Abstract
Dendritic cells (DCs) are specialized leukocytes that orchestrate the adaptive immune response. Mass spectrometry based proteomic study of these cells presents technical challenges, especially when the DCs are human in origin due to the paucity of available biological material. Here, to maximize mass spectrometry coverage of the global human DC proteome, different cell disruption methods, lysis conditions, protein precipitation, and protein pellet solubilisation and denaturation methods were compared. Mechanical disruption of DC cell pellets under cryogenic conditions, coupled with the use of RIPA buffer, was shown to be the method of choice based on total protein extraction and on the solubilisation and identification of nuclear proteins. Precipitation by acetone was found to be more efficient than by 10% TCA/acetone, allowing greater than 28% more protein identifications. Although being an effective strategy to eliminate the detergent residue, the acetone-wash step caused a loss of protein identifications. However, this potential drawback was overcome by adding 1% sodium deoxycholate in the dissolution buffer, which enhanced both solubility of the precipitated proteins and digestion efficiency. This in turn resulted in 6-11% more distinct peptides and 14-19% more total proteins identified than using 0.5M triethylammonium bicarbonate alone with the greatest increase (34%) for hydrophobic proteins.
Keywords: proteomics, monocyte-derived dendritic cells, sample preparation, cell lysis buffer, protein precipitation, protein solubilisation, LC-MS/MS
Introduction
Dendritic cells (DCs) are antigen-presenting cells that function to initiate and modulate the immune response in vertebrates [1]. Given their central role, they have been the subject of much recent research, holding great promise for the prevention of transplant rejection [2], as well as for the treatment of autoimmune disease [3] and of cancer [4]. In addition, new understanding of the molecular mechanisms underlying the interaction of these cells with HIV-1 and the resulting effects on viral spread and pathogenesis [5, 6] as well as their role in the pathogenesis of several other infectious diseases [7], has recently drawn additional attention to DCs as therapeutic targets [8].
Research on DC biology has generally relied upon classical immunological techniques. However, mass spectrometry (MS)-based proteomics, which aims to study biological processes at the protein level in a systematic manner, has recently received considerable attention for the study of the role of human DCs in immune regulation [9]. Proteomics study of human DCs has so far mainly focused on identifying changes in the proteome profile of monocytes during their differentiation into dendritic cells and their subsequent maturation [10-13], comparing the proteomic profiles of different DC model cells [14], as well as evaluating the response to diverse pathogenic challenges [15] and to treatment with different compounds [16-19]. Due to the lack of sufficient quantities of human blood DCs for proteomic analysis, most of these studies have employed monocyte derived dendritic cells (MDDCs), which are generated in vitro from peripheral blood mononuclear cells (PBMCs) obtained from healthy donors [20, 21].
Application of MS-based proteomics techniques usually follows two different approaches. The classical gel-based approach was primarily employed in early DC-related proteomics studies [10-19, 22, 23]. More recently, the solution-based, also referred as gel-free, proteomics sample preparation approach has been used in several DC proteomics studies, in either labelled or label-free modalities [24-26]. Gel-free approaches involve in-solution protein digestion and take advantage of the power of liquid chromatography (LC) to separate the resulting complex peptide mixtures prior to the MS analysis. Compared to gel-based approaches, this solution-based approach has displayed several advantages for the analysis of complex proteomics samples, in terms of sensitivity, accuracy, as well as sample handling and throughput [27].
For a MS-based proteomics study, sample preparation is not only the initial but also a critical step in the workflow. Several factors such as protein solubilisation, the efficiency of proteolytic digestion and chemical composition of the sample solution directly affect the outcome of mass spectrometry analysis. Surfactants which are widely used for protein isolation and solubilisation in protein extraction protocols are usually poorly compatible with mass spectrometry, leading to severe ion competition and ion suppression [28, 29]. Protein sample cleanup by precipitation with organic solvent are generally recognized as a simple and effective approach to eliminate these surfactants. Protein denaturation employed before digestion allows more cleavage sites of the proteins accessible to the proteolytic enzyme, and thus improves digestion efficiency. Sodium dodecyl sulphate (SDS) and urea are two most common reagents to assist in this process and both of them were found to be used in the solution-based sample preparation for DC related proteomic studies [24-26]. However, the sample preparation using SDS suffers from severe trypsin digestion inhibition and incompatibility with reversed-phase LC separation and the subsequent MS analysis. Therefore, its level in the digestion solution and LC-MS sample solution should be controlled [30, 31]. The use of urea is associated with similar protease activity reduction and the other disadvantages such as peptide carbamylation after its decomposition by heat [32]. As a contrast to SDS and urea, Na-DOC, a bile-acid detergent, is considered as a trypsin-friendly and MS-compatible detergent as it has no obvious effect on tryptic digestion at the concentration of 1% and can be easily removed by precipitation at low pH level [33]. Furthermore, it was consistently demonstrated to outperform urea in enhancing the tryptic digestion efficiency by different reach groups [33-35]. Recently, its advantages over urea were comprehensively explored by Leon et al through a series of comparative studies conducted both qualitatively and quantitatively based on in-solution digestion and spin filter-aided digestion [36].
The aim of this study was to establish a simple and efficient in-solution-based sample preparation method suitable for high sample throughput for proteomic profiling of human MDDCs after a variety of treatments. Therefore, we investigated the application of three methods for cellular disruption, coupled with two widely used cell lysis buffer compositions (i.e. Triton X-100 and RIPA) and specifically assessed several different sample preparation workflows by combining solvent-driven protein precipitation strategies with Na-DOC- or SDS-assisted protein solubilisation and denaturation prior to trypsin-based digestion. The outcome of each workflow was evaluated by considering protein solubilisation efficiency, MDDC proteome coverage, the physicochemical properties and subcellular localization of the proteins identified, LC-MS/MS compatibility of individual reagents, the efficiency of surfactant removal and the potential for increasing sample throughput. On the basis of these criteria we developed a procedure that maximizes the desired outcomes and is expected to serve as a standard in the field.
Materials and methods
Materials
NP-40, sodium chloride, acetone, trichloroacetic acid (TCA), triethylammonium bicarbonate (TEAB) (1M, pH 8), SDS, sodium deoxycholate (Na-DOC), Tris-(2-carboxyethyl)phosphine (TCEP), methyl methanethiosulfonate (MMTS), isopropanol, trifluoroacetic acid (TFA), formic acid (FA) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Triton X-100 was purchased from Fisher Scientific (Pittsburgh, PA, USA). Complete protease inhibitor cocktail tablet was purchased from Roche (Basel, Switzerland). HEPES buffer (1M) was from Gibco (Paisley, UK). Sequencing-grade modified trypsin was obtained from Promega (Madison, WI, USA). Acetonitrile (ACN, LC-MS grade) and water (LC-MS grade) were purchased from VWR (Radnor, PA, USA).
Ethics statement
Buffy-coats obtained from anonymous blood donors were provided by the Blood Transfusion Center of the Hematology Service of the University Hospital of Geneva by agreement with the service, after approval of our project by the Ethics Committee of the University Hospital of Geneva (Ref #0704).
MDDC production, protein extraction and sample preparation
MDDCs were generated from Peripheral Blood Mononuclear Cells (PBMCs) as described in the literature [37]. Briefly, PBMCs were isolated from buffy-coats derived from healthy donors by sedimentation over a Ficoll-Plaque Plus (GE Healthcare, Piscataway, NJ) density gradient. The CD14+ monocytes were positively selected from PBMC using CD14-conjugated MicroBeads following the manufacturer’s instructions (Miltenyi Biotec, Auburn, CA). MDDCs were derived by culturing monocytes in the presence of recombinant human granulocyte-macrophage colony-stimulating factor (GM-CSF) and of recombinant human interleukin 4 (IL-4) conditioned medium added at a dilution of 1:50 for 5 days at 37°C in 5% CO2.
