Abstract
There are strong genetic components to cardiorespiratory fitness and its response to exercise training. It would be useful to understand the differences in the genomic profile of highly trained endurance athletes of world class caliber and sedentary controls. An international consortium (GAMES) was established in order to compare elite endurance athletes and ethnicity-matched controls in a case-control study design. Genome-wide association studies were undertaken on two cohorts of elite endurance athletes and controls (GENATHLETE and Japanese endurance runners), from which a panel of 45 promising markers was identified. These markers were tested for replication in seven additional cohorts of endurance athletes and controls: from Australia, Ethiopia, Japan, Kenya, Poland, Russia and Spain. The study is based on a total of 1520 endurance athletes (835 who took part in endurance events in World Championships and/or Olympic Games) and 2760 controls. We hypothesized that world-class athletes are likely to be characterized by an even higher concentration of endurance performance alleles and we performed separate analyses on this subsample. The meta-analysis of all available studies revealed one statistically significant marker (rs558129 at GALNTL6 locus, p = 0.0002), even after correcting for multiple testing. As shown by the low heterogeneity index (I2 = 0), all eight cohorts showed the same direction of association with rs558129, even though p-values varied across the individual studies. In summary, this study did not identify a panel of genomic variants common to these elite endurance athlete groups. Since GAMES was underpowered to identify alleles with small effect sizes, some of the suggestive leads identified should be explored in expanded comparisons of world-class endurance athletes and sedentary controls and in tightly controlled exercise training studies. Such studies have the potential to illuminate the biology not only of world class endurance performance but also of compromised cardiac functions and cardiometabolic diseases.
Introduction
Early studies on the genetic basis of sports performance reported that identical twins who engaged in competitive sports were significantly more likely to participate in the same sports than pairs of dizygotic twins [1]. The first documented attempts to identify genetic markers for sports performance date to the 1968 Mexico and 1976 Montreal Olympic Games and were based on common blood genetic markers. They did not yield any strong positive findings [2–4]. A high maximal oxygen uptake (VO2max) is a necessary condition to reach the level of an endurance athlete of international caliber. A high VO2max can only be achieved if an individual is endowed with a very high level in the sedentary state (intrinsic level) in combination with large increases in response to sustained and demanding exercise training regimens (trainability).
Twin and family studies have revealed that the intrinsic level of VO2max is strongly influenced by a genetic component. For instance, the heritability of VO2max adjusted for age, sex and body composition in sedentary families of European descent reached 51% in the HERITAGE Family Study [5]. Evidence of a significant heritability level has also been documented for exercise training-induced improvements in VO2max. Large individual differences in VO2max gains have been found in sedentary young adults subjected to standardized endurance training programs [6]. A series of exercise training experiments conducted with pairs of identical twins revealed that the differences in trainability were not distributed randomly among the twins, with intraclass correlations in response of VO2max (L O2/min) ranging from 0.44 to 0.77 [7–9], indicating that members of the same twin pair responded similarly to training. In HERITAGE, the increase in VO2max in 481 individuals from 99 two-generation families of Whites of European descent showed 2.5 times more variance between families than within families for VO2max response, with a maximal heritability estimate of 47% [10]. Adjusting the VO2max response data for baseline VO2max did not modify this estimate, suggesting along with other evidence that the familial and genetic factors underlying VO2max in the sedentary state and its response to exercise training are different [11].
In the case-control GENATHLETE study, single nucleotide polymorphisms (SNPs) in several candidate genes were investigated but none have provided strong evidence for differences in allele and genotype frequencies between elite endurance athletes and controls [12–17]. Reports published to date have focused on differences in allele frequencies between athletes and non-athlete controls mainly on angiotensin I converting enzyme (ACE), α-actinin-3 (ACTN3), and peroxisome proliferator-activated receptor-γ coactivator 1α (PPARGC1A) polymorphisms and on mitochondrial DNA (mtDNA) haplogroup distributions [18]. There were several positive findings but most were derived from post hoc subgroup analyses and only a few studies controlled for multiple testing. Most of prior studies have come from elite Japanese runners [19, 20], Spanish endurance athletes [21], Ethiopian and Kenyan endurance runners [22, 23], Russian endurance athletes [24] as well as endurance athletes from Australia [25], Israel [26] and Poland [27].
Unbiased genome-wide approaches have been used in the search for genomic regions, transcripts or DNA variants linked or associated with endurance performance-related traits [28–31]. For instance, in a recent study, the association of 324,611 SNPs with the response of VO2max to endurance training in 473 Whites from HERITAGE was investigated [28]. None of the SNPs reached genome-wide significance even though there were several SNPs moderately associated with VO2max trainability.
It is of scientific interest to understand the differences in the genomic profile of highly trained endurance athletes of world class caliber and sedentary controls from the same ethnic ancestry. Such data could illuminate the biology of cardiorespiratory fitness and human adaptability and would likely have implications for common health problems such as those observed in aging individuals with declining health, diabetic patients with compromised cardiovascular fitness or patients with ischemic heart disease or heart failure to name but a few. Given the heterogeneous results reported in previous studies, we established an international consortium (GAMES) in order to reach larger sample sizes of elite endurance athletes and matched controls in a case-control study design. Genome-wide explorations were undertaken on two cohorts of world-class endurance athletes and controls (GENATHLETE and Japanese endurance runners), which generated a panel of 45 markers that were subsequently used for replication in seven additional cohorts of endurance athletes and controls.
Participants and Methods
The GENATHLETE cohort and a sample of endurance athletes and controls from Japan were used for the discovery phase. The characteristics of the GENATHLETE all male participants are presented in S1 Table and S1 Fig of the Supporting Information. There were 315 elite endurance athletes (national and world-class level) from Germany, Finland, Canada and the USA plus 320 sedentary controls from the same countries. All GENATHLETE participants are males and athletes and controls were closely matched for ethnicity and country of origin. The mean highest recorded VO2max of these athletes was 79.0 mL O2/kg/min with a standard deviation (SD) of 3.4 mL O2 while the mean value for the sedentary controls was 40.0 (SD = 7.1). In the Japanese sample, 60 Japanese elite runners (world-class athletes) and 116 controls were available for the discovery phase. (Additional information on these two cohorts is provided in Supporting Information).
Endurance athletes and controls from 7 countries were used for the replication phase (Table 1). Participants were from Australia, Ethiopia, Japan, Kenya, Poland, Russia and Spain. It should be noted that we used a second Japanese cohort (143 athletes, 692 controls) in the replication phase. Again, in each country, endurance athletes were competing at least at the national level and most of them competed in Olympic or world cup events. A total of 1520 athletes and 2760 controls were involved in the present study. Among the endurance athletes, 1045 (about 69% of total) took part in endurance events at world championships and Olympic Games. We hypothesized that these world-class athletes are likely to be characterized by a higher concentration of “endurance performance alleles” and we performed separate analyses on this subsample (see Supporting Information for details).
Table 1. Number of Athletes and Controls of Each Study Participating in the GAMES Consortium.
| Study | Number of Athletes | Number of World-Class Athletes* | Number of Controls |
|---|---|---|---|
| DISCOVERY PHASE | |||
| GENATHLETE Study | 315 | 168 | 320 |
| (Germany, Finland, Canada, USA) | |||
| Japan Study | 60 | 60 | 116 |
| REPLICATION PHASE | |||
| Australia Study | 207–215 | 160–167 | 252–258 |
| Ethiopia Study | 74–75 | 74–75 | 196–198 |
| Japan | 137–143 | 688–692 | |
| Kenya Study | 269–276 | 66 | 79–83 |
| Poland Study | 98–113 | 50–61 | 136–161 |
| Russia Study | 130–153 | 63–68 | 187–734 |
| Spain Study | 165–170 | 165–170 | 184–198 |
| TOTAL | 1520 | 835 | 2760 |
* World-Class athletes were defined as either having competed at the world championship level and/or in Olympic Games or, in the case of GENATHLETE, as having a maximal oxygen uptake (VO2max) > 78 ml O2/kg/min. The range reflects the smallest and largest numbers of participants available for any given SNP.
Discovery Phase
GAMES was established after the two studies (GENATHLETE and Japanese endurance athlete cohort) had already performed their genome-wide screen and performed their analyses. Thus there are differences in the chips used and the analytical strategies. However, the data of the replication studies were all analyzed centrally at the Pennington site.
In GENATHLETE, genomic DNA was extracted from whole-blood samples by commercial DNA extraction kits (Gentra Systems, Inc., Minneapolis, MN), and the DNA stock samples were diluted to 50 ng/μl concentrations. SNPs for the study were those captured in the Illumina CardioMetabochip (Illumina Inc., San Diego, CA), which contains over 195,000 genetic markers including ~66,000 variants implicated in the aetiology of cardiometabolic traits and disease outcomes from discovery GWAS cohorts, as well as variants around known loci for the purposes of fine-mapping [32]. The SNPs were genotyped using the Illumina Infinium II assay on Illumina iScan platform.
SNPs showing marked deviation from Hardy-Weinberg equilibrium (HWE) (p ≤ 0.00001) were excluded. However, since deviations from HWE may be related to case-control status-related differences of genotype frequencies, identical non-HWE pattern was confirmed both in endurance athletes and non-athlete controls before the SNP was excluded from the database. A total of 143,000 SNPs were polymorphic and passed the quality control filters. In GENATHLETE, we estimate that we an 80% statistical power to detect odds ratios (ORs) of 2.7 and 2.1 for minor allele frequencies of 0.1 and 0.3, respectively, assuming an additive model and an alpha level of 5x10-8.
For the Japanese cohort, total DNA was isolated from saliva or venous blood by use of QIAamp DNA blood Maxi Kit (QIAGEN, Hilden, Germany) or Oragene DNA Collection Kits (DNA genotek, Ontario, Canada), respectively. Total DNA samples were genotyped for more than 700,000 markers using the Illumina® HumanOmniExpress Beadchip. The genotype calls were performed with the Illumina GenomeStudio software. Quality control measures were performed as defined in the Supporting Information. After removing SNPs failing quality control, 541,179 autosomal SNPs in 60 Japanese endurance athletes and 116 Japanese controls were available for association analyses. In the Japanese cohort, we have the ability to detect at 80% statistical power ORs of 6.1 and 4.5 for minor allele frequencies of 0.1 and 0.3, respectively, assuming an additive model and an alpha level of 5x10-8.
Data Analysis
For the GENATHLETE samples, tests of HWE for each SNP were conducted using the exact test implemented in the PEDSTATS software [33]. Allele frequency differences between athletes and controls were tested using a chi-square test as implemented in the PLINK software package [34]. For the Japanese study, standard allelic association analysis was performed by comparing allele-frequency differences between Japanese endurance runners and controls.
None of the SNPs reached genome-wide significance level (p<5 x 10−8) in GENATHLETE or the Japanese Study. Considering that this may be simply the result of the relatively small sample size of the discovery cohorts, we elected to retain the 45 most promising SNPs (all p<1 x 10−4) for further testing in the replication studies. Among these 45 SNPs, 26 came from GENATHLETE CardioMetabochip (13 based on full cohort, 13 from the highest VO2max subgroup analyses) and 19 from the Japanese GWAS.
Replication Phase
The 45 SNPs carried forward were genotyped in athletes and controls from Australia, Ethiopia, Japan, Kenya, Poland, Russia and Spain as described in the Supporting Information. Tests of HWE for each SNP were also performed with the PEDSTATS software [33] and allele-frequency differences between athletes and controls were tested using a chi-square test as implemented in PLINK [34].
Meta-analysis
Results from individual studies were combined using a meta-analysis approach. These analyses were done with the meta-analysis routine of the PLINK software using study-specific association test result files as an input. Three subsets were meta-analyzed: all studies based on athletes of European descent that were part of the replication phase (i.e. Australia, Poland, Russia and Spain), the two studies based on participants from Africa, and all studies combined (including athletes from Ethiopia, Kenya and Japan). In each case, analyses were undertaken based on the results of the comparisons between all athletes and controls from each national cohort.
Further Analyses Performed on World-Class Endurance Athletes
In some of the national cohorts, all endurance athletes were of world-class caliber. However, in other cohorts (GENATHLETE, Poland, Russia and the second Japanese cohort), there was a combination of national level and world-class caliber endurance athletes. We were able to classify the endurance athletes between those who were truly of world-class caliber, as defined by their participation in World Championship competitions or in Olympic Games, and those who competed at the national level (see Supporting Information). In the case of GENATHLETE, the classification was based on the VO2max measurement, with those exhibiting a VO2max value ≥78 mL O2/kg/min being classified as part of the “super elite” contingent. All the analyses described in the previous sections were repeated using only these world-class super elite endurance athletes and controls.
