Summary
The envelope glycoprotein trimer mediates HIV-1 entry into cells. The trimer is flexible, fluctuating between closed and more open conformations and sometimes sampling the fully open, CD4-bound form. We hypothesized that conformational flexibility could hinder the induction of broadly neutralizing antibodies (bNAbs). We therefore modified soluble Env trimers to stabilize their closed, ground states. The trimer variants were indeed stabilized in the closed conformation, with a reduced ability to undergo receptor-induced conformational changes and a decreased exposure of non-neutralizing V3-directed antibody epitopes. In rabbits, the stabilized trimers induced similar autologous Tier-1B or Tier-2 NAb titers to those elicited by the corresponding wild-type trimers, but lower levels of V3-directed Tier-1A NAbs. Stabilized, closed trimers might therefore be useful components of vaccines aimed at inducing bNAbs.
Introduction
The mature, proteolytically cleaved envelope glycoprotein trimer (Env) mediates HIV-1 entry into target cells by undergoing a complex series of conformational changes initiated by binding to the CD4 receptor and the CCR5 or CXCR4 co-receptor. Defining how Env functions during cell entry has major implications for the rational, structure-guided design of trimer-based vaccines aimed at inducing broadly neutralizing antibodies (bNAbs) against highly divergent primary HIV-1 strains. One promising approach is to use recombinant, soluble trimers (Sanders et al., 2013, 2015) as tools to increase our understanding of these coordinated conformational changes, via X-ray and cryo-EM structures and biophysical analyses (Guttman et al., 2014; Julien et al., 2013a; Do Kwon et al., 2015; Lyumkis et al., 2013; Munro et al., 2014; Pancera et al., 2014).
Creating a soluble, native-like trimer is complicated by the instability of the association between the gp120 and gp41 subunits, and between the individual gp120-gp41 protomers. The native trimer is inherently metastable because it must undergo profound conformational rearrangements during virus entry (Sanders and Moore, 2014). One successful stabilization strategy involves introduction of an intermolecular disulfide bond (SOS) to link gp120 and gp41, a point substitution (I559P, i.e. IP) to maintain the gp41 subunits in their prefusion form, and truncation at residue 664 to improve trimer solubility (Binley et al., 2000, 2002; Klasse et al., 2013; Sanders et al., 2002, 2013). The resulting trimers are termed SOSIP.664. Soluble SOSIP trimers based on the clade A BG505 env gene have been used to generate high resolution X-ray and cryo-EM structures (Julien et al., 2013a; Lyumkis et al., 2013; Pancera et al., 2014; Scharf et al., 2015), to isolate new bNAbs that recognize quaternary structure-dependent epitopes, and to characterize known bNAbs (Blattner et al., 2014; Doria-Rose et al., 2014; Huang et al., 2014; Julien et al., 2013b, 2013c; Lee et al., 2015; Sanders et al., 2013; Sok et al., 2014). In addition to BG505, native-like SOSIP.664 trimers have also been produced from the B41, ZM197M and DU422 clade B or C env genes, as well as other sequences (Guenaga et al., 2015; Julien et al., 2015; Pugach et al., 2015; Ringe et al., 2015; Sharma et al., 2015). As immunogens, the BG505 and B41 SOSIP.664 trimers induce NAbs to the neutralization-resistant (Tier-2) autologous virus in rabbits and/or macaques (Sanders et al., 2015).
While the induction of consistent NAb responses against the autologous Tier-2 viruses by the BG505 and B41 SOSIP.664 trimers is an unprecedented achievement, it is the first among several steps towards the induction of bNAbs. It is highly unlikely that any single Env antigen will induce bNAbs. Instead it is probably necessary to devise more sophisticated vaccination regimens that include germline targeting, evolutionary lineages, multivalent immunogens, alone or more likely in combination (Doria-Rose et al., 2014; Dosenovic et al., 2015; Haynes et al., 2012; Jardine et al., 2015; Liao et al., 2013; McGuire et al., 2014; Sliepen et al., 2015).
Limiting the exposure of non-NAb epitopes is also likely to be be necessary for optimal immunogenicity. On BG505 SOSIP.664, non-NAb epitopes in V3 are particularly immunogenic (Sanders et al., 2015). Non-NAbs and narrow specificity NAbs have been proposed to interfere with the induction of bNAbs against many pathogens, including influenza, malaria, HIV-1, and others (Chaudhury et al., 2014; Eggink et al., 2013; Garrity et al., 1997; Hall and Joiner, 1991; Marrack and Kappler, 1994; McGuire et al., 2014; Novotny and Bakaletz, 2003). One mechanistic explanation for this phenomenon is that high affinity non-NAbs may enter germinal centers and block antigen binding to lower affinity B cell receptors with specificity for bNAb epitopes (McGuire et al., 2014; Zhang et al., 2013). In an in vitro study, McGuire et al. showed that when HIV-1 Env antigens were added to a mixture of B cells bearing the germline precursors of bNAbs and non-NAbs, including those for V3 non-NAbs, the latter were preferentially activated. However, modification of the Env antigen to reduce non-NAb epitope presentation (including deletion of the V3 region) facilitated the activation of lower affinity bNAb precursors (McGuire et al., 2014). Refocusing Ab responses away from V3 towards other epitopes has also been reported (Garrity et al., 1997). Moreover, Eggink et al. showed that inhibiting the immunogenicity of the influenza hemagglutinin head domain enhanced responses to the normally subdominant stalk domain, such that more broadly reactive NAbs were elicited (Eggink et al., 2013). Overall, it is highly plausible that limiting the exposure of non-NAb epitopes on native-like Env trimers will improve their use as a platform for inducing bNAbs.
The native Env trimer and its SOSIP.664 mimics are conformationally flexible; they can “breathe” such that they fluctuate between closed and more open forms in a dynamic equilibrium (Guttman et al., 2015; Harris et al., 2013; Munro et al., 2014; Pugach et al., 2015; Scharf et al., 2015). While unliganded BG505 SOSIP.664 trimers have a high propensity to remain in the closed, ground state conformation, the equilibrium for their B41, DU422 and ZM197M counterparts is shifted. Thus, a greater proportion of partially open, but still fully native-like, trimers can be visualized by negative stain electron microscopy (NS-EM) (Julien et al., 2015; Pugach et al., 2015). Another manifestation of this conformational isomerism is the binding of some non-NAbs to a small fraction (presumably the partially open forms) of the SOSIP.664 trimer population; these non-NAbs recognize V3, CD4-induced (CD4i) and a subset of CD4-binding site (CD4bs) epitopes, which are all normally only accessible on neutralization-sensitive (Tier 1) viruses (Sanders et al., 2013). How conformational flexibility influences the immunogenicity of Env trimers is not yet known. However, we hypothesize that a trimer stabilized in the closed, unliganded state would allow the antibody response to be focused on the desired NAb epitopes with minimal induction of unwanted, and perhaps even interfering or distractive, non-NAbs, such as those directed against V3.
Here, we describe the modification of SOSIP.664 trimers to reduce the rate or extent of “breathing” and impede conformational changes involved in the transition to more open conformations that expose non-NAb epitopes.
Results
AMC008 SOSIP.664 trimers as a basis for studying stabilization strategies
For initial studies of how to improve the frequency of closed SOSIP.664 trimers at the expense of partially open or non-native forms, we selected the AMC008 clade B env gene, an early sequence from an elite neutralizer enrolled in the Amsterdam Cohort Studies on HIV/AIDS, as the test case. The parental AMC008 virus was categorized as a Tier-1B virus by the Duke Central Laboratory, suggesting that the native trimer on the virus is relatively open and could expose V3- and/or CD4i-epitopes (not shown). When the AMC008 SOSIP.664 Env proteins were expressed, ~50% migrated as trimers on a BN-PAGE gel (Fig. S1A), but only ~35% of the 2G12/SEC-purified AMC008 trimers were visibly native-like in NS-EM images (Fig. S2C). We have previously described how unliganded native-like SOSIP.664 trimers can exist in two conformations that are distinguishable by NS-EM: a compact “closed” conformation and partially open conformation (Pugach et al., 2015). While the partially open trimers that we have described have a higher degree of loop and/or domain mobility, no major decrease in density at the center of the trimer could be observed, suggesting that the protomers do not become separated at the interface between them. Additionally, the transition from the closed to the partially open conformation is at least partially reversible, and in a thermodynamic equilibrium (Munro et al., 2014). These rapidly interchanging configurations differ from the open conformation described by Scharf et al., which is more analogous to the CD4-induced fully open state that arises when the trimer undergoes a major reorganization and that is likely to be irreversible (Harris et al., 2013; Scharf et al., 2015).
We found that ~25% of the AMC008 SOSIP.664 trimers were in the closed, native-like form, with ~10% partially open, but the remaining ~65% adopted heterogeneous, splayed out shapes that resemble uncleaved, non-native gp140 proteins (Pritchard et al., 2015a; Ringe et al., 2013). The midpoint of thermal denaturation (Tm), determined by differential scanning calorimetry (DSC), for AMC008 SOSIP.664 trimers was 59.6°C, which is much lower than for their BG505 counterparts (67.3°C; Table 1). Finally, the antigenicity profile of the AMC008 SOSIP.664 trimers showed that they bound quaternary structure-dependent bNAbs very poorly, but several non-NAbs efficiently (Data S1A). As such defects are fairly typical of what we have seen with many Env sequences, the AMC008 SOSIP.664 trimers seemed suitable for testing structure-guided stabilization strategies. To assess their general applicability, we also tested the same strategies on the prototype BG505, B41 and ZM197M trimers.