Harvested MDDCs were lysed by combining one of two different cellular disruption methods with one of two different lysis buffers, Triton X-100 (1% Triton X-100, 20 mM HEPES buffer, pH 7.5, 150 mM NaCl and 1 tablet of Roche protease inhibitor cocktail) and Radio Immuno Precipitation Assay (RIPA) buffer (1% NP-40, 0.1% SDS, 0.5% Na-DOC, 20 mM HEPES buffer, pH 7.5, 150 mM NaCl, and 1 tablet of Roche protease inhibitor cocktail). In all cases, cells were harvested by scraping off tissue culture plates using a rubber policeman, before sedimentation to produce a cellular pellet.
Conventional lysis was conducted by suspending cell pellets in either Triton X-100 or RIPA lysis buffer and incubating cell suspensions for 30 min at 4 °C with continual rotation prior to removal of the insoluble fraction by centrifugation at 10000xg for 30 min at 4 °C. Cryogenic lysis was conducted by mechanically disrupting either frozen cell suspensions or pellets. For cell suspension grinding, cell pellets were resuspended in either Triton X-100 or RIPA lysis buffer, transferred to 2 ml Eppendorf tubes and frozen in Liquid N2. A pre-chilled, single 3 mm zirconium oxide bead was added to each tube prior to cryogenic grinding for 2 min at 30 Hz, with a Retsch Mixer Mill 400 grinder equipped with Eppendorf tube holders. After thawing, samples were incubated in the cold and processed as indicated above. For cell pellet grinding, cell pellets obtained as described above were immediately frozen in liquid N2 prior to cryogenic disruption. Frozen cell grindate was thawed directly in lysis buffer and processed as described.
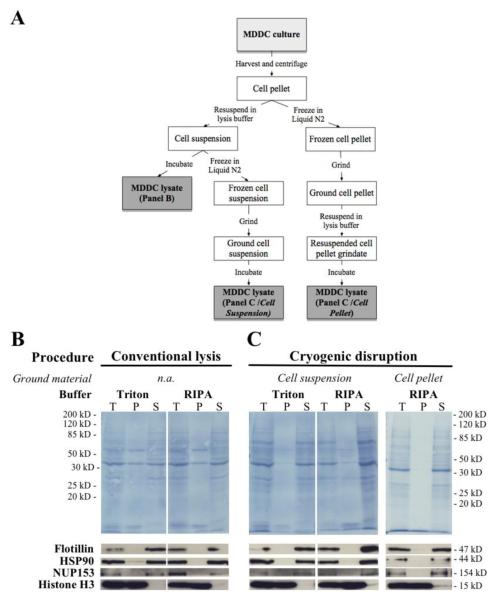
In order to evaluate the extraction and solubilisation efficiencies of the procedures, following cell lysis, total cell lysates (Figure 1, T) were centrifuged for 30 min at 10000 xg at 4° C, to produce insoluble protein pellet (Figure 1, P) and soluble protein suspension (Figure 1, S) fractions. Proportionally equal volumes of each fraction were loaded on SDS-PAGE in order to directly reflect the portion of each protein that partitioned to each fraction. Following transfer onto nitrocellulose, membranes were stained with Amido Black using standard procedures to reveal the total protein content of each fraction. Subsequently, membranes were cut into horizontal strips, and each strip was separately immuno-stained in blocking buffer using standard procedures, in order to analyse the fractionation pattern of key cellular markers. The following antibodies were employed: mouse anti-flotillin (Abcam, Cambridge, MA, USA), rabbit anti-Hsp90 (Cell Signaling, Billerica, MA, USA), rabbit anti-Nup153 (Bethyl Laboratories, Montgomery, TX, USA) rabbit anti-histone H3 (Abcam, Cambridge, MA, USA). After staining, primary antibody binding was detected by incubation with secondary antibody conjugated with Horse Radish Peroxidase (HRP; HRP-anti-Mouse or -Rabbit respectively, Biorad, Hercules, CA).
Figure 1.
Optimization of the procedure for extracting proteins from human monocyte-derived dendritic cells (MDDCs). The efficiency of protein extraction from MDDCs was assessed by comparing three different cellular disruption procedures in combination with two commonly used lysis buffers (as indicated and as described in Materials and Methods). (A) Schematic flowchart depicting the three cell disruption procedures used for this experiment. (B) After harvest, MDDCs were subjected to conventional lysis by incubating cells in the cold with the indicated lysis buffer. (C) Alternatively, MDDCs were subjected to cryogenic grinding either in suspension (Cell suspension) or as cell pellets (Cell pellet), prior to chemical lysis. In all cases, total cell lysates (T) were fractionated by centrifugal sedimentation to give rise to insoluble (P) and soluble (S) protein fractions. After SDS-PAGE and transfer, nitrocellulose membranes were stained with Amido Black to reveal the total protein content of each fraction. Subsequently, individual horizontal strips were subjected to immuno blot to reveal the fractionation pattern of each of the indicated cellular marker. Flotillin is a cell membrane protein associated with lipid rafts, HSP90 is a cytoplasmic and nucleoplasmic marker, Nup153 is a nucleoporin on the nuclear face of the nuclear membrane and Histone H3 is a chromatin protein found exclusively in the nucleus.
The amount of total protein present in the cleared lysate was quantified by Bradford assay according to the manufacturer’s protocol (Pierce, Rockford, IL). The final concentration of the cell lysates (two types and four experimental replicates for each type) was then adjusted to 2 μg/μL with the lysis buffer and aliquots of 50 μL of each cell lysate (equivalent to 100 μg of protein) were used for different sample preparation workflows.
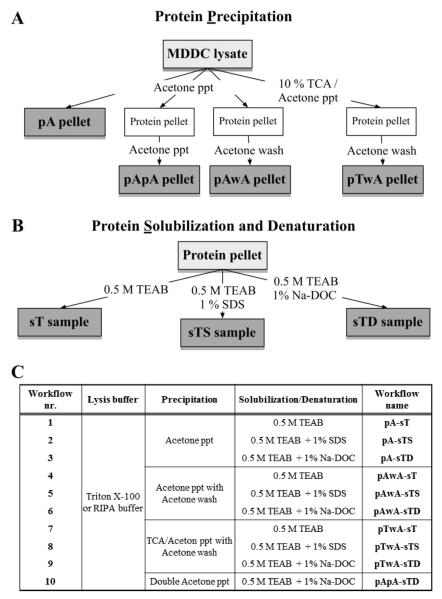
Protein precipitation
Protein precipitation was carried out by four different methods involving two different precipitation reagents (Figure 2): 1) one single precipitation step using six volumes of cold acetone; 2) one-step precipitation with six volumes of cold acetone followed by two pellet-washing steps with 400 μL of cold acetone each; 3) two acetone precipitation steps, where the first pellet was resuspended in 50 μL of 0.5M TEAB (pH 8) dissolution buffer, before further precipitation with 300 μL of acetone; 4) one-step precipitation with six volumes of cold 10% TCA in acetone followed by washing the pellet twice with 400 μL of cold acetone. In all cases, precipitation reagents were pre-cooled at −20°C before use. After the addition of the precipitation reagent, the protein samples were incubated overnight at −20°C and then centrifuged at 13,000 rpm at 4 °C for 30 minutes to collect the protein pellets. For double precipitation methods, the second precipitation step lasted two hours at −20°C. For each washing step, after the addition of acetone, the sample vials were kept at −20°C for 1 hour. After centrifugation, the supernatant in each sample vial after each step was aspirated and the protein pellet was kept for the next step. At the end of the precipitation step, the protein pellets were air-dried for 15 minutes in the ice box.
