Abstract
Herein we develop an approach for optically controlling receptor tension. This is achieved using optomechanical actuator nanoparticles that are controlled with non-invasive near-infrared light. Illumination leads to particle collapse, delivering piconewton forces to specific cell surface receptors with high spatial and temporal resolution. As a proof-of-concept, we applied optomechanical actuation to trigger integrin-based focal adhesion formation, cell protrusion and migration, as well as T cell receptor activation.
By virtue of having a set morphology and architecture, the majority of cells within multicellular organisms experience forces that are highly orchestrated in space and time1. To study the dynamic interplay between chemical and mechanical signals at the cell surface, a number of techniques have been developed to investigate cell response to spatially confined physical perturbations2. For example, micropipettes3 and single molecule techniques4, 5 have been used to prod the apical side of individual cells and record biochemical responses, but such approaches are challenging, low-throughput and highly serial2. Another general approach involves using magnetic actuation of nanoparticles6, 7 and micropillars8 to trigger signaling pathways. Controlling magnetic fields with high spatial resolution requires either a sparse density of magnetic elements or sophisticated micro fabricated magnetic structures that focus an external magnetic field. Therefore, magnetic stimulation of mechanotransduction circuits remains specialized and is not widely employed. In the absence of methods for manipulating forces with molecular specificity and high spatio-temporal resolution, elucidating the local biochemical response to mechanics remains a hurdle2.
In principle, the most desirable approaches for manipulation within biological systems are optical-based. This is evidenced by the rapid proliferation of photo-stimulation techniques employing caged or photoswitchable molecules, and optogenetic constructs9–11. Therefore, the development of methods to harness light for delivering precise physical inputs to biological systems could potentially transform the study of mechanotransduction.
Toward this goal, we develop optomechanical actuator (OMA) nanoparticles to manipulate receptor mechanics with high spatial and temporal resolution using low intensity near-infrared (NIR) illumination (Fig. 1a). OMA nanoparticles are programmed to rapidly shrink upon illumination, thus applying a mechanical load to receptor-ligand complexes decorating the immobilized particle. The NIR optical pulse train controls the amplitude, duration, repetition and loading rate of mechanical input. OMAs are immobilized onto standard glass coverslips allowing cell imaging and manipulation using a conventional fluorescence microscope equipped with an inexpensive NIR laser diode. Therefore, live cell response to mechanical stimulation can be characterized with unprecedented spatial and temporal resolution. Importantly, because mechanical stimulation can be rapidly deployed across arbitrary patterns at the cell surface, we were able to demonstrate the first example of opto-mechanical control of focal adhesion (FA) formation, cell protrusions, cell migration, and T cell activation.
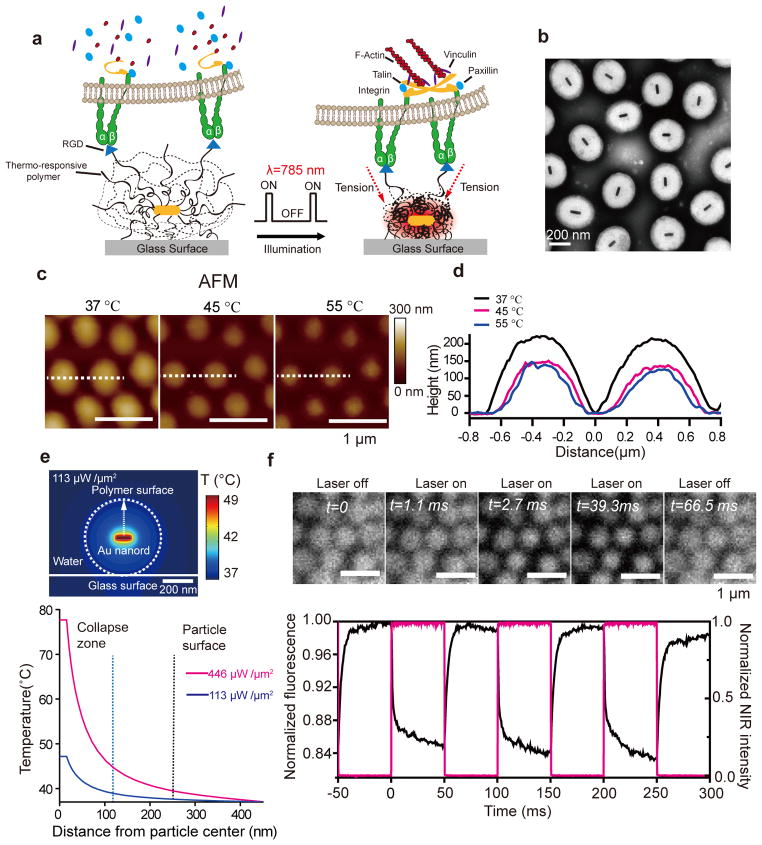
Figure 1. Schematic and characterization of optomechanical actuator (OMA) nanoparticles.
(a) Schematic showing the general design and proposed mechanism of OMA nanoparticles. (b)Representative TEM image of OMA nanoparticles. (c) Representative AFM images of the OMA nanoparticles at T = 37, 45, and 55 °C in DI water. (d) Plot represents the temperature-dependent height profile across two nanoparticles indicated by the dashed white line. (e) Top: Finite element simulation of the heat distribution in a OMA nanoparticle irradiated with 785 nm IR laser beam at a power density of 113 μW μm−2 in water. Bottom: Plot showing the OMA particle temperature as a function of distance from the Au nanorod core irradiated with 785 nm laser for two different power densities (446 and 113 μW μm−2). Note that the power density (113 μW μm−2) is 10 fold greater than that used for cell stimulation because a 100% duty cycle (steady state illumination) is assumed for the energy input calculation. The details of the simulation are included in the Online Methods and Supplementary Figure 4. (f) Top: Representative fluorescence images of OMA nanoparticles labeled with Alexa 488 and captured by a sCMOS camera (Andor Zyla sCMOS) at 1800 fps while chopping the NIR laser source at 10 Hz (50% duty cycle). Data was collected in DI water at 37°C. Bottom: The average normalized fluorescence intensity (black curve) of OMA nanoparticles as the NIR laser (red curve) is modulated with a 10 Hz frequency and 50% duty cycle. The dynamics of fluorescence was used to estimate the time constant of particle collapse and relaxation. The ~15% decrease in fluorescence was primarily due to quenching by the Au nanorod upon particle collapse (Supplementary Figure 5g and h).
OMA nanoparticles are comprised of a Au nanorod (25 nm × 100 nm) coated with a thermo-responsive polymer shell (poly(N-isopropylmethacrylamide, pNIPMAm) (Fig. 1b and Supplementary Figure 1). The Au nanorod functions as a photothermal transducer, converting a NIR pulse to localized heat that drives a transient collapse of the polymer shell. OMA particles can be immobilized onto virtually any type of support and can also be functionalized with a wide variety of small molecule, peptide, and protein ligands specific to a receptor of interest. Thus, mechanical actuation is molecularly selective in that only receptors that are directly engaged to ligands on the OMA nanoparticles experience the mechanical input.
