Abstract
Purpose of review
The interplay between circadian rhythm and cancer has been suggested for more than a decade based on the observations that shift work and cancer incidence are linked. Accumulating evidence implicates the circadian clock in cancer survival and proliferation pathways. At the molecular level, multiple control mechanisms have been proposed to link circadian transcription and cell-cycle control to tumorigenesis.
Recent findings
The circadian gating of the cell cycle and subsequent control of cell proliferation is an area of active investigation. Moreover, the circadian clock is a transcriptional system that is intricately regulated at the epigenetic level. Interestingly, the epigenetic landscape at the level of histone modifications, DNA methylation, and small regulatory RNAs are differentially controlled in cancer cells. This concept raises the possibility that epigenetic control is a common thread linking the clock with cancer, though little scientific evidence is known to date.
Summary
This review focuses on the link between circadian clock and cancer, and speculates on the possible connections at the epigenetic level that could further link the circadian clock to tumor initiation or progression.
Keywords: cancer, cell cycle, circadian clock, epigenome, metabolism
INTRODUCTION
The circadian clock is an endogenous, self-sustaining pacemaker that operates with a periodicity of 24 h, in order to maintain proper rhythms in sleep–wake cycles, behavior, metabolism, hormone secretion, and cell cycle [1,2]. Disruption in proper circadian timekeeping results in detrimental effects on mammalian physiology, and a number of clues from the clinic and laboratory suggest that these disturbances result in uncontrolled cell growth and cancer [3,4]. In humans, circadian disruption found in shift workers puts them at increased risk for breast cancer [5]. Also, mice with an ablation of the central clock located within the suprachiasmatic nucleus (SCN) exhibit increased growth of tumor xenografts compared with mice with an intact circadian pacemaker [6]. Overall, a link exists between cancer and disruption of circadian rhythms, although its extent and molecular mechanisms are not fully elucidated.
At the heart of the circadian machinery are the core DNA-binding transcription factors, CLOCK and BMAL1, that drive the oscillation of approximately 10% of transcripts in the genome in a defined tissue-specific program [7,8]. CLOCK:BMAL1-dependent transcription of core clock and clock-controlled genes (CCGs) peaks during the day, whereas transcription is inhibited by the circadian repressors, Period (PER) and Cryptochrome (CRY), at night [9,10]. In addition to the core transcriptional–translational feedback loop, regulation of circadian transcription is also subject to vast modifications in the epigenetic state that change dynamically over the day–night cycle [9,11].
CIRCADIAN RHYTHMS AND CANCER
A number of genetic mouse models and human clinical studies reveal that the core clock transcriptional machinery is genetically altered in numerous cancer models. The human Clock gene is expressed in colorectal cancer, and its expression level is correlated with hypoxia-inducible factor 1-alpha (HIF-1α), aryl hydrocarbon receptor nuclear translocator, and vascular endothelial growth factor, molecules implicated in hypoxia and tumor angiogenesis [12]. CLOCK expression has also been reported to be upregulated in high-grade human glioma tissue, whereas the glioma suppressor miR-124 has been shown to directly target CLOCK expression and subsequently modulate glioma proliferation [13]. CLOCK has also been involved in breast cancer cell proliferation [14] and CLOCK is reported to interact with estrogen receptor alpha (ERα), whereas estrogen appears to enhance both CLOCK and ERα driven transcription via CLOCK-dependent sumoylation [15]. In addition to CLOCK, BMAL1 has also been reported to be involved in cancer. Bmal1−/− mice display increased salivary gland hyperplasia, and when irradiated, a fraction of these mice exhibit lymphomas in the chest cavity and salivary glands [16].
The PER repressors of the circadian clock have also been linked to cancer. The Per2m/m mice are highly sensitive to gamma-irradiation and display increased rates of salivary gland hyperplasia [17]. Recent studies also demonstrate that low levels of Per1 and Per2 gene expression are associated with poor prognosis in gastric cancer [18]. Per1 mRNA is degraded by IREα, and inhibition of IREα signaling reduces tumorigenesis [19]. Per2 has been implicated in preventing tumor invasion and epithelial–mesenchymal transition (EMT) gene expression profiles [20]. Thus, the core clock machinery has been overall implicated in the regulation of cancer cell growth and survival in mouse models. In addition, clinical data also support a role for clock proteins in cancer.
