Abstract
Hearing loss is the most frequent sensorineural disorder, affecting 1 in 1000 newborns. In more than half of these babies, the hearing loss is inherited. Hereditary hearing loss is a very heterogeneous trait, with about 100 gene localizations and 44 gene identifications for nonsyndromic hearing loss. TMC1 has been identified as the disease-causing gene for autosomal dominant and autosomal recessive nonsyndromic hearing loss at the DFNA36 and DFNB7/11 loci, respectively. To date, two dominant and 18 recessive TMC1 mutations have been reported as the cause of hearing loss in 34 families. In this report, we describe linkage to DFNA36 and DFNB7/11 in one family with dominant and 10 families with recessive nonsyndromic sensorineural hearing loss. In addition, mutation analysis of TMC1 was performed in 51 familial Turkish patients with autosomal recessive hearing loss. TMC1 mutations were identified in seven of the families segregating recessive hearing loss. The pathogenic variants we found included two known mutations, c.100C>T and c.1165C>T, and four new mutations, c.2350C>T, c.776+1G>A, c.767_768del and c.1166G>A. The absence of TMC1 mutations in the remaining six linked families implies the presence of mutations outside the coding region of this gene, or alternatively, at least one additional deafness-causing gene in this region. The analysis of copy number variations in TMC1 as well as DNA sequencing of 15 additional candidate genes did not reveal any proven pathogenic changes, leaving both hypotheses open.
INTRODUCTION
Hearing loss is the most common sensory disorder, affecting one in 1000 newborns. In more than half of these babies, the cause is hereditary (hereditary hearing loss, HHL) (Parving 1999). About 30% of HHL is associated with co-inherited clinical abnormalities and therefore classified as syndromic HL. In the remaining 70% of cases, newborns have nonsyndromic HHL, which is solely characterised by hearing problems and mostly due to cochlear defects. Nonsyndromic HHL is further classified by mode of inheritance. It is almost exclusively monogenic and inherited as an autosomal recessive trait (ARNSHL) in about 70% of cases. Postlingual hearing loss, in contrast, is often multifactorial, the most prevalent example being age-related hearing loss or presbycusis, which affects about half of octogenarians. Families segregating monogenic postlingual autosomal dominant nonsyndromic hearing loss (ADNSHL) are well described but rare compared to presbycusis.
The genetic heterogeneity of HHL is reflected by the mapping of 43 dominant, 52 recessive and 4 X-linked nonsyndromic loci and the identification of 44 genes (Hereditary hearing Loss Homepage; http://webh01.ua.ac.be/hhh/). One example is TMC1 (transmembrane channel-like gene 1) (Genbank ID NT_023935 position 4301249-4615799), mutations of which are a cause of both ADNSHL and ARNSHL at the DFNA36 and DFNB7/11 loci, respectively. The gene has been implicated as the cause of deafness in 34 families: 2 dominant families from North America and 32 recessive families from Pakistan, India, Turkey, Sudan and Tunisia (Kurima et al., 2002; Kalay et al., 2005; Meyer et al., 2005; Santos et al., 2005; Kitajiri et al., 2007; Kitajiri et al., 2007; Tlili et al., 2008) (Table 1). In the mouse ortholog Tmc1, mutations have been identified in both the recessive mutant deafness and the dominant mutant Beethoven (Kurima et al., 2002; Vreugde et al., 2002).
Table 1.
Overview of all TMC1 mutations identified to date.
| Family name | origin | sequence variant | exon | type of sequence variant | inheritance pattern | reference | |
|---|---|---|---|---|---|---|---|
| cDNA level | Protein level | ||||||
| LMG128 | North-America | c.1714 G>A | p.D572N | 19 | Missense mutation | ad | (Kurima et al., 2002) |
| LMG248 | North-America | c.1714 G>C | p.D572H | 19 | Missense mutation | ad | (Kitajiri et al., 2007) |
| PKSR9, PKSN9, PKSN24, PKDF7, PKDF75, PKDF69, PKDF178, PKDF243, PKDF319, PKDF401, 3 Tunisian cases | Pakistan, Tunisia | c.100 C>T | p.R34X | 7 | Nonsense mutation | ar | (Kurima et al., 2002); (Kitajiri et al., 2007); (Tlili et al., 2008) |
| PKDF22 | Pakistan | c.IVS3_IVS5del27kb | / | 5 | Deletion | ar | (Kurima et al., 2002) |
| IN-DKB6 | India | c.295_296delA | / | 8 | Deletion | ar | (Kurima et al., 2002) |
| PKSR25, 4090 | Pakistan | c.IVS10-8T>A | / | Intron 10: −8 | Splice site mutation | ar | (Kurima et al., 2002); (Santos et al., 2005) |
| PKSR1a | Pakistan | c.IVS13+1G>A | / | Intron 13: +1 | Splice site mutation | ar | (Kurima et al., 2002) |
| TR56 | Turkey | c.776 A>G | p.Y259C | 13 | Missense mutation | ar | (Kalay et al., 2005) |
| TR47 | Turkey | c.821 C>T | p.P274L | 13 | Missense mutation | ar | (Kalay et al., 2005) |
| TR50 | Turkey | c.1083_1087delCAGAT | p.R362PfsX6 | 15 | Deletion | ar | (Kalay et al., 2005) |
| TR63 | Turkey | c.1334 G>A | p.R445H | 16 | Missense mutation | ar | (Kalay et al., 2005) |
| PKSR20a | Pakistan | c.1534 C>T | p.R512X | 17 | Nonsense mutation | ar | (Kurima et al., 2002) |
| IN-M17 | India | c.1960 A>G | p.M654V | 20 | Missense mutation | ar | (Kurima et al., 2002) |
| 4049 | Pakistan | c.830 A>G | p.Y277C | 13 | Missense mutation | ar | (Santos et al., 2005) |
| DFNB7/11 Sudanese family Tunisian family |
Sudan Tunisia |
c.1165 C>T | p.R389X | 15 | Nonsense mutation | ar | (Meyer et al., 2005) (Tlili et al., 2008) |
| DFNB7/11 Sudanese family | Sudan | c.IVS19+5G>A | / | Intron 19: +5 | Splice site mutation | ar | (Meyer et al., 2005) |
| PKDF431 | Pakistan | c.1541C>T | p.P514L | 17 | Missense mutation | ar | (Kitajiri et al., 2007) |
| PKDF329, PKDF511 | Pakistan | c.1543T>C | p.C515R | 17 | Missense mutation | ar | (Kitajiri et al., 2007) |
| PKDF274 | Pakistan | c.IVS5+1G>T | / | Intron 5: +1 | Splice site mutation | ar | (Kitajiri et al., 2007) |
| Tunisian family | Tunisia | c.1764G>A | p.W588X | 19 | Nonsense mutation | ar | (Tlili et al., 2008) |
ar=autosomal recessive; ad=autosomal dominant.
