Abstract
Circadian oscillations play a critical role in coordinating the physiology, homeostasis, and behavior of biological systems. Once thought to only be controlled by a master clock, recent high-throughput experiments suggest many genes and metabolites in a cell are potentially capable of circadian oscillations. Each cell can reprogram itself and select a relatively small fraction of this broad repertoire for circadian oscillations, as a result of genetic, environmental, and even diet changes.
Keywords: circadian rhythms, coupled oscillators, clock, oscillatory patterns, circadian transcriptome, circadian metabolome
Role of circadian oscillations: Beyond the “master clock” genes
Circadian rhythms play a key role in ensuring homeostatic balance with the environment and coordinating many aspects of physiology [1, 2], and its disruption has been directly linked to cancer, metabolic disorders, and premature ageing [1, 2, 3]. Research has shown that circadian rhythms are genetically encoded by a molecular clock found in nearly every cell, with a “master clock” located in the supraschiasmatic nucleus (SCN) of the hypothalamus coordinating and interacting with peripheral clocks throughout the body. Species from bacteria to mammals express a 24-hour clock, which is based on a negative transcriptional feedback circuit where core transcriptional activators (Clock and Bmal1) drive the expression of their own negative regulators (Cry1-2, Per1-3) [4, 5]. This attribute of the clock results in a negative transcriptional and translational feedback loop encoded by several genes that perpetuates oscillations in gene expression that occur every 24 hrs. However, recent gene expression experiments reveal that a significant fraction of all transcripts in a cell oscillate in a circadian manner. High-resolution gene expression data from mice liver [6] identified over 3000 transcripts that oscillate with a 24-hour periodicity. In addition, analysis across different tissue types of the same organism reveals little overlap between the molecular species that oscillate in one tissue versus another beyond the core clock genes. Out of 7000 genes studied, 337 transcripts were found oscillating in the SCN compared with 335 in the liver [7]. However, when the oscillating genes in each tissue were compared, only 28 genes were oscillating in both SCN and liver. Likewise, a low amount of genes was identified as circadian in both muscle and liver (57 genes) [8] as well as between heart and liver [9]. Similar results have also been reported with high-throughput metabolomic studies [2, 10]. Furthermore, it has now been established that genetic, environmental, and even diet changes can modify oscillatory patterns by: (i) modifying the phase or amplitude of existing oscillations; (ii) suppressing existing oscillations entirely; and (iii) giving rise to new oscillations that were not observed in the absence of perturbations (Figure 1A). Indeed, comparing gene expression and metabolite levels in liver tissue from high-fat-fed versus normal-chow-fed mice revealed over 2,800 transcripts oscillating in expression across both conditions [2]. Of these, 27.5% (778) did oscillate in both conditions, but often with a change in amplitude and/or phase; 49.5% (1,394) were rhythmic only in the normal-chow-fed condition; and approximately 23% (654) were rhythmic only in the high-fat-fed condition. Similar results were also observed for oscillating metabolites. The results from across tissue and within tissue experiments strongly suggest that a large fraction of all molecular species is capable of oscillating in a circadian manner under some set of conditions.
Figure 1.
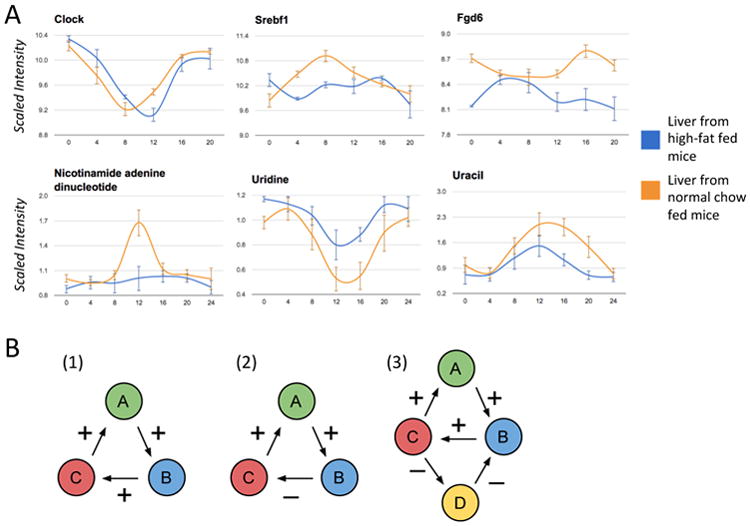
(A) Examples of oscillating transcripts and metabolites from liver comparing high-fat and normal-chow fed mice. Clock, a core circadian regulator, shows a shift in phase. Srebf1, a key transcription factor regulating enzymes involved in lipid synthesis, shows a robust new oscillation only in the high-fat condition. Fgd6 shows a significantly altered oscillatory pattern. Nicotinamide Adenine Dinucleotide (NAD+) oscillates in the normal-chow condition, but not in the high-fat condition. Uracil and Uridine, which metabolically co-react, show a slight change in phase, and a significant dampening in amplitude. (B) 1. A cycle between three molecular species with an even number of negative interactions. Increasing the concentration of A, increases the concentration of B, which increases the concentration of C, which further increases the concentration of A (and vice versa if the concentration of A is decreased). Thus in general such a system does not oscillate and will tend to converge to one of several fixed-point attractors (e.g. all concentrations are high or all concentrations are low). 2. A cycle between three molecular species with an odd number of negative interactions. Increasing the concentration of A, increases the concentration of B, which decreases the concentration of C, which then decreases the concentration of A. Thus such a system will tend to oscillate. 3. Example of two interlocked cycles sharing one positive edge (between B and C) with fixed-point attractors. Changing the sign of the shared interaction creates two oscillatory cycles.