CIRCADIAN CLOCK AND CELL CYCLE
The circadian clock has been previously reported to regulate or ‘gate’ the cell cycle at the G1/S [21–23] and G2/M [24,25] checkpoints, and recent evidence confirms that the clock and cell cycle exhibit phase-locking characteristics and are coordinately synchronized [26]. The expression of a number of cell-cycle regulators is also under the control of the clock: Wee1, c-Myc, p20, p21, and Cyclin D1 all exhibit circadian gene expression [27,28]. Specifically, critical circadian regulators that link the clock to cell-cycle control are the PER1 and PER2 repressors. The expression of a number of cell-cycle genes is abolished in PER2-mutant mice, and PER1 and PER2 interact with checkpoint kinases CHK1/2 [29] and NONO/p16-Ink4A to control cell-cycle progression [30]. In addition, the importance of circadian clock-dependent gating of the cell cycle was recently reported in intestinal stem cells and neural progenitor cells, suggesting circadian control of the cell cycle has far-reaching implications [31,32].
One of the most important features in cancer cells is deregulation of the cell cycle, which can also be linked to the clock. Recent reports revealed that about 30% of circadian transcripts in colon mucosa regulate the cell cycle and specifically mitotic control, and the expression levels of these genes were found to be critical in the response of colon epithelial cells to anticancer agents such as cyclin-dependent kinase (CDK) inhibitors [33]. Also, it is well known that mutations in oncogenes and tumor suppressor genes are critical for tumor development. Two notable examples of cell-cycle regulators that are linked to the circadian clock are the tumor suppressor p53 and c-Myc oncogene. p53 controls multiple checkpoints of the cell cycle [34] and p53 also regulates Per2 expression by blocking CLOCK:-BMAL1-dependent recruitment to the Per2 promoter [35▪▪]. Strikingly, p53-null mice exhibit a shorter circadian period and impaired photo-entrainment [35▪▪]. c-Myc is a known regulator of cell-cycle progression [36,37] and similar to CLOCK:BMAL1, MYC binds E-box sequences to drive its transcriptional program. What has not been determined is the possible interplay between these two transcription networks, and whether, in the case of uncontrolled cell growth, MYC could interfere in the expression of known CLOCK:BMAL1 dependent genes. As an example of this, a recent computational approach linked gene signatures of the circadian clock to oncogenic Ras signaling. Using an inducible system, Ras expression was able to alter the circadian period length, suggesting potential common control mechanisms [38] (Fig. 1). Overall, these examples suggest the possibility that circadian transcriptional networks could overlap with oncogenic signaling pathways, possibly at the level of cell cycle and proliferative control. It could be of critical importance to determine the changes of CLOCK:BMAL1 genome-wide re-distribution in response to loss of p53 expression or activation of oncogenic Myc and Ras signaling in cancer cells.
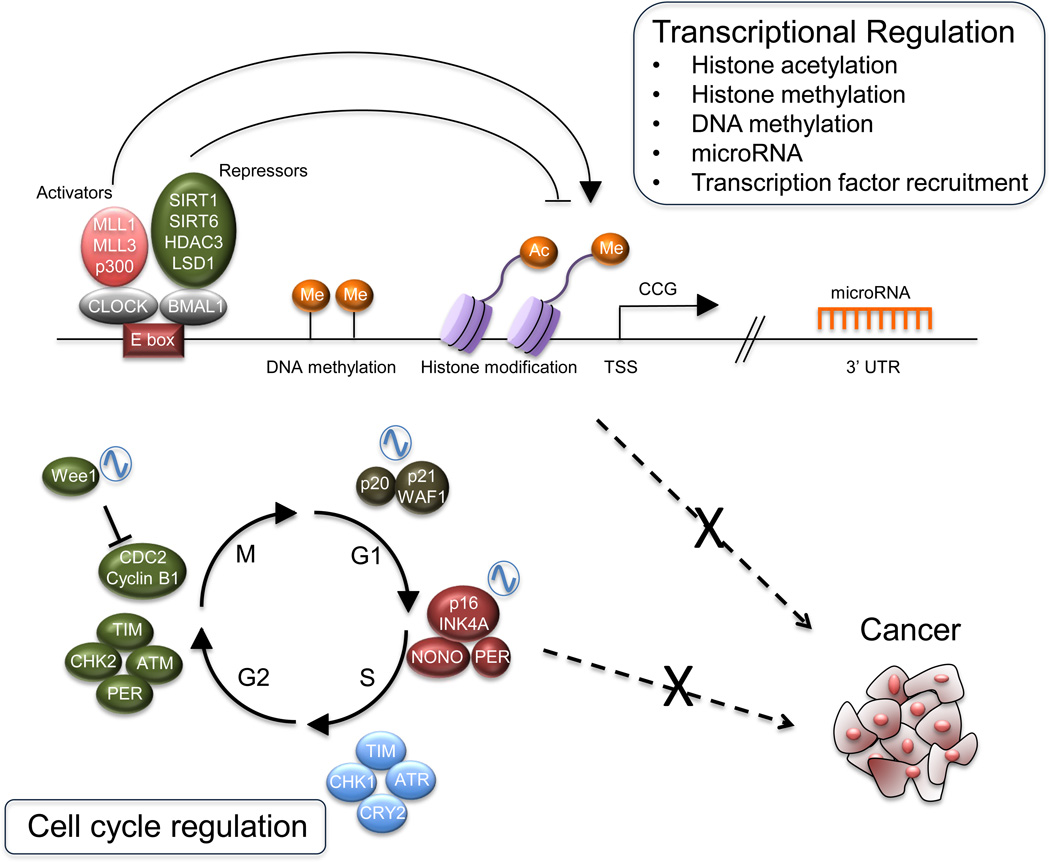
FIGURE 1.