TEM of OMA nanoparticles confirms the core-shell structure, monodispersity, and dimensions of the inorganic core (Fig. 1b and Supplementary Figure 2). Dynamic light scattering showed that the average hydrodynamic diameter of the particles is 480 ± 20 nm at room temperature (RT), shrinking to a 270 ± 10 nm diameter upon heating to great than 42 °C (Supplementary Figure 2e). Vis-NIR spectra of OMA particles as a function of temperature confirmed the phase transition temperature and provided characterization of the NIR absorption band (Supplementary Figure 2f). Temperature-controlled AFM showed that immobilized particles (at 37 °C) displayed a flattened morphology with a mean height and width of 220, and 700 nm, respectively (Fig. 1c and d). The AFM data also revealed that OMA particles collapsed both in the lateral and vertical directions at T > 42 °C, indicating that force vectors point inward toward the particle center with a vertical and horizontal component. Particles displayed a ~70 nm decrease in height and a ~120 nm reduction in particle diameter (Fig. 1d). Structured illumination microscopy (SIM) measurement of particle diameter also showed a ~100 nm decrease following NIR illumination (Supplementary Figure 3a–c and Supplementary Table). The reduction in particle size represents the maximum receptor displacement during NIR illumination. In principle, the magnitude of receptor displacement can be further tuned by changing particle size and the NIR illumination profile.
Key to the optomechanical actuation strategy is that particle heating is transient and confined to the core. Heat is dissipated as a 1/distance function from the Au nanorod core (Online Methods), and thus sufficiently large OMA particles display negligible surface heating. In contrast, mechanical energy is more efficiently transmitted from the particle core to its surface because of the crosslinked nature of the polymer. 3D simulations mapping the heat distribution around the Au nanorod (Fig. 1e and Supplementary Figure 4) confirmed that it is possible to collapse the OMA particle core without significantly altering its surface temperature, thus minimizing thermal effects to cells.
To examine the dynamics of OMAs, we conjugated the particles with Alexa 488, and recorded fluorescence at 1800 frames per second (fps) during pulsed NIR-stimulation at 37 °C in water (Fig. 1f, Supplementary Figure 5 and Supplementary Movie 1). Particles were illuminated at 785 nm with a continuous laser pulsed at a 10 Hz frequency and 50% duty cycle. Under these conditions, both the size of OMA particles and their emission intensity decreased upon illumination (Fig. 1f). Particle actuation was highly reversible and could be sustained for at least 106 cycles (Supplementary Figure 5). The collapse and swelling time constants were 1.3 ± 0.1 msec, and 3.3 ± 0.2 msec, respectively (Supplementary Figure 5e and f). These time constants indicate that one can mechanically stimulate cells with rates 1–2 orders of magnitude greater than can be achieved with magnetic actuation6–8.
To estimate the force applied by OMAs, we used DNA-based fluorescence tension probes that were recently developed in our group (Supplementary Figure 6)12. We observed a reversible increase in fluorescence that coincided with NIR illumination; therefore, particle collapse exerts a force per ligand that is greater than 13 pN, but below ~50 pN (Supplementary Figure 6 and Online Methods).
Given that integrin adhesion receptor forces within FAs are estimated within the ~5 pN to 50 pN range 12–14, OMAs are likely well-suited to trigger integrin activation within FAs. To test this, we transfected NIH/3T3 cells with a plasmid encoding for GFP-paxillin, a widely used surrogate marker for FAs3, 15. Cells were then cultured onto a monolayer of OMA nanoparticles modified with cyclic Arg-Gly-Asp-D-Phe-Lys, c(RGDfK), with a density of 2,000 peptides μm−2 for 12 hr (Supplementary Figure 7–10). We illuminated the sample with a pulsed NIR source while recording GFP-paxillin dynamics (Fig. 2a and Supplementary Movie 2). No detectable increase in surface temperature was observed at these illumination conditions (Supplementary Figure 11). Before stimulation, the GFP-paxillin was localized primarily in punctate regions at the cell edge (Fig. 2a). Representative time-lapse TIRF images showed rapid growth of the total FA size from ~1 μm2 to 5 μm2 after stimulation for t = 17 min within region of interest (ROI) I, coinciding with the NIR illumination area (Fig. 2a and 2b). In contrast, the average FA size at ROI II, adjacent to the region of NIR stimulation, was reduced by 50% during the same time period. FAs in ROI III, 20 μm away from the NIR source, did not display a detectable change in FA size or organization within the 20 min illumination duration. Kymograph analysis and plots of total paxillin intensity over time within these ROIs confirmed this conclusion (Fig. 2c and 2d). Other markers of FAs, such as F-actin and vinculin were also rapidly recruited upon identical NIR illumination (Supplementary Figure 12), demonstrating FA assembly. Dual color imaging of F-actin (mCherry) and paxillin (GFP) upon OMA stimulation showed that F-actin polymerization precedes the recruitment of paxillin and FA maturation, in agreement with existing models (Supplementary Figure 13). 3D-SIM and total internal reflection fluorescence microscopy (TIRFM) nanometry showed that the actin network was mechanically displaced by ~100 nm following the collapse of OMA particles (Supplementary Figure 3 and 14). Taken together, these results indicate that particle collapse in the x-y and z directions mechanically strains integrin receptors and this force is transmitted across the cell membrane activating mechanosensitive proteins, such as talin and vinculin, actin polymerization and triggering FA maturation3, 8.
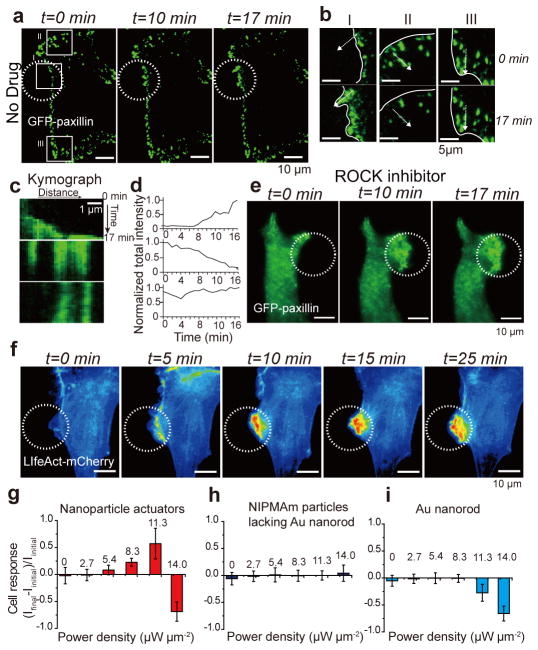
Figure 2. Optomechanical actuation of integrins leads to GFP-paxillin and LifeAct-mCherry recruitment.
