in these cells early in neonatal muscle development. Through its regulation of the ligand ephrinA5, NG2+ cells contact and stimulate myoblasts to migrate towards the end of growing myofibers, where they subsequently fuse, thus providing an increased pool of myonuclei. In support of this notion, both deletion of p65 and EFNA5 from NG2+ cells led to an increase in the number of interstitial cells and a reduction in myonuclei compared to wild type mice. However, we noted that this phenotype did not occur to the same degree in all hindlimb muscles that we examined. This might be due to differences in when migration and accretion of myoblasts are at their maximum, or the ability of specific muscles to compensate for growth defects during neonatal development. It is also possible that failing to migrate to their proper location due to impaired signaling from NG2+ cells, myoblasts may undergo a default pathway such as apoptosis leading to their turn over, which would affect their total numbers in the interstitium. We also observed that loss of EFNA5 in NG2+ cells did not affect the distribution of fiber size to the same extent as when p65 was deleted from these same cells. Although it is possible that a difference in recombination efficiency of flox sites between p65 and EFNA5 alleles accounted for this variance, it is equally probable that, in addition to EFNA5, other NF-κB gene targets in NG2+ cells are utilized to communicate with myoblasts and contribute in muscle growth.
Relatively little is known still how myoblast migrate. Global deletion of scatter factor/hepatocyte growth factor (SC/HGF) had originally been shown to cause a defect in the growth of developing limbs due to a failure in the migration of progenitor myoblasts (Bladt et al., 1995). A further role for the HGF/c-MET axis has been studied in adult muscles undergoing regeneration (Webster and Fan, 2013), but whether this ligand-receptor signaling pathway functions in newborn muscles to stimulate myoblast proliferation or migration is not known. Additional factors, CXCL12 and odorant receptor, MOR23, have also been shown to contribute to myoblast migration during in vitro myogenesis and adult muscle regeneration (Griffin et al., 2010; Griffin et al., 2009), but whether their activities are relevant in promoting myoblasts to the ends of growing fibers in newborn muscles will require additional investigation.
Ephrins/Eph signaling has previously been described in the regulation of axonal guidance, blood vessel formation, and neuromuscular junction maturation (Klein, 2012; Yumoto et al., 2008). Their expression has also been detected in normal and regenerating muscles, and selective ephrins have been shown to stimulate myoblast migration in vitro (Stark et al., 2011). However, in all of these instances, the upstream factors regulating Ephrin/Eph production have not been identified, nor has the in vivo context by which Ephrin/Eph signals regulate myoblasts or other cell types in skeletal muscle been resolved. Our findings demonstrate that one such upstream regulator is NF-κB, which functions as a direct transcriptional activator of EFNA5. Using a series of cell based and genetic approaches in mice, we further revealed that ephrinA5 expression in NG2+ interstitial cells is necessary to promote cell-cell contact with myoblasts, which in turn stimulated myoblast migration. Because we found that NG2+ cells are positioned along the length of a myofiber, whose locations were unaffected by the loss of p65 or ephrinA5, we envision that movement of myoblasts is likely to require multiple interactions with NG2+ cells to reach the end of a developing fiber in newborn mice. The source of these myoblasts is currently unclear, but our results show that sub-laminar Pax7+ cells stain positive for the proliferation marker Ki67 (Figure S4H), suggestive of activated satellite cells. Further immunohistochemical co-staining with dystrophin and laminin confirmed that such activation occurs within the niche where satellite cells reside (Figure S4I). These results indicate that during neonatal muscle growth, satellite cells represent at least one cell type that contribute to the myoblast pool involved in migration and fusion to growing myofibers.
As opposed to adult muscle injury where macrophages and resident mesenchymal progenitor cells have been shown to contribute to a pro-regenerative response (Deng et al., 2012; Joe et al., 2010), the role that stromal cells play in regulating the growth of newborn muscles has been much less appreciated. Only recently has a population of connective tissue fibroblasts expressing the transcription factor, Tcf4, been linked to the maturation of neonatal muscle by regulating the specification of fiber type, potentially through a secreted factor whose cell of origin remains to be elucidated (Mathew et al., 2011). Here, our current work adds to the unique role of a muscle interstitial cell population whose communication with myoblasts plays a key role in neonatal muscle growth.