The genomic structure of TMC1 consists of 24 exons that encode a full-length mRNA of 3201 bp. Its sequence is highly similar to TMC2, and to the corresponding mouse orthologs Tmc1 and Tmc2. These genes, together with 6 other orthologs, belong to the new TMC gene family, which has been created as none of the genes shows nucleotide sequence similarity to other known genes or domains. Two members of the gene family, TMC6 and TMC8, are identical to the long isoforms of EVER1 and EVER2, respectively. Mutations in both genes have been found to be associated with epidermodysplasia verruciformis (MIM 226400). The exact function of the transmembrane proteins encoded by this gene family remains to be determined. Based upon their structure, they may act as ion channels, ion pumps or transporters (Keresztes et al., 2003; Kurima et al., 2003). Studies of the recessive mutant deafness (dn) and the dominant mutant Beethoven (Bth) have given some clues about the possible function of Tmc1 (Marcotti et al., 2006). In mice, the protein is expressed before the onset of hearing in the pericuticular necklace and the endoplasmic reticulum of mature hair cells, as well as during early postnatal development. Therefore, Tmc1 might play a role in normal maturation of the hair cells. It has been suggested that the protein may be responsible for the up- or downregulation of ion channels or molecules of the exocytotic machinery during development. Alternatively, it could be involved in intracellular trafficking. The expression pattern of TMC1 is very specific: apart from its expression in inner and outer hair cells of the cochlea and in neurosensory epithelia of the vestibular end organs, very low levels of transcript are also found in human placenta and testis, but in no other tissues (Kurima et al., 2002).
Here, we report mutation analysis in one DFNA36 family, 10 DFNB7/11 families and 51 Turkish index patients of families with ARNSHL (Table 2). In seven families with ARNSHL, we identified two known and four novel mutations in TMC1. In the remaining five DFNB7/11 families and in the DFNA36 family, no proven disease-causing mutation could be found in TMC1 or in 15 other genes in the overlap of all candidate regions defined by the significantly linked families.
Table 2.
Overview of the families linked to DFNA36 and DFNB7/11, and the Turkish index patients with TMC1 mutations described in this report. For all families, the SLINK score (simulated maximal LOD score) and maximal LOD scores obtained are listed. For families in which sequence variants in TMC1 were identified by DNA sequencing, the variant, its position and the protein change are listed. All variants were homozygous in patients. For families 935, Nas, Fay, GRE, M36 and index patients of families DF139 and DF135, the nucleotide changes found were judged to be pathogenic. The * indicates that the family has previously been reported (Jain et al., 1995).
| Family | Origin | Inheritance pattern | SLINK score | Maximal LOD score | Sequence variant TMC1 | Exon | Type of sequence variant | |
|---|---|---|---|---|---|---|---|---|
| Genomic/cDNA level | Protein level | |||||||
| PE | Guatemala/Mexico | ad | 4,78 | 4,44 | ||||
| GRE | Greece | ar | 4,41 | 3,96 | c.2350C>T | p.R604X | 20 | nonsense mutation |
| TM | India | ar | 3,99 | 3.5 | ||||
| 101* | India | ar | 4,82 | 3.6 | ||||
| G9 | Iran | ar | 2,91 | 2,91 | g.94615A>C | / | 3 | variant in non-coding exon |
| M28 | Iran | ar | 1,78 | 1.66 | ||||
| M13 | Iran | ar | 2,9 | 2,55 | ||||
| M36 | Iran | ar | 2,78 | 2,24 | c.776+1G>A | / | 7 | splice site mutation |
| 935 | Iran | ar | 3,46 | 2,46 | c.100C>T | p.R34X | 7 | nonsense mutation |
| Nas | Lebanon/Jordan | ar | 3,86 | 1,79 | c.100C>T | p.R34X | 7 | nonsense mutation |
| Fay | Lebanon/Jordan | ar | 2,45 | 1,94 | c.1165C>T | p.R389X | 15 | nonsense mutation |
| DF139 | Turkey | ar | / | / | c.767_768del | p.F255FfsX14 | 13 | deletion |
| DF135 | Turkey | ar | / | / | c.1166G>A | p.R389Q | 15 | missense mutation |
ad = autosomal dominant, ar = autosomal recessive.
