Abstract
MicroRNAs, which target mRNAs for post-transcriptional regulation, and heterochromatic siRNAs, which target chromatin causing DNA methylation, make up the majority of the endogenous regulatory small RNA pool in most plant specimens. They both function to guide Argonaute proteins to targeted nucleic acids on the basis of complementarity. Recent work on plant miRNA-target interactions has clarified the general ‘rules’ of complementarity, while also providing several intriguing exceptions to these rules. In addition, emerging evidence suggests that several factors besides miRNA-target complementarity affect plant miRNA function. For heterochromatic siRNAs, recent work has made progress towards comprehensively identifying potential target regions, but numerous fundamental questions remain to be answered.
Introduction
Many 20 to 24 nt non-coding RNAs serve as regulatory molecules in plants. Two major categories are recognized based upon the nature of the RNA precursors: microRNAs (miRNAs), which are processed from stem-loop regions of single-stranded primary RNAs, and endogenous short interfering RNAs (siRNAs), which are processed from double-stranded RNAs [1]. After biogenesis, both miRNAs and siRNAs are bound to Argonaute (AGO) proteins, where they serve to identify ‘target’ RNAs by base-pairing interactions. Successful identification of a target by miRNA/AGO complexes and a minority of siRNA/AGO complexes leads to post-transcriptional repression, while the majority of plant siRNA/AGO complexes instead lead to transcriptional repression via chromatin modifications. The extent and positions of complementarity between targets and AGO-loaded small RNAs clearly has a major influence on target selectivity. Recent evidence also suggests that there may be other factors influencing target selectivity. In this review, we focus on recent work that addresses the factors required for target identification by plant miRNA/AGO and siRNA/AGO complexes.
miRNA-Target Complementarity: Rules and Exceptions
The first experimentally confirmed plant miRNA-target interactions, between miR171 and its SCARECROW-LIKE (SCL) mRNA targets, had perfect complementarity [2]. However, it was soon apparent that perfect complementarity was the exception, rather than the rule: Most of the easily discovered and confirmed plant miRNA targets found in early studies had less than perfect base pairing to their cognate miRNAs [3]. Numerous experimental and computational studies in the mid-2000s (reviewed in [1]) led to a consensus on the ‘rules’ of base-pairing for a functional plant miRNA-target interaction: Little tolerance of mismatches at positions 2–13, with especially little tolerance of mismatches at positions 9–11, and more tolerance of mismatches at positions 1, and 14–21 (Figure 1). Having a set of reliable ‘rules’ for computational prediction of miRNA targets is a critical part of investigating the functions of newly found miRNAs as well as for the design of artificial miRNAs [4,5]. The canonical plant rules are in stark contrast to the general rules for animal miRNA-target complementarity: In animals, pairing at positions 2–7 is sufficient for a functional interaction [6]. This so-called ‘seed pairing’ does not appear to be functional for land plant miRNAs [7], although it has been reported to function in the green alga Chlamydomonas [8].
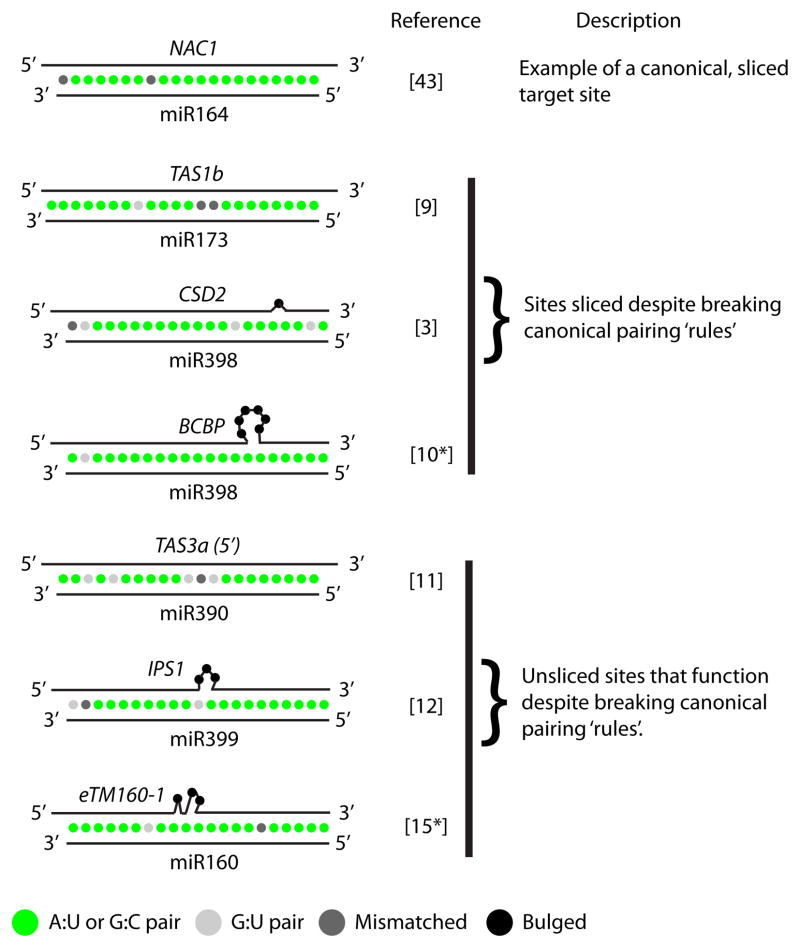
Figure 1.
