Abstract
The spermatogenic process relays in highly regulated gene expression mechanisms at the transcriptional and post-transcriptional levels to generate the male gamete that is needed for the perpetuation of the species. Small non-coding RNA pathways have been determined to participate in the post-transcriptional regulatory processes of germ cells. The most important sncRNA molecules that are critically involved in spermatogenesis belong to the miRNA and piRNAs pathways as illustrated by animal models where ablation of specific protein components displays male infertility. Several elements of these regulatory pathways have been found in the nuage or germ granule, a non-membranous cytoplasmatic structure that can be seen in spermatocytes and spermatids. This notion suggests that germ granules may act as organizer centers for silencing pathways in the germline. In general, miRNAs regulate spermatogenesis through targeting and down-regulation of specific transcripts to eventually promote sperm development. However, piRNAs are powerful repressors of transposon elements expression in the spermatogenic process. Here we describe the suggested functions that miRNA and piRNAs pathways execute in the regulation of spermatogenesis and include some recent studies in the field. Despite major strides on the detailed molecular mechanisms of sncRNAs in relation to spermatogenesis, there is plenty to discover on this fascinating regulatory program.
Keywords: epigenetics, germ cells, germ granules, miRNAs, piRNAs, post-transcriptional regulation, sncRNAs, spermatogenesis
1.1.Introduction
The formation of the male gamete or spermatogenesis is a highly regulated process with the goal of transmitting correct genetic and epigenetic information to next generations [1]. At the embryonic stage, the germ cell linage is specified and segregated early in mammalian development, after implantation, where primordial germ cells (PGCs) migrate and are set aside from other somatic lineages [2-4] (Figure 1). PGCs migrate to the gonadal ridge and initiate germ cell differentiation at around mid-gestation, become gonocytes or prospermatogonia arrested at G1 (G0) phase of the mitotic cell cycle and are localized within cords that are formed by somatic cells that will belong to the future testis. After birth, gonocytes resume mitotic proliferation and become postnatal spermatogonial stem cells (SSC). During embryonic germ cell development, genome-wide epigenetic reprogramming takes places: first somatic epigenetic marks are almost all erased and then novel sex-specific DNA methylation patterns are established in a sex-specific manner [5]. One major feature of this global epigenetic erasure/rewriting is that it also affects selfish transposon sequences.
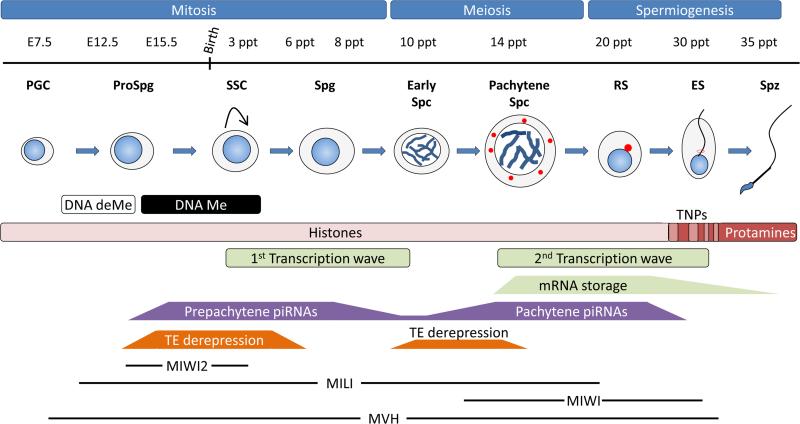
Figure 1.
Spermatogenic process in the mouse testis. During embryogenesis primordial germ cells (PGCs) are specified at embryonic day E7.5. They proliferate and migrate to the future gonads at E11.5 and become arrested at G1 as prospermatogonia (ProSpg). During this phase, a genome-wide epigenetic reprogramming occurs in embryonic germ cells through the near to complete erasure of somatic DNA methylation patterns (DNA deMe) followed by the acquisition of novel sex-specific DNA methylation including differential imprinting of the genes in the male and female germ cells (DNA Me). After birth, the Spermatogonial Stem Cell (SSC) divide mitotically to generate more spermatogonia (Spg) or to differentiate into primary spermatocytes. Early spermatocytes (Early Spc) will enter into meiosis during which synaptonemal complex formation, crossing over and homologous recombination take place in the Pachytene spermatocyte (Pachytene Spc). Round spermatids (RS) are the first haploid cells and undergo a differentiation process or spermiogenesis where elongating spermatids (ES) have their histones replaced first by transition proteins and finally by protamines in the spermatozoa (Spz). Two extensive wave of transcription occurs before and after meiosis and the second lead to the storage of mRNAs that will be needed in the last stages of the spermatogenesis when transcriptions ceases. Unlike Pachytene piRNAs, Prepachytene piRNAs are enriched in sequences that match transposable elements (TEs) and both are expressed at specific spermatogenic stages. Prepachytene piRNAs are expressed simultaneously with MIWI2 (from E15.5 until soon after birth) and MILI (from E12.5 to RS) in foetal ProSpg and during this period there is an extensive derepression of transposable elements (TE) that also occurs in early meiotic spermatocytes. Pachytene piRNAs are expressed together with MIWI (from pachytene Spc until RS) and the presence of the CB (red dots). The MVH, a hallmark of the CB, is exclusively expressed in male germ cells from E10.5 to RS and has been proposed to be involved in both miRNA and piRNA pathways.
Spermatogenesis starts shortly after and takes place in the seminiferous tubules of the testis throughout the male life (Figure 1). SSC located in the base of the seminiferous tubules differentiate into meiotic spermatocytes that will continue the germ cell development process to eventually generate the spermatozoa. The spermatogenic process is typically divided into 3 phases: mitosis, meiosis and differentiation or spermiogenesis. During the first phase, spermatogonia are renewed through mitotic divisions; in the second phase spermatogonia differentiate to generate primary spermatocytes that undergo two meiotic divisions to generate secondary spermatocytes and haploid spermatids; finally, haploid spermatids undergo several morphological changes such as acrosome and flagellum formation, nuclear condensation and cytoplasm reorganization to give rise to the spermatozoa [6]. The proper progression of the spermatogenic process rely on an accurate, spatially and temporally regulated gene expression patterns that takes place at transcriptional, post-transcriptional and epigenetic level [7, 8]. Germ cells have periodically their transcription silenced during spermatogenesis: first at prophase stage during meiosis to allow recombination and then in the transition from round to elongated spermatids [9]. Subsequently, two waves of active transcription occur before and after meiosis prior to nuclear condensation [10, 11]. Thus, epigenetic factors play important roles in the regulation of these processes [12]. A special feature of the sperm cell is its chromatin structure, which is mainly organized by specific proteins called protamines and a limited number of residual histones [11, 13]. In mammals, postmeiotic germ cells differentiate and undergo a chromatin remodeling process where histones are progressively but not completely replaced by protamines. Because spermatid chromatin compaction is incompatible with transcription [11, 14], it is crucial to stabilize the produced mRNAs to allow their translation at later stages. Transcripts are then stored under translational control together with RNA-binding proteins (RBPs) [8, 9, 14, 15].