EXPERIMENTAL PROCEDURES
Mice
All animals were housed, maintained, and used according to protocols approved by the Institutional Animal Care and Use committees (IACUC) at the Ohio State University. NF-κBEGFP/+, Rosa26-Tomato, iNOS−/−, NG2CreER, and p50−/− mice were purchased from Jackson Laboratories. p65−/−;TNFRα−/− and p65fl/fl mice were generated as previously described (Doi et al., 1999; Steinbrecher et al., 2008). The EFNA5 cKO (Efna5fl/fl) mouse was generated using standard Embryonic Stem Cell technology (Tessarollo et al., 2009). The targeting vector was obtained from the International Mouse Knockout Consortium (IKMC). Correctly recombined ES cells were identified by Southern analysis. Germline transmission of the targeted allele (IKMC tm1a allele) was assessed by PCR according to IKMC guidelines. Before use for indicated experiments, Efna5tm1a/+ mice were bred to a ubiquitous Flpe strain (Jackson Laboratories). In the Efna5tm1c/+ (Efna5fl/+) mice, excision of the IRES-LacZ-neo cassette was verified by PCR using the suggested IKMC primers. NG2CreER mice (Jackson Laboratories) were crossed with p65fl/fl or EFNAfl/fl to delete p65 or EFNA5 in NG2+ cells. To induce recombination in early neonatal development, tamoxifen (0.2 mg/pup) dissolved in corn oil was injected subcutaneously to postnatal day 2 (P2) mice.
Immunofluorescence Staining
Muscle tissues were snap frozen with O.C.T. in methyl-butane solutions chilled in liquid nitrogen. To freeze muscle containing EGFP or Tomato reporter, muscle tissues were prefixed with 4% paraformaldehyde, overnight at 4°C followed by a 48h incubation with 30% sucrose at 4°C. For EGFP+ ISC analysis, whole hindlimbs were used from P7 mice, otherwise Quad and Gas muscles were used from mice at P14 or older. 10µm sections were obtained for all staining experiment except for myonuclei number/fiber counting using 30µm-thick sections. Sections were air-dried and incubated with 4% paraformaldehyde for fixation at room temperature, followed by incubation with blocking buffer containing 5% goat serum and 0.2% triton X-100 for 30min at room temperature. Primary antibodies: anti-NG2 (Millipore), anti-CD31 (BD Pharmingen), anti-α laminin (Sigma), anti-MyoD (Dako) were diluted in blocking buffer and incubated for 1h at room temperature. For MyoD, the primary antibody was incubated for overnight at 4°C. After washing three times with PBS, fluorescent-conjugated secondary antibodies (Invitrogen) were incubated for 1h at room temperature. The slides were rinsed with PBS and stained with DAPI solutions. For adherent cell staining, culture plates were washed with PBS and directly fixed with 4% paraformaldehyde. Anti-ki67 (Dako) and anti-troponin T (Sigma) were used as primary antibodies following the same procedure as described for tissue sections. The images were obtained with either an Olympus FV 1000 Spectral or filter confocal laser microscope and analyzed by Image J and FV10-ASW 4.0 viewer. At least three mice were analyzed for each staining.
Statistical Analysis
All quantitative data was shown as mean ± s.e.m. All statistical analysis were performed by using 2-tailed unpaired Student’s t test. (*) p < 0.05, (**) p < 0.01, (***) p< 0.001.
Supplementary Material
Highlights.
NF-κB is activated in NG2+ interstitial cells in neonatal skeletal muscle.
NF-κB functions in NG2+ interstitial cells to promote myoblast migration.
-
-The gene encoding EphrinA5 is a direct target of NF-kB.
- EphrinA5 from NG2+ interstitial cells promotes myoblast migration.
ACKNOWLEDGMENTS
We thank A. Baldwin and A. Beg for generously providing mice, A. Selen Yilmaz for assistance with additional Bioinformatics, the Pollock lab and S. Cole for technical assistance for live cell imaging, and M. Viapiano for assistance with the two-chamber migration system. We also thank G. Leone, M. Ostrowski, the S. Amacher laboratory, and F. Montanaro for their interactions, and M. Campion in the Guttridge laboratory for assistance with whole-mount staining, as well as other members of Guttridge laboratory for engaging discussions throughout the course of this study. This project was supported by an NIH grant, 2R01 AR052787 to D.C.G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
The accession numbers for the data of microarray and RNAseq shown in this paper are GSE75774 and GSE75832, respectively.