(a) Representative time-lapse TIRF images of GFP-paxillin transiently transfected NIH/3T3 cells that were cultured on OMA nanoparticles displaying the RGD peptide, n≥5 cells. Scale bars,10 μm. (a, b) NIR illumination (white dashed circle, 10 Hz, 10% duty cycle, power density = 11.3 μW μm−2) leads to rapid recruitment of GFP-paxillin, ROI I. White line indicates cell edge. Adjacent regions showed a loss of GFP-paxillin, ROI II, while distant regions did not show substantial change in GFP-paxillin, ROI III. (c) The kymograph (measured across dashed white arrow) and total paxillin intensity over time (d) within these regions (I, II, and III) of interest confirm the trends noted above. (e) Cells treated with ROCK inhibitor showed a similar increase in GFP-paxillin following force-driven integrin stimulation. Scale bars,10 μm. (f) Representative time-lapse fluorescence images of LifeAct-mcherry transiently transfected 3T3 cells upon NIR illumination (identical conditions to those used in Fig. 2a), n≥20 cells. Scale bars,10 μm. (g–i) Plots of average cellular response for cells cultured on OMA nanoparticles (g), pNIPMAm particles lacking the gold rod core (h), and gold rods lacking the polymer shell (i), when the surface was excited by different NIR laser power intensities (10Hz, 10% duty cycle). The error bar for each data point represents the standard deviation of cell response from at least n= 6 cells.
Other contributing mechanisms by which OMA collapse could lead to FA maturation may be related to cellular rigidity sensing of the collapsed polymer and increased ligand density in the dehydrated particles. These contributions are likely minimal considering the transient (10% duty cycle) nature of the collapsed particle (Supplementary Note). Moreover, NIR illumination is tuned such that OMA particle are only partially collapsed within the 10 msec illumination window. To further test the role of stiffness sensing and ligand density sensing in cell response, we treated GFP-paxillin expressing cells with the Rho kinase inhibitor Y-27632 for 30 min (blocking myosin II contractility). Based on the literature, ROCK treatment attenuates stiffness sensing mechanisms and ligand density-driven FA maturation3, 16, 17. Within 5 min of particle actuation, cells rapidly protruded toward the source of the stimulus and increased GFP-paxillin density (Fig. 2e and Supplementary Movie 3). Thus, OMA-driven cell stimulation is independent of myosin contractility, and FA growth is primarily attributed to OMA-generated integrin forces, bypassing the requirement for ROCK activity.
To quantify cell response to the NIR intensity and frequency, we transiently transfected cells with LifeAct-mCherry, an F-actin probe, and recorded actin intensity, I=(Ifinal – Iinitial)/Ifinal, where Iinitial is the total actin fluorescence in the ROI prior to stimulation, and Ifinal is the total fluorescence in the same ROI following stimulation at t = 20 min. This readout allowed us to benchmark the response of cells under different conditions. Although appropriate NIR illumination conditions led to a drastic enhancement in F-actin localization (Fig. 2f and Supplementary Movie 4), this was not uniform across all conditions tested. We found that cell response was optimal and similar at frequencies of 10 and 100 Hz (Supplementary Figure 15). In contrast, there was limited cell response at 1 Hz and 0 Hz (Supplementary Figure 15–16), which indicated enhanced integrin activation upon cyclic stimulation. Therefore, for controlling FA formation, OMA nanoparticles require the application of cyclic mechanical stimulation rather than sustained force.
At lower NIR intensities (2.7 μW μm−2), where ~20% of effectively bound ligands experience >13 pN (Supplementary Figure 6), cells did not display a measurable response, indicating that at those force levels, the OMA particles failed to trigger actin polymerization (Fig. 2g). At greater NIR intensities ranging from 5.4 to 11.3 μW μm−2, corresponding to a range from 60% to 100% of effectively bound ligands experiencing >13 pN of tension, we observed an enhancement in F-actin enrichment. In contrast, at average powers equal to or greater than 14.0 μW μm−2, the particle actuation led to cell retraction (Supplementary Figure 17), which is due to thermal heating18. This is supported by thermal measurements showing a 6 ± 1.2 °C increase at this power (Supplementary Figure 11). Importantly, particles that lacked the Au rod core (Fig. 2h), rods lacking the polymer shell (Fig. 2i), or rods coated with a thermally insensitive polymer shell (Supplementary Figure 18) failed to display any increase in actin localization regardless of the NIR intensity tested. These experiments further confirm that cell response is primarily driven by optical manipulation of integrin tension.
Given that FA formation ultimately controls cell protrusion and migration19, we explored whether OMA stimulation could be used to guide cell movement. To test this, we stimulated FA formation in LifeAct-mCherry transfected cells for 2 hrs. Fig. 3a and Supplementary Movie 5 show the results from a representative cell. Initially (within 9 min), new protrusions formed in the illumination area. Subsequently (within 40 min), the cell migrated to maximize its overlap with the region of mechanical actuation. Note that the regions of greatest F-actin intensity always coincided with particle actuation. The NIR illumination area was then moved in a stepwise fashion such that at least one part of the cell maintained exposure to the mechanical stimulus. In this way, the cell was mechanically guided across the field of view. Based on kymograph analysis, the average speed of cell migration was approximately 0.33 μm min−1 (Fig. 3b). A plot of the response of six cells stimulated using this approach is shown in Fig. 3c, and indicates that while the approach is robust, there is cell to cell variability in response to the mechanical stimulus (Supplementary Figure 19). Nonetheless, this experiment represents the first example of controlling cell migration by using active mechanical forces.
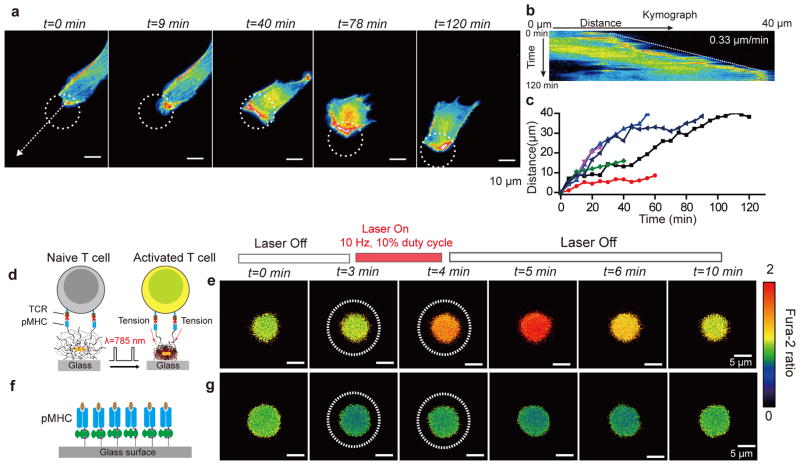
Figure 3. Optical control of cell migration and T cell activation by OMA nanoparticles.