It has recently shown that although the human genome is extensively transcribed, a large fraction of RNAs are not translated to proteins [6]. These transcripts, called non-coding RNAs (ncRNAs), present a high diversity of sizes and have recently been demonstrated to serve as controllers of gene expression. Their role has been shown to be at the transcriptional or post-transcriptional level. Alternatively, the absence of a known protein product despite the presence of the corresponding mRNA, can be due to a regulatory mechanism executed by ncRNAs [16]. Male germ lineages have been found to express a high amount of non-coding RNAs, including both long and small non-coding RNAs (sncRNAs) [3]. In the germline, ncRNAs are required for several mechanisms such as mRNA splicing (snRNPs), protein synthesis (tRNAs and ribosomal RNAs), RNA maturation and modification (snoRNAs) [6]. SncRNAs constitute a well characterized class of ncRNAs [3]. There are three major types of sncRNAs: small interfering RNAs (siRNAs, 21–23 bp), microRNAs (miRNAs, 19–25 bp), and the germline-enriched piwi-interacting RNAs (piRNAs, mostly 24–34 bp) [6]. These sncRNAs bind to evolutionarily conserved proteins of the Piwi/Argonaute family and are characterized by the presence of the PAZ (Piwi-Argonaute-Zwille) and PIWI (P-element induced wimpy testis) domains. This family of proteins is subdivided into two subfamilies: Argonaute (Ago) and Piwi. Ago proteins bind siRNAs and miRNAs and are present in both somatic and germ cells; Piwi proteins, that binds piRNAs, are highly enriched in the germline. MiRNAs, that essentially function by regulating negatively mRNA translation in mammalian cells, are clearly important for spermatogenesis since a global loss of miRNAs through the generation of Dicer knockouts in the supporting or the germ lineage of the testis have detrimental effects on male fertility [8]. In concordance, according to knockout animal models, Piwi proteins (MILI, MIWI2 and MIWI) are essential for the successful completion of spermatogenesis, revealing a crucial role for piRNAs in this process [17]. Since among sncRNAs in mammals, miRNAs and piRNAs are both important regulators of male germ cell differentiation [15], we will discuss their proposed biogenesis and their presence and potential role during the different stages of the spermatogenic process (Figure 2).
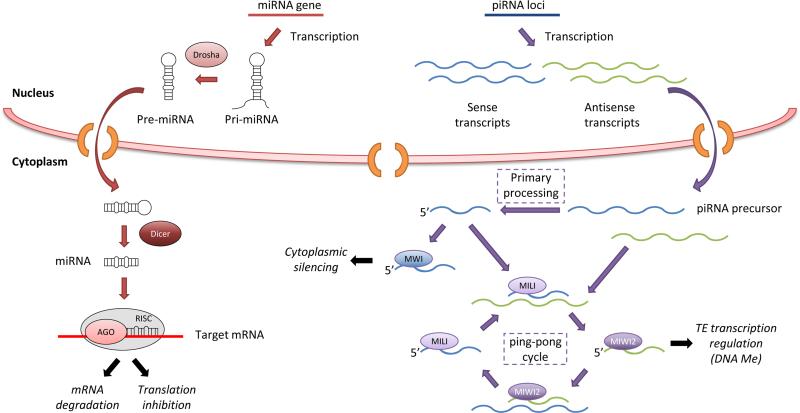
Figure 2.
miRNAs and piRNAs proposed biogenesis pathways and function. Long dsDNA loops from miRNA genes called Pri-miRNAs are cleaved by Drosha in the nucleus and the resulting pre-miRNAs are further exported to the cytoplasm and processed into mature miRNAs by Dicer. Typically, miRNAs mediate translation inhibition or mRNA degradation through their association with Ago proteins in the RISC. In foetal prospermatogonia, prepachytene piRNAs are produced through the primary processing of piRNA precursors by an unknown mechanism and the subsequent ping-pong cycle in where MILI and MIWI2 participate and give rise to piRNAs, although a homotypic mechanism has recently been proposed (not represented in the figure, see [72]). These sense piRNA precursors are likely mRNAs of active transposable elements that preferentially bind MILI. MIWI2 is specifically enriched in secondary antisense piRNAs which may also mediate transposon silencing at the transcriptional level through DNA methylation. After birth, MIWI2 is no longer expressed, MILI continues functioning and MIWI appears to contribute in the cytoplasmatic transposon silencing through primary processing of transposon mRNAs to give rise to pachytene piRNAs. Additionally, MIWI has shown to bind spermiogenic RNAs to potentially cooperate in maintaining the pool of mRNAs that are stored in the round spermatid for later stages. Other protein components have been described to participate in these sncRNA mechanisms in the mouse testis although they have been not included in this representation.
1.2.Germ granules
In the germ cells of many animals it can be found the so called “germ granules”. Germ granules are evolutionarily conserved, non-membranous cytoplasmic structures that contain ribonucleoproteins and are exclusive to the germline [1, 2, 4, 18]. In model animals such as Drosophila, C. elegans and Xenopus, germ granules are involved in germline specification in early embryos; on the contrary, these structures in mammals are first found at later differentiation stages of the germ cells and can be identified as different formations [4, 15, 19]. The most relevant forms are the intermitochondrial cement (IMC) or pi-body, and the chromatoid body (CB) [15, 19]. The IMC is found among clusters of mitochondria in fetal prospermatogonia, postnatal spermatogonia and meiotic spermatocytes (especially abundant in late pachytene) and associated with the nuclear envelope. The CB is more distinct and appears in meiotic spermatocytes and haploid spermatids.
The form and localization of the germ granules are dynamic along the development of the spermatogenic cells in mammals. After birth, the IMC emerges in prospermatogonia and it is clearly visible as an amorphous electron-dense fibrous material in their interstices [20]. Later in the spermatogenic process, the IMC becomes undetectable at the ultrastructural level in B-type spermatogonia and in early spermatocytes from (pre)leptotene, zygotene to early pachytene stages, and reappears at the midpachytene stage with increased size and frequency. The CB starts to be formed at late pachytene stages and by the secondary spermatocyte stage material of the CB aggregates into large dense bodies. Primary spermatocytes display both the CB freely in the cytoplasm and the ICM among mitochondrial clusters [2, 15]. After meiosis, IMC is not anymore detectable and the CB increases in size and is found in haploid spermatids generally as one solitary lobule that is often seen in close contact with nuclear pores. In the murine animal model, the CB is highly mobile around the nucleus and from cell to cell through the cytoplasmic bridges of spermatids [21-24]. As the spermatid develops, the CB assumes various shapes, positions and its size decreases progressively until it disintegrates [15].
As mentioned, one of the basic characteristics of the CB is its variety of localizations: during early spermiogenesis it is localized at the nuclear envelope, associated with the nuclear pores; at later stages it translocates to the annulus area of the tail [23, 25] (Figure 1). Interestingly, the CB contacts perpendicularly to the nuclear envelope revealing a continuity of nuclear material between both organelles, suggesting that the CB could collect material from the nucleus [26]. When the CB makes contact with the nucleus, mRNA and small RNA precursors move towards the CB through the nuclear pores and these RNAs become substrates for the RNA-processing machinery [23, 27]. These movements have been proposed to have a microtubular nature, although how specific components of the CB are transported is still unknown [22, 23]. The implication of the protein eIF4b has been suggested because of its interaction with proteins located in the CB and with RNAs [28]. A recent report also suggested a potential role of the proteins BASP1 and MARCKS, which locate in the CB. Intriguingly, these proteins are involved in non-random movements in neurons [27].
The CB has recently being proposed to operate as a center for RNA storage and metabolism in postmeiotic germ cells and to participate in post-transcriptional processes essential for the completion of spermatogenesis [14, 25, 28]. The several components that have been detected in the CB (Table 1), its high motility in the cytoplasm and its diversity of sizes and form during the spermatogenic process has led to think in the CB as a specialized non-membranous organelle that control germ development through RNA-processing pathways [25, 28]. Among the different elements found in the CB of postmeiotic spermatids, the description of microRNA and RNA decay pathway components was one of the most striking [26]. A detailed description of the components of the mammalian germ granule has recently revealed the full protein and RNA content of the CB [29] increasing the list of new CB components (Table 1). One element that is exclusive for the CB is the ATP-dependent DEAD-box RNA helicase called MVH (Mouse VASA Homolog) and is currently used as a hallmark for the CB [25, 30]. Although the detailed role of MVH remains unknown, its presence within the CB appears crucial for successful progression for spermatogenesis since mouse testes lacking this protein present meiotic arrest and germ cells undergo apoptotic death [25, 31]. Thus, MVH has been proposed to be involved in several RNA processing mechanisms such as miRNA and piRNA pathways [26]. In mouse, the exit of transcripts from the CB in spermatids has been correlated with its translational activation [32]. Importantly, the acetyltransferase HAT1 colocalizes with MVH in the CB and controls MVH RNA-binding activity through direct acetylation of MVH [33]. MVH was hypothesized to function as a regulator of ribonucleoprotein (RNP) complexes in the CB since DEAD-box RNA helicases have been reported to be involved in remodeling of RNA-protein interactions [25] although its molecular mechanism remains unknown. Intriguingly, CLOCK and BMAL1 proteins are other components of the CB that have also recently been described for the first time [34]. These proteins are involved in the circadian clock system and interestingly, when BMAL1 was absent or low expressed the CB of spermatids presented morphological alterations suggesting that both BMAL1 and CLOCK could contribute to CB assembly and physiology. Other very recent studies described novel components of the CB. The RNA methyltransferase NSun2 [36] and the testis-specific serine/threonine kinases TSSK1 and TSSK2, and the substrate TSKS [37], were identified as essential elements for germ cell differentiation in the mouse testis. Moreover, the mitochondrial carrier SCaMC-1L was unexpectedly detected in the CB suggesting a new link between mitochondria and germinal granules [38]. These publications may indicate that many components of the CB and their potential novel functions in the male germ cells are still unveiled.