Informed Consent
The GENATHLETE project was originally approved by the Medical Ethics Committee of Laval University (Quebec, Canada). Approval has also been obtained from the medical ethics committees of all institutions that have contributed participantsto the GENATHLETE cohort. Continuing approval for the study has been granted by the IRB of Pennington Biomedical Research Center. Each GENATHLETE participant has given written informed consent. Written informed consent was obtained from all participants from the Japanese, Ethiopian, and Kenyan cohorts, respectively, and was approved by the Institutional Review Board of Tokyo Metropolitan Institute of Gerontology, National Institute of Health and Nutrition, Japan; the Oxford Tropical Research Ethics Committee, the University of Glasgow Ethics Committee, and a committee from the Ethiopian Athletics Federation; and the University Ethics Committee in Kenya. Written informed consent was obtained for all Australian participants. The study was approved by the institutional review boards and Ethics Committees of the Children’s Hospital at Westmead, the University of Sydney, and the Australian Institute of Sport. The procedures followed in the study from Poland were approved by the Pomeranian Medical University Ethics Committee and all participants gave informed written consent. The elite athlete study from Russia was approved by the Ethics Committee of the Research Institute for Physical-Chemical Medicine. Written informed consent was obtained from each participant. All participants from the Spanish cohort provided written consent and the study protocol was approved by the IRB of Pablo Olavide University (Spain).
Results
Discovery Phase
In GENATHLETE, a total of 143,000 SNPs were polymorphic and passed the quality control filters. A Manhattan plot depicting associations between elite endurance athletes and sedentary controls for all SNPs across the 22 autosomes is shown in Panel A of Fig 1. The strength of the association is shown on the y-axis as a—log10 of the p-value, which represents the statistical significance of the allele frequency chi-square test. None of the SNPs reached the genome-wide significance threshold of 5x10-8. 50 SNPs showed p-values of less than 5.0 x 10−4 and the top 26 SNPs are shown in Table 2. Among the 50 SNPs, 8 were found to be in strong pairwise linkage disequilibrium (r2>0.9) and 2 had a minor allele frequency of <5%. From the 40 SNPs left, the top 26 with the most significant p values in GENATHLETE were retained and are shown in the table.
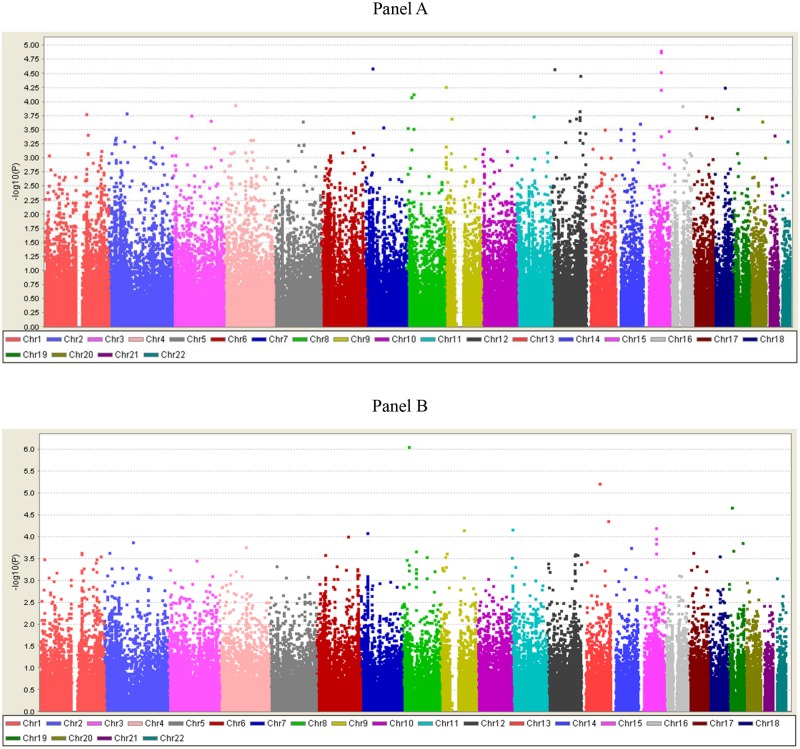
Fig 1. Manhattan Plot of the Association P Values for the CardioMetabochip SNP Differences in Allele Frequencies Between Endurance Athletes and Sedentary Controls in GENATHLETE.
(A) Athletes with VO2max ≥ 75 ml O2/kg/min (N = 315) vs. Controls (N = 320). (B) Elite Athletes with VO2max ≥ 78 ml O2/kg/min (N = 168) vs. Controls (N = 320).
Table 2. Discovery Phase: Most Significant Associations of CardioMetabochip SNPs and All Endurance Athletes as well as World-Class Athletes versus Controls in the GENATHLETE Cohort.
| All Athletes (N = 315) | World-Class Athletes (N = 168) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | CHR | Gene* | Effect Allele | Freq. in Athletes | Freq. in Controls | OR | P | Freq. in Athletes | Freq. in Controls | OR | P |
| rs17459307 | 1 | NOS1AP | A | 0.043 | 0.097 | 0.417 | 0.00016 | 0.060 | 0.097 | 0.590 | 0.04566 |
| rs6726044 | 2 | BMP10 (40 kb) | A | 0.316 | 0.222 | 1.619 | 0.00016 | 0.324 | 0.222 | 1.684 | 0.00050 |
| rs2118908 | 2 | ACOXL | G | 0.129 | 0.083 | 1.634 | 0.00796 | 0.164 | 0.083 | 2.168 | 0.00013 |
| rs6799372 | 3 | CNTN3 | A | 0.189 | 0.278 | 0.604 | 0.00017 | 0.191 | 0.278 | 0.611 | 0.00259 |
| rs10938202 | 4 | ATP8A1 | C | 0.113 | 0.053 | 2.264 | 0.00012 | 0.110 | 0.053 | 2.206 | 0.00113 |
| rs4699824 | 4 | PPP3CA (222 kb) | G | 0.402 | 0.308 | 1.509 | 0.0004771 | 0.429 | 0.308 | 1.687 | 0.00017 |
| rs10499127 | 6 | NKAIN2 | G | 0.129 | 0.069 | 1.999 | 0.00035 | 0.146 | 0.069 | 2.313 | 9.74x10-5 |
| rs17055965 | 8 | ADRA1A | G | 0.097 | 0.041 | 2.532 | 7.37x10-5 | 0.125 | 0.041 | 3.374 | 8.69x10-7 |
| rs7861665 | 9 | LOC101929330, GLIS3 (3 kb) | T | 0.046 | 0.106 | 0.406 | 5.35x10-5 | 0.051 | 0.106 | 0.448 | 0.00339 |
| rs17054974 | 9 | SEMA4D (51 kb) | GADD45G (55 kb) | C | 0.071 | 0.036 | 2.064 | 0.00497 | 0.098 | 0.036 | 2.922 | 7.04x10-5 |
| rs12573965 | 11 | KCNQ1 | C | 0.129 | 0.084 | 1.601 | 0.01062 | 0.170 | 0.084 | 2.217 | 6.70x10-5 |
| rs7947391 | 11 | NPAS4 | A | 0.338 | 0.441 | 0.649 | 0.00018 | 0.354 | 0.441 | 0.696 | 0.00910 |
| rs11613185 | 12 | BCL2L14 | A | 0.125 | 0.214 | 0.526 | 2.61x10-5 | 0.125 | 0.214 | 0.525 | 0.00064 |
| rs12821816 | 12 | NDUFA12 | G | 0.419 | 0.523 | 0.657 | 0.00019 | 0.435 | 0.523 | 0.700 | 0.00829 |
| rs61940911 | 12 | ANKRD13A | T | 0.098 | 0.044 | 2.386 | 0.00015 | 0.104 | 0.044 | 2.542 | 0.00026 |
| rs73195844 | 12 | CCDC63 | MYL2 (4 kb) | T | 0.116 | 0.052 | 2.411 | 3.43x10-5 | 0.116 | 0.052 | 2.415 | 0.00025 |
| rs9543114 | 13 | DIS3 | C | 0.075 | 0.030 | 2.635 | 0.00031 | 0.098 | 0.030 | 3.560 | 5.93x10-6 |
| rs9301108 | 13 | DAOA (653 kb) | EFNB2 (346 kb) | C | 0.067 | 0.033 | 2.105 | 0.00547 | 0.095 | 0.033 | 3.103 | 4.33x10-5 |
| rs214003 | 14 | NRXN3 | G | 0.132 | 0.080 | 1.752 | 0.00253 | 0.158 | 0.080 | 2.163 | 0.00017 |
| rs4776471 | 15 | RPLP1 (245 kb) | TLE3 (348 kb) | T | 0.457 | 0.338 | 1.653 | 1.32x10-5 | 0.464 | 0.338 | 1.701 | 0.00011 |
| rs11856981 | 15 | RPLP1 (288 kb) | TLE3 (305 kb) | T | 0.544 | 0.422 | 1.638 | 1.24x10-5 | 0.557 | 0.422 | 1.720 | 6.17x10-5 |
| rs4288991 | 16 | TOX3 (91 kb) | CHD9 (416 kb) | C | 0.194 | 0.286 | 0.600 | 0.00012 | 0.196 | 0.286 | 0.610 | 0.00231 |
| rs8065364 | 17 | CARD14 | C | 0.268 | 0.366 | 0.636 | 0.00019 | 0.268 | 0.366 | 0.635 | 0.00206 |
| rs578211 | 18 | MYO5B | A | 0.303 | 0.205 | 1.691 | 5.49x10-5 | 0.310 | 0.205 | 1.742 | 0.00027 |
| rs4808571 | 19 | MYO9B | A | 0.229 | 0.155 | 1.619 | 0.00082 | 0.268 | 0.155 | 1.999 | 2.13x10-5 |
| rs62135557 | 19 | MAU2 | G | 0.092 | 0.039 | 2.494 | 0.00013 | 0.074 | 0.039 | 1.977 | 0.01734 |
*The gene located nearest to the SNP. Distance to the gene in kilo bases (1,000 bp) is shown in parentheses. If no distance is shown, the SNP is located within the gene locus.
World-class athletes defined as endurance athletes (N = 168) with VO2max ≥78 ml O2/kg/min. Total number of controls = 320.
OR = Odds ratio.
The strongest evidence of association (1.2x10-5 < p < 6.1x10-5) was detected with a cluster of SNPs located on the long arm of chromosome 15 (15q23) about 69.7 million base pairs from the start of the chromosome (Panel A in Fig 1). While the cluster is located in an intergenic region, the same chromosomal region has been previously reported to be associated with atrial fibrillation in the Framingham Heart Study [35]. Five additional SNPs showed associations with p-values ranging from 2.61x10-5 to 7.37x10-5; these markers were located within or in the vicinity of genes encoding adrenoceptor alpha 1 A (ADRA1A), BCL2-like 14 [apoptosis facilitator] (BCL2L14), GLIS family zinc finger 3 (GLIS3), myosin, light-chain 2, regulatory, cardiac, slow (MYL2), and myosin VB (MYO5B).
To evaluate whether these genetic associations were even more prominent among elite athletes with the highest cardiorespiratory fitness level, the analyses were repeated by comparing only those athletes with a VO2max of ≥78 mL/kg/min (N = 168) versus controls. The rationale was that sequence variants playing a role in maximal endurance capacity should cluster even more tightly in world-class endurance athletes compared to endurance athletes with lower VO2max levels. An overview of the results is shown Panel B of Fig 1. A total of 50 SNPs showed associations with p-values less than 5.0 x 10−4 and eight SNPs with p-values less than 9.7 x 10−5. The strongest evidence of association was detected with a SNP located in the ADRA1A locus (p = 8.69 x 10−7) and the top SNPs are also listed in Table 2.
Japanese Endurance GWAS
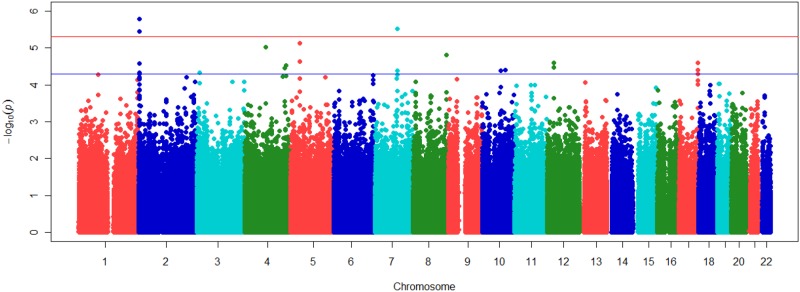
In the Japanese cohort, after removing SNPs and individuals failing quality control, 541,179 autosomal SNPs were available for analysis. A total of 31% of the SNPs were common to the Illumina chips used in the Japanese endurance athlete cohort and GENATHLETE. Allelic association analyses were performed by comparing allele-frequency differences between Japanese endurance athletes and controls. The Quantile-Quantile p-value plot of observed versus expected—log10(p) values is shown in S2 Fig. The genomic inflation factor (λ) value in Japanese was 1.002, indicating that there was no substantial evidence of population stratification. A Manhattan plot of—log10(p) values for associations of elite Japanese endurance status with markers in 22 autosomes is shown in Fig 2.
Fig 2. Manhattan plot of -log10(p) values against genomic position for association of elite endurance status with markers in 22 autosomes in Japanese.
Red line refers to p = 5x10-6; blue line refers to p = 5x10-5.
No SNPs exceeded the threshold for genome-wide significance (p value < 5 x 10−8). The association results for markers with p < 5 x 10−5 plus a few markers as defined next are displayed in Table 3. Out of the total SNPs entered into the association analysis, 21 met this threshold. Among them, 3 SNPs were associated at p < 5 x 10−6. Regional association plots of the top signals were created using LocusZoom Version 1.1 [36], including information on the location and orientation of genes, local estimates of recombination rates and levels of linkage disequilibrium (LD). An individual plot was specified by the SNP of interest and treated as the key marker for the region. Markers within a 500 kb flanking region each side of the index SNP were included. Plots were generated based on Human Genome19 (hg19). Pairwise LD between the index SNP and the surrounding SNPs and recombination rates were estimated in LocusZoom using 1000G Mar 2012 ASN as the reference population.