(a)Representative time-lapse images showing control NIH/3T3 cell migration over long distances by OMA stimulation. The white dashed circle represents the region of NIR illumination. Scale bars,10 μm. (b) Kymograph analysis of cell migration obtained using white dashed arrow shown in (a). (c) Plot showing the cell migration distance as a function of time upon OMA stimulation for n= 6 cells. Cell tracking was stopped when cells ceased to respond to NIR illumination or if cells displayed phototoxicity from fluorescence imaging. Supplementary Figure 19 shows representative images of cell migration from these experiments. (d) Schematic showing OMA nanoparticles functionalized with the OVA peptide-major histocompatibility complex (pMHC) and illuminated to mechanically stimulate T cell receptors on the membrane of OT-I Tlymphocytes. (e) Fluorescence images of the ratiometric intensity of the fura-2 calcium indicator dye. Upon stimulation (t=3 min, dotted white circle), T cells were effectively activated within a 1min as indicated by the increase of fura-2 emission ratio when excited by 340 nm/380 nm. Scale bars, 5 μm. (f) Schematic of control experiments, where OT-I cells were stimulated on a pMHC monolayer that was directly modified on the glass coverslip. (g) Upon stimulation (white circle), no calcium flux was observed and the calcium signal continued to fluctuate at basal levels. Scale bars, 5 μm.
Recent single molecule experiments demonstrate that mechanical stimulation of the T cell receptor leads to T cell activation20, 21, and thus we next aimed to use OMA particles to optically control T cell activation. As a proof-of-concept, we modified OMA particles with a specific peptide-major histocompatibility complex (pMHC) from the OVA model. These pMHC decorated particles were used to mechanically stimulate TCR in naïve OT-I T lymphocytes (Fig. 3d). Upon stimulation (t=3 min, dotted white circle), T cells were effectively activated within one minute as indicated by a calcium flux measured with the fura-2 dye (Fig. 3e, Supplementary Figure 20a and Supplementary Movie 6). OT-I cells optically stimulated on a pMHC monolayer on the glass coverslip showed no response (Fig. 3f and g and Supplementary Figure 20b). Note that T cells rapidly respond to mechanical stimuli (within 1 min) in contrast to cell adhesions, which typically require ~10 min for initial actin polymerization. Therefore, OMA particles can be used to mechanically trigger T cells, demonstrating applications in studying the role of mechanical forces in different biological systems.
In summary, we developed a new class of optomechanical actuator nanoparticles that allow one to optically manipulate receptor tension with optical spatial resolution and 5 msec temporal resolution using low intensity NIR illumination. We demonstrate that nanoparticle-generated forces induce F-actin polymerization, recruitment of paxillin and vinculin into FAs, and ultimately controlling cell migration. Note that control of cell migration using light has been achieved using optogenetic tools9, 10, but unlike these past examples, our approach precludes the need for genetic modification of target cells. Optomechanical stimulation represents a yet unexplored avenue for controlling signaling activity in living cells that may have important applications in a broad range of cell types including neurons, platelets, and T-cells. Moreover, the non-toxic nature of thermo-responsive polymers and gold nanoparticles suggests potential for in vivo applications.
Online Methods
Preparation of OMA nanoparticles
Gold Nanorod preparation
To generate OMA nanoparticles, we first synthesized the gold nanorod (NR) inorganic core using an improved seeded growth synthesis procedure, recently published by Murray and colleagues22. The synthesis of AuNRs employed a binary surfactant mixture composed of hexadecyltrimethylammonium bromide (CTAB) and sodium oleate (NaOL) to grow Au seed crystals into NRs. Briefly, the seed solution was prepared as follows: 5 mL of 0.5 mM HAuCl4 was mixed with 5 mL of 0.2 M CTAB solution in a 20 mL scintillation vial. 0.6 mL of fresh 0.01 M NaBH4 was diluted to 1 mL with water and was then injected into the Au(III)-CTAB solution under vigorous stirring (1,200 rpm). The color of the solution changed from yellow to brownish yellow and the stirring was stopped after 2 min. The seed solution was aged at room temperature for 30 min prior to use.
We adapted the general guidelines presented by Murray et al22. to control the dimensions of the NR such that the NIR plasmon peak was approximately 800 nm. In our protocol, we prepared a growth solution comprised of 3.6 g of CTAB and 0.4936 g of NaOL dissolved in 100 mL of warm water (~50 °C) in a 250 ml Erlenmeyer flask. The solution was allowed to cool down to 30 °C, and then 8.5 mL of 4 mM AgNO3 solution was added. The mixture was kept undisturbed at 30 °C for 15 min after which 100 mL of 1 mM HAuCl4 solution was added. The solution became colorless after 90 min of stirring (700 rpm), and then 0.6 ml of HCl (37 wt. % in water, 12.1 M) was introduced to adjust the pH and acidify the solution. After 15 min of slow stirring at 400 rpm, 0.6 mL of 0.064 M ascorbic acid was added and the solution was vigorously stirred for 30 s. Finally, 80 μL of seed solution was injected into the growth solution. The resultant mixture was stirred for 30 s and left undisturbed at 30 °C for 12 h to complete NR growth. The products were isolated by centrifugation at 5,000 rpm for 60 min followed by removal of the supernatant; the final solution was concentrated to 90 ml and characterized using UV-vis-NIR and TEM.
Polymerization and encapsulation of AuNRs with pNIPMAm
The procedure for polymerization and encapsulation of the AuNRs with pNIPMAm 23 was adapted from methods noted by Contreras-C’aceres et al.24 and F. Tang et al.25 employing Au and Ag spherical nanoparticles, respectively. First, the CTAB and NaOL surfactants coating the AuNRs were replaced with a thiolated and vinyl terminal (C=C) ligand by adding 20 mg of N,N′-bis(acryloyl)cystamine to a 90 ml AuNR solution along with 10 ml ethanol while vigorously stirring at 700 rpm for 12 hours. Subsequently, the AuNR solution was purified by centrifugation at 5,000 rpm for 60 min, removing the supernatant, and then redispersing in 15 mL of deionized water. This solution could be stored at 4 °C for several months without adversely affecting quality of OMAs.