Table 1.
Novel protein components of the mammalian Chromatoid body (see [1] for a list of previously identified CB proteins and [29] for the most updated full RNA and protein composition of the CB).
| Protein | Synonym | Description | Function | Ref |
|---|---|---|---|---|
| BASP1 and MARCKS | NAP-22 for BASP1 | Brain Acid Soluble Protein 1 and Myristoylated Alanin Rich C Kinase Substrate | In neurons; affect actin cytoskeleton dynamics and promote outgrowth | [27] |
| CLOCK and BMAL1 | ARNT-like 1 for BMAL1 | Circadian Locomoter Output Cycles protein Kaput and Brain and Muscle ARNT-Like 1 | CLOCK and BMAL1 are central regulators of the circadian clock. Bmal1 KO and Clock KO mice display morfologic CB alterations due to BMAL1 ablation or low expression | [34] |
| HAT1 and p46 | - | Histone acetyltransferase 1 and Histone-binding protein RBBP7 | Hat1 and its cofactor p46 control the physiology of the CB by acetylation of MVH | [33] |
| MAGO | MAGOH; MGN2 | Protein mago nashi homolog | Its role in normal spermatogenesis of vertebrates is unknown | [29, 104] |
| NANOS1 and PUMILIO2 | - | Nanos homolog 1 and Pumilio homolog 2 | NANOS1 and PUMILIO2 bind to regulate translation of specific mRNAs germ cells of certain model organisms | [105] |
| NONO | - | Non-POU domain containing; octamer binding protein | RNA binding protein. It may participate in various steps of mRNA processing and protein translation | [106] |
| NSun2 | Misu | NOL1/NOP2/Sun domain family member 2 | Cytosine-5 RNA methyltransferase. Nsun2 KO mice have male gametogenesios blocked at the pachytene stage | [107] |
| PABPC1 and PABPC3 | - | Poly(A) Binding Protein; Cytoplasmic 1 and 3 | Bind the poly(A) tail of mRNA. May be involved in cytoplasmic regulatory processes of mRNA metabolism. | [106, 108] |
| SAM68 | - | KH domain-containing; RNA-binding; signal transduction-associated protein 1 | RNA binding protein with potential role in the miRNA pathway during spermatogenesis | [29, 109] |
| SCaMC-1L | - | Small calcium-binding mitochondrial carrier protein 1 like | ATP-Mg/Pi carrier that mediate a reversible electroneutral exchange between ATP-Mg2- and HPO4 2- and control the net transport of adenine nucleotides across the inner mitochondrial membrane | [110] |
| TDRD9 | - | Mouse Tudor repeat-9 | Tudor domain containing protein that is essential for supressessing LINE1 retrotranpsosons. Tdrd9 KO mice have male gametogenesis blocked at zygotene stage | [29, 111] |
| TSSK1; TSSK2 and TSKS | - | Testis-specific serine/threonine-protein kinase 1; 2 and Testis-specific serine kinase substrate | Tssk1 and Tssk2 KO mice have male gametogenesis blocked at elongating spermatids possessing a collapsed mitochondrial sheath | [112] |
1.3.miRNAs
Accumulating evidence points to miRNAs as potentially important elements for the control of gene expression in different cell types and tissues. Specifically in the testis this mechanism has been found to be functional and critical for normal sperm development. MicroRNAs are endogenous RNAs of 22 nucleotides in length that repress the translation of hundreds of mRNAs by binding them in a protein complex [35]. Their role is of great relevance as most of the protein-coding genes appears to be controlled by the miRNA pathway [36]. The regulation mechanisms of miRNAs can be pictured as a cascade of events where a miRNA, in cooperation with other factors or not, can downregulate the expression of a transcription factor and thus, facilitating transcriptional regulation of numerous downstream target genes [37]. Many tissues utilize miRNAs to control gene expression and the expression and localization of the proteins involved in this pathway in the spermatogenic process is still unclear [15]. While miRNAs and miRNA-related proteins have been detected in the CB of round spermatids [26], other components of this pathway have been localized also outside the CB.
Although coding sequences of miRNAs are found all along the genome, approximately half are placed within the introns of genes and their transcription is regulated following the one of their host gene [38]. Precursors of miRNAs (primary RNAs, priRNAs) are mostly transcribed products of the RNA polymerase II that form a long hairpin loop and are cleaved in the nucleus by double-stranded RNA endonuclease Drosha into pre-miRNAs that will be transported to the cytoplasm [15, 17, 30, 39, 40] (Figure 2). In the cytoplasm, the pre-miRNA loop is cleaved by the endoribonuclease Dicer, generating a mature miRNA that will form a complex with AGO proteins called miRNA-induced silencing complex (miRISC) and perform translation regulation of the mRNA targets that are loaded [39]. The fate of the targeted mRNA will depend on its degree and nature of the complementarity to the miRNA and on the type of the AGO proteins found in the RISC complex [39, 41]. For example, in mammals AGO2 is the only AGO protein that cleaves RNA and in conjunction with high miRNA-mRNA complementarity leads to RNA degradation [39, 41, 42]. However, mRNAs with lower or imperfect complementarity to its miRNA partners supposes mRNA sequestration in cytoplasmic granules and/or translational repression [43, 44]. A mechanism for translational repression by miRNAs has been proposed. Several reports have suggested a translational initiation block theory by different processes. MiRNAs can mediate deadenylation of the mRNA polyA tail to avoid binding of the PolyABinding Protein (PABP) required for effective translation [45]. Additionally, AGO2 has been shown to bind the modified mRNA 5′ end and compete with translational initiation factors called Eukaryotic translation Initiation Factor 4D (eIF4E) to finally block translational initiation [46] and eIF6 to prevent the formation of functional 80S subunit of the ribosome [47]. Once translation has been initiated, miRNAs are hypothesized to block protein synthesis [48]. On the other hand, miRNA has been also found to promote translation depending on the proliferation state of cell and the presence of AU-rich elements within the 3′UTR of the mRNA target [39, 49]. Intriguing, a subset of miRNAs called epi-miRNAs can also regulate epigenetic modifications and vice versa [50]. It has been found that epi-miRNAs can control directly or indirectly the expression of the de novo and maintenance DNA methyltransferases as well as histone methylation and acetylation. Intriguingly, same epigenetic modifications regulated by epi-miRNAs can also affect miRNA expression. This regulatory mechanism has been found to be important in tumorigenesis by silencing tumor suppressor genes and activating oncogenes [51, 52].
In the testis, proper functional miRNA mechanisms are required for successful sperm development since spermatogenesis is disrupted at early stages in mice conditionally lacking the enzymes Drosha or Dicer [53-55]. Through expression profile studies using either partially purified germ cells or whole testis, specific miRNAs have been detected to be exclusive of human and mouse testis [56-60] and some of them have been suggested to participate in mammalian spermatogenesis.
1.4.piRNAs
As aforementioned, the function of sncRNAs has been shown to be crucial for the spermatogenic process. Another class of sncRNA consisting of 24 to 30 nucleotides that are generated in a Dicer-independent manner and that interact with Piwi proteins, have been described [61]. Not inactivated by evolution and still competent selfish mobile elements currently pose an ongoing mutagenic threat in mammals. In spermatogenesis, the Piwi pathway has the specific role of silencing retrotransposons [4, 62]. Several evidences have suggested that transposon control is closely associated with proper sperm development, prompting host genomes to present important resources to protect their genetic information [63-65]. Animal models lacking components of the Piwi pathway are male-specific sterile presenting another evidence of the relevance of this sncRNA pathway in spermatogenesis [19, 61].