When a complex physical system is significantly perturbed, in general one does not expect to see a large number of new components oscillating at the same frequency, unless these components were already primed and capable of oscillating at this particular frequency, in which case the perturbation simply reveals a preexisting capability. The important question then is why so many genes and metabolites have the potential to oscillate in a circadian manner, and how does the cell select which molecular species should oscillate in a given situation?
Circadian coupled-oscillator view of molecular networks
Molecular species in isolation cannot oscillate, but feedback loops of interacting molecular species involving various kinds of interactions such as regulatory, protein-protein, and enzymatic interactions can oscillate. Since the beginning of life, the Earth has rotated on its axis well over a trillion times (3.5 × 109 × 365 ≈ 1.3 × 1012)*, inducing a relentless periodic day-night cycle that has deeply impregnated living systems at all levels, from molecular to cellular, to organismal, and beyond. In particular, this relentless cycle has had time to sculpt a large fraction of the feedback loops found in molecular networks and prime them for circadian rhythmicity, which may be the main reason why oscillations are so widespread and their reprogramming is required to maintain homeostasis. By creating compensating oscillations in important transcription factors, enzymes, and metabolites, biological systems can continue to perform important functions and elicit appropriate responses to perturbations. For example, high-fat-fed mice show a robust new oscillation in Srebf1 (Figure 1), a key transcription factor regulating enzymes involved in lipid synthesis.
From a mechanistic point of view, several possible non-exclusive mechanisms may explain the widespread occurrence of oscillations and the ability of cells to reprogram their oscillatory repertoire rapidly. Thus in the coupled-oscillator view of molecular networks, oscillations are not associated with individual molecular species but with directed loops of interacting species. Under fairly general assumptions, it is known from the theory of dynamical systems [11] that activity in loops with an even number of inhibitory interactions tend to converge to one of several possible stable points, whereas activity in loops with an odd number of inhibitory interactions tend to oscillate (Figure 1B). Under this framework, new oscillations could be generated simply by the modification, creation, or suppression of interaction edges. For instance, if the sign of an interaction is changed by a perturbation, a corresponding cycle could flip from stable to oscillatory behavior. Likewise, dynamic changes in the epigenome, like methylation, acetylation, and chromatin remodeling can play a central role in the creation and suppression of interacting edges, thus selecting which loops and corresponding species oscillate. Indeed, recent studies have identified circadian long-range interactions [12] and the role of the Clock gene as a histone acetyltransferase [3]. All these mechanisms could assemble cascades among coupled circadian oscillators by which changes in the phases and amplitudes of key oscillators may possibly propagate to neighboring ones.
In addition, by acting on densely connected molecular hubs, the cell can rapidly and massively reprogram which species oscillate, such as the metabolite NAD+, which plays a central role in regulating circadian rhythms [13]. More importantly, perhaps, the core molecular clock genes (Clock and Bmal1) heterodimerize to bind to a single or pair of E-box sites. E-box sites are short (the canonical sequence is CACGTG), frequent in the genome, and found in promoter regions of many genes, including transcription factors. Indeed, using time-resolved ChIP-seq data, 2,049 Bmal1 binding sites were identified in mouse liver [14]. Among these, approximately 60% showed rhythmic binding and 13% of all Bmal1 sites had a pair of E1-E2 elements with spacers of 6-7 base pairs. In a given environment, again through epigenetic and chromatin reorganization, cells can reveal or hide a fraction of E-box sites thereby controlling which loops are directly or indirectly, affected by Clock and Bmal1.