Table 1. Biophysical properties of stabilized AMC008, BG505, B41 and: ZM197M SOSIP.664 trimers.
| NS-EM | DSC | Glycan analysis | ||||||
|---|---|---|---|---|---|---|---|---|
| Stabilizing mutations / SOSIP version |
Native- like trimers |
Closed Native- like trimers |
Thermo stability (Tm, °C) |
Thermo stability (ΔTm) |
Man8 glycans |
Man9 glycans |
Oligo- mannose glycans |
|
|
AMC 008
SOSIP.664 |
v2,(2G12-SEC) | ~35%a | ~25%a | 59.6a | - | |||
| v2 (PGT145) | >98% | ~15% | 60.2 | - | 14% | 34% | 67% | |
| v3 | >98% | ~15% | 61.6 | 1.4 | ||||
| v4.1 | >98% | ~50% | 64.5 | 4.3 | ||||
| v4.2 | >98% | ~85% | 64.0 | 3.8 | 12% | 39% | 69% | |
| BG505,SOSIP.664 | v2,(2G12-SEC) | >98%b | ~90%b | 67.3c | - | 13%d | 25%d | 56%d |
| v2 (PGT145) | >98% | ~35% | 66.7 | - | 19% | 25% | 66% | |
| E64K | >98% | ~85% | 67.9 | 1.2 | ||||
| H66R | >98% | ~30% | 67.5 | 0.8 | ||||
| A316W | >98% | ~80% | 69.0 | 2.3 | ||||
| v4.1 | >98% | ~70% | 69.5 | 2.8 | 21% | 23% | 64% | |
| v4.2 | >98% | ~55% | 69.3 | 2.5 | ||||
|
B41
SOSIP.664 |
v2,(2G12-SEC) | >98%b | ~60%b | 57.6b | - | |||
| v2 (PGT145) | >98% | ~45% | 58.7 | 15% | 29% | 63% | ||
| v4.1 | >98% | ~55% | 61.7 | 3.0 | 14% | 32% | 64% | |
| v4.2 | >98% | ~50% | ND | |||||
|
ZM197M
SOSIP.664 |
v2,(2G12-SEC) | >90%b | ~85%b | 62.7b | - | |||
| v2 (PGT145) | >95% | ~15% | 62.2 | - | ||||
| v4.1 | >95% | ~45% | ND | |||||
| v4.2 | >90% | ~30% | 62.6 | 0.4 | ||||
Values for AMC008 SOSIP.664 expressed in 293S cells
Values derived from previous studies (Julien et al., 2015; Pugach et al., 2015; Sanders et al., 2013)
Tm for 2G12/SEC purified BG505 SOSIP.664-D7324 expressed in 293F cells. Previously, a Tm of 68.1°C was reported for BG505 SOSIP.664 expressed in 293S cells (Sanders et al., 2013).
Glycan profile of BG505 SOSIP.664 expressed in 293T cells (Pritchard et al., 2015a)
The biophysical properties of 293F cell-expressed, PGT145-purified SOSIP.664-D7324 trimers were assessed using NS-EM to determine native-like trimer formation, DSC to quantify thermostability (Tm), and glycan profiling to measure glycan content. For comparison, previously reported Tm values and glycan profiles for 2G12-SEC purified SOSIP.664 trimers are included (Julien et al., 2015; Pritchard et al., 2015a; Pugach et al., 2015; Sanders et al., 2013). The unprocessed EM, DSC and glycan profiling data are shown in Fig. S2 and S3. The DSC data were fitted using both two state and non-two state models (Fig. S3B). The Tm values based on two-state model fitting are listed here, while values based on the alternative model fitting are in Table S1.
Positive selection of native-like trimers by PGT145 purification
We first investigated whether we could isolate native-like AMC008 SOSIP.664 trimers by affinity chromatography using the quaternary structure-dependent bNAb PGT145, which does not recognize non-native Env proteins (Pugach et al., 2015; Ringe et al., 2015). Even without a subsequent SEC step, PGT145-purification yielded homogeneous, fully cleaved trimers (Fig. S2A&B) that were ~100% native-like when visualized by NS-EM (Fig. S2C). Oligomannose glycoforms dominated (~67%) the glycan profile. The high Man8GlcNAc2 and Man9GlcNAc2 content (~14% and ~34%, respectively) is a hallmark of native trimers (Table 1; Fig. S3D) (Bonomelli et al., 2011; Pritchard et al., 2015a, 2015b). However, NS-EM imaging also showed that most (~85%) of the PGT145-purified trimers were in a partly open conformation (Table 1, Fig. S2C), and their thermal stability was not substantially improved (Tm = 60.2°C; Table 1).
PGT145 purification also yielded a homogeneous population of fully native-like BG505 SOSIP.664 trimers (Fig. S3A). The proportion of trimers in the closed conformation however was reduced to ~35%, compared to >95% after 2G12/SEC purification. Thus, binding to and/or elution from PGT145 causes the BG505 trimers to partially open up, which may be attributable to the known, albeit quite subtle, allosteric effects of PGT145 on the trimer structure (Derking et al., 2015). The stability of these PGT145-purified BG505 trimers (Tm = 66.7°C) was also subtly reduced compared to 2G12/SEC (Tm = 67.3°C; Table 1), but their glycan content (Table 1) (Pritchard et al., 2015a) and antigenicity profiles were both unaltered, compared to 2G12/SEC-purified trimers (Derking et al., 2015). B41 SOSIP.664 trimers, on the other hand, were similar after purification by PGT145 or 2G12/SEC, in that they were partly open (45-60%) in both cases (Pugach et al., 2015). For ZM197M, the closed trimer content was reduced from 80% (2G12/SEC) to ~15% (PGT145) (Table 1, Fig. S2A-C) (Julien et al., 2015).
Mutation of gp41 residues 535 and 543: SOSIP.v3 trimers
A previous comparison of JR-FL, a clade B Env that forms SOSIP trimers inefficiently, with KNH1144, a clade A Env that does so more efficiently, showed that a methionine at position 535 (M535) and glutamine at 543 (Q543) in HR1 of gp41 benefit trimer formation (Fig. 1E, Table S2) (Dey et al., 2008). Similar findings were made when HIV-1 was cultured under harsh conditions (Leaman and Zwick, 2013). We noted that the BG505 sequence contains M535 and N543. Based on the high quality of BG505 trimers, we considered that N543 might have the same stabilizing effect as Q543 on the gp41 prefusion structure. In contrast, the AMC008 sequence contains neither M535 nor N/Q543, but instead the unfavorable I535 and L543 (Fig. 1E, Table S2). We therefore constructed AMC008 SOSIP.664 trimer mutants containing one or more of the I535M, L543N and L543Q substitutions (Fig. S1A). All three individual changes improved trimer formation, which was also the case for the I535M+L543N combination (Fig. S1A). When this double mutant was PGT145-purified, NS-EM analysis showed the percentage of closed vs. open trimers was unchanged (~15% closed; Table 1), but its thermostability was slightly increased from 60.2°C (wild-type) to 61.6°C (Table 1). The I535M+L543N substitutions also modestly improved binding of bNAbs PGT145, 35O22 and PGT151, but otherwise did not affect trimer antigenicity (Table 2, Fig. Data S1A&E).
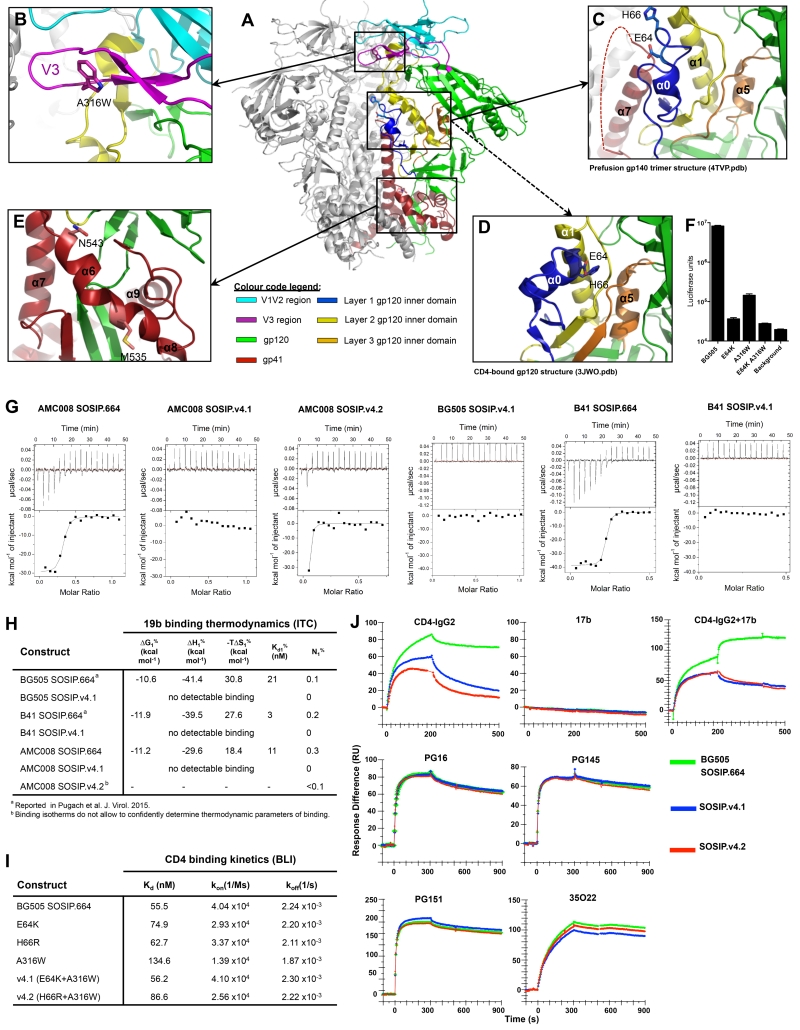
Figure 1. Design of amino-acid substitutions to stabilize SOSIP.664 trimers.