Figure 2.
Structure of different sample preparation workflows used to prepare MDDC tryptic digest for LC-MS/MS analysis. (A) Protein precipitation method; (B) Protein solubilisation and denaturation method; (C) Different workflows constructed by combining one protein precipitation method and one protein solubilisation and denaturation method.
Protein solubilisation and denaturation and in-solution digestion
For comparison reason, the air-dried pellets were reconstituted in either 20 μL of dissolution buffer (0.5M TEAB, pH 8) alone or in the same dissolution buffer but containing different surfactants (i.e. 0.1% SDS or 1% Na-DOC). After that, proteins were reduced with 5 mM TCEP at 60°C for 1 hour, followed by alkylation with 10mM MMTS at room temperature for 10 minutes. Modified sequencing-grade trypsin was added to the reduced and alkylated samples at a 1:40 enzyme-to-protein ratio and the mixtures were incubated at 37°C for 16 hours. The tryptic digestion was stopped by acidifying the samples with 2 μL of 50% formic acid, followed by centrifugation at 13,000 rpm at 4 °C for 30 minutes. For LC-MS/MS analysis, the supernatants of all the protein samples were diluted to 0.2 μg/μL with 0.1% TFA in water.
LC-MS/MS analysis
Tryptic digests of proteins (0.2 μg/μL) were analysed by reverse-phase HPLC- MS/MS using a NanoLC-Ultra 2D plus system (Eksigent, Dublin, CA, USA) coupled to a TripleTOF 5600 mass spectrometer (AB SCIEX, Concord, ON, Canada) with a nanoelectrospray ionisation source equipped by an uncoated fused silica emitter tip (20 μm inner diameter, 10 μm tip, New Objective, Woburn, MA). The samples were desalted and concentrated with 0.1% TFA in water at a flow rate of 5μL/min on a C18 nanoscale trapping cartridge (Acclaim PepMap100, 5 μm, 100 Å, 300 μm i.d. × 1 mm, Dionex, Sunnyvale, CA, USA) for 5 minutes, followed by LC separation on a nanoscale LC column (Acclaim PepMap100, C18, 3 μm, 100 Å, 75 μm i.d. × 15 cm, Dionex, Sunnyvale, CA, USA). Mobile phase A consisted of 0.1% FA in water, and mobile phase B consisted of 0.1% FA in ACN. The elution gradient was set as follows: 0-2 min, 2% B; 2-5 min, 2-5% B; 5-65 min, 5-35% B; 65-72 min, 35-60% B; 72-73 min, 60-90% B; 73-80 min, 90% B; 80-82 min, 90-2% B; 82-85 min, 2% B at a flow rate of 300 nL/min. The column oven was maintained at 40°C. The injection volume was 5 μL for all experiments. The mass spectrometer was operated in the positive ion mode. Information dependent acquisition (IDA) was used by selecting the top 20 most intense precursor ions in each MS scan for the subsequent MS/MS scans over the m/z range of 50-1500. The total cycle time was 1150 ms with accumulation time for MS survey scan (100 ms) and dependant scans (50 ms each). The dynamic exclusion window for MS/MS was set as 7 s. The CID energy was automatically adjusted by the rolling CID function of Analyst TF 1.5.1. After acquisition of 4 samples, TOF MS spectra and TOF MS/MS spectra were automatically calibrated by injecting 25 fmol beta-galactosidase. For each sample preparation workflow, the peptide digests were analysed twice as technical replicates.
Protein identification and data analysis
The raw files from two technical replicates of the same sample were concatenated and searched by ProteinPilot software (v 4.2, AB SCIEX) using the Paragon algorithm [38] with False Discovery Rate (FDR) analysis. The following sample parameters were used: trypsin digestion, cysteine alkylation set to MMTS, and specie Homo sapiens. Processing parameters were set to “Biological modifications” and “Amino acid substitutions” together with a thorough ID search effort. All data files were searched against the UniProt database (updated on 10-03-2012) with a total of 40,468 human protein sequences. A global FDR of 1% for protein identification and local FDR of 5% for peptide identification were applied to filter the searching results. Extracted ion chromatograms (XICs) were generated and processed by PeakView software (v1.1.0.0, AB SCIEX). The average hydrophobicity expressed as grand average of hydropathy (GRAVY) value [39] for the identified proteins was calculated by online tool available at http://www.bioinformatics.org/sms2/protein_gravy.html. The pI values of the proteins were calculated by on-line tool available at http://web.expasy.org/compute_pi/. Identified proteins were mapped to cellular component Gene Ontology (GO) terms via the generic GOTermMapper tool (http://go.princeton.edu/cgi-bin/GOTermMapper) [40].
Results and Discussion
Optimization of cellular extract preparation
In order to maximize the extraction and consequent MS detection of proteins derived from Monocyte Derived Dendritic Cells (MDDCs), we initially concentrated our efforts on the development of an efficient method for cellular disruption and protein extract preparation (Figure 1A). In particular, we compared a standard cell lysis method (Figure 1B) with two alternative cryogenic disruption procedures (Figure 1C). In the first of such procedures, cells were resuspended in the indicated lysis buffer, frozen in liquid N2 and ground as described in Materials and Methods. In the second cryogenic disruption method, solid cell pellets were first frozen and ground and then they were thawed in the indicated lysis buffer. In addition to this we also compared the effect of using two commonly used lysis solutions, i.e. a buffer containing 1% Triton X-100 and the conventional Radio Immuno Precipitation Buffer (RIPA) buffer. Both buffers contain a mild non-ionic detergent, Triton X-100 or NP-40. The RIPA buffer additionally contains a small amount of two other ionic detergents, SDS (0.1%) and Na-DOC (0.5%). When protein recovery was evaluated by SDS-PAGE followed by Amido-Black staining, and by immuno blotting, we observed that combining mechanical disruption with chemical lysis allowed better solubilisation of both the bulk of cellular proteins (Figure 1B and 1C, top panels), and of specific cellular markers (Figure 1B and 1C, bottom panels). The difference between Triton X-100 and RIPA was less obvious, even though RIPA appeared to be marginally better in the extraction of the chaperone protein HSP90, found in both cytoplasm and in the nucleus, and the chromatin protein Histone H3. In addition, results indicated that protein extraction could be further improved by directly grinding cell pellets (Figure 1C, Cell pellet) rather than cellular suspensions (Figure 1C, Cell suspension). In particular, mechanical disruption of frozen cellular pellets, followed by thawing in RIPA for chemical lysis, significantly reduced the presence of bulk proteins in the insoluble fraction and further increased the fraction of Histone H3 that could be released in the supernatant. Based on the combination of these initial observations, we subsequently concentrated our efforts on the combined use of mechanical disruption of frozen cell pellets followed by chemical lysis.