The polymerization of pNIPMAm on the Au nanorods surface was carried out as follows: 0.1 g of N-isopropylmethacrylamide, 0.01 g of the crosslinking agent N,N′-methylenebisacrylamide were dissolved in 15 ml Milli-Q water in a three neck flask. The mixture was heated to 70 °C under continuous stirring and purged with continuous N2 flow. Subsequently, 1 ml of the AuNR solution modified with the thiolated vinyl group (described above) was added to the three-neck flask. After 1 min, pNIPMAm polymerization was initiated with the addition of 80 μl (0.1 M) of the free radical initiator 2,2′azobis(2-methylpropionamidine) dihydrochloride (AAPH) and the polymerization was allowed to proceed for 2 hr at 70 °C. In some cases, we aimed to decorate the OMA particles with the alkyne functional group. This was achieved by adding 30 μl of propargyl methacrylate monomer dissolved in 1 mL ethanol at the 100 min time point of reaction and allowing the free-radical polymerization to proceed for the full 2 hr. Please note that vigorous stirring (1,200 rm) was used in every step in the OMA particle synthesis, and no additional detergent was used in the synthetic protocol. After the reaction was complete, the sample was allowed to gradually cool to room temperature while stirring. To remove gold-free nanoparticles, the dispersion was diluted with water, centrifuged, then the supernatant was removed and the resulting pellet was redispersed in water. The procedure was repeated at least three times, yielding OMAs with ~95% purity based on TEM analysis.
The synthesis of the control pNIPMAm particles with 50% crosslinking density used 40 mg of N-isopropylmethacrylamide and 40 mg of the crosslinking agent N,N′-methylenebisacrylamide, however, all other steps were identical to the protocol described above for the 10% crosslinking density pNIPMAm-coated particles. The synthesis of the control pNIPMAm particles lacking the inorganic Au NR core was identical to the protocol described above, with a single exception in that the 1 mL AuNR solution was not added.
Characterization of OMAs
Temperature controlled dynamic light scattering (DLS) was performed on a Zetasizer Nano ZS90 (Malvern Instruments, Malvern UK). Negative staining transmission electron microscopy(TEM) was carried out on a Hitachi H-7500 transmission electron microscope at an accelerating voltage of 75 kV. AFM measurements were performed on a “FastScan Bio” AFM (Bruker, USA) in a closed liquid cell with the ability to control the temperature of the solution from room temperature to 55 °C. Silicon AFM tips (Bruker)with a tip radius of curvature = 12 nm were used to image the sample in peak-force tapping mode.
Determination of Dynamics of OMA nanoparticles
To determine the collapse and relaxation dynamics of OMA nanoparticles in response to light, we functionalized the particles with a fluorescent dye and used pulsed NIR excitation coupled with fluorescence imaging to monitor particle dynamics. Note that Alexa488 was selected to record particle dynamics because of its known temperature independence, which was verified in Supplementary Figure 5g and h. Therefore, changes in fluorescence quantum yield (intensity) could be ascribed to OMA particle collapse rather than to local heating. The OMA nanoparticles were immobilized onto azide modified coverslips (as described in the surface preparation section). However, rather than modifying the remaining alkyne functional groups with the cRGDfK peptide, the OMA nanoparticles were conjugated with an azide-modified fluorescent dye. Briefly, a 50 μL solution containing 2.5 μL of 2 nM Alexa Fluor 488-cRGDfK- azide (Note: the synthesis of this molecule was adapted from Liu et al26). 2.5 μL of 10 mM ascorbic acid, 5 μL of 10 mM Cu(II)-TBTA (55% DMSO), and 40 μL of 55% (v/v) DMSO was placed between two OMA-modified coverslips. The sandwiched coverslips were sealed in a secondary petri dish, and the reaction was allowed to proceed for 12 hr at room temperature. The coverslips were then rinsed with ethanol and dried under a stream of nitrogen. The samples with Alexa488-OMA particles were then simultaneously imaged using epifluorescence microscopy while chopping the NIR laser source at 10 Hz (50% duty cycle). A standard FITC cube was used for fluorescence imaging. In addition, two filters (product number: 87-745,47-291, Edmund optics, USA) were included in the optical path to block the NIR illumination. The fluorescence images were captured by a sCMOS camera (Zyla 4.2 sCMOS, Andor Technologies Ltd., UK) at 1800 fps.
Determination of cRGDfK peptide density on OMA nanoparticles
The value of the extinction coefficient for the cRGDfK peptide was too small for absorption spectrometry to quantify the peptide loading density. Instead, to estimate the peptide density, azide dye was coupled to the alkyne terminal group of the OMA nanoparticles using click chemistry. Based on the assumption that the dye and the cRGDfK peptide have similar coupling efficiencies, we quantified the dye loading density to approximate the peptide loading density.
Quantitative fluorescence calibration curve using supported lipid bilayers
In order to quantify the dye loading density, a quantitative fluorescence supported lipid bilayer calibration curve was constructed to relate fluorescence to dye density27. To establish the linear relationship between the observed fluorescence intensity and the fluorophore density, we prepared lipid bilayers with predefined concentrations of fluorescent lipid molecules. Non-fluorescent DOPC supported lipid membranes were doped with DPPE-fluoresce in lipids with ratios ranging from 0 to 1 mol% and then imaged under the same conditions used in live cell imaging (Supplementary Figure 9a and b). The number of fluorophore molecules per unit area was directly calculated from the footprint of a DOPC lipid, which was reported at 0.72 nm2.27
Determination of the F factor
The scaling factor F represents the ratio of FAM emission versus the DPPE-fluorescein emission and is defined as: F = Ibulk(FAM–azide)/Ibulk(DPPE–fluorescein), where Ibulk(FAM-azide) and Ibulk(DPPE-fluorescein) are the intensity of the ligand and lipid molecules in solution after being normalized for concentration, respectively. This value was measured on the fluorescence microscope by moving the focal plane (~200 μm) into the center of the sample (Supplementary Figure 9d).
Determination of FAM density on OMA nanoparticles surface
To exclude fluorescence quenching by the Au NR core, 50 mM KCN was used to dissolve the gold NR core prior to covalent immobilization of the OMAs nanoparticles onto the glass substrate. After gold dissolution, OMA nanoparticles were washed 5 times with DI water by centrifugation (8000 rpm, 10 min) to wash away the KCN. Subsequently, OMA nanoparticles without the AuNR core were covalently immobilized onto the azide functionalized glass substrates using the click reaction. In order to exclude fluorescence self-quenching, the FAM-azide concentration was ten times lower than the cRGDfK peptide concentration when functionalizing OMA particles. Briefly, a 50 μL solution containing 2.5 μL of 30 mM FAM-azide, 2.5 μL of 10 mM ascorbic acid, 5 μL of 10 mM Cu (II)-TBTA (55% DMSO, 45% H2O), and 40 μL of 55% by volume DMSO were added to an OMA(Au NR free)-functionalized coverslip and sandwiched with a second OMA(Au NR free)-functionalized glass coverslip. This reaction was allowed to proceed at 50 °C for 3 h prior to rinsing with ethanol and drying with nitrogen. The surface was imaged under the same conditions used in live cell imaging. Using the calibration curve, F factor and fluorescence intensity from FAM labeled OMA (Au NR free)-functionalized coverslips (Supplementary Figure 9d), FAM density was estimated to be ~2,000 molecules μm−2. Note, because the FAM-azide reaction concentration is ten times lower than the cRGDfK peptide concentration used for cell experiments, the absolute RGDfK peptide density should be much higher than 2,000 molecules μm−2, and this value represents the lower bound estimate.