Rule-breakers. Examples of functionally validated miRNA-target interactions from Arabidopsis thaliana. The miR164/NAC1 interaction on top is representative of the canonical ‘rules’ for plant miRNA targeting: Extensive pairing to positions 2–13 of the miRNA, with most or all mismatches occurring to the 3′ end of the miRNA. The other six target sites all violate these rules, but have nonetheless been experimentally proven to function in vivo.
Several exceptions to the canonical rules have been observed (Figure 1). The Arabidopsis non-coding RNA TRANS-ACTING SIRNA 1B (TAS1b) is targeted by miR173 despite the presence of two consecutive mismatches at positions 9 and 10 [9]. The Arabidopsis miR398-COPPER/ZINC SUPEROXIDE DISMUTASE 2 (CSD2) interaction has two G-U wobble pairs and a bulged nucleotide in the critical 2–13 region [3]. In isolation, however, the efficacy of this target site is quite weak [7]. Another Arabidopsis miR398 target, BLUE COPPER BINDING PROTEIN (BCBP), is regulated despite a six nucleotide bulge between positions 6–7 of the miRNA [10]*. Like the miR398 target site in CSD2, the BCBP miR398 site functions, albeit weakly, outside of the context of its native mRNA. These exceptions show that the canonical ‘rules’ are not absolute. It also seems possible that miR398 is especially tolerant of unusual target sites. It will be important to determine whether there are indeed miRNA-specific variations in complementarity requirements, as this could have a major influence on designing better artificial miRNAs, and on reducing false negatives in miRNA target predictions.
Another major set of exceptions to the canonical plant miRNA-target complementarity rules are the target-mimics (Figure 1). The natural target-mimic sites found to date are in non-coding RNAs and have G-U wobbles and/or mismatches at positions 9–11 [11] or bulged target nucleotides between positions 9–11 [12], but are otherwise almost perfectly paired. These sites prevent AGO-catalyzed ‘slicing’, which normally occurs between positions 10–11. They also function as competitive inhibitors of the regulation of canonical miRNA targets, and as such can be exploited to create reduction-in-function phenotypes for miRNAs [13,14]. Importantly, naturally occurring target-mimics embedded in long non-coding RNAs appear to be frequent and conserved in plants, suggesting that they may be an important aspect of miRNA function [15*]. Target-mimic sites also function when placed in the context of protein-coding mRNAs, and several patterns of central mismatches or bulges are allowed [7,13–17]. However, there are conflicting data on whether such sites can also direct repression of their host transcripts. Our group found that single target-mimic sites in transiently expressed reporters were not themselves repressed [7]. In contrast, a single target-mimic site in an over-expressed ENHANCED YELLOW FLUORESCENT PROTEIN (eYFP) transgene was sufficient to repress eYFP fluorescence in transgenic plants [13], and 2–8 tandem target-mimic sites located in the 3′-untranslated region (UTR), or a single site located in the 5′-UTR, repressed translation of reporter mRNA in an in vitro AGO-repression system [18]. It is possible that these discrepancies can be accounted for by the ratio of miRNA to target: Li et al. [19**] observed that a target-mimic-like site only became able to cause repression of its host mRNA when the ratio of miRNA to target was high.