Proteomic sample preparation for LC-MS/MS analysis
Since only marginal differences could be observed between the use of Triton X-100 and of RIPA buffer, subsequent LC-MS/MS based sample analysis experiments were conducted to compare the results we would obtain with both of these two buffers. Thus, both Triton X-100 cell lysate (TCL) and RIPA cell lysate (RCL) were subjected to one of four protein precipitation methods (Figure 2A) in combination with one of three protein solubilisation and denaturation methods (Figure 2B), prior to trypsin digestion. In total, ten different combinations (Figure 2C), of protein precipitation and protein solubilisation and denaturation methods were screened by LC-MS/MS analysis. An overview of the results obtained from two experimental replicates for each combination (hereafter referred to as workflow) is provided in Supplementary Table S1. Such table includes number of MS/MS spectra (total and identified), number of distinct peptides (total, fully cleaved and with missed cleavages) and number of unique proteins. A detailed comparison of the effects of each of the sample preparation workflows, mainly focusing on the distinct peptides and proteins identified in each case, is presented in individual sections below. Results are presented as pairwise comparisons to directly examine the effect of one variable at a time.
Evaluation of acetone and TCA/acetone protein precipitation methods
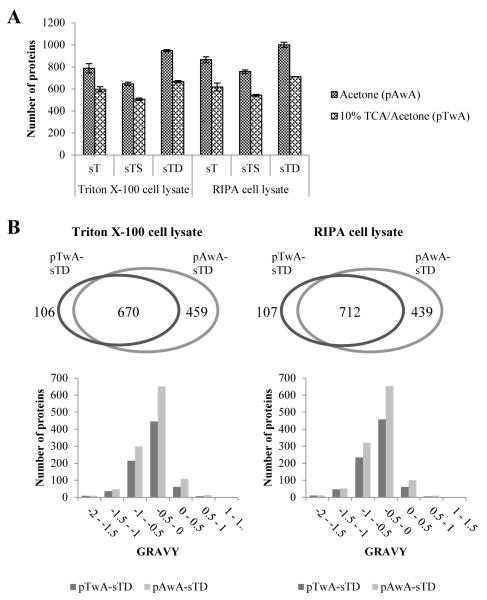
Protein precipitation is generally used to concentrate proteins and to remove surfactants (i.e. Triton X-100 and NP-40), which are not compatible with downstream steps such as tryptic digestion and MS analysis. Here, the whole MDDC proteome was precipitated by employing either one of two commonly used precipitation reagents, acetone versus 10% TCA/acetone both in combination with an acetone wash (procedures hereafter, referred to as pAwA and as pTwA respectively; see also Figure 2), and the results obtained were compared (Figure 3A and Supplementary Table S1). Direct inspection of the protein pellet size together with assessment of the number of peptides and of proteins identified allowed us to conclude that the pAwA procedure was better than the alternative method. In particular, using this method 19% - 51% more distinct peptides and 28% - 42% more proteins were identified for either TCL or RCL.
Figure 3.
Comparative study of protein profiles identified by different precipitation methods. pAwA: protein precipitation by acetone plus acetone-wash step; pTwA: protein precipitation by 10% TCA/acetone; sT, sTS and sTD represent protein solubilisation and denaturation using 0.5M TEAB, 0.1% SDS in 0.5M TEAB and 1% Na-DOC in 0.5M TEAB respectively. (A) Number of proteins identified by the workflows combining different protein precipitation methods but the same protein solubilisation and denaturation option. Since pTwA method included an actone-wash step, the comparative study was performed based on the workflows using pAwA as the protein precipitation method. The bar chart shows the means of two experimental replicates for each workflow with error bars representing two individual values. (B) Comparative study of the proteins identified by the workflow pTwA-sTD and pAwA-sTD with the venn diagrams shown on the top panel and the histograms of GRAVY value distribution shown on the bottom panel. For both panels, the results for the Triton X-100 cell lysate are shown on the left side while RIPA cell lysate shown on the right side. Data were obtained by merging two experimental replicates for each workflow.
When protein pellets produced by either pAwA or pTwA were resuspended using the solubilisation with TEAB and Na-DOC (sTD) method (Figure 2), the results obtained by aggregating two independent experimental replicates produced a total of 1129 (pAwA) vs. 776 (pTwA) identified proteins for the case of Triton X-100 cell lysate and 1151 (pAwA) vs. 819 (pTwA) for the case of RIPA cell lysate. Comparative studies of the resulting proteins revealed that for both TCL and RCL more than 85% of the protein identifications obtained by pTwA method were also identified by the pAwA method, whereas only about 60% of the acetone-precipitated proteins could be identified by the pTwA method (Figure 3B, top panel). In addition, all of the identified proteins where subdivided into six groups according to their hydrophobicity characteristics, as estimated based on their calculated GRAVY index (Figure 3B, bottom panel). Proteins with positive GRAVY values are generally accepted as hydrophobic, whereas those with negative values are considered hydrophilic. Compared with the pTwA method, the pAwA method showed an increased recovery of proteins characterized by higher hydrophobicity (Supplementary Table S2), resulting in a higher number of proteins in all groups except for the most hydrophilic group (i.e. GRAVY value ranges from −2 to −1.5) where a similar number of proteins was recovered. Notably there were no significant differences in the pI and MW distribution profiles obtained with either pAwA or with pTwA as indicated by the fact that the additional proteins identified by pAwA were evenly distributed along the pI and MW range (Supplementary Figure S1).
Detergent removal by different acetone precipitation methods
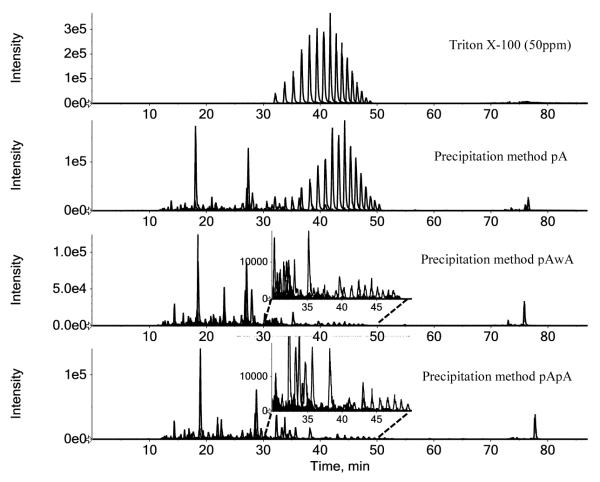
Detergents are usually poorly compatible with reversed-phase chromatography and MS analysis, causing severe ion competition and suppression. As a consequence, proper sample preparation procedures should eliminate these contaminants from the protein sample before LC-MS analysis. Having established the superiority of acetone based methods for MDDC protein recovery, we next evaluated the effectiveness of three different acetone-pellet treatment options for the removal of 1% Triton X-100 from TCL and 1% NP-40 from RCL , while at the same time maintaining high protein recovery. We therefore compared 1) single acetone precipitation (pA), 2) acetone precipitation followed by acetone-wash (pAwA), and 3) double acetone precipitation (pApA). The amount of the detergents remaining in protein extracts after these pellet treatment options was estimated by comparing the XIC-derived peak areas of their typical ions detected during MDDC sample analysis with those observed from a standard solution of detergent, i.e. 50 ppm of Triton X-100 (Figure 4 and Supplementary Table S3) or NP-40 (Supplementary Figure S2 and Supplementary Table S3). Typical total ion chromatograms (TICs) of sample solutions prepared by these three acetone precipitation methods were also displayed in Supplementary Figure S3.
Figure 4.
Extracted ion chromatograms (XICs) of typical ions derived from detergent residues as found in TCL protein extracts after different acetone pellet treatments. In all cases protein pellets were solubilised by using the sTD method (i.e. 1% Na-DOC in 0.5M TEAB dissolution buffer). From top to bottom: Triton X-100 at 50 ppm used for comparison sake; single precipitation by acetone (pA); acetone precipitation plus acetone-wash step (pAwA); double precipitation by acetone (pApA).