Circadian epigenome and cell-cycle regulation in cancer. Schematic representation of the core clock machinery is shown. The activating heterodimer CLOCK:BMAL1 binds to the E-box elements on the genome, controlling a large number of genes. CLOCK:BMAL1 action is counteracted by the PER and CRY repressor proteins. Additional regulators and chromatin remodelers contribute to circadian gene expression. Among the genes controlled by the clock, a number of them are key cell-cycle regulators. The molecular clock has also been shown to interplay with oncogenes and tumor suppressor genes. CRY, Cryptochrome; MLL, mixed lineage leukemia; PER, Period.
CIRCADIAN EPIGENOME AND CANCER
The vast genomic reprogramming that occurs in tumors has prompted in-depth investigations on the epigenetic changes and chromatin transitions occurring in cancer cells. The emerging view is that key chromatin remodelers involved in histone modifications play a central role in the development and establishment of a cancerous state for a given cell or tissue, directing a specialized set of histone modifications [39]. The remarkable role played by chromatin remodeling in circadian control begs the question on whether the epigenetic reprogramming that occurs during tumorigenesis could involve the clock system.
Circadian transcription is coupled with rhythmic chromatin modifications that regulate oscillations in gene expression (Fig. 1). The promoter regions of CCGs exhibit rhythmic histone acetylation at H3K9 and H3K14 [9,11,40], which has been attributed to the histone acetyltransferase (HAT) p300 [41] and the intrinsic HAT activity of CLOCK [40,42]. Conversely, circadian acetylation of histone and nonhistone proteins is counterbalanced by the NAD+-dependent deacetylases sirtuin 1 (SIRT1) [43,44] and SIRT6 [45]. Moreover, histone methylation is known to be rhythmic at H3K4, H3K9, H3K27 and H3K36, and these marks are mediated by a number of histone methyltransferases (HMTs) and demethylases that will be discussed below [9,46–48].
A number of histone-modifying enzymes that regulate the circadian clock have also been implicated in cancer. The mammalian sirtuins, SIRT1 and SIRT6, have been shown to participate in circadian control [44,45], but are also involved in aging and cancer. The role of SIRT1 in cancer is currently unclear as SIRT1 targets a number of acetylated proteins including Myc, p53, HIF, TGF-β, and Wnt signaling, apparently acting both as tumor suppressor and tumor promoter depending on the biological system studied [49–51]. Striking evidence illustrates that SIRT1 expression is elevated in leukemia stem cells, and a cross-talk exists between SIRT1 and MYC oncogenic signaling that is responsible for driving FLT3 receptor tyrosine kinase resistance in acute myeloid leukemia (AML) [52▪]. SIRT6 was recently described as a tumor suppressor and a potent regulator of aerobic glycolysis in cancer cells, which is a key mechanism for energy production upon which cancer cells are reliant for growth [53]. The role of the sirtuins in cancer could be regulated at the level of the circadian clock, and further work is needed to validate this idea experimentally. In addition to the sirtuins, liver-specific deletion of histone deacetylase 3 (HDAC3) results in hepatocellular carcinoma (HCC) [54] and the HDAC3–nuclear receptor corepressor 1 (NcoR1)–Rev-Erb axis has been implicated in the circadian control of gene expression [55,56].