Plant miRNA Targeting: Factors Beyond Complementarity
Emerging evidence suggests that there are other determinants of targeting beyond miRNA-target complementarity (Table 1). In a large study of artificial miRNAs, all with approximately similar patterns of high complementarity, Li et al. [20] observed a trend toward higher efficacies when target sites were closer to the 5′ end of the target mRNA. The trend was especially apparent when measurements of miRNA efficacy were made at the level of protein accumulation, rather than at the level of mRNA accumulation. Consistent with this, Iwakawa and Tomari [18] observed that, in vitro, target sites in the 5′-UTR and open reading frame (ORF) blocked elongation of translation, while those in the 3′-UTR blocked translation initiation. However, there are some caveats to these conclusions. Random variations in artificial miRNA biogenesis and/or AGO loading, or in mRNA secondary structures at the various target sites, may have contributed to the variable effects of artificial miRNAs reported by Li et al. [20]. Iwakawa and Tomari’s [18] experiments depended on a mutated AGO1 protein that was incapable of target cleavage, which of course does not occur in vivo. Further work is required to more rigorously test whether the position of a target site within an mRNA has a consistent, predictable effect on miRNA target efficacy in vivo.
Table 1.
Factors other than miRNA-target complementarity that may affect plant miRNA repression efficacies.
| Factor | Possible Molecular Explanation(s) | Key Reference(s) |
|---|---|---|
| Relative location of target site within protein-coding mRNA | Distinct mechanisms of target repression based on target site location? | [18],[20] |
| Target mRNA sequences flanking the target site | Occurrence of inhibitory target mRNA secondary structures? Recruitment of RNA-binding proteins? | [19**],[21],[24] |
| Ratio of miRNA levels to target mRNA levels | Target sites with sub-optimal complementarity are not detectably repressed in the absence of overwhelming amounts of miRNA? | [19**] |
Recent advances in methodology have allowed transcriptome-wide views of mRNA accessibility in plants [21–23]. These data collectively reveal a great deal of variability in mRNA accessibility due to a combination of mRNA secondary structures and mRNA-binding protein occupancies. Conceptually, miRNA/AGO complexes may have a harder time finding target sites that are occluded by RNA-binding proteins or by locally stable mRNA secondary structures. Consistent with this notion, sites flanking plant miRNA target sites tend to be depleted in GC residues, which could indicate selection for less robust local secondary structures [24]. Direct analysis of mRNA accessibility using a high-throughput approach supports this conclusion: miRNA target sites generally are more accessible than their flanking regions in Arabidopsis [21]. This hypothesis was directly tested for Arabidopsis MYELOBLASTOSIS33 (MYB33), a conserved target of miR159: Simultaneous mutation of five nucleotides immediately upstream of the target site and six nucleotides immediately downstream of the target site caused over-accumulation of both MYB33 mRNAs and MYB33 proteins [19**]. This result strongly suggests that the nucleotides flanking the MYB33 miR159 site have been selected to somehow allow optimal exposure of the target site. However, because this experiment made three amino acid substitutions in the MYB33 protein, it remains possible that the results were instead due to aberrant MYB33 protein behavior. Nonetheless, taken as a whole, several different observations together suggest that local sequence context affects plant miRNA target site function. Future work should endeavor towards a deeper understanding of how these effects work. This will allow better design of effective artificial miRNA target sites.
A final mechanism by which plant miRNA-targeting may be influenced is the ratio of miRNA to target mRNA levels. Li et al. [19**] observed that over-expressed MIR159a-based artificial miRNAs with two central mismatches suppressed the phenotype of miR159ab mutant plants and repressed miR159’s targets. However, native-promoter driven MYB33 transgenes with one or two central mismatches were not able to be substantially repressed by native miR159. These observations are consistent with a hypothesis where sub-optimal patterns of complementarity (i.e., those with central mismatches), can cross a threshold into detectable repression when there are high miRNA/target ratios. Indeed, such central mismatch sites are known to function as target mimics [7,16], and thus can likely stably bind miRNA/AGO complexes. One interpretation of the available data is that when there is enough miRNA present, interactions that normally would simply be target-mimics become repressed in their own right. If this is true, it has general implications for artificial miRNA design, because one would need to consider ‘mimic-like’ patterns of complementarity as potential targets (or avoid inadvertently repressing them as off-targets).