As observed, the detergent (i.e. Triton X-100 or NP-40) residues remaining in the protein extracts were below 50 ppm after the first acetone precipitation step, which were further reduced to less than 1ppm by either washing the pellet with acetone or by subjecting re-solubilised proteins to a second acetone precipitation step. In addition to the detergent, a large amount of very hydrophobic contaminants from the cell lysates could also be removed (Supplementary Figure S3). Since both the detergent and those contaminants generated a large amount of non-identifiable MS/MS spectra, removing this material increased the percentage of the MS/MS spectra that could be assigned with confident peptide identifications (i.e. 48.9% vs. 54.4% and 57.0% for workflow pA-sTD vs. pAwA-sTD and pApA-sTD, respectively, for Triton X-100 cell lysate and the corresponding number obtained for RIPA cell lysate were 54.4% vs. 58.6% and 57.4%). Although the pAwA and the pApA methods demonstrated comparable sample clean-up capacities, the latter was less favoured as more steps were required while at the same time fewer distinct peptides (both total peptides and fully-cleaved peptides) as well as proteins were identified (i.e. pAwA-sTD vs. pApA-sTD in Supplementary Table S1).
Evaluation of protein identification efficiency in the presence of Na-DOC
After the MDDC proteins were precipitated from the cell lysates, the recovery of these proteins was generally affected by the extent of their resolubilisation in the dissolution buffer. Since the presence of detergent residues might facilitate this process, we reasoned that it might be more difficult for acetone-washed protein pellets to be solubilised than un-washed pellets, due to relatively lower amount of detergent residues present in the former. In turn this would help explain the discrepancy between the numbers of proteins identified by pA versus pAwA for the method screening results (Supplementary Table S1). In order to obtain further confirmation of these results, we repeated the sample preparation workflows with more experimental replicates. As shown in Table 1 and Supplementary Figure S4, an average drop of 9% (TCL) and of 5% (RCL) of distinct peptides and accordingly a drop of 10% (TCL) and of 8% (RCL) of protein identifications, was observed by applying this wash step to the protein pellets prior to protein solubilisation in 0.5M TEAB (i.e.workflow pA-sT vs. pAwA-sT). Moreover, hydrophobic rather than hydrophilic proteins were more likely to be affected, resulting in an overall loss of 17% and 8% for hydrophobic proteins and hydrophilic proteins respectively (Supplementary Table S4).
Table 1.
LC-MS/MS analysis results of the tryptic digests prepared by different sample preparation workflows for MDDC Triton X-100 cell lysate and RIPA cell lysate. Data reported here are the average results over four experimental replicates for each workflow (two technical replicates of each experimental replicate were merged). Identification FDR threshold for MS/MS spectra is 5% (local FDR), for distinct peptides is 5% (local FDR) and for proteins is 1% (global FDR). Only the distinct peptides that are associated with the confidently identified proteins are included in the results.
|
Cell
lysate |
Sample preparation workflow (n=4) |
MS/MS spectra | Distinct peptides |
Protein (CV%) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total (CV%) |
Identified (CV%) |
% of identified |
Total (CV%) |
Fully cleaved (CV%) |
% of fully cleaved |
Missed cleavage |
||||
| 1 | ≥2 | |||||||||
| Triton X-100 (TCL) |
pA-sT | 52748 (3.4) |
26227 (4.1) |
49.7 | 5563 (5.4) |
5332 (5.7) | 95.8 | 222 | 9 | 880 (4.0) |
| pAwA-sT | 42864 (5.4) |
21791 (8.1) |
50.8 | 5045 (4.0) |
4898 (3.9) | 97.1 | 144 | 4 | 791 (6.2) |
|
| pAwA-sTS | 52485 (7.1) |
19948 (14.9) |
38.0 | 3770 (6.7) |
3658 (6.4) | 97.0 | 110 | 1 | 687 (6.9) |
|
| pAwA-sTD | 42880 (4.1) |
23969 (6.0) |
55.9 | 5612 (3.0) |
5378 (3.1) | 95.8 | 228 | 7 | 941 (2.0) |
|
| RIPA (RCL) |
pA-sT | 49978 (6.0) |
27125 (4.8) |
54.3 | 5675 (4.3) |
5449 (4.7) | 96.0 | 220 | 7 | 908 (1.8) |
| pAwA-sT | 43297 (1.9) |
23354 (3.9) |
53.9 | 5372 (4.8) |
5196 (5.6) | 96.7 | 172 | 4 | 838 (5.4) |
|
| pAwA-sTS | 47590 (7.6) |
21734 (10.7) |
45.7 | 4117 (5.3) |
3985 (5.2) | 96.8 | 130 | 2 | 771 (4.9) |
|
| pAwA-sTD | 43274 (1.6) |
25908 (2.0) |
59.9 | 5676 (2.9) |
5442 (3.0) | 95.9 | 227 | 7 | 959 (4.7) |
|
In order to enhance the solubility of the acetone-precipitated protein pellet while preserving LC-MS/MS compatibility, the 0.5M TEAB dissolution buffer was supplemented with 1% (w/v) Na-DOC. This reagent can be easily removed by precipitation at low pH levels and is considered as a trypsin- and MS-compatible detergent. We therefore introduced this step into our sample preparation workflow and referred to it as Na-DOC assisted protein solubilisation and denaturation. LC-MS/MS analyses of the MDDC proteomic samples prepared from the workflow pAwA-sT, where 0.5M TEAB dissolution buffer was used, and workflow pAwA-sTD, where the 1% Na-DOC was added to the dissolution buffer, generated almost equal numbers of MS/MS spectra (<0.1% difference). However, database search results showed that with 10% (TCL) and 11% (RCL) more spectra that could be confidently matched to distinct peptides, which in turn produced 11% (TCL) and 6% (RCL) more distinct peptides or alternatively 10% (TCL) and 5% (RCL) more fully-cleaved distinct peptides, the addition of Na-DOC in the dissolution buffer increased the number of proteins detectable from pAwA MDDC protein pellets by an average of 19% or 14% , for TCL and RCL respectively, under otherwise the same conditions (Table 1). The identification enhancement was more pronounced for hydrophobic (GRAVY index >0) rather than hydrophilic proteins (GRAVY index <0), yielding an overall increase of 34% and 14% respectively for both types of cell lysates (Supplementary Table S5). Furthermore, as shown in Table 2, the overall sequence coverage of proteins found with both workflows was increased from 20.4% to 21.6% for TCL and from 20.5% to 21.1% for RCL, by using pAwA-sTD workflow. These results are in agreement with our expectation that enhanced tryptic digestion efficiency could be achieved by the utilization of Na-DOC assisted protein dissolution and denaturation.
Table 2.