MATERIALS AND METHODS
Family data
In this study, different approaches were used to collect families and perform mutation analysis. Eleven families of different origin segregating ADNSHL or ARNSHL were collected and analysed. (Table 2) (Supplementary Fig. 1). Family 101 has been reported before (Jain et al., 1995). In addition, genetic analysis was performed in 51 Turkish index patients from families with ARNSHL containing two or more affecteds. All these patients were seen personally and completed a questionnaire to exclude syndromic hearing loss. All participants signed an informed consent form. For family PE, audiometry was performed by measuring air conduction thresholds at frequencies ranging from 125 to 8000 Hz. In family GRE, auditory evoked potentials were measured in two affected individuals.
Linkage analysis
DNA was isolated from blood samples of participating subjects using a standard salting-out protocol. In a first part of the study, the information content for linkage of eleven hearing loss families was estimated, based on SLINK simulations using the program Easylinkage (version 4.01) (Lindner and Hoffmann 2005). Fixed linkage parameters were used for all LOD score calculations with an allele frequency of 0.001 and a phenocopy rate of 0%. For the dominant family, the penetrance was 0% for the wild-type/wild-type genotype (wt/wt) and 100% for the wild-type/mutant and mutant/mutant genotypes (wt/mt, mt/mt). For the recessive families, penetrance was 0% for wt/wt and wt/mt and 100% for mt/mt.
Different strategies were used for linkage analysis. If the SLINK score had a value of 3.3 or higher, the family was considered informative enough for genome-wide linkage analysis (Lander and Kruglyak 1995). For families in which the LOD score was below 3.3, linkage analysis was performed for known deafness loci and LOD score calculations were combined with haplotype analysis to confirm or exclude linkage. For families 935, Nas and Fay, DFNB1 was excluded by direct sequencing. Next, a genome-wide scan was performed for these families, as well as for families PE, TM and 101. Linkage analysis on family 101 has been reported before (Jain et al., 1995). In families GRE, G9 and M28, nine of the more common recessive loci were checked for linkage (DFNB1, DFNB2, DFNB3, DFNB4, DFNB7/11, DFNB9, DFNB12, DFNB21 and DFNB23). In families M13 and M36, only DFNB1 and DFNB7/11 were screened in order to find additional TMC1-families. Linkage analysis was performed by calculating two-point and multipoint LOD scores using the Easylinkage program. All genotyping was done by PCR amplification of fluorescently labelled microsatellite markers and fragment analysis on an ABI 3130 automated DNA sequencer (Applied Biosystems, California, USA), using standard procedures.
DNA sequence analysis
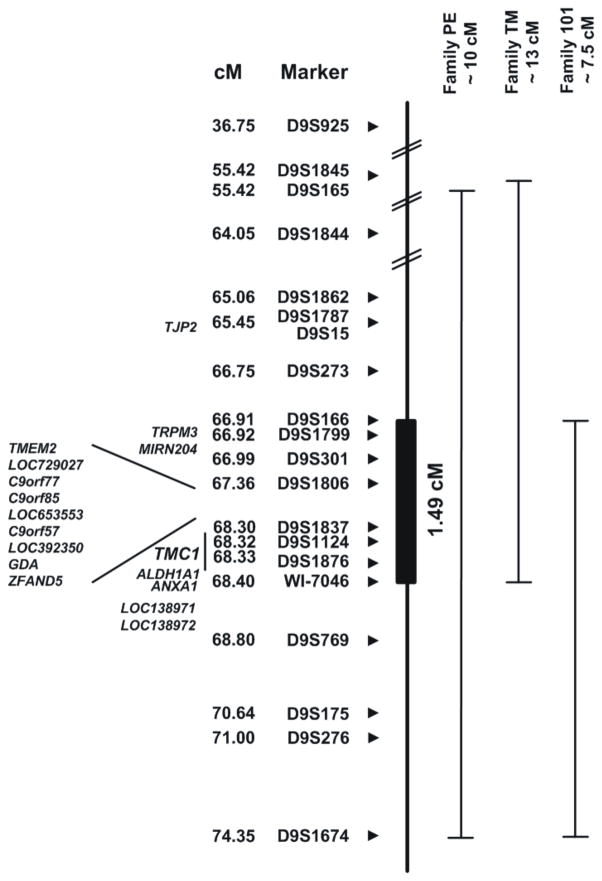
To screen candidate genes, non-coding and coding exons including the intron-exon boundaries were PCR amplified. Subsequent DNA sequencing of both strands was carried out on an ABI 3130 automated DNA sequencer (Applied Biosystems) using the big-Dye terminator cycle sequencing kit Version 3.1 (Applied Biosystem). DNA sequencing of TMC1 was performed in two patients of eleven hearing loss families and in 51 Turkish index patients of ARNSHL families. In affected subjects from larger families in which no TMC1 mutation was identified, additional candidate genes were sequenced. Most of these genes localize to the minimal shared candidate region defined by the three significantly linked families negative for TMC1 mutations and include TJP2, MIRN204, TMEM2, LOC729027, C9ORF77, C9ORF85, LOC653553, C9ORF57, LOC392350, GDA, ZFAND5, ALDH1A1, ANXA1, LOC138971 and LOC138972 (Fig. 2). In addition, DNA sequencing of TRPM3 was performed in families TM and 101. In all Turkish index patients, the coding exon of GJB2 was sequenced.
Fig. 2.
Candidate regions of 3 families significantly linked to DFNA36 or DFNB7/11, without TMC1 mutation identified. All analysed markers and their corresponding positions from the Decode genetic map are indicated. At the left, all candidate genes located in the common linked region are listed, containing 8 genes and 6 gene predictions. The common region is 1.49 cM large and includes the TMC1-gene.