A high diversity of piRNAs have been described, suggesting the presence of million individual piRNAs, a remarkable notion when compared to a few hundred miRNAs. The genes that express piRNAs are not conserved but their genomic location can be seen as clusters suggesting that piRNAs are processed from long primary transcripts [63, 66]. piRNAs, whose sequence dramatically change during sperm development, are classified in mammals mainly into two categories, prepachytene and pachytene piRNAs [15, 19] (Figure 1). The piRNA sequences can contain repeat-derived sequences, intergenic and genic regions [17, 19, 66]. Prepachytene piRNAs are enriched in repeat-derived sequences and associate with MIWI2 and MILI in prospermatogonia; however, pachytene piRNAs are enriched in intergenic, unannotated sequences and bind MILI and MIWI proteins in pachytene spermatocytes and round spermatids [63, 66-68]. Just few days after birth, the majority of the piRNAs in the mouse testis are prepachytene; however, at 14.5 days postpartum (dpp), pachytene piRNAs appears in the developing spermatocytes when they enter the pachytene phase of meiotic prophase I. Interestingly, these pachytene piRNas will correspond to >95% of piRNAs in the adult mouse testis. The transcription factor A-MYB contributes to initiating pachytene piRNA production through directly regulating piRNAs transcription together with the regulation of genes that encode for protein components of the piRNA pathway [69].
Due to its complexity and singularity, a detailed pathway of the piRNA biogenesis is still incomplete. It is however known that the piRNA biogenesis pathway differs from miRNAs's mainly because its independency from Dicer's activity [4, 70]. Two pathways have been proposed: the primary processing to produce primary piRNAs and the ping-pong mechanism to amplify piRNAs and requires slicer activity of Piwi proteins [71] (Figure 2). In pachytene mouse spermatocytes, piRNAs are only generated by primary processing, however, in prospermatogonia and premeiotic spermatogonia, the ping-pong mechanism further amplifies specific sequences generated by the primary biogenesis pathway [63, 66, 71]. In the primary processing, it has been suggested that precursors of piRNAs correspond to long single-stranded RNAs just transcribed from the piRNA cluster genes that have already determined their 5’ end and consequently loaded onto Piwi proteins [4, 19, 71, 72]. Therefore, the 3’ end is cleaved giving rise to a pool of different piRNAs. In the ping-pong mechanism, Piwi proteins (MILI in mice) recognize complementarity between RNAs and primary piRNAs, then, the targeted RNAs are cleaved through the slicer activity of the Piwi protein at the position 10th from the 5’ end of the primary piRNA, producing secondary piRNA precursors [4, 72]. At this point, secondary piRNA are specifically amplified by another mouse Piwi protein named MIWI2. Interestingly, these two Piwi proteins, MILI, preferentially associated to primary piRNAs, and MIWI2, to secondary piRNAs, have non-redundant roles in the ping-pong mechanism [67], although it has recently proposed a homotypic ping-pong mechanism where MILI is the only Piwi protein involved [73]. In any case, this biological process relies on the complementarity of sequences to specifically amplify piRNAs that are antisense to TE mRNAs and consequently piRNAs are able to target them [4, 62, 71]. At the same time as piRNAs are produced through the ping-pong cycle, transposon transcripts are cleaved by piRISCs [72, 74]. This mechanism has been seen in primordial mouse testis but not in the adult, where MIWI2 in no longer expressed, suggesting that MILI and MIWI are involved in piRNAs biogenesis through predominantly primary processing [75]. Additionally, piRNAs are also suggested to regulate directly transposon element (TE) transcription since MILI and MIWI2 mutants fail to establish de novo DNA methylation of TE in fetal male germ cells when de novo DNA methylation takes place [67, 76]. Recently, the Piwi pathway was reported to be responsible for methylation of a paternally imprinted gene locus [77] suggesting that this pathway may have further functions beyond TE silencing.
Several protein components of the piRNA pathway have also been described and some of them been localized in the CB (Table 1). In mice, one of the first protein identified to be associated to Piwi proteins are the Tudor family proteins [4]. Tudor proteins are suggested to associate with Piwi proteins through their recognition of arginine methylated N-terminal conserved residues and promote their localization to the nuage [78] and to form effector RNP complexes with piRNAs [19]. Thus, Tudor proteins have been suggested to function as scaffolds to congregate macromolecular complexes [4]. The RNA helicase MVH, a hallmark of the CB, is another important and conserved component of the Piwi pathway. MVH is required for piRNA production and transposon silencing since mice lacking functional MVH present a clear upregulation of TE expression, reduction of piRNAs and impairment of DNA methylation [64]. Other protein elements like MOV10L1, MAELSTROM, GASZ/ASZ1, MITOPLD and FKBP6 are also essential components of the Piwi pathway and crucial for the spermatogenic process [17, 79-82].
Nonetheless, it is clear that the crucial participation of Piwi pathway in germline development highlights the importance and intimate integration of transposon control in this process. The detailed molecular mechanism of the biogenesis and function of piRNAs is still uncertain.
1.5.ncRNAs in the spermatogenic process
Both miRNA and piRNA pathways function along the sperm development and execute crucial roles to ensure the proper formation of the male gamete. During mammalian early embryonic development, sncRNA pathways are functional, although germinal granules are not observed in the prospective germ cells and their components cannot be detected [2, 83]. The regulation of a variety of pluripotency genes needed for germ cell-specification has been found to require miRNAs, such as miR-145 that totally and partially suppress the expression of OCT4 and SOX2 respectively in human embryonic stem (ES) cells and hence promote differentiation to germ cells [39, 84].
Spermatogonial stem cells can decide for self-renewal or differentiation; the first option lasts until the male old age in human males and the last generates a spermatozoa through a meiotic division and a differentiation pathway [11]. miRNAs have been suggested to interpret and transduce cellular signals to allow the maintenance of the undifferentiated stem cell population as well as allowing cell differentiation during spermatogenesis [39]. Recent studies have demonstrated the involvement of several miRNAs in maintaining the SSC population. These include miR-21 [85], Mir-17-92 (Mirc1) and its paralog Mir-106b-25 (Mirc3) clusters [86], Mir146 [87] and miR-221/222 [88]. Similarly, a study of cryptorchid testes in rats found that the function of miR-135a contributed to stem cell maintenance and its target, the transcription factor FoxO1, is known to be essential for SSC maintenance [89]. On the other hand, other miRNAS were found to potentially have a role in promoting spermatogonial differentiation such as Mirlet7 family miRNAs [90].
It is noteworthy to mention that, concurrently with the two waves of active transcription, germ cells from the meiotic phase and haploid spermatids have been found to express most of the identified miRNAs in the testis [57, 58, 91]. Despite the suggested specificity of several miRNAs in distinct stages of the sperm development, several miRNAs are enriched in both meiotic and postmeiotic spermatogenic cells. As an example, the expression of miR-184 was restricted to the male germ cells from spermatogonia to round spermatids and was found to be involved in the post-transcriptional regulation of mRNAs of nuclear receptor corepressor 2 (Ncor2) through inhibiting NCOR2 protein translation in mammalian spermatogenesis [92]. However, other miRNAs were found enriched in premeiotic germ cells such as miR-383, the expression of which is restricted to spermatogonia and early spermatocytes, and has been associated with male infertility and with inducing testicular embryonal carcinoma cell proliferation [93]. Thus, it is suggested that miR-383 may act on early stages of spermatogenesis regulating germ cell proliferation or death. At the same time, a potential feedback loop between FMRP and miR-383 during spermatogenesis was also proposed, suggesting that impairment of the FMRP-miR-383 pathway may partially contribute to human spermatogenic failure with maturation arrest [94].
Other miRNAs have been proposed to be specifically functional in meiosis such as miR-214, possibly through interaction with members of the heat shock family, and miR-24 potentially through its binding with Methyl-CpG binding domain protein 6 (MBD6) and histone 2A family member X (H2AX) [91].