The Elusive Targets of Heterochromatic siRNAs
Heterochromatic siRNAs (hc-siRNAs) are the other major class of endogenous plant small RNAs. They are usually derived from intergenic and/or repetitive regions, and unlike the ~21 nt miRNAs, tend to be ~24 nts in length. After biogenesis in the nucleus (recently reviewed in [25*]), 24 nt hc-siRNA duplexes are transported to the cytoplasm and loaded into an AGO4-clade protein. After slicing the passenger strand of the hc-siRNA duplex, the guide-strand assembled hc-siRNA/AGO complex is transported back to the nucleus [26]. The nuclear-cytoplasmic-nuclear shuttling during hc-siRNA biogenesis may reflect the fact that hc-siRNAs are mobile within the plant [27], such that the recipient nucleus may often be distinct from the source nucleus. Once in the recipient nucleus, the hc-siRNA/AGO complex identifies specific target regions in the chromatin. Successful targeting is accompanied by DNA methylation at the target locus, especially in the CHG and CHH sequence contexts.
Transcription by the plant-specific DNA-dependent RNA polymerase complex, Pol V, is essential for hc-siRNA-mediated DNA methylation [28], and a key hc-siRNA associated Argonaute, AGO4, can co-immunoprecipitate with Pol V-dependent RNAs [29–30]. AGO4 physically associates with a repetitive C-terminal domain of the Pol V largest sub-unit [31]. These and similar observations have led to the predominant model for hc-siRNA targeting: hc-siRNA/AGO complexes identify complementary sites on nascent Pol V transcripts (Figure 2 - model 1), and successful identification leads to recruitment of DNA methyltransferases to the vicinity of the locus. To date, genome-wide inventories of Pol V RNAs have not been reported. This is likely due to their very low abundance; the handful of known Arabidopsis Pol V RNAs reported to date have only been detected by reverse transcriptase polymerase chain reaction (RT-PCR) assays, which are highly sensitive. Obtaining complete, genome-wide inventories of Pol V RNAs will be critical to better understand targeting by hc-siRNAs.
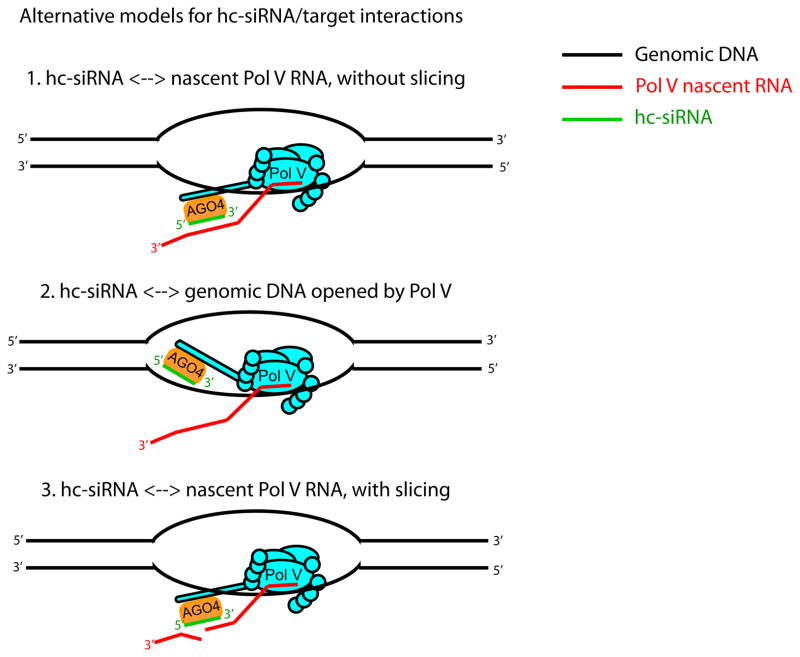
Figure 2.
Alternative models for hc-siRNA/target interactions. Model 1: AGO4-bound hc-siRNAs identify target sites on nascent Pol V RNAs, but don’t cleave them. Model 2: AGO4-bound hc-siRNAs identify target sites in genomic DNA within the Pol V transcription bubble. Model 3: Like model 1, except that AGO4 catalyzes slicing of the Pol V RNA.