Mixed lineage leukemia 1 (MLL1) operates as a critical regulator of rhythmic H3K4 tri-methylation and recruitment of CLOCK:BMAL1 to CCG promoters [46], and recent reports show that also MLL3 contributes to histone methylation and circadian gene expression in the liver [57]. The MLL protein family has been long implicated in leukemia through multiple mechanisms pertaining to loss of MLL expression, mutation, amplification, or translocation events, whereby oncogenic fusion proteins are formed between MLL and multiple partners [58]. The enzymatic HMT activity of MLL is critically involved in the epigenetic regulation in cancer and recent work has identified a small-molecule inhibitor for MLL1 that inhibits its interaction with WD repeat-containing protein 5 (WDR5) to repress H3K4 methylation, gene expression, cell-cycle arrest, and apoptosis in leukemia cells [59▪]. Furthermore, MLL3 is frequently mutated in multiple human cancers and was recently identified as a tumor suppressor located at a commonly deleted chromosomal locus in AML. MLL3-dependent changes in gene signatures corresponding to cellular differentiation and immune response suggest that H3K4 methylation profiles are also altered in these AML cancers [60]. As the HMT activity of MLL proteins controls rhythmic histone methylation, it could be envisioned that this circadian regulation of histone methylation is altered or abolished in cancer cells, potentially linking the circadian clock to tumorigenesis at the epigenetic level. In addition to the HMT activity of the MLL family of proteins, MLL is targeted by microRNAs [61–63], which could also be a common control mechanism with the circadian clock, as microRNAs are known to be involved in the circadian output and gene expression [64–66].
The histone demethylase LSD1 targets H3K4 and H3K9 methylation, and was recently reported to be a key component of the circadian machinery and regulator of CCG expression [67]. Interestingly, LSD1 functions as a histone demethylase in Sox2-expressing lung squamous cell carcinomas [68] and also controls proliferation and metastasis of colon cancer cells [69]. Another H3K4 demethylase, Jarid1a, demethylates H3K4 and has been implicated in circadian control by interacting directly with CLOCK:BMAL1 and regulating circadian gene expression [70]. Some clues suggest that Jarid1a could be involved in the tumor suppressor network of Retinoblastoma (Rb) [71], though how this is linked to the circadian clock remains unclear.
Finally, DNA methylation could also play an important role in linking the clock to cancer. Striking evidence for dynamic DNA methylation in the central clock region of the SCN identified light-induced changes in DNA methylation at specific promoters that correspond to circadian gene expression [72▪▪]. Yet, what is currently unclear is the extent to which changes in DNA methylation are rhythmic, considering CpG methylation is believed to exert long-lasting suppressive effects on gene expression. Global alterations in DNA methylation have been reported in cancer and not surprisingly, the Per and Cry promoters have been reported to be methylated in specific cancers [73,74]. Also, DNA methyltransferases and histone-modifying enzymes, such as HDACs, HMTs, and histone demethylases, are known to cross-talk and regulate the recruitment of one another, suggesting major complexity in this epigenetic control axis [75].
CIRCADIAN CONTROL OF TUMOR METABOLISM
Cancer cells have adapted alternative methods of energy production, such that the rates of aerobic glycolysis are dramatically elevated to maintain the energetic needs of the tumor, termed the ‘Warburg effect’ [76]. Additionally, a revised view of cancer cell metabolic reprogramming beyond glycolysis also includes enhanced glutamine metabolism for lipogenesis, which is needed for membrane synthesis and proliferation, fatty acid beta-oxidation for energy production, and methionine dependency which is needed for methylation reactions and polyamine synthesis [77,78▪]. Not surprisingly, the circadian clock is heavily implicated in metabolic control and a number of metabolites, like glucose and fatty acids, display diurnal rhythms [79–81]. It is conceivable that as cancer cells are highly energy-consuming, a possible loss or deregulation of circadian metabolic control can lead to exacerbated cell proliferation in cancer cells. The details of this remain fully unexplored, but factors such as p53, mechanistic target of rapamycin (mTOR), Ras, and c-Myc, which are implicated in clock control, are known to also regulate metabolic processes in cancer cells.
CIRCADIAN RHYTHMS AND CANCER TREATMENT
Targeting the circadian machinery may be valuable in the treatment or intervention of human cancers. The administration of chemotherapeutic agents at optimal times (chronotherapy) has been considered to maximize the antitumor effects and minimize the toxic influence on other organs. For instance, evaluation of Bmal1 and Rev-erbα transcription profiles could help minimize the toxicity of chemotherapy [82]. Also, Bmal1 overexpression reduces cancer cell proliferation and improves the sensitivity to oxaliplatin in colorectal cancer [83]. Furthermore, mTOR protein expression is rhythmic, and administration of mTOR inhibitors in renal cell carcinoma during the peak expression phase improves survival [84]. At a time when personalized medicine is vastly valued, this set of findings suggests that the timing of chemotherapy should be adjusted to each patient, depending on the cyclic expression of clock genes and sensitivity of drug target – thereby following the internal circadian rhythm of the patient.
CONCLUSION
The elevated incidence of cancer in modern society parallels the epidemiology data, showing elevated cancer risk in shift workers. Regarding this point, the detrimental effects of light pollution on the circadian clock are a growing concern. A number of clinical and laboratory results suggest that genetic and epigenetic changes in cell-cycle control and cell proliferation link the circadian pacemaker to cancer. In addition to the accumulating evidence discussed in this review, an area that remains completely unexplored is the connection between tumor metabolism and the circadian clock. The link between the circadian clock, cancer, epigenetics, and possibly metabolism opens up multiple avenues that require further exploration and could result in potential therapeutic intervention.