We used the ConSeq Server to check the conservation of amino acids affected by the identified TMC1 mutations (Berezin et al., 2004). A BLAST analysis of the human TMC1 protein sequence showed 61 sequences with an e-value below 0.001, of which all hits with a sequence identity below 20% were excluded. With the remaining 27 sequences, a multiple sequence alignment was made, which was subsequently used as an input at the ConSeq Server. The same strategy was used for calculating the conservation scores for TJP2. ConSeq-scores that are obtained using this procedure vary from 1 (variable) to 9 (conserved).
The I-Mutant2.0 program was used to predict the effect of the TJP2 variant on the protein stability (Capriotti et al., 2005). A ΔΔG value is calculated from the unfolding Gibbs free energy value of the mutated protein minus the unfolding Gibbs free energy value of the wild type protein (Kcal/mol). The calculation was performed at pH 7.0 and at temperatures of 25°C (in vitro conditions) and 37°C (in vivo conditions). ΔΔG values below zero indicate a reduced stability of the mutant protein. A reliability index (RI) is computed if a negative ΔΔG value is present, indicating how reliable the prediction is.
Multiplex Amplicon Quantification (MAQ)
TMC1 was screened for copy number variations (CNVs) using the Multiplex Amplicon Quantification (MAQ) technique (Sleegers et al., 2006; Suls et al., 2006). This technique consists of a multiplex PCR amplification of several fluorescently labelled test and reference amplicons, followed by fragment analysis. Twelve test amplicons located in TMC1 and eight reference amplicons located at randomly selected genomic positions outside known CNVs were simultaneously PCR amplified in samples from two affected subjects and one control from each of six families (PE, G9, M13, M28, TM and 101). The primer sequences of the test and reference amplicons are given in Table 3. Unlabelled and FAM-labelled primers were ordered from Eurogentec (Belgium). The multiplex PCR reactions were performed on 40 ng genomic DNA in a 15 μl reaction containing 1×Titanium™Taq PCR buffer (Clontech, Palo Alto, CA, USA) with a final concentration of 2.5 mM for each dNTP (Invitrogen, Carlsbad, CA, USA) and a total of 0.075 μl of Titanium™Taq DNA Polymerase (Clontech). PCR cycle conditions were 2′ at 98 °C, followed by 23 cycles of 45″ at 95 °C, 45″ at 60 °C and 2′ at 68 °C. After a final extension step of 10′ at 72 °C, samples were cooled to 8 °C. Subsequently, fragment analysis was performed on an Applied Biosystems 3730 DNA analyzer with the GeneScan ™ 500 LIZ™ Size Standard (Applied Biosystems, Foster City, CA). The experiment was performed in duplicate. Peak areas of the test amplicons were normalized to those of the reference amplicons. Chromatogram files were analyzed with the MAQs software (http://www.vibgeneticservicefacility.be/soft/maqs.php). The dosage quotients (DQ) of every amplicon is calculated as described by Suls et al. (2006) (Suls et al., 2006). DQ values below 0.75 were considered indicative of a deletion and values above 1.25 indicative of a duplication.
Table 3.
Primer sequences of the test and reference amplicons used for MAQ analysis of TMC1.
| Amplicon name | Forward primer sequence (3′-5′) | Reversed primer sequence (3′-5′) |
|---|---|---|
| TMC1-Amp01 | CTTCAATCAAGTCCCAGTTTCCT | TCAAACACACAGTAGTGCCTTCTA |
| TMC1-Amp02 | GATTCAGTTTCAATAAATGCTTCCT | CTGTATCAGCCCAGCTTCCT |
| TMC1-Amp03 | TATCTCTTCTTGGATTTCCTTTGCT | CTCATACCATTCTCACATTCATTCC |
| TMC1-Amp04 | GGGTAGTTTCCCTTTGTTTCCT | CAACAATAGGGTTTGATGTCTCCT |
| TMC1-Amp05 | TTGAGGGTAACTTATGTGTCAACAAC | TAAACCCAGTGCTCAAAGTACACTAA |
| TMC1-Amp06 | TGCCACATTCTCATTCTTCCT | CTGTCTTGAAAGCCTTCTGATCTA |
| TMC1-Amp07 | TCACTGGCCCACTCTTCC | CAGTCAGGTCAACCACATTCC |
| TMC1-Amp08 | TAAATCAAAGGGCATTTCACG | CAAACCCTGAAATCCAACAAC |
| TMC1-Amp09 | CGTCCACTTGATCAGATTCCT | GATCTTGCTGTGCAAATTCCT |
| TMC1-Amp10 | TTGGTCAGTCTCCTCTGATTCTCTA | ATTCAATGTCCAGTCTCCATGTC |
| TMC1-Amp11 | GCCAATAACTGTGTGTTCACG | GAACCAAATCCTTTGCATCAAC |
| TMC1-Amp12 | GTGAAAGGGTGAAAGTTCAATTC | TTCAACCACCTCATCTTCTGC |
| TMC1-Contr01 | TTCAATATGTATACCCAACCTTCG | TAAACTTCAAGGCTACGCTTCTC |
| TMC1-Contr02 | CAGTATCTAAGACCAGGGTGATTCT | AAAGATTTCTTCTTCCCAGGCTA |
| TMC1-Contr04 | CTTTGTATCCGAGCTCCATTCT | TCATCCTGTCTGCTTTCACAAC |
| TMC1-Contr05 | TGAAATTCTCCAAACACCTGTC | TTTCCAAAGCCAGATTATTCCTAA |
| TMC1-Contr06 | CATACCCTTAATGGCTCTTCTTTCT | TTCTGGGTTCTCAGCTCTGC |
| TMC1-Contr08 | TCAGGGTAAACAAGGGCAAC | TGACTGCCACCATCTTTCG |
| TMC1-Contr09 | CAAGCTCCTCCTCTCCTTCC | TGCACACCCATGCATAATAAC |