Table 3. Association Results for Markers Chosen for Replication from the Japanese World-Class Endurance Athlete Cohort.
| SNP | Chr | Gene* | Minor allele | Freq. in athletes | Freq. in controls | OR | 95% CI | p-value |
|---|---|---|---|---|---|---|---|---|
| rs10874242 | 1 | LPHN2 (153 kb) | T | 0.30 | 0.53 | 0.39 | 0.24–0.62 | 5.38x10-5 |
| s12047209 | 1 | AKT3 | C | 0.42 | 0.22 | 2.60 | 1.61–4.20 | 7.29x10-5 |
| rs921665 | 2 | TSSC1 (18 kb) | G | 0.30 | 0.57 | 0.32 | 0.20–0.52 | 1.68x10-6 |
| rs2694093 | 2 | TSSC1 | G | 0.17 | 0.36 | 0.35 | 0.20–0.61 | 0.0001 |
| rs6548153 | 2 | TSSC1 | C | 0.28 | 0.51 | 0.37 | 0.23–0.59 | 2.69x10-5 |
| rs2361506 | 2 | MROH2A | T | 0.12 | 0.31 | 0.30 | 0.16–0.56 | 8.32x10-5 |
| rs7650685 | 3 | VGLL4 | G | 0.63 | 0.40 | 2.54 | 1.61–3.99 | 4.73x10-5 |
| rs10007111 | 4 | MEPE | A | 0.45 | 0.22 | 2.87 | 1.79–4.62 | 9.72x10-6 |
| rs558129 | 4 | GALNTL6 | T | 0.06 | 0.24 | 0.20 | 0.09–0.45 | 3.01x10-5 |
| rs2910756 | 5 | GDNF-AS1 | G | 0.14 | 0.37 | 0.28 | 0.16–0.50 | 7.58x10-6 |
| rs9355947 | 6 | PARK2 | G | 0.57 | 0.34 | 2.52 | 1.60–3.96 | 5.46x10-5 |
| rs6959675 | 7 | CFAP69 | C | 0.27 | 0.49 | 0.38 | 0.24–0.62 | 6.82x10-5 |
| rs11975386 | 7 | BET1 (70 kb) | A | 0.42 | 0.19 | 3.14 | 1.92–5.13 | 3.07x10-6 |
| rs16906888 | 8 | FAM135B (898 kb) | G | 0.38 | 0.17 | 2.97 | 1.79–4.92 | 1.58x10-5 |
| rs3780169 | 9 | PAX5 | C | 0.28 | 0.50 | 0.39 | 0.24–0.62 | 7.17x10-5 |
| rs17690338 | 10 | ZNF503-AS1 | A | 0.45 | 0.24 | 2.63 | 1.65–4.21 | 4.21x10-5 |
| rs2761291 | 10 | MYOF | A | 0.24 | 0.08 | 3.54 | 1.89–6.64 | 4.03x10-5 |
| rs9580890 | 13 | SPATA13 | G | 0.18 | 0.38 | 0.35 | 0.20–0.60 | 8.50x10-5 |
| rs4541108 | 17 | RBFOX3 | A | 0.22 | 0.06 | 4.00 | 2.02–7.90 | 2.51x10-5 |
OR = Odds ratio; CI = 95% confidence interval.
For athletes, N = 60 and for controls, N = 116.
*The gene located nearest to the SNP. Distance to the gene in kilo bases (1,000 bp) is shown in parentheses. If no distance is shown, the SNP is located within the gene locus.
Regional association plots of the 21 SNPs with a p < 5 x 10−5 were further inspected, leading to the exclusion of 10 SNPs, which were considered as redundant. The regional association plots for the remaining 11 SNPs (see Table 3) are presented in S3–S13 Figs of the Supporting Information. Another set of 7 SNPs with a p < 10−4 were added to the panel after examination of regional association plots (S14–S20 Figs). Furthermore, rs2694093 (p = 0.00014) was independently associated with athlete status and is in a region near the strongest signal rs921665 (chr2:3174321, hg19/1000 Genomes Mar 2012 ASN; S21 Fig). It was also included in the discovery set. All 19 SNPs retained for the replication phase based on the results on the Japanese athletes are summarized in Table 3. These 19 SNPs were combined with the 26 retained from GENATHLETE to constitute the panel of 45 SNPs that was used in the replication cohorts.
Replication Phase
Table 4 depicts the association results for the 42 SNPs that could be tested in at least two of the four replication cohorts of Caucasians, namely Australian, Polish, Russian and Spanish endurance athletes and controls. Meta-analysis showed that none of the markers summarized in Table 4 reached statistical significance after accounting for multiple testing (Bonferroni-corrected statistical significance p<0.0011), although one marker (rs558129 in GALNT6) showed a nominal p-value of P = 0.02. Five SNPs were nominally (p<0.05) associated with endurance athlete status in the Australian study, as well as one in the Polish cohort, four in the Spanish cohort, and five in the Russian cohort. One SNP (rs7947391) showed nominally significant associations in three replication cohorts (Australians, Polish and Spanish). However, as is evident from the meta-analysis results, the direction of the association was not the same across the three studies; the minor allele frequency was lower in athletes than in controls in the Spanish cohort (as well as in the GENATHLETE discovery cohort), whereas Australian and Polish athletes had greater minor allele frequency than their non-athlete controls.
Table 4. Results for Replication Meta-Analysis and for Individual Cohorts for All Athletes and Controls of European Descent; Results in bold type are for p≤0.05.
| Meta-analysis | Australia | Poland | Spain | Russia | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Chr | MAP | Gene* | Effect allele | OR | P | Q | I2 | OR | P | OR | P | OR | P | OR | P |
| rs10874242 | 1 | 81,426,166 | LPHN2 (153 kb) | T | 0.95 | 0.61 | 0.41 | 0.0 | 1.16 | 0.37 | 0.86 | 0.53 | 0.97 | 0.88 | 0.77 | 0.17 |
| rs17459307 | 1 | 162,245,878 | NOS1AP | A | 1.01 | 0.95 | 0.69 | 0.0 | 1.08 | 0.77 | 1.19 | 0.61 | 0.83 | 0.53 | NA | NA |
| rs12047209 | 1 | 243,791,105 | AKT3 | C | 1.19 | 0.18 | 0.23 | 30.5 | 1.16 | 0.44 | 1.07 | 0.83 | 0.89 | 0.63 | 1.60 | 0.009 |
| rs921665 | 2 | 3,170,550 | TSSC1 (18 kb) | A | 1.29 | 0.07 | 0.27 | 23.0 | 0.90 | 0.64 | 1.56 | 0.12 | 1.38 | 0.20 | 1.56 | 0.05 |
| rs2694093 | 2 | 3,254,403 | TSSC1 | G | 0.87 | 0.34 | 0.01 | 74.1 | 1.03 | 0.82 | 0.93 | 0.71 | 1.04 | 0.81 | 0.58 | 0.00008 |
| rs6548153 | 2 | 3,322,274 | TSSC1 | A | 1.08 | 0.68 | 0.06 | 63.7 | 0.76 | 0.13 | 1.36 | 0.22 | NA | NA | 1.28 | 0.16 |
| rs2118908 | 2 | 111,067,015 | ACOXL | G | 1.05 | 0.75 | 0.08 | 55.7 | 0.92 | 0.70 | 0.68 | 0.17 | 1.11 | 0.64 | 1.51 | 0.02 |
| rs2361506 | 2 | 233,830,694 | MROH2A | T | 1.13 | 0.16 | 0.84 | 0.0 | 1.17 | 0.25 | 1.02 | 0.90 | NA | NA | 1.17 | 0.33 |
| rs7650685 | 3 | 11,660,982 | VGLL4 | G | 0.96 | 0.57 | 0.47 | 0.0 | 0.84 | 0.23 | 0.84 | 0.38 | 1.04 | 0.80 | 1.13 | 0.45 |
| rs6799372 | 3 | 74,481,495 | CNTN3 | A | 0.90 | 0.44 | 0.17 | 44.0 | 0.92 | 0.56 | 1.17 | 0.45 | 0.69 | 0.049 | NA | NA |
| rs10938202 | 4 | 42,650,320 | ATP8A1 | C | 1.18 | 0.38 | 0.22 | 31.7 | 1.83 | 0.04 | 0.87 | 0.69 | 1.37 | 0.36 | 0.86 | 0.61 |
| rs10007111 | 4 | 87,828,549 | MEPE | A | 0.95 | 0.49 | 0.96 | 0.0 | 0.95 | 0.70 | 0.99 | 0.96 | 0.89 | 0.45 | 0.98 | 0.90 |
| rs4699824 | 4 | 100,801,328 | EMCN (283 kb), PPP3CA (222 kb) | G | 1.01 | 0.94 | 0.07 | 58.5 | 0.74 | 0.03 | 1.02 | 0.90 | 1.19 | 0.28 | 1.20 | 0.26 |
| rs558129 | 4 | 171,829,960 | GALNTL6 | T | 0.84 | 0.02 | 0.38 | 2.1 | 0.69 | 0.01 | 0.82 | 0.31 | 0.99 | 0.96 | 0.89 | 0.39 |
| rs2910756 | 5 | 37,859,972 | GDNF-AS1 | G | 0.92 | 0.50 | 0.18 | 38.2 | 1.03 | 0.88 | 0.95 | 0.84 | 0.60 | 0.02 | 1.08 | 0.65 |
| rs10499127 | 6 | 124,594,509 | NKAIN2 | G | 1.16 | 0.21 | 0.60 | 0.0 | 1.11 | 0.62 | 0.87 | 0.61 | 1.38 | 0.17 | 1.29 | 0.30 |
| rs9355947 | 6 | 161,887,402 | PARK2 | A | 0.94 | 0.50 | 0.24 | 28.5 | 0.87 | 0.27 | 0.74 | 0.10 | 1.15 | 0.36 | 1.03 | 0.86 |
| rs6959675 | 7 | 90,253,815 | CFAP69 | T | 0.78 | 0.49 | 0.32 | 11.9 | 0.36 | 0.10 | 0.71 | 0.69 | NA | NA | 1.16 | 0.73 |
| rs11975386 | 7 | 94,075,721 | BET1 (70 kb) | A | 1.24 | 0.25 | 0.42 | 0.0 | 1.68 | 0.07 | 0.71 | 0.46 | 1.27 | 0.55 | 1.00 | 0.99 |
| rs17055965 | 8 | 26,769,562 | ADRA1A | G | 1.25 | 0.13 | 0.68 | 0.0 | 1.43 | 0.20 | 0.76 | 0.53 | 1.31 | 0.26 | 1.26 | 0.44 |
| rs16906888 | 8 | 137,231,375 | FAM135B (898 kb) | G | 1.17 | 0.17 | 0.50 | 0.0 | NA | NA | NA | NA | 1.27 | 0.15 | 1.09 | 0.61 |
| rs7861665 | 9 | 4,302,988 | LOC101929330, GLIS3 (3 kb) | T | 0.96 | 0.77 | 0.26 | 25.1 | 0.82 | 0.38 | 0.70 | 0.28 | 1.41 | 0.15 | 0.92 | 0.77 |
| rs3780169 | 9 | 36,979,398 | PAX5 | T | 1.02 | 0.76 | 0.68 | 0.0 | 0.90 | 0.44 | 1.09 | 0.63 | 1.15 | 0.40 | 1.04 | 0.78 |
| rs17690338 | 10 | 75,357,798 | ZNF503-AS1 | A | 0.94 | 0.54 | 0.23 | 31.2 | 1.08 | 0.56 | 1.00 | 0.98 | 1.00 | 0.99 | 0.70 | 0.03 |
| rs2761291 | 10 | 93,328,423 | MYOF | C | 0.90 | 0.32 | 0.44 | 0.0 | 1.04 | 0.78 | 0.76 | 0.18 | NA | NA | 0.86 | 0.37 |
| rs12573965 | 11 | 2,660,402 | KCNQ1 | C | 0.76 | 0.06 | 0.60 | 0.0 | 0.87 | 0.50 | 0.75 | 0.37 | 0.62 | 0.06 | NA | NA |
| rs7947391 | 11 | 66,419,411 | NPAS4 | A | 1.13 | 0.57 | 0.00 | 87.4 | 1.37 | 0.02 | 1.85 | 0.0008 | 0.63 | 0.003 | 1.06 | 0.74 |
| rs11613185 | 12 | 12,075,733 | BCL2L14 | A | 0.91 | 0.62 | 0.19 | 41.5 | 1.07 | 0.70 | 0.72 | 0.18 | NA | NA | NA | NA |
| rs61940911 | 12 | 110,029,339 | ANKRD13A | T | 1.21 | 0.24 | 0.95 | 0.0 | 1.29 | 0.31 | 1.14 | 0.70 | 1.17 | 0.58 | NA | NA |
| rs73195844 | 12 | 110,906,768 | CCDC63, MYL2 (4 kb) | T | 0.86 | 0.41 | 0.57 | 0.0 | 0.78 | 0.33 | 0.96 | 0.87 | NA | NA | NA | NA |
| rs9580890 | 13 | 24,165,281 | SPATA13 | G | 0.87 | 0.35 | 0.76 | 0.0 | 0.80 | 0.26 | 0.89 | 0.70 | NA | NA | 1.03 | 0.92 |
| rs9543114 | 13 | 72,767,956 | DIS3 | C | 0.94 | 0.77 | 0.61 | 0.0 | 0.75 | 0.38 | 1.01 | 0.99 | 1.29 | 0.57 | NA | NA |
| rs9301108 | 13 | 106,143,743 | DAOA (653 kb), EFNB2 (346 kb) | C | 1.14 | 0.45 | 0.36 | 6.2 | 0.75 | 0.33 | 1.61 | 0.21 | 1.17 | 0.56 | 1.44 | 0.33 |
| rs214003 | 14 | 78,336,687 | NRXN3 | G | 0.89 | 0.55 | 0.11 | 50.6 | 0.59 | 0.03 | 0.80 | 0.51 | 1.52 | 0.17 | 0.95 | 0.83 |
| rs4776471 | 15 | 69,700,119 | RPLP1 (245 kb), TLE3 (348 kb) | T | 1.14 | 0.25 | 0.08 | 55.1 | 1.17 | 0.25 | 1.17 | 0.39 | 1.47 | 0.01 | 0.86 | 0.29 |
| rs11856981 | 15 | 69,743,207 | RPLP1 (288 kb), TLE3 (305 kb) | T | 1.11 | 0.15 | 0.77 | 0.0 | 1.08 | 0.57 | 1.09 | 0.65 | 1.27 | 0.11 | 0.96 | 0.76 |
| rs4288991 | 16 | 52,639,252 | TOX3 (91 kb), CHD9 (416 kb) | C | 0.86 | 0.09 | 0.96 | 0.0 | 0.89 | 0.43 | 0.81 | 0.38 | 0.82 | 0.23 | 0.91 | 0.58 |
| rs4541108 | 17 | 79,332,527 | RBFOX3 | A | 1.05 | 0.73 | 0.99 | 0.0 | 1.03 | 0.90 | 1.07 | 0.83 | NA | NA | 1.06 | 0.78 |
| rs8065364 | 17 | 80,189,160 | CARD14 | C | 0.96 | 0.62 | 0.65 | 0.0 | 1.00 | 0.98 | 1.16 | 0.45 | 0.85 | 0.31 | 0.92 | 0.58 |
| rs578211 | 18 | 49,868,025 | MYO5B | A | 1.00 | 0.98 | 0.08 | 59.9 | 0.83 | 0.19 | 0.88 | 0.56 | 1.38 | 0.08 | NA | NA |
| rs4808571 | 19 | 17,115,419 | MYO9B | A | 1.03 | 0.85 | 0.06 | 59.9 | 0.74 | 0.08 | 1.17 | 0.49 | 1.49 | 0.05 | 0.94 | 0.70 |
| rs62135557 | 19 | 19,355,484 | MAU2 | G | 0.93 | 0.75 | 0.24 | 30.7 | 0.77 | 0.40 | 0.61 | 0.28 | 1.31 | 0.30 | NA | NA |
Effect allele (i.e., minor allele).