KEY POINTS.
Mouse models as well as human clinical studies have suggested that disruption of circadian rhythms facilitates tumorigenesis.
The circadian clock has been shown to ‘gate’ the cell cycle: the clock and cell cycle display phase-locking characteristics and are coordinately synchronized.
Circadian transcription is coupled to rhythmic changes of histone modifications and a number of histone-modifying enzymes are also implicated in cancer.
Acknowledgements
None.
Financial support and sponsorship
Work in the Sassone-Corsi laboratory is supported by the National Institute of Health (NIH), INSERM (Institut National de la Sante et la Recherche Medicale, France), KAUST (King Abdullah University of Science and Technology, Saudi Arabia), and Merieux Pharmaceuticals (France). S.M. is supported by the UC Irvine Chao Family Cancer Center and K.K. is supported by a JSPS postdoctoral fellowship.
Footnotes
Conflicts of interest
The authors declare no conflicts of interest.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
▪ of special interest
▪▪ of outstanding interest
- 1.Feng D, Lazar MA. Clocks, metabolism, and the epigenome. Mol Cell. 2012;47:158–167. doi: 10.1016/j.molcel.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gamble KL, Berry R, Frank SJ, et al. Circadian clock control of endocrine factors. Nat Rev Endocrinol. 2014;10:466–475. doi: 10.1038/nrendo.2014.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fu L, Lee CC. The circadian clock: pacemaker and tumour suppressor. Nat Rev Cancer. 2003;3:350–361. doi: 10.1038/nrc1072. [DOI] [PubMed] [Google Scholar]
- 4.Sahar S, Sassone-Corsi P. Metabolism and cancer: the circadian clock connection. Nat Rev Cancer. 2009;9:886–896. doi: 10.1038/nrc2747. [DOI] [PubMed] [Google Scholar]
- 5.Schernhammer ES, Laden F, Speizer FE, et al. Rotating night shifts and risk of breast cancer in women participating in the Nurses’ Health Study. J Natl Cancer Inst. 2001;93:1563–1568. doi: 10.1093/jnci/93.20.1563. [DOI] [PubMed] [Google Scholar]
- 6.Filipski E, King VM, Li X, et al. Host circadian clock as a control point in tumor progression. J Natl Cancer Inst. 2002;94:690–697. doi: 10.1093/jnci/94.9.690. [DOI] [PubMed] [Google Scholar]
- 7.Masri S, Sassone-Corsi P. Plasticity and specificity of the circadian epigenome. Nat Neurosci. 2010;13:1324–1329. doi: 10.1038/nn.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Partch CL, Green CB, Takahashi JS. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 2014;24:90–99. doi: 10.1016/j.tcb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koike N, Yoo SH, Huang HC, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rey G, Cesbron F, Rougemont J, et al. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011;9:e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ripperger JA, Schibler U. Rhythmic CLOCK–BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet. 2006;38:369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- 12.Wang L, Chen B, Wang Y, et al. hClock gene expression in human colorectal carcinoma. Mol Med Rep. 2013;8:1017–1022. doi: 10.3892/mmr.2013.1643. [DOI] [PubMed] [Google Scholar]
- 13.Li A, Lin X, Tan X, et al. Circadian gene Clock contributes to cell proliferation and migration of glioma and is directly regulated by tumor-suppressive miR-124. FEBS Lett. 2013;587:2455–2460. doi: 10.1016/j.febslet.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 14.Xiao L, Chang AK, Zang MX, et al. Induction of the CLOCK gene by E2-ERalpha signaling promotes the proliferation of breast cancer cells. PLoS One. 2014;9:e95878. doi: 10.1371/journal.pone.0095878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li S, Wang M, Ao X, et al. CLOCK is a substrate of SUMO and sumoylation of CLOCK upregulates the transcriptional activity of estrogen receptor-alpha. Oncogene. 2013;32:4883–4891. doi: 10.1038/onc.2012.518. [DOI] [PubMed] [Google Scholar]
- 16.Lee S, Donehower LA, Herron AJ, et al. Disrupting circadian homeostasis of sympathetic signaling promotes tumor development inmice. PLoS One. 2010;5:e10995. doi: 10.1371/journal.pone.0010995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu L, Pelicano H, Liu J, et al. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 18.Zhao H, Zeng ZL, Yang J, et al. Prognostic relevance of Period1 (Per1) and Period2 (Per2) expression in human gastric cancer. Int J Clin Exp Pathol. 2014;7:619–630. [PMC free article] [PubMed] [Google Scholar]