Estimation of OMA nanoparticle collapse-driven forces using molecular tension sensor
To estimate the force applied during OMA nanoparticle collapse, we immobilized OMA nanoparticles onto a surface functionalized with DNA-based molecular tension probes that were recently developed in our lab12, 26, 28. The DNA tension sensor consists of three components: 1) a lower ssDNA strand modified with a molecular quencher (BHQ1) at the 3′ end, 2) an upper ssDNA strand modified with a Cy3B dye as fluorescence reporter at the 5′ end, 3) and a bridging hairpin duplex flanked by two complementary ssDNA arms that brings the BHQ1 and Cy3B into close proximity. Due to the high quenching efficiency (> 95%) between BHQ1 and Cy3B dye, minimal fluorescence is detected when the hairpin is folded. However, when sufficient external forces are applied to the two ends of DNA, the hairpin structure becomes unfolded due to force, which effectively results in a physical separation of Cy3B dye away from BHQ1. This increased distance between dye and quencher is accompanied by a significant amount of fluorescence increase (~10–30 fold) due to de-quenching of the dye. Therefore, this method enables force measurements in real-time. Here, we used a DNA hairpin that has been experimentally calibrated to have a F1/2 of ~13 pN, which indicates that 50% of harpins will unfold at a force=13 pN.
To immobilize the DNA tension sensor to the coverslip, we adapted a previously published protocol for AuNP tension sensor26. Initially, 9 nm spherical AuNPs were randomly anchored to a lipoic acid modified glass coverslip. To efficiently attach the DNA tension sensors onto the AuNP coverslip with high coverage, we modified the sensor with a thiol group at the 5′ end of the lower ssDNA and directly incubated the sensor construct to the AuNP surface. Typically, 300 nM thiolated DNA hairpins in 1 M NaCl solution were incubated with the AuNP surface overnight. Note, all hairpin samples were pre-folded by applying an annealing cycle in PCR thermocycler. On the 3′ end of the upper ssDNA, we added an additional biotin group available to bind to streptavidin, which provides a link for the tension sensor to ultimately bind to the biotinylated OMA particles.
Upon NIR illumination of particles, we observed a reversible increase in fluorescence that coincided with NIR illumination. DNA unfolding could be repeated for hundreds of cycles until photobleaching of the force-reporting dye occurred. This suggested that the particle collapse exerts a force that is greater than 13 pN (force response threshold), but below the ~50 pN force required to shear DNA duplexes (Supplementary Figure 6).
Simulations
Calculations of the OMA particle temperature and heat distribution were performed by finite element simulation (Comsol Multiphysics, Burlington MA). In all the simulations, the NIR laser power was held constant at 35 mW but with different radius of the laser spot (3, 5, 7, 10 μm, respectively). In these simulations, we assume that during the transient laser heating period (10 msec), the temperature of the OMA particle reaches steady state conditions. Thus, we calculate the temperature distribution in the nanorod and polymer at steady state to determine the maximum temperatures that would be observed during the illumination cycle. We also assume that the nanorod is located exactly at the center of polymer and the laser illumination provides a constant and uniform heat generation that dissipates uniformly from the gold nanorod. The absorption cross section of Au NR was estimated at 20,000 nm2 based on literature precedent 29. The heat capacity and thermal conductivity of the polymer hydrogel is assumed to be equivalent to that of water 29. The value of thermal conductivity of gold nanorod was set to 317 W mK−1 30. Note that the power density of 113 μW μm−2 (35 mW, the radius of the laser spot was 10 μm, 100% duty cycle) is 10 fold greater than that used for cell stimulation because a 100% duty cycle (steady state illumination) is assumed for the energy input calculation.
Surface Preparation
Glass coverslips (No. 2–25mm diameter, VWR) were sonicated in Nanopure water (18.2 mΩ) for 10 min and then etched in piranha for 10 min - please take caution - piranha is extremely corrosive and may explode if exposed to organics. The glass coverslips were then washed twice in a beaker of Nanopure water (18.2 mΩ) and placed into three successive wash beakers containing ethanol and left in a final fourth beaker containing 1% 6-azidosulfonylhexyl-triethoxysilane (Gelest, Inc.) in ethanol for 6 hours. The azide functionalized substrates were then immersed in the ethanol three times and subsequently rinsed with ethanol and dried under N2. To covalently immobilize the OMAs nanoparticles (or pNIPMAm particles lacking the Au NR core)onto the azide functionalized glass substrates, a 50 μL solution containing 10 μL of 1 nM OMA nanoparticles,10 μL of 10 mM ascorbic acid, 10 μL of 10 mM Cu (II)-TBTA(55% DMSO), and 20 μL of DMSO were added onto one azide modified glass coverslip and then covered using a second azide functionalized glass coverslip. The sandwiched coverslips were sealed in a secondary petri dish, and the reaction was allowed to proceed for 12 hr at room temperature. The coverslips were then rinsed with ethanol and dried under a stream of N2. This procedure generated coverslips coated with a monolayer of covalently immobilized OMA particles. To further modify OMA particles with the cyclic Arg-Gly-Asp-d-Phe-Lys c(RGDfK) peptide, a 50 μL solution containing 2 μL of 318 mM RGD-N3, 5 μL of 10 mM ascorbic acid, 5 μL of 10 mM Cu (II)-TBTA (55% DMSO), and 40 μL of 55% (by volume) DMSO was sandwiched between two OMA-functionalized substrates. This reaction was allowed to proceed at 50 °C for 3 h prior to rinsing with ethanol and drying with nitrogen.
To prepare Au NRs lacking the polymer shell, the CTAB and NaOL surfactants coating the AuNRs were exchanged with 1 mM alkyne terminated PEG thiol (Catalog number: PG2-AKTH-1k, 1000 g/mol, Nanocs, USA) in a 1 ml AuNR solution (described in Gold nanorod preparation). To allow for more complete ligand exchange, the solution was sonicated for 3 min and left at room temperature for 12 hr. Subsequently, the AuNR solution was purified by centrifugation at 5,000 rpm for 10 min, removing the supernatant, and then redispersing in 25 μl of deionized water. These Au NRs were immobilized onto azide-functionalized glass coverslips by placing a 70 μL solution containing 25 μl of Au NRs modified with the alkyne terminal group, 5 μL of 10 mM ascorbic acid, 5 μL of 10 mM Cu(II)-TBTA(55% DMSO), and 35 μL of DMSO between two coverslips. The sandwiched coverslips were sealed in a secondary petri dish, and the reaction was allowed to proceed for 12 hr at room temperature. The coverslips were then rinsed with ethanol and dried under a stream of nitrogen. The coverslips were subsequently incubated with 500 μL of 100 μg ml−1 human fibronectin (Sigma-Adrich) for 1 hr. Fibronectin enhanced cell binding to the bare AuNR surface to allow for control experiments. Coverslips were rinsed with sterile PBS prior to seeding cells and conducting experiments.