It is striking that despite intense research on hc-siRNAs, there has not yet been a single experimentally confirmed example of a discrete, single hc-siRNA target site on a discrete Pol V RNA. This may be due to the difficulty of identifying Pol V RNAs in the first place (see above). However, an alternative hypothesis of hc-siRNA targeting is also possible. Matzke et al. [25*] suggest that hc-siRNA/AGO complexes target genomic DNA instead of Pol V-transcribed nascent RNA. In this model, Pol V transcription is required not to produce RNA per se, but to open a transcription bubble that allows hc-siRNA/AGO complexes the opportunity to scan for sequence matches in the genomic DNA (Figure 2 - model 2). The demonstrated associations between AGO4 and Pol V RNAs could be interpreted as indirect consequences of their proximity, rather than as evidence of a direct hc-siRNA/Pol V RNA interaction. That AGO4 is in close proximity to chromatin at Pol V-occupied loci is not in doubt, as AGO4 chromatin immunoprecipitation-sequencing (ChIP-Seq) has been shown to produce useful results [32]. Further study is required to determine whether the alternative hypothesis of a direct hc-siRNA/genomic DNA interaction is true.
Tracking Pol V-bound Chromatin
Regardless of whether hc-siRNAs target nascent RNA or genomic DNA, it seems clear that the sites of Pol V transcription or occupancy delineate eligible target regions. Several studies have reported on (ChIP-Seq) experiments that collectively provide genome-wide Pol V occupancy profiles in Arabidopsis [33–34,35**]. Somewhat surprisingly, Pol V occupancy is enriched at gene promoters and ‘young’ transposons [33], and loss of Pol V function causes a global shift in DNA methylation in the CHH context away from euchromatic regions and into pericentromeric regions [34]. One interpretation of these data is that Pol V functions to patrol protein-coding regions of the genome for infiltration by new transposons by making them eligible targets of hc-siRNAs. Pol V occupancy, and presumably transcription, depends upon a complex comprised of the DEFECTIVE IN RNA-DIRECTED DNA METHYLATION1 (DRD1), DEFECTIVE IN MERISTEM SILENCING3 (DMS3), and RNA-DIRECTED DNA METHYLATION1 (RDM1) proteins [33]. Pol V occupancy also depends upon the SU(VAR)3-9 HOMOLOG2 (SUVH2) and SU(VAR)3-9 HOMOLOG9 (SUVH9) proteins [35**,36**]. SUVH2 and SUVH9 both bind methylated DNA, indicating a requirement of already-methylated DNA for SUVH2 and SUVH9 to direct Pol V occupancy.
Why does Pol V, a factor required for initiation and maintenance of DNA methylation, require its binding sites to be already methylated? An emerging consensus is that the 24 nt hc-siRNA/Pol V pathway represents a steady-state, self-reinforcing ‘surveillance’ function for genomic elements that have already been identified in previous generations as silencing targets. Consistent with this hypothesis, Nuthikattu et al. [37] found that initiation of de novo epigenetic silencing of re-activated transposable elements requires 21–22 nt hc-siRNAs, not the ‘canonical’ 24 nt hc-siRNAs. These TE derived 21–22 nt siRNAs can be loaded into AGO6, an Argonaute in the AGO4-clade, and mediate Pol V-dependent DNA methylation [38*]. Similarly, a re-activated Arabidopsis transposon, EVD, was initially targeted by 21–22 nt hc-siRNAs that primarily functioned at the post-transcriptional level [39]. After several generations, the 21–22 nt hc-siRNAs were replaced with 24 nt hc-siRNAs, and DNA methylation became apparent. This transition from initiation of silencing by 21–22 nt siRNAs to maintenance by 24 nt siRNAs does not require transposition, as it also occurs during virus-induced gene silencing of epigenetically sensitive protein-coding genes [40].
Unknowns of hc-siRNA Targeting
hc-siRNAs presumably fulfill the same basic molecular function as miRNAs and other types of siRNAs: Guiding AGO complexes to targets based on siRNA - target complementarity. However, a single discrete, naturally occurring Pol V-enabled target site (on DNA or RNA) of a single known hc-siRNA has yet to be conclusively identified. This is a difficult experiment, both because natural Pol V targets are currently rather obscure, and because hc-siRNAs tend to be made as diverse populations of multiple siRNAs, as opposed to the discrete, single species typical of miRNAs. Because of this, the amount of hc-siRNA/target complementarity required for function is currently unknown. It stands to reason that perfect matches will function. But it also may be that imperfect pairing also functions, as it does for plant miRNAs. If so, this would require a re-assessment of the scope of hc-siRNA targeting in plant genomes. Determining the base-pairing requirements for hc-siRNA targeting of Pol V RNAs and/or Pol V-opened chromatin should be a high priority for future research.