OR = odds ratio from a random-effects meta-analysis.
P = p-value for OR from a random-effects meta-analysis.
Q = p-value for Cochrane's Q statistic (tests heterogeneity in effects across individual studies).
I2 (I-squared) = heterogeneity index (0–100; 0 = no heterogeneity, 100 = max. heterogeneity).
MAP based on GRCh38 (hg38).
*The gene located nearest to the SNP. Distance to the gene in kilo bases (1,000 bp) is shown in parentheses. If no distance is shown, the SNP is located within the gene locus.
The meta-analysis of the subgroup of athletes classified as world class and truly elite endurance athletes did not reveal any significant associations. Five SNPs had nominal p-values less than 0.05 in Australian world-class athletes, one in Polish world-class athletes, four SNPs in Spanish world-class athletes, and three in Russian world-class athletes (Table 5). However, these associations were generally not directionally consistent among cohorts.
Table 5. Results for Meta-Analysis of World-Class Athletes and Controls of European Descent; Results in bold type are for p≤0.05.
| Meta-analysis | Australia | Poland | Spain | Russia | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Chr | MAP | Gene* | Effect allele | OR | P | Q | I2 | OR | P | OR | P | OR | P | OR | P |
| rs10874242 | 1 | 81,426,166 | LPHN2 (153 kb) | T | 1.00 | 1.00 | 0.33 | 11.9 | 1.19 | 0.32 | 1.13 | 0.68 | 0.97 | 0.88 | 0.67 | 0.13 |
| rs17459307 | 1 | 162,245,878 | NOS1AP | A | 1.02 | 0.91 | 0.60 | 0.0 | 1.25 | 0.43 | 1.00 | 0.99 | 0.83 | 0.53 | NA | NA |
| rs12047209 | 1 | 243,791,105 | AKT3 | C | 1.26 | 0.31 | 0.02 | 69.4 | 1.16 | 0.46 | 0.96 | 0.92 | 0.89 | 0.63 | 2.29 | 0.0002 |
| rs921665 | 2 | 3,170,550 | TSSC1 (18 kb) | A | 1.19 | 0.21 | 0.58 | 0.0 | 0.93 | 0.76 | 1.13 | 0.76 | 1.38 | 0.20 | 1.46 | 0.19 |
| rs2694093 | 2 | 3,254,403 | TSSC1 | G | 0.91 | 0.33 | 0.32 | 14.0 | 0.98 | 0.91 | 0.89 | 0.63 | 1.04 | 0.81 | 0.67 | 0.04 |
| rs6548153 | 2 | 3,322,274 | TSSC1 | A | 1.31 | 0.28 | 0.03 | 71.4 | 0.85 | 0.40 | 1.65 | 0.08 | NA | NA | 1.71 | 0.01 |
| rs2118908 | 2 | 111,067,015 | ACOXL | G | 1.00 | 1.00 | 0.28 | 20.9 | 0.98 | 0.92 | 0.50 | 0.08 | 1.11 | 0.64 | 1.23 | 0.41 |
| rs2361506 | 2 | 233,830,694 | MROH2A | T | 1.12 | 0.27 | 0.80 | 0.0 | 1.13 | 0.41 | 0.99 | 0.97 | NA | NA | 1.22 | 0.32 |
| rs7650685 | 3 | 11,660,982 | VGLL4 | G | 0.93 | 0.40 | 0.77 | 0.0 | 0.82 | 0.20 | 0.96 | 0.86 | 1.04 | 0.80 | 0.91 | 0.68 |
| rs6799372 | 3 | 74,481,495 | CNTN3 | A | 0.86 | 0.30 | 0.19 | 39.3 | 0.85 | 0.30 | 1.23 | 0.42 | 0.69 | 0.049 | NA | NA |
| rs10938202 | 4 | 42,650,320 | ATP8A1 | C | 1.35 | 0.08 | 0.52 | 0.0 | 1.90 | 0.03 | 1.02 | 0.96 | 1.37 | 0.36 | 1.03 | 0.94 |
| rs10007111 | 4 | 87,828,549 | MEPE | A | 0.92 | 0.36 | 0.99 | 0.0 | 0.93 | 0.61 | 0.96 | 0.85 | 0.89 | 0.45 | 0.95 | 0.82 |
| rs4699824 | 4 | 100,801,328 | EMCN (283 kb), PPP3CA (222 kb) | G | 1.08 | 0.63 | 0.04 | 64.7 | 0.73 | 0.04 | 1.24 | 0.35 | 1.19 | 0.28 | 1.36 | 0.14 |
| rs558129 | 4 | 171,829,960 | GALNTL6 | T | 0.87 | 0.13 | 0.78 | 0.0 | 0.80 | 0.15 | 0.80 | 0.37 | 0.99 | 0.96 | 0.86 | 0.47 |
| rs2910756 | 5 | 37,859,972 | GDNF-AS1 | G | 0.88 | 0.36 | 0.22 | 32.4 | 1.10 | 0.60 | 0.89 | 0.73 | 0.60 | 0.02 | 0.95 | 0.83 |
| rs10499127 | 6 | 124,594,509 | NKAIN2 | G | 1.25 | 0.09 | 0.62 | 0.0 | 1.26 | 0.28 | 0.83 | 0.59 | 1.38 | 0.17 | 1.43 | 0.24 |
| rs9355947 | 6 | 161,887,402 | PARK2 | A | 0.95 | 0.57 | 0.29 | 19.1 | 0.80 | 0.11 | 0.83 | 0.40 | 1.15 | 0.36 | 1.05 | 0.82 |
| rs6959675 | 7 | 90,253,815 | CFAP69 | T | 0.79 | 0.55 | 0.54 | 0.0 | 0.46 | 0.23 | 0.67 | 0.72 | NA | NA | 1.18 | 0.76 |
| rs11975386 | 7 | 94,075,721 | BET1 (70 kb) | A | 1.35 | 0.14 | 0.50 | 0.0 | 1.82 | 0.045 | 1.01 | 0.99 | 1.27 | 0.55 | 0.75 | 0.61 |
| rs17055965 | 8 | 26,769,562 | ADRA1A | G | 1.20 | 0.27 | 0.60 | 0.0 | 1.39 | 0.27 | 0.91 | 0.86 | 1.31 | 0.26 | 0.67 | 0.44 |
| rs16906888 | 8 | 137,231,375 | FAM135B (898 kb) | G | 1.10 | 0.58 | 0.20 | 38.7 | NA | NA | NA | NA | 1.27 | 0.15 | 0.90 | 0.62 |
| rs7861665 | 9 | 4,302,988 | LOC101929330, GLIS3 (3 kb) | T | 1.00 | 0.99 | 0.12 | 48.3 | 0.71 | 0.19 | 0.62 | 0.27 | 1.41 | 0.15 | 1.40 | 0.34 |
| rs3780169 | 9 | 36,979,398 | PAX5 | T | 1.07 | 0.48 | 0.71 | 0.0 | 0.98 | 0.88 | 1.27 | 0.28 | 1.15 | 0.40 | 0.97 | 0.88 |
| rs17690338 | 10 | 75,357,798 | ZNF503-AS1 | A | 0.95 | 0.62 | 0.25 | 27.6 | 1.13 | 0.41 | 0.91 | 0.69 | NA | NA | 0.66 | 0.06 |
| rs2761291 | 10 | 93,328,423 | MYOF | C | 0.84 | 0.36 | 0.12 | 52.8 | 1.13 | 0.48 | 0.72 | 0.23 | NA | NA | 0.66 | 0.07 |
| rs12573965 | 11 | 2,660,402 | KCNQ1 | C | 0.79 | 0.13 | 0.50 | 0.0 | 0.89 | 0.60 | 0.95 | 0.89 | 0.62 | 0.06 | NA | NA |
| rs7947391 | 11 | 66,419,411 | NPAS4 | A | 1.17 | 0.51 | 0.00 | 85.5 | 1.35 | 0.04 | 1.86 | 0.007 | 0.63 | 0.003 | 1.23 | 0.30 |
| rs11613185 | 12 | 12,075,733 | BCL2L14 | A | 0.97 | 0.89 | 0.24 | 28.9 | 1.13 | 0.53 | 0.74 | 0.31 | NA | NA | NA | NA |
| rs61940911 | 12 | 110,029,339 | ANKRD13A | T | 1.12 | 0.54 | 0.83 | 0.0 | 1.17 | 0.56 | 0.87 | 0.76 | 1.17 | 0.58 | NA | NA |
| rs73195844 | 12 | 110,906,768 | CCDC63, MYL2 (4 kb) | T | 0.73 | 0.16 | 0.94 | 0.0 | 0.74 | 0.29 | 0.71 | 0.35 | NA | NA | NA | NA |
| rs9580890 | 13 | 24,165,281 | SPATA13 | G | 0.78 | 0.16 | 0.72 | 0.0 | 0.78 | 0.25 | 0.60 | 0.24 | NA | NA | 0.96 | 0.92 |
| rs9543114 | 13 | 72,767,956 | DIS3 | C | 1.03 | 0.90 | 0.82 | 0.0 | 0.98 | 0.94 | 0.86 | 0.77 | 1.29 | 0.57 | NA | NA |
| rs9301108 | 13 | 106,143,743 | DAOA (653 kb), EFNB2 (346 kb) | C | 1.07 | 0.80 | 0.08 | 55.2 | 0.50 | 0.06 | 2.06 | 0.09 | 1.17 | 0.56 | 1.21 | 0.71 |
| rs214003 | 14 | 78,336,687 | NRXN3 | G | 0.82 | 0.43 | 0.07 | 57.4 | 0.51 | 0.01 | 0.82 | 0.65 | 1.52 | 0.17 | 0.73 | 0.34 |
| rs4776471 | 15 | 69,700,119 | RPLP1 (245 kb), TLE3 (348 kb) | T | 1.09 | 0.59 | 0.02 | 68.3 | 1.23 | 0.15 | 1.03 | 0.89 | 1.47 | 0.01 | 0.70 | 0.07 |
| rs11856981 | 15 | 69,743,207 | RPLP1 (288 kb), TLE3 (305 kb) | T | 1.09 | 0.30 | 0.53 | 0.0 | 1.10 | 0.50 | 0.88 | 0.59 | 1.27 | 0.11 | 0.98 | 0.91 |
| rs4288991 | 16 | 52,639,252 | TOX3 (91 kb), CHD9 (416 kb) | C | 0.85 | 0.09 | 0.85 | 0.0 | 0.85 | 0.33 | 0.70 | 0.25 | 0.82 | 0.23 | 0.98 | 0.92 |
| rs4541108 | 17 | 79,332,527 | RBFOX3 | A | 1.16 | 0.34 | 0.87 | 0.0 | 1.06 | 0.77 | 1.22 | 0.56 | NA | NA | 1.27 | 0.37 |
| rs8065364 | 17 | 80,189,160 | CARD14 | C | 0.95 | 0.59 | 0.83 | 0.0 | 1.03 | 0.84 | 0.93 | 0.77 | 0.85 | 0.31 | 1.02 | 0.94 |
| rs578211 | 18 | 49,868,025 | MYO5B | A | 0.94 | 0.77 | 0.04 | 69.2 | 0.77 | 0.10 | 0.74 | 0.30 | 1.38 | 0.08 | NA | NA |
| rs4808571 | 19 | 17,115,419 | MYO9B | A | 1.05 | 0.73 | 0.16 | 42.6 | 0.80 | 0.22 | 0.97 | 0.92 | 1.49 | 0.05 | 1.06 | 0.78 |
| rs62135557 | 19 | 19,355,484 | MAU2 | G | 0.89 | 0.71 | 0.15 | 47.1 | 0.79 | 0.46 | 0.33 | 0.12 | 1.31 | 0.30 | NA | NA |
Effect allele (i.e., minor allele).