(A) Crystal structure of BG505 SOSIP.664 trimer (Pancera et al., 2014), showing the locations of amino-acid substitutions relevant to this study. One protomer is colored according to sub-regions: gp41 in red; V1V2 in cyan; V3 in purple; gp120 inner domain layer 1 in blue; layer 2 in yellow; layer 3 in orange; outer domain and N- and C- termini of gp120 in green. (B) Detailed view of V3 and surrounding regions showing the A316W substitution. (C) Detailed view of layers 1 and 2 of the gp120 inner domain, showing how E64 and H66 face the unstructured region of gp41 (residues 548-568) between α6 and α7. (D) Detailed view of a CD44-liganded gp120 monomer structure (Pancera et al., 2010) highlighting the same region as in (C) and showing how the E64 and H66 side chains face gp120 layer 2. (E) Depiction of how the α6-α9 helices of gp41 encircle the N- and C-termini of gp120, with M535 and N543 located in the middle and at the C-terminus of α6, respectively. (F) Infection of TZM-bl cells by BG505.T332N Env-pseudoviruses with an A316W, E64K or H66R substitution. (G) V3 Fab 19b binding to AMC008, BG505 and B41 SOSIP.664 and SOSIP.v4 trimers was assessed by ITC. The enthalpy changes (ΔH), dissociation constants (KD) and stoichiometries of binding (molar ratio; N) are listed in panel (H). (I) Biolayer interferometry analysis of CD4 binding to AviB-tagged mutant wild-type BG505 SOSIP.664 trimers. (J) SPR analysis of ligand binding to BG505 SOSIP.664-His trimers and both versions of SOSIP.v4-His trimers. Top row: CD4- IgG2 (left panel), 17b (middle panel), CD4-IgG2 followed by 17b (right panel); bottom row, PG16, PGT145, PGT151 and 35O22. Antibodies and CD4-IgG2 were injected at 500 nM. See also figure S1, table S2 and S3.
Table 2. Antigenicity of stabilized AMC008, BG505, B41 and ZM197M SOSIP trimers.
| bNAbs | Non-NAbs | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| V1V2 Apex | V3 glycan |
OD glycan |
CD4 binding site |
Gp120-41 interface |
CD4i | V3 | CD4 bs |
||||||||||||
| PG9 | PG16 | PGT 145 |
PGT 121 |
PGT 126 |
2G12 | PGT 135 |
VRC 01 |
CH 103 |
CD4 IgG2 |
PGT 151 |
35 O22 |
17b | 17b +sCD4 |
14e | 19b | b6 | |||
| SOSIP.664 constructs | AMC008 | WT | ND | ND | 0.19 | 0.25 | 0.02 | 0.03 | 0.56 | 0.06 | 0.05 | 0.06 | 0.40 | 0.47 | 0.11 | 0.01 | 0.01 | 0.01 | 0.04 |
| v3.2 | ND | ND | 137 | 140 | 110 | 105 | 60 | 129 | 69 | 119 | 166 | 185 | 105 | 104 | 95 | 109 | 113 | ||
| v4.1 | ND | ND | 194 | 180 | 111 | 100 | 84 | 174 | 91 | 81 | 236 | 254 | 0 | 12 | 21 | 42 | 35 | ||
| v4.2 | ND | ND | 133 | 116 | 79 | 70 | 26 | 117 | 74 | 68 | 154 | 158 | 0 | 7 | 15 | 20 | 25 | ||
| BG505 | WT | 0.18 | 0.61 | 0.03 | 0.01 | 0.01 | 0.02 | 0.06 | 0.02 | 0.06 | 0.02 | 0.01 | 0.1 | 0.18 | 0.004 | 0.02 | 0.07 | 0.13 | |
| E64K | 179 | 206 | 167 | 70 | 131 | 97 | 117 | 137 | 150 | 65 | 102 | 85 | 0 | 23 | 96 | 102 | 114 | ||
| H66R | 145 | 182 | 216 | 78 | 92 | 98 | 169 | 179 | 162 | 68 | 112 | 137 | 0 | 31 | 112 | 102 | 123 | ||
| A316W | 161 | 197 | 137 | 56 | 82 | 98 | 171 | 126 | 116 | 81 | 132 | 142 | 73 | 63 | 22 | 55 | 97 | ||
| v4.1 | 100 | 143 | 145 | 66 | 94 | 90 | 81 | 90 | 77 | 26 | 90 | 173 | 0 | 7 | 13 | 67 | 34 | ||
| v4.2 | 114 | 116 | 329 | 74 | 74 | 121 | 76 | 87 | 89 | 28 | 91 | 243 | 0 | 8 | 14 | 71 | 22 | ||
| B41 | WT | 0.37 | 1.1 | 0.9 | 1.31 | 0.06 | 0.05 | 0.09 | 0.04 | 0.24 | 0.03 | ND | 1.72 | 0.16 | 0.01 | 0.01 | 0.01 | 0.05 | |
| v4.1 | 69 | 95 | 141 | 122 | 44 | 73 | 27 | 89 | 72 | 38 | ND | 168 | 0 | 7 | 7 | 13 | 11 | ||
| v4.2 | 132 | 108 | 159 | 132 | 98 | 101 | 44 | 119 | 88 | 38 | ND | 218 | 0 | 11 | 8 | 17 | 10 | ||
| ZM19M | WT | ND | 0.47 | 0.34 | 0.04 | ND | 0.49 | ND | 0.03 | ND | 0.05 | 0.3 | ND | 0.04 | 0.01 | 0.04 | 0.65 | 0.87 | |
| v4.1 | ND | 96 | 76 | 72 | ND | 72 | ND | 61 | ND | 62 | 78 | ND | 0 | 11 | 10 | 81 | 65 | ||
| v4.2 | ND | 132 | 143 | 90 | ND | 70 | ND | 65 | ND | 46 | 216 | ND | 0 | 11 | 8 | 68 | 53 | ||
Binding of bNAbs and non-NAbs to PGT145-purified SOSIP.v4 and SOSIP.664 trimers was assessed by D7324-capture ELISA (Data S1E-H). Half maximal binding concentrations (EC50, in μg/ml) or area under the curve (AUC) values are shown. AUC values were used for some bNAbs and non-Nab 17b because plateau values differed substantially (>20%) between trimer variants or binding curves did not reach plateau levels (17b). Values determined with AUC values are underlined. Ab binding to each SOSIP.v4 trimer is expressed as a percentage of binding to the corresponding SOSIP.664 trimer (= 100%). Whenever possible, the comparison was based on EC50 values. OD glycan is a reference to outer domain glycans. The experimental error is within ± 20% of the recorded value.
The B41 and ZM197M sequences also both lack one of the beneficial amino acids at positions 535 and 543, so we introduced the L543N substitution into B41 and V535M into ZM197M. The L543N substitution improved PGT145 binding to B41 SOSIP.664 trimers in ELISA (Data S1C). From here on, we refer to SOSIP.664 proteins containing the optimal amino acids at position 535 (i.e. methionine) and 543 (i.e. asparagine or glutamine) as SOSIP version 3 (SOSIP.v3). Specifically, SOSIP.v3.1 contains the 543Q change, and SOSIP.v3.2 contains 543N. For more details on nomenclature, see Table S3.
Reducing V3 exposure by enhancing hydrophobic packing
Stabilizing the closed conformation of SOSIP.664 trimers requires the V3 region to be sequestered and the exposure of non-NAb epitopes (for Tier-2 viruses), including CD4i epitopes, to be reduced. Although the V3 region is tucked beneath the V1V2 domain in the BG505 SOSIP.664 structure, it can become transiently exposed on some or all trimers as they breathe in vitro (Sanders et al., 2013). V3 is also immunogenic in vivo (Sanders et al., 2015). One way to gauge V3 exposure is MAb reactivity with D7324-tagged SOSIP.664 trimers under ELISA conditions in which V3 epitopes become readily accessible (Sanders et al., 2013).
We designed an A316W substitution to strengthen hydrophobic interactions at the apex, thereby decreasing the propensity for V3 to flip out of its ground-state location (Fig. 1A&B). For AMC008 and BG505 SOSIP.664 proteins, the A316W change improved trimer formation and also increased the thermostability of the BG505 trimers as assessed by an assay that can be used with unpurified Env proteins (Fig. S1D-G). The V3 non-NAbs 447-52D, 39F, 14e and 19b bound less well to all the A316W variant trimers compared to wild-type (Data S1E-H). For example, 14e and 19b binding to PGT145-purified BG505 SOSIP.664 A316W trimers was reduced by ~80% and ~50%, respectively; similarly, binding of the CD4i non-NAbs 17b and 412d was ~50% lower (Table 2, Data S1F). As the A316W substitution did not affect binding of the same non-NAbs to isolated V3 peptides, we conclude that it works indirectly on the trimer by impeding exposure of V3 epitopes (Fig. S1H). In contrast, the A316W substitution had no adverse effect on the binding of multiple bNAbs to the AMC008, BG505, B41 and ZM197M SOSIP.664 trimers (Data S1E-H). Other bulky hydrophobic amino acids (Tyr/Phe/Val/Ile) also decreased V3 exposure (>3-fold), but other substitutions had little effect (Fig. S1I).