Protein sequence coverage of the MDDC proteins commonly identified by two sample preparation workflows. MDDC proteins were precipitated by acetone with wash (pAwA) method and the acetone-washed protein pellets were solubilised by 0.5M TEAB dissolution buffer (sT) or 0.5M TEAB with additional 1% Na-DOC (sTD). Proteins were identified with global FDR 1% and associated distinct peptides were identified with local FDR 5%.
| Experimental replicate |
Triton X-100 cell lysate (TCL) | RIPA cell lysate (RCL) | ||||
|---|---|---|---|---|---|---|
| # of proteins | pAwA-sT | pAwA-sTD | # of proteins | pAwA-sT | pAwA-sTD | |
| #1 | 657 | 20.0 | 21.7 | 793 | 20.3 | 20.8 |
| #2 | 725 | 20.7 | 21.6 | 739 | 20.4 | 21.0 |
| #3 | 715 | 20.0 | 21.3 | 751 | 20.3 | 20.4 |
| #4 | 638 | 20.9 | 21.9 | 666 | 21.0 | 22.2 |
| Average | - | 20.4 | 21.6 | - | 20.5 | 21.1 |
The introduction of Na-DOC to the protein solubilisation and denaturation step in the workflow pAwA-sTD compensated for the sample loss caused by acetone wash, and therefore increased the MDDC proteome coverage to a level even higher than what we obtained without the acetone-wash step (pA-sT) (Table 1 and Supplementary Figure S4). The use of Na-DOC also improved the solubility and digestion efficiency of protein pellets that were not subjected to acetone-wash, giving rise to 8% (TCL) or 4% (RCL) and 11% (TCL) or 4% (RCL) more distinct peptides and protein identifications, respectively, compared with the workflow using 0.5M TEBA alone (pA-sT vs. pA-sTD in Supplementary Table S1). Compared with the workflow pAwA-sTD, however, the workflow pA-sTD could not further improve the MDDC proteome coverage even though the protein resolubilisation step in pA-sTD was also facilitated by the higher amount of residual detergent besides the use of Na-DOC in the dissolution buffer. On the contrary, its higher percentage of the non-identifiable MS/MS spectra might hinder the peptide identification rate and thus decrease the number of identified proteins.
The impact of SDS on the MDDC proteomics analysis
SDS is generally used not only to increase the protein solubility, but also to denature proteins for enhanced enzymatic digestion. In this experiment, 0.1% SDS was added to the solubilisation buffer (0.5M TEAB) to assist in the solubilisation and denaturation process. According to the manufacturer’s documentation, this concentration of SDS is compatible with trypsin activity. For comparison purposes, the tryptic digests of MDDC protein extracts prepared by all the different workflows (Figure 2C) were diluted to the same concentration before LC-MS/MS analysis. In these ready-to-inject sample solutions, the concentration of SDS, when present, was approximately 0.004%. Even though, as illustrated in Supplementary Table S1 (e.g. pAwA-sTS vs. pAwA-sT), more MS/MS spectra were obtained from sample solutions resulting from workflows involving this SDS-assisted protein solubilisation and denaturation, the number of spectra leading to confident peptide identifications were comparatively fewer. Since we did observe notable intensity suppression in the TIC traces of SDS-containing sample solutions, the low MS/MS spectrum identification rate could be attributed to the decreased quality of the MS/MS spectra. Data collected from four experimental replicates, as shown in Table 1, indicated that the addition of SDS in the 0.5M TEAB dissolution buffer to dissolve the acetone-washed protein pellet after the protein precipitation step caused an average loss of 25% (TCL) or 23% (RCL) and 13% (TCL) or 8% (RCL) identifications for the distinct peptides and proteins, respectively. Therefore, the potential benefit brought by 0.1% SDS to the protein solubilisation and denaturation process could not compensate for the drawback of interfering with the subsequent LC-MS/MS analysis even when SDS was present in the final sample solution at a level below 0.01%.
Triton X-100 or RIPA as the cell lysis buffer
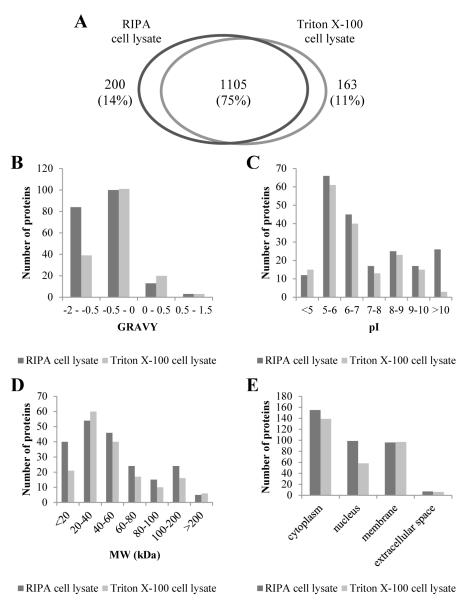
With the exception of a marginal difference in the extraction of the nuclear marker Histone H3 and of the HSP90 chaperone (Figure 1), comparable results were obtained with either Triton X-100 or RIPA lysis buffers with respect to all parameters monitored up to this point, demonstrating highly similar performances of these two lysis buffers in MDDC protein extraction. In order to select one of the two buffers for MDDC proteomic profiling experiments, we therefore concentrated our efforts in the evaluation of the identity of the proteins identified by each of the buffers. To do so we compared the outcome of the pAwA-sTD workflow on four experimental replicates from either TCL or RCL. In addition to 1105 (75% of total) proteins, which were identified in both cell lysates, 163 proteins were found to be uniquely present in the TCL and 200 were unique to RCL respectively (Figure 5A).
Figure 5.
Comparison of the number, biophysical characteristics and expected cellular location of MDDC proteins identified from RIPA cell lysate and Triton X-100 cell lysate. Both cell lysates were precipitated by acetone and the acetone-washed pellets were solubilised by 1% Na-DOC in 0.5M TEAB dissolution buffer (workflow pAwA-sTD). (A) Distribution of all proteins after merging four experimental replicates for each workflow; Uniquely identified proteins in each sample type distributed according to their (B) GRAVY index, (C) calculated isoelectric point (pI), (D) average molecular weight (MW) and (E) expected subcellular localisation based on associated cellular component GO terms.
These unique proteins were grouped according to their GRAVY indexes (Figure 5B), calculated isoelectric points (pI) (Figure 5C), average molecular weights (MW) (Figure 5D) and their expected subcellular localisation in MDDCs based on GO term mapping (Figure 5E). The proteins that were uniquely identified from RCL were found to be mostly hydrophilic proteins with GRAVY values lower than −0.5, very basic with pI values higher than 10 or proteins of relatively smaller size, which is consistent with the behaviour of histones. This represented a 2-, 9- or 2-fold, increase respectively, compared to the proteins identified by Triton X-100 buffer. This result is consistent with what we observed in Figure 1. As a further confirmation of the greater nuclear extraction ability of RIPA with respect to Triton X-100, RIPA buffer provided 71% more predicted nuclear proteins (41 proteins) than Triton X-100 buffer (Figure 5E). The efficient solubilisation of nuclear proteins is highly desirable in proteomic profiling experiments, due to the nuclear localization of several regulatory proteins such as transcription factors. For this reason RIPA buffer was selected for MDDC profiling.