Apoptosis in spermatocytes is a frequent event since meiotic cells have to undergo crossover/homologous recombination through double strand breaks that must be repaired after recombination is finished. During this process any chromosomal mistake can lead the meiotic checkpoint mechanism that promotes spermatocyte apoptosis [95]. Several miRNAs have also been shown to control apoptotic signaling of different cellular types. The miR-449 cluster and miR-34b/c were highly detected in meiotic and postmeiotic germ cells and their functions were found to be redundant in the regulation of male germ cell development in mice by targeting the E2F-pRb pathway in the early meiotic phase [60]. This represents a mechanism for the suppression of E2F activity during meiotic and postmeiotic male germ cell to potentially prevent massive meiotic male germ cells apoptosis. Similarly, the involvement of miR-17-92 cluster in the regulation of E2F-1 transcription factor during normal spermatogenesis and in carcinoma was also investigated [96]. When pri-mir-17-5p expression was strongest, in pachytene spermatocytes, E2F-1 protein was lowest, proposing that the physiological inhibition of E2F1 mRNA translation may be another important mechanism to prevent apoptosis during meiotic recombination. In a study intended to determine the specific function of miR-34c in germline differentiation, a miRNA well documented to be pro-apoptotic, it was found that miR-34c enhanced murine male germ cell apoptosis through targeting ATF1 (activating transcription factor 1) and providing a novel mechanism of the involvement of miRNAs in the regulation of germ cell apoptosis [97].
Several miRNAs have been described to participate in gene expression regulation of postmetiotic spermatogenic cells. MiR-122a, which is mainly expressed in late-stage male germ cells and specifically on polysomes, promotes the cleavage of Tnp2 transcripts, a nuclear protein that is involved in chromatin remodeling assisting the histone-to-protamine transition in mouse spermatogenesis [56]. In mice lacking the testis-specific member of the DEAD-box family (GRTH, Table 1), which is an essential post-transcriptional regulator of spermatogenesis, miR-469 was found overexpressed. This miRNA was also observed to repress Tnp2 and Prm2 translation leading to infertility due to failure of spermatids to elongate. Thus, GRTH was suggested to act as a negative regulator of miR-469 biogenesis and consequently repress its function during spermatogenesis [98]. It has also been recently shown that miR-18 directly targets heat shock factor 2 (HSF2), a transcription factor involved in spermatogenesis [37]. This miRNA was found at highest intensity in the spermatocytes and was suggested to regulate HSF2 activity in spermatogenesis and link miR-18 to HSF2-mediated postmeiotic physiological processes such as correct chromatin organization and sperm maturation.
While miRNAs are expressed abundantly during all stages of spermatogenesis, piRNAs are mainly present in pachytene spermatocytes and round spermatids. During mouse embryonic testis development, MILI and MIWI2 are co-expressed and localized in the prospermatogonia in two distinct types of germ granules that are often in contact, the pi-bodies and the piP-bodies respectively. MILI is found in the pi-bodies together with a Tudor protein (TDRD1), MVH and GASZ and MIWI2 in the piP-bodies with another Tudor protein (TDRD9) and MAEL [64, 65, 81]. The correct formation of these germ granules is crucial for the proper function of the piRNA pathway in TE silencing [71]. In postnatal male germ cells, MIWI2 expression ceases and MIWI appears at the same time of the CB, suggesting that pachytene piRNAs associated with MIWI and MILI may perform their role in this germ. Additionally, it has been indicated that pachytene piRNAs correspond to the end products of RNA processing that mostly belong to non-coding transcripts with potential meiotic functions [99]. It was also demonstrated that Miwi directly binds and stabilize spermiogenic mRNAs to participate in the formation of mRNP complexes. This study established novel roles of the piRNA pathway that shed light on this mechanism in relation to the mammalian male germ cell development.
The relevance of piRNAs at several distinct stages of spermatogenesis and the absence of redundant functions are illustrated by the fact that mouse mutants for components of the piRNA pathway are male-specific sterile. These defects are observed in two different sperm developmental stages: during meiosis in spermatocytes and post-meiotically in haploid spermatids [4, 19]. In mouse, mutations in Mili and Miwi2 block spermatogenic progression through pachytene spermatocytes, whereas mutations in Miwi led to round spermatid arrest [100-102]. In Mili and Miwi2 mutants, their phenotype can also be seen in fetal prospermatogonia where the expression of TE is upregulated and their corresponding genes are hypomethylated. On the other hand, the molecular link between the piRNAs and the spermatid development has not been unveiled in Miwi mutants. Interestingly, MIWI activity was found to directly cleave transposon RNAs providing an explanation for the continued maintenance of repeat-derived piRNAs [68]. In addition, in a recent study MILI and the repressive histone H3 dimethylated K9 modification in meiotic pachytene and mitotic germ cells respectively were also required for post-transcriptionally silence of TEs in the adult [103]. All this suggests a scenario in which MILI and MIWI2 establish both transcriptional and post-transcriptional repression of TEs in germ cells from embryos; on the other hand, MILI and MIWI together with other epigenetic mechanisms silence TEs post-transcriptionally after birth enforcing genomic stability in those cells.
1.6. Conclusion
The successful transmission of genetic and epigenetic information to next generations depends on the correct formation of sperm cells through the protection of their genomes and the proper establishment and maintenance of epigenetic marks. Many studies have demonstrated that sncRNAs are crucial regulators of gene expression controlling the fate of their target RNAs, mainly at the post-transcriptional level. Interestingly, sncRNA pathways such as miRNAs and piRNAs have been shown to play an important role in spermatogenesis. Dysregulation of miRNAs during spermatogenesis leads to male infertility and, although not discussed in this review, it has also been related to testicular cancer. The specific role of many miRNAs in each stage of the spermatogenic process is still unknown and requires future studies. On the other hand, the germ cell specific piRNA pathway is still poorly understood compared to other small RNA silencing processes. While major strides have been achieved in the understanding piRNA physiology, many enigmas are still unveiled such as how piRNA genes are transcribed and their molecular mechanism of action. It is now clear that piRNAs provide protection of the germline genome against detrimental expression of TEs. The understanding of the molecular mechanism that both the miRNA and piRNA pathways execute in spermatogenesis still needs further investigation. This issue is of high interest since it will potentially allow researchers to control these pathways for male contraception as well as for diagnostic or treatment proposes in the cases of testicular cancer or male infertility.
Highlights.
Small non-coding RNAs are crucial for the development of the male germ cell.
sncRNAs operate through post-transcriptional regulatory mechanisms.
miRNA and piRNAs are the most important sncRNAs in regulating spermatogenesis.
Components of the sncRNAs pathway are found in the germ granule localized in the cytoplasm of germ cells.
Acknowledgements
NIH grants GM081634 and AG033888, Merieux Research Grant, and Sirtris Pharmaceuticals SP-48984 (PSC).