OR = odds ratio from a random-effects meta-analysis.
P = p-value for OR from a random-effects meta-analysis.
Q = p-value for Cochrane's Q statistic (tests heterogeneity in effects across individual studies).
I2 (I-squared) = heterogeneity index (0–100; 0 = no heterogeneity, 100 = max. heterogeneity).
MAP based on GRCh38 (hg38).
*The gene located nearest to the SNP. Distance to the gene in kilo bases (1,000 bp) is shown in parentheses. If no distance is shown, the SNP is located within the gene locus.
Next, we report on the associations between 35 SNPs identified in the discovery phase and endurance athlete status in Ethiopian and Kenyan athletes and controls (Table 6). The lower number of SNPs available for these analyses is due to the fact that several SNPs selected for the replication phase were monomorphic or very rare in African populations. The meta-analysis of the Ethiopian and Kenyan athletes did not reveal any associations that were significant after accounting for multiple testing, although four SNPs had nominal p-value less than 0.05. Five and three SNPs had nominal p-value less than 0.05 in the Kenyan sample (276 athletes versus 83 controls) and in the Ethiopian study (75 athletes versus 198 controls), respectively, but none of these SNPs overlapped between the two cohorts. In fact, the most significant SNPs observed in the Kenyan cohort showed associations in the opposite direction in the Ethiopian cohort.
Table 6. Results for the Replication in Athletes and Controls from Kenya and Ethiopia; Results in bold type are for p ≤0.05.
| Meta-analysis | Kenya | Ethiopia | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Chr | MAP | Gene* | Effect allele | OR | P | Q | I2 | OR | P | OR | P |
| rs10874242 | 1 | 81,426,166 | LPHN2 (153 kb) | T | 1.39 | 0.11 | 0.22 | 32.9 | 1.14 | 0.58 | 1.70 | 0.02 |
| rs12047209 | 1 | 243,791,105 | AKT3 | C | 0.74 | 0.67 | 0.02 | 82.5 | 0.37 | 0.01 | 1.52 | 0.33 |
| rs921665 | 2 | 3,170,550 | TSSC1 (18 kb) | A | 0.75 | 0.03 | 0.33 | 0.0 | 0.67 | 0.02 | 0.87 | 0.51 |
| rs2694093 | 2 | 3,254,403 | TSSC1 | G | 0.94 | 0.70 | 0.84 | 0.0 | 0.97 | 0.89 | 0.91 | 0.68 |
| rs6726044 | 2 | 68,911,860 | BMP10 (40 kb) | A | 1.07 | 0.85 | 0.02 | 81.7 | 1.57 | 0.04 | 0.73 | 0.19 |
| rs2118908 | 2 | 111,067,015 | ACOXL | G | 0.98 | 0.94 | 0.98 | 0.0 | 0.98 | 0.94 | 0.99 | 0.98 |
| rs7650685 | 3 | 11,660,982 | VGLL4 | G | 0.98 | 0.90 | 0.61 | 0.0 | 1.05 | 0.80 | 0.92 | 0.65 |
| rs6799372 | 3 | 74,481,495 | CNTN3 | A | 1.39 | 0.04 | 0.92 | 0.0 | 1.37 | 0.16 | 1.42 | 0.13 |
| rs10938202 | 4 | 42,650,320 | ATP8A1 | C | 0.57 | 0.07 | 0.21 | 35.4 | 0.74 | 0.31 | 0.40 | 0.02 |
| rs10007111 | 4 | 87,828,549 | MEPE | A | 1.03 | 0.95 | 0.02 | 82.2 | 1.51 | 0.05 | 0.68 | 0.14 |
| rs4699824 | 4 | 100,801,328 | EMCN (283 kb), PPP3CA (222 kb) | A | 1.00 | 0.98 | 0.64 | 0.0 | 1.07 | 0.74 | 0.94 | 0.73 |
| rs558129 | 4 | 171,829,960 | GALNTL6 | T | 0.80 | 0.15 | 0.44 | 0.0 | 0.72 | 0.11 | 0.91 | 0.69 |
| rs2910756 | 5 | 37,859,972 | GDNF-AS1 | G | 0.74 | 0.04 | 0.35 | 0.0 | 0.82 | 0.27 | 0.62 | 0.04 |
| rs10499127 | 6 | 124,594,509 | NKAIN2 | G | 0.92 | 0.67 | 0.37 | 0.0 | 0.75 | 0.34 | 1.08 | 0.78 |
| rs9355947 | 6 | 161,887,402 | PARK2 | G | 0.93 | 0.60 | 0.73 | 0.0 | 0.89 | 0.54 | 0.98 | 0.90 |
| rs11975386 | 7 | 94,075,721 | BET1 (70 kb) | A | 1.05 | 0.83 | 0.83 | 0.0 | 1.09 | 0.76 | 0.99 | 0.98 |
| rs17055965 | 8 | 26,769,562 | ADRA1A | G | 1.27 | 0.37 | 0.70 | 0.0 | 1.44 | 0.39 | 1.17 | 0.64 |
| rs16906888 | 8 | 137,231,375 | FAM135B (898 kb) | G | 1.12 | 0.40 | 0.46 | 0.0 | 1.23 | 0.27 | 1.01 | 0.98 |
| rs7861665 | 9 | 4,302,988 | LOC101929330, GLIS3 (3 kb) | T | 0.81 | 0.13 | 0.70 | 0.0 | 0.78 | 0.16 | 0.87 | 0.51 |
| rs3780169 | 9 | 36,979,398 | PAX5 | T | 0.80 | 0.12 | 0.32 | 0.0 | 0.90 | 0.59 | 0.68 | 0.08 |
| rs17054974 | 9 | 89,549,371 | SEMA4D (51 kb), GADD45G (55 kb) | C | 1.08 | 0.87 | 0.13 | 57.4 | 1.75 | 0.21 | 0.71 | 0.36 |
| rs17690338 | 10 | 75,357,798 | ZNF503-AS1 | A | 1.02 | 0.92 | 0.19 | 42.4 | 1.24 | 0.31 | 0.84 | 0.40 |
| rs12573965 | 11 | 2,660,402 | KCNQ1 | C | 0.82 | 0.90 | 0.01 | 86.3 | 0.19 | 0.005 | 4.02 | 0.10 |
| rs7947391 | 11 | 66,419,411 | NPAS4 | G | 0.75 | 0.03 | 0.71 | 0.0 | 0.71 | 0.06 | 0.79 | 0.21 |
| rs61940911 | 12 | 110,029,339 | ANKRD13A | T | 1.23 | 0.57 | 0.69 | 0.0 | 0.98 | 0.98 | 1.34 | 0.48 |
| rs73195844 | 12 | 110,906,768 | CCDC63, MYL2 (4 kb) | T | 1.43 | 0.29 | 0.93 | 0.0 | 1.51 | 0.59 | 1.41 | 0.35 |
| rs9543114 | 13 | 72,767,956 | DIS3 | C | 1.37 | 0.56 | 0.92 | 0.0 | 1.51 | 0.71 | 1.33 | 0.65 |
| rs9301108 | 13 | 106,143,743 | DAOA (653 kb), EFNB2 (346 kb) | C | 0.82 | 0.69 | 0.00 | 87.9 | 0.50 | 0.0008 | 1.38 | 0.26 |
| rs214003 | 14 | 78,336,687 | NRXN3 | G | 0.86 | 0.38 | 0.21 | 36.3 | 0.74 | 0.09 | 1.04 | 0.86 |
| rs4776471 | 15 | 69,700,119 | RPLP1 (245 kb), TLE3 (348 kb) | T | 1.12 | 0.38 | 0.85 | 0.0 | 1.10 | 0.62 | 1.15 | 0.46 |
| rs11856981 | 15 | 69,743,207 | RPLP1 (288 kb), TLE3 (305 kb) | T | 1.13 | 0.37 | 0.98 | 0.0 | 1.13 | 0.54 | 1.14 | 0.51 |
| rs4288991 | 16 | 52,639,252 | TOX3 (91 kb), CHD9 (416 kb) | C | 0.96 | 0.78 | 0.72 | 0.0 | 1.03 | 0.92 | 0.91 | 0.66 |
| rs8065364 | 17 | 80,189,160 | CARD14 | C | 1.24 | 0.13 | 0.74 | 0.0 | 1.30 | 0.20 | 1.19 | 0.39 |
| rs578211 | 18 | 49,868,025 | MYO5B | A | 0.80 | 0.15 | 0.40 | 0.0 | 0.88 | 0.51 | 0.67 | 0.12 |
| rs4808571 | 19 | 17,115,419 | MYO9B | A | 0.94 | 0.89 | 0.53 | 0.0 | 1.30 | 0.68 | 0.77 | 0.61 |
OR = odds ratio from a random-effects meta-analysis.
P = p-value for OR from a random-effects meta-analysis.
Q = p-value for Cochrane's Q statistic (tests heterogeneity in effects across individual studies).
I2 (I-squared) = heterogeneity index (0–100; 0 = no heterogeneity, 100 = max. heterogeneity).
MAP based on GRCh38 (hg38).
*The gene located nearest to the SNP. Distance to the gene in kilo bases (1,000 bp) is shown in parentheses. If no distance is shown, the SNP is located within the gene locus.
In an attempt to identify lead SNPs based on all the cohorts contributing to the GAMES project, meta-analyses were performed using all contributing studies. The results are summarized in Table 7. Finally, the summary meta-analyses were repeated with all subsamples of world-class endurance athletes and these results are depicted in Table 8. Among all available athletes and controls (9 studies, including discovery cohorts), the meta-analysis revealed one statistically significant SNP (rs558129 at GALNTL6 locus, p = 0.0002), even after correcting for multiple testing (Bonferroni-corrected statistical significance p<0.0011). As shown by the low heterogeneity index (I2 = 0), all eight cohorts showed the same direction of association with rs558129, even though p-values varied considerably across the individual studies. When the meta-analysis was repeated without the discovery cohorts, the association persisted near the multiple testing-adjusted significance threshold (p = 0.0019).