Another outstanding question is the role of AGO4 ‘slicing’ during targeting. Like most plant Argonautes, AGO4 possesses a conserved catalytic motif. AGO4 slicing of the passenger strand of the initial hc-siRNA duplex is clearly essential for proper loading of hc-siRNAs in the cytoplasm [26]. In vitro, hc-siRNA/AGO4 complexes can slice target RNA [41], suggesting that AGO4 slicing might also be required at the targeting stage. However, It is currently unknown whether AGO4 actually slices target RNAs in vivo. A long-standing but unproved hypothesis is that hc-siRNA assembled AGO4 slices nascent Pol V transcripts (Figure 2 - model 3), and the slicing products are then used as substrates to produce more hc-siRNAs, amplifying the silencing. However, as described above, Pol V RNAs can be detected by AGO4 immunoprecipitation [29–30], which suggests stable binding between AGO4 and targets in vivo. Stable binding to target RNAs would not be expected for interactions that result in AGO4-catalyzed target slicing; for plant miRNAs, whose targeting often involves slicing, only catalytically inactive AGOs can co-immunoprecipitate target RNAs [42]. Further experiments are needed to more fully describe the role of AGO4-catalyzed slicing, if any, during the targeting phase of the hc-siRNA pathway.
Conclusion
The basic parameters of miRNA-target interactions in plants are now well known. However, several factors beyond the base-pairing pattern are now implicated in modulating miRNA targeting efficiencies. For hc-siRNAs, the specific details of targeting are much less well-known at present. Better understanding of these interactions will facilitate improved target identification and artificial small RNA design, which will be valuable enabling knowledge for both research and industry.
Acknowledgments
We thank Andrzej Wierzbicki for discussion of this manuscript. Seth Polydore is supported by the NIH-PSU funded Computation, Bioinformatics and Statistics (CBIOS) Predoctoral Training Program (1T32GM102057-0A1). This work was supported by NSF award 1121438 to MJA.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Axtell MJ. Classification and comparison of small RNAs from plants. Annu Rev Plant Biol. 2013;64:137–159. doi: 10.1146/annurev-arplant-050312-120043. [DOI] [PubMed] [Google Scholar]
- 2.Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 3.Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 4.Schwab R, Ossowski S, Warthmann N, Weigel D. Directed gene silencing with artificial microRNAs. Methods Mol Biol. 2010;592:71–88. doi: 10.1007/978-1-60327-005-2_6. [DOI] [PubMed] [Google Scholar]
- 5.Carbonell A, Takeda A, Fahlgren N, Johnson SC, Cuperus JT, Carrington JC. New generation of artificial MicroRNA and synthetic trans-acting small interfering RNA vectors for efficient gene silencing in Arabidopsis. Plant Physiol. 2014;165:15–29. doi: 10.1104/pp.113.234989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Q, Wang F, Axtell MJ. Analysis of complementarity requirements for plant MicroRNA targeting using a Nicotiana benthamiana quantitative transient assay. Plant Cell. 2014;26:741–753. doi: 10.1105/tpc.113.120972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamasaki T, Voshall A, Kim E-J, Moriyama E, Cerutti H, Ohama T. Complementarity to an miRNA seed region is sufficient to induce moderate repression of a target transcript in the unicellular green alga Chlamydomonas reinhardtii. Plant J. 2013;76:1045–1056. doi: 10.1111/tpj.12354. [DOI] [PubMed] [Google Scholar]
- 9.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 10*.Brousse C, Liu Q, Beauclair L, Deremetz A, Axtell MJ, Bouché N. A non-canonical plant microRNA target site. Nucleic Acids Res. 2014;42:5270–5279. doi: 10.1093/nar/gku157. This paper demonstrates that Arabidopsis BCBP has a highly unusual miR398 target site, containing a six-nucleotide target bulge between positions six and seven, in its 5′-UTR. The site is sliced in vivo and functional in a heterologous context. It does not, however, appear to be a wide-spread pattern of miRNA targets. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Axtell MJ, Jan C, Rajagopalan R, Bartel DP. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006;127:565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 12.Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 13.Todesco M, Rubio-Somoza I, Paz-Ares J, Weigel D. A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet. 2010;6:e1001031. doi: 10.1371/journal.pgen.1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan J, Gu Y, Jia X, Kang W, Pan S, Tang X, Chen X, Tang G. Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell. 2012;24:415–427. doi: 10.1105/tpc.111.094144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15*.Wu H-J, Wang Z-M, Wang M, Wang X-J. Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant Physiol. 2013;161:1875–1884. doi: 10.1104/pp.113.215962. Demonstrated that target-mimic RNAs are widespread in plants. Several conserved examples were noted, and two (for miR160 and miR166) were experimentally confirmed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ivashuta S, Banks IR, Wiggins BE, Zhang Y, Ziegler TE, Roberts JK, Heck GR. Regulation of gene expression in plants through miRNA inactivation. PLoS ONE. 2011;6:e21330. doi: 10.1371/journal.pone.0021330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reichel M, Li Y, Li J, Millar AJ. Inhibiting plant microRNA activity: molecular SPONGEs, target MIMICs, and STTMs all display variable efficacies against target microRNAs. Plant Biotechnol J. 2015 doi: 10.1111/pbi.12327. [DOI] [PubMed] [Google Scholar]
- 18.Iwakawa H, Tomari Y. Molecular insights into microRNA-mediated translational repression in plants. Mol Cell. 2013;52:591–601. doi: 10.1016/j.molcel.2013.10.033. [DOI] [PubMed] [Google Scholar]
- 19**.Li J, Reichel M, Millar AA. Determinants beyond both complementarity and cleavage govern microR159 efficacy in Arabidopsis. PLoS Genet. 2014;10:e1004232. doi: 10.1371/journal.pgen.1004232. This study demonstrates that several factors besides just the complementarity pattern affect plant miRNA targeting. These additional factors include the ratio of miRNA to target, and the identities of nucleotides immediately flanking the miRNA target site. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J-F, Chung HS, Niu Y, Bush J, McCormack M, Sheen J. Comprehensive protein-based artificial microRNA screens for effective gene silencing in plants. Plant Cell. 2013;25:1507–1522. doi: 10.1105/tpc.113.112235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li F, Zheng Q, Vandivier LE, Willmann MR, Chen Y, Gregory BD. Regulatory impact of RNA secondary structure across the Arabidopsis transcriptome. Plant Cell. 2012;24:4346–4359. doi: 10.1105/tpc.112.104232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, Assmann SM. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505:696–700. doi: 10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- 23.Gosai SJ, Foley SW, Wang D, Silverman IM, Selamoglu N, Nelson ADL, Beilstein MA, Daldal F, Deal RB, Gregory BD. Global analysis of the RNA-protein interaction and RNA secondary structure landscapes of the Arabidopsis nucleus. Mol Cell. 2015;57:376–388. doi: 10.1016/j.molcel.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu W, Wang X, Zhai C, Xie X, Zhou T. Selection on synonymous sites for increased accessibility around miRNA binding sites in plants. Mol Biol Evol. 2012;29:3037–3044. doi: 10.1093/molbev/mss109. [DOI] [PubMed] [Google Scholar]
- 25*.Matzke MA, Kanno T, Matzke AJM. RNA-Directed DNA Methylation: The Evolution of a Complex Epigenetic Pathway in Flowering Plants. Annu Rev Plant Biol. 2015;66:243–267. doi: 10.1146/annurev-arplant-043014-114633. In addition to summarizing the current knowledge in hc-siRNA functions with special attention to evolutionary aspects, this review proposes an alternative mechanism of hc-siRNA targeting: Direct targeting of genomic DNA at sites made accessible by Pol V activity. [DOI] [PubMed] [Google Scholar]
- 26.Ye R, Wang W, Iki T, Liu C, Wu Y, Ishikawa M, Zhou X, Qi Y. Cytoplasmic assembly and selective nuclear import of Arabidopsis Argonaute4/siRNA complexes. Mol Cell. 2012;46:859–870. doi: 10.1016/j.molcel.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 27.Melnyk CW, Molnar A, Bassett A, Baulcombe DC. Mobile 24 nt small RNAs direct transcriptional gene silencing in the root meristems of Arabidopsis thaliana. Curr Biol. 2011;21:1678–1683. doi: 10.1016/j.cub.2011.08.065. [DOI] [PubMed] [Google Scholar]