Abbreviations
- Ago
Argonaute
- CB
Chromatoid body
- IMC
Intermitochondiral cement
- miRNAs
microRNAs
- mRNPs
mRNA ribonucleoproteins
- MVH
mouse vasa homolog protein
- ncRNAs
non-coding RNAs
- PGCs
Primordial germ cells
- piRNAs
piwi-interacting RNAs
- PIWI
P-element induced wimpy testis
- RBP
RNA-binding proteins
- RISC
RNA-induced silencing complex
- siRNAs
Small interfering RNAs
- sncRNAs
small non-coding RNAs
- SSC
spermatogonial stem cells
- TE
Transposone Element
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Voronina E, Seydoux G, Sassone-Corsi P, Nagamori I. RNA granules in germ cells. Csh Perspect Biol. 2011:3. doi: 10.1101/cshperspect.a002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chuma S, Hosokawa M, Tanaka T, Nakatsuji N. Ultrastructural characterization of spermatogenesis and its evolutionary conservation in the germline: Germinal granules in mammals. Mol Cell Endocrinol. 2009;306:17–23. doi: 10.1016/j.mce.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 3.Kotaja N. Small RNAs in spermatogenesis. Int J Androl. 2010;33:35. [Google Scholar]
- 4.Chuma S, Nakano T. piRNA and spermatogenesis in mice. Philos T R Soc B. 2013:368. doi: 10.1098/rstb.2011.0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet. 2008;9:129–40. doi: 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- 6.Saxe JP, Lin HF. Small Noncoding RNAs in the Germline. Csh Perspect Biol. 2011:3. doi: 10.1101/cshperspect.a002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kimmins S, Sassone-Corsi P. Chromatin remodelling and epigenetic features of germ cells. Nature. 2005;434:583–9. doi: 10.1038/nature03368. [DOI] [PubMed] [Google Scholar]
- 8.Papaioannou MD, Nef S. microRNAs in the Testis: Building Up Male Fertility. J Androl. 2010;31:26–33. doi: 10.2164/jandrol.109.008128. [DOI] [PubMed] [Google Scholar]
- 9.Paronetto MP, Sette C. Role of RNA-binding proteins in mammalian spermatogenesis. Int J Androl. 2010;33:2–12. doi: 10.1111/j.1365-2605.2009.00959.x. [DOI] [PubMed] [Google Scholar]
- 10.Geremia R, Boitani C, Conti M, Monesi V. RNA synthesis in spermatocytes and spermatids and preservation of meiotic RNA during spermiogenesis in the mouse. Cell Differ Dev. 1977;5:343–55. doi: 10.1016/0045-6039(77)90072-0. [DOI] [PubMed] [Google Scholar]
- 11.Sassone-Corsi P. Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science. 2002;296:2176–8. doi: 10.1126/science.1070963. [DOI] [PubMed] [Google Scholar]
- 12.Rajender S, Avery K, Agarwal A. Epigenetics, spermatogenesis and male infertility. Mutat Res. 2011;727:62–71. doi: 10.1016/j.mrrev.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Oliva R. Protamines and male infertility. Hum Reprod Update. 2006;12:417–35. doi: 10.1093/humupd/dml009. [DOI] [PubMed] [Google Scholar]
- 14.Idler RK, Yan W. Control of Messenger RNA Fate by RNA-Binding Proteins: An Emphasis on Mammalian Spermatogenesis. J Androl. 2012;33:309–37. doi: 10.2164/jandrol.111.014167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meikar O, Da Ros M, Korhonen H, Kotaja N. Chromatoid body and small RNAs in male germ cells. Reproduction. 2011;142:195–209. doi: 10.1530/REP-11-0057. [DOI] [PubMed] [Google Scholar]
- 16.Novotny GW, Nielsen JE, Sonne SB, Skakkebaek NE, Rajpert-De Meyts E, Leffers H. Analysis of gene expression in normal and neoplastic human testis: new roles of RNA. Int J Androl. 2007;30:316–26. doi: 10.1111/j.1365-2605.2007.00773.x. discussion 26-7. [DOI] [PubMed] [Google Scholar]
- 17.Watanabe T, Chuma S, Yamamoto Y, Kuramochi-Miyagawa S, Totoki Y, Toyoda A, et al. MITOPLD Is a Mitochondrial Protein Essential for Nuage Formation and piRNA Biogenesis in the Mouse Germline. Dev Cell. 2011;20:364–75. doi: 10.1016/j.devcel.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eddy EM. Germ Plasm and Differentiation of Germ-Cell Line. Int Rev Cytol. 1975;43:229–80. doi: 10.1016/s0074-7696(08)60070-4. [DOI] [PubMed] [Google Scholar]
- 19.Pillai RS, Chuma S. piRNAs and their involvement in male germline development in mice. Dev Growth Differ. 2012;54:78–92. doi: 10.1111/j.1440-169X.2011.01320.x. [DOI] [PubMed] [Google Scholar]
- 20.Eddy EM. Fine-Structural Observations on Form and Distribution of Nuage in Germ-Cells of Rat. Anat Rec. 1974;178:731–57. doi: 10.1002/ar.1091780406. [DOI] [PubMed] [Google Scholar]
- 21.Fawcett DW, Eddy EM, Phillips DM. Observations on the fine structure and relationships of the chromatoid body in mammalian spermatogenesis. Biol Reprod. 1970;2:129–53. doi: 10.1095/biolreprod2.1.129. [DOI] [PubMed] [Google Scholar]
- 22.Ventela S, Toppari J, Parvinen M. Intercellular organelle traffic through cytoplasmic bridges in early spermatids of the rat: mechanisms of haploid gene product sharing. Mol Biol Cell. 2003;14:2768–80. doi: 10.1091/mbc.E02-10-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parvinen M. The chromatoid body in spermatogenesis. Int J Androl. 2005;28:189–201. doi: 10.1111/j.1365-2605.2005.00542.x. [DOI] [PubMed] [Google Scholar]
- 24.Onohara Y, Fujiwara T, Yasukochi T, Himeno M, Yokota S. Localization of mouse vasa homolog protein in chromatoid body and related nuage structures of mammalian spermatogenic cells during spermatogenesis. Histochem Cell Biol. 2010;133:627–39. doi: 10.1007/s00418-010-0699-5. [DOI] [PubMed] [Google Scholar]
- 25.Nagamori I, Sassone-Corsi P. The chromatoid body of male germ cells Epigenetic control and miRNA pathway. Cell Cycle. 2008;7:3503–8. doi: 10.4161/cc.7.22.6977. [DOI] [PubMed] [Google Scholar]
- 26.Kotaja N, Bhattacharyya SN, Jaskiewicz L, Kimmins S, Parvinen M, Filipowicz W, et al. The chromatoid body of male germ cells: Similarity with processing bodies and presence of Dicer and microRNA pathway components. P Natl Acad Sci USA. 2006;103:2647–52. doi: 10.1073/pnas.0509333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mosevitsky MI, Snigirevskaya ES, Komissarchik YY. Immunoelectron microscopic study of BASP1 and MARCKS location in the early and late rat spermatids. Acta Histochem. 2012;114:237–43. doi: 10.1016/j.acthis.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 28.Kotaja N, Sassone-Corsi P. The chromatoid body: a germ-cell-specific RNA-processing centre. Nat Rev Mol Cell Bio. 2007;8:85–90. doi: 10.1038/nrm2081. [DOI] [PubMed] [Google Scholar]
- 29.Meikar O, Vagin VV, Chalmel F, Sostar K, Lardenois A, Hammell M, et al. An atlas of chromatoid body components. Rna. 2014;20:483–95. doi: 10.1261/rna.043729.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yokota S. Historical survey on chromatoid body research. Acta Histochem Cytoc. 2008;41:65–82. doi: 10.1267/ahc.08010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka SS, Toyooka Y, Akasu R, Katoh-Fukui Y, Nakahara Y, Suzuki R, et al. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev. 2000;14:841–53. [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen Chi M, Chalmel F, Agius E, Vanzo N, Khabar KS, Jegou B, et al. Temporally regulated traffic of HuR and its associated ARE-containing mRNAs from the chromatoid body to polysomes during mouse spermatogenesis. Plos One. 2009;4:e4900. doi: 10.1371/journal.pone.0004900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagamori I, Cruickshank VA, Sassone-Corsi P. Regulation of an RNA granule during spermatogenesis: acetylation of MVH in the chromatoid body of germ cells. J Cell Sci. 2011;124:4346–55. doi: 10.1242/jcs.096461. [DOI] [PubMed] [Google Scholar]
- 34.Peruquetti RL, De Mateo S, Sassone-Corsi P. Circadian Proteins CLOCK and BMAL1 in the Chromatoid Body, a RNA Processing Granule of Male Germ Cells. Plos One. 2012:7. doi: 10.1371/journal.pone.0042695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas M, Lieberman J, Lal A. Desperately seeking microRNA targets. Nat Struct Mol Biol. 2010;17:1169–74. doi: 10.1038/nsmb.1921. [DOI] [PubMed] [Google Scholar]
- 36.Friedman RC, Farh KKH, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bjork JK, Sandqvist A, Elsing AN, Kotaja N, Sistonen L. miR-18, a member of Oncomir-1, targets heat shock transcription factor 2 in spermatogenesis. Development. 2010;137:3177–84. doi: 10.1242/dev.050955. [DOI] [PubMed] [Google Scholar]