Synthesis of RGDfK-N3
500 μg of cRGDfK peptide (Peptides International, MW: 603.68) was reacted with 1.2 mg azide-NHS linker (Thermo Scientific. MW: 198.14) in 20 μL DMF. To this reaction mixture, 0.1 μL of neat triethylamine was added as an organic base and the reaction was allowed to proceed for 12 hr at room temperature. The product was purified by reverse phase HPLC (flow rate 1 ml/min; solvent A: 99.5% DI water, 0.5% TFA; solvent B: 99.5% acetonitrile 0.5% TFA; initial condition was 10% B with a gradient of 1% per min). The final RGDfK-N3 product was verified by MALDI-TOF.
Cell culture and transfection
NIH/3T3 fibroblast cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% Cosmic Calf Serum (Mediatech), HEPES (9.9 mM, Sigma), sodium pyruvate (1 mM, Sigma), L-glutamine (2.1 mM, Mediatech), penicillin G (100 IU ml−1, Mediatech) and streptomycin (100 μg ml−1, Mediatech) and were incubated at 37 °C with 5% CO2. Cells were passaged at 60–80% confluency and plated at a density of 10% using standard cell culture procedures. All cell transfection was performed in a 24-well plate. All procedures are based on a standard protocol provided by Life Technologies (Carlsbad, CA). Briefly, 4×104 cells were plated in each well one day before transfection. During transfection, 0.5–1g DNA was mixed with Lipofectamine LTX with Plus™ Reagent for each well and incubated for 24–48 hr before imaging.
Microscopy and optomechanics experiments
Transfected NIH 3T3 cells were cultured on a monolayer of OMA nanoparticles modified with cyclic Arg-Gly-Asp-d-Phe-Lys c(RGDfK) with a density of 2,000 peptides per μm2 for 12 hr. Live cells were imaged in standard cell imaging buffer (Hank’s balanced salt, pH 7.4, 10 mM HEPES without phenol red) at 37 °C. During imaging, physiological temperature was maintained with a warming apparatus consisting of a sample warmer and an objective warmer (Warner Instruments 641674D and 640375). The microscope was Nikon Eclipse Ti driven by the Elements software package. The microscope features an Evolve electron multiplying charge coupled device (EMCCD; Photometrics), an Intensilight epifluorescence source (Nikon), a CFI Apo 100× (numerical aperture (NA) 1.49) objective (Nikon) and a TIRF launcher with three laser lines: 488 nm (10 mW), 561 (40 mW), and 638 nm (20 mW). The experiments used the following Chroma filter cubes: TIRF 488, TRITC, FITC and reflection interference contrast microscopy (RICM). For optomechanics experiments, a near-infrared diode laser (Schäfter+Kirchhoff, Hamburg, Germany) was mounted using the standard dark-field condenser accessory available from Nikon. The NIR laser wavelength was 785 nm ± 10 nm, while the laser output maximum power was 53 mW. The minimum size (diameter) of the focused spot was 6 μm. The laser modulation frequency was controlled by a frequency generator following the instructions provided by the manufacturer. Two additional NIR cutoff filters (product number: 87-745, 47-291, Edmund optics, USA) were included into the optical path to eliminate NIR blead through to the EMCCD. Using this setup, we simultaneously illuminated the sample with a pulsed NIR source (10 Hz, 10% duty cycle) while recording FAs dynamics for the cell.
Calcium imaging
To modify OMA surface with peptide-major histocompatibility complex (pMHC) from the OVA model. The amine functionalized OMA particles were incubated with NHS-biotin (Thermo Fisher) at 2 mg ml−1 in DMSO (Sigma) overnight. Subsequently, the substrates were washed with EtOH and dried under a stream of nitrogen. This was then followed by incubation with streptavidin (1 μg ml−1, 1 hour and room temperature). After rinsing with PBS, the substrates were incubated with the biotinylated H-2K(b) monomers (80 μg ml−1, 1 hour, room temperature). After final rinsing with PBS, the substrates were assembled into the cell imaging chamber (Life Technologies), replaced with hank’s balanced salt imaging buffer (Sigma) and used immediately for cell experiment. Here, biotinylated H-2K(b) monomers were provided by the National Institutes of Health Tetramer Core Facility at Emory University. Monomers are comprised of the cognate chicken ovalbumin 257-264 epitope SIINFEKL (N4) peptide loaded in the alpha chain of mouse H-2K(b) that is complexed with beta-2-microglobulin (b2m) from mouse origin.
Freshly purified OT1 cells (n = 1×106) were centrifuged to remove the RPMI media and re-suspended in 3 ml of hank’s imaging buffer. 10 ml of 1 mM of fura-2/Am in DMSO was added into the cell suspension. The solution was kept at 37 °C for 30 min. Afterwards, cells were pelleted by centrifugation at 1,200 rpm for 4 min and re-suspended in 3 ml of imaging buffer for an additional 15 min. This step is to ensure full de-esterification of the fura-2/Am. Then, the cells were again pelleted and re-suspended in 1 ml of imaging buffer and prepared for seeding on the OMA surface. The cells were incubated for 20 mins before stimulation.
For the calcium imaging, the microscope employed a Nikon CFI S Fluor 100x oil objective, a Chroma 340 excitation filter set (ET340x, T400lp and ET510/80m) and Chroma 380 excitation filter set (ET380x, T400lp and ET510/80m). During the experiment, the fluorescence images were acquired by using the 340 nm and 380 nm filter sets sequentially. Afterwards, ImageJ was used to generate a mask for both channels excited by 340 nm and 380 nm. Finally, the fura-2 ratio (I340/I380) was calculated by using the image calculator function in ImageJ.