Conclusion
In this study, we examined multiple protein extraction methods as well as multiple LC-MS/MS proteomic sample preparation workflows applied to MDDCs with a focus on the maximization of MDDC proteome coverage, with particular attention to the extraction of nuclear factors. Mechanical disruption of frozen cell pellets followed by immediate thawing and incubation in RIPA buffer gave the overall best results in term of both bulk cellular protein extraction and the solubilisation of proteins representing critical cellular compartments, such as the plasma membrane and the nuclear interior. This result was consistent with the subsequent LC-MS/MS analysis results, which showed that more protein identifications were detected in RIPA cell lysate with substantially increased nuclear proteome coverage if compared to the proteins identified in Triton X-100 cell lysate. The results of LC-MS/MS analysis of MDDC samples prepared using workflows differing in protein precipitation, dissolution and denaturation approaches, revealed that the number of identified distinct peptides could range between 4006 and 5837, while the number of protein identifications could vary from and 506 to 1000. The acetone precipitation method exhibited higher efficiency as compared to the 10% TCA/acetone method, as determined by at least 28% larger MDDC proteome coverage. In addition, the data suggested that the acetone precipitation method led to the recovery of more hydrophobic proteins. The acetone-wash step applied to the precipitated protein pellet further removed the detergent residues, leaving a cleaner protein mixture for downstream LC-MS/MS analysis. However, executing this step resulted in reduced proteome coverage most likely due to the fact that the protein pellet after being washed was less soluble in the 0.5 M TEAB solubilisation buffer. This issue was overcome by the addition of Na-DOC, a trypsin- and MS-compatible surfactant. As an additive used in the dissolution buffer, Na-DOC not only increased the protein identification rate by promoting solubilisation, especially for hydrophobic proteins, but also improved the protein sequence coverage by enhanced digestion efficiency. Since the drawback of protein pellet wash step was compensated by the use of Na-DOC, the workflow including this step was ultimately preferred in order to minimize surfactant contamination (Figure 4) and the detrimental effect it might have on the LC-MS instrument and on sample analysis. On the other hand, the addition of SDS to enhance protein solubilisation and denaturation was not desirable in our hands due to the fact that it interfered with the subsequent LC-MS/MS analysis by generating many more non-identifiable MS/MS spectra and would presumably lead to a reduction in the peptide and protein identification rate. In conclusion, taking all results into account, we propose that the method of choice to prepare MDDC whole cell lysates for LC-MS/MS analysis is cryogenic disruption of cell pellets followed by RIPA protein extraction. For subsequent MS-friendly sample preparation, a precipitation step with acetone should be followed by acetone precipitation, acetone wash of the pellet and by Na-DOC assisted protein solubilisation and denaturation prior to the tryptic digestion.
Supplementary Material
Acknowledgements
This work was supported by the Swiss National Science Foundation Sinergia grant CRSII3_136282 and by National Institutes of Health (USA) grants RO1111809 and DP1DA034990.
Abbreviations
- MS
mass spectrometry
- MDDC
monocyte-derived dendritic cells
- DCs
dendritic cells
- RIPA
Radio Immuno Precipitation Assay
- PBMCs
peripheral blood mononuclear cells
- LC
liquid chromatography
- TCA
trichloroacetic acid
- TEAB
triethylammonium bicarbonate
- TCEP
Tris-(2-carboxyethyl)phosphine
- Na-DOC
sodium deoxycholate
- SDS
sodium dodecyl sulfate
- MMTS
methyl methanethiosulfonate
- TFA
trifluoroacetic acid
- FA
formic acid
- TCL
Triton X-100 cell lysate
- RCL
RIPA cell lysate
- GRAVY
grand average of hydropathy
- IDA
information dependent acquisition
- FDR
false discovery rate
- GO
Gene Ontology
- XIC
extracted ion chromatogram
- TIC
total ion chromatogram
Footnotes
Competing interests statement
The authors declare that they have no competing financial interests.
References
- [1].Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- [2].Vassalli G. Dendritic cell-based approaches for therapeutic immune regulation in solid-organ transplantation. Journal of transplantation. 2013;2013:761429. doi: 10.1155/2013/761429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gross CC, Wiendl H. Dendritic cell vaccination in autoimmune disease. Current opinion in rheumatology. 2013;25:268–274. doi: 10.1097/BOR.0b013e32835cb9f2. [DOI] [PubMed] [Google Scholar]
- [4].Palucka K, Banchereau J. Dendritic-Cell-Based Therapeutic Cancer Vaccines. Immunity. 2013;39:38–48. doi: 10.1016/j.immuni.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Luban J. Innate immune sensing of HIV-1 by dendritic cells. Cell host & microbe. 2012;12:408–418. doi: 10.1016/j.chom.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wu L, KewalRamani VN. Dendritic-cell interactions with HIV: infection and viral dissemination. Nat Rev Immunol. 2006;6:859–868. doi: 10.1038/nri1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Rahman S, Khan ZK, Jain P. The tug-of-war between dendritic cells and human chronic viruses. International reviews of immunology. 2011;30:341–365. doi: 10.3109/08830185.2011.561506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sehgal M, Khan ZK, Talal AH, Jain P. Dendritic Cells in HIV-1 and HCV Infection: Can They Help Win the Battle? Virology: Research and Treatment. 2013;4:1–25. doi: 10.4137/VRT.S11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ferreira GB, Mathieu C, Overbergh L. Understanding dendritic cell biology and its role in immunological disorders through proteomic profiling. Proteomics. Clinical applications. 2010;4:190–203. doi: 10.1002/prca.200900162. [DOI] [PubMed] [Google Scholar]
- [10].Ferreira GB, Overbergh L, van Etten E, Lage K, D'Hertog W, Hansen DA, Maris M, Moreau Y, Workman CT, Waelkens E, Mathieu C. Protein-induced changes during the maturation process of human dendritic cells: A 2-D DIGE approach. Proteomics. Clinical applications. 2008;2:1349–1360. doi: 10.1002/prca.200800110. [DOI] [PubMed] [Google Scholar]
- [11].Le Naour F, Hohenkirk L, Grolleau A, Misek DE, Lescure P, Geiger JD, Hanash S, Beretta L. Profiling changes in gene expression during differentiation and maturation of monocyte-derived dendritic cells using both oligonucleotide microarrays and proteomics. The Journal of biological chemistry. 2001;276:17920–17931. doi: 10.1074/jbc.M100156200. [DOI] [PubMed] [Google Scholar]
- [12].Pereira SR, Faca VM, Gomes GG, Chammas R, Fontes AM, Covas DT, Greene LJ. Changes in the proteomic profile during differentiation and maturation of human monocyte-derived dendritic cells stimulated with granulocyte macrophage colony stimulating factor/interleukin-4 and lipopolysaccharide. Proteomics. 2005;5:1186–1198. doi: 10.1002/pmic.200400988. [DOI] [PubMed] [Google Scholar]
- [13].Rivollier A, Perrin-Cocon L, Luche S, Diemer H, Strub JM, Hanau D, van Dorsselaer A, Lotteau V, Rabourdin-Combe C, Rabilloud T, Servet-Delprat C. High expression of antioxidant proteins in dendritic cells: possible implications in atherosclerosis. Molecular & cellular proteomics : MCP. 2006;5:726–736. doi: 10.1074/mcp.M500262-MCP200. [DOI] [PubMed] [Google Scholar]