- 19.Pluquet O, Dejeans N, Bouchecareilh M, et al. Posttranscriptional regulation of PER1 underlies the oncogenic function of IREalpha. Cancer Res. 2013;73:4732–4743. doi: 10.1158/0008-5472.CAN-12-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwang-Verslues WW, Chang PH, Jeng YM, et al. Loss of corepressor PER2 under hypoxia up-regulates OCT1-mediated EMT gene expression and enhances tumor malignancy. Proc Natl Acad Sci USA. 2013;110:12331–12336. doi: 10.1073/pnas.1222684110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dekens MP, Santoriello C, Vallone D, et al. Light regulates the cell cycle in zebrafish. Curr Biol. 2003;13:2051–2057. doi: 10.1016/j.cub.2003.10.022. [DOI] [PubMed] [Google Scholar]
- 22.Grechez-Cassiau A, Rayet B, Guillaumond F, et al. The circadian clock component BMAL1 is a critical regulator of p21WAF1/CIP1 expression and hepatocyte proliferation. J Biol Chem. 2008;283:4535–4542. doi: 10.1074/jbc.M705576200. [DOI] [PubMed] [Google Scholar]
- 23.Laranjeiro R, Tamai TK, Peyric E, et al. Cyclin-dependent kinase inhibitor p20 controls circadian cell-cycle timing. Proc Natl Acad Sci USA. 2013;110:6835–6840. doi: 10.1073/pnas.1217912110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuo T, Yamaguchi S, Mitsui S, et al. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 25.Plikus MV, Vollmers C, de la Cruz D, et al. Local circadian clock gates cell cycle progression of transient amplifying cells during regenerative hair cycling. Proc Natl Acad Sci USA. 2013;110:E2106–E2115. doi: 10.1073/pnas.1215935110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feillet C, Krusche P, Tamanini F, et al. Phase locking and multiple oscillating attractors for the coupled mammalian clock and cell cycle. Proc Natl Acad Sci USA. 2014;111:9828–9833. doi: 10.1073/pnas.1320474111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelleher FC, Rao A, Maguire A. Circadian molecular clocks and cancer. Cancer Lett. 2014;342:9–18. doi: 10.1016/j.canlet.2013.09.040. [DOI] [PubMed] [Google Scholar]
- 28.Masri S, Cervantes M, Sassone-Corsi P. The circadian clock and cell cycle: interconnected biological circuits. Curr Opin Cell Biol. 2013;25:730–734. doi: 10.1016/j.ceb.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunt T, Sassone-Corsi P. Riding tandem: circadian clocks and the cell cycle. Cell. 2007;129:461–464. doi: 10.1016/j.cell.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 30.Kowalska E, Ripperger JA, Hoegger DC, et al. NONO couples the circadian clock to the cell cycle. Proc Natl Acad Sci USA. 2013;110:1592–1599. doi: 10.1073/pnas.1213317110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bouchard-Cannon P, Mendoza-Viveros L, Yuen A, et al. The circadian molecular clock regulates adult hippocampal neurogenesis by controlling the timing of cell-cycle entry and exit. Cell Rep. 2013;5:961–973. doi: 10.1016/j.celrep.2013.10.037. [DOI] [PubMed] [Google Scholar]
- 32.Karpowicz P, Zhang Y, Hogenesch JB, et al. The circadian clock gates the intestinal stem cell regenerative state. Cell Rep. 2013;3:996–1004. doi: 10.1016/j.celrep.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siffroi-Fernandez S, Dulong S, Li XM, et al. Functional genomics identify Birc5/survivin as a candidate gene involved in the chronotoxicity of cyclin-dependent kinase inhibitors. Cell Cycle. 2014;13:984–991. doi: 10.4161/cc.27868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Agarwal ML, Agarwal A, Taylor WR, et al. p53 controls both the G2/M and the G1 cell cycle checkpoints and mediates reversible growth arrest in human fibroblasts. Proc Natl Acad Sci USA. 1995;92:8493–8497. doi: 10.1073/pnas.92.18.8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Miki T, Matsumoto T, Zhao Z, et al. p53 regulates Period2 expression and the circadian clock. Nat Commun. 2013;4:2444. doi: 10.1038/ncomms3444. This study illustrates a direct control mechanism for p53 on the circadian machinery, suggesting bidirectional control of the clock and cell cycle.