When BG505 SOSIP.664 A316W trimers were purified on PGT145 columns and viewed by NS-EM, ~80% were in the closed conformation, an increase compared to the ~35% value for the wild-type BG505 trimers. Their thermostability was also greater (Tm = 69.0°C vs. 66.7°C; Table 1). When introduced into the BG505 Env-pseudovirus, the A316W substitution reduced infectivity by ~98%, implying either that conformational flexibility is important during entry or that residue A316 plays a direct role in co-receptor binding (Fig. 1F).
Reducing spontaneous sampling of the CD4-bound conformation
We next evaluated two substitutions (E64K and H66R) in layer 1 of the gp120 inner domain that were identified in a study of how HIV-1 becomes resistant to, and eventually dependent on, the entry inhibitor VIR165 (Fig. 1A, C, D) (D.E., S.W.dT., I.B., P.J.K., B.B., R.W.S., unpublished data). In brief, the resistance changes appear to impede native trimers from assuming the CD4-bound conformation, but when VIR165 is present the trimers can undergo CD4-induced conformational changes. In the unliganded BG505 SOSIP.664 trimer, the side chains of residues 64 and 66 are positioned to interact with HR1 of gp41 (Finzi et al., 2010; Do Kwon et al., 2015; Pancera et al., 2014), but the high crystallographic B-values around residues 64 and 66 imply some local flexibility (Fig. 1C) (Pancera et al., 2014). Hydrogen-deuterium exchange (HDX) studies confirmed that this region does not adopt a stable secondary structure, but becomes ordered after CD4-binding (Guttman et al., 2014). Indeed, in the structure of a CD4-liganded gp120 core protein, the side chains of residues 64 and 66 have reoriented to interact with gp120 layer 2 (Fig. 1D) (Pancera et al., 2010). Thus, these residues may be part of a switch that transduces CD4-induced conformational changes from the CD4bs and layers 3 and 2 of gp120 to gp41 via layer 1. We hypothesize that the E64K and H66R substitutions impede this allosteric process.
Exploratory studies on unpurified culture supernatants indicated that the E64K and H66R changes each substantially reduced, or even eliminated, the binding of the CD4i non-NAbs to AMC008, BG505, B41 and ZM197M Env proteins (Fig. S1J&K, Data S1A-D). When mutant BG505 SOSIP.664 trimers were PGT145-purified and visualized by NS-EM, the percentage of closed trimers was substantially increased for the E64K mutant (~85% closed vs. 35% for wild-type), but not H66R (~30% closed; Table 1). Each substitution had only a minor effect on thermostability (Tm = 67.9°C for E64K, 67.5°C for H66R vs. 66.7°C for wild-type). In an ELISA, bNAb binding was unaltered or sometimes enhanced (e.g., for the quaternary structure-dependent bNAbs PG16 and PGT145), whereas the 17b and 412d non-NAbs no longer bound (Table 2, Data S1F).
Combining A316W with E64K or H66R substitutions: SOSIP.v4 trimers
The E64K or H66R substitutions were combined with A316W to make double mutant AMC008, BG505, B41 and ZM197M variants based on the SOSIP.v3 design (Table S3). In each case, pilot studies on unpurified Env proteins (culture supernatants) indicated that the E64K+A316W and H66R+A316W double mutants from all four genotypes had acquired the beneficial properties associated with each individual change. Each double mutant was expressed efficiently, formed fully cleaved trimers and was slightly more thermostable than wild-type (Fig. S1A-G). Moreover, in all four cases, the binding of non-NAbs to CD4i-epitopes and V3 epitopes was diminished compared to wild type, or even abolished, while bNAb epitopes were mostly unaffected (Data S1A-D).
For simplicity, we here introduce the following nomenclature: SOSIP.v4 trimers contain the optimal amino acids at position 535 and 543 (SOSIP.v3; see above) and, in addition, the A316W change and either the E64K (designated SOSIP.v4.1) or the H66R (SOSIP.v4.2) substitution (Table S3).
Biophysical properties of stabilized SOSIP.v4 trimers
The various PGT145-purified SOSIP.v4 trimers were fully cleaved and highly homogeneous, as assessed by reducing and non-reducing SDS-PAGE and BN-PAGE analysis (Fig. S2A&B). The proportion of closed trimers was increased in each case compared to wild-type, most notably for AMC008 SOSIP.v4.2 (~85% closed vs. ~15%; Table 1, Fig. S2C). Except for ZM197M SOSIP.v4 (no change), the SOSIP.v4 trimers were also more thermostable by 2-4°C, compared to wild-type (Table 1, Fig. S3B). The glycosylation profiles differed little from their wild-type counterparts, with oligomannose glycans again predominating (Table 1, Fig. S3D). Dynamic light scattering (DLS) experiments showed that, like the BG505 wild-type trimers, both SOSIP.v4 variants were monodisperse and had the same hydrodynamic radius (Rh) of ~69 Å (Fig. S3C). Furthermore, in small angle x-ray scattering (SAXS) studies, these trimers were all well folded, with a radius of gyration (Rg) of ~53 Å (Fig. S3C).
Antigenicity of stabilized SOSIP.v4 trimers
D7324-tagged versions of wild-type (i.e., SOSIP.664) and stabilized trimer variants were PGT145 affinity-purified and studied by ELISA. The AMC008, BG505, B41, ZM197M SOSIP.v4 trimers all retained the ability to bind bNAbs (2G12, PGT135, PGT121, PGT126, PGT145, 35O22, VRC01 and CH103) (Table 2, Data S1E-H). This outcome was confirmed by surface plasmon resonance (SPR) for BG505 SOSIP.v4 and the quaternary structure-dependent bNAbs PG16, PGT145, 35O22 and PGT151 (Fig. 1J). However, in ELISA, there was a marked reduction in binding of the V3 non-NAbs 14e and 19b and the CD4bs non-NAb b6, while the CD4i non-NAbs 17b and 412d bound to a negligible extent (Table 2, Data S1E-H). Furthermore, although the SOSIP.v4 trimers still bound CD4-IgG2, they did not undergo CD4-induced conformational changes efficiently, as judged by the lack of induction of the 17b and 412d CD4i epitopes (Table 2, Data S1E-H).
In previous isothermal titration calorimetry (ITC) studies, the V3 non-NAb 19b IgG bound minimally to the BG505 SOSIP.664 trimers, with a stoichiometry of 0.2 (i.e., on average one IgG per 5 trimers), which suggest that a subpopulation of the trimers exposes V3, or that all trimers expose V3 transiently (Pugach et al., 2015; Sanders et al., 2013). In contrast, 19b binding to the BG505 SOSIP.v4.1 mutant was abolished (Fig. 1G&H). Reactivity of 19b was also decreased for AMC008 SOSIP.v4 compared to wild-type; the stoichiometry of the 19b-trimer complex was 0.3 for wild-type (i.e., ~1 IgG per 3 trimers), but 0 (i.e., no complexes were detected) for SOSIP.v4.1 and negligible (~0.03; i.e., ~1 IgG per 30 trimers) for SOSIP.v4.2 (Fig. 1G&H). 19b binding to B41 SOSIP.v4 was also abolished (no detectable binding, compared to a stoichiometry of 0.2 for B41 SOSIP.664; Fig. 1G&H). Hence, the V3 region is effectively sequestered on SOSIP.v4 trimers.
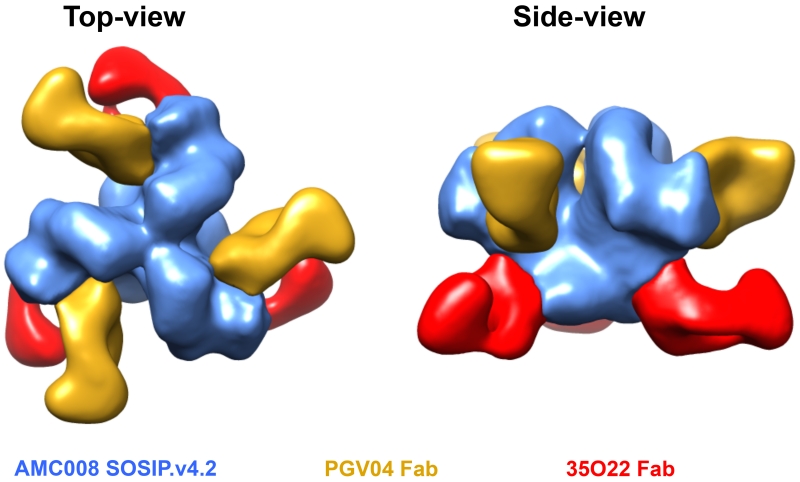
To confirm their overall native-like structure and bNAb epitope presentation, we prepared complexes of the AMC008 SOSIP.v4.2 trimers with bNAbs PGV04 and 35O22 (added as Fabs), and visualized them by NS-EM. The stabilized trimers were all compact entities that were indistinguishable from previously published low-resolution reconstructions of BG505 and B41 SOSIP.664 trimers (Pugach et al., 2015; Sanders et al., 2013) (Fig. 3, Fig. S5).
Figure 3. EM reconstruction of AMC008 SOSIP.v4.2 trimers in complex with PGV04 and 35O22 Fabs.
Top and side view of AMC008.v4.2 trimers (blue) in complex with PGV04 (yellow) and 35O22 (red) Fabs. The reconstruction shows that AMC008.v4.2 trimers are compact and native-like. At the resolution obtained here, there is a very high degree of structural similarity with BG505 SOSIP.664 trimers in complex with 35O22 [EMDB-2672). See also figure S5.