Table 7. Results for the Meta-analysis of All Studies of All Athletes and Controls (GENATHLETE, Japan, Australia, Poland, Russia, Spain, Kenya, and Ethiopia); Results in bold type are for p ≤ 0.05.
| SNP | Chr | MAP | Gene* | Effect Allele | N | OR | P | Q | I2 |
|---|---|---|---|---|---|---|---|---|---|
| rs10874242 | 1 | 81,426,166 | LPHN2 (153 kb) | T | 8 | 1.01 | 0.94 | 0.02 | 57.9 |
| rs17459307 | 1 | 162,245,878 | NOS1AP | A | 6 | 0.80 | 0.29 | 0.03 | 60.8 |
| rs12047209 | 1 | 243,791,105 | AKT3 | C | 8 | 1.09 | 0.49 | 0.01 | 60.5 |
| rs921665 | 2 | 3,170,550 | TSSC1 (18 kb) | A | 8 | 1.07 | 0.57 | 0.00 | 67.2 |
| rs2694093 | 2 | 3,254,403 | TSSC1 | G | 8 | 0.85 | 0.04 | 0.07 | 47.4 |
| rs6548153 | 2 | 3,322,274 | TSSC1 | A | 3 | 1.08 | 0.68 | 0.06 | 63.7 |
| rs6726044 | 2 | 68,911,860 | BMP10 (40 kb) | A | 5 | 1.17 | 0.41 | 0.00 | 84.5 |
| rs2118908 | 2 | 111,067,015 | ACOXL | G | 8 | 1.16 | 0.20 | 0.14 | 36.3 |
| rs2361506 | 2 | 233,830,694 | MROH2A | T | 5 | 0.97 | 0.76 | 0.09 | 50.3 |
| rs7650685 | 3 | 11,660,982 | VGLL4 | G | 8 | 1.01 | 0.78 | 0.66 | 0.0 |
| rs6799372 | 3 | 74,481,495 | CNTN3 | A | 7 | 0.95 | 0.66 | 0.00 | 70.1 |
| rs10938202 | 4 | 42,650,320 | ATP8A1 | C | 8 | 1.06 | 0.78 | 0.00 | 70.4 |
| rs10007111 | 4 | 87,828,549 | MEPE | A | 8 | 1.02 | 0.76 | 0.13 | 37.3 |
| rs4699824 | 4 | 100,801,328 | EMCN (283 kb), PPP3CA (222 kb) | G | 8 | 1.08 | 0.35 | 0.02 | 59.0 |
| rs558129 | 4 | 171,829,960 | GALNTL6 | T | 8 | 0.81 | 0.0002 | 0.66 | 0.0 |
| rs2910756 | 5 | 37,859,972 | GDNF-AS1 | G | 8 | 0.89 | 0.23 | 0.02 | 58.8 |
| rs10499127 | 6 | 124,594,509 | NKAIN2 | G | 8 | 1.10 | 0.48 | 0.01 | 63.4 |
| rs9355947 | 6 | 161,887,402 | PARK2 | A | 8 | 0.94 | 0.22 | 0.58 | 0.0 |
| rs6959675 | 7 | 90,253,815 | CFAP69 | T | 3 | 0.78 | 0.49 | 0.32 | 11.9 |
| rs11975386 | 7 | 94,075,721 | BET1 (70 kb) | A | 8 | 1.15 | 0.12 | 0.61 | 0.0 |
| rs17055965 | 8 | 26,769,562 | ADRA1A | G | 7 | 1.43 | 0.008 | 0.21 | 28.0 |
| rs16906888 | 8 | 137,231,375 | FAM135B (898 kb) | G | 6 | 1.06 | 0.39 | 0.46 | 0.0 |
| rs7861665 | 9 | 4,302,988 | LOC101929330, GLIS3 (3 kb) | T | 8 | 0.84 | 0.22 | 0.01 | 62.4 |
| rs3780169 | 9 | 36,979,398 | PAX5 | T | 8 | 1.02 | 0.81 | 0.22 | 26.4 |
| rs17054974 | 9 | 89,549,371 | SEMA4D (51 kb), GADD45G (55 kb) | C | 5 | 1.32 | 0.13 | 0.11 | 47.4 |
| rs17690338 | 10 | 75,357,798 | ZNF503-AS1 | A | 8 | 0.94 | 0.24 | 0.36 | 9.3 |
| rs2761291 | 10 | 93,328,423 | MYOF | C | 3 | 0.90 | 0.32 | 0.44 | 0.0 |
| rs12573965 | 11 | 2,660,402 | KCNQ1 | C | 7 | 0.87 | 0.52 | 0.00 | 69.9 |
| rs7947391 | 11 | 66,419,411 | NPAS4 | A | 8 | 1.11 | 0.46 | 0.00 | 85.2 |
| rs11613185 | 12 | 12,075,733 | BCL2L14 | A | 2 | 0.91 | 0.62 | 0.19 | 41.5 |
| rs12821816 | 12 | 94,976,701 | NDUFA12 | G | 3 | 0.88 | 0.44 | 0.00 | 82.0 |
| rs61940911 | 12 | 110,029,339 | ANKRD13A | T | 6 | 1.45 | 0.01 | 0.29 | 19.5 |
| rs73195844 | 12 | 110,906,768 | CCDC63, MYL2 (4 kb) | T | 5 | 1.29 | 0.33 | 0.01 | 70.5 |
| rs9580890 | 13 | 24,165,281 | SPATA13 | G | 3 | 0.87 | 0.35 | 0.76 | 0.0 |
| rs9543114 | 13 | 72,767,956 | DIS3 | C | 6 | 1.32 | 0.26 | 0.09 | 48.1 |
| rs9301108 | 13 | 106,143,743 | DAOA (653 kb), EFNB2 (346 kb) | C | 7 | 1.14 | 0.55 | 0.00 | 74.3 |
| rs214003 | 14 | 78,336,687 | NRXN3 | G | 8 | 0.98 | 0.89 | 0.01 | 63.7 |
| rs4776471 | 15 | 69,700,119 | RPLP1 (245 kb), TLE3 (348 kb) | T | 8 | 1.18 | 0.04 | 0.01 | 59.9 |
| rs11856981 | 15 | 69,743,207 | RPLP1 (288 kb), TLE3 (305 kb) | T | 8 | 1.17 | 0.02 | 0.09 | 42.9 |
| rs4288991 | 16 | 52,639,252 | TOX3 (91 kb) | CHD9 (416 kb) | C | 8 | 0.84 | 0.007 | 0.31 | 14.9 |
| rs4541108 | 17 | 79,332,527 | RBFOX3 | A | 3 | 1.05 | 0.73 | 0.99 | 0.0 |
| rs8065364 | 17 | 80,189,160 | CARD14 | C | 8 | 0.95 | 0.53 | 0.03 | 53.8 |
| rs578211 | 18 | 49,868,025 | MYO5B | A | 7 | 1.00 | 1.00 | 0.00 | 74.2 |
| rs4808571 | 19 | 17,115,419 | MYO9B | A | 8 | 1.14 | 0.30 | 0.03 | 56.1 |
| rs62135557 | 19 | 19,355,484 | MAU2 | G | 4 | 1.18 | 0.59 | 0.01 | 76.3 |
N = Number of studies used in meta-analysis.
OR = odds ratio from a random-effects meta-analysis.
P = p-value for OR from a random-effects meta-analysis.
Q = p-value for Cochrane's Q statistic (tests heterogeneity in effects across individual studies).
I2 (I-squared) = heterogeneity index (0–100; 0 = no heterogeneity, 100 = max. heterogeneity).
MAP based on GRCh38 (hg38).
*The gene located nearest to the SNP. Distance to the gene in kilo bases (1,000 bp) is shown in parentheses. If no distance is shown, the SNP is located within the gene locus.
Table 8. Results for the Meta-analysis of All Studies of World-class Athletes and Controls (GENATHLETE, Japan, Australia, Poland, Russia, Spain); Results in bold type are for p ≤ 0.05.
| SNP | Chr | MAP | Gene* | Effect Allele | N | OR | P | Q | I2 |
|---|---|---|---|---|---|---|---|---|---|
| rs10874242 | 1 | 81,426,166 | LPHN2 (153 kb) | T | 6 | 0.91 | 0.50 | 0.03 | 59.1 |
| rs17459307 | 1 | 162,245,878 | NOS1AP | A | 5 | 0.83 | 0.30 | 0.25 | 25.7 |
| rs12047209 | 1 | 243,791,105 | AKT3 | C | 6 | 1.29 | 0.17 | 0.00 | 75.8 |
| rs921665 | 2 | 3,170,550 | TSSC1 (18 kb) | A | 6 | 1.17 | 0.31 | 0.05 | 55.1 |
| rs2694093 | 2 | 3,254,403 | TSSC1 | G | 6 | 0.86 | 0.04 | 0.36 | 9.1 |
| rs6548153 | 2 | 3,322,274 | TSSC1 | A | 3 | 1.31 | 0.28 | 0.03 | 71.4 |
| rs6726044 | 2 | 68,911,860 | BMP10 (40 kb) | A | 3 | 1.25 | 0.35 | 0.00 | 82.2 |
| rs2118908 | 2 | 111,067,015 | ACOXL | G | 5 | 1.15 | 0.48 | 0.02 | 63.6 |
| rs2361506 | 2 | 233,830,694 | MROH2A | T | 5 | 0.88 | 0.41 | 0.01 | 72.9 |
| rs7650685 | 3 | 11,660,982 | VGLL4 | G | 6 | 1.06 | 0.56 | 0.06 | 53.1 |
| rs6799372 | 3 | 74,481,495 | CNTN3 | A | 5 | 0.79 | 0.02 | 0.19 | 35.4 |
| rs10938202 | 4 | 42,650,320 | ATP8A1 | C | 5 | 1.56 | 0.005 | 0.30 | 17.9 |
| rs10007111 | 4 | 87,828,549 | MEPE | A | 6 | 1.05 | 0.68 | 0.03 | 58.6 |
| rs4699824 | 4 | 100,801,328 | EMCN (283 kb), PPP3CA (222 kb) | G | 6 | 1.10 | 0.53 | 0.00 | 77.6 |
| rs558129 | 4 | 171,829,960 | GALNTL6 | T | 6 | 0.78 | 0.04 | 0.05 | 54.5 |
| rs2910756 | 5 | 37,859,972 | GDNF-AS1 | G | 6 | 0.88 | 0.49 | 0.00 | 74.5 |
| rs10499127 | 6 | 124,594,509 | NKAIN2 | G | 5 | 1.24 | 0.25 | 0.01 | 67.1 |
| rs9355947 | 6 | 161,887,402 | PARK2 | A | 6 | 0.84 | 0.11 | 0.02 | 61.7 |
| rs6959675 | 7 | 90,253,815 | CFAP69 | T | 3 | 0.79 | 0.55 | 0.54 | 0.0 |
| rs11975386 | 7 | 94,075,721 | BET1 (70 kb) | A | 6 | 1.23 | 0.40 | 0.02 | 64.1 |
| rs17055965 | 8 | 26,769,562 | ADRA1A | G | 5 | 1.44 | 0.18 | 0.01 | 69.4 |
| rs16906888 | 8 | 137,231,375 | FAM135B (898 kb) | G | 4 | 1.15 | 0.52 | 0.00 | 84.5 |
| rs7861665 | 9 | 4,302,988 | LOC101929330, GLIS3 (3 kb) | T | 6 | 0.94 | 0.78 | 0.01 | 69.8 |
| rs3780169 | 9 | 36,979,398 | PAX5 | T | 6 | 1.12 | 0.19 | 0.22 | 28.1 |
| rs17054974 | 9 | 89,549,371 | SEMA4D (51 kb), GADD45G (55 kb) | C | 3 | 1.70 | 0.08 | 0.04 | 68.3 |
| rs17690338 | 10 | 75,357,798 | ZNF503-AS1 | A | 6 | 1.03 | 0.84 | 0.01 | 67.5 |
| rs2761291 | 10 | 93,328,423 | MYOF | C | 3 | 0.84 | 0.36 | 0.12 | 52.8 |
| rs12573965 | 11 | 2,660,402 | KCNQ1 | C | 5 | 1.05 | 0.84 | 0.00 | 77.4 |
| rs7947391 | 11 | 66,419,411 | NPAS4 | A | 6 | 1.16 | 0.45 | 0.00 | 86.5 |
| rs11613185 | 12 | 12,075,733 | BCL2L14 | A | 2 | 0.97 | 0.89 | 0.24 | 28.9 |
| rs12821816 | 12 | 94,976,701 | NDUFA12 | G | 2 | 0.95 | 0.76 | 0.03 | 71.4 |
| rs61940911 | 12 | 110,029,339 | ANKRD13A | T | 4 | 1.39 | 0.16 | 0.07 | 57.1 |
| rs73195844 | 12 | 110,906,768 | CCDC63, MYL2 (4 kb) | T | 3 | 1.11 | 0.81 | 0.00 | 84.6 |
| rs9580890 | 13 | 24,165,281 | SPATA13 | G | 3 | 0.78 | 0.16 | 0.72 | 0.0 |
| rs9543114 | 13 | 72,767,956 | DIS3 | C | 4 | 1.47 | 0.30 | 0.01 | 73.1 |
| rs9301108 | 13 | 106,143,743 | DAOA (653 kb), EFNB2 (346 kb) | C | 4 | 1.37 | 0.33 | 0.00 | 75.8 |
| rs214003 | 14 | 78,336,687 | NRXN3 | G | 5 | 1.07 | 0.77 | 0.00 | 76.4 |
| rs4776471 | 15 | 69,700,119 | RPLP1 (245 kb), TLE3 (348 kb) | T | 5 | 1.24 | 0.08 | 0.01 | 69.5 |
| rs11856981 | 15 | 69,743,207 | RPLP1 (288 kb), TLE3 (305 kb) | T | 5 | 1.23 | 0.04 | 0.05 | 54.6 |
| rs4288991 | 16 | 52,639,252 | TOX3 (91 kb), CHD9 (416 kb) | C | 6 | 0.79 | 0.003 | 0.57 | 0.0 |
| rs4541108 | 17 | 79,332,527 | RBFOX3 | A | 3 | 1.16 | 0.34 | 0.87 | 0.0 |
| rs8065364 | 17 | 80,189,160 | CARD14 | C | 6 | 0.87 | 0.10 | 0.26 | 22.6 |
| rs578211 | 18 | 49,868,025 | MYO5B | A | 5 | 1.12 | 0.53 | 0.00 | 76.4 |
| rs4808571 | 19 | 17,115,419 | MYO9B | A | 5 | 1.24 | 0.22 | 0.01 | 69.2 |
| rs62135557 | 19 | 19,355,484 | MAU2 | G | 4 | 1.11 | 0.71 | 0.05 | 60.7 |
N = Number of studies used in meta-analysis.