| TMC1-Contr11 | ACCGGATTCACACACTACCAC | AACCACAGCGAGGGATTCT |
RESULTS
Clinical data
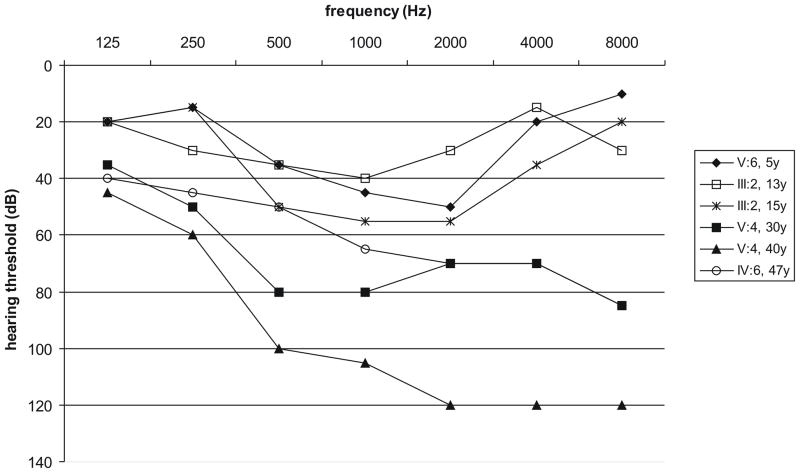
The Guatemalan family PE segregates ADNSHL characterized by hearing impairment that starts in the mid frequencies during the first decade of life and progresses to involve all frequencies. The rate of progression is faster in the higher frequencies, leading to a downward-sloping audiogram after several decades (Fig. 1). Five subjects in the Greek family GRE were diagnosed as affected. For the two youngest patients (3 and 7 years old), auditory evoked potentials were available, which showed no response at equipment limits, consistent with profound prelingual ARNSHL. Affected subjects in the other families as well as the Turkish index patients were reported to have congenital severe-to-profound sensorineural hearing impairment. By completing questionnaires, syndromic hearing loss could be excluded in all cases.
Fig. 1.
Air conduction thresholds of the better ear of 4 patients of family PE at different ages. There is clear progression of the hearing loss for individuals III:2 and V:4. For all patients, mainly the mid and high frequencies are affected.
Linkage analysis
Genome-wide linkage analysis in family PE revealed linkage to DFNA36 with a maximal LOD score of 4.44. 80% of the total genome was excluded with LOD scores below −2. Apart from the region on chromosome 9q, no other linked regions were found. In the recessive family GRE, linkage to locus DFNB7/11 was identified by linkage analysis of known loci, showing a LOD score of 3.96. We tried to identify additional families that link to this genomic region in different ways. A series of Iranian families segregating ARNSHL had been collected before and although their SLINK scores were not above the genome wide significance level, we chose to perform linkage analysis for a set of known loci. This screening revealed suggestive linkage at DFNB7/11 in four families, G9, M28, M13 and M36, all having LOD scores between 1.66 and 2.91. Three more families, 935, Nas and Fay, were identified with probable linkage, having LOD scores between 1.79 and 2.46. All other genomic regions were excluded. For one additional family TM, significant DFNB7/11 linkage was found with a LOD score of 3.5, while no linkage to other genomic regions was identified. Family 101 was previously reported to show significant linkage to DFNB7/11 and was also included in our analysis (Jain et al., 1995). For all families, candidate regions were defined by polymorphic marker analysis and linkage was checked by both haplotype reconstruction (Supplementary Fig. 1) and LOD score calculation (Table 2). The linked regions of the families included TMC1.
Candidate gene analysis
DNA sequencing of TMC1
Mutation screening of TMC1 was completed by direct DNA sequencing in two patients of each family with suggestive and significant linkage and in 51 Turkish index patients with ARNSHL. All sequence variants segregating with the hearing loss phenotype are listed in Table 2. In families 935 and Nas, the known mutation c.100C>T was identified in homozygous condition in all affected subjects. This change predicts the nonsense mutation p.R34X. Similarly, in family Fay, affected subjects were homozygous for the known mutation c.1165C>T, which causes the nonsense mutation p.R389X. In the Greek family GRE, a new nonsense mutation was identified in exon 20, which segregates with the hearing loss. The identified nucleotide change c.2350C>T causes a nonsense mutation p.R604X, leading to a premature termination codon (PTC). In the Iranian family M36, a new splice site mutation was identified in all affected subjects. The mutation c.776+1G>A is located at the splice donor site of exon 7 and changes the 5′ splice site from GT to AT. This change was not identified in 100 Iranian control samples. In an index patient of family DF139, a 1-bp deletion c.767_768del was identified in exon 13 of TMC1. The deletion changes the highly conserved leucine at position 255 (ConSeq-score 9) and leads to a frameshift p.F255FfsX14. In a patient of family DF135, a new missense mutation c.1166G>A was identified, causing the amino acid change p.R389Q. The mutation was not found in 100 ethnically matched control samples and the amino acid at position 389 has a ConSeq-score of 8. In family G9, a nucleotide change g.94615A>C was identified in exon 3 (non-coding) of TMC1 and was found to segregate with the hearing loss. The variant was not found in 100 Iranian and 100 Belgian control samples.