CD4 binding and CD4A-induced conformational changes
In ELISA, CD4-IgG2 binding to AMC008, B41 and ZM197 SOSIP.v4 trimers was slightly reduced compared to SOSIP.664, and more markedly so for the BG505 trimers, while there was no significant change in binding of CD4bs bNAbs CH103 and VRC01. An SPR analysis showed that CD4-IgG2 reached lower plateaus earlier for the SOSIP.v4 trimers than for the wild-type trimers, and the ligand dissociated markedly faster from the SOSIP.v4 trimers (Fig. 1J). This finding is consistent with a report that the introduction of the H66N substitution into a gp120 core protein leads to a faster rate of sCD4 dissociation but no change in the association kinetics (Kassa et al., 2009a). Biolayer interferometry data confirmed that the affinity of sCD4 for various stabilized, BG505 SOSIP.664 trimers was similar or slightly lower, with the greatest reduction (2.4-fold) seen with the A316W single mutant and the SOSIP.v4.2 double mutant (Fig. 1I).
The ELISA studies also showed that the sCD4-induction of the 17b and 412d CD4i epitopes was negligible for the AMC008, B41, ZM197M and BG505 SOSIP.v4 trimers (5-11% compared to 100% for their SOSIP.664 counterparts; Table 2, Data S1E-H). When 17b was added to CD4-IgG2-trimer complexes in an SPR study, it did not bind detectably to the BG505 SOSIP.v4 trimers, in marked contrast to its strong binding to the wild-type BG505 SOSIP.664 trimers under the same conditions (Fig. 1J). Thus, the stabilizing substitutions in the SOSIP.v4 trimers impede CD4-induced conformational changes.
Conformational flexibility assessed by hydrogen-deuterium exchange
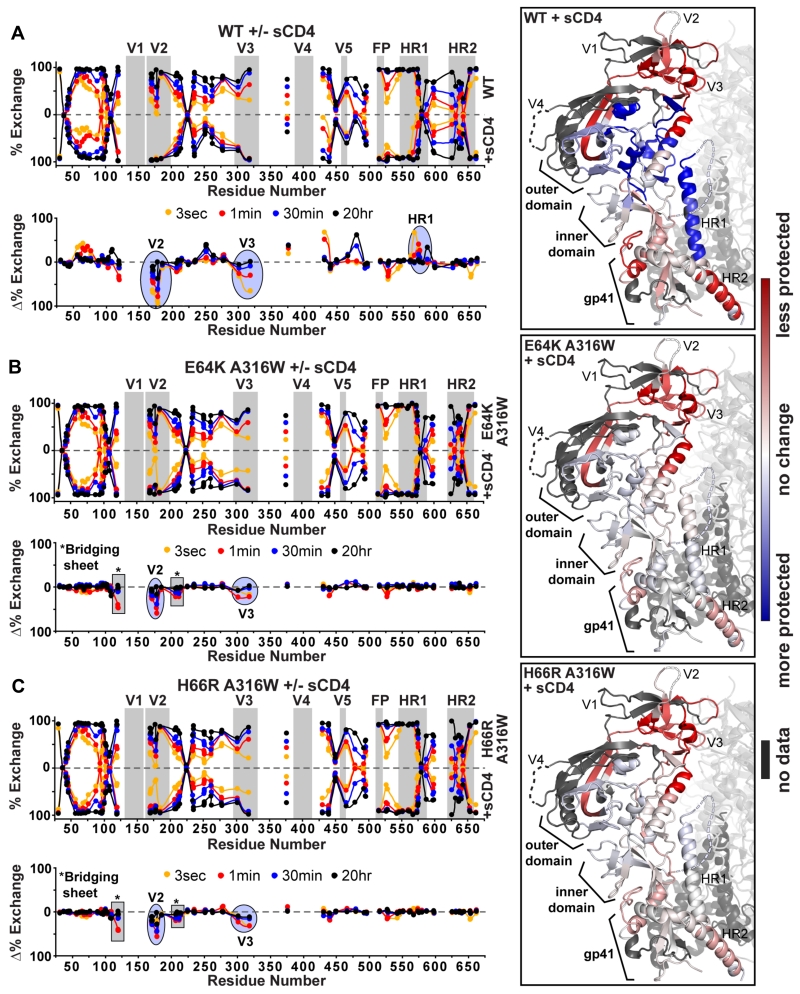
We used hydrogen-deuterium exchange coupled with mass spectrometry (HDX-MS) to study PGT145-purified BG505 SOSIP.664 and SOSIP.v4 trimers. The profiles for all three trimers were very similar to those obtained previously for 2G12/SEC-purified BG505 SOSIP.664 trimers with only minor changes in exchange patterns proximal to the stabilizing mutations (Fig. 2, Fig. S4A&B) (Guttman et al., 2014). Hence, the E64K+A316W and H66R+A316W substitutions did not alter the overall structure and conformational dynamics of the trimers.
Figure 2. HDX-MS analysis of BG505 SOSIP.664 and SOSIP.v4 trimers.
Butterfly plots of PGT145-purified BG505 (A) SOSIP.664, (B) SOSIP.v4.1, (C) SOSIP.v4.2 trimers, comparing deuterium exchange levels in the presence and absence of sCD4. The percent exchange at each time point for each peptide is shown at its position along the primary sequence of the protein. Regions that are less (red) or more (blue) protected upon CD4-binding are mapped on the BG505 SOSIP.664 crystal structure (Pancera et al., 2014), to depict CD4-induced conformational changes. The differences in CD4-induction between the SOSIP.664 and SOSIP.v4.1 or SOSIP.v4.2 trimers were also plotted on the crystal structure (rightmost panels of B and C). The exchange kinetics of individual peptides are shown in Fig. S4 and Data S1.
When sCD4 was added in molar excess, the deuterium-exchange patterns now differed markedly for SOSIP.v4 trimers compared to wild-type SOSIP.664 (Fig. 2, Data S1I). sCD4-binding to wild-type trimers resulted in greater protection of the CD4bs, but also of layers 1-3 in the gp120 inner domain and HR1 of gp41 (Fig. 2A). Thus, sCD4-binding orders and/or buries these regions, as reported previously (Guttman et al., 2014). Conversely, sCD4-binding led to decreased protection of the V2 and V3 loops as well as in several gp41 regions, which is consistent with a sCD4-induced opening of the trimer apex that eventually propagates and increases the accessibility of the gp41 fusion machinery (Fig. 2A). Regions of the trimer that are less (red) or more (blue) protected upon sCD4 binding are mapped onto the BG505 SOSIP.664 crystal structure (Fig. 2A-C).
In contrast, the above sCD4-induced changes were greatly attenuated or entirely abrogated when the SOSIP.v4 trimers were studied. The stabilizing substitutions appear, therefore, to block the ability of sCD4 to induce ordering of residues 370-382 (CD4bs), 245-256 and 476-483 (layer 3), 206-226 (layer 2), 53-92 (layer 1) and 566-592 (HR2; α7) (Fig. 2B&C, Data S1I). Furthermore, the substitutions reduced the sCD4-induced disorder of residues 165-181 (V2), 286-320 (V3), 520-537 (α6) and 593-628 (gp41 disulfide loop). Thus, the E64K+A316W and H66R+A316W substitutions restrain the sCD4-induced opening of the trimer apex, as well as the conformational cascade that spreads from the CD4bs via layer 3, layer 2 and layer 1 to gp41 HR1 (Fig. 2B&C), described as the allosteric “priming” network (Guttman et al., 2014).
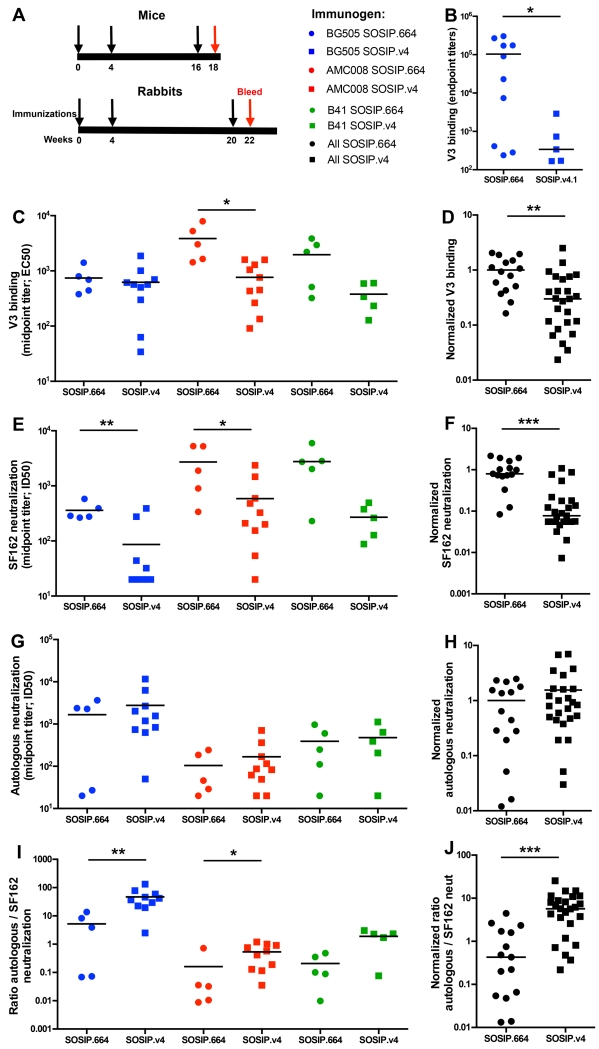
Reduced V3 antibody and Tier-1A neutralizing antibody responses to SOSIP.v4 trimers in mice and rabbits
Our principal goal was to reduce the exposure, and by inference the immunogenicity, of non-NAb epitopes such as V3. Accordingly, we immunized mice with BG505 SOSIP.664 or SOSIP.v4.1 trimers and analyzed the trimer- and V3-specific Ab binding titers two weeks after the third immunization (week 18; Fig. 4A). While trimer binding titers were similar (Fig. S6A), V3 binding Ab responses, as measured by a direct V3-peptide ELISA, were approximately 100-fold lower in SOSIP.v4.1-immunized mice compared to the SOSIP.664 recipients (p=0.039, Fig. 4B).