- 36.Bouchard C, Thieke K, Maier A, et al. Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27. EMBO J. 1999;18:5321–5333. doi: 10.1093/emboj/18.19.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perez-Roger I, Kim SH, Griffiths B, et al. Cyclins D1 and D2 mediate mycinduced proliferation via sequestration of p27(Kip1) and p21(Cip1) EMBO J. 1999;18:5310–5320. doi: 10.1093/emboj/18.19.5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Relogio A, Thomas P, Medina-Perez P, et al. Ras-mediated deregulation of the circadian clock in cancer. PLoS Genet. 2014;10:e1004338. doi: 10.1371/journal.pgen.1004338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chi P, Allis CD, Wang GG. Covalent histone modifications – miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 41.Etchegaray JP, Lee C, Wade PA, et al. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421:177–182. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- 42.Hirayama J, Sahar S, Grimaldi B, et al. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature. 2007;450:1086–1090. doi: 10.1038/nature06394. [DOI] [PubMed] [Google Scholar]
- 43.Asher G, Gatfield D, Stratmann M, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 44.Nakahata Y, Kaluzova M, Grimaldi B, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Masri S, Rigor P, Cervantes M, et al. Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism. Cell. 2014;158:659–672. doi: 10.1016/j.cell.2014.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Katada S, Sassone-Corsi P. The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nat Struct Mol Biol. 2010;17:1414–1421. doi: 10.1038/nsmb.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Etchegaray JP, Yang X, DeBruyne JP, et al. The polycomb group protein EZH2 is required for mammalian circadian clock function. J Biol Chem. 2006;281:21209–21215. doi: 10.1074/jbc.M603722200. [DOI] [PubMed] [Google Scholar]
- 48.Duong HA, Weitz CJ. Temporal orchestration of repressive chromatin modifiers by circadian clock Period complexes. Nat Struct Mol Biol. 2014;21:126–132. doi: 10.1038/nsmb.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roth M, Chen WY. Sorting out functions of sirtuins in cancer. Oncogene. 2014;33:1609–1620. doi: 10.1038/onc.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Menssen A, Hydbring P, Kapelle K, et al. The c-MYC oncoprotein, the NAMPT enzyme, the SIRT1-inhibitor DBC1, and the SIRT1 deacetylase form a positive feedback loop. Proc Natl Acad Sci USA. 2012;109:E187–E196. doi: 10.1073/pnas.1105304109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin Z, Fang D. The roles of SIRT1 in cancer. Genes Cancer. 2013;4:97–104. doi: 10.1177/1947601912475079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li L, Osdal T, Ho Y, et al. SIRT1 activation by a c-MYC oncogenic network promotes the maintenance and drug resistance of human FLT3-ITD acute myeloid leukemia stem cells. Cell Stem Cell. 2014;15:431–446. doi: 10.1016/j.stem.2014.08.001. A critical role for SIRT1 and Myc signaling in regulating leukemia stem cells is shown in this study.
- 53.Sebastian C, Zwaans BM, Silberman DM, et al. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell. 2012;151:1185–1199. doi: 10.1016/j.cell.2012.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bhaskara S, Knutson SK, Jiang G, et al. Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell. 2010;18:436–447. doi: 10.1016/j.ccr.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alenghat T, Meyers K, Mullican SE, et al. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature. 2008;456:997–1000. doi: 10.1038/nature07541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feng D, Liu T, Sun Z, et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Valekunja UK, Edgar RS, Oklejewicz M, et al. Histone methyltransferase MLL3 contributes to genome-scale circadian transcription. Proc Natl Acad Sci USA. 2013;110:1554–1559. doi: 10.1073/pnas.1214168110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 59. Cao F, Townsend EC, Karatas H, et al. Targeting MLL1 H3K4 methyltransferase activity in mixed-lineage leukemia. Mol Cell. 2014;53:247–261. doi: 10.1016/j.molcel.2013.12.001. This study illustrates the effective therapeutic strategy for targeting the MLL-dependent global epigenetic landscape in leukemia.