Multiplex Amplicon Quantification (MAQ) of TMC1
In the 6 families in which no pathogenic TMC1 changes were found, MAQ analysis was completed using 12 amplicons covering TMC1. Family G9 was also analysed as we could not prove that the variant we found was pathogenic. No differences in peak area were observed between normalized chromatograms of affected subjects and controls. This result indicates that no deletion or duplication is present at the regions of these amplicons.
DNA sequencing of other candidate genes
Other candidate genes were also selected for DNA sequencing under the hypothesis that mutations in another gene in this region also cause HHL. To narrow the interval, we assumed that the same gene was mutated in the three families that were significantly linked to the TMC1-locus (PE, TM and 101), thereby defining a shared interval of 1.49 cM (2,596 Mb), common to all families (Fig. 2). This region contains 8 genes and 6 gene predictions, including TMC1. The coding exons and intron-exon boundaries of all 13 candidate genes in this region were sequenced in the three families with significant linkage as well as in the remaining three families with suggestive linkage, but no mutations could be identified. LOC138971 and LOC138972 were also sequenced because they were located in the shared region in an earlier phase of the project when fewer families had been collected, but no possible pathogenic change was found in these two genes. In addition to these genes chosen on the basis of location, TJP2 (ZO-2) was considered a good candidate due to its interesting function as a tight junction protein and its expression in the cochlea. In exon 19 of the gene, a new sequence variant c.2971A>T was identified, segregating with the hearing loss phenotype in family PE and causing the amino acid change p.D924V. This aspartic acid residue has a ConSeq-score of 7 and belongs to a conserved acidic domain of the protein. The Grantham score of the amino acid change was 152, while scores above 100 indicate radical amino acid changes (Grantham 1974). The I-Mutant2.0 program predicted a decreased stability of the mutant protein (ΔΔG = −1.84 at 25°C and −1.65 at 37°C with RI=2). The change was not found in any of the non-affected family members, nor in 104 independent Belgian control samples and 103 ethnically matched controls. The gene was sequenced in two affected subjects from 26 additional small families segregating deafness but no variants were identified (data not shown). The DNA sequencing of GJB2 in all Turkish index patients revealed the presence of 35delG in heterozygous state in the index patient of family DF139.
DISCUSSION
Twenty different mutations in TMC1 have been reported as the cause of hearing loss in two families segregating ADNSHL and 32 families segregating ARNSHL. A literature search allowed making an estimate of the most frequent causes of ARNSHL, based on the reported numbers of mutations. GJB2 is without any doubt the most frequent cause of ARNSHL, carrying over 220 different mutations. The other more frequent genes, ranked according to their reported frequency, are SLC26A4, MYO15A, OTOF, CDH23 and TMC1. The literature data, together with the data from this report indicate that TMC1 is one of the more frequent causes of ARNSHL. In this study, we have reported one additional dominant family linked to the DFNA36 locus and 10 additional recessive families linked to the DFNB7/11 locus. We were able to identify disease-causing TMC1 mutations in five of these 11 families. In two families, putative mutations were identified, one in TMC1 and one in TJP2, but for none of them, the pathogenicity could be confirmed. In addition, we found two new TMC1 mutations in an index patient of two Turkish families segregating ARNSHL.
In families 935, Nas and Fay, known recessive mutations c.100C>T and c.1165C>T were identified in TMC1. Both are nonsense mutations leading to a premature termination codon (PTC). As the mRNA contains a PTC, it may be detected and degraded by nonsense-mediated decay (NMD), a eukaryotic mRNA surveillance mechanism (Linde et al., 2007). Including this study, c.100C>T has been identified in 16 families segregating ARNSHL, accounting for 47% of all recessive TMC1 mutations.
In four families with ARNSHL, we detected new pathogenic changes in TMC1. In family GRE, a nonsense mutation c.2350C>T was found to cause a PTC at nucleotide position 47 of exon 20. In the Iranian family M36, a novel splice site mutation c.776+1G>A was found to change the 5′ splice donor site of exon 7 from GT to AT. The effect of this splice site mutation depends on the adjacent sequence. Use of a cryptic splice site will lead to partial exon skipping or partial intron retention. Alternatively, if no cryptic splice site is used, then either exon skipping or a greatly reduced production of normal transcript will be the predominant outcome (Krawczak et al., 1992). Experimental study of this effect using patient mRNA is not possible, as TMC1 is not expressed in lymphocytes. In two Turkish families with ARNSHL, new TMC1 mutations were identified in an index patient. The first mutation c.767_768del is a 1-bp deletion causing a frameshift. As a result, 14 amino acids are altered and followed by a stop codon. The second mutation, c.1166G>A, causes the highly conserved amino acid arginine to change into glutamine at position 389 of the protein, causing a shift from a positively charged into a neutral residu. In addition, the variant was not found in 100 Turkish control samples. Therefore, we believe that this new variant may be pathogenic.
In family G9, a putative pathogenic change was found in the third non-coding exon of TMC1, segregating with the hearing loss and absent in 200 control samples. The variant may be located in the promoter region or in a regulatory region of the gene, but none of these regions have been identified so far. Functional studies should be performed to further investigate the effect of the variant on the protein.
In family PE, a putative mutation in TJP2 was found to segregate with the hearing loss. Different arguments support the hypothesis that the variation may be a true pathogenic change. The amino acid is highly conserved and belongs to a conserved acidic protein domain, it has a high Grantham score, the mutant protein is predicted to have a decreased stability and the variant was absent in 207 control samples. However, no conclusive evidence could be given as it is a missense mutation and no TJP2 mutations were identified in other hearing loss families. A mutation in TJP2 has already been suggested to be associated with familial hypercholanemia (OMIM 607709) (Carlton et al., 2003). However, the inheritance may be oligogenic, only causing the disease in combination with a mutation in a second gene. It is possible that the TJP2 variant is a pathogenic variant involved in familial hypercholanemia or another autosomal recessive disease and that the occurrence in the current family is correlated to the hearing impairment.