Figure 4. Immunogenicity of AMC008, BG505 and B41 SOSIP.v4 trimers.
(A) Immunization schedules. Mice were immunized at weeks 0, 4 and 16 (black arrows) and the Ab responses were analyzed at week 18 (red arrow). Rabbits were immunized at weeks 0, 4 and 20 (black arrows) and the Ab responses were analyzed at week 22 (red arrow). Blue symbols represent BG505 trimer-immunized animals, red symbols AMC008 and green symbols B41. The SOSIP.664 recipients are shown by closed circles, SOSIP.v4 by squares. Statistical comparisons between groups were performed using a two-tailed Mann-Whitney U test (*p>0.05, **p>0.01, ***p>0.001). (B) V3 peptide ELISA endpoint binding Ab titers in mouse sera are plotted. (C, D) V3 peptide binding responses were determined by ELISA. The midpoint binding Ab titers in rabbit sera are plotted in (C), and the normalized values of all SOSIP.664 and SOSIP.v4 values, compared to the mean of the corresponding SOSIP.664 group, are shown in (D). The midpoint (E, F) SF162 (Tier-1A), and (G, H) autologous Tier-2 (BG505 and B41) or Tier-1B (AMC008) neutralization titers were determined using the TZM-bl assay (Tables S4 and S5). Specifically, the serum dilutions at which HIV-1 infectivity is inhibited by 50% (ID50) are plotted in (E) and (G), and the normalized values of all SOSIP.664 and SOSIP.v4 values, compared to the mean of the corresponding SOSIP.664 group, are shown in (F) and (H). (I) The ratios of autologous/SF162 NAb titers are shown for sera from the various SOSIP.664 and SOSIP.v4 immunized rabbits. (J) The normalized values derived from the data in (I) are plotted. See also figure S6 and table S4 and S5.
We also immunized rabbits with AMC008, BG505 and B41 SOSIP.664 trimers and their SOSIP.v4 counterparts, and quantified the Ab responses by ELISA two weeks after the third immunization (week 22; Fig. 4A). All the rabbit sera contained high titers of binding Abs against the corresponding SOSIP.664 and SOSIP.v4 trimers (Fig. S6B-D). When we used the direct V3 peptide ELISA we found that the V3 Ab responses were significantly reduced in the SOSIP.v4-immunized rabbits compared to SOSIP.664 (p=0.0015 for the comparison of all SOSIP.664 vs. SOSIP.v4; Fig. 4C&D). Similar results were obtained with a V3 peptide-competition ELISA (Fig. S6E).
Since Tier-1A NAb responses to BG505 SOSIP.664 trimers are dominated by V3-specificities (Sanders et al., 2015), we expected that SOSIP.v4 trimers would induce lower, or less V3-dependent, titers of such NAbs. That was indeed the case; the titers of NAbs against the Tier-1A virus SF162 were significantly lower in SOSIP.v4 immunized rabbits compared to SOSIP.664 (by 14-, 8- and 10-fold for BG505, AMC008 and B41, respectively; p=0.0001 for the combined datasets; Fig. 4E&F; Tables S4 and S5). In contrast, there was no such reduction in the autologous NAb titers. Overall, strong autologous neutralization (ID50 > 500) was observed in 9 of 10 BG505 SOSIP.v4-immunized rabbits, with a weaker response in the 10th, and in 3 of 5 BG505 SOSIP.664 recipients (Fig. 4G&H; Table S5). Autologous NAb titers were observed in 4 out of 5 rabbits from each of the B41 SOSIP.664 and SOSIP.v4.1 immunized groups (Fig. 4G&H; Table S5). An autologous NAb response was also seen in the majority of the AMC008 trimer recipients. However, the AMC008 NAb titers were lower than the corresponding autologous responses to the BG505 and B41 trimers, despite AMC008 being the more sensitive virus (i.e., Tier-1B vs. Tier-2) (Fig. 4G&H; Table S5). Autologous neutralization strongly correlated with BG505 and AMC008 SOSIP.v4 trimer binding titers (Fig. S6F-I). For each of the trimer genotypes, the ratio of the SF162 (Tier-1A) and autologous NAb titers show that the Ab responses in the SOSIP.v4 immunized rabbits were directed away from the V3 region, compared to the responses elicited by the SOSIP.664 trimers (Fig. 4I&J; Table S4 &5). We also tested all the rabbit sera against a large panel of heterologous viruses and found that cross-neutralization of Tier-2 viruses was weak and sporadic, which is consistent with our findings using BG505 SOSIP.664 trimers (Sanders et al., 2015).
Discussion
We re-designed native-like SOSIP.664 trimers to reduce their intrinsic “breathing”. The E64K/H66R and A316W substitutions stabilized the trimers in the closed, pre-fusion ground state, impeding or even preventing their spontaneous sampling of the CD4-induced conformation, and reducing the accessibility of V3. Two gp41 substitutions, I/V535M and L543N/Q, increased trimer formation. Together, these four substitutions improve the current SOSIP.664 design (Sanders et al., 2013) and the probability of generating stable, native-like closed trimers for a range of Env sequences. These trimer improvement strategies allowed us to generated native-like trimers from a new Env sequence derived from a patient that developed a bNAb response. The AMC008 SOSIP.v4 trimers might be suitable starting points for lineage vaccines designed to mimic the development of neutralization in the patient.
Other mutation strategies, such as an I201C-A433C intra-gp120 disulfide bond and a A433P point substitution, also reduce non-NAb binding to BG505 SOSIP.664 trimers, increase their thermostability and constrain their conformational flexibility (Do Kwon et al., 2015). The immunogenicity of these modified trimers has not yet been reported. It is also not yet known whether these strategies are effective for trimers from other genotypes, or whether they can improve intrinsically lower quality trimers such as AMC008 SOSIP.664.
Our results provide insights into the functions of the Env trimer. The A316W, E64K and H66R changes do not affect the folding or overall structure of soluble, SOSIP.664 trimers as the resulting structures are very similar to wild-type, but they do ablate infectivity when introduced into viral Env. The likely explanation is that residues E64 and H66 are critical for facilitating CD4-induced conformational changes and the formation of the CD4i epitopes associated with the co-receptor binding site that are essential for virus-cell entry but not important in the soluble trimer context. Other substitutions at position 66 (H66N and H66A) are known to increase the resistance of HIV-1 to low-temperature inactivation by decreasing spontaneous sampling of the CD4-induced conformation, which was previously described as “intrinsic Env reactivity” (Haim et al., 2011; Kassa et al., 2009a, 2009b).
In the context of an infectious virus, V3 needs to be accessible and dissociate from the V1V2 domain, perhaps as an integral part of the entry process. Native-like SOSIP.664 trimers appear to retain this natural plasticity, such that their V3 regions become exposed under certain conditions (Sanders et al., 2013, 2015). The same flexibility in the orientation of the V1, V2 and V3 loops at the trimer apex may underpin the transitions between fully closed and partially open versions of some native-like SOSIP.664 trimers (e.g., B41) that we have described elsewhere (Pugach et al., 2015). We now show that these spontaneous conformational transitions in soluble trimers can be impeded by the introduction of stabilizing changes.
In conclusion, by preventing the conformational changes normally required for Env function, we have further stabilized recombinant, native-like trimers now designated SOSIP.v4. The principal rationale for the stabilization strategy was that it might be beneficial for generating protective humoral immune responses. Antigenicity studies showed that bNAb epitopes, in particular those that are dependent on quaternary structure, were better presented on the SOSIP.v4 trimers while non-NAb epitopes such as V3, which might serve as immunological decoys, were occluded. The reduced antigenicity of the V3 region had an immunological correlate, in that the SOSIP.v4 trimers induced weaker anti-V3 responses in rabbits and, as a result, lower titers of V3-dependent NAbs against the Tier-1A virus SF162. This outcome was achieved without compromising the autologous NAb response. As noted earlier, a single native-like trimer of any genotype will probably not be sufficient to induce bNAbs. However, vaccination regimens that include germline targeting, evolutionary lineages, and/or multivalent trimers are strategies towards bNAb generation that merit pursuing (Doria-Rose et al., 2014; Dosenovic et al., 2015; Haynes et al., 2012; Jardine et al., 2015; Liao et al., 2013; McGuire et al., 2014; Sliepen et al., 2015). In all of these vaccination scenarios, it is likely that reducing the immunogenicity of immunodominant non-NAb epitopes in favor of the subdominant bNAb epitopes, such as through the generalizable trimer-stabilizing mutations we describe here, will be beneficial.
The improved stability of SOSIP.v4 trimers might also improve their in vivo halfclife or how they are affected by certain adjuvants, thereby maximizing the opportunity for naïve B cells to encounter bNAb epitopes. The added resistance to CD4-induced opening, which may occur in vivo when macaques or humans (but not rabbits) are immunized, would further help maintain the closed, immunologically relevant conformation of the stabilized trimer. Overall, we propose that the stabilized SOSIP.v4 trimers are promising Env vaccine candidates for evaluation in various strategies intended to induce heterologous Tier-2 NAb responses.