- 38.Shomron N, Levy C. MicroRNA-Biogenesis and Pre-mRNA Splicing Crosstalk. J Biomed Biotechnol. 2009 doi: 10.1155/2009/594678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McIver SC, Roman SD, Nixon B, McLaughlin EA. miRNA and mammalian male germ cells. Hum Reprod Update. 2012;18:44–59. doi: 10.1093/humupd/dmr041. [DOI] [PubMed] [Google Scholar]
- 40.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–39. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 41.Macfarlane LA, Murphy PR. MicroRNA: Biogenesis, Function and Role in Cancer. Curr Genomics. 2010;11:537–61. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–41. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 43.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 44.Perron MP, Provost P. Protein interactions and complexes in human microRNA biogenesis and function. Front Biosci. 2008;13:2537–47. doi: 10.2741/2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wakiyama M, Takimoto K, Ohara O, Yokoyama S. Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes Dev. 2007;21:1857–62. doi: 10.1101/gad.1566707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kiriakidou M, Tan GS, Lamprinaki S, De Planell-Saguer M, Nelson PT, Mourelatos Z. An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell. 2007;129:1141–51. doi: 10.1016/j.cell.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 47.Chendrimada TP, Finn KJ, Ji X, Baillat D, Gregory RI, Liebhaber SA, et al. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447:823–8. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- 48.Liu J. Control of protein synthesis and mRNA degradation by microRNAs. Curr Opin Cell Biol. 2008;20:214–21. doi: 10.1016/j.ceb.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 49.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–4. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 50.Iorio MV, Piovan C, Croce CM. Interplay between microRNAs and the epigenetic machinery: an intricate network. Biochim Biophys Acta. 2010;1799:694–701. doi: 10.1016/j.bbagrm.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 51.Chuang JC, Jones PA. Epigenetics and microRNAs. Pediatr Res. 2007;61:24R–9R. doi: 10.1203/pdr.0b013e3180457684. [DOI] [PubMed] [Google Scholar]
- 52.Valeri N, Vannini I, Fanini F, Calore F, Adair B, Fabbri M. Epigenetics, miRNAs, and human cancer: a new chapter in human gene regulation. Mamm Genome. 2009;20:573–80. doi: 10.1007/s00335-009-9206-5. [DOI] [PubMed] [Google Scholar]
- 53.Maatouk DM, Loveland KL, McManus MT, Moore K, Harfe BD. Dicer1 is required for differentiation of the mouse male germline. Biol Reprod. 2008;79:696–703. doi: 10.1095/biolreprod.108.067827. [DOI] [PubMed] [Google Scholar]
- 54.Romero Y, Meikar O, Papaioannou MD, Conne B, Grey C, Weier M, et al. Dicer1 depletion in male germ cells leads to infertility due to cumulative meiotic and spermiogenic defects. Plos One. 2011;6:e25241. doi: 10.1371/journal.pone.0025241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu Q, Song R, Ortogero N, Zheng H, Evanoff R, Small CL, et al. The RNase III enzyme DROSHA is essential for microRNA production and spermatogenesis. The Journal of biological chemistry. 2012;287:25173–90. doi: 10.1074/jbc.M112.362053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu Z, Raabe T, Hecht NB. MicroRNA Mirn122a reduces expression of the posttranscriptionally regulated germ cell transition protein 2 (Tnp2) messenger RNA (mRNA) by mRNA cleavage. Biol Reprod. 2005;73:427–33. doi: 10.1095/biolreprod.105.040998. [DOI] [PubMed] [Google Scholar]
- 57.Ro S, Park C, Sanders KM, McCarrey JR, Yan W. Cloning and expression profiling of testis-expressed microRNAs. Dev Biol. 2007;311:592–602. doi: 10.1016/j.ydbio.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yan N, Lu Y, Sun H, Tao D, Zhang S, Liu W, et al. A microarray for microRNA profiling in mouse testis tissues. Reproduction. 2007;134:73–9. doi: 10.1530/REP-07-0056. [DOI] [PubMed] [Google Scholar]
- 59.Smorag L, Zheng Y, Nolte J, Zechner U, Engel W, Pantakani DV. MicroRNA signature in various cell types of mouse spermatogenesis: evidence for stage-specifically expressed miRNA-221, -203 and - 34b-5p mediated spermatogenesis regulation. Biol Cell. 2012;104:677–92. doi: 10.1111/boc.201200014. [DOI] [PubMed] [Google Scholar]
- 60.Bao J, Li D, Wang L, Wu J, Hu Y, Wang Z, et al. MicroRNA-449 and microRNA-34b/c function redundantly in murine testes by targeting E2F transcription factor-retinoblastoma protein (E2F-pRb) pathway. The Journal of biological chemistry. 2012;287:21686–98. doi: 10.1074/jbc.M111.328054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development. 2008;135:3–9. doi: 10.1242/dev.006486. [DOI] [PubMed] [Google Scholar]
- 62.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–4. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 63.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 64.Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Takamatsu K, Chuma S, Kojima-Kita K, et al. MVH in piRNA processing and gene silencing of retrotransposons. Genes Dev. 2010;24:887–92. doi: 10.1101/gad.1902110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reuter M, Chuma S, Tanaka T, Franz T, Stark A, Pillai RS. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat Struct Mol Biol. 2009;16:639–46. doi: 10.1038/nsmb.1615. [DOI] [PubMed] [Google Scholar]
- 66.Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–7. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 67.Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785–99. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reuter M, Berninger P, Chuma S, Shah H, Hosokawa M, Funaya C, et al. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature. 2011;480:264–7. doi: 10.1038/nature10672. [DOI] [PubMed] [Google Scholar]
- 69.Li XZ, Roy CK, Dong X, Bolcun-Filas E, Wang J, Han BW, et al. An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol Cell. 2013;50:67–81. doi: 10.1016/j.molcel.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–4. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 71.Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12:246–58. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- 72.Ishizu H, Siomi H, Siomi MC. Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines. Genes Dev. 2012;26:2361–73. doi: 10.1101/gad.203786.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.De Fazio S, Bartonicek N, Di Giacomo M, Abreu-Goodger C, Sankar A, Funaya C, et al. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature. 2011;480:259–63. doi: 10.1038/nature10547. [DOI] [PubMed] [Google Scholar]
- 74.Luteijn MJ, Ketting RF. PIWI-interacting RNAs: from generation to transgenerational epigenetics. Nature reviews Genetics. 2013;14:523–34. doi: 10.1038/nrg3495. [DOI] [PubMed] [Google Scholar]
- 75.Beyret E, Liu N, Lin HF. piRNA biogenesis during adult spermatogenesis in mice is independent of the ping-pong mechanism. Cell Res. 2012;22:1429–39. doi: 10.1038/cr.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–17. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Watanabe T, Tomizawa S, Mitsuya K, Totoki Y, Yamamoto Y, Kuramochi-Miyagawa S, et al. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science. 2011;332:848–52. doi: 10.1126/science.1203919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vagin VV, Wohlschlegel J, Qu J, Jonsson Z, Huang X, Chuma S, et al. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev. 2009;23:1749–62. doi: 10.1101/gad.1814809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zheng K, Xiol J, Reuter M, Eckardt S, Leu NA, McLaughlin KJ, et al. Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. P Natl Acad Sci USA. 2010;107:11841–6. doi: 10.1073/pnas.1003953107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Soper SF, van der Heijden GW, Hardiman TC, Goodheart M, Martin SL, de Boer P, et al. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev Cell. 2008;15:285–97. doi: 10.1016/j.devcel.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ma L, Buchold GM, Greenbaum MP, Roy A, Burns KH, Zhu H, et al. GASZ is essential for male meiosis and suppression of retrotransposon expression in the male germline. PLoS Genet. 2009;5:e1000635. doi: 10.1371/journal.pgen.1000635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xiol J, Cora E, Koglgruber R, Chuma S, Subramanian S, Hosokawa M, et al. A role for Fkbp6 and the chaperone machinery in piRNA amplification and transposon silencing. Mol Cell. 2012;47:970–9. doi: 10.1016/j.molcel.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 83.Eddy EM, Clark JM, Gong D, Fenderson BA. Origin and Migration of Primordial Germ-Cells in Mammals. Gamete Res. 1981;4:333–62. [Google Scholar]