- 60.Chen C, Liu Y, Rappaport AR, et al. MLL3 is a haploinsufficient 7q tumor suppressor in acute myeloid leukemia. Cancer Cell. 2014;25:652–665. doi: 10.1016/j.ccr.2014.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kotani A, Ha D, Hsieh J, et al. miR-128b is a potent glucocorticoid sensitizer in MLL-AF4 acute lymphocytic leukemia cells and exerts cooperative effects with miR-221. Blood. 2009;114:4169–4178. doi: 10.1182/blood-2008-12-191619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jiang X, Huang H, Li Z, et al. MiR-495 is a tumor-suppressor microRNA downregulated in MLL-rearranged leukemia. Proc Natl Acad Sci USA. 2012;109:19397–19402. doi: 10.1073/pnas.1217519109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen P, Price C, Li Z, et al. miR-9 is an essential oncogenic microRNA specifically overexpressed in mixed lineage leukemia-rearranged leukemia. Proc Natl Acad Sci USA. 2013;110:11511–11516. doi: 10.1073/pnas.1310144110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luo W, Sehgal A. Regulation of circadian behavioral output via a MicroRNAJAK/ STAT circuit. Cell. 2012;148:765–779. doi: 10.1016/j.cell.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gatfield D, Le Martelot G, Vejnar CE, et al. Integration of microRNA miR-122 in hepatic circadian gene expression. Genes Dev. 2009;23:1313–1326. doi: 10.1101/gad.1781009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cheng HY, Papp JW, Varlamova O, et al. microRNA modulation of circadianclock period and entrainment. Neuron. 2007;54:813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nam HJ, Boo K, Kim D, et al. Phosphorylation of LSD1 by PKCalpha is crucial for circadian rhythmicity and phase resetting. Mol Cell. 2014;53:791–805. doi: 10.1016/j.molcel.2014.01.028. [DOI] [PubMed] [Google Scholar]
- 68.Zhang X, Lu F, Wang J, et al. Pluripotent stem cell protein Sox2 confers sensitivity to LSD1 inhibition in cancer cells. Cell Rep. 2013;5:445–457. doi: 10.1016/j.celrep.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ding J, Zhang ZM, Xia Y, et al. LSD1-mediated epigenetic modification contributes to proliferation and metastasis of colon cancer. Br J Cancer. 2013;109:994–1003. doi: 10.1038/bjc.2013.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.DiTacchio L, Le HD, Vollmers C, et al. Histone lysine demethylase JARID1a activates CLOCK-BMAL1 and influences the circadian clock. Science. 2011;333:1881–1885. doi: 10.1126/science.1206022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chicas A, Kapoor A, Wang X, et al. H3K4 demethylation by Jarid1a and Jarid1b contributes to retinoblastoma-mediated gene silencing during cellular senescence. Proc Natl Acad Sci USA. 2012;109:8971–8976. doi: 10.1073/pnas.1119836109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Azzi A, Dallmann R, Casserly A, et al. Circadian behavior is lightreprogrammed by plastic DNA methylation. Nat Neurosci. 2014;17:377–382. doi: 10.1038/nn.3651. The first demonstration of dynamic light-induced DNA methylation in the SCN.
- 73.Ripperger JA, Merrow M. Perfect timing: epigenetic regulation of the circadian clock. FEBS Lett. 2011;585:1406–1411. doi: 10.1016/j.febslet.2011.04.047. [DOI] [PubMed] [Google Scholar]
- 74.Joska TM, Zaman R, Belden WJ. Regulated DNA methylation and the circadian clock: implications in cancer. Biology (Basel) 2014;3:560–577. doi: 10.3390/biology3030560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 77.Cavuoto P, Fenech MF. A review of methionine dependency and the role of methionine restriction in cancer growth control and life-span extension. Cancer Treat Rev. 2012;38:726–736. doi: 10.1016/j.ctrv.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 78. Carracedo A, Cantley LC, Pandolfi PP. Cancer metabolism: fatty acid oxidation in the limelight. Nat Rev Cancer. 2013;13:227–232. doi: 10.1038/nrc3483. A revised view on tumor metabolism, beyond the Warburg effect.
- 79.Minami Y, Kasukawa T, Kakazu Y, et al. Measurement of internal body time by blood metabolomics. Proc Natl Acad Sci USA. 2009;106:9890–9895. doi: 10.1073/pnas.0900617106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Eckel-Mahan KL, Patel VR, Mohney RP, et al. Coordination of the transcriptome and metabolome by the circadian clock. Proc Natl Acad Sci USA. 2012;109:5541–5546. doi: 10.1073/pnas.1118726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dallmann R, Viola AU, Tarokh L, et al. The human circadian metabolome. Proc Natl Acad Sci USA. 2012;109:2625–2629. doi: 10.1073/pnas.1114410109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li XM, Mohammad-Djafari A, Dumitru M, et al. A circadian clock transcription model for the personalization of cancer chronotherapy. Cancer Res. 2013;73:7176–7188. doi: 10.1158/0008-5472.CAN-13-1528. [DOI] [PubMed] [Google Scholar]
- 83.Zeng ZL, Luo HY, Yang J, et al. Overexpression of the circadian clock gene Bmal1 increases sensitivity to oxaliplatin in colorectal cancer. Clin Cancer Res. 2014;20:1042–1052. doi: 10.1158/1078-0432.CCR-13-0171. [DOI] [PubMed] [Google Scholar]
- 84.Okazaki H, Matsunaga N, Fujioka T, et al. Circadian regulation of mTOR by the ubiquitin pathway in renal cell carcinoma. Cancer Res. 2014;74:543–551. doi: 10.1158/0008-5472.CAN-12-3241. [DOI] [PubMed] [Google Scholar]