- [14].Horlock C, Shakib F, Mahdavi J, Jones NS, Sewell HF, Ghaemmaghami AM. Analysis of proteomic profiles and functional properties of human peripheral blood myeloid dendritic cells, monocyte-derived dendritic cells and the dendritic cell-like KG-1 cells reveals distinct characteristics. Genome biology. 2007;8:R30. doi: 10.1186/gb-2007-8-3-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Sanarico N, Colone A, Grassi M, Speranza V, Giovannini D, Ciaramella A, Colizzi V, Mariani F. Different Transcriptional Profiles of Human Monocyte-Derived Dendritic Cells Infected with Distinct Strains of Mycobacterium tuberculosis and Mycobacterium bovis Bacillus Calmette-Guérin. Clinical and Developmental Immunology. 2011;2011:14. doi: 10.1155/2011/741051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Boukli NM, Saiyed ZM, Ricaurte M, Rodriguez JW, Rios Olivares E, Cubano LA, Nair MP. Implications of ER stress, the unfolded protein response, and pro- and anti-apoptotic protein fingerprints in human monocyte-derived dendritic cells treated with alcohol. Alcoholism, clinical and experimental research. 2010;34:2081–2088. doi: 10.1111/j.1530-0277.2010.01304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Reynolds JL, Mahajan SD, Aalinkeel R, Nair B, Sykes DE, Schwartz SA. Proteomic analyses of the effects of drugs of abuse on monocyte-derived mature dendritic cells. Immunological investigations. 2009;38:526–550. doi: 10.1080/08820130902874110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Reynolds JL, Mahajan SD, Sykes DE, Schwartz SA, Nair MP. Proteomic analyses of methamphetamine (METH)-induced differential protein expression by immature dendritic cells (IDC) Biochimica et biophysica acta. 2007;1774:433–442. doi: 10.1016/j.bbapap.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wang CY, Staniforth V, Chiao MT, Hou CC, Wu HM, Yeh KC, Chen CH, Hwang PI, Wen TN, Shyur LF, Yang NS. Genomics and proteomics of immune modulatory effects of a butanol fraction of echinacea purpurea in human dendritic cells. BMC genomics. 2008;9:479. doi: 10.1186/1471-2164-9-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Fearnley DB, Whyte LF, Carnoutsos SA, Cook AH, Hart DNJ. Monitoring Human Blood Dendritic Cell Numbers in Normal Individuals and in Stem. Cell Transplantation. 1999 [PubMed] [Google Scholar]
- [21].Zhou LJ, Tedder TF. CD14+ blood monocytes can differentiate into functionally mature CD83+ dendritic cells. Proceedings of the National Academy of Sciences. 1996;93:2588–2592. doi: 10.1073/pnas.93.6.2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Duclos S, Clavarino G, Rousserie G, Goyette G, Boulais J, Camossetto V, Gatti E, LaBoissiere S, Pierre P, Desjardins M. The endosomal proteome of macrophage and dendritic cells. Proteomics. 2011;11:854–864. doi: 10.1002/pmic.201000577. [DOI] [PubMed] [Google Scholar]
- [23].Gundacker NC, Haudek VJ, Wimmer H, Slany A, Griss J, Bochkov V, Zielinski C, Wagner O, Stockl J, Gerner C. Cytoplasmic proteome and secretome profiles of differently stimulated human dendritic cells. J Proteome Res. 2009;8:2799–2811. doi: 10.1021/pr8011039. [DOI] [PubMed] [Google Scholar]
- [24].Ge J, Yan H, Li S, Nie W, Dong K, Zhang L, Zhu W, Fan F, Zhu J. Changes in proteomics profile during maturation of marrow-derived dendritic cells treated with oxidized low-density lipoprotein. Proteomics. 2011;11:1893–1902. doi: 10.1002/pmic.201000658. [DOI] [PubMed] [Google Scholar]
- [25].Nicholas D, Tang H, Zhang Q, Rudra J, Xu F, Langridge W, Zhang K. Quantitative Proteomics Reveals a Role for Epigenetic Reprogramming During Human Monocyte Differentiation. Molecular & cellular proteomics : MCP. 2014 doi: 10.1074/mcp.M113.035089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Schlatzer DM, Sugalski J, Dazard JE, Chance MR, Anthony DD. A quantitative proteomic approach for detecting protein profiles of activated human myeloid dendritic cells. Journal of immunological methods. 2012;375:39–45. doi: 10.1016/j.jim.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Abdallah C, Dumas-Gaudot E, Renaut J, Sergeant K. Gel-Based and Gel-Free Quantitative Proteomics Approaches at a Glance. International Journal of Plant Genomics. 2012;2012:17. doi: 10.1155/2012/494572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Rundlett KL, Armstrong DW. Mechanism of Signal Suppression by Anionic Surfactants in Capillary Electrophoresis–Electrospray Ionization Mass Spectrometry. Analytical Chemistry. 1996;68:3493–3497. doi: 10.1021/ac960472p. [DOI] [PubMed] [Google Scholar]
- [29].Zhang N, Li L. Effects of common surfactants on protein digestion and matrix-assisted laser desorption/ionization mass spectrometric analysis of the digested peptides using two-layer sample preparation. Rapid communications in mass spectrometry : RCM. 2004;18:889–896. doi: 10.1002/rcm.1423. [DOI] [PubMed] [Google Scholar]
- [30].Botelho D, Wall MJ, Vieira DB, Fitzsimmons S, Liu F, Doucette A. Top-Down and Bottom-Up Proteomics of SDS-Containing Solutions Following Mass-Based Separation. Journal of Proteome Research. 2010;9:2863–2870. doi: 10.1021/pr900949p. [DOI] [PubMed] [Google Scholar]
- [31].Lin Y, Liu H, Liu Z, Liu Y, He Q, Chen P, Wang X, Liang S. Development and evaluation of an entirely solution-based combinative sample preparation method for membrane proteomics. Analytical biochemistry. 2013;432:41–48. doi: 10.1016/j.ab.2012.09.023. [DOI] [PubMed] [Google Scholar]
- [32].Chen EI, Cociorva D, Norris JL, Yates JR., 3rd Optimization of mass spectrometry-compatible surfactants for shotgun proteomics. J Proteome Res. 2007;6:2529–2538. doi: 10.1021/pr060682a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lin Y, Zhou J, Bi D, Chen P, Wang X, Liang S. Sodium-deoxycholate-assisted tryptic digestion and identification of proteolytically resistant proteins. Analytical biochemistry. 2008;377:259–266. doi: 10.1016/j.ab.2008.03.009. [DOI] [PubMed] [Google Scholar]
- [34].Proc JL, Kuzyk MA, Hardie DB, Yang J, Smith DS, Jackson AM, Parker CE, Borchers CH. A Quantitative Study of the Effects of Chaotropic Agents, Surfactants, and Solvents on the Digestion Efficiency of Human Plasma Proteins by Trypsin. Journal of Proteome Research. 2010;9:5422–5437. doi: 10.1021/pr100656u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lin Y, Liu H, Liu Z, Wang X, Liang S. Shotgun analysis of membrane proteomes using a novel combinative strategy of solution-based sample preparation coupled with liquid chromatography-tandem mass spectrometry. Journal of chromatography. B, Analytical technologies in the biomedical and life sciences. 2012;901:18–24. doi: 10.1016/j.jchromb.2012.05.035. [DOI] [PubMed] [Google Scholar]
- [36].Leon IR, Schwammle V, Jensen ON, Sprenger RR. Quantitative assessment of in-solution digestion efficiency identifies optimal protocols for unbiased protein analysis. Molecular & cellular proteomics : MCP. 2013;12:2992–3005. doi: 10.1074/mcp.M112.025585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Reinhard C, Bottinelli D, Kim B, Luban J. Vpx rescue of HIV-1 from the antiviral state in mature dendritic cells is independent of the intracellular deoxynucleotide concentration. Retrovirology. 2014;11:12. doi: 10.1186/1742-4690-11-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Shilov IV, Seymour SL, Patel AA, Loboda A, Tang WH, Keating SP, Hunter CL, Nuwaysir LM, Schaeffer DA. The Paragon Algorithm, a Next Generation Search Engine That Uses Sequence Temperature Values and Feature Probabilities to Identify Peptides from Tandem Mass Spectra. Molecular & Cellular Proteomics. 2007;6:1638–1655. doi: 10.1074/mcp.T600050-MCP200. [DOI] [PubMed] [Google Scholar]
- [39].Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. Journal of Molecular Biology. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- [40].Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. GO::TermFinder--open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.