Methods and Experimental Procedures
Trimer design, expression and purification
The AMC008 env gene is derived from a subtype B Tier-1B virus, isolated 8 months post-seroconversion from an elite neutralizer in the Amsterdam Cohort Studies on HIV/AIDS (patient H18818 in reference (Euler et al., 2010)). The design of AMC008 SOSIP.664 trimers is identical to the BG505, B41 and ZM197M SOSIP.664 constructs described elsewhere (Julien et al., 2015; Pugach et al., 2015; Sanders et al., 2013). For more details see SI Methods
Antigenicity assays
Antigenicity was determined using tagged SOSIP.664 trimers for SPR or D7324-capture ELISA, as described elsewhere (Derking et al., 2015; Sanders et al., 2013; Yasmeen et al., 2014). In ELISA, the D7324 antibody was attached to the solid phase and used to capture the epitope-tagged trimers, which were then recognized by various solution-phase MAbs. ITC was used to generate data on MAb-trimer interactions in solution (Julien et al., 2013b, 2013c).
Thermostability assays
As a screening assay, we used a PCR machine-based system to expose unpurified SOSIP.664-D7324 trimers to a range of temperatures, before using D7324-capture ELISA to assess when the 2G12 epitope had been lost. A conventional DSC assay was used to quantify the melting of purified trimers, as described previously (Pugach et al., 2015). The two methods yielded data on multiple trimers that correlated well.
Biophysical techniques
Multiple biophysical assays were used to analyze the properties of various SOSIP.664 trimers, including DLS, SAXS and HDX-MS as described elsewhere (Guttman and Lee, 2013; Guttman et al., 2014). Images of purified SOSIP.664 trimers, either alone or as Fab complexes, were generated by NS-EM following previously described procedures (Sanders et al., 2013). A more detailed description of the EM process can be found in SI Methods.
Mouse and rabbit immunizations
Mice and rabbits were immunized with 10 or 22 μg of trimer, respectively, formulated with ISCOMATRIX™, at weeks 0, 4 and 16 (mice) or 0, 4, and 20 (rabbits). For details see SI Methods
Supplementary Material
Acknowledgments
We thank Hermann Katinger, Mark Connors, James Robinson, Dennis Burton, John Mascola, Peter Kwong and William Olson for donating antibodies and reagents directly or through the NIH AIDS Research and Reference Reagent Program. The Amsterdam Cohort Studies on HIV infection and AIDS, a collaboration between the Amsterdam Health Service, the Academic Medical Center of the University of Amsterdam, Sanquin Blood Supply Foundation, Medical Center Jan van Goyen and the HIV Focus Center of the DC-Clinics, are part of the Netherlands HIV Monitoring Foundation and financially supported by the Center for Infectious Disease Control of the Netherlands National Institute for Public Health and the Environment. The EM reconstruction data reported in this paper have been deposited in the Electron Microscopy Data Bank, www.emdatabank.org (EMDB ID code EMDB-6500).
Funding
This work was supported by National Institutes of Health Grants P01 AI82362 and P01 AI110657, R21 AI112389, UM1 AI100663 (Scripps CHAVI-ID) and NIAID Contract # HHSN27201100016C (to DCM); by the International AIDS Vaccine Initiative (IAVI); by the Bill and Melinda Gates Foundation through the Collaboration for AIDS Vaccine Discovery (CAVD) and by the Aids fonds Netherlands, Grant #2012041. R.W.S. is a recipient of a Vidi grant from the Netherlands Organization for Scientific Research (NWO) and a Starting Investigator Grant from the European Research Council (ERC-StG-2011-280829-SHEV). The electron microscopy data were collected at The Scripps Research Institute Electron Microscopy Facility.
Footnotes
Author Contributions
Conceived and designed the experiments: SWdT, GO, ATdP, TvdK, PP, IB, JH, HA, JS, WCK, HS, DE, BB, HD, CL, SC, MC, DCM, PJK, KKL, JPM, IAW, ABW, RWS
Performed the experiments: SWdT, GO, ATdP, MG, JPJ, JLT, LKP, JAB, AY, CL, JC
Analyzed the data: SWdT, GO, ATdP, MG, JPJ, SC, PJK, JPM, IAW, ABW, RWS
Wrote the paper: SWdT, GO, JPM, IAW, ABW, RWS
Edited the paper: SWdT, GO, JPM, IAW, ABW, RWS, MG, KKL, MC, LKP, PJK, JPJ, WCK, BB, CL
References
- Binley JM, Sanders RW, Clas B, Schuelke N, Master A, Guo Y, Kajumo F, Anselma DJ, Maddon PJ, Olson WC, et al. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 2000;74:627–643. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley JM, Sanders RW, Master A, Cayanan CS, Wiley CL, Schiffner L, Travis B, Kuhmann S, Burton DR, Hu S, et al. Enhancing the proteolytic maturation of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 2002;76:2606–2616. doi: 10.1128/JVI.76.6.2606-2616.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner C, Lee J, Sliepen K, Derking R, Falkowska E, DelaPeña A, Cupo A, Julien JP, VanGils M, Lee PS, et al. Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 env trimers. Immunity. 2014;40:669–680. doi: 10.1016/j.immuni.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonomelli C, Doores KJ, Dunlop DC, Thaney V, Dwek RA, Burton DR, Crispin M, Scanlan CN. The glycan shield of HIV is predominantly oligomannose independently of production system or viral clade. PLoS One. 2011;6:1–7. doi: 10.1371/journal.pone.0023521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury S, Reifman J, Wallqvist A. Simulation of B cell affinity maturation explains enhanced antibody cross-reactivity induced by the polyvalent malaria vaccine AMA1. J. Immunol. 2014;193:2073–2086. doi: 10.4049/jimmunol.1401054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derking R, Ozorowski G, Sliepen K, Yasmeen A, Cupo A, Torres JL, Julien J, Lee JH, van Montfort T, de Taeye SW, et al. Comprehensive antigenic map of a cleaved soluble HIV-1 envelope trimer. PLoS Pathog. 2015;11:e1004767. doi: 10.1371/journal.ppat.1004767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey AK, David KB, Ray N, Ketas TJ, Klasse PJ, Doms RW, Moore JP. N-terminal substitutions in HIV-1 gp41 reduce the expression of non-trimeric envelope glycoproteins on the virus. Virology. 2008;372:187–200. doi: 10.1016/j.virol.2007.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, et al. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 2014;509:55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosenovic P, von Boehmer L, Escolano A, Jardine J, Freund NT, Gitlin AD, McGuire AT, Kulp DW, Oliveira T, Scharf L, et al. Immunization for HIV-1 broadly neutralizing antibodies in human Ig knockin mice. Cell. 2015;161:1505–1515. doi: 10.1016/j.cell.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggink D, Goff PH, Palese P. Guiding the immune response against Influenza virus hemagglutinin toward the conserved stalk domain by hyperglycosylation of the globular head domain. J. Virol. 2013;88:699–704. doi: 10.1128/JVI.02608-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euler Z, van Gils MJ, Bunnik EM, Phung P, Schweighardt B, Wrin T, Schuitemaker H. Cross-reactive neutralizing humoral immunity does not protect from HIV type 1 disease progression. J. Infect. Dis. 2010;201:1045–1053. doi: 10.1086/651144. [DOI] [PubMed] [Google Scholar]
- Finzi A, Xiang S-H, Pacheco B, Wang L, Haight J, Kassa A, Danek B, Pancera M, Kwong PD, Sodroski J. Topological layers in the HIV-1 gp120 inner domain regulate gp41 interaction and CD4-triggered conformational transitions. Mol. Cell. 2010;37:656–667. doi: 10.1016/j.molcel.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity RR, Rimmelzwaan G, Minassian A, Tsai W, Lin G, de Jong J, Goudsmit J, Nara PL. Refocusing neutralizing antibody response. J. Immunol. 1997;159:279–289. [PubMed] [Google Scholar]
- Guenaga J, de Val N, Tran K, Feng Y, Satchwell K, Ward AB, Wyatt RT. Well-ordered trimeric HIV-1 subtype B and C soluble spike mimetics generated by negative selection display native-like properties. PLoS Pathog. 2015;11:e1004570. doi: 10.1371/journal.ppat.1004570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Lee KK. A functional interaction between gp41 and gp120 is observed for monomeric but not oligomeric, uncleaved HIV-1 Env gp140. J. Virol. 2013;87:11462–11475. doi: 10.1128/JVI.01681-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Garcia NK, Cupo A, Matsui T, Julien JP, Sanders RW, Wilson IA, Moore JP, Lee KK. CD4-induced activation in a soluble HIV-1 Env trimer. Structure. 2014;22:974–984. doi: 10.1016/j.str.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Cupo A, Julien J-P, Sanders RW, Wilson I. a., Moore JP, Lee KK. Antibody potency relates to the ability to recognize the closed, pre-fusion form of HIV Env. Nat. Commun. 2015;6:6144. doi: 10.1038/ncomms7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haim H, Strack B, Kassa A, Madani N, Wang L, Courter JR, Princiotto A, McGee K, Pacheco B, Seaman MS, et al. Contribution of intrinsic reactivity of the HIV-1 envelope glycoproteins to CD4-independent infection and global inhibitor sensitivity. PLoS Pathog. 2011;7:e1002101. doi: 10.1371/journal.ppat.1002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BF, Joiner KA. Strategies of obligate intracellular parasites for evading host defences. Immunol. Today. 1991;12:A22–A27. doi: 10.1016/S0167-5699(05)80007-6. [DOI] [PubMed] [Google Scholar]