OR = odds ratio from a random-effects meta-analysis.
P = p-value for OR from a random-effects meta-analysis.
Q = p-value for Cochrane's Q statistic (tests heterogeneity in effects across individual studies).
I2 (I-squared) = heterogeneity index (0–100; 0 = no heterogeneity, 100 = max. heterogeneity).
MAP based on GRCh38 (hg38).
*The gene located nearest to the SNP. Distance to the gene in kilo bases (1,000 bp) is shown in parentheses. If no distance is shown, the SNP is located within the gene locus.
When the meta-analysis was restricted to the world class athletes versus all controls, none of the SNPs retained for replication reached multiple testing-corrected statistical significance, although SNPs rs4288991 (p = 0.0028) and rs10938202 (p = 0.0053) were trending close. SNP rs558129 was characterized by a nominal p value of 0.04 in this meta-analysis.
Discussion
It is commonly recognized that to perform at a world-class level in endurance athletic events, one has to be intrinsically well endowed in terms of cardiorespiratory and skeletal muscle potential to exercise at high intensity for sustained periods of time in combination with the ability to respond very favorably to exercise training regimens. Observational genetic epidemiology studies and experimental studies in humans as well as in rodents have shown that there are substantial genetic components to intrinsic cardiorespiratory fitness and its trainability [5, 9, 10]. However, the exact genomic features responsible for these genetic effects have not been identified despite many years of candidate gene driven research, a topic that has been reviewed in recent times [18, 37–39]. A limited number of studies have used an unbiased genomic search to identify genomic regions harboring allelic markers of athlete status [40] or response to exercise training of physiological traits [30, 31, 41–43] or GWAS-identified loci associated with determinants of aerobic fitness or its trainability [28–30] but most findings have not been subjected to replication studies. The present report constitutes the largest effort to date designed to identify in an unbiased manner common variants that could begin to define the world-class endurance performance genotype. Even though the total sample size was substantially higher than any other published effort in this field, it is recognized that it was not optimal for the identification of common genomic variants with small effect size that could discriminate between elite endurance athletes and sedentary controls. The main conclusion from the present study is that common genetic variants do not appear to be strong determinants of elite endurance athlete status.
From the GWAS performed on a panel of Japanese endurance athletes and controls plus the CardioMetabochip screen on the endurance athletes from four countries and their matched controls of the GENATHLETE cohort, a panel of 45 SNPs was retained and carried forward for replication in seven other cohorts. Importantly, none of these 45 SNPs reached genome-wide significance in the two discovery studies. But these studies were small and were expected to generate targets at the genome-wide level of significance only if SNPs with large effect sizes were contributing to elite endurance athlete status. There was no overlap among the top SNPs identified in both discovery panels.
When the results from all cohorts were pooled together in meta-analyses, SNP rs558129 located in the GALNTL6 locus on chromosome 4q34.1 was statistically significant (p = 0.0002) even after multiple testing was taken into account. The nominal p-values of the individual cohorts ranged from 0.011 (Australia) to 0.9572 (Spain), but the direction of the association was uniform across all cohorts, that is the T allele was less frequent in athletes than in controls. The association remained robust after excluding the discovery cohort from the meta-analysis (p = 0.0071). The same SNP in GALNTL6 was also nominally significant (p = 0.037) when the meta-analysis was undertaken on the subsets of world-class endurance athletes. This illustrates how random effects meta-analysis helps to identify consistent trends across several relatively small cohorts which individually would not allow for the documentation of such an association. The GALNTL6 gene encodes N-acetylgalactosaminyltransferase-like 6 but the functional role of the peptide is not fully elucidated at this time. It is expressed primarily in the testes but also in the brain and to some extent in skeletal muscle [44]. The rs558129 polymorphic site is in the last intron of the gene.
A few weak leads may also be of interest in future research. Two SNPs in or near TSSC1, the gene encoding tumor suppressing substransferable candidate 1, were associated with endurance athlete status in the cohort from Russia and one of these SNPs (rs2694093) was also shown to be nominally significant and directionally consistent in the global meta-analysis of all cohorts for all endurance athletes as well as the world-class athletes. Of potential interest could also be SNPs near TOX3/CHD9 on chr 16 and RPLP1/TLE3 on chr 15 which were nominally associated with endurance athlete status in the global meta-analyses of all endurance athletes and in the subset of world-class athletes as well. None of these genes have been implicated in exercise biology before.
Interestingly, SNPs in three candidate genes (CKM, ACTN3 and GNB3) selected on the basis of prior results in Japanese endurance athletes were not associated with endurance athlete status in any of the cohorts or in meta-analyses. Along the same line, 161 SNPs related to 13 candidate genes for endurance performance derived from the human gene map for physical performance [18] were available among the CardioMetabochip markers genotyped in GENATHLETE. These candidate genes were: ACE, ACSL1, ACTN3, ADRB1, ADRB2, AMPD1, BDKRB2, GH1, IL6, KDR, NOS3, PPARA and PPARGC1A. None of the 161 SNPs reached the Bonferroni-adjusted significance threshold of p = 3.5 x 10−7. Two SNPs in the vicinity of PPARGC1A were the most strongly associated with endurance athlete status at p≤0.0012. None of the SNP associations were close to significance when the subsample of the GENATHLETE world-class endurance athletes (VO2max ≥78 mL O2/kg/min) was considered.
Over all, there is no convincing evidence for the contribution of common genomic variants to elite endurance performance in the present report. However, since we were underpowered to identify contributing alleles with small effect sizes, one may be justified in proposing that a few leads, even though supported by very modest levels of evidence, be explored in subsequent studies with adequate statistical power. For instance, the joint contribution from gene-members of a pathway or a network module could be significant, even if individual gene contributions are small and non-significant. We have used this approach successfully in other GWA studies, for example on the analysis of genetic associations to maximal oxygen uptake in response to exercise [45]. An alternative approach employs unsupervised bioinformatics data analysis techniques to generate functional hypotheses on how some of the top candidates from a study may be related to a trait. Graph based methods, such as those based on stochastic random walks [46], are well suited for this type of analysis and can provide useful information for candidate gene prioritization. In this regard, it may be useful to expand the comparison of world-class endurance athletes and sedentary controls to DNA sequence variants in or in the vicinity of a number of genes (for instance: GALNTL6 but also perhaps ADRA1A, ATP8A1, CHD9, CNTN3, RPLP1, TLE3, TOX3, TSSC1, AKT3, BET1, BMP10) in order to shed some light on the genetics, biology and the highly demanding selection process leading to world class endurance performance. Thus, using the 12 genes listed above in a Biograph analysis, in which the query was on athletic performance, we have found additional support for the suggestion that sequence variants in these genes should be further investigated (results not shown). Even when SNPs and genes are characterized by small effect size and marginal nominal significance, the joint contributions from gene-members of a pathway or a network may provide useful information.
The present report has several limitations. The SNP chips used for the discovery phase differed between GENATHLETE and the Japanese cohort of endurance athlete. These discovery studies were performed independently and later used to identify the panel of the most promising 45 SNPs. It is also recognized that the CardioMetabochip used in GENATHLETE offers less than a comprehensive coverage of the genome and it may provide less than optimal inclusion of markers and genes that could be important for the peripheral determinants of endurance performance. In some of the replication studies, participants in the control group were from the general population or recreationally active subjects while in others they were confirmed sedentary participants. However, even though the endurance athletes were exposed to variable training regimens, which are undoubtedly heterogeneous by athletic event and country, these factors should not have an influence on the results as they all achieved national and international levels of competition requiring an extraordinary level of cardiorespiratory endurance. Obviously sample size continues to be a major issue for this type of studies. This is a challenge that is very difficult to overcome as the number of world-class endurance athletes on the planet is limited and those who reach this level of performance are hesitant to give consent to participate in genetic studies. Finally, one has also to consider that a number of the athletes who gave consent to participate in these genetic studies are using performance enhancement drugs. However, it is unlikely that doping of any kind could have influenced the results as the use of these drugs is not by itself responsible for an athlete reaching world-class level but rather their uses come more into play when athletes of world-class caliber are engaged competitively against one another.
In all likelihood, attaining the required sample size of world class caliber endurance athletes, for an adequate statistical power in the search for critical sequence variants, will require a buy-in from the relevant world sports federations and the International Olympic Committee. With their support and active participation, thousands of world-class endurance athletes could be enrolled in genomics studies aimed at understanding the fundamentals of inherited biological traits that are necessary to perform at the world class level. Such an effort, particularly if it relied on whole genome sequencing, would allow for the exploration of not only common polymorphisms but also rare variants and copy number variants and could be complemented by the investigation of epigenomic signatures in accessible tissues. In summary, we found that the T allele in GALNTL6 was less frequent in endurance athletes of all studies compared to ethnicity-matched controls. However, we could not find evidence for a detailed genomic signature that differentiates endurance athletes from controls.
Supporting Information
(TIFF)
(TIFF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIFF)
(TIFF)
(TIFF)
(TIFF)
(TIFF)
(TIFF)
(TIFF)
(TIFF)
(DOCX)
(TIFF)
Acknowledgments
The contributions of Monique Chagnon and Anne-Marie Bricault from Laval University, Quebec, Canada, Kathryn Cooper and Jessica Watkins from Pennington Biomedical Research Center, Baton Rouge, LA, USA, to the GENATHLETE study are gratefully acknowledged. Gratitude is expressed to Dr. Vladimir A. Naumov from the Research Institute for Physical-Chemical Medicine, Russia, and to Dr. Leysan J. Gabdrakhmanova, Dr. Emiliya S. Egorova and Dr. Albina A. Galeeva from the Volga Region State Academy of Physical Culture, Sport and Tourism, Russia.
Data Availability
All relevant data are provided within the paper and its Supporting Information files. Regulations regarding confidentiality and access to original and confidential data on athletes vary from country to country. The original athlete data are controlled by the Principle Investigator of each study. Interested researchers may access the data of the two discovery cohorts (GENATHLETE and Japanese cohort) at Figshare (Rankinen, Tuomo (2015): GAMES discovery data sets http://dx.doi.org/10.6084/m9.figshare.1619893) and at www.athlomeconsortium.org. For the data of the various replication studies, the principal investigator of each contributing study should be contacted. If problems arise, interested scientists may contact Claude Bouchard (claude.bouchard@pbrc.edu) who will facilitate the interactions with the investigators of the contributing studies.