Although there are not enough data currently to model these mechanisms on a large-scale, progress has been made in producing comprehensive network maps of which molecular species interact and which oscillate under particular sets of conditions [10, 15]. These maps might provide the basis for developing coarse models that could be refined in time as more data becomes available.
Concluding remarks
Since the world is drastically different during the night versus during the day, over a trillion night-and-day cycles during the course of evolution have deeply sculpted the molecular networks of the cell and made 24h oscillations pervasive. High-throughput transcriptomic and metabolomics experiments across tissue types and conditions are starting to reveal that a large fraction of the molecular network of a cell is primed for and potentially capable of oscillating in a circadian manner. Only a relatively small fraction of this broad repertoire actually oscillates characteristically in a given cell, as a result of many factors, such as epigenetic modifications and environmental conditions broadly construed. Thus from a circadian position, the cell could be viewed as an intricate network of coupled-oscillators with complex couplings. Circadian oscillations are quite different from the oscillations of a simple physical system, such as a spring, because they are non-rigid and highly plastic with respect to perturbations, allowing for instance an organism to change its diet or its time of sleep. Perturbations not only modify existing oscillations, but can also induce new oscillations through these couplings, revealing the underlying oscillatory fabric that enables flexible cellular reprogramming. Thus the complement of transcripts and other species that oscillate in a circadian manner provides a characteristic signature describing a cell and its physiological condition. Ongoing and future work should provide the data to better model and understand circadian oscillator networks, predict how they respond to perturbations, and use these responses to explain biology and direct therapeutic intervention.
Acknowledgments
We thank Nicholas Ceglia, Yuzo Kanomata, Yu Liu, and Paul Rigor for helping develop and maintain the CircadiOmics system and web site. The work of VP and PB has been supported by the following grants: NSF IIS-1321053, NIH LM010235, and NIHNLM T15 LM07443 to PB.
Footnotes
Footnote: The actual number is slightly larger since the period of rotation of the Earth has been increasing over evolutionary times primarily as a result of the attraction by the moon and the corresponding tidal acceleration.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Takahashi JS, et al. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9(10):764–75. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eckel-Mahan KL, et al. Reprogramming of the Circadian Clock by Nutritional Challenge. Cell. 2013;155(7):1464–1478. doi: 10.1016/j.cell.2013.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Froy O. Circadian rhythms, aging, and life span in mammals. Physiology (Bethesda) 2011;26(4):225–35. doi: 10.1152/physiol.00012.2011. [DOI] [PubMed] [Google Scholar]
- 4.Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–49. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 5.Yan J, et al. Analysis of gene regulatory networks in the mammalian circadian rhythm. PLoS Comput Biol. 2008;4(10):e1000193. doi: 10.1371/journal.pcbi.1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hughes ME, et al. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009;5(4):e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109(3):307–20. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 8.Miller BH, et al. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci U S A. 2007;104(9):3342–7. doi: 10.1073/pnas.0611724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Storch KF, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 10.Eckel-Mahan KL, et al. Coordination of the transcriptome and metabolome by the circadian clock. Proc Natl Acad Sci U S A. 2012;109(14):5541–6. doi: 10.1073/pnas.1118726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baldi P, Atiya A. Oscillations and Synchronizations in Neural Networks: an Exploration of the Labeling Hypothesis. International Journal of Neural Systems. 1989;1(2):103–124. [Google Scholar]
- 12.Aguilar-Arnal L, et al. Cycles in spatial and temporal chromosomal organization driven by the circadian clock. Nature Structural & Molecular Biology. 2013;20:1206–1213. doi: 10.1038/nsmb.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakahata Y, et al. Circadian control of the NAD+ salvage pathway by CLOCK- SIRT1. Science. 2009;324(5927):654–7. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rey G, et al. Genome-Wide and Phase-Specific DNA-Binding Rhythms of BMAL1 Control Circadian Output Functions in Mouse Liver. PLoS Biol. 2011;9(2):e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patel VR, Eckel-Mahan KL, Sassone-Corsi P, Baldi P. Integrating Circadian Genomics, Transcriptomics, Proteomics, and Metabolomics. Nature Methods. 2012;9(8):772–773. doi: 10.1038/nmeth.2111. [DOI] [PubMed] [Google Scholar]