In previous reports, patients of six families with significant linkage to DFNB7/11 did not carry TMC1 mutations (Santos et al., 2005). In addition, we found 3 significantly linked families and 3 families with possible linkage that also lack pathogenic changes in TMC1. To screen for possible disease-causing mutations we tested three hypotheses.
First, we hypothesized that TMC1 is the disease-causing gene in these families but that the pathogenic change could not be detected by DNA sequencing. Therefore, MAQ of TMC1 was performed in six hearing loss families to determine whether CNVs could be observed. While this screen failed to identify any deletions or duplication of the amplicons we used, it should be noted that smaller CNVs could have been missed, as TMC1 is large and only 12 amplicons were studied. Complete CNV analysis of TMC1 may allow detecting smaller rearrangements. This could be done either by using a fine-tiling array-CGH with oligonucleotides or using a high density SNP microarray. Assaying for other pathogenic changes in TMC1 at the transcription level using mRNA from blood is not feasible since TMC1 is not expressed in white blood cells. It should be noted, however, that this type of change is infrequent, making it somewhat implausible that all families would carry this type of pathogenic mutation. For family PE, we compared the hearing loss phenotype with two other families, LMG128 and LMG248, segregating ADNSHL and carrying TMC1 mutations (Fig. 1) (Kurima et al., 2002; Kitajiri et al., 2007). The hearing loss of patients from all three families is comparable, starting in the first to second decade and affecting mainly the mid-to-high frequencies. Later on, the hearing loss becomes profound across most frequencies, showing some variation in the low-frequency thresholds. In family LMG128, the rate of progression seems to be faster compared to families LMG248 and PE. This phenotypic similarity may be an additional support to the hypothesis that TMC1 is the disease-causing gene in family PE.
The second hypothesis we tested stated that a second deafness gene lies near TMC1 and is the cause of the hearing loss in the families lacking a TMC1 mutation. The common candidate region shared by all families with significant linkage includes only 14 genes. No single pathogenic change could be identified in any of these genes. However, our screen does not definitively exclude these genes as a possible new deafness gene. We may have missed mutations not detectable by DNA sequence analysis of exons. In addition, the LOC-genes and c9orf-genes might have additional exons which have not been annotated yet.
In a third hypothesis we considered that some families carry an undetected TMC1 mutation, while others carry mutations in one or more new deafness genes. As a corollary to this hypothesis, the candidate region that contains this new gene(s) is much larger and many more candidate genes must therefore be screened for mutations.
In conclusion, we have shown that TMC1 mutations are one of the more frequent causes of ARNSHL. Our data also show that a subset of families linked to the DFNA36-DFNB7/11 loci do not carry mutations in TMC1 exons, suggesting either a remarkably high proportion of mutations outside of the exons or an additional deafness gene in this region.
Supplementary Material
Pedigrees of all families analysed, with the linked haplotypes at loci DFNA36 and DFNB7/11. All individuals with hearing loss are indicated with black symbols, the open symbols are individuals with normal hearing and symbols with questions marks indicate individuals with an uncertain phenotype. The analysed markers are listed at the left and the linked haplotype is indicated with a box.
Acknowledgments
We are very grateful to Gilad Wainreb for helping us to score the conservation of amino acids using the ConSeq Server. This study was supported by grants from EUROHEAR (LSHG-CT-2004-512063), the Fund for Scientific Research Flanders (FWO-F, Grant G.0138.07), the National Institutes of Health (1RO1DC02842, RJHS) and the Oticon Foundation (MBP). Nele Hilgert is a fellow of the Fund for Scientific Research Flanders (FWO-F).
References
- Berezin C, Glaser F, Rosenberg J, Paz I, Pupko T, Fariselli P, Casadio R, Ben-Tal N. ConSeq: the identification of functionally and structurally important residues in protein sequences. Bioinformatics. 2004;20:1322–4. doi: 10.1093/bioinformatics/bth070. [DOI] [PubMed] [Google Scholar]
- Capriotti E, Fariselli P, Casadio R. I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005;33:W306–10. doi: 10.1093/nar/gki375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton VE, Harris BZ, Puffenberger EG, Batta AK, Knisely AS, Robinson DL, Strauss KA, Shneider BL, Lim WA, Salen G, et al. Complex inheritance of familial hypercholanemia with associated mutations in TJP2 and BAAT. Nat Genet. 2003;34:91–6. doi: 10.1038/ng1147. [DOI] [PubMed] [Google Scholar]
- Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–4. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- Jain PK, Fukushima K, Deshmukh D, Ramesh A, Thomas E, Lalwani AK, Kumar S, Plopis B, Skarka H, Srisailapathy CR, et al. A human recessive neurosensory nonsyndromic hearing impairment locus is potential homologue of murine deafness (dn) locus. Hum Mol Genet. 1995;4:2391–4. doi: 10.1093/hmg/4.12.2391. [DOI] [PubMed] [Google Scholar]