AUTHOR CONTRIBUTION
J-M.G. designed, performed, and analyzed the experiments. J.W., J.M.P, and J.S. assisted in optimizing FACS conditions and in vivo mouse experiments. S.L. performed the bioinformatics analysis for the array data. V.C. generated Efna5tm1a/+ mice. B.K. and A.F. helped to optimize the live cell imaging experiment. D.D.C. contributed to discussion. J-M.G. wrote and helped with the editing phase of the manuscript. D.C.G supervised, designed experiments, and contributed to the editing of the manuscript.
REFERENCES
- Bladt F, Riethmacher D, Isenmann S, Aguzzi A, Birchmeier C. Essential role for the c-met receptor in the migration of myogenic precursor cells into the limb bud. Nature. 1995;376:768–771. doi: 10.1038/376768a0. [DOI] [PubMed] [Google Scholar]
- Braun T, Gautel M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat Rev Mol Cell Biol. 2011;12:349–361. doi: 10.1038/nrm3118. [DOI] [PubMed] [Google Scholar]
- Britto PR, Perez-Escamilla R. No second chances? Early critical periods in human development. Introduction. Social science & medicine. 2013;97:238–240. doi: 10.1016/j.socscimed.2013.09.001. [DOI] [PubMed] [Google Scholar]
- Cappellari O, Cossu G. Pericytes in development and pathology of skeletal muscle. Circ Res. 2013;113:341–347. doi: 10.1161/CIRCRESAHA.113.300203. [DOI] [PubMed] [Google Scholar]
- Dahlman JM, Bakkar N, He W, Guttridge DC. NF-kappaB functions in stromal fibroblasts to regulate early postnatal muscle development. J Biol Chem. 2010;285:5479–5487. doi: 10.1074/jbc.M109.075606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis TA, Fiorotto ML. Regulation of muscle growth in neonates. Curr Opin Clin Nutr Metab Care. 2009;12:78–85. doi: 10.1097/MCO.0b013e32831cef9f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellavalle A, Maroli G, Covarello D, Azzoni E, Innocenzi A, Perani L, Antonini S, Sambasivan R, Brunelli S, Tajbakhsh S, et al. Pericytes resident in postnatal skeletal muscle differentiate into muscle fibres and generate satellite cells. Nature communications. 2011;2:499. doi: 10.1038/ncomms1508. [DOI] [PubMed] [Google Scholar]
- Deng B, Wehling-Henricks M, Villalta SA, Wang Y, Tidball JG. IL-10 triggers changes in macrophage phenotype that promote muscle growth and regeneration. J Immunol. 2012;189:3669–3680. doi: 10.4049/jimmunol.1103180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi TS, Marino MW, Takahashi T, Yoshida T, Sakakura T, Old LJ, Obata Y. Absence of tumor necrosis factor rescues RelA-deficient mice from embryonic lethality. Proc Natl Acad Sci. 1999;96:2994–2999. doi: 10.1073/pnas.96.6.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehler E, Gautel M. The sarcomere and sarcomerogenesis. Adv Exp Med Biol. 2008;642:1–14. doi: 10.1007/978-0-387-84847-1_1. [DOI] [PubMed] [Google Scholar]
- Griffin CA, Apponi LH, Long KK, Pavlath GK. Chemokine expression and control of muscle cell migration during myogenesis. Journal of cell science. 2010;123:3052–3060. doi: 10.1242/jcs.066241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin CA, Kafadar KA, Pavlath GK. MOR23 promotes muscle regeneration and regulates cell adhesion and migration. Dev Cell. 2009;17:649–661. doi: 10.1016/j.devcel.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. NF-kappaB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 2012;26:203–234. doi: 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joe AW, Yi L, Natarajan A, Le Grand F, So L, Wang J, Rudnicki MA, Rossi FM. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat Cell Biol. 2010;12:153–163. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitiyakara A, Angevine DM. A Study of the Pattern of Postembryonic Growth of M. Gracilis in Mice. Dev Biol. 1963;8:322–340. doi: 10.1016/0012-1606(63)90033-2. [DOI] [PubMed] [Google Scholar]