- 84.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–58. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 85.Niu Z, Goodyear SM, Rao S, Wu X, Tobias JW, Avarbock MR, et al. MicroRNA-21 regulates the self-renewal of mouse spermatogonial stem cells. P Natl Acad Sci USA. 2011;108:12740–5. doi: 10.1073/pnas.1109987108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tong MH, Mitchell DA, McGowan SD, Evanoff R, Griswold MD. Two miRNA clusters, Mir-17-92 (Mirc1) and Mir-106b-25 (Mirc3), are involved in the regulation of spermatogonial differentiation in mice. Biol Reprod. 2012;86:72. doi: 10.1095/biolreprod.111.096313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huszar JM, Payne CJ. MicroRNA 146 (Mir146) modulates spermatogonial differentiation by retinoic acid in mice. Biol Reprod. 2013;88:15. doi: 10.1095/biolreprod.112.103747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang QE, Racicot KE, Kaucher AV, Oatley MJ, Oatley JM. MicroRNAs 221 and 222 regulate the undifferentiated state in mammalian male germ cells. Development. 2013;140:280–90. doi: 10.1242/dev.087403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moritoki Y, Kojima Y, Mizuno K, Shibata Y, Kamisawa H, Imura M, et al. The Role of Mir-135a Via Regulation of Foxo1 Gene Expression in Maintenance of Spermatogonial Stem Cells. J Urology. 2013;189:E106–E7. [Google Scholar]
- 90.Tong MH, Mitchell D, Evanoff R, Griswold MD. Expression of Mirlet7 family microRNAs in response to retinoic acid-induced spermatogonial differentiation in mice. Biol Reprod. 2011;85:189–97. doi: 10.1095/biolreprod.110.089458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Marcon E, Babak T, Chua G, Hughes T, Moens PB. miRNA and piRNA localization in the male mammalian meiotic nucleus. Chromosome Res. 2008;16:243–60. doi: 10.1007/s10577-007-1190-6. [DOI] [PubMed] [Google Scholar]
- 92.Wu J, Bao J, Wang L, Hu Y, Xu C. MicroRNA-184 downregulates nuclear receptor corepressor 2 in mouse spermatogenesis. BMC Dev Biol. 2011;11:64. doi: 10.1186/1471-213X-11-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lian J, Tian H, Liu L, Zhang XS, Li WQ, Deng YM, et al. Downregulation of microRNA-383 is associated with male infertility and promotes testicular embryonal carcinoma cell proliferation by targeting IRF1. Cell Death Dis. 2010;1:e94. doi: 10.1038/cddis.2010.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tian H, Cao YX, Zhang XS, Liao WP, Yi YH, Lian J, et al. The targeting and functions of miRNA-383 are mediated by FMRP during spermatogenesis. Cell Death Dis. 2013;4:e617. doi: 10.1038/cddis.2013.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hamer G, Roepers-Gajadien HL, van Duyn-Goedhart A, Gademan IS, Kal HB, van Buul PP, et al. DNA double-strand breaks and gamma-H2AX signaling in the testis. Biol Reprod. 2003;68:628–34. doi: 10.1095/biolreprod.102.008672. [DOI] [PubMed] [Google Scholar]
- 96.Novotny GW, Sonne SB, Nielsen JE, Jonstrup SP, Hansen MA, Skakkebaek NE, et al. Translational repression of E2F1 mRNA in carcinoma in situ and normal testis correlates with expression of the miR-17-92 cluster. Cell Death Differ. 2007;14:879–82. doi: 10.1038/sj.cdd.4402090. [DOI] [PubMed] [Google Scholar]
- 97.Liang X, Zhou D, Wei C, Luo H, Liu J, Fu R, et al. MicroRNA-34c enhances murine male germ cell apoptosis through targeting ATF1. Plos One. 2012;7:e33861. doi: 10.1371/journal.pone.0033861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dai L, Tsai-Morris CH, Sato H, Villar J, Kang JH, Zhang J, et al. Testis-specific miRNA-469 up-regulated in gonadotropin-regulated testicular RNA helicase (GRTH/DDX25)-null mice silences transition protein 2 and protamine 2 messages at sites within coding region: implications of its role in germ cell development. The Journal of biological chemistry. 2011;286:44306–18. doi: 10.1074/jbc.M111.282756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vourekas A, Zheng Q, Alexiou P, Maragkakis M, Kirino Y, Gregory BD, et al. Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis. Nat Struct Mol Biol. 2012;19:773–81. doi: 10.1038/nsmb.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Deng W, Lin HF. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell. 2002;2:819–30. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- 101.Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–49. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- 102.Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–14. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 103.Di Giacomo M, Comazzetto S, Saini H, De Fazio S, Carrieri C, Morgan M, et al. Multiple epigenetic mechanisms and the piRNA pathway enforce LINE1 silencing during adult spermatogenesis. Mol Cell. 2013;50:601–8. doi: 10.1016/j.molcel.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 104.Zhao W, Zhou F, Zhou X, Hou Y, He Y, Cheng H, et al. Mago, a vertebrate homolog of Drosophila Mago nashi protein, is a component of the chromatoid body in the cytoplasm of the postmeiotic spermatid. J Exp Zool B Mol Dev Evol. 2010;314:232–41. doi: 10.1002/jez.b.21331. [DOI] [PubMed] [Google Scholar]
- 105.Ginter-Matuszewska B, Kusz K, Spik A, Grzeszkowiak D, Rembiszewska A, Kupryjanczyk J, et al. NANOS1 and PUMILIO2 bind microRNA biogenesis factor GEMIN3, within chromatoid body in human germ cells. Histochem Cell Biol. 2011;136:279–87. doi: 10.1007/s00418-011-0842-y. [DOI] [PubMed] [Google Scholar]
- 106.Xu K, Yang L, Zhao D, Wu Y, Qi H. AKAP3 Synthesis Is Mediated by RNA Binding Proteins and PKA Signaling During Mouse Spermiogenesis. Biol Reprod. 2014 doi: 10.1095/biolreprod.113.116111. [DOI] [PubMed] [Google Scholar]
- 107.Hussain S, Tuorto F, Menon S, Blanco S, Cox C, Flores JV, et al. The mouse cytosine-5 RNA methyltransferase NSun2 is a component of the chromatoid body and required for testis differentiation. Mol Cell Biol. 2013;33:1561–70. doi: 10.1128/MCB.01523-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Meikar O, Da Ros M, Liljenback H, Toppari J, Kotaja N. Accumulation of piRNAs in the chromatoid bodies purified by a novel isolation protocol. Exp Cell Res. 2010;316:1567–75. doi: 10.1016/j.yexcr.2010.02.023. [DOI] [PubMed] [Google Scholar]
- 109.Messina V, Meikar O, Paronetto MP, Calabretta S, Geremia R, Kotaja N, et al. The RNA binding protein SAM68 transiently localizes in the chromatoid body of male germ cells and influences expression of select microRNAs. Plos One. 2012;7:e39729. doi: 10.1371/journal.pone.0039729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Amigo I, Traba J, Satrustegui J, del Arco A. SCaMC-1Like a member of the mitochondrial carrier (MC) family preferentially expressed in testis and localized in mitochondria and chromatoid body. Plos One. 2012;7:e40470. doi: 10.1371/journal.pone.0040470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shoji M, Tanaka T, Hosokawa M, Reuter M, Stark A, Kato Y, et al. The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev Cell. 2009;17:775–87. doi: 10.1016/j.devcel.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 112.Shang P, Baarends WM, Hoogerbrugge J, Ooms MP, van Cappellen WA, de Jong AA, et al. Functional transformation of the chromatoid body in mouse spermatids requires testis-specific serine/threonine kinases. J Cell Sci. 2010;123:331–9. doi: 10.1242/jcs.059949. [DOI] [PubMed] [Google Scholar]