- Kalay E, Karaguzel A, Caylan R, Heister A, Cremers FP, Cremers CW, Brunner HG, de Brouwer AP, Kremer H. Four novel TMC1 (DFNB7/DFNB11) mutations in Turkish patients with congenital autosomal recessive nonsyndromic hearing loss. Hum Mutat. 2005;26:591. doi: 10.1002/humu.9384. [DOI] [PubMed] [Google Scholar]
- Keresztes G, Mutai H, Heller S. TMC and EVER genes belong to a larger novel family, the TMC gene family encoding transmembrane proteins. BMC Genomics. 2003;4:24. doi: 10.1186/1471-2164-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajiri S, Makishima T, Friedman T, Griffith A. A novel mutation at the DFNA36 hearing loss locus reveals a critical function and potential genotype-phenotype correlation for amino acid-572 of TMC1. Clin Genet. 2007;71:148–52. doi: 10.1111/j.1399-0004.2007.00739.x. [DOI] [PubMed] [Google Scholar]
- Kitajiri SI, McNamara R, Makishima T, Husnain T, Zafar AU, Kittles RA, Ahmed ZM, Friedman TB, Riazuddin S, Griffith AJ. Identities, frequencies and origins of TMC1 mutations causing DFNB7/B11 deafness in Pakistan. Clin Genet. 2007;72:546–50. doi: 10.1111/j.1399-0004.2007.00895.x. [DOI] [PubMed] [Google Scholar]
- Krawczak M, Reiss J, Cooper DN. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum Genet. 1992;90:41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- Kurima K, Peters LM, Yang Y, Riazuddin S, Ahmed ZM, Naz S, Arnaud D, Drury S, Mo J, Makishima T, et al. Dominant and recessive deafness caused by mutations of a novel gene, TMC1, required for cochlear hair-cell function. Nat Genet. 2002;30:277–84. doi: 10.1038/ng842. [DOI] [PubMed] [Google Scholar]
- Kurima K, Yang Y, Sorber K, Griffith AJ. Characterization of the transmembrane channel-like (TMC) gene family: functional clues from hearing loss and epidermodysplasia verruciformis. Genomics. 2003;82:300–8. doi: 10.1016/s0888-7543(03)00154-x. [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–7. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Linde L, Boelz S, Neu-Yilik G, Kulozik AE, Kerem B. The efficiency of nonsense-mediated mRNA decay is an inherent character and varies among different cells. Eur J Hum Genet. 2007;15:1156–62. doi: 10.1038/sj.ejhg.5201889. [DOI] [PubMed] [Google Scholar]
- Lindner TH, Hoffmann K. easyLINKAGE: a PERL script for easy and automated two-/multi-point linkage analyses. Bioinformatics. 2005;21:405–7. doi: 10.1093/bioinformatics/bti009. [DOI] [PubMed] [Google Scholar]
- Marcotti W, Erven A, Johnson SL, Steel KP, Kros CJ. Tmc1 is necessary for normal functional maturation and survival of inner and outer hair cells in the mouse cochlea. J Physiol. 2006;574:677–98. doi: 10.1113/jphysiol.2005.095661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer CG, Gasmelseed NM, Mergani A, Magzoub MM, Muntau B, Thye T, Horstmann RD. Novel TMC1 structural and splice variants associated with congenital nonsyndromic deafness in a Sudanese pedigree. Hum Mutat. 2005;25:100. doi: 10.1002/humu.9302. [DOI] [PubMed] [Google Scholar]
- Parving A. The need for universal neonatal hearing screening--some aspects of epidemiology and identification. Acta Paediatr Suppl. 1999;88:69–72. doi: 10.1111/j.1651-2227.1999.tb01163.x. [DOI] [PubMed] [Google Scholar]
- Santos RL, Wajid M, Khan MN, McArthur N, Pham TL, Bhatti A, Lee K, Irshad S, Mir A, Yan K, et al. Novel sequence variants in the TMC1 gene in Pakistani families with autosomal recessive hearing impairment. Hum Mutat. 2005;26:396. doi: 10.1002/humu.9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleegers K, Brouwers N, Gijselinck I, Theuns J, Goossens D, Wauters J, Del-Favero J, Cruts M, van Duijn CM, Van Broeckhoven C. APP duplication is sufficient to cause early onset Alzheimer’s dementia with cerebral amyloid angiopathy. Brain. 2006;129:2977–83. doi: 10.1093/brain/awl203. [DOI] [PubMed] [Google Scholar]
- Suls A, Claeys KG, Goossens D, Harding B, Van Luijk R, Scheers S, Deprez L, Audenaert D, Van Dyck T, Beeckmans S, et al. Microdeletions involving the SCN1A gene may be common in SCN1A-mutation-negative SMEI patients. Hum Mutat. 2006;27:914–20. doi: 10.1002/humu.20350. [DOI] [PubMed] [Google Scholar]
- Tlili A, Rebeh IB, Aifa-Hmani M, Dhouib H, Moalla J, Tlili-Chouchene J, Said MB, Lahmar I, Benzina Z, Charfedine I, et al. TMC1 but Not TMC2 Is Responsible for Autosomal Recessive Nonsyndromic Hearing Impairment in Tunisian Families. Audiol Neurootol. 2008;13:213–218. doi: 10.1159/000115430. [DOI] [PubMed] [Google Scholar]
- Vreugde S, Erven A, Kros CJ, Marcotti W, Fuchs H, Kurima K, Wilcox ER, Friedman TB, Griffith AJ, Balling R, et al. Beethoven, a mouse model for dominant, progressive hearing loss DFNA36. Nat Genet. 2002;30:257–8. doi: 10.1038/ng848. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pedigrees of all families analysed, with the linked haplotypes at loci DFNA36 and DFNB7/11. All individuals with hearing loss are indicated with black symbols, the open symbols are individuals with normal hearing and symbols with questions marks indicate individuals with an uncertain phenotype. The analysed markers are listed at the left and the linked haplotype is indicated with a box.