Determination of F-actin displacement using TIRF-based nanometry
The evanescent field intensity in TIRF exponentially decays with a predictable function31,
where I0 is the intensity at z = 0, n1 and n2 are the indices of refraction of the glass coverslip and the sample, respectively, λ is the excitation wavelength, and θ is the angle of incidence. Therefore, one can infer the height changes of cellular structures, as long as the number of fluorophores remains fixed31. In our experiments, we assumed that n1 (glass) = 1.515, n2 (cell) =1.37, λ= 488 nm, θ= 70° ~78°. Experimentally, we found that the TIRF intensity of F-actin increased by 70% ±12% under steady-state NIR illumination (Supplementary Figure 14). Using these parameters, and assuming an initial height of the F-actin at 220 nm, and fixed number of GFP emitters, we were able to estimate the change in height for the F-actin network. The height changes of the F-actin network are estimated to range between 95–115 nm in response to OMA particle collapse. Note that the n2 is assumed to remain constant. However, the index of refraction for pNIPMAm is known to increase following collapse29. An increase in n2 will reduce the value of I. Therefore, the calculated decrease in z, represents the minimum displacement.
Supplementary Material
Acknowledgments
The authors are grateful for support from the US National Institutes of Health (R01-GM097399), the Alfred P. Sloan Research Fellowship, the Camille-Dreyfus Teacher-Scholar Award, the National Science Foundation (NSF) EAGER Award (1362113) and the NSF CAREER Award (1350829). This research project was also supported in part by the Emory University Integrated Cellular Imaging Microscopy Core. We thank M. Zhang for collecting DLS data and S. Nie (Biomedical Engineering at Emory University and the Georgia Institute of Technology, Atlanta, U.S.A.) for providing access to the DLS instrument. We thank B. Evavold and L. Blanchfield (Emory University, Atlanta, U.S.A.) for providing pMHC and T cells. We also thank C. Hill (Emory University, Atlanta, U.S.A.) for access to the temperature-controlled Vis-IR spectrophotometer.
Footnotes
Author contributions:
K.S. and Z.L. devised the overall experimental strategy. Z.L. and Y.L. performed cell experiments, and analyzed data. Z.L. synthesized and characterized the OMAs and developed the near infrared laser illumination system for investigating the kinetics of OMAs and for single cell stimulation experiments. Y.L. synthesized molecular tension sensors, Y.C. and Y.L. synthesized the RGD-N3 molecule. Z.L., Y.L., K.Y. and Y.Z. performed OMAs modifications. H.S. and A.H. performed the 3D steady state thermal conduction simulations. A.M., Y.L. and Z. L. performed the 3D-SIM experiments and TIRF nanometry analysis. Z.H. performed temperature-controlled AFM imaging in liquid. The manuscript was prepared by Z.L. and K.S. with input from all authors.
References
- 1.Vogel V, Sheetz M. Nature Rev Mol Cell Biol. 2006;7:265–275. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- 2.Dufrene YF, et al. Nat Methods. 2011;8:123–127. doi: 10.1038/nmeth0211-123. [DOI] [PubMed] [Google Scholar]
- 3.Riveline D, et al. J Cell Biol. 2001;153:1175–1186. doi: 10.1083/jcb.153.6.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charras GT, Horton MA. Biophys J. 2002;82:2970–2981. doi: 10.1016/S0006-3495(02)75638-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang YX, et al. Nature. 2005;434:1040–1045. doi: 10.1038/nature03469. [DOI] [PubMed] [Google Scholar]
- 6.Tseng P, Judy JW, Di Carlo D. Nat Methods. 2012;9:1113–1119. doi: 10.1038/nmeth.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Etoc F, et al. Nat Nanotech. 2013;8:193–198. doi: 10.1038/nnano.2013.23. [DOI] [PubMed] [Google Scholar]
- 8.Sniadecki NJ, et al. Proc Natl Acad Sci USA. 2007;104:14553–14558. doi: 10.1073/pnas.0611613104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levskaya A, Weiner OD, Lim WA, Voigt CA. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu YI, et al. Nature. 2009;461:104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pastrana E. Nat Methods. 2011;8:24–25. [Google Scholar]
- 12.Zhang Y, Ge C, Zhu C, Salaita K. Nat Commun. 2014;5:5167. doi: 10.1038/ncomms6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Ha T. Science. 2013;340:991–994. doi: 10.1126/science.1231041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jurchenko C, Chang Y, Narui Y, Zhang Y, Salaita KS. Biophys J. 2014;106:1436–1446. doi: 10.1016/j.bpj.2014.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oakes PW, Beckham Y, Stricker J, Gardel ML. J Cell Biol. 2012;196:363–374. doi: 10.1083/jcb.201107042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coyer SR, et al. J Cell Sci. 2012;125:5110–5123. doi: 10.1242/jcs.108035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mih JD, Marinkovic A, Liu F, Sharif AS, Tschumperlin DJ. J Cell Sci. 2012;125:5974–5983. doi: 10.1242/jcs.108886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang X, El-Sayed IH, Qian W, El-Sayed MA. J Am Chem Soc. 2006;128:2115–2120. doi: 10.1021/ja057254a. [DOI] [PubMed] [Google Scholar]
- 19.Iskratsch T, Wolfenson H, Sheetz MP. Nat Rev Mol Cell Biol. 2014;15:825–833. doi: 10.1038/nrm3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim ST, et al. J Biol Chem. 2009;284:31028–31037. doi: 10.1074/jbc.M109.052712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu BY, Chen W, Evavold BD, Zhu C. Cell. 2014;157:357–368. doi: 10.1016/j.cell.2014.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye X, Zheng C, Chen J, Gao Y, Murray CB. Nano Lett. 2013;13:765–771. doi: 10.1021/nl304478h. [DOI] [PubMed] [Google Scholar]
- 23.Das M, Sanson N, Fava D, Kumacheva E. Langmuir. 2007;23:196–201. doi: 10.1021/la061596s. [DOI] [PubMed] [Google Scholar]
- 24.Contreras-Caceres R, et al. Adv Mater. 2008;20:1666–1670. [Google Scholar]
- 25.Tang F, Ma N, Wang X, He F, Li L. J Mater Chem. 2011;21:16943–16948. [Google Scholar]
- 26.Liu Y, Yehl K, Narui Y, Salaita K. J Am Chem Soc. 2013;135:5320–5323. doi: 10.1021/ja401494e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galush WJ, Nye JA, Groves JT. Biophys J. 2008;95:2512–2519. doi: 10.1529/biophysj.108.131540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stabley DR, Jurchenko C, Marshall SS, Salaita KS. Nat Methods. 2012;9:64–67. doi: 10.1038/nmeth.1747. [DOI] [PubMed] [Google Scholar]
- 29.Rodriguez-Fernandez J, Fedoruk M, Hrelescu C, Lutich AA, Feldmann J. Nanotechnology. 2011;22:245708. doi: 10.1088/0957-4484/22/24/245708. [DOI] [PubMed] [Google Scholar]
- 30.Ekici O, et al. J Phys D: Appl Phys. 2008;41:185501. doi: 10.1088/0022-3727/41/18/185501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saffarian S, Kirchhausen T. Biophys J. 2008;94:2333–2342. doi: 10.1529/biophysj.107.117234. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.