- Klein R. Eph/ephrin signalling during development. Development. 2012;139:4105–4109. doi: 10.1242/dev.074997. [DOI] [PubMed] [Google Scholar]
- Li J, Johnson SE. Ephrin-A5 promotes bovine muscle progenitor cell migration before mitotic activation. J Anim Sci. 2013;91:1086–1093. doi: 10.2527/jas.2012-5728. [DOI] [PubMed] [Google Scholar]
- Magness ST, Jijon H, Van Houten Fisher N, Sharpless NE, Brenner DA, Jobin C. In vivo pattern of lipopolysaccharide and anti-CD3-induced NF-kappa B activation using a novel gene-targeted enhanced GFP reporter gene mouse. Journal of immunology. 2004;173:1561–1570. doi: 10.4049/jimmunol.173.3.1561. [DOI] [PubMed] [Google Scholar]
- Mathew SJ, Hansen JM, Merrell AJ, Murphy MM, Lawson JA, Hutcheson DA, Hansen MS, Angus-Hill M, Kardon G. Connective tissue fibroblasts and Tcf4 regulate myogenesis. Development. 2011;138:371–384. doi: 10.1242/dev.057463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie RJ, Doyle AD, Yamada KM. Random versus directionally persistent cell migration. Nature reviews Molecular cell biology. 2009;10:538–549. doi: 10.1038/nrm2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliakov A, Cotrina M, Wilkinson DG. Diverse roles of eph receptors and ephrins in the regulation of cell migration and tissue assembly. Dev Cell. 2004;7:465–480. doi: 10.1016/j.devcel.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Siegel AL, Atchison K, Fisher KE, Davis GE, Cornelison DD. 3D timelapse analysis of muscle satellite cell motility. Stem cells. 2009;27:2527–2538. doi: 10.1002/stem.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark DA, Karvas RM, Siegel AL, Cornelison DD. Eph/ephrin interactions modulate muscle satellite cell motility and patterning. Development. 2011;138:5279–5289. doi: 10.1242/dev.068411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbrecher KA, Harmel-Laws E, Sitcheran R, Baldwin AS. Loss of epithelial RelA results in deregulated intestinal proliferative/apoptotic homeostasis and susceptibility to inflammation. J Immunol. 2008;180:2588–2599. doi: 10.4049/jimmunol.180.4.2588. [DOI] [PubMed] [Google Scholar]
- Swartz ME, Eberhart J, Pasquale EB, Krull CE. EphA4/ephrin-A5 interactions in muscle precursor cell migration in the avian forelimb. Development. 2001;128:4669–4680. doi: 10.1242/dev.128.23.4669. [DOI] [PubMed] [Google Scholar]
- Tessarollo L, Palko ME, Akagi K, Coppola V. Gene targeting in mouse embryonic stem cells. Methods Mol Biol. 2009;530:141–164. doi: 10.1007/978-1-59745-471-1_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster MT, Fan CM. c-MET regulates myoblast motility and myocyte fusion during adult skeletal muscle regeneration. PloS one. 2013;8:e81757. doi: 10.1371/journal.pone.0081757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RB, Bierinx AS, Gnocchi VF, Zammit PS. Dynamics of muscle fibre growth during postnatal mouse development. BMC Dev Biol. 2010;10:21. doi: 10.1186/1471-213X-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams PE, Goldspink G. Longitudinal growth of striated muscle fibres. J Cell Sci. 1971;9:751–767. doi: 10.1242/jcs.9.3.751. [DOI] [PubMed] [Google Scholar]
- Yumoto N, Wakatsuki S, Kurisaki T, Hara Y, Osumi N, Frisen J, Sehara-Fujisawa A. Meltrin beta/ADAM19 interacting with EphA4 in developing neural cells participates in formation of the neuromuscular junction. PloS one. 2008;3:e3322. doi: 10.1371/journal.pone.0003322. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.