- Harris AK, Bartesaghi A, Milne JLS, Subramaniam S. HIV-1 envelope glycoprotein trimers display open quaternary conformation when bound to the gp41 membrane-proximal external-region-directed broadly neutralizing antibody Z13e1. J. Virol. 2013;87:7191–7196. doi: 10.1128/JVI.03284-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, Kelsoe G, Harrison SC, Kepler TB. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat. Biotechnol. 2012;30:423–433. doi: 10.1038/nbt.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Kang BH, Pancera M, Lee JH, Tong T, Feng Y, Imamichi H, Georgiev IS, Chuang G-Y, Druz A, et al. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature. 2014;515:138–142. doi: 10.1038/nature13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine JG, Ota T, Sok D, Pauthner M, Kulp DW, Kalyuzhniy O, Skog PD, Thinnes TC, Bhullar D, Briney B, et al. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science. 2015;349:156–161. doi: 10.1126/science.aac5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien J-P, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse P-J, Burton DR, Sanders RW, Moore JP, et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013a;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien J-P, Lee JH, Cupo A, Murin CD, Derking R, Hoffenberg S, Caulfield MJ, King CR, Marozsan AJ, Klasse PJ, et al. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proc. Natl. Acad. Sci. U. S. A. 2013b;110:4351–4356. doi: 10.1073/pnas.1217537110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien J-P, Lee JH, Ozorowski G, Hua Y, Torrents de la Peña A, de Taeye SW, Nieusma T, Cupo A, Yasmeen A, Golabek M, et al. Design and structure of two HIV-1 clade C SOSIP.664 trimers that increase the arsenal of native-like Env immunogens. Proc. Natl. Acad. Sci. U. S. A. 2015;112:11947–11952. doi: 10.1073/pnas.1507793112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien JP, Sok D, Khayat R, Lee JH, Doores KJ, Walker LM, Ramos A, Diwanji DC, Pejchal R, Cupo A, et al. Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS Pathog. 2013c;9:e1003342. doi: 10.1371/journal.ppat.1003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassa A, Madani N, Schön A, Haim H, Finzi A, Xiang S, Wang L, Princiotto A, Pancera M, Courter J, et al. Transitions to and from the CD4-bound conformation are modulated by a single-residue change in the human immunodeficiency virus type 1 gp120 inner domain. J. Virol. 2009a;83:8364–8378. doi: 10.1128/JVI.00594-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassa A, Finzi A, Pancera M, Courter JR, Smith AB, Sodroski J. Identification of a human immunodeficiency virus type 1 envelope glycoprotein variant resistant to cold inactivation. J. Virol. 2009b;83:4476–4488. doi: 10.1128/JVI.02110-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasse PJ, Cupo A, Cocco N, Korzun J, Yasmeen A, Ward AB, Wilson IA, Sanders RW, Moore JP. Influences on trimerization and aggregation of soluble , cleaved HIV-1 SOSIP envelope glycoprotein. J. Virol. 2013;87:9873–9885. doi: 10.1128/JVI.01226-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do Kwon Y, Pancera M, Acharya P, Georgiev IS, Crooks ET, Gorman J, Joyce MG, Guttman M, Ma X, Narpala S, et al. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat. Struct. Mol. Biol. 2015;22:522–531. doi: 10.1038/nsmb.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaman DP, Zwick MB. Increased functional stability and homogeneity of viral envelope spikes through directed evolution. PLoS Pathog. 2013;9:e1003184. doi: 10.1371/journal.ppat.1003184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Leaman DP, Kim AS, Torrents de la Peña A, Sliepen K, Yasmeen A, Derking R, Ramos A, de Taeye SW, Ozorowski G, et al. Antibodies to a conformational epitope on gp41 neutralize HIV-1 by destabilizing the Env spike. Nat. Commun. 2015;6:8167. doi: 10.1038/ncomms9167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H-X, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyumkis D, Julien J, de Val N, Cupo A, Potter CS, Klasse P, Burton DR, Sanders RW, Moore JP, Carragher B, et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrack P, Kappler J. Subversion of the immune system by pathogens. Cell. 1994;76:323–332. doi: 10.1016/0092-8674(94)90339-5. [DOI] [PubMed] [Google Scholar]
- McGuire AT, Dreyer AM, Carbonetti S, Lippy A, Glenn J, Scheid JF, Mouquet H, Stamatatos L. HIV antibodies. Antigen modification regulates competition of broad and narrow neutralizing HIV antibodies. Science. 2014;346:1380–1383. doi: 10.1126/science.1259206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro JB, Gorman J, Ma X, Zhou Z, Arthos J, Burton DR, Koff WC, Courter JR, Smith AB, 3rd, Kwong PD, et al. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science. 2014;346:759–763. doi: 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotny LA, Bakaletz LO. The fourth surface-exposed region of the outer membrane protein P5-homologous adhesin of nontypable haemophilus influenzae is an immunodominant but nonprotective decoying epitope. J. Immunol. 2003;171:1978–1983. doi: 10.4049/jimmunol.171.4.1978. [DOI] [PubMed] [Google Scholar]
- Pancera M, Majeed S, Ban YA, Chen L, Huang C, Kong L, Kwon Y. Do, Stuckey J, Zhou T, Robinson JE, et al. Structure of HIV-1 gp120 with gp41- interactive region reveals layered envelope architecture and basis of conformational mobility. Proc. Natl. Acad. Sci. U. S. A. 2010;107:1166–1171. doi: 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang G, Ofek G, et al. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014;514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard LK, Vasiljevic S, Ozorowski G, Seabright GE, Cupo A, Ringe R, Kim HJ, Sanders RW, Doores KJ, Burton DR, et al. Structural constraints determine the glycosylation of HIV-1 envelope trimers. Cell Rep. 2015a;11:1604–1613. doi: 10.1016/j.celrep.2015.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard LK, Harvey DJ, Bonomelli C, Crispin M, Doores KJ. Cell-and protein-directed glycosylation of native cleaved HIV-1 envelope. J. Virol. 2015b;89:8932–8944. doi: 10.1128/JVI.01190-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugach P, Ozorowski G, Cupo A, Ringe R, Yasmeen A, de Val N, Derking R, Kim HJ, Korzun J, Golabek M, et al. A native-like SOSIP.664 trimer based on a HIV-1 subtype B env gene. J. Virol. 2015;89:3380–3395. doi: 10.1128/JVI.03473-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringe RP, Sanders RW, Yasmeen A, Kim HJ, Lee JH, Cupo A, Korzun J, Derking R, van Montfort T, Julien J-P, et al. Cleavage strongly influences whether soluble HIV-1 envelope glycoprotein trimers adopt a native-like conformation. Proc. Natl. Acad. Sci. U. S. A. 2013;110:18256–18261. doi: 10.1073/pnas.1314351110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringe RP, Yasmeen A, Ozorowski G, Go EP, Pritchard LK, Guttman M, Ketas TA, Cottrell CA, Wilson IA, Sanders RW, et al. Influences on the design and purification of soluble, recombinant native-like HIV-1 envelope glycoprotein trimers. J. Virol. 2015;89:12189–12210. doi: 10.1128/JVI.01768-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, Moore JP. HIV: A stamp on the envelope. Nature. 2014;514:437–438. doi: 10.1038/nature13926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, Vesanen M, Schuelke N, Master A, Schiffner L, Kalyanaraman R, Paluch M, Berkhout B, Maddon PJ, Olson WC, et al. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol. 2002;76:8875–8889. doi: 10.1128/JVI.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Peña AT, Korzun J, et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, Ozorowski G, Cupo A, Simonich C, Goo L, et al. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science. 2015;349:aac4223. doi: 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf L, Wang H, Gao H, Chen S, McDowall AW, Bjorkman PJ. Broadly neutralizing antibody 8ANC195 recognizes closed and open states of HIV-1 Env. Cell. 2015;162:1379–1390. doi: 10.1016/j.cell.2015.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SK, de Val N, Bale S, Guenaga J, Tran K, Feng Y, Dubrovskaya V, Ward AB, Wyatt RT. Cleavage-independent HIV-1 Env trimers engineered as soluble native spike mimetics for vaccine design. Cell Rep. 2015;11:539–550. doi: 10.1016/j.celrep.2015.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliepen K, Medina-Ramírez M, Yasmeen A, Moore JP, Klasse PJ, Sanders RW. Binding of inferred germline precursors of broadly neutralizing HIV-1 antibodies to native-like envelope trimers. Virology. 2015;486:116–120. doi: 10.1016/j.virol.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sok D, van Gils MJ, Pauthner M, Julien J, Saye-Francisco KL, Hsueh J, Briney B, Lee JH, Le KM, Lee PS, et al. Recombinant HIV envelope trimer selects for quaternary-dependent antibodies targeting the trimer apex. Proc. Natl. Acad. Sci. 2014;111:17624–17629. doi: 10.1073/pnas.1415789111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasmeen A, Ringe R, Derking R, Cupo A, Julien J-P, Burton DR, Ward AB, Wilson I. a, Sanders RW, Moore JP, et al. Differential binding of neutralizing and non-neutralizing antibodies to native-like soluble HIV-1 Env trimers, uncleaved Env proteins, and monomeric subunits. Retrovirology. 2014;11:41. doi: 10.1186/1742-4690-11-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Meyer-Hermann M, George LA, Figge MT, Khan M, Goodall M, Young SP, Reynolds A, Falciani F, Waisman A, et al. Germinal center B cells govern their own fate via antibody feedback. J. Exp. Med. 2013;210:457–464. doi: 10.1084/jem.20120150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.