- 28.Stroud H, Greenberg MVC, Feng S, Bernatavichute YV, Jacobsen SE. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell. 2013;152:352–364. doi: 10.1016/j.cell.2012.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wierzbicki AT, Ream TS, Haag JR, Pikaard CS. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet. 2009;41:630–634. doi: 10.1038/ng.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Böhmdorfer G, Rowley MJ, Kuciński J, Zhu Y, Amies I, Wierzbicki AT. RNA-directed DNA methylation requires stepwise binding of silencing factors to long non-coding RNA. Plant J. 2014;79:181–191. doi: 10.1111/tpj.12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.El-Shami M, Pontier D, Lahmy S, Braun L, Picart C, Vega D, Hakimi M-A, Jacobsen SE, Cooke R, Lagrange T. Reiterated WG/GW motifs for functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev. 2007;21:2539–2544. doi: 10.1101/gad.451207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng Q, Rowley MJ, Böhmdorfer G, Sandhu D, Gregory BD, Wierzbicki AT. RNA polymerase V targets transcriptional silencing components to promoters of protein-coding genes. Plant J. 2012 doi: 10.1111/tpj.12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhong X, Hale CJ, Law JA, Johnson LM, Feng S, Tu A, Jacobsen SE. DDR complex facilitates global association of RNA polymerase V to promoters and evolutionarily young transposons. Nat Struct Mol Biol. 2012;19:870–875. doi: 10.1038/nsmb.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wierzbicki AT, Cocklin R, Mayampurath A, Lister R, Rowley MJ, Gregory BD, Ecker JR, Tang H, Pikaard CS. Spatial and functional relationships among Pol V-associated loci, Pol IV-dependent siRNAs, and cytosine methylation in the Arabidopsis epigenome. Genes Dev. 2012;26:1825–1836. doi: 10.1101/gad.197772.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35**.Johnson LM, Du J, Hale CJ, Bischof S, Feng S, Chodavarapu RK, Zhong X, Marson G, Pellegrini M, Segal DJ, et al. SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation. Nature. 2014;507:124–128. doi: 10.1038/nature12931. Along with [36], this study demonstrates that the SUVH2 and SUVH9 proteins are required for Pol V positioning. Importantly, both SUVH2 and SUVH9 specifically bind to methylated DNA, implying that Pol V occupancy requires pre-existing DNA methylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36**.Liu Z-W, Shao C-R, Zhang C-J, Zhou J-X, Zhang S-W, Li L, Chen S, Huang H-W, Cai T, He X-J. The SET domain proteins SUVH2 and SUVH9 are required for Pol V occupancy at RNA-directed DNA methylation loci. PLoS Genet. 2014;10:e1003948. doi: 10.1371/journal.pgen.1003948. Along with [35], this study demonstrates that the SUVH2 and SUVH9 proteins are required for Pol V positioning. Importantly, both SUVH2 and SUVH9 specifically bind to methylated DNA, implying that Pol V occupancy requires pre-existing DNA methylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nuthikattu S, McCue AD, Panda K, Fultz D, DeFraia C, Thomas EN, Slotkin RK. The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21–22 nucleotide small interfering RNAs. Plant Physiol. 2013;162:116–131. doi: 10.1104/pp.113.216481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38*.McCue AD, Panda K, Nuthikattu S, Choudury SG, Thomas EN, Slotkin RK. ARGONAUTE 6 bridges transposable element mRNA-derived siRNAs to the establishment of DNA methylation. EMBO J. 2015;34:20–35. doi: 10.15252/embj.201489499. Demonstrates that the initiation phase of transposon silencing uses 21–22 nt hc-siRNAs to direct DNA methylation via the AGO6 protein in Arabidopsis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marí-Ordóñez A, Marchais A, Etcheverry M, Martin A, Colot V, Voinnet O. Reconstructing de novo silencing of an active plant retrotransposon. Nat Genet. 2013;45:1029–1039. doi: 10.1038/ng.2703. [DOI] [PubMed] [Google Scholar]
- 40.Bond DM, Baulcombe DC. Epigenetic transitions leading to heritable, RNA-mediated de novo silencing in Arabidopsis thaliana. Proc Natl Acad Sci US A. 2015;112:917–922. doi: 10.1073/pnas.1413053112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qi Y, He X, Wang X-J, Kohany O, Jurka J, Hannon GJ. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature. 2006;443:1008–1012. doi: 10.1038/nature05198. [DOI] [PubMed] [Google Scholar]
- 42.Carbonell A, Fahlgren N, Garcia-Ruiz H, Gilbert KB, Montgomery TA, Nguyen T, Cuperus JT, Carrington JC. Functional analysis of three Arabidopsis ARGONAUTES using slicer-defective mutants. Plant Cell. 2012;24:3613–3629. doi: 10.1105/tpc.112.099945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mallory AC, Dugas DV, Bartel DP, Bartel B. MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr Biol. 2004;14:1035–1046. doi: 10.1016/j.cub.2004.06.022. [DOI] [PubMed] [Google Scholar]