Funding Statement
Partial funding for the study has been received from the Prince Faisal Prize awarded to Drs. C. Bouchard, T. Rankinen, M. Sarzynski and B. Wolfahrt. CB is partially funded by the John W. Barton Jr Chair in Genetics and Nutrition. The study in Russia was supported by a grant from the Federal Medical-Biological Agency (“Sportgen project”), http://fmbaros.ru/en/fmba/infor/. The Spanish group (AL, CAM, CS, TY) was funded by Fondo de Investigaciones Sanitarias (grant # PI12/00914). Research in sports genetics to A. Lucia in Spain was funded by grants from the Consejo Superior de Deportes (CSD). This work in Japan was supported in part by grants from the programs Grants-in-Aid for Challenging Exploratory Research (24650414 to NF) from the Ministry of Education, Culture, Sports, Science and Technology; and by a grant-in-aid for scientific research from the Ministry of Health, Labor, and Welfare of Japan (to MM). No specific funding for this work was received for the studies performed in Australia, Poland, Kenya and Ethiopia. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Gedda L. Sports and genetics. A study on twins (351 pairs). Acta Genet Med Gemellol (Roma). 1960;9:387–406. Epub 1960/10/01. . [DOI] [PubMed] [Google Scholar]
- 2.Chagnon YC, Allard C, Bouchard C. Red blood cell genetic variation in Olympic endurance athletes. J Sport Sci. 1984;2(2):121–9. [PubMed] [Google Scholar]
- 3.Couture L, Chagnon M, Allard C, Bouchard C. More on red blood cell genetic variation in Olympic athletes. Can J Appl Sport Sci. 1986;11(1):16–8. Epub 1986/03/01. . [PubMed] [Google Scholar]
- 4.de Garay A, Levine L, Carter JEL. Genetic and Anthropological Studies of Olympic Athletes. Press A, editor. New York: Academic Press; 1974. [Google Scholar]
- 5.Bouchard C, Daw EW, Rice T, Perusse L, Gagnon J, Province MA, et al. Familial resemblance for VO2max in the sedentary state: the HERITAGE family study. Medicine and science in sports and exercise. 1998;30(2):252–8. Epub 1998/03/21. . [DOI] [PubMed] [Google Scholar]
- 6.Lortie G, Bouchard C, Leblanc C, Tremblay A, Simoneau JA, Theriault G, et al. Familial similarity in aerobic power. Hum Biol. 1982;54(4):801–12. Epub 1982/12/01. . [PubMed] [Google Scholar]
- 7.Bouchard C, Tremblay A, Despres JP, Theriault G, Nadeau A, Lupien PJ, et al. The response to exercise with constant energy intake in identical twins. Obes Res. 1994;2(5):400–10. Epub 1994/09/01. . [DOI] [PubMed] [Google Scholar]
- 8.Hamel P, Simoneau JA, Lortie G, Boulay MR, Bouchard C. Heredity and muscle adaptation to endurance training. Medicine and science in sports and exercise. 1986;18(6):690–6. Epub 1986/12/01. . [PubMed] [Google Scholar]
- 9.Prudhomme D, Bouchard C, Leblanc C, Landry F, Fontaine E. Sensitivity of maximal aerobic power to training is genotype-dependent. Medicine and science in sports and exercise. 1984;16(5):489–93. [DOI] [PubMed] [Google Scholar]
- 10.Bouchard C, An P, Rice T, Skinner JS, Wilmore JH, Gagnon J, et al. Familial aggregation of VO(2max) response to exercise training: results from the HERITAGE Family Study. Journal of applied physiology (Bethesda, Md: 1985). 1999;87(3):1003–8. Epub 1999/09/14. . [DOI] [PubMed] [Google Scholar]
- 11.Skinner JS, Jaskolski A, Jaskolska A, Krasnoff J, Gagnon J, Leon AS, et al. Age, sex, race, initial fitness, and response to training: the HERITAGE Family Study. Journal of applied physiology (Bethesda, Md: 1985). 2001;90(5):1770–6. Epub 2001/04/12. . [DOI] [PubMed] [Google Scholar]
- 12.Rankinen T, Wolfarth B, Simoneau JA, Maier-Lenz D, Rauramaa R, Rivera MA, et al. No association between the angiotensin-converting enzyme ID polymorphism and elite endurance athlete status. Journal of applied physiology (Bethesda, Md: 1985). 2000;88(5):1571–5. Epub 2000/05/08. . [DOI] [PubMed] [Google Scholar]
- 13.Rivera MA, Dionne FT, Simoneau JA, Perusse L, Chagnon M, Chagnon Y, et al. Muscle-specific creatine kinase gene polymorphism and VO2max in the HERITAGE Family Study. Medicine and science in sports and exercise. 1997;29(10):1311–7. Epub 1997/11/05. [DOI] [PubMed] [Google Scholar]
- 14.Rivera MA, Perusse L, Gagnon J, Dionne FT, Leon AS, Rao DC, et al. A mitochondrial DNA D-loop polymorphism and obesity in three cohorts of women. Int J Obes Relat Metab Disord. 1999;23(6):666–8. Epub 1999/07/20. . [DOI] [PubMed] [Google Scholar]
- 15.Wolfarth B, Rankinen T, Muhlbauer S, Ducke M, Rauramaa R, Boulay MR, et al. Endothelial nitric oxide synthase gene polymorphism and elite endurance athlete status: the Genathlete study. Scand J Med Sci Sports. 2008;18(4):485–90. Epub 2007/12/11. SMS717 [pii] 10.1111/j.1600-0838.2007.00717.x . [DOI] [PubMed] [Google Scholar]
- 16.Wolfarth B, Rankinen T, Muhlbauer S, Scherr J, Boulay MR, Perusse L, et al. Association between a beta2-adrenergic receptor polymorphism and elite endurance performance. Metabolism. 2007;56(12):1649–51. Epub 2007/11/14. S0026-0495(07)00268-5 [pii] 10.1016/j.metabol.2007.07.006 . [DOI] [PubMed] [Google Scholar]
- 17.Wolfarth B, Rivera MA, Oppert JM, Boulay MR, Dionne FT, Chagnon M, et al. A polymorphism in the alpha2a-adrenoceptor gene and endurance athlete status. Medicine and science in sports and exercise. 2000;32(10):1709–12. Epub 2000/10/20. . [DOI] [PubMed] [Google Scholar]
- 18.Bray MS, Hagberg JM, Perusse L, Rankinen T, Roth SM, Wolfarth B, et al. The human gene map for performance and health-related fitness phenotypes: the 2006–2007 update. Medicine and science in sports and exercise. 2009;41(1):35–73. Epub 2009/01/06. . [DOI] [PubMed] [Google Scholar]
- 19.Mikami E, Fuku N, Takahashi H, Ohiwa N, Pitsiladis YP, Higuchi M, et al. Polymorphisms in the control region of mitochondrial DNA associated with elite Japanese athlete status. Scand J Med Sci Sports. 2013;23(5):593–9. Epub 2012/02/01. 10.1111/j.1600-0838.2011.01424.x . [DOI] [PubMed] [Google Scholar]
- 20.Mikami E, Fuku N, Takahashi H, Ohiwa N, Scott RA, Pitsiladis YP, et al. Mitochondrial haplogroups associated with elite Japanese athlete status. Br J Sports Med. 2011;45(15):1179–83. Epub 2010/06/17. [pii]. . [DOI] [PubMed] [Google Scholar]
- 21.Buxens A, Ruiz JR, Arteta D, Artieda M, Santiago C, Gonzalez-Freire M, et al. Can we predict top-level sports performance in power vs endurance events? A genetic approach. Scand J Med Sci Sports. 2011;21(4):570–9. Epub 2010/05/13. [pii]. . [DOI] [PubMed] [Google Scholar]
- 22.Ash GI, Scott RA, Deason M, Dawson TA, Wolde B, Bekele Z, et al. No association between ACE gene variation and endurance athlete status in Ethiopians. Medicine and science in sports and exercise. 2011;43(4):590–7. Epub 2010/08/28. . [DOI] [PubMed] [Google Scholar]
- 23.Scott RA, Georgiades E, Wilson RH, Goodwin WH, Wolde B, Pitsiladis YP. Demographic characteristics of elite Ethiopian endurance runners. Medicine and science in sports and exercise. 2003;35(10):1727–32. Epub 2003/10/03. . [DOI] [PubMed] [Google Scholar]
- 24.Ahmetov II, Druzhevskaya AM, Astratenkova IV, Popov DV, Vinogradova OL, Rogozkin VA. The ACTN3 R577X polymorphism in Russian endurance athletes. Br J Sports Med. 2010;44(9):649–52. Epub 2008/08/23. [pii]. . [DOI] [PubMed] [Google Scholar]
- 25.Yang N, MacArthur DG, Gulbin JP, Hahn AG, Beggs AH, Easteal S, et al. ACTN3 genotype is associated with human elite athletic performance. Am J Hum Genet. 2003;73(3):627–31. Epub 2003/07/25. [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eynon N, Meckel Y, Alves AJ, Yamin C, Sagiv M, Goldhammer E. Is there an interaction between PPARD T294C and PPARGC1A Gly482Ser polymorphisms and human endurance performance? Exp Physiol. 2009;94(11):1147–52. Epub 2009/08/12. [pii]. . [DOI] [PubMed] [Google Scholar]
- 27.Zarebska A, Sawczyn S, Kaczmarczyk M, Ficek K, Maciejewska-Karlowska A, Sawczuk M, et al. Association of rs699 (M235T) polymorphism in the AGT gene with power but not endurance athlete status. J Strength Cond Res. 2013;27(10):2898–903. Epub 2013/01/05. . [DOI] [PubMed] [Google Scholar]
- 28.Bouchard C, Sarzynski MA, Rice TK, Kraus WE, Church TS, Sung YJ, et al. Genomic predictors of the maximal O(2) uptake response to standardized exercise training programs. Journal of applied physiology (Bethesda, Md: 1985). 2011;110(5):1160–70. Epub 2010/12/25. [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rankinen T, Sung YJ, Sarzynski MA, Rice TK, Rao DC, Bouchard C. Heritability of submaximal exercise heart rate response to exercise training is accounted for by nine SNPs. Journal of applied physiology (Bethesda, Md: 1985). 2012;112(5):892–7. Epub 2011/12/17. [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Timmons JA, Knudsen S, Rankinen T, Koch LG, Sarzynski M, Jensen T, et al. Using molecular classification to predict gains in maximal aerobic capacity following endurance exercise training in humans. Journal of applied physiology (Bethesda, Md: 1985). 2010;108(6):1487–96. Epub 2010/02/06. [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Argyropoulos G, Stutz AM, Ilnytska O, Rice T, Teran-Garcia M, Rao DC, et al. KIF5B gene sequence variation and response of cardiac stroke volume to regular exercise. Physiol Genomics. 2009;36(2):79–88. Epub 2008/11/06. [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Voight BF, Kang HM, Ding J, Palmer CD, Sidore C, Chines PS, et al. The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet. 2012;8(8):e1002793 Epub 2012/08/10. [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet. 2005;76(5):887–93. Epub 2005/03/25. S0002-9297(07)60735-6 [pii] 10.1086/429864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. Epub 2007/08/19. S0002-9297(07)61352-4 [pii] 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Larson MG, Atwood LD, Benjamin EJ, Cupples LA, D'Agostino RB Sr., Fox CS, et al. Framingham Heart Study 100K project: genome-wide associations for cardiovascular disease outcomes. BMC Med Genet. 2007;8 Suppl 1:S5 Epub 2007/10/16. 1471-2350-8-S1-S5 [pii] 10.1186/1471-2350-8-S1-S5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–7. Epub 2010/07/17. 10.1093/bioinformatics/btq419 ; PubMed Central PMCID: PMCPmc2935401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pitsiladis Y, Wang G, Wolfarth B, Scott R, Fuku N, Mikami E, et al. Genomics of elite sporting performance: what little we know and necessary advances. Br J Sports Med. 2013;47(9):550–5. Epub 2013/05/02. [pii]. . [DOI] [PubMed] [Google Scholar]
- 38.Rankinen T, Sarzynski MA, Bouchard C. Genes and response to training In: Bouchard C, Hoffman EP, editors. Genetic and Molecular Aspects of Sport Performance. Volume XVIII of the Enclyclopaedia of Sports Medicine. An IOC Medical Commission Publication. West Sussex, UK: Wiley-Blackwell; 2011. p. 177–84. [Google Scholar]
- 39.Wolfarth B. Genes and Endurance Performance In: Bouchard C, Hoffman EP, editors. Genetic and Molecular Aspects of Sport Performance. Volume XVIII of the Enclyclopaedia of Sports Medicine. An IOC Medical Commission Publication. West Sussex, UK: Wiley-Blackwell; 2011. p. 151–8. [Google Scholar]
- 40.De Moor MH, Spector TD, Cherkas LF, Falchi M, Hottenga JJ, Boomsma DI, et al. Genome-wide linkage scan for athlete status in 700 British female DZ twin pairs. Twin Res Hum Genet. 2007;10(6):812–20. Epub 2008/01/09. 10.1375/twin.10.6.812 . [DOI] [PubMed] [Google Scholar]
- 41.Rice TK, Sarzynski MA, Sung YJ, Argyropoulos G, Stutz AM, Teran-Garcia M, et al. Fine mapping of a QTL on chromosome 13 for submaximal exercise capacity training response: the HERITAGE Family Study. Eur J Appl Physiol. 2012;112(8):2969–78. Epub 2011/12/16. 10.1007/s00421-011-2274-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spielmann N, Leon AS, Rao DC, Rice T, Skinner JS, Rankinen T, et al. Genome-wide linkage scan for submaximal exercise heart rate in the HERITAGE family study. Am J Physiol Heart Circ Physiol. 2007;293(6):H3366–71. Epub 2007/10/09. 00042.2007 [pii] 10.1152/ajpheart.00042.2007 . [DOI] [PubMed] [Google Scholar]
- 43.Stutz AM, Teran-Garcia M, Rao DC, Rice T, Bouchard C, Rankinen T. Functional identification of the promoter of SLC4A5, a gene associated with cardiovascular and metabolic phenotypes in the HERITAGE Family Study. Eur J Hum Genet. 2009;17(11):1481–9. Epub 2009/04/23. [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peng C, Togayachi A, Kwon YD, Xie C, Wu G, Zou X, et al. Identification of a novel human UDP-GalNAc transferase with unique catalytic activity and expression profile. Biochemical and biophysical research communications. 2010;402(4):680–6. Epub 2010/10/28. 10.1016/j.bbrc.2010.10.084 . [DOI] [PubMed] [Google Scholar]
- 45.Ghosh S, Vivar JC, Sarzynski MA, Sung YJ, Timmons JA, Bouchard C, et al. Integrative pathway analysis of a genome-wide association study of (V)O(2max) response to exercise training. Journal of applied physiology (Bethesda, Md: 1985). 2013;115(9):1343–59. 10.1152/japplphysiol.01487.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liekens AM, De Knijf J, Daelemans W, Goethals B, De Rijk P, Del-Favero J. BioGraph: unsupervised biomedical knowledge discovery via automated hypothesis generation. Genome biology. 2011;12(6):R57 10.1186/gb-2011-12-6-r57 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIFF)
(TIFF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIFF)
(TIFF)
(TIFF)
(TIFF)
(TIFF)
(TIFF)
(TIFF)
(TIFF)
(DOCX)
(TIFF)
Data Availability Statement
All relevant data are provided within the paper and its Supporting Information files. Regulations regarding confidentiality and access to original and confidential data on athletes vary from country to country. The original athlete data are controlled by the Principle Investigator of each study. Interested researchers may access the data of the two discovery cohorts (GENATHLETE and Japanese cohort) at Figshare (Rankinen, Tuomo (2015): GAMES discovery data sets http://dx.doi.org/10.6084/m9.figshare.1619893) and at www.athlomeconsortium.org. For the data of the various replication studies, the principal investigator of each contributing study should be contacted. If problems arise, interested scientists may contact Claude Bouchard (claude.bouchard@pbrc.edu) who will facilitate the interactions with the investigators of the contributing studies.